ABSTRACT
Male circumcision reduces female-to-male HIV transmission. Hypothesized mechanisms for this protective effect include decreased HIV target cell recruitment and activation due to changes in the penis microbiome. We compared the coronal sulcus microbiota of men from a group of uncircumcised controls (n = 77) and from a circumcised intervention group (n = 79) at enrollment and year 1 follow-up in a randomized circumcision trial in Rakai, Uganda. We characterized microbiota using16S rRNA gene-based quantitative PCR (qPCR) and pyrosequencing, log response ratio (LRR), Bayesian classification, nonmetric multidimensional scaling (nMDS), and permutational multivariate analysis of variance (PerMANOVA). At baseline, men in both study arms had comparable coronal sulcus microbiota; however, by year 1, circumcision decreased the total bacterial load and reduced microbiota biodiversity. Specifically, the prevalence and absolute abundance of 12 anaerobic bacterial taxa decreased significantly in the circumcised men. While aerobic bacterial taxa also increased postcircumcision, these gains were minor. The reduction in anaerobes may partly account for the effects of circumcision on reduced HIV acquisition.
IMPORTANCE
The bacterial changes identified in this study may play an important role in the HIV risk reduction conferred by male circumcision. Decreasing the load of specific anaerobes could reduce HIV target cell recruitment to the foreskin. Understanding the mechanisms that underlie the benefits of male circumcision could help to identify new intervention strategies for decreasing HIV transmission, applicable to populations with high HIV prevalence where male circumcision is culturally less acceptable.
Introduction
Male circumcision (MC) reduces the risk of HIV acquisition in men by 50 to 60% (1–3) and decreases the incidence and prevalence of herpes simplex virus 2 (HSV-2) (4) and human papillomavirus (HPV) (4, 5). The impact of MC on classical bacterial sexually transmitted infections (STIs), such as Chlamydia trachomatis, Neisseria gonorrhoeae, Treponema pallidum, and Trichomonas vaginalis infection, is more equivocal (4, 6–8). Women with circumcised male partners are at lower risk for STIs ranging from HPV to Trichomonas vaginalis infection (6, 9). This suggests that MC reduces the risk of viral STIs in men and of STI transmission to their female partners (10).
MC is hypothesized to reduce HIV risk in men by changing the penile anatomy and by altering the genital microbiology (11). With respect to the anatomic changes, MC removes the prepuce, which decreases the number of available HIV target cells on the penis (11, 12). It remains unclear whether decreases in viral STIs post-MC contribute to HIV risk reduction. HSV-2 infection increases the risk of HIV in observational studies (13, 14), but trials aimed at controlling viral and classical bacterial STIs have largely failed to reduce HIV transmission (15, 16). Removal of the preputial tissue also eliminates the moist subpreputial environment, which can modify the genital bacterial communities (i.e., the microbiota) and may have a broad impact on the genital microbiology (17).
Recently, genital epithelial inflammation associated with bacterial antigens has emerged as a possible factor in increasing susceptibility of genital HIV target cells (18–23). These findings suggest that specific groups of genital bacteria, including those not associated with classical STIs, could elicit local immune responses that promote epithelial inflammation and recruitment of HIV target cells. Thus, changes in the genital bacterial microbiota could be linked to HIV acquisition.
Previously, we reported the impact of MC on the coronal sulcus microbiota composition in 12 men (17). However, this study lacked uncircumcised controls. In the current study, we assessed the effect of MC on the genital microbiota using absolute abundance. In addition, we applied novel analyses to assess the microbiota changes attributable to MC. We hypothesized that MC would significantly decrease coronal sulcus bacterial abundance and modify the microbiota in participants randomly assigned to receive MC but not in those who remained uncircumcised. Here, we report a study of penile coronal sulcus microbiota in 77 control and 79 intervention-arm participants from the Rakai MC randomized controlled trial in Uganda.
RESULTS
Study participant profile at enrollment.
At enrollment, men from the control and intervention arms had similar sociodemographic characteristics, sexual practices, sexually transmitted infections, and symptoms (Table 1).
TABLE 1 .
Demographic characteristics, sexual behaviors, and symptoms of sexually transmitted infections for the control and intervention arms at enrollment
| Characteristic | No. (%) in group |
|
|---|---|---|
| Control (n = 77) | Intervention (n = 79) | |
| Age (yr) | ||
| 15–19 | 1 (1.3) | 1 (1.3) |
| 20–24 | 13 (16.9) | 15 (19.0) |
| 25–29 | 21 (27.3) | 25 (31.6) |
| 30–49 | 42 (54.5) | 38 (48.1) |
| Marital status | ||
| Currently married, monogamous | 70 (91.0) | 71 (89.9) |
| Currently married, polygamous | 7 (9.0) | 8 (10.1) |
| No. of sexual partners in past yr | ||
| 1 | 45 (58.4) | 45 (57.0) |
| 2 | 23 (29.9) | 26 (32.9) |
| ≥3 | 9 (11.7) | 8 (10.1) |
| Nonmarital sexual relationships | ||
| No | 66 (85.7) | 67 (84.8) |
| Yes | 11 (14.3) | 12 (15.2) |
| Condom use in past yr | ||
| None | 46 (59.7) | 54 (68.3) |
| Inconsistent use | 30 (39.0) | 24 (30.4) |
| Consistent use | 1 (1.3) | 1 (1.3) |
| Syphilis infection | ||
| No | 72 (93.5) | 73 (92.4) |
| Yes | 4 (5.2) | 5 (6.3) |
| Not tested | 1 (1.3) | 1 (1.3) |
| HSV-2 infection | ||
| No | 36 (46.8) | 37 (46.8) |
| Yes | 32 (41.5) | 32 (40.5) |
| Indeterminate | 9 (11.5) | 9 (11.4) |
| Not tested | 1 (1.3) | 1 (1.3) |
| Self-reported symptoms of sexually transmitted infection | ||
| Genital ulcer disease | 0 (0.0) | 0 (0.0) |
| Urethral discharge | 0 (0.0) | 1 (1.3) |
| Dysuria | 0 (0.0) | 0 (0.0) |
Coronal sulcus bacteria in the uncircumcised penis at enrollment. (i) Prevalence.
At enrollment, the prevalences of coronal sulcus bacterial were comparable between the two study arms (Table 2). Some of the most common coronal sulcus bacteria seen at enrollment included those from the Prevotellaceae, Veillonellaceae, Clostridiales family XI, Actinomycetaceae, Coriobacteriaceae, and Porphyromonadaceae. Two groups of bacteria from the order Clostridiales were highly prevalent but could not be assigned with sufficient confidence to known lower taxa and are referred to as unclassified Clostridiales family XI and unclassified Clostridiales (Table 2).
TABLE 2 .
