Abstract
Approximately 40 single nucleotide polymorphisms (SNPs) that are associated with prostate cancer (PCa) risk have been identified through genome-wide association studies (GWAS). However, these GWAS-identified PCa risk-associated SNPs can explain only a small proportion of heritability (~13%) of PCa risk. Gene–gene interaction is speculated to be one of the major factors contributing to the so-called missing heritability. To evaluate the gene–gene interaction and PCa risk, we performed a two-stage genome-wide gene–gene interaction scan using a novel statistical approach named “Boolean Operation-based Screening and Testing”. In the first stage, we exhaustively evaluated all pairs of SNP–SNP interactions for ~500,000 SNPs in 1,176 PCa cases and 1,101 control subjects from the National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) study. No SNP–SNP interaction reached a genome-wide significant level of 4.4E–13. The second stage of the study involved evaluation of the top 1,325 pairs of SNP–SNP interactions (Pinteraction < 1.0E–08) implicated in CGEMS in another GWAS population of 1,964 PCa cases from the Johns Hopkins Hospital (JHH) and 3,172 control subjects from the Illumina iControl database. Sixteen pairs of SNP–SNP interactions were significant in the JHH population at a Pinteraction cutoff of 0.01. However, none of the 16 pairs of SNP–SNP interactions were significant after adjusting for multiple tests. The current study represents one of the first attempts to explore the high-dimensional etiology of PCa on a genome-wide scale. Our results suggested a list of SNP–SNP interactions that can be followed in other replication studies.
Introduction
Prostate cancer (PCa) ranks top in incidence of cancers affecting men in western countries. Among the components that contribute to PCa etiology, genetic factor is a major risk factor, with a heritability estimate of 42% (Lichtenstein et al. 2000). Genome-wide association study (GWAS) is a powerful approach to identify genetic loci that are associated with diseases or QTL traits. Until now, approximately forty PCa risk-associated single nucleotide polymorphisms (SNPs) have been identified using GWAS approaches in European populations (Amundadottir et al. 2006; Duggan et al. 2007; Eeles et al. 2008, 2009; Gudmundsson et al. 2007, 2008, 2009; Halldorsson et al. 2007; Hsu et al. 2009; Kote-Jarai et al. 2011; Sun et al. 2008, 2009; Thomas et al. 2008; Yeager et al. 2007, 2009; Zheng et al. 2009).
Although this essential single-gene approach is successful in identifying the genetic loci that are individually associated with PCa risk, only a small percentage of heritability can be explained, with an estimate of ~13% of heritability accounted for by combining ~30 GWAS-identified PCa risk-associated SNPs (Kim et al. 2010). One major factor that may contribute to the so-called missing heritability is the joint genetic effect, or epistasis (Manolio et al. 2009). Epitasis is generally defined as the interaction among genetic variants of multiple genes (or loci). In fact, gene–gene interaction has been the interest of investigation in the genetic field for a long time (Phillips 2008). Gene– gene interaction is considered to play an important roles in the pathogenesis of many complex diseases, including diabetes, obesity, hypertension, asthma, and cancer (Cordell 2009). However, very few studies have performed assessment on gene–gene interaction on a genome-wide scale. One of the largest difficulties preventing from the genome-wide search for gene–gene interactions was the computation burden. An exhaustive search for pair-wise interactions for 500,000 SNPs for thousands of samples will take 1.25 × 1011 statistical tests. Several methods have been developed recently to tackle this challenging task, including PLINK (Purcell et al. 2007), MDR (Ritchie et al. 2001), Random Jungle (Schwarz et al. 2010), RuningReliefFm (Moore 2007), BEAM (Zhang and Liu 2007), and BOOST (Wan et al. 2010).
Among these methods, BOOST, which stands for “Boolean Operation-based Screening and Testing”, has been reported to be the fastest approach to exhaustively search for the gene–gene interaction in the genome-wide data. Evaluation of gene–gene interaction in GWAS dataset based on the other methods suffers either from a relative low power by requiring a pre-screening stage (Cordell 2009; Marchini et al. 2005; Ritchie et al. 2001), or from a relatively slow speed and thus requiring a parallel computation system (Cordell 2009; Ma et al. 2009). In contrast, BOOST uses Boolean values to represent genotype data, which greatly facilitates the calculation of the contingency tables based on fast logic operations, and thus dramatically improves the CPU efficiency and computation speed.
In the current study, we performed a two-stage genome-wide gene–gene interaction scan using the BOOST approach. In the first stage, we evaluated all pairs of SNP– SNP interaction for ~500,000 SNP in 1,176 PCa cases and 1,101 control subjects in the National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) study. The second stage of study involved evaluation of the top SNP–SNP interactions (P < 1e1E–08) in another GWAS population of 1,964 PCa cases from the Johns Hopkins Hospital (JHH) and 3,172 control subjects from the Illumina iControlDB (iControl) database.
Materials and methods
Study populations
The first population was obtained from Stage 1 of the CGEMS study. It included 1,176 PCa cases and 1,101 control subjects, selected from the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial (Thomas et al. 2008; Yeager et al. 2009). The genotype and phenotype data of the study are publicly available and our use of the data was approved by CGEMS.
The second population was from a JHH PCa GWAS which included 1,964 PCa cases and 3,172 control subjects. The cases are Caucasian PCa patients who underwent radical prostatectomy for the treatment of PCa at JHH from 1 January 1999, through 31 December 2008 (Xu et al. 2010). The clinical characteristics of these patients are presented in the Supplementary Table 1. The control subjects for this population were an independent group of Caucasian individuals from the iControls dataset (https://www.illumina.com/science/icontrodb.ilmn). No demographic information on the iControls was available and both men and women were included in the iControls. The iControl subjects were matched to the cases by ancestry based on the principal component (PC) analysis, implemented by EIGENSOFT software (Price et al. 2006). Subjects excluded from the analysis were the outliers with deviation of six standard errors in the PC analysis (Price et al. 2006).
