Abstract
The vascular endothelial growth factor (VEGF)-VEGF Receptor (VEGFR) system is an important pathway for regulation of angiogenesis. However, its evolutionary development, particularly the step from invertebrates to vertebrates, is still largely unknown. Here, we molecularly cloned the VEGFR-like gene from Halocynthia roretzi, a species belonging to the Tunicata, the chordate subphylum recently considered the sister group of vertebrates. The cDNA encoded a homolog of human VEGFR, including the transmembrane domain, and the tyrosine kinase domain with a kinase-insert region, which was designated S. sq VEGFR (GenBank AB374180). Similar to Tunicates including ascidians in the phylogenetic tree, the Amphioxus, another chordate, is located close to vertebrates. However, S. sq VEGFR has a higher homology than the Amphioxus VEGFR-like molecule (GenBank AB025557) to human VEGFR in the kinase domain-2 region. The S. sq VEGFR mRNA was expressed at highest levels in circulatory system-containing tissues, suggesting that S. sq VEGFR plays an important role in the formation or maintenance of circulatory system in Tunicates, Halocynthia roretzi.
Keywords: vascular endothelial growth factor receptor, Halocynthia roretzi, sea squirt, circulatory system
Abbreviation
- VEGF
Vascular endothelial growth factor
- VEGFR
Vascular endothelial growth factor receptor
- S. sq.
Sea squirt
- B. schlosseri
Botryllus schlosseri
- H. roretzi
Halocynthia roretzi
- C. intestinalis
Ciona intestinalis
1. Introduction
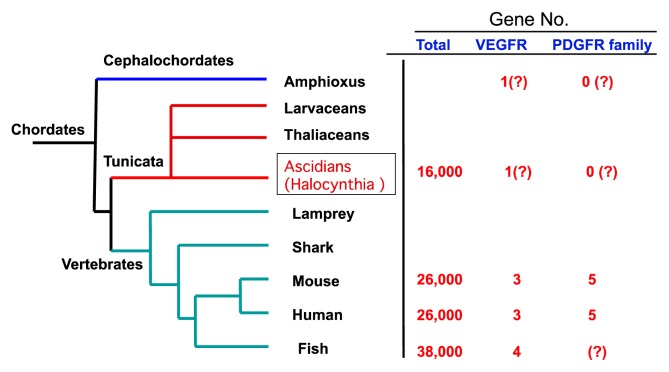
By comparing invertebrates’ genomes with those of humans and other animals, we could find new insights into the evolutionary origins of human tissues, as well as a better understanding of chordate and vertebrate development in general. The chordates have given rise to three groups: the vertebrates, Cephalochordates (Amphioxus), and Tunicates, which include the ascidians such as Halocynthia roretzi (H. roretzi) (Figure 1). The tunicates have recently emerged as a model system for understanding the organization of genomes. Because of their relative simplicity and position as an out-group to vertebrates, tunicates have a unique potential to illuminate the molecular mechanisms underlying the primitive body plan from which modern chordates diversified [1–6]. H. roretzi are bag-like gelatinous creatures that its body appears more primitive than creatures with backbones and spinal columns. Its genome is about 160 Mb in size, or one twentieth the size of the human genome, coding for about 16,000 genes [1,5]. The small size of the ascidian genome provides a distinct advantage for understanding genome organization and gene function [7,8]. Finding those genetic signals in the relatively simple animal could add to our knowledge of development such as heart and circulatory system in humans [9–14]. In Ascidians, several animals such as Botryllus schlosseri (B. schlosseri) and Ciona intestinalis (C. intestinalis) have been extensively studied on their development, morphogenesis and phylogeny. Particularly, B. schlosseri is unique in terms of their asexual growth of zooid and their circulatory system [10–12]. The circulatory system of B. schlosseri is divided into two: one is connected to heart and lacunae in various tissues in each zooid, and another consists of multiple ampullae and vessel system which is located at the peripheral area of a group of zooids.
Figure 1.
Phylogenic tree of the chordate and the number of genes for Vascular Endothelial Growth Factor receptor (VEGFR) and Platelet-derived growth factor receptor (PDGFR) family.
Recent studies have identified several signaling pathways and regulatory factors governing angiogenesis in vertebrates. In 1990 we isolated a gene for novel receptor-type tyrosine kinase containing seven Immunoglobulin-like domains in the ligand-binding region from human tissue, and designated it as Fms-like tyrosine kinase-1 (Flt-1) (GenBank X51602) [15–17]. A few years later, de Vries et al. reported the binding of Flt-1 with Vascular Endothelial Growth Factor (VEGF), thus, Flt-1 is also known as VEGFR1 [18]. Two sequences distantly related to Flt-1/VEGFR1 were isolated from mammals; VEGFR2 (KDR/Flk-1) and VEGFR3 (Flt-4). We and others have demonstrated that these VEGFR family gene products have unique signaling pathways within the cell, and play crucial roles in angiogenesis and lymphangiogenesis [16,17,19]: Flt-1/VEGFR1 has very strong affinity to VEGF, but does a weak tyrosine kinase activity, showing dual (positive and negative) roles in angiogenesis. On the other hand, VEGFR2 has about one order-lower affinity to VEGF compared to Flt-1/VEGFR1, but has a 10-fold stronger tyrosine kinase activity, and transduces major angiogenic signals using Phospholipase Cγ-C-kinase-MAP kinase pathway for cell proliferation via the autophosphorylation of a single tyrosine (Y)-containing motif at the 1175Y site of carboxy terminal region [20,21].
