ABSTRACT
Pneumocystis jirovecii is a fungus that causes Pneumocystis pneumonia in immunosuppressed patients and has been closely associated with AIDS since the beginning of the AIDS epidemic. Because in vitro cultivation of P. jirovecii is not possible, progress has been hindered in our understanding of its life cycle, mode of transmission, metabolic function, and genome. Limited amounts of P. jirovecii can be obtained from infected patients, but the occurrence of bacteria, other fungi, and human cells in clinical samples presents new challenges for whole-genome sequencing and downstream bioinformatic analysis. In a recent article, Cissé et al. used cell immunoprecipitation enrichment together with whole-genome amplification to generate sufficient quantities of DNA for Roche 454 and Illumina sequencing [O. H. Cissé, M. Pagni, and P. M. Hauser, mBio 4(1):e00428-12, 2012, doi:10.1128/mBio.00428-12]. In addition, a bioinformatic pipeline was devised to sort and assemble lung microbiome reads, thereby generating an 8.1-Mb P. jirovecii genome comprised of 356 contigs with an N50 (median length of all contigs) of 41.6 kb. Knowledge of this genome will open new avenues of research, including the identification of nutritional requirements for in vitro cultivation as well as the identification of new and novel drug and vaccine targets.
Commentary
Next-generation sequencing (NGS) provides the technology necessary to probe the genomes of microbes that have resisted cultivation outside their host species. The sequencing and annotation of the human pathogen Pneumocystis jirovecii using Roche 454 and Illumina paired-end reads represent a significant advancement in mycological genomics research (1). This wealth of data, together with the newly released Pneumocystis murina genome (http://www.broadinstitute.org/annotation/genome/Pneumocystis_group/MultiHome.html) and the Pneumocystis carinii genome (http://pgp.cchmc.org), now makes it possible to address many previously unapproachable questions, including the conserved syntenic relationships among these closely related, though host-specific, fungal species. As it has been postulated that each Pneumocystis species coevolved with its specific mammalian host, it will now be possible to understand how each species diverged and what core genetic components were retained. The identification of new and novel drug targets may now be a possibility, or at least structural and functional comparisons of the proteins will reveal whether the rodent models of pneumonia provide accurate and reliable predictors for drug development. It is a given that a better understanding of the basic biology of these fungi will be attained.
STRUCTURES OF THE GENOMES
The assembly of P. jirovecii at 8.1 Mb, with a predicted gene inventory of 3,878 genes and a gene density of 481 genes per Mb (or 1 gene/2,029 bp), is similar to predictions of the assembly of P. carinii, namely, 1 gene/2,139 bp, with an estimate of 3,740 total genes for the genome (1, 2). Although the assembly size of P. carinii was stated to be 6.3 Mb (see Table 1 in reference 1), based on the summation of chromosome-sized DNA bands from electrophoretic karyotypes, the estimated genome size is 8.2 Mb (3). The estimated sizes of other Pneumocystis genomes based on electrophoretic karyotyping are 8.2 Mb for P. murina and 7.0 Mb for a single isolate of P. jirovecii, considerably smaller than the genomic sequencing data (3). As the karyotype of the P. jirovecii strain used in the present study is not available, it is not possible to determine where the discrepancy may lie at this time. It should also be noted that the genome size inferred by electrophoretic methods is an estimate based on migration of the DNA bands in a gel, which may introduce some variability.