Genotype data, imputation, and quality control
GWAS of the CGEMS population was performed using HumanHap300 and HumanHap240 assays from Illumina Corp. GWAS of the JHH case population was performed using the Illumina 610K chip (24). GWAS of the iControls population was performed using Illumina Hap300 and Hap550 Chips. For SNP–SNP interactions that were significant at a Pinteraction cutoff of 1.0E–08 in the CGEMS population, we further performed the in silico replication in the JHH GWAS population.
To maximize the number of SNPs that were available for analysis in the JHH population, we imputed all the known SNPs that are cataloged in HapMap Phase II (www.hapmap.org) using the IMPUTE computer program (Marchini et al. 2007). A posterior probability of 0.90 was used as a threshold to call genotypes.
Individuals with a call rate below 0.95 were removed from GWAS analysis. The following quality control criteria were used to filter SNPs: MAF < 0.01, HWE < 0.001 and call rate < 0.95.
Statistical analysis
We implemented the BOOST approach to identify gene– gene interaction on the genome-wide scale (Wan et al. 2010). BOOST utilizes log-linear model to evaluate a two-locus interaction, which is much faster than the logistic model since it does not require iteration. It has been shown that using log-linear model is equivalent with the logistic model (Agresti 2002). Since there are three genotypes for each SNP, a 3 × 3 × 2 contingency table was constructed to test the interaction effect of SNP pair (XP, XQ). The log-linear models for the homogeneous association model (MH) with main effect terms and the saturated model (MS) with an interaction term are as following:
where µijk is the mean of the cell count of the contingency table corresponding to SNP Xp and Xq and disease status Y, λ is the overall mean of the natural log of the expected frequencies, and λs are the corresponding effects which SNPs and disease status have on the cell frequencies. Here, measures the gene–gene interaction and is the term of interest, if we use LH and LS to denote the log-likelihood of MH and MS. The significance of interaction term is tested by the difference of the maximum likelihood estimations: LS – LH.
A two-stage approach was then implemented to improve the computation efficiency while keeping the interactions in check. In the screening stage, BOOST transformed the genotype data into Boolean values and stored it to a contingency table. The difference of maximum likelihood estimation (MLE) of these two models was used to test interaction significance of a SNP pair. To overcome the computation difficulty that there is no close form estimate of MLE, BOOST approximates the likelihood ratio statistics L for the homogeneous association model using Kirkwood superposition approximation (KSA). As LS – LKSA is an upper bound of LS – LH, this method filters out most non-significant interactions and also guarantees survival of significant ones. A SNP pair would be removed from consideration, if the difference of MLE between these two models [2(LS – LKSA)] is less than a specified threshold t. In our analysis, we set t to be 43 for CGEMS and 9.49 for the confirmation population (JHH), respectively. This values correspond to an unadjusted Pinteraction of 1E–08, and a Pinteraction of 0.05, respectively. In the testing stage, likelihood ratio test statistics 2(LS – LH) was used to test the interaction of remaining SNP pairs. The test statistic is evaluated by a χ2 test with four degrees of freedom. The Pinteraction can be further adjusted by Bonferroni correction, if needed. Ancestral proportions obtained based on EIGENSOFT software (Price et al. 2006) were included as covariates to minimize the impact of potential population stratification in the JHH population. No age adjustment was performed due to the incomplete information in the iControls.
Results
After applying the quality control (QC) criteria, 509,916 SNPs remained in the analysis in the CGEMS GWAS population. A total of 1,325 pairs of SNP–SNP interactions reached a Pinteraction cutoff of 1.0E–08 (Supplementary Figure 1). None of the SNP pairs reached a genome-wide significant level of 4.4E–13, considering 1.25 × 1011 statistical tests. Specifically, 17 pairs of SNP–SNP interaction were found to be significant at a Pinteraction cutoff of 1.0E–10, and 131 pairs were significant at a Pinteraction cutoff of 1.0E–09. The most significant hit was observed between rs1178517 located on CNTN4 gene at chromosome 3 and rs1355096 located on FAM173B gene at chromosome 5, with a Pinteraction of 2.3E–11.
We then examined the interaction effects for the top list of SNP–SNP interactions in another independent GWAS population of JHH. Among the 1,325 pairs of SNP–SNP interactions that were significant at a Pinteraction of 1.0E–08 in CGEMS, 96 pairs of SNP–SNP interactions were significant at a nominal Pinteraction of 0.05 in JHH (Supplementary Table 2). Sixteen pairs of SNP–SNP interactions were significant at a more stringent Pinteraction cutoff of 0.01 (Table 1). However, no SNP pairs reached a Bonferroni corrected Pinteraction of 3.8E–05 in JHH population (0.05/1,325).
Table 1.
