Abstract
Translational researchers conduct research in a highly data-intensive and continuously changing environment and need to use multiple, disparate tools to achieve their goals. These researchers would greatly benefit from meta-composite software development or the ability to continuously compose and recompose tools together in response to their ever-changing needs. However, the available tools are largely disconnected, and current software approaches are inefficient and ineffective in their support for meta-composite software development. Building on the composite services development approach, the de facto standard for developing integrated software systems, we propose a concept-map and agent-based meta-composite software development approach. A crucial step in composite services development is the modeling of users’ needs as processes, which can then be specified in an executable format for system composition. We have two key innovations. First, our approach allows researchers (who understand their needs best) instead of technicians to take a leadership role in the development of process models, reducing inefficiencies and errors. A second innovation is that our approach also allows for modeling of complex user interactions as part of the process, overcoming the technical limitations of current tools. We demonstrate the feasibility of our approach using a real-world translational research use case. We also present results of usability studies evaluating our approach for future refinements.
Keywords: Data integration, systems integration, meta-composite, translational research, informatics
INTRODUCTION
The National Institutes of Health categorizes translational research into two areas of translation: one is the process of applying discoveries generated during research in the laboratory and in preclinical studies to the development of trials and studies in humans. The second area of translation concerns research aimed at enhancing the adoption of best practices in the community [1]. A core need for translational researchers is the ability to connect data bidirectionally across these scientific domains, from laboratory to clinical to community and back. In a needs assessment conducted at the University of Alabama at Birmingham [2], researchers reported having to interact with multiple tools to conduct their research, which resulted in a loss of efficiency, as well as potentially introducing errors. Over the last decade or more, several infrastructure tools that are largely disconnected and disjointed have been created [3]. Thus, meta-composite software tools — tools that would support connecting data across multiple tools and simplify user interactions — would greatly increase the productivity of researchers. Elaborations of research use cases with individual research groups further illustrated this need for us. An example use case is below.
Use Case Highlighting Need for Meta-Composite Software Development
This use case was developed in collaboration with genomic researchers at the University of Alabama at Birmingham. A task of genomic researchers is to discover candidate genes for chromosome regions of interest identified in genetic linkage studies. This task of identifying candidate genes is cumbersome and time-consuming despite the availability of sophisticated tools aiding this effort. Currently, the National Center of Biotechnology (NCBI) MapViewer tool allows downloading lists of genes for chromosome regions of interest, while PubMed and Online Mendelian Inheritance in Man (OMIM) websites facilitate the searching and downloading of relevant articles for particular genes. The researchers first search for information for all genes in the chromosome region. They then organize and iteratively search through the information using keywords of interest. An example [4] highlights the difficulty of this effort: for a region from 60 to 120 cM in Chromosome 18, there are 230+ annotations out of which 136 are genes, 70 are hypothetical genes, 16 are open reading frames (ORFs), 10 are pseudo genes, 3 are micro RNAs, and 2 are anticodons. The number of PubMed articles: 4181, OMIM citations: 121. Thus, despite the availability of supportive tools, this use case is time-consuming, sometimes taking months to complete.
The above use case can be thought of as a series of steps that requires passing data through several programs in sequence. The data needs to be formatted to conform to application-dependent file formats and then passed through selected scientific applications or services, which yield a handful of results or generate new data. This new data in turn requires reformatting and passing through other services. Often, researchers have to transfer results manually between tools by noting these values and re-keying them into a new interface or by cutting and pasting. In other words, simply providing mechanisms to access biological data is not sufficient. Researchers also need the ability to integrate data across software tools and research methods.
Meta-composite software tools that connect multiple systems and simplify user interactions would make the above use case more efficient. The tool we developed — GeneRECQuest (GENome REgion Characterization QUEST) tool [4] — automates and simplifies the manual steps involved in the use case. However, this use case is just one instance of the several approaches using multiple tools and databases utilized by these translational researchers. These use cases are also constantly evolving. It took us almost a year to develop GeneRECQuest despite utilizing techniques such as rapid prototyping and simplifying development using service-oriented design and web services. Our software development effort was an example of the inefficiencies in the development of meta-composite software tools.
In this paper, we describe a concept-map and agent-based modeling approach to develop meta-composite systems building on the work of Gurupur et al. [5–6]. Instead of technicians leading the modeling effort as is currently done, we focus on providing a structured modeling approach, in which domain experts who best understand their processes can take a leadership role in the modeling of their needs, and then generating executable specifications from these models for meta-composite software development. We demonstrate how our modeling approach can be used to model a real-world translational research use case and generate executable specifications to develop a meta-composite software system.
2. CHALLENGES PREVENTING META-COMPOSITE SOFTWARE DEVELOPMENT
Our approach uses the composite services model, which includes the development of large systems using service-oriented approaches [7–9]. Composite services development is currently the de facto standard for developing large and integrated software systems. Composite services development focuses on specifying, discovering, selecting, and integrating services. Below, we highlight the limitations of current software development that prevent efficient and effective meta-composite software development.