Prevalences and proportional abundances of the 40 most common coronal sulcus bacteria in the control and intervention arms at enrollmenta
| Family | Genus | Prevalence (%) in group |
Avg proportional abundance [% (SD)] in group |
||
|---|---|---|---|---|---|
| Control (n = 77) | Intervention (n = 79) | Control | Intervention | ||
| Clostridiales family XIa | Peptoniphilus spp. | 74 (96.1) | 72 (91.1) | 5.4 (5.9) | 5.1 (5.8) |
| Clostridiales family XIa | Anaerococcus spp. | 71 (92.2) | 68 (86.1) | 5.1 (8.1) | 4.3 (6.2) |
| NA | Unclassified Clostridiales | 69 (89.6) | 70 (88.6) | 15.9 (16.0) | 14.3 (14.5) |
| Prevotellaceae | Prevotella spp. | 69 (89.6) | 67 (84.8) | 21.4 (17.0) | 23.1 (20.5) |
| Clostridiales family XIa | Finegoldia spp. | 63 (81.8) | 64 (81.0) | 6.5 (8.3) | 7.1 (10.6) |
| Clostridiales family XIa | Murdochiella spp. | 62 (80.5) | 58 (73.4) | 3.2 (4.7) | 5.2 (8.6) |
| Porphyromonadaceae | Porphyromonas spp. | 61 (79.2) | 55 (69.6) | 5.4 (6.1) | 4.8 (8.4) |
| Corynebacteriaceae | Corynebacterium spp. | 57 (74.0) | 52 (65.8) | 12.2 (21.2) | 8.5 (18.8) |
| Clostridiales family XI | Unclassified Clostridiales Family XI | 54 (70.1) | 53 (67.1) | 0.7 (0.8) | 0.8 (1.5) |
| Veillonellaceae | Dialister spp. | 53 (68.8) | 43 (54.4) | 1.6 (1.9) | 1.0 (1.6) |
| Veillonellaceae | Negativicoccus spp. | 40 (51.9) | 36 (45.6) | 1.0 (2.4) | 0.8 (1.8) |
| Peptostreptococcaceae | Peptostreptococcus spp. | 31 (40.3) | 36 (45.6) | 0.9 (2.1) | 1.3 (2.7) |
| Actinomycetaceae | Mobiluncus spp. | 38 (49.4) | 26 (32.9) | 1.5 (5.2) | 1.1 (3.8) |
| Bifidobacteriaceae | Gardnerella spp. | 33 (42.9) | 25 (31.6) | 1.8 (5.9) | 0.9 (3.2) |
| Lactobacillaceae | Lactobacillus spp. | 26 (33.8) | 28 (35.4) | 2.4 (10.7) | 8 (21.0) |
| Staphylococcaceae | Staphylococcus spp. | 29 (37.7) | 21 (26.6) | 1.4 (3.9) | 1.1 (6.0) |
| Ruminococcaceae | Saccharofermentans spp. | 28 (36.4) | 21 (26.6) | 0.3 (0.6) | 0.5 (1.9) |
| Streptococcaceae | Streptococcus spp. | 26 (33.8) | 19 (24.1) | 1.3 (6.1) | 0.5 (2.8) |
| Actinomycetaceae | Actinomyces spp. | 26 (33.8) | 17 (21.5) | 0.1 (0.5) | 0.1 (0.2) |
| Veillonellaceae | Veillonella spp. | 26 (33.8) | 14 (17.7) | 0.5 (1.5) | 0.6 (3.1) |
| Peptococcaceae 1 | Peptococcus spp. | 25 (32.5) | 14 (17.7) | 0.1 (0.1) | 0.04 (0.1) |
| Coriobacteriaceae | Olsenella spp. | 20 (26.0) | 19 (24.1) | 0.1 (0.2) | 0.1 (0.2) |
| Actinomycetaceae | Arcanobacterium spp. | 23 (29.9) | 14 (17.7) | 0.1 (0.2) | 0.1 (0.2) |
| Lachnospiraceae | Howardella spp. | 19 (24.7) | 9 (11.4) | 0.1 (0.1) | 0.03 (0.1) |
| Clostridiales family XIa | Parvimonas spp. | 17 (22.1) | 10 (12.7) | 0.2 (0.5) | 0.3 (1.2) |
| Coriobacteriaceae | Atopobium spp. | 14 (18.2) | 12 (15.2) | 0.1 (0.2) | 0.2 (1.1) |
| Leptotrichiaceae | Sneathia spp. | 13 (16.9) | 13 (16.5) | 0.2 (0.8) | 0.3 (1.4) |
| Sutterellaceae | Sutterella spp. | 13 (16.9) | 12 (15.2) | 0.1 (0.2) | 0.03 (0.1) |
| Lachnospiraceae | Moryella spp. | 14 (18.2) | 7 (8.9) | 0.1 (0.2) | 0.03 (0.1) |
| Peptostreptococcaceae | Peptostreptococcaceae family | 12 (15.6) | 9 (11.4) | 0.1 (0.5) | 0.03 (0.1) |
| Spirochaetaceae | Treponema spp. | 10 (13.0) | 11 (13.9) | 0.2 (0.6) | 0.2 (0.5) |
| Fusobacteriaceae | Fusobacterium spp. | 8 (10.4) | 13 (16.5) | 0.2 (0.8) | 0.8 (3.8) |
| Synergistaceae | Pyramidobacter spp. | 13 (16.9) | 7 (8.9) | 0.2 (0.8) | 0.2 (0.8) |
| Aerococcaceae | Facklamia spp. | 12 (15.6) | 8 (10.1) | 0.1 (0.3) | 0.1 (0.5) |
| Clostridiales family XIa | Anaerosphaera spp. | 9 (11.7) | 11 (13.9) | 0.02 (0.1) | 0.1 (0.2) |
| Micrococcaceae | Kocuria spp. | 10 (13.0) | 8 (10.1) | 0.05 (0.2) | 0.1 (0.2) |
| Veillonellaceae | Megasphaera spp. | 10 (13.0) | 8 (10.1) | 0.2 (0.6) | 0.1 (0.4) |
| Micrococcaceae | Micrococcus spp. | 8 (10.4) | 10 (12.7) | 0.04 (0.2) | 0.04 (0.1) |
| Bacillales family XI | Gemella spp. | 10 (13.0) | 6 (7.6) | 0.04 (0.1) | 0.03 (0.1) |
| Burkholderiaceae | Ralstonia spp. | 13 (16.9) | 2 (2.5) | 0.1 (0.3) | 0.01 (0.1) |
*, false discovery rate (FDR)-adjusted P value < 0.05.
(ii) Relative abundance.
Most coronal sulcus bacteria were observed in relatively low abundances (Table 2). Prevotella spp. were the most dominant, followed by unclassified members of the Clostridiales and Corynebacterium spp. Six others—Peptoniphilus spp., Anaerococcus spp., Finegoldia spp., Murdochiella spp., Porphyromonas spp., and Lactobacillus spp.—were found at relative abundances of approximately 5%. The remaining coronal sulcus bacteria were detected at lower than 1% (Table 1).