Results for top list of SNP–SNP interactions in the CGEMS and JHH populations (Pinteraction <1.0E–08 in CGEMS and Pinteraction < 0.01 in JHH)
| Study | SNP A |
SNP B |
Interaction | Annotation for SNP A |
Annotation for SNP B |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | CHR | BP | A1 | MAF | Main effect P |
rsID | CHR | BP | A1 | MAF | Main effect P |
BOOST P | Location | Gene | Relative position |
Location | Gene | Relative position |
|
| CGEMS | rs7514217 | 1 | 13,795,386 | G | 0.42 | 0.75 | rs7934426 | 11 | 37,270,065 | G | 0.46 | 0.38 | 5.3E–09 | Intron | PDPN | – | Intergenic | RAG2 | −693,660 |
| JHH | G | 0.45 | 0.16 | G | 0.44 | 0.37 | 1.9E–03 | ||||||||||||
| CGEMS | rs2503220 | 1 | 66,272,145 | G | 0.08 | 0.80 | rs2579790 | 10 | 77,725,730 | C | 0.38 | 0.32 | 9.3E–09 | Intron | PDE4B | – | Intron | C10orf11 | – |
| JHH | G | 0.08 | 0.75 | C | 0.39 | 0.23 | 3.1E–03 | ||||||||||||
| CGEMS | rs13402702 | 2 | 29,033,906 | G | 0.2 | 0.05 | rs7329899 | 13 | 105,276,784 | A | 0.26 | 0.99 | 6.4E–09 | Intergenic | SNORD53 | 30,394 | Intergenic | DAOA | 335,400 |
| JHH | G | 0.18 | 0.15 | A | 0.25 | 0.33 | 9.0E–03 | ||||||||||||
| CGEMS | rs4314028 | 2 | 51,990,169 | C | 0.25 | 0.41 | rs11980379 | 7 | 50,437,475 | C | 0.26 | 0.49 | 5.6E-09 | Intergenic | CHAC2 | −1,858,264 | 3′UTR | IKZF1 | – |
| JHH | C | 0.26 | 0.99 | C | 0.26 | 0.18 | 3.4E–03 | ||||||||||||
| CGEMS | rs4314028 | 2 | 51,990,169 | C | 0.25 | 0.41 | rs4132601 | 7 | 50,438,098 | G | 0.26 | 0.51 | 3.4E–09 | Intergenic | CHAC2 | −1,858,264 | 3′UTR | IKZF1 | – |
| JHH | C | 0.26 | 0.99 | G | 0.26 | 0.14 | 5.1E–03 | ||||||||||||
| CGEMS | rs4664789 | 2 | 156,575,782 | C | 0.49 | 0.97 | rs8019172 | 14 | 51,715,669 | A | 0.1 | 0.48 | 8.8E–09 | Intergenic | NR4A2 | 313,408 | Intergenic | PTGDR | −88,512 |
| JHH | A | 0.49 | 0.17 | A | 0.1 | 0.35 | 5.8E–03 | ||||||||||||
| CGEMS | rs4973194 | 2 | 229,945,721 | G | 0.37 | 0.49 | rs1949403 | 3 | 6,070,643 | C | 0.28 | 0.26 | 9.4E–09 | Intron | DNER. | – | Intergenic | EDEM1 | 833,993 |
| JHH | G | 0.36 | 0.91 | C | 0.27 | 0.65 | 8.9E–03 | ||||||||||||
| CGEMS | rs6772801 | 3 | 140,979,161 | G | 0.29 | 0.16 | rs6955437 | 7 | 137,547,448 | T | 0.11 | 0.77 | 3.9E–09 | Intergenic | NMNAT3 | −99,631 | Intergenic | AKR1D1 | 93,858 |
| JHH | G | 0.27 | 3.3E–04 | T | 0.09 | 0.31 | 8.9E–03 | ||||||||||||
| CGEMS | rs6878100 | 5 | 129,370,204 | A | 0.33 | 0.77 | rs2960753 | 7 | 141,386,546 | T | 0.4 | 0.47 | 3.6E–09 | Intron | CHSY3 | – | Intron | MGAM | – |
| JHH | A | 0.33 | 0.44 | T | 0.39 | 0.39 | 9.7E–03 | ||||||||||||
| CGEMS | rs6948622 | 7 | 145,043,041 | A | 0.36 | 0.85 | rs1154140 | 14 | 40,456,756 | G | 0.3 | 0.23 | 8.2E–09 | Intergenic | TPK1 | −878,962 | Intergenic | LRFN5 | −689,758 |
| JHH | A | 0.34 | 0.28 | G | 0.32 | 0.14 | 9.2E–03 | ||||||||||||