However, understanding these interactions in details has been complicated by extensive gene multiplication events in vertebrates, and it is difficult to clarify their basal characteristics in circulatory system in the phylogenetic development [9–11,14,22–25]. To circumvent this genetic redundancy, we have focused on VEGFR in basal chordates. VEGFR/platelet-derived growth factor receptor (PDGFR)-related tyrosine kinase has been identified in Drosophila and other invertebrates including Cephalochordate (amphioxus) and Tunicates (ascidians) such as B. schlosseri, and C. intestinalis[12,26–29]. Therefore, we attempted to molecularly clone a receptor related to mammalian VEGFR from another popular ascidian, H. roretzi.
2. Results and Discussion
2.1. Isolation of VEGFR-Like Tyrosine Kinase Gene in H. roretzi by Using Degeneration-Primer Method
Total RNA from H. roretzi tissue was prepared by the AGPC method [30] (see Experimental Section). To test the quality of dsDNA, we selected Eph-Receptor as a positive control [31] well conserved among various species. By sequential PCR, we obtained a cDNA product of expected size and with the expected patterns of digestion by restriction enzymes.
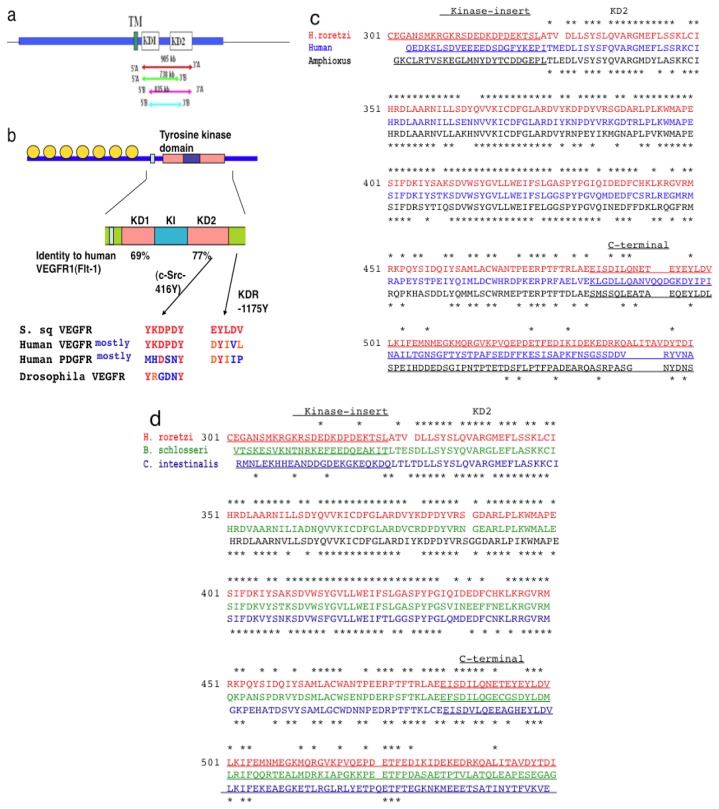
To test for the possible existence of VEGFR in H. roretzi, we made four degenerated oligonucleotide primers based on the sequence of the tyrosine kinase domain in VEGFR genes highly conserved among various species (Figure 2a). Once we obtained RT-PCR bands, we attempted a second PCR using another degenerated primer set at the inner sequences. We finally isolated a DNA sequence which encodes for tyrosine kinase with a kinase insert closely related to VEGFR (Figure 2b,c).
Figure 2.
Amino acid sequence of S. sq VEGFR, and its strong homology with human VEGFR. (a) Isolation of cDNA related to VEGFR gene from ascidian, H. roretzi; (b) Schematic diagram of VEGFR of H. roretzi in comparison to human Flt-1 (VEGFR1). A high degree of conservation of critical tyrosine-containing residues in S. sq VEGFR and human VEGFR; (c) Sequence homology of S. sq VEGFR with human VEGFR1 in the KD2 region (157 amino acid-long sequence; ATVD—RLAE in S. sq VEGFR). [*]: identical amino acids with the VEGFR-related sequence in the middle; (d) Sequence homology among ascidian VEGFRs (S. sq VEGFR, B. schlosseri VEGFR, and C. intestinalis VEGFR) in the KD2 region. [*]: identical amino acids with the VEGFR-related sequence in the middle.