Male circumcision reduces coronal sulcus bacterial load.
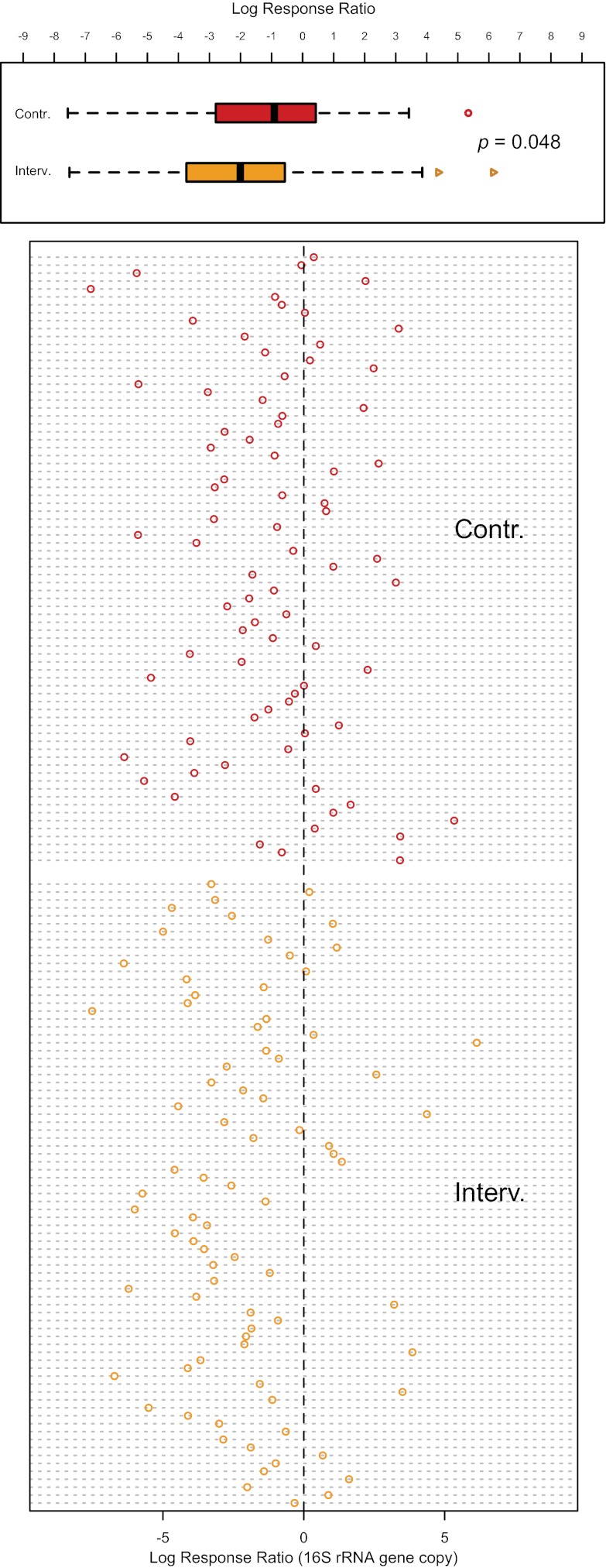
At enrollment, similar mean bacterial loads were seen in the two study groups based on measurements of the bacterial 16S rRNA gene, with an average of 1.4 × 105 copies (standard deviation [SD] = 3.1 × 105) in the control arm and 2.0 × 105 copies (SD = 4.8 × 105) in the intervention arm. At year 1, the total bacterial load decreased significantly in both arms. In the uncircumcised men, the average bacterial load decreased to 5.7 × 104 copies (SD = 1.19 × 105), but the circumcised men had an average of 3.8 × 104 copies (SD = 1.80 × 105) (log response ratio P = 0.048) (Fig. 1). Thus, MC significantly decreased the coronal sulcus bacterial load relative to changes in uncircumcised men.
FIG 1 .
Changes in the coronal sulcus bacterial load as measured by the log response ratio for the uncircumcised (Contr.; in red) versus the circumcised (Interv.; in orange) men, shown by group (top panel) and by individual (bottom panel). In the group comparison, the box of each box plot denotes the interquartile range (IQR) (quartile 1 [Q1] to Q3) and the corresponding median, whereas the whiskers signify the upper and lower 1.5× IQR. Outliers are shown as open symbols in each box plot. There was a statistically significant reduction in bacterial load for the circumcised men compared to that for the uncircumcised men (P = 0.048). As shown in the scatter plot in the bottom panel, although a decrease was observed for many individuals from both groups, more circumcised men showed decreases (i.e., negative log response ratios) (62/79, 78.5%) than did those that remained uncircumcised (51/77, 66.2%).
Male circumcision significantly altered prevalences of coronal sulcus bacteria.
Fifteen coronal sulcus bacteria significantly decreased in prevalence post-MC (P < 0.05), among which 12 were strict anaerobes, including Porphyromonas spp. (ΔΔPrevalence = −43.10%), Prevotella spp. (ΔΔPrevalence = −34.21%), Negativicoccus spp. (ΔΔPrevalence = −28.95%), Dialister spp. (ΔΔPrevalence = −30.18%), Mobiluncus spp. (ΔΔPrevalence = −13.69%), and six genera from Clostridiales family XI, among others (Table 3). The reductions in anaerobe prevalence due to MC were often substantial, but MC did not significantly reduce all anaerobes; notably, Atopobium spp., Sneathia spp., and Megasphaera spp. showed no statistically significant decrease post-MC.
TABLE 3 .