| CGEMS | rs12682543 | 8 | 29,135,477 | G | 0.34 | 0.04 | rs11231168 | 11 | 62,158,196 | T | 0.42 | 0.23 | 2.2E–09 | Intron | KIF13B | – | Intron | GANAB | – |
| JHH | G | 0.32 | 0.12 | T | 0.4 | 0.34 | 4.7E–03 | ||||||||||||
| CGEMS | rs10810961 | 9 | 18,361,966 | G | 0.10 | 0.11 | rs643853 | 21 | 43,656,244 | A | 0.18 | 0.05 | 3.7E–09 | Intergenic | MIR3152 | −201,338 | Intergenic | SIK1 | 2,583 |
| JHH | G | 0.11 | 0.26 | A | 0.21 | 0.22 | 5.2E–03 | ||||||||||||
| CGEMS | rs4837960 | 9 | 124,163,792 | T | 0.15 | 0.98 | rs275769 | 12 | 123,687,425 | T | 0.31 | 0.45 | 9.8E–09 | Intergenic | PTGS1 | −9,258 | Intergenic | SCARB1 | 140,702 |
| JHH | T | 0.14 | 0.45 | T | 0.3 | 0.24 | 8.9E–03 | ||||||||||||
| CGEMS | rs1038972 | 10 | 31,243,098 | T | 0.11 | 0.17 | rs2022896 | 14 | 27,074,470 | A | 0.24 | 0.65 | 4.2E–09 | Intron | ZNF438 | – | Intergenic | MIR4307 | 626,699 |
| JHH | T | 0.12 | 0.33 | A | 0.25 | 0.70 | 7.8E–03 | ||||||||||||
| CGEMS | rs12861843 | 13 | 35,638,422 | C | 0.42 | 0.06 | rs3862743 | 13 | 41,208,633 | C | 0.39 | 0.34 | 4.1E-09 | Intergenic | SOHLH2 | 1,923 | Intron | KIAA0564. | – |
| JHH | C | 0.45 | 0.91 | C | 0.4 | 0.28 | 3.2E–03 | ||||||||||||
| CGEMS | rs2136267 | 13 | 107,332,783 | T | 0.25 | 0.37 | rs1884393 | 20 | 1,404,079 | A | 0.11 | 0.46 | 9.3E–09 | Intergenic | LIG4 | 325,010 | 3′UTR | SIRPB2 | – |
| JHH | T | 0.27 | 0.33 | A | 0.11 | 0.95 | 6.5E–03 | ||||||||||||
Main effect P refers to the single-locus P value based on the two-degree of freedom test; BOOST P refers to the P values for the multiplicative interaction term, as calculated by BOOST approach. Relative position is the distance of SNPA/SNPB relative to the nearest gene if SNPA/SNPB is located in the intergenic region
SNP A the first interacting SNP, SNP B the second interacting SNP, CHR chromosome, MAF minor allele frequency, CGEMS the Cancer Genetic Markers of Susceptibility, JHH Johns Hopkins Hospital
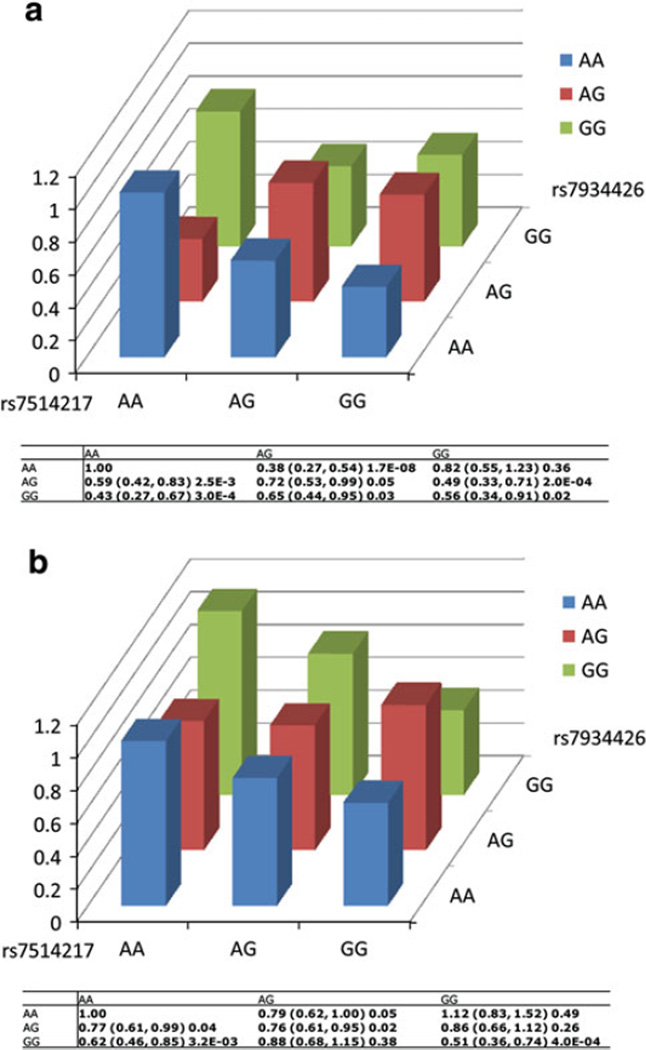
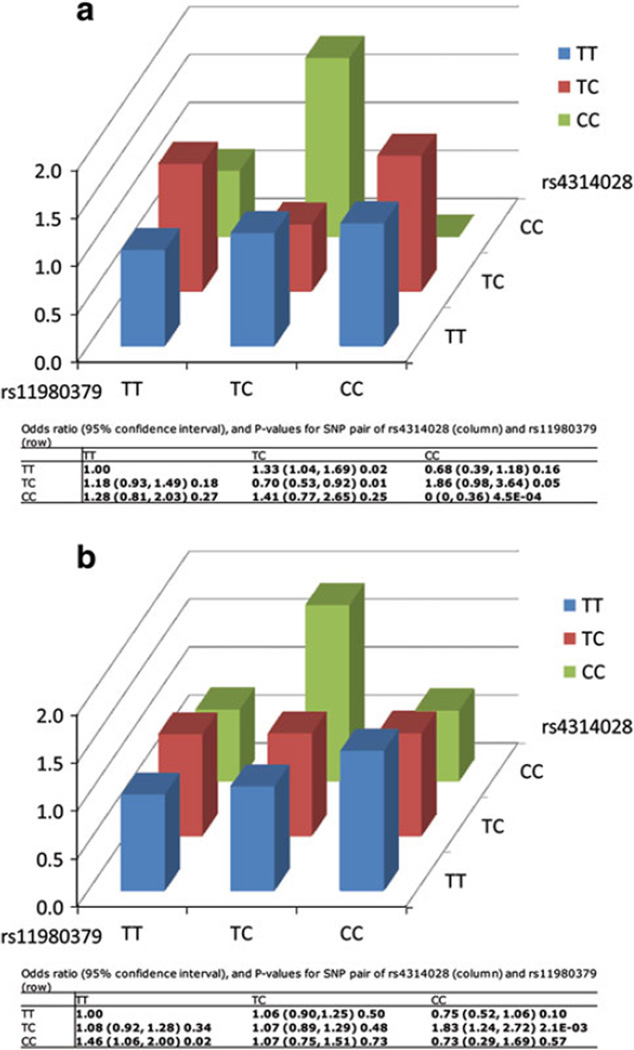
Among the 16 SNP–SNP interactions that reached the significance level at Pinteraction < 1.0E–08 for the CGEMS population and Pinteraction < 0.01 for the JHH population, three were found between SNPs that are both located within the intragenic regions, seven between an intragenic SNP and an intergenic SNP, and six between SNPs both located in intergenic regions. Two interactions deserved to be emphasized because they involve two cancer-related genes. One interaction (Pinteraction = 5.3E–09 for CGEMS and 1.9E–03 for JHH) was between rs7514217 (within the intron of PDPN at 1p36) and rs7934426 (intergenic, at 11p12). The second interaction was between rs11980379, located within the 3′UTR of IKZF1 at 7p12.2, rs4314028 at 2p16.3 (Pinteraction = 5.6E–09 and 3.4E–03, respectively (Table 1).