Composite services development is mainly comprised of three stages: 1) Process modeling, in which user needs are captured and represented as process models for composition. Included in these process models are the various tools with which users need to connect to complete their tasks. Examples are models created using Business Process Execution Language (BPEL) and Business Process Modeling (BPM). 2) Process composition, in which the process models are composed, including linking with the various tools that users need to realize software systems. 3) Process analysis and optimization, in which the composite services are analyzed and optimized.
Process modeling is a crucial first step as the rest of the software development is dependent on it. It can be thought of as two phases: the abstract and the executable. Abstract process modeling starts with a high-level representation of user needs with few details [10]. Abstract processes are developed until models with details sufficient to generate executable processes are reached. Usually, tools such as Microsoft Visio or Unified Modeling Language are used to develop abstract process models. The abstract process models are then used to develop executable process models. Executable process models provide an executable representation of the process that can be directly composed using a composition engine. Examples are models developed using tools like BPEL or BPM.
A number of limitations exist that prevent meta-composite software development. First, many of the tools that we need have disparate interfaces that make it difficult to integrate them. However, a number of web services and semantic markup techniques have been developed to address this issue [9, 11–13]. Thus, an infrastructure of resources that can be continuously composed and recomposed as part of meta-composite software development can be developed.
A second limitation is that while a number of modeling techniques, including semantic modeling approaches, have been developed [8, 14–15], none provide adequate support for meta-composite software development. There are two main reasons for this shortcoming. First, three main types of interactions exist in a workflow: 1) mechanistic or computer-computer (e.g., computer-database, software-software), 2) user-computer (e.g., people interacting with software), and 3) user-user (e.g., people interacting with one another either directly or through software) [16]. Current process modeling tools are generally designed to model mechanistic interactions [17]. However, very few real processes are simply a sequence of mechanistic connections; real processes usually involve steps in which the task is dependent on user interaction. Thus, the process models developed using these tools are limited representations of the actual processes. This problem is often referred to as a semantic gap between the user needs and the implemented process [18]. Consequently, additional support for the process steps involving user interaction has to be developed ad hoc, outside the process composition, limiting the ability to compose or recompose the process models for meta-composite software development. The second reason the current modeling techniques do not provide adequate support for meta-composite software development is that these tools are designed to support process models developed by technicians [17]. Thus, a domain expert (e.g., a translational researcher), who best understands the process, has difficulty contributing to the development of these models [17]. As noted above, our GeneRECQuest software development took nearly a year to complete, and the majority of the time was spent trying to understand the research process. What would be ideal is a modeling tool that allows researchers to model their needs, including steps to interact with particular tools, and generates the executable specifications for composition. Technicians then would not have to spend considerable time trying to understand the researchers’ processes and instead could focus their efforts in more productive activities. We address this second limitation in this paper.
3. A CONCEPT-MAP BASED APPROACH
Building on the work of Gurupur et al. [5–6], we have developed a concept-map approach to model researchers’ needs that can then be transformed into executable models. A key difference between our modeling approach and others is that we focused on providing a structured approach for domain experts to lead the modeling effort. As described in the above section, other modeling approaches have limitations that prevent domain experts from contributing to the development of process models. Our concept-map approach enables modeling of complex user interactions as part of the process, overcoming the technical limitations of current tools.
Concept maps support our approach in multiple ways. Concept maps are powerful tools for eliciting and communicating knowledge about a complex domain [19–20]. They are graphical tools for organizing and representing knowledge [21]. Concept maps include concepts, usually enclosed in circles or boxes of some type, with relationships between concepts designated by lines linking concepts. Words on the lines, referred to as linking words or linking phrases, specify the relationship between the concepts [21]. Concept maps provide a concise representation of information for human use, since these diagrams provide a means of representation between that of traditional representations (which are hard to capture and require intervention by knowledge engineers) and text (which may obscure the structure of a domain) [22]. Concept maps are easily understandable by domain experts, not just by software developers [21, 23–24], thus allowing domain users to actively participate in process modeling, which can result in better representation of the users’ needs. Because of the intuitive nature of concept maps, domain experts can even lead the creation of models [25]. Finally, the conversion of concept maps to executable specifications can be automated [5–6, 26].
To guide concept-map developers, we have created a “service agents and resources” concept model based on the task-system model developed by Coffman and Denning [27]. The task-system model was originally developed to model the processes of an operating system [27], but later adapted to model the processes of large organizations [28–30]. A process is made up of tasks and associated resources [27]. A task constitutes the unit of compositional activity in a process. The task is specified in terms of its external behavior, such as the input it requires, the output it generates, its action or function, and its execution time. Resources are any (not necessarily physical) device, which is used by tasks.