Prevalences and changes in prevalence of the 40 most common coronal sulcus bacteria for uncircumcised and circumcised men at year 1
| Bacterial group | Oxygen tolerancea | Prevalence (%)b |
ΔΔPrevalence (%)c | |
|---|---|---|---|---|
| Uncircumcised | Circumcised | |||
| Peptoniphilus spp. | AN | 71 (92.2) | 37 (46.8)** | −40.41 |
| Anaerococcus spp. | AN | 71 (92.2) | 59 (74.7) | −11.39 |
| Unclassified Clostridiales spp. | NA | 65 (84.4) | 38 (48.1)** | −35.31 |
| Prevotella spp. | AN | 70 (90.9) | 41 (51.9)** | −34.21 |
| Finegoldia spp. | AN | 61 (79.2) | 38 (48.1)** | −30.31 |
| Murdochiella spp. | AN | 51 (66.2) | 12 (15.2)** | −43.94 |
| Porphyromonas spp. | AN | 62 (80.5) | 23 (29.1)** | −43.10 |
| Corynebacterium spp. | FAN | 71 (92.2) | 77 (97.5)** | 13.46 |
| Unclassified Clostridiales family XI | NA | 55 (71.4) | 20 (25.3)** | −43.07 |
| Dialister spp. | AN | 47 (61.0) | 13 (16.5)** | −30.18 |
| Negativicoccus spp. | AN | 35 (45.5) | 8 (10.1)** | −28.95 |
| Peptostreptococcus spp. | AN | 32 (41.6) | 13 (16.5)* | −30.41 |
| Mobiluncus spp. | AN | 31 (40.3) | 8 (10.1)* | −13.69 |
| Gardnerella spp. | FAN | 34 (44.2) | 32 (40.5) | 7.56 |
| Lactobacillus spp. | FAN/AN/MAE | 32 (41.6) | 35 (44.3) | 1.07 |
| Staphylococcus spp. | FAN | 48 (62.3)* | 69 (87.3)** | 36.08 |
| Saccharofermentans spp. | AN | 21 (27.3) | 10 (12.7) | −4.83 |
| Streptococcus spp. | FAN | 34 (44.2) | 28 (35.4) | 1.00 |
| Actinomyces spp. | FAN | 27 (35.1) | 8 (10.1) | −12.69 |
| Veillonella spp. | AN | 26 (33.8) | 16 (20.3) | 2.53 |
| Peptococcus spp. | AN | 14 (18.2) | 7 (8.9) | 5.42 |
| Olsenella spp. | AN | 19 (24.7) | 5 (6.3) | −16.42 |
| Arcanobacterium spp. | FAN | 15 (19.5) | 4 (5.1) | −2.27 |
| Howardella spp. | AN | 18 (23.4) | 5 (6.3) | −3.76 |
| Parvimonas spp. | AN | 13 (16.9) | 12 (15.2) | 7.73 |
| Atopobium spp. | AN | 18 (23.4) | 11 (13.9) | −6.46 |
| Sneathia spp. | AN | 15 (19.5) | 9 (11.4) | −7.66 |
| Sutterella spp. | AN | 15 (19.5) | 2 (2.5)* | 15.26 |
| Moryella spp. | AN | 11 (14.3) | 4 (5.1) | 0.10 |
| Peptostreptococcaceae family | NA | 4 (5.2) | 1 (1.3) | 0.26 |
| Treponema spp. | AN | 8 (10.4) | 4 (5.1) | −6.26 |
| Fusobacterium spp. | AN | 17 (22.1) | 11 (13.9) | −14.22 |
| Pyramidobacter spp. | AN | 7 (9.1) | 3 (3.8) | 2.73 |
| Facklamia spp. | FAN | 17 (22.1) | 21 (26.6) | 9.96 |
| Anaerosphaera spp. | AN | 9 (11.7) | 7 (8.9) | −5.06 |
| Kocuria spp. | AE | 20 (26.0) | 36 (45.6)** | 22.46 |
| Megasphaera spp. | AN | 11 (14.3) | 10 (12.7) | 1.23 |
| Micrococcus spp. | AE | 22 (28.6) | 39.24 | 8.40 |
| Gemella spp. | FAN | 15 (19.5) | 13 (16.5) | 2.37 |
| Ralstonia spp. | AE | 16 (20.8) | 2 (2.5) | −3.90 |
AN, strictly anaerobic; AE strictly aerobic; FAN, facultative anaerobic; MAE, microaerophilic; NA, no data.
**, FDR-adjusted P value < 0.0001 for change in prevalence over time (i.e., ΔPrevalence); *, FDR-adjusted P value < 0.05 for ΔPrevalence.
ΔΔPrevalence, shown as a percentile, is the change in prevalence seen for the circumcised men over time compared to that for men that remain uncircumcised.
Seven coronal sulcus bacteria became more prevalent post-MC. Among these, five also increased in prevalence in the uncircumcised men over time, suggesting either an effect of time or changes in behavior with participation in the trial. Nevertheless, a greater number of the circumcised than of the uncircumcised men acquired these specific bacteria, as shown by the positive ΔΔPrevalence values (Table 3). The aerobic Kocuria spp. and the facultative anaerobic Facklamia spp. were the two types of bacteria that became more prevalent exclusively in the circumcised men. Other bacteria were uncommon in the uncircumcised penis but increased in prevalence post-MC (see Table S1 in the supplemental material).
Male circumcision modified coronal sulcus microbiota biodiversity and composition. (i) Microbiota biodiversity.
MC significantly reduced the evenness of the microbiota, indicating a general decrease in the number of dominant coronal sulcus bacteria post-MC (E treatment effect = −0.053; 95% CI = −0.101 to −0.005). In addition, MC also significantly decreased the structural diversity of the microbiota (D treatment effect = −1.26; 95% CI = −2.04 to −0.52)
(ii) Microbiota composition.
MC reshaped the composition of the coronal sulcus microbiota, producing a more homogeneous post-MC profile (Fig. 2A and B). While both study groups show significant temporal changes in microbiota composition, the change was more marked in the circumcised men (PerMANOVA F statistic = 13.1; P < 0.001) (Fig. 2A; see also Fig. S1 in the supplemental material) than in the uncircumcised men (PerMANOVA F statistic = 3.1l P = 0.02) (Fig. 2B; see also Fig. S1).
FIG 2 .
The nonmetric multidimensional scaling (nMDS) ordination plots enable the visualization of individuals’ microbiota over time. In nMDS plots, each data point represents an individual’s microbiota at one time point. The centroids and 95% confidence ellipses for each study group are as shown. Here, the coronal sulcus microbiota in men that remained uncircumcised showed minor variations from enrollment (in blue) to year-1 (in orange) (Fig. 2A). In contrast, significant shifts were seen in the circumcised men (Fig. 2B).
Circumcision significantly reduced previously abundant coronal sulcus bacteria.
To quantify the impact of MC on coronal sulcus bacteria, we determined the MC effect size. This was performed for genera that either significantly decreased (i.e., “negative responders”) or increased (i.e., “positive responders”) after MC (Table 4). Among the negative responders, Prevotella spp., Porphyromonas spp., Finegoldia spp., and Peptostreptococcus spp. decreased in both prevalence and absolute abundance, with effective load reductions ranging from −1,157 to −25,327 16S rRNA gene copies (Table 4). Other negative responders decreased significantly in either prevalence (n = 8) or absolute abundance (n = 2). Several negative responders had substantial effective load reductions that were also highly variable, such as unclassified Clostridiales, Peptoniphilus spp., and Murdochiella spp. As a result, they have large but non-statistically significant effect sizes (Table 4).
TABLE 4 .