At the time of writing of this commentary, sequences of the mitochondrial (mt) genomes of P. jirovecii and P. murina (4) and a resequencing of the previously published P. carinii mt genome (5) were published. As in the previous report by Sesterhenn et al. (5), the P. carinii mt genome was found to be linear in structure, with telomere-like repeats at the ends. The authors of the P. murina mt genome suggested that it is also linear, while the P. jirovecii mt genome was found to be circular. The presence of linear and circular mt genomes within a single fungal genus is not without precedent, and since linear and circular mt genomes use distinct modes of replication, this knowledge could present opportunities to develop drugs that specifically target one type or the other (6). While Cissé et al. do not mention the structural configuration of their mitochondrial assembly, the mt genome size of P. jirovecii (27 kb) was considerably smaller than that reported by Ma et al. (33.7 kb) (4). The GC contents of the whole mt genome differed in the studies of Cissé et al. and Ma et al. as well (29.5% versus 25.7% across the mt genome and 32.5% versus 14% in the coding regions, respectively). Moreover, while both studies identified 2 rRNA genes per mt genome, Ma et al. identified 25 tRNA genes, while Cissé et al. found only 12. Such discrepancies will need to be resolved before accurate comparative analyses are conducted.
BIOLOGICAL APPLICATIONS
A significant finding of the report by Cissé et al. was the paucity of genes related to amino acid biosynthesis, which may provide clues to the supplementation of in vitro media that could eventually lead to sustainable ex vivo growth. Other missing genes detailed in the study may also provide clues for nutritional additives. Looking to therapies, identification of P. jirovecii’s receptors and transporters offers potential parasite-specific drug targets.
Cissé et al. noted the lack of a glyoxylate cycle in P. jirovecii, which was previously found to be absent in P. carinii as well (7). The importance of the glyoxylate pathway has been stressed as a potential drug target in those fungi that maintain this cycle, which serves as a shortcut across the citric acid cycle and is not found in humans. It is a potential virulence factor as well, since fungi that can survive after phagocytosis appear to induce this cycle, while those that do not have these genes are unable to germinate and cause disease. The transcriptome of P. carinii and the data from the P. jirovecii genome suggest that these apparently obligate fungal parasites have adapted to their specific mammalian hosts and are able to achieve a careful balance without killing immunologically intact hosts, although they do require nutritional supplementation. This adaptation is important, as their entire life cycle, except during transmission to the next host, appears to take place within mammalian lungs. Analysis of the Pneumocystis genomes should provide a better understanding of the survival strategies used by these fungi.
rDNA COPY NUMBER
Early studies of Pneumocystis electrophoretic karyotypes showed only a single band of hybridization to a ribosomal DNA (rDNA) probe, with a hybridization signal consistent with a single-copy gene, suggesting that Pneumocystis had very few copies of the locus (8). This is in contrast with what occurs in other fungi, which typically have hundreds of rDNAs located on multiple chromosomes. High-resolution restriction fragment length analysis of lambda phages containing rRNA genes and genomic DNA, as well as quantitative PCR (qPCR), later confirmed that P. carinii and P. jirovecii genomes have a single rDNA copy (9–12). In addition, it was shown that Pneumocystis rDNA evolved at a rate typical for eukaryotes (13).
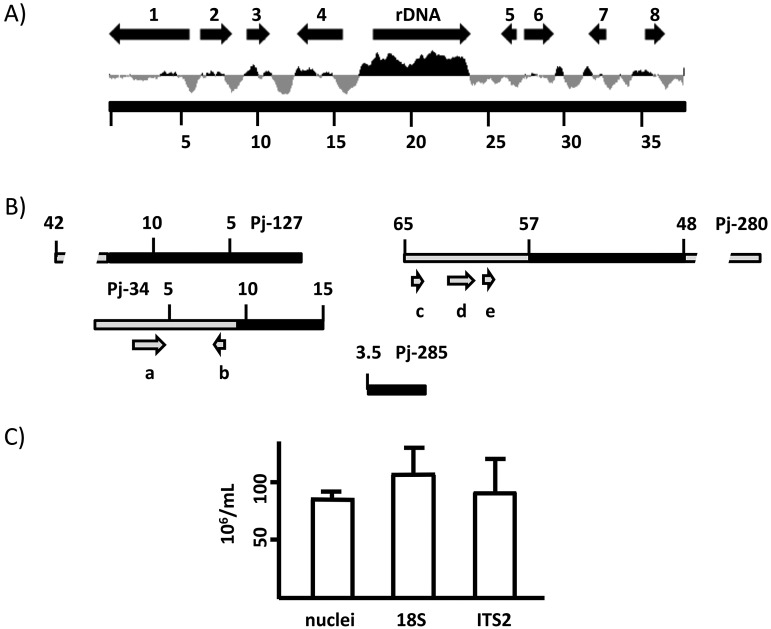