The central concept in our approach is the service-agents concept, which maps to a task in a process. Service agents can be either a mechanistic service agent or a people service agent. Mechanistic service agents imply the interaction in the task is between computers. They are abstractions of composable resources, such as services, methods, and programs (mechanistic interactions in a process). People service agents are abstractions of people, implying that the task involves interactions between an individual or a group of users and service functions (user interactions in a process). We have also defined six resource concepts to model the resources that a service agent might need to perform its task: Knowledge, Rule, Role, User, Infrastructure, and Communication. Knowledge-Concept (KC) provides specific information that is needed to interact with a task. For example, in the use-case discussed in this paper, researchers need to enter a series of input to the NCBI MapViewer tool to download a list of gene files from it. Researchers need to understand what input to provide or what information is available in the gene’s list file to execute the task and move forward to the next. Rules-Concept (RuC) provides specific conditions to execute a task. For example, there are decision points at which a researcher might opt to ignore a particular gene. We have included the RuC to capture such information. Roles-Concept (RoC) provides a representation of the user or group of users interacting with a task and their access permissions. For example, certain tasks may be or need to be accessible to certain user groups. In this case, the RoC is used to specify that the task is only accessible to that group. User-Concept (UC) provides a profile of the users associated with a task and their preferences. The UC could be simple information collected during a login or more sophisticated preferences. Users are considered as a resource in our model because we consider them as part of the process. For a task to execute, users may have to push a button, make a decision, and perform a service. In this view, their function is similar to the function of other resources (e.g., a web service). Considering a user as a service allows us to model them as part of the process, overcoming limitations of other modeling tools. This distinction is a key difference between our modeling approach and others that consider users only in the context of using a resource. Infrastructure-Concept (InC) provides a representation of the mechanistic resources and tools that are used to perform a task. An example is a particular database from which the system needs to access data. Communication-Concept (CoC) provides a representation of the input and output messages that are involved in performing a task.
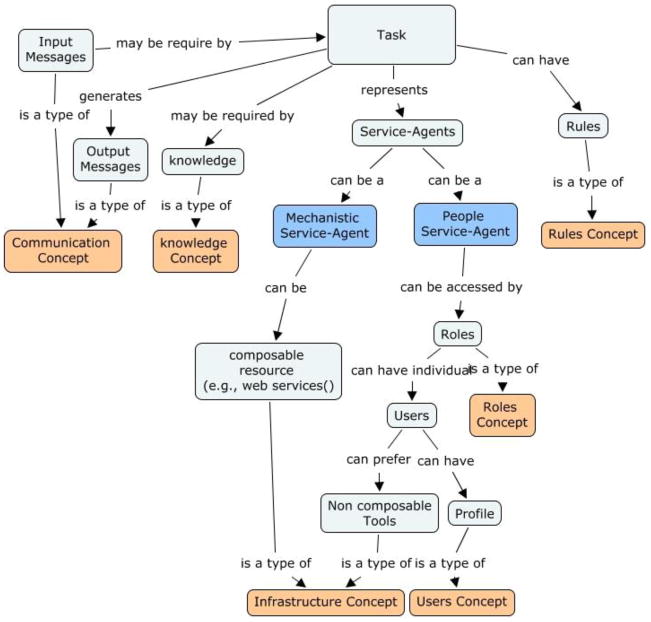
Figure 1 is a concept-map representation of service agents and connected resources. The color codes in the Figure are used only to differentiate between a task and its resources, service agents, and concepts. A task represents a service agent. As described, the service agents can be of two types: mechanistic or people. Mechanistic service agents are abstractions of composable resources, such as services, methods, and programs. Mechanistic service agents specify that the interaction is a computer interaction. People service agents are abstractions of human activity. People service agents specify that the interaction is a user interaction. Every task requires input messages for operation and generates output messages. The input and output messages are types of CoC. A service agent may require KC for performing a task. RuC specify the conditions necessary to carry out a task. The concept map also shows that a people service agent can be accessed by certain roles of users that are specified by RoC. Each role can have a set of users specified by UC. Users can use certain tools, a type of InC, to perform their tasks. These describe non-composable tools that are used by users, such as Microsoft Word or Excel. Users can also have a profile, which can be used to personalize their interaction with the process. The representation of the tasks of a process using the relationships described in the concept map is demonstrated in the case study examples.
Figure 1.
