Abstract
Human uracil DNA glycosylase (hUNG) plays a central role in DNA repair and programmed mutagenesis of Ig genes, requiring it to act on sparsely or densely spaced uracil bases located in a variety of contexts, including U/A and U/G base pairs, and potentially uracils within single stranded DNA. An interesting question is whether the facilitated search mode of hUNG, which includes both DNA sliding and hopping, changes in these different contexts. Here we find that hUNG uses an enhanced local search mode when it acts on uracils in ssDNA, and also, in a context where uracils are densely clustered in duplex DNA. In the context of ssDNA hUNG performs an enhanced local search by sliding with a larger mean sliding length as compared to dsDNA. In the context of duplex DNA, insertion of high-affinity abasic product sites between two uracil lesions serves to significantly extend the apparent sliding length on dsDNA from 4 to 20 base pairs, and in some cases, leads to directionally biased 3′→5′ sliding. The presence of intervening abasic product sites mimics the situation where hUNG acts iteratively on densely spaced uracils. The findings suggest that intervening product sites serve to increase the amount of time the enzyme remains associated with DNA as compared to nonspecific DNA, which in turn increases the likelihood of sliding as opposed to falling off the DNA. These findings illustrate how the search mechanism of hUNG is not predetermined, but instead, depends on the context in which the uracils are located.
Human uracil DNA glycosylase (hUNG) is an extremely versatile catalyst that excises uracils in a wide variety of genomic DNA contexts (1). For example, during chemotherapy with antifolate and fluoropyrimidine drugs dUTP levels rise and replicative DNA polymerases frequently incorporate dUTP opposite to adenine (2, 3). Within this framework hUNG is likely confronted with densely spaced uracils in the context of U/A base pairs. The enzyme must also act on uracils that are generated by the enzyme activation induced cytosine deaminase (AID) during the process of adaptive immunity, which includes the two distinct processes of somatic hypermutation (SHM) (4) and class switch recombination (CSR) (5). Somatic hypermutation involves the iterative action of AID on multiple cytosines localized in the hypervariable regions of Ig genes. These cytosines are generated during the transient period where these regions are present as single stranded R loops during active transcription (4, 6). Thus, depending on the timing of hUNG with respect to transcription-coupled hypermutation, the enzyme might encounter either single-stranded uracils or uracils that are paired with guanine. Finally, to initiate CSR, AID must deaminate closely spaced cytosines on opposite strands of duplex DNA (generating U/G mismatches) such that recombinogenic double-stranded breaks are introduced after hUNG acts at such sites (5). Given the diverse contexts of these genomic uracils we wondered whether the facilitated search mechanism of hUNG might be altered from that observed with sparsely spaced uracils in duplex DNA (7, 8).
What specific aspects of a uracil’s environment might influence the search mechanism of hUNG? In the case of DNA sliding to a uracil site, the most critical factors are the bound lifetime (τbind), which provides the upper limit time frame for sliding, and the 1D diffusion constant (D1), that sets the speed limit for sliding (9). Together, these parameters define the sliding length, . Thus, if D1 or τbind are increased in a given DNA context, hUNG will search longer stretches of DNA by a sliding mode. In the case of DNA hopping between uracil sites (i.e. short range dissociation and reassociation events), such events will become more likely if the persistence length of DNA is decreased (as in ssDNA), because the probability (P) for successful hopping between sites is inversely related to the distance (r) between the sites (P ~ 1/r) (10). In addition, hopping is a required pathway to locate clustered uracils that are located on opposite strands of DNA (7, 8). From these considerations, it is reasonable to envisage that features such as single strand DNA bubbles, U/G base mismatches, and clustered uracils or abasic sites could change the hopping or sliding efficiency.
Here we explore how the search mechanism of hUNG is affected by the context in which the uracil sites are found. These studies show that the enzyme has an enhanced ability to slide along linear ssDNA substrates as compared to helical duplex DNA. Additionally, the sliding length of hUNG2 can also be significantly extended in duplex DNA when high affinity product abasic sites are inserted between two uracil target sites. These findings provide a window into the flexibility of the search mechanism of hUNG, which can be tuned to optimally locate densely spaced uracils that occur during adaptive immunity as well as sparse uracils that arise during spontaneous cytosine deamination or infrequent incorporations of dUTP.
Materials and Methods
Protein and Oligonucleotide Reagents
hUNG was purified as previously described (8). Protein concentrations were determined by absorbance measurements at 280 nm using an extinction coefficient of 33.68 mM−1 cm−1. All 90 mer uracil containing oligonucleotides were ordered from Integrated DNA Technologies (www.IDTDNA.com) in the crude desalted form and purified by denaturing PAGE. The sequences of these oligonucleotides are reported in the Supplemental Methods.
Experimental conditions
All measurements in this paper and the accompanying paper (insert reference upon publication) were made at 37 °C in a standard reaction buffer consisting of 20 mM HEPES pH 7.5, 0.002% Brij 35 detergeant (Sigma Aldrich), 3 mM EDTA (added from a 0.5 M pH 8.0 stock), and 1 mM DTT unless otherwise noted.
Intramolecular Site Transfer Assay
For all substrates used here, oligonucleotides were labeled at the 5′ and 3′ ends by incubation with [γ32P] ATP and 3′-deoxyadenosine 5′-triphosphate (cordycepin 5′-triphosphate),-[α-32P] and polynucleotide kinase and terminal transferase (New England Biolabs) respectively and excess radioactivity and salt was removed by gel filtration as described in the accompanying paper. Duplex substrates containing tetrahydrofuran (F) abasic site mimics (S5F, S11F, and S19F) were hybridized to the complement oligonucleotide by heating to 95 °C for 10 min in a dry heat block and allowed to slowly cool to room temperature.