We then carefully examined the two-locus interaction pattern of the above two SNP pairs in CGEMS and JHH populations. Figure 1a showed the odds ratios for the × combinations of the genotypes of rs7514217 and rs7934426. Men who carried “GG/GG” double homozygotes for both SNPs had a significantly decreased risk of developing PCa (OR = 0.56, 95% CI 0.34–0.91; P = 0.02) in CGEMS compared with men who carried homozygous “A” allele (major allele) for both rs7514217 and rs7934436 (reference group) (Fig. 1a). A similar pattern of interaction was observed in JHH population. Particularly, men who carry GG/GG genotypes also had a decreased risk of developing PCa (OR = 0.51, 95% CI 0.36–0.74; P = 4.0E–04, Fig. 1b) compared with men who carried homozygous “A” allele (major allele) for both of the SNPs. Figure 2a and b showed the interaction pattern between rs11980379 and rs4314028 in CGEMS and JHH, respectively. Men who carried the “TC/CC” genotype for rs11980379 and rs4314028 had a marginal significantly increased risk for PCa (OR = 1.86, 95% CI 0.98–3.64; P = 0.05) in CGEMS, compared with men with homozygous “T” allele (major allele) (Fig. 2a). Similar in JHH, men who carry the “TC/CC” genotype combination also had a significantly increased risk for PCa (OR = 1.83, 95% CI 1.24–2.72; P = 2.1E–03) (Fig. 2b). However, the interaction pattern was not consistent with a dominant model since “TC/CC” genotype for rs11980379 and rs4314028 showed an increase in risk, while the “CC/CC” genotype shows a reduced risk (both compared to “TT/TT”) (Fig. 2b).
Fig. 1.
Interaction between rs7514217 and rs7934426 in CGEMS (a) and JHH (b). X axis represent the genotype for rs7514217, Y axis represent the genotype for rs7934426, the Z axis showed the odds ratio of the SNP pair (rs7514217 and rs7934426). Odds ratios are estimated relative to the baseline AA/AA double homozygote
Fig. 2.
Interaction between rs11980379 and rs4314028 in CGEMS (a) and JHH (b). X axis represent the genotype for rs11980379, Y axis represent the genotype for rs4314028 and the Z axis showed the odds ratio of the SNP pair (rs11980379 and rs4314028). Odds ratios are estimated relative to the baseline AA/AA double homozygote
Discussion
In this current study, we performed a two-stage genome-wide search for gene–gene interactions that were associated with PCa risk. It represents one of the first attempts to exhaustively evaluate all possible pairs of SNP–SNP interactions associated with PCa risk on a genome-wide scale. We identified 1,325 pairs of SNP–SNP interactions with a Pinteraction cutoff of 1.0E–08 in the discover population of CGEMS. Among the 1,325 pairs of interactions, 16 pairs of interactions were also significant in an independent population from JHH, with a Pinteraction cutoff of 0.01.
Interestingly, the SNP–SNP interactions revealed by our study involved loci with no evidence of main effect or weak marginal effect. Previous studies also showed that all the SNPs that were implicated in the top hits of interactions in Type 1 Diabetes and Type 2 Diabetes displayed weak main effects (Bell et al. 2011; Wan et al. 2010).Culverhouse et al. (2002) also reported large interaction effects in the complete absence of marginal effect. Thus these findings, combined with ours, highlight the need for exhaustive search when evaluating epistasis in a genome-wide scale. To reduce the computation burden, many methods have been developed with pre-screening algorithms built in (Cordell 2009; Marchini et al. 2005; Ritchie et al. 2001). However, most of the pre-screening algorithms are based on evaluating the marginal effects of single-locus. The SNPs with weak or no marginal effects and with significant interaction effects will be screened out based on such algorithms. In contrast, BOOST was able to evaluate all pair-wise interactions on the genome-wide scale with a relatively fast speed, which allows for exhaustive search of interaction effects across the entire genome.
Interaction, or epistasis, has been defined in multiple ways. In a recently review, Phillips (2008) highlights three major forms of epistasis: functional epistasis is a functional definition that describes the molecular interactions; compositional epistasis describes the phenomena of blocking of one allelic effect by another allele at a different locus (Bateson and Mendel 1909); statistical interaction refers as the presence of non-additive statistical deviation of two loci on the phenotype (Fisher 1918). Statistical interaction, as detected by BOOST, provides an approximation of measurement to reflect the biological implication that one locus’s effect on the phenotype depends on another locus (Phillips 2008). Although the extent to which statistical interaction relates to biological or functional interaction remains in extensive debate, the statistical interaction may still provide valuable insights on the potential biological interaction.
