Summary
International efforts to test gene function in the mouse by the systematic knockout of each gene are creating many lines in which embryonic development is compromised. These homozygous lethal mutants represent a potential treasure trove for the biomedical community. Developmental biologists could exploit them in their studies of tissue differentiation and organogenesis; for clinical researchers they offer a powerful resource for investigating the origins of developmental diseases that affect newborns. Here, we outline a new programme of research in the UK aiming to kick-start research with embryonic lethal mouse lines. The ‘Deciphering the Mechanisms of Developmental Disorders’ (DMDD) programme has the ambitious goal of identifying all embryonic lethal knockout lines made in the UK over the next 5 years, and will use a combination of comprehensive imaging and transcriptomics to identify abnormalities in embryo structure and development. All data will be made freely available, enabling individual researchers to identify lines relevant to their research. The DMDD programme will coordinate its work with similar international efforts through the umbrella of the International Mouse Phenotyping Consortium [see accompanying Special Article (Adams et al., 2013)] and, together, these programmes will provide a novel database for embryonic development, linking gene identity with molecular profiles and morphology phenotypes.
Knockout mice: the potential of embryonic lethal lines
A global 10-year effort by the International Knockout Mouse Consortium (IKMC) to create targeted knockout strains for every gene in the mouse genome (∼20,000) is now well underway. This ambitious scheme will create an unparalleled resource for uncovering the role of individual genes in maintaining normal physiological function in the mouse and will be invaluable for identifying genes that might contribute to human disease (Brown and Moore, 2012). Standardised high-throughput phenotypic screens have been established to characterise adult mutant lines, coordinated by the International Mouse Phenotyping Consortium (IMPC), and these are already identifying the impact of individual gene mutations on adult mouse growth, metabolism, physiology and behaviour.
However, research has shown that a strikingly large proportion (about one-third) of all targeted knockouts in the mouse result in embryonic or perinatal (EP) death and cannot therefore be studied using existing pipelines, except as heterozygotes. Yet these lines could be of tremendous value to developmental biologists seeking to identify genes that are essential for embryogenesis. If characterised further, these lines could provide a unique window into the genetic regulation of tissue differentiation, organ formation and embryo morphogenesis. The insights gained from such studies will also shed light on the origins of human developmental disorders and could provide an important boost for research into new diagnostic and therapeutic approaches for these debilitating, and sometimes lethal, conditions.
Initiating work to identify and characterise knockout mutants that result in EP death is a priority for the IMPC [see accompanying Special Article on the April 2012 IMPC meeting on embryo phenotyping (Adams et al., 2013)]. In the UK, a consortium of developmental biologists and clinician scientists funded by the Wellcome Trust has now been formed to set the wheels in motion. ‘Deciphering the Mechanisms of Developmental Disorders’ (DMDD) is a 5-year programme, commencing in the spring of 2013, that aims to provide systematic characterisation of all EP lethal mouse lines generated in the UK by the Wellcome Trust Sanger Institute (WTSI). By combining comprehensive imaging with detailed transcriptomics, the DMDD programme aims to provide a systematic and unbiased study of EP lethals at both the anatomical and transcriptional levels. This will yield a novel and rich public resource that should transform our understanding of embryo tissue differentiation and organ morphogenesis.
Careful and standardised analysis of embryos from EP lethal lines will also enable identification of mouse null alleles that mimic aspects of human congenital syndromes and disease. Through such work, the DMDD programme will provide new opportunities to identify and investigate the underlying molecular and developmental pathways that are disrupted in human developmental disorders. Its work will complement existing clinical genetic programmes [such as the Deciphering Developmental Disorders (DDD; http://www.ddduk.org) and UK10K [http://www.uk10k.org] projects] that are currently exploiting the latest generation sequencing methods in efforts to identify previously unknown disease-associated alleles.
Outline of the DMDD phenotyping pipeline
Experience gained over more than a decade of mouse transgenics work has revealed that approximately half of all mouse gene knockouts demonstrating recessive EP lethality complete the major period of organogenesis (E14.5–E15.5) and reach a point when organ arrangement resembles that of the adult. Another third of the knockouts develop at least to the mid-gestation stage but fail to complete organogenesis. Furthermore, existing phenotyping efforts show that at least 60% of all EP lethals studied to date have readily detectable structural defects in one or more organ systems. (The true figure is likely to be significantly higher because existing studies have rarely been systematic, nor have they used consistent or comparable imaging methods.) Taken together, these observations indicate that simple screening of embryo morphology provides a remarkably efficient way to identify genes that are important in embryo development and organogenesis.