By using rapid amplification of cDNA and further extension of cDNA, we isolated a H. roretzi cDNA which corresponds to 557 amino acids (GenBank AB374180) (Figure 2b). The cDNA encodes the transmembrane domain, the entire tyrosine kinase domain with a 120-amino-acid-long kinase insert region, and the carboxyl tail (Figure 2b). Although we used a variety of degenerate primers, we could not find any other VEGFR-like sequences in H. roretzi. This strongly suggests that only one member of the VEGFR family exists in H. roretzi. Since “Sea squirt” is a popular name for ascidians, we designated the H. roretzi VEGFR-like sequence as S. sq VEGFR. The S. sq VEGFR showed a high homology with VEGFRs obtained from other ascidians, B. schlosseri and C. intestinalis (Figure 2d).
2.2. The Structure of S. sq VEGFR Is Closer to VEGFR Family than PDGFR Family in Humans
Although the S. sq VEGFR gene is not completely homologous with that of humans, the basic structure of tyrosine kinase relevant to its function is very well conserved. A sequence alignment of S. sq VEGFR versus human VEGFR1 [15] in different parts, i.e., the kinase domain-1 (KD1), kinase-insert (KI) and kinase domain-2 (KD2), showed amino acid identity of 69%, 17%, and 77%, respectively (Figure 2c, data was not shown). As we found, the similarity was greatest at KD2. The sequence also shows homology with mammalian PDGFR, but less than with mammalian VEGFR (Figure 3). The higher homology of S. sq VEGFR with human VEGFRs than PDGFRs is consistent with the idea that this sequence encodes for a 7-Ig VEGFR-like molecule in ascidians, H. roretzi (Figure 2b).
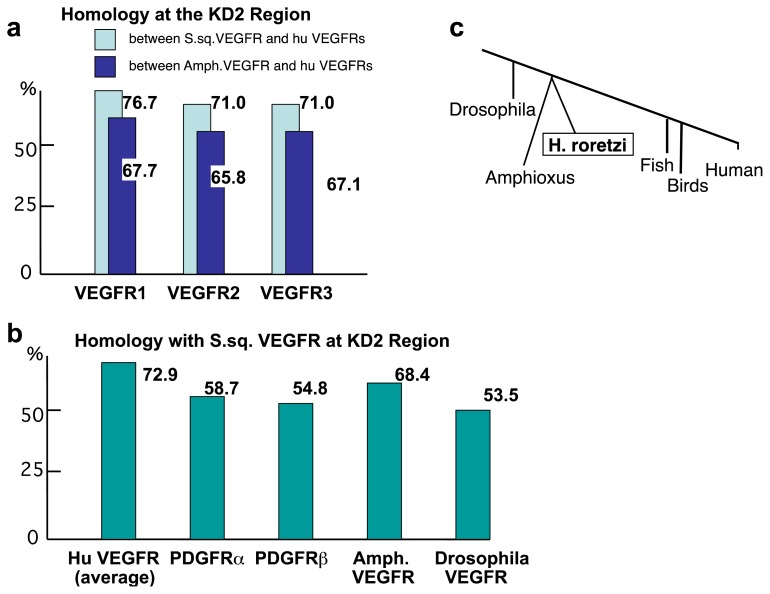
Figure 3.
Human VEGFR is more homologous to S. sq VEGFR than to Amphioxus VEGFR-like protein. (a) Comparison of the KD2 domain among S. sq VEGFR, Amphioxus VEGFR-like molecule and human VEGFRs; (b) Comparison of KD2 domain in S. sq VEGFR with invertebrate VEGFRs and human VEGFRs as well as PDGFRs; (c) Diagram of Phylogenic tree (the order of ascidians such as H. roretzi and amphioxus is not completely fixed).
Recently, the Amphioxus VEGFR-like sequence was published at the regions of a part of KI, KD2, and carboxyl tail (GenBank AB025557). The KD2 of human VEGFR1 shares 77%, 68% and 56%-identity with those of H. roretzi, Amphioxus, and Drosophila, respectively. Human VEGFR2 and VEGFR3 at the KD2 region also share more homology with S. sq VEGFR than with Amphioxus VEGFR (hVEGFR2 to S. sq VEGFR, 71% vs. hVEGFR2 to Amphioxus VEGFR, 65.8%; hVEGFR3 to S. sq VEGFR, 71% vs. hVEGFR3 to Amphioxus VEGFR, 67.1%) (Figure 3a). In the phylogenetic tree, based on the sequence homology of 18S rDNA, Amphioxus was previously suggested to locate closer than tunicates to vertebrates (Figure 3c) [32]. However, S. sq VEGFR has a higher homology in KD (Figure 3a,b) than the Amphioxus PDGFR/VEGFR-like molecule to human VEGFR.