For post-reaction processing of the single and double stranded uracil substrates, the abasic sites generated by hUNG were cleaved with either hot piperidine (20 minutes at 90 °C), or by the addition of 0.25 M ethylenediamine (pH 8.0) (see accompanying paper). Samples were then loaded onto a 10% denaturing gel (19:1 bis:acrylamide) that was preheated in order to denature any residual structure.
For ssDNA substrates where no site preference was observed, the data was analyzed as previously using eq 1 (insert reference to the companion manuscript upon publication). However in the case of substrates containing F sites, the data was analyzed using the method of Stanford et al. (see Results section) (11).
| (1) |
Determination of the Efficiency of Uracil Excision
The efficiency (E) with which hUNG excises a uracil when it encounters a site as opposed to falling off the site, is defined by eq 2.
| (2) |
The efficiency for uracil in a ssDNA and in the context of the F containing substrate S19F was determined as previously described for duplex DNA using a pulse-chase kinetic partitioning experiment (7, 8). Briefly for a substrate uracil within ssDNA, using a three syringe rapid mixing device (Kintek RQF3), 20 μL of a 4 μM solution of hUNG was rapidly mixed with 20 μL of 5′ 32P labeled 1XUss substrate at a concentration of 0.5 or 1 μM. The reaction was quenched at a 2 ms aging time by the addition of either 0.5 M HCl, or chased with a duplex DNA (60 μM or 30 μM) containing a high affinity F site (chDNA, Supplemental Methods). The concentrations of the quench listed are that in the quench syringe of the rapid mixer resulting in approximately a 2/3 dilution in the final quenched solution. Identical results were observed in experiments varying the DNA/Enzyme ratio and when varying chase DNA concentration (Supplemental Fig. S2, Fig. 2). For the samples machine-quenched by the chDNA, subsequent time points were taken between 5 and 30 seconds and manually quenched with an equal volume of 0.5 M HCl. To all samples an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, Invitrogen) was added and the samples were vortexed. The layers were allowed to separate by gravity and an equal volume of 2 M piperidine followed by centrifugation for 10 minutes at 13,000 × g. The aqueous layer was then transferred to another tube and heated to 90 °C for 20 minutes to cleave the abasic sites and then dried to completion to remove the piperidine. The samples were resuspended in 50% formamide gel loading buffer and substrate and product were separated by electrophoresis on a 10% denaturing gel. The gels were dried and imaged as describe above. Detailed explanation and kinetic simulations validating the approach are described in Porecha et al. and the corresponding Supplemental Methods (7).
Figure 2.
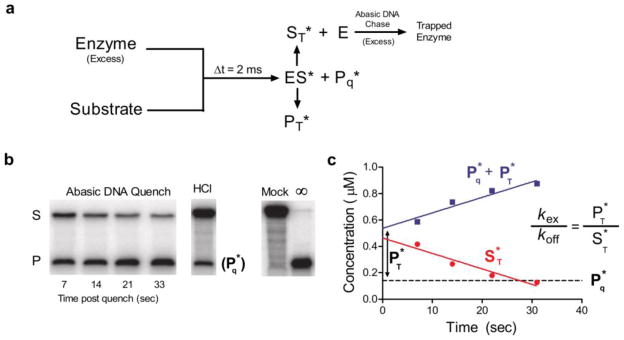
Determination of the excision efficiency (E) for uracil cleavage by hUNG on ssDNA. (a) Rapid mixing scheme where hUNG (4 μM) was reacted with 1.0 or 0.5 μM 90mer DNA substrate containing a single uracil site (1XUss 90mer, Supplemental Methods. Both DNA concentrations gave identical results). After mixing, the reaction was either quenched with 0.5 M HCl or chased with 60 μM duplex DNA containing a tetrahydrofuran abasic site mimic (chDNA). Following the chase, the reaction mix was then manually quenched with HCl at the indicated times in panels b and c. (b) Separation of product and substrate bands by denaturing gel electrophoresis in the quenched and chased samples. Mock is a control reaction where the DNA was subjected to the exact processing procedure without the addition of enzyme. Note that the amount of trapped product at the time of chase mixing (PT*) must be obtained from Ptot by subtracting the amount of product already present within the 2 ms aging time as determined by the acid quenched samples. Thus, when 2 μM UNG was mixed with 1 μM substrate for 2 ms and the reaction was quenched with 0.5 M HCl, 0.13 ± 0.01 μM excision product was formed (P*q) and 0.86 ± 0.01 μM bound substrate was left unreacted (ES*). Linear extrapolation to zero time is used determine the amount of total product (Ptot = Pq* + PT*) formed and substrate (ST*) remaining at the time of addition of the chase DNA. (c) When the acid quench was replaced with 60 μM F containing DNA duplex (chDNA) to serve as a trap for hUNG after it dissociated from the ES* complex, the 0.86 μM ES* present at 2 ms was converted to 0.53 ± 0.08 μM free substrate (S*T), and 0.47 ± 0.08 μM was excised to form product (P*T). Because P*T/S*T = kex/koff = 0.34/0.53 = 0.64, then the average excision efficiency E = kex/(koff + kex) = PT*/(PT* + ST*) may be directly calculated as be 0.39 ± 0.14 from nine independent trials. Values are reported as the average ± 1 s.d. Control experiments varying the enzyme/substrate ratio as well as the chase DNA concentration gave identical results and are shown in Supplemental Fig. S2.