Specifically, we want to emphasize on two pairs of interactions implicated in our study, because they involve two cancer-related genes. One interaction was between rs7514217 (within the intron of PDPN at 1p36) and rs7934426 (intergenic, at 11p12). PDPN encodes a mucin-type transmembrane glycoprotein (podoplanin) that reportedly performs diverse functions including regulating actin cytoskeleton organization and cell migration(Navarro et al. 2011; Wicki et al. 2006), inducing platelet aggregation(Kaneko et al. 2004), and modulating lymphatic vasculature formation(Schacht et al. 2003). The crucial role of PDPN in tumorigenesis is indicated by the observations that PDPN is frequently overexpressed in various human cancer types (Martin-Villar et al. 2005; Mishima et al. 2006; Rahadiani et al. 2010; Schacht et al. 2005), and that podoplanin facilitates tumor cell migration and invasion (Wicki and Christofori 2007) and promotes lymphangio-genesis and lymph node metastasis (Raica et al. 2008). Expression of PDPN was also reported in prostatic tumors (Kanner et al. 2010), suggesting a role in PCa as well. The second pair of interaction was between rs11980379, located within the 3′UTR of IKZF1 at 7p12.2, and rs4314028, located on an intergenic region at 2p16.3. IKZF1 encodes a Ikaros family zinc finger transcription factor that normally directs hematopoietic lineage commitment and pituitary neuroendocrine cell expansion by regulating differentiation, proliferation, and apoptosis of these cell lineages (Ezzat et al. 2006; Georgopoulos et al. 1994). IKZF1 is considered a hematological and pituitary tumor suppressor, such that abnormalities in its splicing have been associated with leukemias (Sun et al. 1999) and pituitary tumors (Ezzat et al. 2003). Recently, expression of IKZF1 was also found in a variety of other human tissues including prostate and was associated with prognosis of breast, lung, ovarian and skin cancers (Yang et al. 2010), suggesting a potentially important role of IKZF1 in other cancer types such as PCa. However, given the “gene desert” localizations of the partner SNPs that interact with these PDPN/IKZF1-harbored SNPs, it is hard to make biological inferences as what molecular mechanisms might account for these two interactions. Nonetheless, as revealed by the ongoing ENCODE project and several genome-wide cistrome studies on important transcription factors such as Forkhead box A1 (FOXA1) (Wang et al. 2009), androgen receptor (AR) (Wang et al. 2009) and estrogen receptor (ER) (Carroll et al. 2006), the DNA sequences whereby certain functionally critical transcription factors bind and regulate expression of their target genes are extensively localized throughout the genome, which sometimes are several hundred kbps away from their target genes and may reside in intergenic regions (enhancers or suppressors). Thus, it is possible that these intergenic SNPs, or those in linkage disequilibrium (LD) with them, may lie within the DNA sequences containing enhancer or suppressor activities that distantly regulate one or several target genes. These genes may collaborate with PDPN/IKZF1 in a common signaling network that combinatorially determines the well-being of prostatic epithelial cells. A certain combination of genetic variants in these interacting intergenic and intragenic loci, though insufficient by themselves alone, may cause the synthetic deficiency of the crucial signaling network and result in explicitly increased risk for PCa. This enhancer/suppressor-involved mechanism may potentially provide explanations for the remaining interactions as well, especially for the six interactions which involve two intergenic SNPs. However, it should be pointed out these contemplations are largely speculative and may require in-depth mechanistic and functional studies to prove.
Our results need to be interpreted with caution. First, in this study we performed a total of 1.25 × 1011 statistical tests, in which a Pinteraction ≤4.4E–13 would be corresponding to a genome-wide significant level (0.05/1.25 × 1011). However, simply applying a Bonferroni correction here is very conservative, considering the LD structure existing among SNPs. The number of dependent tests exponentially increases when evaluating pair-wise gene– gene interactions. For example, the ~500,000 SNPs assessed in our GWAS analysis could be collapsed into ~50,000 independent LD clusters using a r2 criteria of 0.2 using the clumping function of PLINK (Purcell et al. 2007). In this case, we roughly estimated the number of independent interaction tests to be 1.9 × 109 (50,000 × 49,999/2 = 1.2 × 109). Then, a Pinteraction cutoff of 4.2E–11 would be used to claim a genome-wide significance level. One pair of interaction could be claimed to exceed a genome-wide significance level (Pinteraction = 2.3E–11, between rs1178517 and rs1355096). However, this SNP pair was not replicated in the JHH study (Pinteraction > 0.05), which demonstrated the challenge of selecting an appropriate threshold to claim a statistically conclusive finding. Second, another potential factor which may prevent interaction pairs from reaching the genome-wide significance level was the limited sample size in the GWAS discovery population. Specifically, with ~1,200 cases and ~1,200 controls, we have limited power to detect interactions with small to modest interaction effects (OR < 2.0) at a Pinteraction level of either 4.4E–13 or 4.2E–11. Third, we chose to use a relatively stringent P value cutoff of 1E–08 to select SNP pairs to be followed in the 2nd stage. We understand that using a stringent cutoff and with a small sample size in the GWAS stage lowers the power to detect true interactions with modest effects, which comes with the price that any true interaction that did not make this stringent cutoff can no longer be identified in this study. A more detailed power calculation can be found in the Supplementary Table 2, assuming a P value cutoff of 1E–08, a range of ORs and minor allele frequencies (MAFs). Using a less stringent P value cutoff, i.e., 1E–07, slightly increases the power for the first stage. However, it also significantly increases the number of SNPs pairs that need to be evaluated in the 2nd stage, which decreases the power for the 2nd stage. Therefore, the use of 1E–08 as a cutoff to select SNP pairs to be followed was to balance the power of stage 1 and stage 2. Third, the inclusion of over 60% of women as controls in the iControl populations may lead to potential bias when studying a male disease. However, the concern is minimal, since we limited the controls to male only when analyzing SNPs on X chromosome. Also, as shown in our previous finding, we have used the same iControl population as controls to successfully discover a novel PCa risk loci. (Feng et al. 2011).