Effect size of MC, measured as the change in absolute abundances of coronal sulcus bacteria that significantly decreased (i.e., “negative responders”) or increased (i.e., “positive responders”) post-MC, adjusted by changes in abundance among uncircumcised men over timea
| Category and bacterial groupb | Indicator value (FDR-adjusted P value) | MC effect size [mean (90% CI)] |
|---|---|---|
| Negative responders to MC | ||
| Prevotella spp.*** | 0.18 (0.06) | −25,327 (−48,812 to −3,988) |
| Porphyromonas spp.*** | 0.27 (<0.01) | −14,232 (−28,698 to −2,358) |
| Unclassified Clostridiales spp.* | 0.24 (0.01) | −10,087 (−32,278 to 12,802) |
| Unclassified Clostridiales family XI* | 0.22 (0.03) | −3,299 (−8,715 to 647) |
| Murdochiella spp.* | 0.30 (<0.01) | −3,207 (−10,314 to 5,319) |
| Peptoniphilus spp.* | 0.29 (<0.01) | −1,349 (−8,121 to 5,388) |
| Finegoldia spp.*** | 0.24 (0.07) | −1,343 (−2,385 to −438) |
| Anaerococcus spp.** | 0.27 (0.03) | −1,284 (−2,292 to −337) |
| Peptostreptococcus spp.*** | 0.21 (<0.01) | −1,157 (−2,415 to −188) |
| Mobiluncus spp.* | 0.16 (0.02) | −568 (−1,480 to 290) |
| Actinomyces spp. | 0.20 (0.01) | −286 (−780 to 13) |
| Saccharofermentans spp. | 0.16 (0.03) | −281 (−1,545 to 1,032) |
| Negativicoccus spp.* | 0.28 (<0.01) | −244 (−785 to 321) |
| Sutterella spp.* | 0.13 (0.01) | −110 (−258 to 12) |
| Howardella spp. | 0.14 (0.01) | −44 (−175 to 75) |
| Olsenella spp.* | 0.17 (0.01) | −37 (−270 to 171) |
| Ralstonia spp. | 0.14 (0.01) | 0 (0 to 1) |
| Dialister spp.* | 0.24 (<0.01) | 1,434 (−1,259 to 4,618) |
| Positive responders to MC | ||
| Rothia spp. | 0.12 (<0.01) | 1 (0 to 2) |
| Roseomonas spp. | 0.11 (0.04) | 1 (0 to 2) |
| Kocuria spp.*** | 0.32 (<0.01) | 8 (5 to 11) |
| Brevibacterium*** | 0.30 (<0.01) | 13 (5 to 22) |
| Eremococcus** | 0.26 (0.06) | 45 (8 to 94) |
| Bifidobacterium | 0.16 (<0.01) | 101 (−7 to 243) |
| Helcococcus*** | 0.21 (0.02) | 161 (22 to 345) |
| Staphylococcus*** | 0.56 (<0.01) | 259 (168 to 353) |
| Corynebacterium*** | 0.50 (<0.01) | 2,857 (1,440 to 4,722) |
The negative and positive responders to MC were identified using indicator analysis, which also produced the indicator values.
*, significant change in prevalence only; **, significant change in absolute abundance (i.e., load) only; ***, significant change in both prevalence and absolute abundance.
In contrast, many positive responders had smaller MC effect sizes that were statistically significant, a finding that indicated a more uniform bacterial gain among circumcised men. On average, Corynebacterium spp. increased by 2,860 and Staphylococcus spp. by 249 16S rRNA gene copies per individual (Table 4). The third-highest mean MC effect size was seen in Helcococcus spp., which belong to Clostridiales family XI, and that response contrasts with the broadly negative impact of MC on other Clostridiales family XI members. Overall, the relatively larger MC effect sizes in negative responders indicate that MC primarily reduced previously abundant coronal sulcus bacteria, accompanied by other minor abundance gains.
DISCUSSION
In a randomized trial of MC, we showed that MC significantly reduced the bacterial load by reducing both the prevalence and the absolute abundance of many coronal sulcus bacteria. The two study groups had comparable coronal sulcus microbiota at enrollment that consisted of multiple microbiota types, but MC profoundly altered the composition of the microbiota and reduced its biodiversity. Over time, changes in the coronal sulcus microbiota were observed in the uncircumcised men. However, after adjusting for these temporal changes, we found that there were significantly greater decreases in the total bacterial load, microbiota biodiversity, and microbiota composition in the circumcised men that were attributable to MC.
The role of coronal sulcus bacteria in heterosexual HIV acquisition remains unknown. Recent studies suggest that the non-STI genital bacteria may affect the susceptibility of foreskin HIV target cells (22, 24). Of the HIV target cell types found in the foreskin, Langerhans cells (LCs) have been hypothesized to play a key role in mediating HIV infection (24). Located proximal to the epithelial surface, naive LCs bind, internalize, and degrade HIV particles; however, when activated by a high HIV load, active STIs, or bacterium-associated inflammatory mediators, such as lipopolysaccharide (LPS) and tumor necrosis factor alpha (TNF-α), LCs bind and present HIV particles to CD4+ T cells (24, 25).
As already mentioned, the changes in penile microbiota and STI incidence after MC may be attributable not only to the anatomic alteration itself but also to behavioral changes in circumcised men or men enrolled in a clinical trial in general. However, analysis of the Rakai data showed that MC did not significantly alter behavior during the trial (1). Likewise, the natural dynamics of the circumcised coronal sulcus microbiota are unknown, but sampling performed 1 year postcircumcision likely represents a persistent change. Our use of novel analysis metrics, such as the log response ratio and MC effect size, permitted adjustment for the impact of time and trial participation, which allowed us to quantify microbiota changes attributable solely to MC.
MC has been associated with reduction of bacterial vaginosis (BV) in female sexual partners, but the sharing of genital microbiota between sexual partners is not well understood (6, 26). We found that a subset of bacteria associated with BV decreased after MC, including Prevotella spp., Fusobacterium spp., and Mobiluncus spp., while others, such as Gardnerella spp., Sneathia spp., Actinomyces spp., Atopobium spp., Megasphaera spp., and Veillonella spp., were not significantly altered.
MC selected for bacteria capable of surviving in the aerated circumcised microenvironment. At enrollment, the microbiota types were comparable in the two study groups. However, nearly all of the bacteria that decreased after MC were strict anaerobes, except for Actinomyces spp. and Arcanobacterium spp., which are facultative anaerobes. We also show that as a facultative anaerobe (27), Helcococcus spp. constituted the single positive responder to MC in Clostridiales family XI. The large competitive advantage of a single genus from a previously diverse and abundant bacterial family illustrates the strong and functionally cohesive selective pressure exerted by MC through changes to the coronal sulcus microenvironment.
One of the largest positive responders to MC was the Staphylococcus species group. Although we did not attempt to perform species-level analysis, the most abundant Staphylococcus bacteria on the post-MC coronal sulcus included S. haemolyticus, S. hominis, S. epidermidis, S. xylosus, and their genetic near neighbors. It is important to note that S. aureus and S. epidermidis are common commensals on exposed human epithelial and mucosal surfaces. Thus, their increase post-MC is unlikely to affect the pathogenic potential.