For determining the efficiency of cleavage and the single turnover rate of single uracil containing substrates analogous to S19F (S19F 5′ site and S19F 3′ site) an identical procedure was employed, however after reaction with hUNG, quenching and phenol-chloroform extraction, the DNA containing solution was instead neutralized with an appropriate volume of 3M Tris base. Formamide was then added to 65% final concentration and the sample was subsequently heated to 90 °C for 3 hours to cleave the abasic sites and immediately run on a 10% denaturing polyacrylamide gel as described above.
Results
Site Transfer Mechanism on ssDNA
Previous site transfer measurements were made on ssDNA substrates with closely spaced uracils at 5 and 10 ntds (8). In order to further understand hUNG transfer on ssDNA we made site transfer measurements at lengths out to 40 ntds (S5ss, S10ss, S20ss and S40ss). These ssDNA sequences were designed to minimize any propensity for intramolecular hydrogen bonding that might give rise to secondary structures and anomalous site transfer results (Supplemental Fig. S1). Representative data for S20ss in the absence and presence of the uracil trap shows a significant degree of intramolecular transfer (revealed by excess A and C fragments) that is diminished, but not entirely removed, by the presence of uracil (Fig. 1a). Furthermore, the plateau region of trapping has been reached because identical data were obtained in the presence of 10 and 15 mM uracil. Extrapolation of Ptransobs (eq 1) to zero time shows that Ptrans = 0.44 ± 0.03 and Pslide = 0.14 ± 0.03 for S20ss (Fig. 1b). Similar measurements were made on the 40 ntd ssDNA substrates and the results are summarized in Fig. 1c–e.
Figure 1.
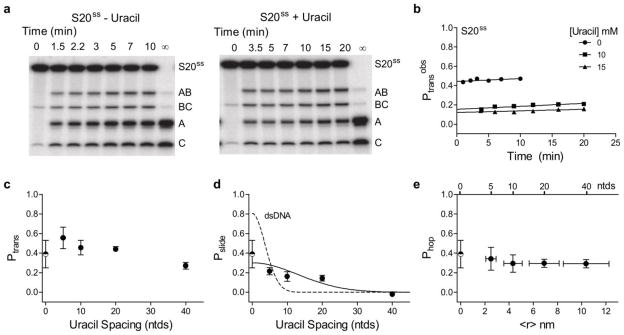
Facilitated transfer of hUNG on ssDNA as determined by the ‘molecular clock’ approach (8). (a) Gel images of the site transfer assay performed using S20ss in the presence and absence of the uracil trap. We note the presence of a very small percentage (<1%) of the 3′ labeled BC and C bands in the zero time lane. These bands result from a very small amount of uracil DNA glycosylase activity (<1% cleavage in >2 hrs reaction at 37 °C) in the commercially available 3′ terminal deoxynucleotidyl transferase enzyme used in the 3′ 32P labeling of the DNA substrates. These background bands were determined to have a neglible effect on the calculations of site transfer probabilities and initial rates. (b) Determination of Ptrans for S20ss by linear extrapolation of the Ptransobs (eq 1) to zero time. (c) Total transfer (Ptrans) as determined by the site transfer assay without the addition of the trap (uracil). (d) Transfer by sliding (Pslide) by determination of site transfer at high trap concentrations. The solid line is a least squares fit to a random walk sliding model (Pslide = E × [kslide/(kslide+koff)]N where N = ntds2 is the number of stochastic steps taken during sliding transfers and E is the efficiency of uracil excision (see Fig. 2 and eq 2). The dotted line is the same fit previously obtained for dsDNA (8). (e) Spacing dependence of the hopping probability as determined by the difference between Ptrans and Pslide. The mean distance between uracil sites was determined from the worm like chain model (33). The x-axis error bars for the mean square distance (<r>) are maximum and minimum values calculated using various experimental estimates of the persistence and contour lengths for ssDNA (13, 14) (see Supplemental Methods). The shown data for ssDNA at 5 and 10 bp spacing were previously reported (8). Pslide was calculated as the average plateau value for all data points performed at 10 or 15 mM uracil. For data obtained at 20 and 40 bp uracil spacings, Pslide was determined from the average of 3 replicate trials at each uracil concentration. The half-filled circle at zero site spacing in all panels is the efficiency of uracil excision when hUNG has found a target site as determined in Fig. 2 (E = 0.39 ± 0.14 for ssDNA). All transfer probability errors represent the mean plus or minus one standard deviation obtained from at least three replicate measurements at 0 mM uracil and six replicates at high uracil concentrations (three replicates each at 10 and 15 mM uracil).
A unique feature of the transfer data for ssDNA substrates as compared to previous results with dsDNA is the flat dependence of Ptrans on uracil spacing (Fig. 1c). In fact, extrapolation of the data in Fig. 1c to zero site spacing suggests that the maximal transfer efficiency is only around 50% for single stranded DNA. One possible explanation for this limiting value is that once a uracil site is encountered, hUNG then partitions evenly between falling off the DNA and moving forward along the reaction coordinate to excise the base (i.e. kex/koff ~ 1). This ratio will serve to limit excision events at the second site even if intramolecular transfer is very efficient (7, 8). Previous measurements of this partitioning ratio for cleavage of uracil sites in dsDNA by both the human and E. coli UNG enzymes established that uracil sites were processed efficiently when they were encountered (kex/koff ~ 5/1) (7, 8). Here a similar pulse-chase rapid kinetic approach was used to measure a much lower kex/koff = 0.64 for uracil within a ssDNA context (Fig. 2). This ratio indicates that the efficiency (E, see eq 2) of excising a uracil site once it is encountered in ssDNA is only 0.39 ± 0.14 (Fig. 2 and denoted by the half-filled circle in Fig. 1c–e). Correcting the Ptrans values in Fig. 1c for this efficiency (i.e. Ptrans/E) yields the true site transfer probability for ssDNA, which is in the remarkably high range of ~0.6 to 1.0 for spacings from 40 to 5 ntds.