2.3. Amino Acid Sequence Critical for the Intracellular Signaling of Human VEGFR Is Conserved in S. sq VEGFR
The “Y-K-D/N-P-D-Y” amino acid residue at KD2 of human VEGFR (residue 1048–1053 in human VEGFR1) is highly conserved among VEGFRs in vertebrates. VEGFR1 in mammals and birds have the sequence “Y-K-N-P-D-Y” whereas VEGFR2 and VEGFR3 in essentially all the vertebrates reported so far have “Y-K-D-P-D-Y”. The last “Y” corresponds to the 416-tyrosine in c-Src tyrosine kinase which is critical for activation of Src-kinase (Figure 2b). PDGFR family members have “M-R/H-D-S-N-Y” at this position, which is much diverged from the sequence in VEGFR. Interestingly, “Y-K-D-P-D-Y” is not well conserved in Drosophila VEGFR which has “Y-R-G-D-N-Y”. These results strongly suggest that the S. sq VEGFR has evolutionally developed to become closer to VEGFRs in vertebrates (Figure 2b).
We have recently shown 1175-Y in human VEGFR2, which corresponds to 1169-Y in VEGFR1, to be a site of autophosphorylation essential for the PhospholipaseCγ-PKC-MAP kinase pathway towards endothelial cell growth [20,21]. At the “D-Y-I-V-L” site in VEGFR2, S. sq VEGFR but not Drosophila VEGFR carries a similar sequence motif, “E-Y-L-D-V” (Figure 2b). This suggests that before vertebrates, S. sq VEGFR had already acquired a similar intracellular signaling system. The “Y-K-D-P-D-Y” motif in KD2, and “D-Y-I-V-L” motif in the 1175Y region are highly conserved in VEGFRs of other ascidians, B. schlosseri and C. intestinalis, with minor modifications (Figure 2d).
2.4. An Intimate Relationship between Expression of S. sq VEGFR mRNA and the Circulatory System in H. roretzi
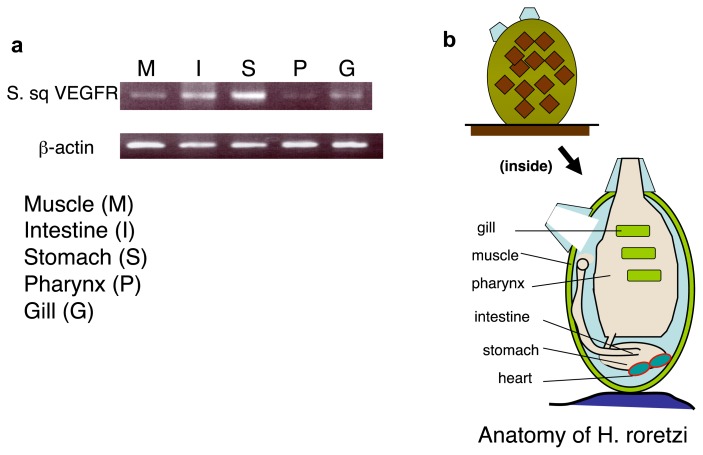
To see whether the levels of the S. sq. VEGFR transcript are tissue-specific or not in H. roretzi, we examined the expression patterns of S. sq VEGFR mRNA by using RT-PCR. The level of mRNA was highest in the stomach (heart-localized tissue), followed by the intestine and muscle, and lower in the pharynx and gill (Figure 4).
Figure 4.
The expression of the S. sq VEGFR gene in the H. roretzi tissues is correlated with the existence of circulatory system. (a) RT-PCR analysis of S. sq VEGFR gene expression in the different H. roretzi tissues. Upper panel: PCR was performed using a S. sq VEGFR-specific primer set and reverse-transcribed cDNA from the total RNA of each tissue as the template. Lower panel: PCR was preformed using the H. roretzi β-actin-specific primer set and reverse-transcribed cDNA from total RNA of each tissue as the template (positive control); (b) Anatomical scheme of H. roretzi.
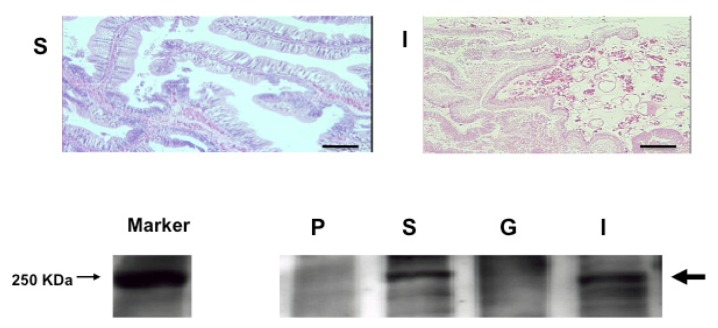
A high level of expression of the S. sq VEGFR gene in the stomach (heart-localized tissue) suggests an intimate relationship between S. sq VEGFR mRNA and the circulatory system in this animal. Western blot analysis also revealed that the S. sq VEGFR is highly expressed in the stomach (including heart), and then intestine and significantly less abundant in gill, and pharynx (Figure 5). These results suggest that S. sq VEGFR plays a crucial role in the formation or maintenance of heart and vessel-like structure as well as blood cells in the latest stage of invertebrates.