Additionally, we calculated the potential power gain if all existing PCa GWAS would be available. So far, a total of four PCa GWAS have been reported for Caucasians. Those four studies include one study conducted in Iceland with 1,453 cases and 3,064 controls (Gudmundsson et al. 2007), one study conducted in UK with 1,854 cases and 1,894 controls (Eeles et al. 2009), two studies conducted in US, including the CGEMS’s study with 1,176 cases and 1,101 controls (Yeager et al. 2009), and our JHH study with 1,964 cases and 3,172 controls. With a total of 6,447 cases and 9,231 controls, we would have 80% power to detect an interaction OR of 1.4 at a Pinteraction level of either 4.4E–13 or 4.2E–011, assuming a MAF of 0.3 for both SNPs. Therefore, future studies with larger samples are needed to detect modest interaction effects that are significant at a genome-wide level.
In summary, although the SNP–SNP interactions implicated in this study did not reach a genome-wide significant level, we hope the publication of the list will encourage more replication studies. More importantly, it represents one of the first application studies which implemented a novel statistical method of BOOST to detect interactions on a genome-wide scale. The pairs of SNP–SNP interactions suggested in our study represent the first step towards obtaining further biological insight into the high-dimensional etiology of PCa. Additional studies are needed to further evaluate the interactions suggested in this current study.
Supplementary Material
Acknowledgments
We thank all of the study subjects who participated in the JHH study and the urologists who provided their patients to the JHH study. We acknowledge the contribution of multiple physicians and researchers in designing and recruiting study subjects. We also acknowledge the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) for making the data publicly available. We also want to thank Dr. Can Yang for kindly responding to all our questions related to BOOST software. This work was supported by a DOD grant to J.S (W81XWH-09-1-0488), an intramural funding from the Van Andel Research Institute to J.X, and a R01 grant from the National Cancer Institute (CA129684 J.X).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-012-1148-4) contains supplementary material, which is available to authorized users.
Contributor Information
Sha Tao, Center for Genetic Epidemiology and Prevention, Van Andel Research Institute, Grand Rapids, MI, USA.
Junjie Feng, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Timothy Webster, Center for Genetic Epidemiology and Prevention, Van Andel Research Institute, Grand Rapids, MI, USA.
Guangfu Jin, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Fang-Chi Hsu, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Department of Biostatistical Sciences, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Shyh-Huei Chen, Department of Biostatistical Sciences, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Seong-Tae Kim, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Zhong Wang, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Zheng Zhang, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Siqun L. Zheng, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
William B. Isaacs, Department of Urology, Johns Hopkins Medical Institutions, Baltimore, MD, USA
Jianfeng Xu, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genetic Epidemiology and Prevention, Van Andel Research Institute, Grand Rapids, MI, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA; Department of Urology, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
Jielin Sun, Email: jisun@wakehealth.edu, Center for Cancer Genomics, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Center for Genomics and Personalized Medicine Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA.
References
- Agresti A. Wiley Series in Probability and Statistics. 2nd edn. New York: Wiley; 2002. Categorical data analysis. [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Bateson W, Mendel G. Mendel’s principles of heredity. Cambridge: Cambridge University Press; 1909. [Google Scholar]
- Bell JT, Timpson NJ, Rayner NW, Zeggini E, Frayling TM, Hattersley AT, Morris AP, McCarthy MI. Genome-wide association scan allowing for epistasis in type 2 diabetes. Ann Hum Genet. 2011;75:10–19. doi: 10.1111/j.1469-1809.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Balter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Gronberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat S, Yu S, Asa SL. Ikaros isoforms in human pituitary tumors: distinct localization, histone acetylation, and activation of the 50 fibroblast growth factor receptor-4 promoter. Am J Pathol. 2003;163:1177–1184. doi: 10.1016/S0002-9440(10)63477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat S, Zhu X, Loeper S, Fischer S, Asa SL. Tumor-derived Ikaros 6 acetylates the Bcl-XL promoter to up-regulate a survival signal in pituitary cells. Mol Endocrinol. 2006;20:2976–2986. doi: 10.1210/me.2006-0265. [DOI] [PubMed] [Google Scholar]
- Feng J, Sun J, Kim ST, Lu Y, Wang Z, Zhang Z, Gronberg H, Isaacs WB, Zheng SL, Xu J. A genome-wide survey over the ChIP-on-chip identified androgen receptor-binding genomic regions identifies a novel prostate cancer susceptibility locus at 12q13.13. Cancer Epidemiol Biomarkers Prev. 2011;20:2396–2403. doi: 10.1158/1055-9965.EPI-11-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The correlations between relatives on the supposition of Mendelian inheritance. Philos Trans Royal Soc Edinburgh. 1918;52:35. [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldorsson JG, Flekkoy KM, Gudmundsson KR, Arnkelsson GB, Arnarson EO. Urban–rural differences in pediatric traumatic head injuries: a prospective nationwide study. Neuropsychiatr Dis Treat. 2007;3:935–941. doi: 10.2147/ndt.s2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Sun J, Wiklund F, Isaacs SD, Wiley KE, Purcell LD, Gao Z, Stattin P, Zhu Y, Kim ST, Zhang Z, Liu W, Chang BL, Walsh PC, Duggan D, Carpten JD, Isaacs WB, Gronberg H, Xu J, Zheng SL. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69:2720–2723. doi: 10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Kato Y, Kunita A, Fujita N, Tsuruo T, Osawa M. Functional sialylated O-glycan to platelet aggregation on Aggrus (T1alpha/Podoplanin) molecules expressed in Chinese hamster ovary cells. J Biol Chem. 2004;279:38838–38843. doi: 10.1074/jbc.M407210200. [DOI] [PubMed] [Google Scholar]