Do ssDNA site transfers in the presence of uracil correspond to chain sliding? It would be anticipated that the probability of successful intramolecular transfer by sliding would follow a site spacing dependence according to eq 3, where E is the site excision efficiency at zero site spacing and S is the kinetic partitioning ratio (S = ksl/(ksl + koff) that describes the likelihood that the enzyme will slide along the DNA strand as opposed to dissociating after each sliding step (n) during transfer (9, 12).
| (3) |
The square in eq 3 results from the fundamental stochastic property of diffusion where the total number of steps taken to traverse a given distance varies with the square of the separation in step length units (i.e. traversing a site separation of 10 ntd step lengths would require an average of 100 total steps) (9, 12). We used eq 3 to fit the observed transfer probabilities in the presence of uracil as a function of nucleotide spacing between the two sites, and compared the results obtained with ssDNA to that of duplex DNA (Fig. 1d, dashed line). Within experimental error the site transfer probabilities on ssDNA decrease with increasing site spacing according to the expectations of a sliding mechanism. Using this model, the mean sliding length (defined as the uracil spacing where Pslide is diminished by 50%) is calculated as 19 ntds, compared to only 4–5 bp for duplex DNA (Fig. 1d, dashed line) (8). A discussion of whether the term “sliding” is an appropriate descriptor for a 1-dimensional random walk on a flexible polymer like ssDNA is deferred to the Discussion.
The probability of successfully hopping between two sites (Phop = PoffPreturn, see Fig. 1 of accompanying paper)(insert reference to the companion manuscript upon publication) separated by a linear distance r should follow the relationship of eq 4, where a is the diameter of an idealized spherical target (9, 12). For single stranded DNA, the average distances between target sites (<r>) must be obtained using the worm like chain model and experimental estimates for the persistence and contour lengths of ssDNA at the salt concentration used in these experiments (Supplemental Methods) (13, 14).
| (4) |
Although eq 4 describes very well the transfer probability of human and E. coli UNG for site spacings in duplex DNA between 20 bp and 800 bp (7, 8), this relationship fails to account for the flat distance dependence of Phop in ssDNA (Fig. 1e). This result is not unexpected because the persistence length of ssDNA is very short compared to dsDNA (1–3 nm versus 50 nm) (13, 14). Thus, the largest uracil spacing of 40 ntds only results in an average target site separation of about 10 nanometers (Fig. 1e). Moreover, eq 4 breaks down when a ~ r because the enzyme engages a length of DNA that is similar to the site spacing (hUNG contacts at least 5 ntds of single stranded DNA) (15).
Intervening Abasic Sites Extend the Sliding Length of hUNG
We used a modified method to analyze the transfer data for the DNA constructs that contained abasic sites due to a large apparent site preference with some of these constructs (Fig. 3). This method (initial rates method) differs in that the initial rates for formation of the individual fragments are calculated first (vAB, vBC, vA, vC) and Ptrans is calculated using eq 5 (11), whereas previously we had calculated the site transfer using eq 1 at each time point and linearly extrapolated back to zero time (extrapolation method).
Figure 3.
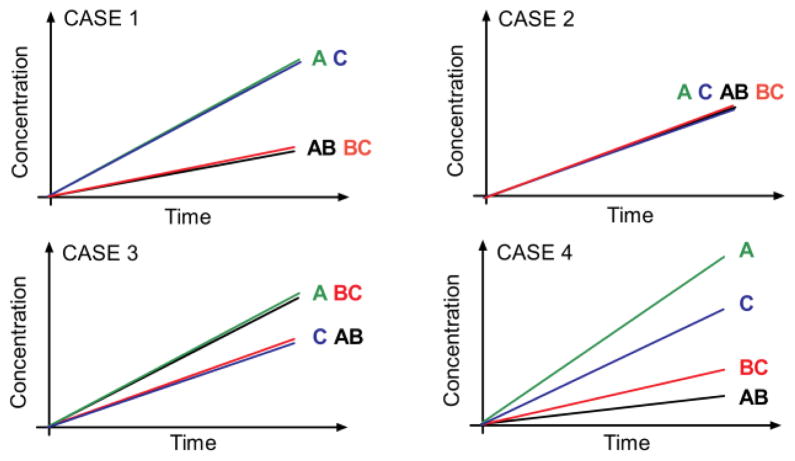
Possible outcomes for site transfer measurements as determined using the method of initial rates [adapted from (11)]. The cartoon panels depict the initial time courses for formation of the cleavage products AB, BC, A and C. Case I: hUNG is processive with an equal preference for both sites resulting in a larger initial velocity for the products A and C compared to AB and BC. Case II: hUNG is fully distributive in its reaction and the enzyme dissociates from the DNA after each single excision event, after which it becomes in equilibrium with all DNA substrate molecules. Case III: hUNG is fully distributive, but reacts preferentially at site 1 as compared to site 2, generating more of fragments A and BC. Case IV: hUNG is processive but also has a site preference. As shown in the Results and Supplemental Materials, an apparent site preference may result from either an excision preference or biased transfer in one direction.
| (5) |
Both methods are equivalent, however we find that using the initial rates in the site transfer equation was more reliable and intuitive when a site excision or directional biases to transfer were present (vAB ≠ vBC or vC ≠ vA). As previously described (11), the initial rates for formation of the individual fragments describe four possible mechanistic scenarios as follows (Fig. 3): (Case 1) when vAB = vBC = vC = vA, there is no site preference and only primary excision events occur, (Case 2) when vA = vc > vAB = vBC, there is no site preference, but directionally unbiased intramolecular transfers lead to consumption of the AB and BC intermediates, (Case 3) when vBC = vA and vAB = vc, a site preference exists, but only primary excision events occur, and (Case 4) when vA ≠ vC > vAB ≠ vBC, a site preference or directionally biased transfer is indicated, which cannot be distinguished unless other information is available.