We integrated culture-independent bacterial identification, an ecological analytical framework, and a randomized study design to reveal the impact of MC on the penis microbiome. Combining bacterial quantification with parallel sequencing showed that circumcision resulted in significant decreases in the absolute abundances of several anaerobic bacterial taxa that defined the uncircumcised penis microbiome. Currently, we know little about the role of these fastidious anaerobes in the male urogenital tract or the broader context of human health. Future studies are required to determine if a decreased anaerobic bacterial load modifies foreskin inflammation and HIV target cell recruitment/susceptibility, which may play a role in HIV risk reduction conferred by MC.
MATERIALS AND METHODS
Study design and subjects.
We conducted a randomized trial of MC for HIV prevention in 2004 to 2006 (1). In this study, HIV-negative, uncircumcised men of ages 15 to 49 were randomized to either immediate circumcision (intervention group) or circumcision delayed for 24 months (control group), as described previously (1, 4). All circumcision procedures were performed in one surgical facility by the same team of urologists and trained medical officers, using a single surgical procedure, the “sleeve method” of circumcision. Prophylactic antibiotics were not used for these procedures, and antibiotic use in this geographic region was minimal. Study participants were provided access to regular reproductive health services and followed at 6, 12, and 24 months to assess HIV and sexually transmitted infection (STI) acquisition, as described in detail elsewhere (1). Men with a diagnosis of syphilis or symptoms suggestive of an STI were treated, but this was uncommon. Specifically, there were three new cases of syphilis in the control group and one new case in the intervention group who were treated with intramuscular benzathine penicillin. In addition, five control group men but none from the intervention group were treated for STI symptoms at month 6, and three control group men and three intervention group men were treated at year 1. Symptomatic patients were given azithromycin, ciprofloxacin, or metronidazole. HSV-2 and HPV data were not available at the time of the trial, and thus no treatment was given.
At each visit, clinicians collected penile swabs from the coronal sulcus as follows. Sterile cotton-tipped applicators (Thermo, Fisher Scientific, Waltham, MA) were premoistened with sterile saline and rolled over the coronal sulcus twice in a nontraumatic fashion. The swabs were immediately placed in 1 ml of Amplicor specimen transport medium (Roche Diagnostics, Indianapolis, IN) and stored at −80°C until analysis. In this analysis, we evaluated the enrollment and year 1 swabs from 77 control and 79 intervention arm participants, selected at random from among all married men who, together with their spouses, remained persistently HIV negative during the trial. The study was approved by four institutional review boards: the Science and Ethics Committee of the Uganda Virus Research Institute (Entebbe, Uganda), the HIV subcommittee of the National Council for Science and Technology (Kampala, Uganda), the Committee for Human Research at Johns Hopkins University’s Bloomberg School of Public Health (Baltimore, MD), and the Western Institutional Review Board (Olympia, WA).
Sample processing.
We processed samples from each participant in the same batch to control for interrun variation. For each sample, we lysed 100 µl of eluted transport medium using enzyme-free chemical and mechanical lysis. We purified the lysate using a Qiagen AllPrep DNA/RNA minikit (Qiagen, Valencia, CA) and performed DNA elution using 100 µl of buffer EB. Additional methodological details can be found in Text S1 in the supplemental material.
Bacterial load quantification and 16S rRNA gene-based pyrosequencing analysis.
We quantified the bacterial load, measured as the bacterial 16S rRNA gene copies per μl of coronal sulcus swab eluent, using a broad-coverage qPCR assay (28). We also generated bar-coded V3-V6 amplicons using broad-coverage fusion PCR primers, which were pooled and sequenced on the Genome Sequencer FLX instrument (Roche Applied Science, Branford, CT). Resultant pyrosequences were chimera checked (29), demultiplexed, and quality checked (30). We performed taxonomic classification using the Ribosomal Database Project Naïve Bayesian Classifier (RDP release 10, update 28) (31). Detailed description of the bioinformatics analyses can be found in Text S1 in the supplemental material.
After stringent filtering, pyrosequencing yielded a total of 104,425 16S rRNA gene sequences for samples from control men at enrollment and 90,560 at year 1; for the intervention group, there were 88,834 16S rRNA gene sequences at enrollment and 66,265 at year 1. These sequences represented 18 phyla, 31 classes, 49 orders, 121 families, and 306 genera at a ≥80% bootstrap confidence level after excluding taxonomic groups with only a single sequence detected from the full sample set. For Clostridiales and Clostridiales family XI, many sequences could not be further classified at a ≥80% bootstrap confidence level. These were included in the data set as unclassified Clostridiales and unclassified Clostridiales family XI, respectively.
Bacterial load comparison.
We expressed the bacterial load change in each participant over time as a log response ratio (LRR) using the following: ln[(bacterial load at year 1)/(bacterial load at baseline)] (32). LRR quartiles and means for participants from each group were plotted in the R (version 2.13.1) software environment (33) and compared used a two-tailed t test with unequal variance at an α value of 0.05.
16S rRNA gene-based microbiota comparative analysis.
We analyzed the coronal sulcus microbiota based on operational taxonomic unit (OTU), i.e., the unique bacterial groups detected at each taxonomic level. We converted the per-participant OTU data into four metrics: prevalence, relative abundance, absolute abundance, and log-transformed absolute abundance.
We calculated each OTU’s prevalence as (total number of participants positive for the OTU in group X)/(total number of participants in group X) and the relative abundance as (number of sequences assigned to the OTU in participant A)/(total number of sequences from participant A). We calculated absolute abundance using the formula (relative abundance of each OTU in participant A) × (bacterial load in participant A) and the log-transformed absolute abundance as ln(absolute abundance + 1).
For the 40 most common genera at enrollment, we compared the baseline prevalences and relative abundances between the study arms using the chi-square test and two-tailed t test, respectively. We assessed the change in prevalence (i.e., ΔPrevalence) at year 1 in each arm using a presence-absence data matrix and a two-tailed paired t test. All P values were adjusted for false discovery. The ΔPrevalences between the circumcised and uncircumcised men were further compared based on the following: ΔΔPrevalence = [(ΔPrevalenceintervention) − (ΔPrevalencecontrol)].
We compared the change in overall microbiota composition visually based on family-level log-transformed absolute abundance data using nonmetric multidimensional scaling (nMDS) and Bray-Curtis distance (34–36). The resultant nMDS plots were annotated with centroids and 95% confidence ellipses (34). We assessed the microbiota change over time for each study group using permutational multivariate analysis of variance (PerMANOVA) (34) based on the log-transformed absolute abundance data in Euclidean distance.