Concept-Map depicting the relationships between a task, service-agents, and resources
4. CASE STUDY – MODELING THE GENETIC LINKAGE PROCESS
Our case study is an expansion of the use case described in Section 1. To identify candidate genes for genetic linkage studies, the researchers with whom we worked typically undertake several steps:
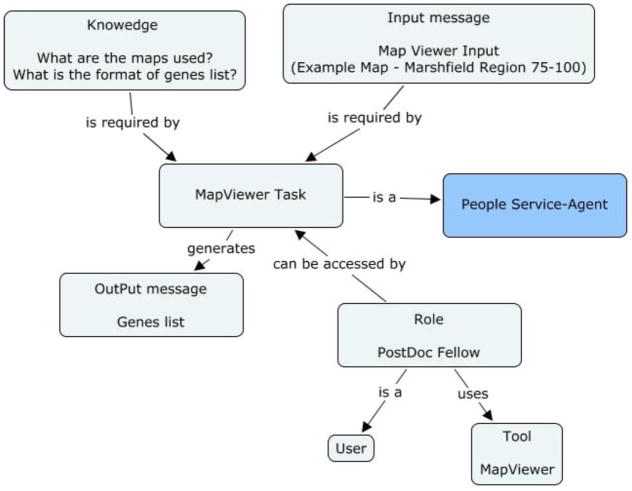
The researcher visits the NCBI Web site and uses a tool called MapViewer. MapViewer shows integrated views of chromosome maps for many organisms, including human and numerous other vertebrates, invertebrates, fungi, protozoa, and plants [31]. The researcher uses MapViewer to obtain the chromosome maps and then to study the genes in a specific region of the chromosome. After making a series of selections through several Web pages, the researcher can select the specific region of the chromosome under investigation and also a few other details on the viewing style. The tool finally produces a page from which the genes on the sequence in the selected region of the chromosome can be downloaded. This downloaded document is a tab-delimited file and is most readable in spreadsheet applications, such as Microsoft Excel.
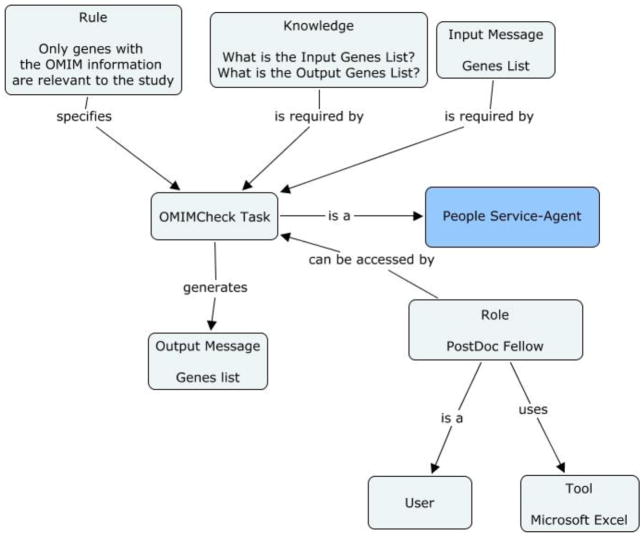
On studying this document downloaded from MapViewer, the researcher identifies the genes that have associated OMIM information. OMIM is a NCBI database that lists all the known diseases and the related genes in the human genome [31]. The researcher then manually goes to the NCBI OMIM search interface and looks up the OMIM information for each gene. Similarly, each gene in the list can be associated with one or multiple PubMed documents. PubMed is an NCBI database that stores citations for biomedical articles [31]. When the researcher types a gene into the search interface of PubMed, it displays a list of all documents from the database that cite that particular gene. Each document will have summaries or abstracts of the article.
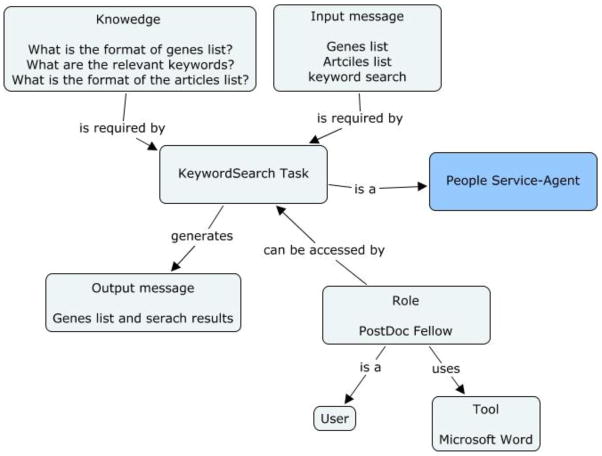
The OMIM and the PubMed information associated with each gene offers vital information for the researcher to discover candidate genes for further research. This step is done by carrying out a key-word search on the retrieved PubMed and OMIM information. The genes that yield “hits” during the search are those genes that are identified as candidate genes. Other genes are discarded.
The tasks and resources of the genetic linkage process were identified and then categorized as service agents and resource concepts (Table 1). The concept maps in Figures 2a–e depict the relationship between the service agents and resources for each task. The IMHC COE knowledge modeling kit (version 5.0.03) was used to develop the concept-map models. The concept map of the articles task in Web ontology language (OWL) format is provided (Figure 3).
Table 1.