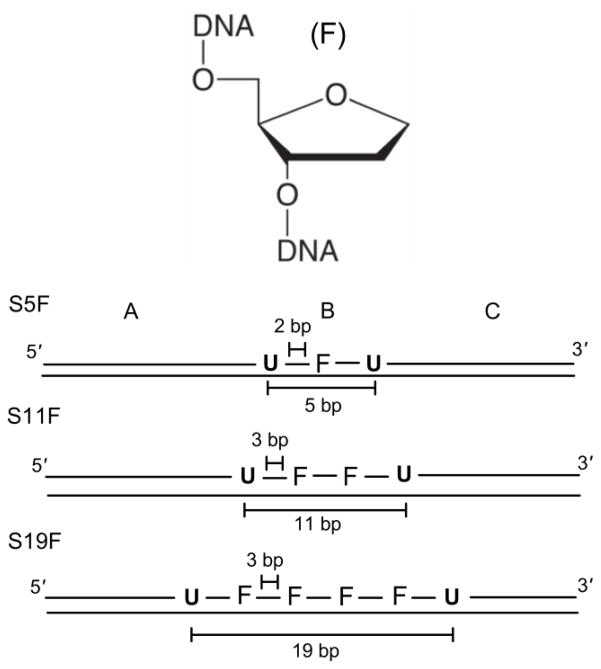
Substrates containing one or more intervening F residues were designed with five, eleven and nineteen bp uracil site spacings (Fig. 4). These uracil spacings are equal to or greater than the previously determined sliding length of ~4 bp for dsDNA and allow investigation of the effects of intervening abasic sites on both hopping and sliding efficiencies. The substrates S5F, S11F and S19F were designed to have identical two or three bp sequences surrounding the uracil target sites and the F residues were located no closer than two base pairs from the uracil sites to minimize possible direct effects on the catalytic complex. In addition, with the aim of increasing the probability of successful sliding between F sites, each intervening site was separated by three bp, which is less than the sliding length on duplex DNA. Thus the substrate with the five bp uracil spacing contained one F site, the substrate with an 11 bp spacing contained two intervening F sites, and the 19 bp site spacing contained four F sites (S5F, S11F and S19F, Fig. 4). The effect of multiple abasic site substitutions on the structure of the local intervening DNA is not obtainable, but it is reasonable to expect that a dynamic equilibrium between structures that resemble locally unpaired and paired strands might exist. Although it is certainly desirable to understand the structural effects of these pseudo-abasic site constructs, measurement of the transfer effects does not require knowledge of structure. Of course, interpretation of the observed effects must be made with this uncertainty in mind.
Figure 4.
Structure of the tetrahydrofuran abasic site mimic (F) and design of the uracil substrates containing intervening F residues.
We collected site transfer data for S5F, S11F, and S19F in the presence and absence of uracil and the collective data are provided as Supplemental Fig. S3 and the individual analyses of the velocity data are shown in Fig. 5. An interesting aspect of the transfer reactions using the F substrates was that the apparent initial velocities for primary excision events at site 1 (vBC) and 2 (vAB) became increasingly divergent as the site spacing increased, indicating a site preference. Indeed, a plot of vBC/vAB against site spacing shows that the ratio is essentially unity at the five bp uracil spacing and increases to almost 10 at the 19 bp spacing (Fig. 5a). For substrate SF5 (Fig. 5b and e), the transfer measurements in the absence and presence of uracil correspond to Case 1 in Fig. 3 (i.e. no site preference with facilitated transfers by hopping and sliding). For substrates S11F (Fig. 5c and f) and S19F (Fig. 5d and g), the measurements correspond to Case 4 (a small or large site preference, with facilitated transfers). It is notable that no previously investigated duplex or single stranded substrates for hUNG have displayed a site preference, and that the large preference for site 1 appears only as the site spacing is increased.
Figure 5.
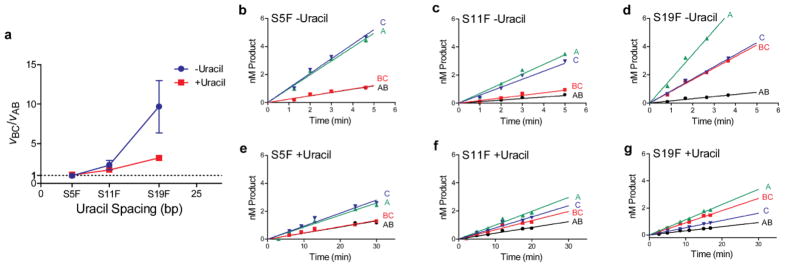
Site transfer measurements as determined using the method of initial rates for substrates S5F, S11F, and S19F. (a) The site preference (vBC/vAB) as a function of uracil site spacing in the presence and absence of uracil. Panels (b) and (e); velocities for formation of individual fragments derived from substrate S5F in the presence (b) and absence (e) of uracil. Panels (c) and (f); velocities for formation of individual fragments derived from substrate S11F in the presence (c) and absence (f) of uracil. Panels (d) and (g); velocities for formation of individual fragments derived from substrate S19F in the presence (d) and absence (f) of uracil. Reported errors are 1 SD as determined from at least 3 trials at 0 mM uracil trap and 6 trials at high uracil trap concentrations (3 trials each at 10 and 15 mM). Raw comparison showing equivalence of the initial rates and extrapolation method are presented in Supplemental Table S1.