We assessed the change in microbiota biodiversity in each group using two biodiversity metrics: diversity (D), calculated as D = Simpson’s diversity index, and evenness (E), calculated as E = D/S, where S is richness (37). Evenness reflects the dominance by many (i.e., high evenness) versus few (i.e., low evenness) OTUs, whereas richness is a measurement of the total number of unique OTUs present. We calculated ΔE and ΔD for each individual and applied bootstrapping to generate random control-intervention pairs (i = 1,000). We estimated the mean E and D effect sizes (ES) as follows: mean EES = mean (ΔEintervention Xi − ΔEcontrol Yi) and mean DES = mean (ΔDintervention Xi − ΔDcontrol Yi), as well as the accompanying 95% confidence intervals.
We identified indicator bacterial genera impacted significantly by MC with indicator species analysis using log-transformed data (38). For these indicator genera, we quantified the mean MC effect sizes and the 90% confidence intervals. Detailed description of the statistical analyses can be found in Text S1 in the supplemental material.
Literature review.
We performed a literature review of the oxygen tolerance of the 40 most common genera in the uncircumcised-group microbiota. Bergey’s Manual of Determinative Bacteriology (39) was used for Corynebacterium spp., Lactobacillus spp., Staphylococcus spp., and Streptococcus spp. For others, we performed a search of the MEDLINE database via the PubMed tool with a cutoff date of April 2012 using a combined term of the applicable genus name and “nov” to identify publications defining new species within the genus. Detailed results from the literature review can be found in Table S2 in the supplemental material.
SUPPLEMENTAL MATERIAL
Detailed laboratory protocols and bioinformatic methods. Download
An nMDS plot comparing changes in the coronal sulcus microbiota composition from enrollment (in orange) to year 1 (in blue) in the two study groups (see key, top left). Each data point represents an individual’s microbiota at a given time point. The centroids and 95% confidence ellipses for each group are as shown. The microbiota composition at enrollment was similar between the controls and the intervention groups and remained largely unchanged among the uncircumcised men at year 1. In contrast, microbiota composition for the circumcised men changed significantly post-MC and formed a cluster distinct from the uncircumcised-group microbiota profiles. Download
Comparison of year-1 prevalence, as well as changes in prevalence from enrollment to year 1 for uncircumcised versus circumcised men for all detected coronal sulcus bacterial genera.
References for oxygen tolerance designations.
ACKNOWLEDGMENTS
Funding for this work was provided by grants R01AI087409-01A1, U01AI51171, and 1U01AI075115-01A1 from the National Institutes of Health. C.M.L. was supported by the Northern Arizona University Technology and Research Initiative Fund (TRIF) and the Cowden Endowment in Microbiology at Northern Arizona University. A.A.R.T. was supported by NIH 1K23AI093152-01A1 and the Doris Duke Charitable Foundation Clinician Scientist Development Award (no. 22006.02).
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Footnotes
CitationLiu CM, Hungate BA, Tobian AAR, Serwadda D, Ravel J, Lester R, Kigozi G, Aziz M, Galiwango RM, Nalugoda F, Contente-Cuomo TL, Wawer MJ, Keim P, Gray RH, Price LB. 2013. Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. mBio 4(2):e00076-13. doi:10.1128/mBio.00076-13.
REFERENCES
- 1. Gray RH, Kigozi G, Serwadda D, Makumbi F, Watya S, Nalugoda F, Kiwanuka N, Moulton LH, Chaudhary MA, Chen MZ, Sewankambo NK, Wabwire-Mangen F, Bacon MC, Williams CF, Opendi P, Reynolds SJ, Laeyendecker O, Quinn TC, Wawer MJ. 2007. Male circumcision for HIV prevention in men in Rakai, Uganda: a randomised trial. Lancet 369:657–666 [DOI] [PubMed] [Google Scholar]
- 2. Auvert B, Taljaard D, Lagarde E, Sobngwi-Tambekou J, Sitta R, Puren A. 2005. Randomized, controlled intervention trial of male circumcision for reduction of HIV infection risk: the ANRS 1265 trial. PLoS Med. 2:e298 http://dx.doi.org/10.1371/journal.pmed.0020298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey RC, Moses S, Parker CB, Agot K, Maclean I, Krieger JN, Williams CF, Campbell RT, Ndinya-Achola JO. 2007. Male circumcision for HIV prevention in young men in Kisumu, Kenya: a randomised controlled trial. Lancet 369:643–656 [DOI] [PubMed] [Google Scholar]
- 4. Tobian AA, Serwadda D, Quinn TC, Kigozi G, Gravitt PE, Laeyendecker O, Charvat B, Ssempijja V, Riedesel M, Oliver AE, Nowak RG, Moulton LH, Chen MZ, Reynolds SJ, Wawer MJ, Gray RH. 2009. Male circumcision for the prevention of HSV-2 and HPV infections and syphilis. N. Engl. J. Med. 360:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gray RH, Serwadda D, Kong X, Makumbi F, Kigozi G, Gravitt PE, Watya S, Nalugoda F, Ssempijja V, Tobian AA, Kiwanuka N, Moulton LH, Sewankambo NK, Reynolds SJ, Quinn TC, Iga B, Laeyendecker O, Oliver AE, Wawer MJ. 2010. Male circumcision decreases acquisition and increases clearance of high-risk human papillomavirus in HIV-negative men: a randomized trial in Rakai, Uganda. J. Infect. Dis. 201:1455–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gray RH, Kigozi G, Serwadda D, Makumbi F, Nalugoda F, Watya S, Moulton L, Chen MZ, Sewankambo NK, Kiwanuka N, Sempijja V, Lutalo T, Kagayii J, Wabwire-Mangen F, Ridzon R, Bacon M, Wawer MJ. 2009. The effects of male circumcision on female partners’ genital tract symptoms and vaginal infections in a randomized trial in Rakai, Uganda. Am. J. Obstet. Gynecol. 200:42.e1–42.e7 http://dx.doi.org/10.1016/j.ajog.2008.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sobngwi-Tambekou J, Taljaard D, Nieuwoudt M, Lissouba P, Puren A, Auvert B. 2009. Male circumcision and Neisseria gonorrhoeae, Chlamydia trachomatis and Trichomonas vaginalis: observations after a randomised controlled trial for HIV prevention. Sex. Transm. Infect. 85:116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mehta SD, Moses S, Agot K, Parker C, Ndinya-Achola JO, Maclean I, Bailey RC. 2009. Adult male circumcision does not reduce the risk of incident Neisseria gonorrhoeae, Chlamydia trachomatis, or Trichomonas vaginalis infection: results from a randomized, controlled trial in Kenya. J. Infect. Dis. 200:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wawer MJ, Tobian AA, Kigozi G, Kong X, Gravitt PE, Serwadda D, Nalugoda F, Makumbi F, Ssempiija V, Sewankambo N, Watya S, Eaton KP, Oliver AE, Chen MZ, Reynolds SJ, Quinn TC, Gray RH. 2011. Effect of circumcision of HIV-negative men on transmission of human papillomavirus to HIV-negative women: a randomised trial in Rakai, Uganda. Lancet 377:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tobian AA, Quinn TC, Gray RH. 2011. Male circumcision for prevention of oncogenic HPV infection. Lancet 378:314–315; author reply, 315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinh MH, Fahrbach KM, Hope TJ. 2011. The role of the foreskin in male circumcision: an evidence-based review. Am. J. Reprod. Immunol. 65:279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson KE, Redd AD, Quinn TC, Collinson-Streng AN, Cornish T, Kong X, Sharma R, Tobian AA, Tsai B, Sherman ME, Kigozi G, Serwadda D, Wawer MJ, Gray RH. 2011. Effects of HIV-1 and herpes simplex virus type 2 infection on lymphocyte and dendritic cell density in adult foreskins from Rakai, Uganda. J. Infect. Dis. 203:602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glynn JR, Biraro S, Weiss HA. 2009. Herpes simplex virus type 2: a key role in HIV incidence. AIDS 23:1595–1598 [DOI] [PubMed] [Google Scholar]