Figure 5.
Strong expression of S. sq VEGFR protein in H. roretzi stomach (heart-localized tissue) and intestine. Upper part, Histology of the different tissues of H. roretzi. Lower part, Western blot analysis of S. sq VEGFR in different H. roretzi tissues. The tissues examined are: Intestine (I), Stomach (S), Pharynx (P) and Gill (G). Scale bar indicates 120 m.
The tunicate occupies a pivotal position in the animal kingdom for understanding the evolution of vertebrates. The ascidian genome provides an insight into the basic set of genes available at the very beginning of vertebrate evolution since the tunicates diverged just prior to the widespread gene duplication processes that are thought to have shaped and transformed the vertebrate genome [1–8,33].
Recently VEGFR mRNA of the C. intestinalis was reported (GenBank XM_002126535, in 2005). This sequence shows a high homology at amino acid levels in tyrosine kinase domain (KD1 and KD2), except for the kinase insert region, with that of the sequence S. sq VEGFR (H. roretzi) (Figure 2d). However, the C. intestinalis VEGFR-like sequence reported has very short, almost a half of, extracellular domain compared to those of human VEGFR and Drosophila VEGFR. This short sequence cannot contain seven Immunoglobulin-like domains which are important characteristics of VEGFR family. Thus, we suggest that this C. intestinalis VEGFR-like sequence is not for a typical VEGFR but a variant form.
More recently, Tiozzo et al. isolated a B. schlosseri cDNA encoding VEGFR-related molecule [12]. By using this sequence, they showed that the VEGFR mRNA is expressed in the epithelial cells covering the ampullae, and suppression of the gene expression via siRNA or human VEGFR-kinase inhibitor significantly attenuated the angiogenesis occurred after ampullaectomy and disorganized the whole morphology. These results suggest that VEGFR function is involved in a unique angiogenic process and morphogenesis in B. schlosseri.
In this study, we showed a higher homology of S. sq VEGFR (KD2 region) to human VEGFR compared to that of Amphioxus VEGFR to humans. This might indicate a strong phylogenetic affinity between tunicates and vertebrates. However, the information on the homology derived from a single gene product such as VEGFR is limited, and it could be modified with the structure of interacting molecule(s). Thus, the views on the relationships between the chordates should be carefully studied both at genome-wide level and each gene product level [13,14,34–37].
Both VEGFs and PDGFs and their receptors have been implicated in mammalian vascular development, and the prototype VEGF (VEGF-A) also regulates the development of hematopoietic cells. A VEGFR was found in Drosophila; however, there is no blood vessel structure in this animal. The VEGFR in Drosophila was shown to regulate the migration of border-cells, hemocytes, and thoracic epithelial cells [27,38,39].
To the authors’ knowledge, this is the first report for a VEGFR in Halocynthia roretzi that express significantly in the tissues containing hearts and circulatory system. However, it is not confirmed yet whether the S. sq VEGFR is expressed in the epithelial cells surrounding vessel-like structure and hematopoietic cells in this animal. Further analysis of this molecule in Halocynthia roretzi (S. sq VEGFR) should shed more light on the function of VEGFR in angiogenesis.
3. Experimental Section
3.1. Animals
H. roretzi was selected as the ascidian in our experiment. After the surface of each individual was washed with Ca2+ and Mg2+-free artificial sea water (460 mM NaCl, 9 mM KCl, 32 mM Na2SO4, 5 mM HEPES (pH 7.0), 5 mM EDTA-2Na, and 6 mM NaHCO3), the tunicate was removed and the exposed inner walls were opened. We mainly picked up heart (with part of stomach), intestine, gill, and pharynx for histological analysis.
3.2. The Acid Guanidium-Phenol-Chloroform (AGPC) Method
The AGPC method (Chomczynski and Sacchi 1987) [31] was used to isolate total RNA from different tissues of H. roretzi. Fresh tissue from different parts of H. roretzi was minced on ice and then homogenized at room temperature with 1 mL of denaturing solution (4 M guanidium thiocynate, 25 mM sodium citrate, pH 7; 0.5% sarcosyl, and 0.1 M 2-mercaptoethanol; solution D) in a glass-Teflon.
3.3. cDNA Synthesis
First-Strand cDNA was synthesized according to the protocol of the SMART™ cDNA Library Construction Kit (Clontech, CA, USA). Next, we amplified the cDNA by long-distance PCR (LD PCR). Then we purified the PCR product according to the protocol of the Qiaquick PCR Purification Kit (Qiagen, Hilden, Germany). Finally, cDNA size fractionation was carried out using CHROMA SPIN-400 columns according to the manufacturer’s protocol.