Numerical simulations were used to explore possible interpretations for the large preference for excision at site 1 in S19F (Supplemental Methods). These simulations confirmed that the data cannot distinguish between three scenarios (i) a greater rate of cleavage at site 1 as compared to site 2, resulting in low levels of fragment AB compared to BC, (ii) a preference for transfer in the site 2→1 direction (which would consume AB efficiently to make A), or (iii) a combination of both mechanisms.
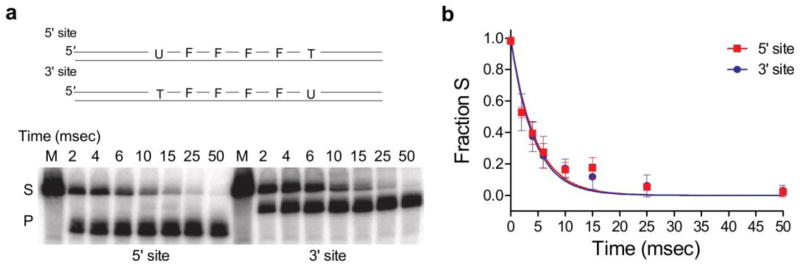
To distinguish between these possibilities, substrates were designed that were identical to S19F but contain only a single uracil site (Fig. 6a). Single turnover measurements confirmed that the rates of uracil cleavage (kex) at each uracil site (5′ or 3′) to the intervening F residues were identical (Fig. 6). Another possibility for these results would be a difference in the excision efficiency between the two sites as described above for single stranded DNA. Using an identical trapping approach as described above for the ssDNA substrate, we determined that the efficiency (E) of cleaving a uracil once the site is located is identical for each site (0.92 ± 0.12 for the 5′ site and 0.86 ± 0.04) (Supplemental Fig. S4). These results establish that there is no off-rate difference once hUNG has landed on either uracil site. Therefore, the only reasonable explanation for the observed site preference is preferential transfer in the site 2 → site 1 direction.
Figure 6.
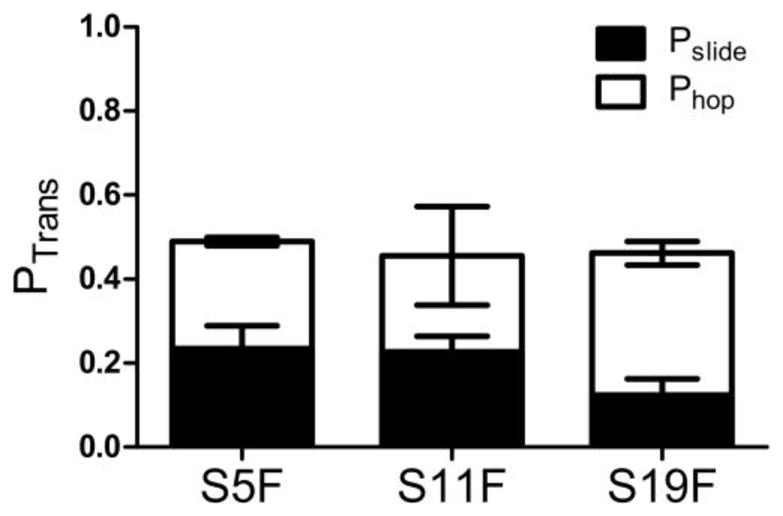
Facilitated site transfer properties of hUNG on uracil DNA constructs containing intervening F sites. The total transfer (Ptrans) is the sum of the trappable pathway or uracil sensitive (Phop) and the untrappable or uracil insensitive pathway (Pslide). Errors represent the mean plus or minus 1 s.d. determined from at least 3 trials at 0 mM uracil and 6 at high uracil (3 replicates each at 10 and 15 mM uracil).
A summary of the overall site transfer properties for these substrates is presented in Fig. 7. Notably the uracil insensitive or sliding pathway (Pslide) persists at eleven and nineteen base pairs which is considerably longer than that of duplex DNA where sliding persists over only 4–5 bp (8). Although, the molecular origin of the increased site transfer in the 5→3′ direction is not fully discernible, it is clear that the site transfer properties, including the apparent sliding length of hUNG, are very much context dependent.
Figure 7.
Single-turnover kinetic measurements of uracil cleavage from each site in the F containing DNA substrate S19F. (a) Substrate design and representative gel showing separation of substrate and product as a function of aging time in the rapid mixer using a final concentration of 4 μM hUNG and 280 nM substrate DNA (5′ or 3′ site). (b) Fraction of substrate as function of aging time. The least squares fit is to a single exponential that provides the cleavage rate of uracil from DNA. The determined rates were independent of enzyme concentration and identical within error, kex (5′ site) = 239 ± 19 s−1 and kex (3′ site) = 226 ± 17 s−1 where the error is the standard error of the least squares fit as determined using Graphpad Prism. The data points shown are the average ± SD (n = 3) of data points determined using 4 and 8 μM hUNG and 280 nM DNA substrate.