Tasks, service agents and resources of the genetic linkage process
| Tasks | Concepts | Description |
|---|---|---|
| RuC | N/A | |
| RoC | The post-doctoral fellows of the Department of Epidemiology are assigned to the task. | |
| UC | Dr. XXX, a post-doctoral fellow of the Department of Epidemiology, is a user of the task. She uses the NCBI Map View tool to perform the task. | |
| InC | NCBI Map Viewer tool. | |
| CoC | Input message: The map and region required to invoke the Map Viewer tool. Output message: The genes list. |
|
| OMIMCheck task | KC |
|
| RuC | In the input genes list, the “LINKS” column indicates the presence or absence of OMIM information. Textually, it is represented as “OMIM” in the “LINKS” column to indicate the presence of OMIM information. An empty field in the “LINKS” column indicates the absence of OMIM information. | |
| RoC | The post-doctoral fellows of the Department of Epidemiology are assigned to the task. | |
| UC | Dr. XXX, a post-doctoral fellow of the Department of Epidemiology, is a user of the task. She uses the Microsoft Excel tool to perform the task. | |
| InC | The Microsoft Excel tool, which is used to view and manipulate the genes list. | |
| CoC | Input message: The genes list. Output message: The modified genes list created after the genes without OMIM information are removed. |
|
| Articles task | KC | N/A |
| RuC |
|
|
| RoC | N/A | |
| UC | N/A | |
| InC |
|
|
| CoC | Input message: The modified genes list created after the genes without OMIM information are removed. Output message: the articles retrieved from the OMIM and PubMed database service. |
|
| KeywordSearch task | KC |
|
| RuC | N/A | |
| RoC | The post-doctoral fellows of the Department of Epidemiology are assigned to the task. | |
| UC | Dr. XXX, a post-doctoral fellow of the Department of Epidemiology, is a user of the task. She uses the Microsoft Word tool to perform the task. | |
| InC | Microsoft Word tool that is used to view the articles and store the results of the keyword of interest search. | |
| CoC | Input message: The keywords of interest. Output message: The search results for the keyword of interest search. |
|
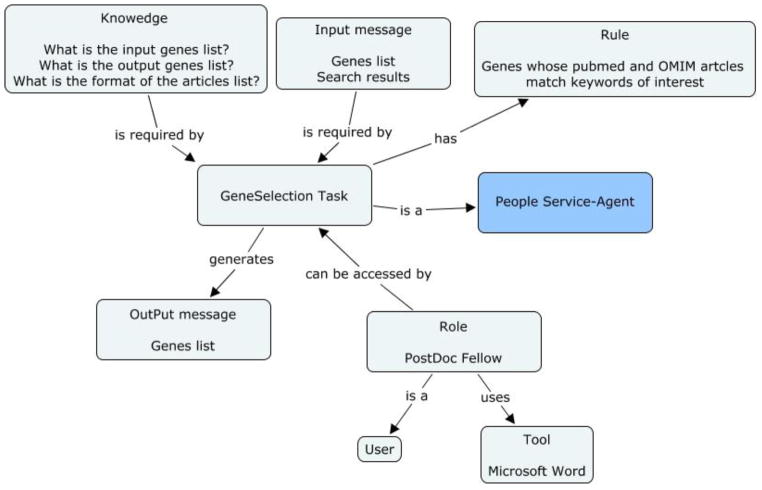
| GeneSelection task | KC |
|
| RuC | Genes whose PubMed and OMIM summary matched the keyword of interest relevant to the study are the set of genes for continued study. | |
| RoC | The post-doctoral fellows of the Department of Epidemiology are assigned to the task. | |
| UC | Dr. XXX, a post-doctoral fellow of the Department of Epidemiology, is a user of the task. She uses the Microsoft Word tool to perform the task. | |
| InC | Microsoft Word tool is the tool that is used to view the articles and store the results of the keyword of interest search. | |
| CoC | Input message: 1) The input genes list. 2) The matched articles, which is the results of the keyword search task. Output message: The output genes list. |
Figure 2.
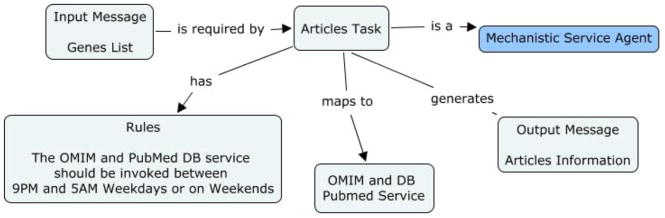
Figure 2a: Concept map of MapViewer task in the genetic linkage use case
Figure 2b: Concept map of OMIMCheck task in the genetic linkage use case
Figure 2c: Concept map of Articles task in the genetic linkage use case
Figure 2d: Concept map of KeywordSearch task in the genetic linkage use case
Figure 2e: Concept map of GeneSelection task in the genetic linkage use case
Figure 3.
Concept map of the articles task specified in Web ontology format
5. Usability Evaluation
In a pilot usability study, we evaluated our approach in individual sessions (n=4) with researchers at the University of Alabama at Birmingham with the goal of understanding whether users will be able to understand the service agents and resource concepts and be able to use them to model a research process. We adapted the Think Aloud approach as detailed by Kushniruk [32–33]. In this approach, as participants review the content, they are asked to vocalize thoughts, feelings, and opinions. Think Aloud allows understanding of how users might approach the interface and what considerations they may have in their minds when utilizing the interface.