The single-copy hypothesis for P. carinii was further investigated by determining the sequence for a 38-kb pWEB cosmid (3C5) containing a single rDNA locus (Fig. 1A). BLAST analysis of 3C5 showed that it contains a single GC-rich 37S rDNA locus flanked by several protein-encoding genes (Fig. 1A). DNA alignments between 3C5 and P. jirovecii contigs indicated that there was limited conserved synteny between P. jirovecii and P. carinii (Fig. 1B). For example, the gene order of approximately 13 kb of contig Pj-127 matched, in order, arrows 1 to 4 (Fig. 1A) of the left flank of 3C5. Approximately 6 kb of Pj-34 also matched the left flank of 3C5 (arrows 3 and 4 in Fig. 1A), but 8 kb contained an unknown open reading frame (ORF) and rad13 (arrows a and b in Fig. 1B) and did not match 3C5 or Pj-127. It is unclear why contigs Pj-127 and Pj-34 shared 5 kb of DNA (>99% nucleotide identity) but contained different adjacent regions. This might be due to gene duplication, or it may represent software assembly artifacts. Similarly, the gene order of 9 kb of contig Pj-280 matched the right flank of 3C5; however, 8 kb of this contig (5′ region, 57 to 65 kb) did not match rDNA, as expected, but rather contained an unknown ORF, rad13, and orc4 (arrows c, d and e in Fig. 1B). Unfortunately, contig Pj-285 contained only 37S rDNA sequences, so it was not possible to determine what genes were associated with the locus (Fig. 1B).
FIG 1 .
Pneumocystis contains one copy of rDNA. (A) Genetic map of the 38-kb cosmid 3C5 containing P. carinii rDNA. The black arrows represent approximate nucleotide locations and sizes of rDNA and BLASTx (16) genes in the flanking regions of 3C5. The number above each black arrow indicates a BLASTx hit, as follows: arrow 1, a cell morphogenesis protein (PAG1); arrow 2, NADPH cytochrome P450 reductase (CprA); arrow 3, sporulation-specific protein 5 (Spo5); arrow 4, a 26S proteasome subunit (Rpn1); arrow 5, U3 small nucleolar RNA-associated protein 6; arrow 6, a developmental regulator (FlbA); arrow 7, a hypothetical zinc finger protein; and arrow 8, damage-inducible protein 1. The black arrow for rDNA contains the entire 37S ribosomal DNA locus, which includes the 18S, internal transcribed spacer 1 (ITS1), 5.8S, ITS2, and 26S genes. Below the black arrows is the GC content of 3C5; black and gray plots represent above- and below-average GC contents, respectively. The numbers indicate nucleotide positions of 3C5. (B) P. jirovecii contigs similar to 3C5. Five P. jirovecii contigs had significant alignments with 3C5. They were Pj-127 (length, 42 kb), Pj-34 (15 kb), Pj-285 (3.5 kb), Pj-280 (65 kb), and Pj-200 (22 kb, not shown). Black-filled rectangles represent regions of similarity between P. jirovecii contigs and 3C5 (cf. panel A), whereas gray-filled rectangles represent dissimilarity. The overall percentages of nucleotide identity from the BLAST alignments between corresponding matching regions of 3C5 and each P. jirovecii contig were 87.2% ± 4.5% (Pj-127), 86.2% ± 4.2% (Pj-280), 87.6% ± 4.1% (Pj-34), 83.2% ± 3% (Pj-200), and 95.7% ± 2.1% (Pj-285). The gray arrows indicate the approximate locations and sizes of BLASTx hits in regions of P. jirovecii contigs that do not match 3C5, as follows: arrow a, an unknown protein product; arrow b, a DNA repair protein (Rad13) (b); arrow c, an unknown protein product; arrow d, origin recognition complex subunit 4 (Orc4); and arrow e, DNA-directed RNA polymerase subunit 4 (Rpb4). Contig Pj-200 matched only arrow 5 of 3C5 (i.e., the U3 small nucleolar RNA-associated protein 6 gene [comparison not shown]). (C) There is one copy of rDNA in P. murina. Microscope slides containing P. murina organisms were stained with Diff-Quik (Baxter Scientific, McGraw Park, IL), and nuclei were counted as described previously (17) and are expressed as numbers of nuclei per ml. Genomic DNA was extracted from P. murina as previously described (17). A 5′ region of the P. murina 18S rDNA locus (GenBank accession no. AY532651) was amplified with primers Pm-18sFor (5′ ATGCATGTCTAAGTATAAGCAAG 3′) and Pm-18Srev (5′ CTAATAAATACATCCCTTCCATAA 3′). The second transcribed spacer locus in AY532651 was amplified with primers ITS2For (5′ AGGTTCGTGTTGGGCTATGC 3′) and ITS2Rev (5′ ATTCAAAAAATCAGCTTAACATTTC 3′). Forty cycles of PCR were performed in the presence of SYBR green under the following conditions: 95°C for 15 s, 50°C for 15 s, and 72°C for 15 s. DNA plasmids containing these 18S rDNA and ITS2 targets were used as standards to establish a linear relationship between log-transformed numbers of copies of the target and PCR threshold cycles as previously described (17).
These DNA sequence comparisons suggest that P. carinii and P. jirovecii have divergent chromosomes, which is consistent with the 100 million years of evolution of their respective lineages (14). It will be of interest to comprehensively compare the syntenic relationships among these two species as well as their sequences to the recently released P. murina genome. A probable hypothesis is that chromosomes of the closely related rodent species P. carinii and P. murina are more similar to each other than either is to human-derived P. jirovecii. There is new evidence in support of this prediction. Analysis of the architecture of Pneumocystis mt genomes has revealed a remarkable level of conserved synteny between P. carinii and P. murina but much less synteny between both of their genomes and that of P. jirovecii (4). Consistent with this was our sequence comparison of 3C5 and a P. murina supercontig containing 37S rDNA, which showed a conserved gene order of the entire 38-kb region of 3C5. Close inspection of the sequences in the supercontig indicated the presence of a single 37S rDNA locus, which was consistent with our quantitative PCR twice targeting the 37S rDNA of P. murina (Fig. 1C).
DRUG DEVELOPMENT
There is a critical need for new and novel approaches to the treatment of Pneumocystis pneumonia (PcP) and, potentially, for those patients who are colonized by P. jirovecii or who have comorbidities associated with its presence, as the latter patients may require a treatment regimen different than that of patients with frank disease. Chemotherapeutics has been the mainstay of anti-PcP therapy, and the standard therapy has been and remains trimethoprim-sulfamethoxazole (TMP-SMX). Second-line treatment includes pentamidine isethionate, atovaquone, and clindamycin primaquine, none of which are as effective as TMP-SMX. It has been known for over a decade that P. jirovecii has evolved mutations in the DHPS (dihydropteroate synthase) gene that lead to increased resistance in other microbial pathogens, but the role of these mutations in the clinical outcome of P. jirovecii infection is not clear. Prophylaxis with atovaquone has been associated with resistance by mutation of the cytochrome bc1 gene (15). Other new antifungal compounds, like the echinocandins, reduce the formation of asci, but the more numerous, asexually dividing trophic forms are largely left intact. Detailed analysis of the P. jirovecii genome and identification of Pneumocystis-specific metabolic requirements, transporters, and unique enzyme functions should provide the scientific community with new avenues for drug development.
The availability of genomic data promises to level the scientific playing field for these heretofore intractable fungi.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Citation Cushion MT, Keely SP. 2013. Assembly and annotation of Pneumocystis jirovecii from the human lung microbiome. mBio 4(2):e00224-13. doi:10.1128/mBio.00224-13.