Discussion
hUNG is unique among DNA glycosylases in that it has the catalytic flexibility to remove uracils from both duplex and single stranded DNA with almost equivalent efficiency, and thus provides a valuable system to understand intramolecular site transfer within a variety of DNA contexts. Although the action of hUNG on uracils in ssDNA has not been directly established in vivo, the process of somatic hypermutation of Ig genes in B cells involves enzymatic deamination of cytosines by AID in single stranded DNA that forms transiently during active transcription of these genes (16). Indeed AID has been shown to be highly processive in deaminating neighboring cytosines leaving behind clusters of closely but not uniformly spaced uracils, similar to the spacing in our assays here (17–19). Thus, it seems hUNG likely acts on uracils positioned within a variety of contexts including ssDNA and uracils positioned among neighboring abasic sites.
Does hUNG “Slide” on ssDNA?
The present data with ssDNA substrates unambiguously show that hUNG can efficiently transfer between uracil sites in ssDNA and that transfers persist even in the presence of the uracil trap, which is at least phenomenologically consistent with “sliding” (see Results and Fig. 1d). Despite this apparent sliding behavior on the ssDNA platform, it should be noted that rigorous interpretation of the observed transfer behavior with ssDNA is inherently more complicated than dsDNA because of several intrinsic properties of ssDNA. Boundary estimates for one-dimensional sliding of hUNG on duplex DNA have been previously determined (8)(insert reference to companion paper upon publication) and it is of interest to perform a similar analysis for ssDNA to gain insights into the possible nature of sliding on ssDNA as compared to dsDNA. Calculation of a lower limit 1D diffusion constant for sliding transfers requires knowledge of the mean sliding length on ssDNA (Lslide) and the binding lifetime to nontarget ssDNA (τbind = 1/koff). The required value for Lslide = 19 ntds may be obtained from Fig. 1d, and a value for τbind = 1/KDns × kon = 4 ms may be estimated from (i) measurements of the nonspecific ssDNA binding affinity of hUNG as measured by fluorescence anisotropy (KDns 2.0 ± 0.3 μM, Supplemental Fig. S5) and, (ii) the diffusion-controlled on-rate for reaction of ssDNA substrate DNA (kon = 1.1 × 108 M−1 s−1, Supplemental Fig. S6). Insertion of these parameters into eq 6 (9), gives a value for D1 = 8 × 104 ntd2 s−1.
| (6) |
This value may be converted to standard distance units using a contour length for ssDNA of 0.6 nm under the low salt conditions employed here (13, 14), which gives D1 (ssDNA) = 3 × 10−2 μm2 s−1 (Supplemental Methods). This value may be compared with the boundary limits for duplex DNA which were calculated using two limiting cases (i) that hUNG during its entire bound lifetime is in a conformation that is competent for sliding (D1 = 0.07 × 10−2 μm2 s−1), and (ii) that hUNG is in a conformational state that is competent for sliding only 5% of its total bound lifetime (D1 = 1.4 × 10−2 μm2 s−1) (8). As discussed in the previous paper in this issue, these boundary conditions were estimated based on NMR studies of the conformational dynamics of hUNG bound to nonspecific DNA (20)(insert reference to companion paper upon publication).
The above calculations indicate that the apparent D1 for ssDNA (calculated using a sliding time equal to the total bound lifetime) is 40 times larger than the corresponding value for duplex DNA. Thus, substantial differences in the interactions or mechanism of sliding between ssDNA and duplex DNA are clearly apparent. The smaller diffusion constant for sliding on duplex DNA is not likely to arise from its higher charge density as compared to ssDNA because methylphosphonate substitution does not reveal any evidence for a strong electrostatic component to sliding (insert reference to companion paper upon publication). Since there are no structures of long ssDNA molecules bound to hUNG (21–24), it is quite possible that the flexible polymer nature of ssDNA may allow interactions over a more extended binding surface of hUNG than the more rigid duplex DNA polymer. Such an extended surface for ssDNA, and even “scrunching” of the polymer, could lead to longer apparent sliding lengths and correspondingly larger calculated diffusion constants (see above). Another possible explanation for the smaller diffusion constant for duplex DNA is that sliding on duplex DNA involves the increased frictional resistance arising from rotation-coupled diffusion along the helical DNA chain while sliding on single stranded DNA does not (25, 26). Additionally, we note that facilitated diffusion on ssDNA has been previously observed in bulk solution and single-molecule measurements of the AID family member cytidine deaminase APOBEC3G (27, 28).
Mechanism of Directional Bias During Reaction at Clustered Uracils
Although some enzymes such as helicases and DNA polymerases can use the free energy of nucleotide hydrolysis to move directionally along DNA, the movement of DNA glycosylases is driven only by thermal energy and thus would not be expected to have directional bias. Indeed, we have always found that there is no 5′ or 3′ directional bias for transfer of hUNG between uracil sites in duplex or single stranded DNA (7, 8). However, the expectation of no directional bias for a thermally driven transfer process might be negated if the intervening DNA chain connecting the sites contained high affinity regions that served as thermodynamic sinks to pull the enzyme in a biased direction.
Here we have shown that insertion of high affinity and flexible abasic site mimics between two uracil target sites can increase the average apparent sliding length of hUNG by 5-fold. This finding suggests that when hUNG acts on clustered uracils (resulting in clustered abasic sites) its search strategy is modified to increase the local search efficiency by sliding. An unexpected result in these studies was the observed 3′→5′ directional bias observed for S19F (Fig. 5d). While the exact mechanism of this directionally biased transfer is not easily discernible, the data would seem to require asymmetric interactions of hUNG with the DNA, because the only difference between the two uracil target sites is whether the F sites lie 3′ or 5′ distal to the uracil. Recent H/D exchange mass spectrometry experiments have provided evidence for an asymmetry in the interaction of hUNG with a 30 mer duplex DNA containing a F site (29). This data suggested a previously undetected DNA binding surface of the enzyme that could interact with the DNA 3′ to the product site. Thus one reason that directionality may appear in these abasic constructs and not normal DNA is that binding to the newly detected DNA binding region is favored by the introduction of duplex destabilizing lesions which allowing more facile DNA bending. This is also consistent with previous studies showing that hUNG favors binding to destabilized base pairs (30, 31). It is not known whether the 3′→5′ bias is merely related to an inherent asymmetry in the hUNG DNA binding mode, or if there exists a larger functional significance for this behavior in a biological context.