- 14. Celum C, Levine R, Weaver M, Wald A. 2004. Genital herpes and human immunodeficiency virus: double trouble. Bull. World Health Organ. 82:447–453 [PMC free article] [PubMed] [Google Scholar]
- 15. Gray RH, Wawer MJ. 2008. Reassessing the hypothesis on STI control for HIV prevention. Lancet 371:2064–2065 [DOI] [PubMed] [Google Scholar]
- 16. Gray RH, Serwadda D, Tobian AA, Chen MZ, Makumbi F, Suntoke T, Kigozi G, Nalugoda F, Iga B, Quinn TC, Moulton LH, Laeyendecker O, Reynolds SJ, Kong X, Wawer MJ. 2009. Effects of genital ulcer disease and herpes simplex virus type 2 on the efficacy of male circumcision for HIV prevention: analyses from the Rakai trials. PLoS Med. 6:e1000187 http://dx.doi.org/10.1371/journal.pmed.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, Ravel J, Keim PS, Serwadda D, Wawer MJ, Gray RH. 2010. The effects of circumcision on the penis microbiome. PLoS One 5:e8422 http://dx.doi.org/10.1371/journal.pone.0008422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flacher V, Bouschbacher M, Verronèse E, Massacrier C, Sisirak V, Berthier-Vergnes O, de Saint-Vis B, Caux C, Dezutter-Dambuyant C, Lebecque S, Valladeau J. 2006. Human Langerhans cells express a specific TLR profile and differentially respond to viruses and Gram-positive bacteria. J. Immunol. 177:7959–7967 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Li G, Bafica A, Pantelic M, Zhang P, Broxmeyer H, Liu Y, Wetzler L, He JJ, Chen T. 2005. Neisseria gonorrhoeae enhances infection of dendritic cells by HIV type 1. J. Immunol. 174:7995–8002 [DOI] [PubMed] [Google Scholar]
- 20. Donoval BA, Landay AL, Moses S, Agot K, Ndinya-Achola JO, Nyagaya EA, MacLean I, Bailey RC. 2006. HIV-1 target cells in foreskins of African men with varying histories of sexually transmitted infections. Am. J. Clin. Pathol. 125:386–391 [PubMed] [Google Scholar]
- 21. Patterson BK, Landay A, Siegel JN, Flener Z, Pessis D, Chaviano A, Bailey RC. 2002. Susceptibility to human immunodeficiency virus-1 infection of human foreskin and cervical tissue grown in explant culture. Am. J. Pathol. 161:867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ogawa Y, Kawamura T, Kimura T, Ito M, Blauvelt A, Shimada S. 2009. Gram-positive bacteria enhance HIV-1 susceptibility in Langerhans cells, but not in dendritic cells, via Toll-like receptor activation. Blood 113:5157–5166 [DOI] [PubMed] [Google Scholar]
- 23. McGowin CL, Ma L, Martin DH, Pyles RB. 2009. Mycoplasma genitalium-encoded MG309 activates NF-kappaB via Toll-like receptors 2 and 6 to elicit proinflammatory cytokine secretion from human genital epithelial cells. Infect. Immun. 77:1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Jong MA, Geijtenbeek TB. 2009. Human immunodeficiency virus-1 acquisition in genital mucosa: Langerhans cells as key-players. J. Intern. Med. 265:18–28 [DOI] [PubMed] [Google Scholar]
- 25. de Witte L, Nabatov A, Geijtenbeek TB. 2008. Distinct roles for DC-SIGN+-dendritic cells and Langerhans cells in HIV-1 transmission. Trends Mol. Med. 14:12–19 [DOI] [PubMed] [Google Scholar]
- 26. Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. 2011. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One 6:e26732 http://dx.doi.org/10.1371/journal.pone.0026732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collins MD, Facklam RR, Rodrigues UM, Ruoff KL. 1993. Phylogenetic analysis of some Aerococcus-like organisms from clinical sources: description of Helcococcus kunzii gen. nov., sp. nov. Int. J. Syst. Bacteriol. 43:425–429. [DOI] [PubMed]
- 28. Liu CM, Aziz M, Kachur S, Hsueh PR, Huang YT, Keim P, Price LB. 2012. BactQuant: an enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 12:56 http://dx.doi.org/10.1186/1471-2180-12-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 http://dx.doi.org/10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lajeunessei MJ. 2011. On the meta-analysis of response ratios for studies with correlated and multi-group designs. Ecology 92:2049–2055 [DOI] [PubMed] [Google Scholar]
- 33. R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 34. Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens HH, Wagner H. 2011. Vegan: community ecology package. R package version 1.17-8. [Google Scholar]
- 35. Kindt RC. 2005. Tree diversity analysis. In A manual and software for common statistical methods for ecological and biodiversity studies. World Agroforestry Centre; (ICRAF; ), Nairobi, Kenya [Google Scholar]
- 36. Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 7:1–19 [Google Scholar]
- 37. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 38. Roberts DW. 2010. labdsv: ordination and multivariate analysis for ecology. http://CRAN.R-project.org/package=labdsv
- 39. Buchanan RE, Gibbons NE. 1974. Bergey’s manual of determinative bacteriology, 8th ed, vol 26 Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed laboratory protocols and bioinformatic methods. Download
An nMDS plot comparing changes in the coronal sulcus microbiota composition from enrollment (in orange) to year 1 (in blue) in the two study groups (see key, top left). Each data point represents an individual’s microbiota at a given time point. The centroids and 95% confidence ellipses for each group are as shown. The microbiota composition at enrollment was similar between the controls and the intervention groups and remained largely unchanged among the uncircumcised men at year 1. In contrast, microbiota composition for the circumcised men changed significantly post-MC and formed a cluster distinct from the uncircumcised-group microbiota profiles. Download
Comparison of year-1 prevalence, as well as changes in prevalence from enrollment to year 1 for uncircumcised versus circumcised men for all detected coronal sulcus bacterial genera.
References for oxygen tolerance designations.