3.4. Degenerated Oligonucleotide Primers
(A) Primers for isolation of a VEGFR-like tyrosine kinase gene in H. roretzi
Degenerated oligonucleotide primers (5′A: 5′-TGGCCATGGLGCLTTYGG-3′; 5′B: 5′-ACAGTATGCMGCMGTLAASATG-3′; 3′A: 5′-GTCCAACATGSTYTMSTANA-3′, and 3′B: 5′-CAAAAAGCCARACSTCSCT-3′, wherein R is G or A; S is C or G; Y is C or T; L is A,C,G or T; M is A or C; N is C or G) were designed based on the sequences of VEGFR genes highly conserved among various species.
(B) Primers for detection of β-actin mRNA in H. roretzi
Degenerated oligonucleotide primers (5′A: 5′-TGAAGCCAGAGCAAGAGAGG-3′; 5′B: 5′-GTATCGTAACCAACTGGGACG-3′; 3′A: 5′-TTTGCTGATCCACATCTGTTG-3′; 3′B: 5′-TTAGAAGCATTTGCGGTGAAC-3′) were designed based on the sequence of β-actin genes highly conserved among various species. Rapid amplification of cDNA ends of the 5′-or 3′-end was performed with these degenerate primers and anchor primers corresponding to the anchor sequence combined with the 3′-end of the oligo(dT) primer or the sequence involved in the 5′-end of the SMART IV oligonucleotide. The nucleotide sequences were determined using a BigDye Terminator cycle sequencing kit and an ABI310 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). To further extend the S. sq VEGFR sequence, we generated several primers based on partial sequences of S. sq VEGFR determined as the original, in both the 5′ and 3′ direction.
3.5. PCR
PCR was performed in a 20 μL reaction mixture containing 1 μL of cDNA, 5 μL of 10+PCR buffer, 0.2 mM of each dNTP, 1 mM of MgSO4, and 15 pmol of each primer. PCR was performed with one cycle of incubation at 94 °C for 2 min, followed by 35 cycles at 94 °C for 15 s and 50 °C for 30 s, with a final cycle at 68 °C for 5 min. The PCR products were separated by electrophoresis in a 1.5% agarose gel and then purified by gel extraction. Sequencing was performed with a PE-Biosystems BigDye terminator kit and an ABI Prism 377 XL sequenator.
3.6. Restriction Enzymes
HaeIII, Saw3AI, HinfI, and SspI as restriction enzymes have been used for checking the existence of Eph receptors as a positive control in H. roretzi.
3.7. Histological Analysis
Tissue samples were fixed in 4% paraformaldehyde (PFA). For Hematoxylin and Eosin staining, paraffin-embedded specimens were sectioned at a thickness of 5 μm and stained with hematoxylin and eosin using a standard protocol.
3.8. RT-PCR Detection of S. sq VEGFR mRNA
Total RNA samples were exacted from the different H. roretzi tissues (Heart with stomach tissue, intestine, pharynx, gill and muscle) using the AGPC method [31]. Reverse transcription was performed using the SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and dT 15 primer. PCR assay was carried out to detect the expression of the S. sq VEGFR gene in tissues. The PCR mixture contained reverse-transcribed DNA from the total RNA samples of ascidian tissues, Fw primer (50 pmol), Rw primer (50 pmol), dNTPs (20 pmol), PCR buffer, and rTaq DNA polymerase (TaKaRa) in 100 μL. The PCR conditions used were as follows: 94 °C for 4 min; 30 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 5 min. PCR with the β-actin Fw primer (5′TGAAGCCCAGAGCAAGAGAGG-3′) and β-actin Rw primer (5′CGTTCTTGATTCTGGCGATGG-3′) was used to detect the expression of the β-actin gene in the ascidian tissues, as the positive control. After finding the sequence of β-actin in H. roretzi, we generated a pair of primers from the β-actin sequence for RT-PCR (Fw, 5′TGAAGCCCAGAGCAAGAGAGG-3′, Rw, 5′CGTTCTTGATTCTGGCGATGG-3′).
3.9. Generation of Antibody
To generate antibody against the S. sq VEGFR protein, the 20-amino-acid peptide “CEGANSMKRGKRSDEDKDPD” which corresponds to a part of the kinase-insert region within the tyrosine kinase domain, was synthesized and conjugated with the KLH carrier. Rabbits were immunized several times with the conjugated protein, and antiserum was obtained.
3.10. Western Blotting
Proteins separated by SDS-PAGE were transferred to Immobilon P membrane. The membrane was blocked with 5% BSA in PBST buffer and incubated with purified primary antibody for 2 h. Corresponding horseradish peroxidase-conjugated secondary antibodies were used for detection of the primary antibody with chemiluminescent reagent.