Conclusion
This report establishes that hUNG can change its search mode in the direction of longer DNA sliding events when it is confronted with ssDNA and clustered lesions such as abasic sites. This property is likely to be relevant during adaptive immunity, as selective patterning of uracil cleavage events has been shown to be important in the controlled mutagenesis of immunoglobin hypervariable sequences (32). Additionally, it is envisaged that facilitated sliding will be important in other regions of the genome where destabilized or ssDNA persists such as replication foci or regions of high negative supercoiling.
Supplementary Material
Acknowledgments
This work was supported by US National Institute of Health grant GM056834.
Abbreviations
- hUNG
catalytic domain of human uracil DNA glycosylase
- ntds
nucleotides
- bps
base pairs
- CSR
class switch recombination
- SHM
somatic hypermutation
- F
tetrahydrofuran abasic site
Footnotes
This work was supported by NIH grant GM056834 (J.T.S).
Supporting Information Available. Supplemental methods and six supporting figures: (1) mFold output showing the lack of secondary structure for the single strand substrate S20ss. (2) Control experiments for determination of the excision efficiency of ssDNA. (3) Site transfer assay gel images of F containing substrates. (4) Determination of the excision efficiency for the 5′ and 3′ uracil sites of S19F. (5) Non-specific binding of hUNG and ssDNA. (6) Steady-state kinetic parameters of hUNG cleavage of a single uracil site within ssDNA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stivers JT. Extrahelical damaged base recognition by DNA glycosylase enzymes. Chemistry. 2008;14:786–793. doi: 10.1002/chem.200701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 3.Van Triest B, Pinedo HM, Giaccone G, Peters GJ. Downstream molecular determinants of response to 5-fluorouracil and antifolate thymidylate synthase inhibitors. Ann Oncol. 2000;11:385–391. doi: 10.1023/a:1008351221345. [DOI] [PubMed] [Google Scholar]
- 4.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 5.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 7.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci USA. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schonhoft JD, Stivers JT. Timing facilitated site transfer of an enzyme on DNA. Nat Chem Biol. 2012;8:205–210. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg HC. Random walks in biology. Princeton University Press; 1993. [Google Scholar]
- 11.Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three- dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg OG, Winter RB, Von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1 Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 13.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc Natl Acad Sci USA. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg HC. Random walks in biology. Princeton University Press; 1993. [Google Scholar]
- 16.Maul RW, Maul RW, Gearhart PJ, Gearhart PJ. AID and Somatic Hypermutation. In: Alt FW, Alt FW, editors. Advances in Immunology. Academic Press; 2010. pp. 159–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 18.Pham P, Chelico L, Goodman MF. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair. 2007;6:689–92. doi: 10.1016/j.dnarep.2007.01.001. author reply 693–4. [DOI] [PubMed] [Google Scholar]
- 19.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 20.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker JB, Stivers JT. Uracil DNA glycosylase: revisiting substrate-assisted catalysis by DNA phosphate anions. Biochemistry. 2008;47:8614–8622. doi: 10.1021/bi800854g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang YL, Ichikawa Y, Song F, Stivers JT. Powering DNA Repair through Substrate Electrostatic Interactions. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 24.Xiao G, Tordova M, Jagadeesh J, Drohat AC, Stivers JT, Gilliland GL. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins. 1999;35:13–24. [PubMed] [Google Scholar]
- 25.Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA: Hydrodynamic considerations. Biophysical chemistry. 1979;9:413–414. [PubMed] [Google Scholar]
- 26.Bagchi B, Blainey PC, Xie XS. Diffusion Constant of a Nonspecifically Bound Protein Undergoing Curvilinear Motion along DNA. J Phys Chem B. 2008;112:6282–6284. doi: 10.1021/jp077568f. [DOI] [PubMed] [Google Scholar]
- 27.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 28.Senavirathne G, Jaszczur M, Auerbach PA, Upton TG, Chelico L, Goodman MF, Rueda D. Single-stranded DNA scanning and deamination by APOBEC3G cytidine deaminase at single molecule resolution. J Biol Chem. 2012;287:15826–15835. doi: 10.1074/jbc.M112.342790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, Jamison JW, Liu T, Lee JH, Tainer JA, Eyck, Ten LF, Woods VL. Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase. Nucleic Acids Res. 2012;40:6070–6081. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krosky DJ, Schwarz FP, Stivers JT. Linear Free Energy Correlations for Enzymatic Base Flipping: How Do Damaged Base Pairs Facilitate Specific Recognition? Biochemistry. 2004;43:4188–4195. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
- 31.Krosky DJ, Song F, Stivers JT. The Origins of High-Affinity Enzyme Binding to an Extrahelical DNA Base. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Durán P, Belver L, de Yébenes VG, Delgado P, Pisano DG, Ramiro AR. UNG shapes the specificity of AID-induced somatic hypermutation. J Exp Med. 2012;209:1379–1389. doi: 10.1084/jem.20112253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazur AK. Evaluation of elastic properties of atomistic DNA models. Biophys J. 2006;91:4507–4518. doi: 10.1529/biophysj.106.091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.