- Kanner WA, Galgano MT, Atkins KA. Podoplanin expression in basal and myoepithelial cells: utility and potential pitfalls. Appl Immunohistochem Mol Morphol. 2010;18:226–230. doi: 10.1097/PAI.0b013e3181c65141. [DOI] [PubMed] [Google Scholar]
- Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, Isaacs WB, Xu J, Sun J. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic Variation. Prostate. 2010;70:1729–1738. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dork T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Roder MA, Tybjaerg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer— analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Ma L, Runesha H, Dvorkin D, Garbe J, Da Y. Parallel and serial computing tools for testing single-locus and epistatic SNP effects of quantitative traits in genome-wide association studies. BMC Bioinformatics. 2009;9 doi: 10.1186/1471-2105-9-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Martin-Villar E, Scholl FG, Gamallo C, Yurrita MM, Munoz-Guerra M, Cruces J, Quintanilla M. Characterization of human PA2.26 antigen (T1alpha-2, podoplanin), a small membrane mucin induced in oral squamous cell carcinomas. Int J Cancer. 2005;113:899–910. doi: 10.1002/ijc.20656. [DOI] [PubMed] [Google Scholar]
- Mishima K, Kato Y, Kaneko MK, Nakazawa Y, Kunita A, Fujita N, Tsuruo T, Nishikawa R, Hirose T, Matsutani M. Podoplanin expression in primary central nervous system germ cell tumors: a useful histological marker for the diagnosis of germinoma. Acta Neuropathol. 2006;111:563–568. doi: 10.1007/s00401-006-0033-4. [DOI] [PubMed] [Google Scholar]
- Moore JWB. Tuning reliefF for genome-wide genetic analysis. Lect Notes Comput Sci. 2007;4447:10. [Google Scholar]
- Navarro A, Perez RE, Rezaiekhaligh MH, Mabry SM, Ekekezie II. Polarized migration of lymphatic endothelial cells is critically dependent on podoplanin regulation of Cdc42. Am J Physiol Lung Cell Mol Physiol. 2011;300:L32–L42. doi: 10.1152/ajplung.00171.2010. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis: the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahadiani N, Ikeda J, Makino T, Tian T, Qiu Y, Mamat S, Wang Y, Doki Y, Aozasa K, Morii E. Tumorigenic role of podoplanin in esophageal squamous-cell carcinoma. Ann Surg Oncol. 2010;17:1311–1323. doi: 10.1245/s10434-009-0895-5. [DOI] [PubMed] [Google Scholar]
- Raica M, Cimpean AM, Ribatti D. The role of podoplanin in tumor progression and metastasis. Anticancer Res. 2008;28:2997–3006. [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht V, Ramirez MI, Hong YK, Hirakawa S, Feng D, Harvey N, Williams M, Dvorak AM, Dvorak HF, Oliver G, Detmar M. T1alpha/podoplanin deficiency disrupts normal lymphatic vasculature formation and causes lymphedema. EMBO J. 2003;22:3546–3556. doi: 10.1093/emboj/cdg342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht V, Dadras SS, Johnson LA, Jackson DG, Hong YK, Detmar M. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. Am J Pathol. 2005;166:913–921. doi: 10.1016/S0002-9440(10)62311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DF, Konig IR, Ziegler A. On safari to random jungle: a fast implementation of random forests for high-dimensional data. Bioinformatics. 2010;26:1752–1758. doi: 10.1093/bioinformatics/btq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Heerema N, Crotty L, Wu X, Navara C, Vassilev A, Sensel M, Reaman GH, Uckun FM. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 1999;96:680–685. doi: 10.1073/pnas.96.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Purcell L, Gao Z, Isaacs SD, Wiley KE, Hsu FC, Liu W, Duggan D, Carpten JD, Gronberg H, Xu J, Chang BL, Partin AW, Walsh PC, Isaacs WB, Zheng SL. Association between sequence variants at 17q12 and 17q24.3 and prostate cancer risk in European and African Americans. Prostate. 2008;68:691–697. doi: 10.1002/pros.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, Kim ST, Zhu Y, Zhang Z, HsuFC, Turner AR, Stattin P, LiuW,KimJW,Duggan D, Carpten J, Isaacs W, Gronberg H, Xu J, Chang BL. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Wan X, Yang C, Yang Q, Xue H, Fan X, Tang NL, Yu W. BOOST: a fast approach to detecting gene–gene interactions in genome-wide case–control studies. Am J Hum Genet. 2010;87:325–340. doi: 10.1016/j.ajhg.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicki A, Christofori G. The potential role of podoplanin in tumour invasion. Br J Cancer. 2007;96:1–5. doi: 10.1038/sj.bjc.6603518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Isaacs SD, Wiley KE, Wiklund F, Sun J, Kader AK, Li G, Purcell LD, KimST, HsuFC, Stattin P, Hugosson J, Adolfsson J, Walsh PC, Trent JM, Duggan D, Carpten J, Gronberg H, Isaacs WB. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci USA. 2010;107:2136–2140. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luo Y, Wei J. Integrative genomic analyses on Ikaros and its expression related to solid cancer prognosis. Oncol Rep. 2010;24:571–577. doi: 10.3892/or_00000894. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF, Jr, Hunter DJ, Thomas G, Chanock SJ. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case–control studies. Nat Genet. 2007;39:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, Smith S, Pruett K, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Johansson JE, Liu W, Kim JW, Chang BL, Duggan D, Carpten J, Rodriguez C, Isaacs W, Gronberg H, Xu J. Two independent prostate cancer risk-associated loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.