To our participants, we provided an overview of our approach and gave a description of the different resources. We also provided screenshots of the various tasks we modeled of the above use case. We asked users to review the screenshots and vocalize any thoughts they may have. We prompted them if we needed elaborations of any particular point. Overall, the researchers liked our approach and could understand our models. They thought it was a promising approach and that such a tool would be very helpful. However, they did ask for clarification of some of the resources. For example, they asked for clarification on what information would be included in the KC versus the RuC and provided suggestions on how we can better define these resources, such as KC as being something that a researcher would need to know to interact with the tools, while RuC would be conditional logic that a tool or the researcher would need to make a branching decision. One researcher was not sure that KC was the right term and wondered whether “tips” would be more appropriate as the examples we provided sounded more like tips than pieces of knowledge. He suggested that clarifying the concepts with training videos would improve the usability of the tool. Another researcher asked whether a batch file with a series of inputs would be classified as an input CoC or an InC because the batch file would need to be composed into the system. Examples of additional resources were also suggested. For example, researchers talked about the need to connect gene information with tools, such as GeneNote (to check whether the genes are expressed in tissues related to interest). Researchers talked about their potentially being multiple tools for the same function and different researchers preferring different tools. For example, Ingenuity, WebGestalt, or GeneSpring can be used check whether each gene is enriched or functional. We are incorporating these inputs into our future versions.
6. DISCUSSION AND FUTURE WORK
Translational researchers conduct research in highly changable environments [17]. They are problem solvers. They need to adapt to constant change and use several tools in their efforts. Their work cannot be accomplished following guidelines or completing individual tasks; they interact with multiple systems to perform their tasks. Many researchers rely on writing perl-scripts to integrate systems together and simplify their workflow. More sophisticated research groups have access to software engineers to help them build Web services and workflow tools to connect systems together. However, this model is not sustainable as most research groups do not have these resources to develop in-house solutions.
Two challenges must be met when developing meta-composite tools for biomedical researchers [34]. First, retrieval of data alone is not sufficient. The challenge is to manipulate the retrieved data to investigate specific biomedical problems. Secondly, merely providing a library package that interfaces to a large number of databases and computation software is not useful if it requires long and tedious programming.. Existing software development approaches fall short of addressing the needs of integrated systems [34–38]. Mainly, these approaches address problems in discovering and accessing biological data only. They ignore the problem of integration of data sources together with computation tools based on workflows to solve biological problems (i.e., meta-composition).
While current software development approaches do not fully support meta-composite software development, they do provide building blocks toward it. Web services and service-oriented architecture tools have simplified the life-cycle of system development [9]. Semantic web technologies even promise to enable the automated composition of systems [7–8, 13]. Process-oriented technologies (e.g., BPEL) allow us to drive systems development from a workflow perspective.
In this paper, we have described a concept-map approach that will facilitate meta-composite software development and build systems addressing the integration needs of translational researchers quickly and effectively. We have demonstrated how our approach can be used to model and generate executable process specifications for a real-world translational use case. Our overall goal is to develop a user-friendly tool that allows researchers to define tasks and service-agents and drag and drop different resources that are needed by the tasks. As an extension of the work described in this paper, we are further refining our parsers to map the different concepts into executable process entities so that meta-composite systems can be composed from them. We are also further refining our resources concepts based on the results of our usability evaluation.
As described above, our approach has two major innovations. First, it allows researchers to actively contribute and even lead the development of process models, reducing inefficiencies in the development of process models. Since the researchers understand their processes best, their resulting models may have fewer errors. In addition, technicians can focus on the more worthwhile effort of developing infrastructure that can be used as a resource for composition. Second, because our approach enables the representation of user interactions in the process model, our models provide a comprehensive view of the “real” process, enabling the ability to compose or recompose them for meta-composite software development.
Our ultimate goal is to develop a meta-composite software development tool that allows researchers to drag and drop services-agents and resources and develop their models. As we develop this tool, we will conduct further usability evaluation studies to validate our tool, i.e., to ensure that the “right” system is developed for the user with respect to meeting real-world needs [39]. Because of the inherent complexities in developing a meta-composite software development tool, our usability goals cannot be met by using a single approach. Thus, we will use a combination of quantitative and qualitative approaches. As described, the Think Aloud approach provides an understanding of how a user might approach the tool interface and what considerations they may have in their minds when utilizing the interface [32–33]. This approach is useful in gathering users’ perspectives during the design of the tool.
A more comprehensive usability evaluation includes assessing the tool for its efficiency, effectiveness, and satisfaction [40]. Effectiveness has been defined as the accuracy and completeness with which users achieve specified goals [41]. Accuracy measures quantify the number of errors users make either during the process of completing tasks or in the solution to the tasks, while completeness refers to measures of the extent to which tasks are solved. Efficiency has been defined as the resources expended in relation to the accuracy and completeness with which users achieve goals. Satisfaction is both freedom from discomfort and positive attitudes toward the user of the product. A more detailed explanation of these measures is provided by Hornbaek [40].