REFERENCES
- 1. Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4(1):e00428-12 http://dx.doi.org/10.1128/mBio.00428-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smulian AG, Sesterhenn T, Tanaka R, Cushion MT. 2001. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157:991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cushion MT. 2005. Pneumocystis pneumonia, p 763–806 In Merz WG, Hay RJ, Topley and Wilson’s microbiology and microbial infections, 10th ed. ASM Press, Washington, DC [Google Scholar]
- 4. Ma L, Huang DW, Cuomo CA, Sykes S, Fantoni G, Das B, Sherman BT, Yang J, Huber C, Xia Y, Davey E, Kutty G, Bishop L, Sassi M, Lempicki RA, Kovacs JA. 7 February 2013. Sequencing and characterization of the complete mitochondrial genomes of three Pneumocystis species provide new insights into divergence between human and rodent Pneumocystis. FASEB J. [Epub ahead of print.] http://dx.doi.org/10.1096/fj.12-224444 [DOI] [PMC free article] [PubMed]
- 5. Sesterhenn TM, Slaven BE, Keely SP, Smulian AG, Lang BF, Cushion MT. 2010. Sequence and structure of the linear mitochondrial genome of Pneumocystis carinii. Mol. Genet. Genomics 283:63–72 [DOI] [PubMed] [Google Scholar]
- 6. Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. 2006. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 34:2472–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2(5):e423 http://dx.doi.org/10.1371/journal.pone.0000423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cushion MT, Zhang J, Kaselis M, Giuntoli D, Stringer SL, Stringer JR. 1993. Evidence for two genetic variants of Pneumocystis carinii coinfecting laboratory rats. J. Clin. Microbiol. 31:1217–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giuntoli D, Stringer SL, Stringer JR. 1994. Extraordinarily low number of ribosomal RNA genes in P. carinii. J. Eukaryot. Microbiol. 41:88S [PubMed] [Google Scholar]
- 10. Tang X, Bartlett MS, Smith JW, Lu JJ, Lee CH. 1998. Determination of copy number of rRNA genes in Pneumocystis carinii f. sp. hominis. J. Clin. Microbiol. 36:2491–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nahimana A, Francioli P, Blanc DS, Bille J, Wakefield AE, Hauser PM. 2000. Determination of the copy number of the nuclear rDNA and beta-tubulin genes of Pneumocystis carinii f. sp. hominis using PCR multicompetitors. J. Eukaryot. Microbiol. 47:368–372 [DOI] [PubMed] [Google Scholar]
- 12. Xu Z, Lance B, Vargas C, Arpinar B, Bhandarkar S, Kraemer E, Kochut KJ, Miller JA, Wagner JR, Weise MJ, Wunderlich JK, Stringer J, Smulian G, Cushion MT, Arnold J. 2003. Mapping by sequencing the Pneumocystis genome using the ordering DNA sequences V3 tool. Genetics 163:1299–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fischer JM, Keely SP, Stringer JR. 2006. Evolutionary rate of ribosomal DNA in Pneumocystis species is normal despite the extraordinarily low copy-number of rDNA genes. J. Eukaryot. Microbiol. 53:S156–S158 [DOI] [PubMed] [Google Scholar]
- 14. Keely SP, Fischer JM, Cushion MT, Stringer JR. 2004. Phylogenetic identification of Pneumocystis murina sp. nov., a new species in laboratory mice. Microbiology 150:1153–1165 [DOI] [PubMed] [Google Scholar]
- 15. Walker DJ, Meshnick SR. 1998. Drug resistance in Pneumocystis carinii: an emerging problem. Drug Resist. Updat. 1:201–204 [DOI] [PubMed] [Google Scholar]
- 16. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 17. Keely SP, Linke MJ, Cushion MT, Stringer JR. 2007. Pneumocystis murina MSG gene family and the structure of the locus associated with its transcription. Fungal Genet. Biol. 44:905–919 [DOI] [PMC free article] [PubMed] [Google Scholar]