Such a screen lies at the heart of the DMDD pipeline (Figs 1, 2) which will use state-of-the-art imaging methods to phenotype EP lethal embryos based on their external and internal embryo morphology. Where possible, embryos will be analysed after organogenesis (E14.5–E15.5). Lines presenting no homozygous mutant embryos at E14.5 (or those in which homozygous mutants are evidently dying or dead) will be re-assessed at mid-gestation (E9.5–E10.5), a stage late enough to identify structural defects in several major organ systems, including cardiovascular and neural tissue. Data collated by The Jackson Laboratory on existing mouse mutants (http://www.informatics.jax.org) indicates that, by studying embryos at these two time points, the DMDD programme can expect to capture more than three-quarters of all structural abnormalities in the EP embryos [see table 1 of the accompanying Special Article (Adams et al., 2013)].
Fig. 1.
Flow diagram of the DMDD programme. Embryo viability of homozygous nulls will be assessed 2 weeks after birth (P14) to identify EP lethal lines. Viability of EP embryos will then be checked at 14 days of gestation (E14.5) and viable embryos will be comprehensively imaged for malformations. If embryos are absent (resorbed), embryo litters will be examined after 9 or 10 days of gestation (E9.5–E10.5) and homozygous null embryos identified for imaging. All possible EP lines will be used for transcriptomics analysis at early stages of development. EP lines that appear normal at E14.5 will be reanalysed shortly before birth (E18.5) in order to detect neurological abnormalities. Data that will be presented on the DMDD web portal is indicated by pink boxes.
Fig. 2.
Overall DMDD workflow. Mouse lines identified as EP lethal will undergo analysis in the DMDD pipeline, with all anatomical and histological data scored for defects. Collective data for each line will be reviewed and published in full via the web portal.
Current UK plans envisage the creation of ∼270 targeted, null lines in the UK annually [approximately 160 at the WTSI and 110 at the UK Medical Research Council (MRC) Mary Lyon Centre], which can be expected to yield ∼400 EP lethal lines over the next 5 years. At least 240 of these lines will provide viable embryos, either at mid-gestation or after organogenesis, and will therefore be amenable to analysis. This will dramatically extend the pool of mouse genes known to be critical for normal embryo development, enhancing research into the genetic networks that are responsible for tissue organisation and organogenesis. It will also provide experimental testing of candidate developmental disease genes suggested from clinical studies.
Of course, embryonic lethality can arise not only through direct disruption of embryonic tissue formation or organ function; mid-gestational lethality or stunted growth prior to later-stage death can also result from compromised placental development. For this reason, the DMDD programme includes a comprehensive screen of EP lines for placental abnormalities.
Around 15% of EP mutants are expected to show peri- or postnatal, rather than earlier embryonic, lethality, and such embryos are likely to exhibit no obvious morphological abnormalities at E14.5–E15.5. For a large proportion of these mutants, the gene knockout will affect later aspects of neural development, and will not be easily detected using simple morphological criteria. Nonetheless, these mutations result in severe defects (for example, in movement, breathing, cognition, vision or suckling) that render the pups unviable. Lines falling into this category will therefore be assayed immediately prior to birth, initially triaged by a range of simple tests, including assessment of appearance and the response to stimuli. Histological analysis will be used to detect late-appearing phenotypes such as craniosynostosis, hindbrain herniation (Chiari II defect) and submucous cleft palate, whereas immunohistochemistry will identify defects in synaptogenesis and neuronal differentiation or morphology.
Optimising the pipeline
Evidently, any screen of this scale must balance effectiveness against cost. This is a difficult calculus because the bulk of costs arise simply from animal husbandry, constraining the scale of any screening. By contrast, we can expect considerable variations in phenotypic penetrance, despite the use of a single genetic background, with the result that some lines will require analysis of many more null embryos than others in order to detect the resulting abnormalities. For the DMDD programme, we have chosen to screen three homozygous knockout embryos, together with one wild-type littermate, for structural defects. Assuming phenotype penetrance of 0.5, this sample size gives a 0.875 probability of finding at least one phenotypically abnormal embryo for each EP line. In pilot studies, this approach has proved effective in identifying a range of specific abnormalities in virtually every line examined.