A popular approach to evaluate usability is task analysis [40, 42], which allows for assessment of completeness, accuracy, and efficiency of a tool. In this approach, users are asked to complete certain tasks using the tool being evaluated. An example task could be to model a research workflow. Completeness can be assessed by documenting the number of tasks users are able to complete, accuracy by documenting the number of errors committed during completion of these tasks, and efficiency by the time taken to complete the tasks. One challenge for our evaluation is that we do not have a gold standard measurement with which to compare our tool. However, we can complement the quantitative measures of task analysis evaluation with a qualitative evaluation assessing the users’ acceptance and satisfaction with the tool. We may also be able to generate more objective completeness and accuracy measures for the models developed during the task completion evaluation by simulating them under different conditions [30, 43–44]. The simulation parameters for these tests would be based on input gathered from users.
7. CONCLUSION
Translational researchers need meta-composite software development or the ability to compose multiple tools together to integrate data across these tools. Current software development approaches have limitations that prevent meta-composite software development. We have presented a concept-map and agent-based modeling approach that address limitations of current software development approaches. We demonstrated the feasibility of our approach using a real research case study, and we have conducted usability evaluations for future refinements.
Acknowledgments
We would like to thank our colleagues, especially Dr. Donna Arnett at the University of Alabama at Birmingham School of Public Health Epidemiology Department for providing the problem and supporting the development and evaluation of the modeling approach. We also thank Dr. Chittoor V. Ramamoorthy for directing our research in the area of meta-composition.
References
- 1.National Institutes of Health. Institutional Clinical and Translational Science Award (U54) Mar2007. Dec 3; Available: http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-07-007.html.
- 2.England B, et al. Insight Findings and Recommendations for University of Alabama at Birmingham. Mar 21, 2007. [Google Scholar]
- 3.Buetow KH. Cyberinfrastructure: empowering a “third way” in biomedical research. Science. 2005 May 6;308:821–4. doi: 10.1126/science.1112120. [DOI] [PubMed] [Google Scholar]
- 4.Sadasivam RS, et al. Genetic region characterization (Gene RECQuest) - software to assist in identification and selection of candidate genes from genomic regions. BMC Res Notes. 2009;2:201. doi: 10.1186/1756-0500-2-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurupur VP, et al. Enhancing medical research efficiency by using concept maps. Adv Exp Med Biol. 2011;696:581–8. doi: 10.1007/978-1-4419-7046-6_59. [DOI] [PubMed] [Google Scholar]
- 6.Gurupur VP, Tanik MM. A System for Building Clinical Research Applications using Semantic Web-Based Approach. J Med Syst. 2010 Feb 24; doi: 10.1007/s10916-010-9445-8. [DOI] [PubMed] [Google Scholar]
- 7.Alesso HP, Smith CF. Developing Semantic Web services. Natick, Mass: A K Peters; 2005. [Google Scholar]
- 8.Mandell DJ, McIlraith SA. Adapting BPEL4WS for the semantic web: The bottom-up approach to web service interoperation. Semantic Web - Iswc 2003. 2003;2870:227–241. [Google Scholar]
- 9.Newcomer E, Lomow G. Understanding SOA with Web services. Upper Saddle River, NJ: Addison-Wesley; 2005. [Google Scholar]
- 10.Duan Z, et al. A model for abstract process specification, verification and composition. New York, NY, USA: 2004. pp. 232–241. [Google Scholar]
- 11.Fensel D. Semantic web services. 1. New York: Springer; 2011. [Google Scholar]
- 12.Studer R, et al. Semantic Web Services: Concepts, Technologies, and Applications. Springer; 2010. [Google Scholar]
- 13.Rao JH, Su XM. A survey of automated web service composition methods. Semantic Web Services and Web Process Composition. 2005;3387:43–54. [Google Scholar]
- 14.WS-BPEL Extension for People. 2005 Aug 15; Available: http://www.ibm.com/developerworks/webservices/library/specification/ws-bpel4people/
- 15.Ludäscher B, et al. Scientific workflow management and the Kepler system: Research Articles. Concurr Comput: Pract Exper. 2006;18:1039–1065. [Google Scholar]
- 16.Ramamoorthy CV. A study of the service industry - functions, features, and control. ICICE Transactions Communications. 2000 May;E83-B:885–903. [Google Scholar]
- 17.Harrison-Broninski K. Human Interactions: The Heart and Soul of Business Process Management. Tampa, FL: Meghan-Kiffer Press; 2005. [Google Scholar]