DNA Data Base Accession number for S. sq VEGFR: GenBank AB374180.
4. Conclusions
For the first time, we molecularly cloned the VEGFR-like gene in Halocynthia roretzi, one of three basal chordates. S. sq VEGFR has a higher homology than the Amphioxus VEGFR-like molecule to human VEGFR. The S. sq VEGFR mRNA was expressed at higher levels in heart-localized tissue (stomach) and circulatory system-containing tissue (intestine), suggesting that S. sq VEGFR plays an important role in the formation or maintenance of circulatory system in tunicates, H. roretzi.
Acknowledgments
This work was supported by Grant-in-Aid Special Project Research on Cancer-Bioscience 17014020 from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and the program “Promotion of Fundamental Research in Health Science” of the Organization for Pharmaceutical Safety and Research (OPSR).
Conflict of Interest
There is no conflict of interest.
References
- 1.Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., de Tomaso A., Davidson B., di Gregorio A., Gelpke M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 2.Holland L.Z., Gibson-Brown J.J. The Ciona intestinalis genome: When the constraints are off. BioEssays. 2003;25:529–532. doi: 10.1002/bies.10302. [DOI] [PubMed] [Google Scholar]
- 3.Johnson D.S., Davidson B., Brown C.D., Smith W.C., Sidow A. Noncoding regulating sequences of ciona exhibit strong coreesespondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leveugle M., Prat K., Popovici C., Birnbaum D., Coulier F. Phylogenetic analysis of Ciona intestinalis gene super-families supports the hypothesis of successive gene expansion. J. Mol. Evol. 2004;58:168–181. doi: 10.1007/s00239-003-2538-y. [DOI] [PubMed] [Google Scholar]
- 5.Satou Y., Mineta K., Ogasawara M., Sasakura Y., Shoguchi E., Ueno K., Yamada L., Matsumoto J., Wasserscheid J., Dewar K., et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhan A., Macisaac H.J., Cristescu M.E. Invasion genetics of the Ciona intestinalis species complex: From regional endemism to global homogeneity. Mol. Ecol. 2010;19:4678–4694. doi: 10.1111/j.1365-294X.2010.04837.x. [DOI] [PubMed] [Google Scholar]
- 7.Wada S., Tokuoka M., Shoguchi E., Kobayashi K., di Gregorio A., Spagnuolo A., Branno M., Kohara Y., Rokhsar D., Levine M., et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev. Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- 8.Shoguchi E., Hamada M., Fujie M., Satoh N. Direct examination of chromosomal clustering of organ-specific genes in the chordate Ciona intestinalis. Genesis. 2011;49:662–672. doi: 10.1002/dvg.20730. [DOI] [PubMed] [Google Scholar]
- 9.Munoz-Chapuli R., Carmona R., Guadix J.A., Macias D., Perez-Pomares J.M. The origin of the endothelial cells: An evo-devo approach for the invertebrate/vertebrate transition of the circulatory system. Evol. Dev. 2005;7:351–358. doi: 10.1111/j.1525-142X.2005.05040.x. [DOI] [PubMed] [Google Scholar]
- 10.Gasparini F., Longo F., Manni L., Burighel P., Zaniolo G. Tubular sprouting as a mode of vascular formation in a colonial ascidian (Tunicata) Dev. Dyn. 2007;236:719–731. doi: 10.1002/dvdy.21073. [DOI] [PubMed] [Google Scholar]
- 11.Gasparini F., Burighel P., Manni L., Zaniolo G. Vascular regeneration and angiogenic-like sprouting mechanism in a compound ascidian is similar to vertebrates. Evol. Dev. 2008;10:591–605. doi: 10.1111/j.1525-142X.2008.00274.x. [DOI] [PubMed] [Google Scholar]
- 12.Tiozzo S., Voskoboynik A., Brown F.D., de Tomaso A.W. A conserved role of the VEGF pathway in angiogenesis of an ectodermally-derived vasculature. Dev. Biol. 2008;315:243–255. doi: 10.1016/j.ydbio.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolfi A., Gainous T.B., Young J.J., Mori A., Levine M., Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz-Chapuli R. Evolution of angiogenesis. Int. J. Dev. Biol. 2011;55:345–351. doi: 10.1387/ijdb.103212rm. [DOI] [PubMed] [Google Scholar]
- 15.Shibuya M., Yamaguchi S., Yamane A., Ikeda T., Tojo A., Matsushime H., Sato M. Nucleotide sequence and expression of a novel human receptor-type tyrosine kinase gene (flt) closely related to the fms family. Oncogene. 1990;5:519–524. [PubMed] [Google Scholar]
- 16.Shibuya M., Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp. Cell Res. 2006;312:549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Shibuya M. Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J. Biochem. Mol. Biol. 2006;39:469–478. doi: 10.5483/bmbrep.2006.39.5.469. [DOI] [PubMed] [Google Scholar]