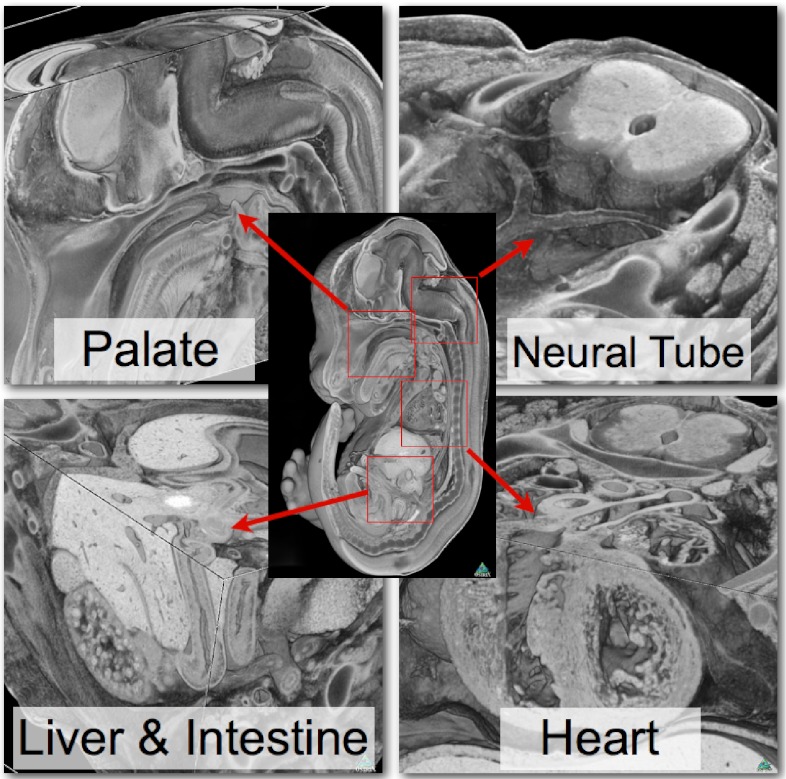
With relatively small embryo numbers and a limited amount of time before the lines will be archived, it is essential that embryo structure examination is as effective as possible. Furthermore, such data must lend itself readily to computer analysis to allow the power of digital resectioning and three-dimensional (3D) modelling to be harnessed in the detection of abnormal phenotypes. The DMDD programme will utilise high-resolution episcopic microscopy (HREM) to image embryos at the highest levels of resolution currently feasible without histological sectioning (Fig. 3). This will maximise our ability to identify the largest range of phenotypes by qualitative analysis (Geyer et al., 2009).
Fig. 3.
Phenotyping embryos with high-resolution episcopic microscopy (HREM). Volume-rendered 3D models from HREM data allow the visualisation of embryo morphology at remarkably high resolution.
Considerable progress has also been made in recent years using computer-based methods for automatic registration of multiple image data sets, paving the way for quantitative comparison of defined embryo organs and organ regions (Dorr et al., 2008; Zamyadi et al., 2010; Zhang et al., 2010). This technique, which has been applied to magnetic resonance imaging (MRI) and X-ray computed tomography (μCT) data from embryos, has the potential to complement qualitative phenotyping. In addition to being automated rather than manual, this approach can identify volumetric changes that are difficult to detect in 2D analysis (Spring et al., 2007; Spring et al., 2010). Furthermore, it can assist expert phenotyping by highlighting regions of mutant embryos that display the greatest variance compared with reference controls. Because the bulk of embryo imaging in the DMDD programme will occur after organogenesis (E14.5–E15.5), we plan to image embryos non-invasively by μCT prior to HREM analysis. By implementing this sequential imaging pipeline, we will accumulate a repository of image data that has the greatest utility for future studies, while also facilitating the use of emerging automated analysis methods.
The value of systematic transcriptomics
As a first step towards identifying the cellular processes and molecular pathways that are disrupted by gene deletion, the DMDD programme will complement morphological analysis by analysing the transcriptome of knockout embryos, using RNA sequencing (RNA-seq). By adopting a transcript counting methodology (Anders and Huber, 2010) developed for efficient and cost-effective high-throughput analysis, we will be able to detect differential expression of known protein-coding genes, non-coding RNAs (ncRNAs) and newly identified transcripts compared with sibling controls (Airik et al., 2005; Rodriguez-Zas et al., 2008; Tsutsumi et al., 2008). Bioinformatic analysis could then be used to identify candidate pathways and downstream targets that might be affected (Lu et al., 2009). Of course, gene expression data will only reveal the genetic consequences of gene knockout at the stage chosen for analysis and cannot, in isolation, distinguish between the primary molecular lesion and its downstream consequences. However, by choosing the most appropriate early time point for transcriptomic analysis, DMDD’s goal is to obtain the most useful pointers to guide in-depth studies of each mutant line (Park and Choi, 2011).