- 18.Yeh R, et al. A systemic approach to process modeling. Journal of Systems Integration. 1991;1:265–282. [Google Scholar]
- 19.Novak JD. Learning, Creating, and Using Knowledge: Concept Maps(tm) As Facilitative Tools in Schools and Corporations. Routledge: 1998. [Google Scholar]
- 20.Novak JD, Gowin DB. Learning How to Learn. Cambridge University Press; 1984. [Google Scholar]
- 21.Novak JD, Cañas AJ. Florida Inst for Human and Machine Cognition, Tech Rep: IHMC CmapTools. Jan, 2006. The theory underlying concept maps and how to construct them. [Google Scholar]
- 22.Leake DB, et al. Aiding knowledge capture by searching for extensions of knowledge models. presented at the K-CAP; Sanibel Island, Florida. 2003. [Google Scholar]
- 23.Richardson R, et al. Using Concept Maps as a Tool for Cross-Language Relevance Determination. 2007. [Google Scholar]
- 24.Richardson R, et al. Evaluating concept maps as a cross-language knowledge discovery tool for NDLTD. Proceedings ETD; Sydney. 2005. [Google Scholar]
- 25.Feinman A, et al. Using Formalized Concept Maps To Model Role-based Workflows. Concept Maps: Theory, Methodology, Technology: Second Int. Conference on Concept Mapping; San José, Costa Rica. 2006. [Google Scholar]
- 26.Institute for Human and Machine Cognition COE tool. 2011 Jun 13; Available: http://www.ihmc.us/groups/coe/
- 27.Coffman EG, Denning PJ. Operating Systems Theory. Englewood Cliffs, NJ: Prentice Hall; 1973. [Google Scholar]
- 28.Delcambre S, Tanik MM. Using task system templates to support process description and evolution. Journal of Systems Integration. 1998;8:83–111. [Google Scholar]
- 29.Mills SF. PhD dissertation. Eng. and Appl. Sci., Southern Methodist Univ; Dallas, TX: 1997. A resource-focused framework for process engineering. [Google Scholar]
- 30.Sadasivam RS. An Architecture Framework for Process-PersonalizedComposite Services: Service-oriented Architecture, Web Services, Business-Process Engineering, and Human Interaction Management. VDM Verlag; 2008. [Google Scholar]
- 31.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004 Jan 1;32:35–40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kushniruk AW, Patel VL. Cognitive computer-based video analysis: its application in assessing the usability of medical systems. Medinfo. 1995;8(Pt 2):1566–9. [PubMed] [Google Scholar]
- 33.Kushniruk AW. Analysis of complex decision-making processes in health care: cognitive approaches to health informatics. J Biomed Inform. 2001 Oct;34:365–76. doi: 10.1006/jbin.2001.1021. [DOI] [PubMed] [Google Scholar]
- 34.Wong L. Technologies for integrating biological data. Brief Bioinform. 2002 Dec;3:389–404. doi: 10.1093/bib/3.4.389. [DOI] [PubMed] [Google Scholar]
- 35.Jagadish HV, Olken F. Report of the NSF/NLM Workshop on the Data Management for Molecular and Cell Biology, Workshop Report LBNL-52767. Nov 4, 2003. Data Management for the BioSciences. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez T, Kambhampati S. Integration of biological sources: current systems and challenges ahead. SIGMOD Rec. 2004;33:51–60. http://doi.acm.org/10.1145/1031570.1031583 “. [Google Scholar]
- 37.Venkatesh TV, Harlow HB. Integromics: challenges in data integration. Genome Biol. 2002 Jul 17;3:REPORTS4027. doi: 10.1186/gb-2002-3-8-reports4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein LD. Integrating biological databases. Nat Rev Genet. 2003 May;4:337–45. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 39.Thomas H, Pedro M. VVT terminology: a proposal. IEEE Expert: Intelligent Systems and their Applications. 1993;8:48–55. [Google Scholar]
- 40.Hornbaek K. Current practice in measuring usability: Challenges to usability studies and research. International Journal of Human-Computer Studies. 2006;64:79–102. [Google Scholar]
- 41.ISO. Ergonomic requirements for office work with visual display terminals (VDTs) -- Part 11: Guidance on usability (ISO 9241-11:1998) 1998 Available: http://www.iso.org/iso/home/store/catalogue_tc/catalogue_detail.htm?csnumber=16883.
- 42.Fr E, et al. Measuring usability: are effectiveness, efficiency, and satisfaction really correlated?. presented at the Proceedings of the SIGCHI conference on Human Factors in Computing Systems; The Hague, The Netherlands. 2000. [Google Scholar]
- 43.Zhang H, et al. Reflections on 10 Years of Software Process Simulation Modeling: A Systematic Review. In: Wang Q, et al., editors. Making Globally Distributed Software Development a Success Story. Vol. 5007. Springer; Berlin Heidelberg: 2008. pp. 345–356. [Google Scholar]
- 44.Sierhuis M, Clancey WJ. Modeling and simulating practices, a work method for work systems design. Intelligent Systems, IEEE. 2002;17:32–41. [Google Scholar]