- 18.De Vries C., Escobedo J.A., Ueno H., Houck K., Ferrara N., Williams L.T. The fms-like tyrosine kinase, a receptor for vascular endothelial growth factor. Science. 1992;255:989–991. doi: 10.1126/science.1312256. [DOI] [PubMed] [Google Scholar]
- 19.Alitalo K., Carmeliet P. Molecular mechanisms of lymphangiogenesis in health and disease. Cancer Cell. 2002;1:219–227. doi: 10.1016/s1535-6108(02)00051-x. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi T., Yamaguchi S., Chida K., Shibuya M. A single autophosphorylation site on KDR/Flk-1 is essential for VEGF-A-dependent activation of PLC-γ and DNA synthesis in vascular endothelial cells. EMBO J. 2001;20:2768–2778. doi: 10.1093/emboj/20.11.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakurai Y., Ohgimoto K., Kataoka Y., Yoshida N., Shibuya M. Essential role of Flk-1 (VEGF receptor 2) tyrosine residue 1173 in vasculogenesis in mice. Proc. Natl. Acad. Sci. USA. 2005;102:1076–1081. doi: 10.1073/pnas.0404984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland P.W., Garcia-Fernandez J., Williams N.A., Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994;1994:125–133. [PubMed] [Google Scholar]
- 23.Hirano T., Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi. I. Origin of mesodermal tissues of the juvenile. Dev. Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- 24.Hoch R.V., Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann F.G., Opazo J.C. Evolution of the relaxin/insulin-like gene family in placental mammals: Implications for its early evolution. J. Mol. Evol. 2011;72:72–79. doi: 10.1007/s00239-010-9403-6. [DOI] [PubMed] [Google Scholar]
- 26.Suga H., Hoshiyama D., Kuraku S., Katoh K., Kubokawa K., Miyata T. Protein tyrosine kinase cDNAs from amphioxus, hagfish, and lamprey: Isoform duplications around the divergence of cyclostomes and gnathostomes. J. Mol. Evol. 1999;49:601–608. doi: 10.1007/pl00006581. [DOI] [PubMed] [Google Scholar]
- 27.Duchek P., Somogyi K., Jekely G., Beccari S., Rorth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 28.Duloquin L., Lhomond G., Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134:2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y., Brock A.R., Wang Y., Fujitani K., Ueda R., Galko M.J. A blood-borne PDGF/VEGF-like ligand initiates wound-induced epidermal cell migration in Drosophila larvae. Curr. Biol. 2009;19:1473–1477. doi: 10.1016/j.cub.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 31.Satou Y., Imai K.S., Satoh N. FGF genes in the basal chordate Ciona intestinalis. Dev. Genes Evol. 2002;212:432–438. doi: 10.1007/s00427-002-0266-8. [DOI] [PubMed] [Google Scholar]
- 32.Wada H., Satoh N. Details of the evolutionary history from invertebrates to vertebrates, as deduced from the sequences of 18S rDNA. Proc. Natl. Acad. Sci. USA. 1994;91:1801–1804. doi: 10.1073/pnas.91.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsagkogeorga G., Turon X., Galtier N., Douzery E.J., Delsuc F. Accelerated evolutionary rate of housekeeping genes in tunicates. J. Mol. Evol. 2010;71:153–167. doi: 10.1007/s00239-010-9372-9. [DOI] [PubMed] [Google Scholar]
- 34.Oda H., Wada H., Tagawa K., Akiyama-Oda Y., Satoh N., Humphreys T., Zhang S., Tsukita S. A novel amphioxus cadherin that localizes to epithelial adherens junctions has an unusual domain organization with implications for chordate phylogeny. Evol. Dev. 2002;4:426–434. doi: 10.1046/j.1525-142x.2002.02031.x. [DOI] [PubMed] [Google Scholar]
- 35.Cañestro C., Bassham S., Postlethwait J.H. Seeing chordate evolution through the Ciona genome sequence. Genome Biol. 2003;4:208. doi: 10.1186/gb-2003-4-3-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gee H. Evolution: Careful with that amphioxus. Nature. 2006;439:923–924. doi: 10.1038/439923a. [DOI] [PubMed] [Google Scholar]
- 37.Lemaire P. Evolutionary crossroads in developmental biology: The tunicates. Development. 2011;138:2143–2152. doi: 10.1242/dev.048975. [DOI] [PubMed] [Google Scholar]
- 38.Ishimaru S., Ueda R., Hinohara Y., Ohtani M., Hanafusa H. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J. 2004;23:3984–3994. doi: 10.1038/sj.emboj.7600417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heino T.I., Karpanen T., Wahlstrom G., Pulkkinen M., Eriksson U., Alitalo K., Roos C. The Drosophila VEGF receptor homolog is expressed in hemocytes. Mech. Dev. 2001;109:69–77. doi: 10.1016/s0925-4773(01)00510-x. [DOI] [PubMed] [Google Scholar]