The systematic accumulation of transcriptomic data linked to phenotypic data would open many new opportunities for basic and translational research. The accumulated data from a large number of mutant lines and their controls will provide an unparalleled resource for bioinformatic analysis of likely gene networks driving normal tissue and organ development. By hierarchically combining information across different gene knockouts, it will be possible to begin constructing and refining gene regulatory networks (GRNs) that can inform our understanding of complex phenotypes (Aijö and Lähdesmäki, 2009). In addition, this resource will be invaluable for elucidating possible mechanisms underlying gene-based developmental abnormalities.
Focusing on developmental disease
A key goal of DMDD is to exploit mouse studies to advance understanding of human congenital diseases. High-throughput sequencing studies are already transforming our knowledge of the aetiology of such diseases, and have pinpointed homozygous and heterozygous loss-of-function mutations as a major cause of childhood developmental disorders. Over 300 haploinsufficient genes have so far been identified (Huang et al., 2010; Zhang et al., 2009), and establishing the full extent to which loss-of-function among the 22,000 human genes causes developmental disorders is a pressing challenge. A major obstacle is the observation that every patient genome in fact harbours many loss-of-function mutations; thus, distinguishing pathogenic from benign loss-of-function variants is not easy with clinical studies alone (Huang et al., 2010; MacArthur et al., 2012). Here, the DMDD programme could be invaluable, by producing the appropriate mouse lines to study loss-of-function in both heterozygote and homozygote animals. These could be used to directly test the putative association between loss-of-function variants and developmental disease phenotypes in vivo.
To help achieve this goal, the DMDD programme will prioritise the analysis of mutants for mouse genes that are orthologous to high-confidence causal alleles associated with human developmental disorders. Not only will this provide an experimental test of such linkage, the associated systematic phenotyping could unveil other associated phenotypes that merit further investigation in a clinical context. The UK10K and DDD projects have also shown the predictive value of gene-gene and protein-protein interaction networks for identifying additional disease alleles. DMDD will therefore also prioritise genes that, on the basis of network analysis, are predicted to interact or cooperate with genes known to cause developmental disease.
Challenges and outlook
The potential of EP lethal lines for advancing our understanding of embryo development and the genetics of developmental disease is easy to grasp; but the sheer scale of the opportunity afforded by a knockout screen covering the entire mouse genome is potentially daunting. The burden of mouse husbandry costs enforces the need for strict selection of lines by individual researchers, generally based on pre-existing genetic or phenotypic data, yet this is entirely absent for many of the genes now being identified in EP lethal lines. The DMDD programme is designed to fill this gap, providing the molecular and structural phenotype data to enable researchers to identify mutant lines that are relevant to their research interests. To encourage such engagement, all data obtained by the DMDD programme will therefore be made freely and immediately available for review and analysis by the research community.
Achieving this will be a considerable challenge. However, pilot studies have demonstrated that, despite the enormous size of individual embryo image sets, it is perfectly possible to make these data freely accessible, using a web-based platform that can provide real-time access to image data and its associated annotations (http://embryoimaging.org). Systematic, qualitative phenotyping by dedicated, expert anatomists has also proved feasible in pilot screens. By employing a standardised ontology, phenotypers will be able to record over 200 different abnormalities in embryo structure that will be incorporated in searchable form into web interface. Combined with links to the associated transcriptomics data, the DMDD web resource will provide an unparalleled level of phenotypic description, all of which can be reviewed and re-assessed by individual researchers.
The consortium of scientists brought together in the DMDD programme are drawn from across the UK and its leading research centres, bringing together a unique combination of developmental biology, genetics and clinical expertise. We hope that our efforts will herald a new level of cooperation and engagement that can help transform our understanding of embryo development and developmental disease.
REFERENCES
- Adams D., Baldock R., Bhattacharya S., Copp A. J., Dickinson M., Greene N. D. E., Henkelman M., Justice M., Mohun T., Murray S. A., et al. (2013). Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis. Model. Mech. 6, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aijö T., Lähdesmäki H. (2009). Learning gene regulatory networks from gene expression measurements using non-parametric molecular kinetics. Bioinformatics 25, 2937–2944 [DOI] [PubMed] [Google Scholar]
- Airik R., Kärner M., Karis A., Kärner J. (2005). Gene expression analysis of Gata3−/− mice by using cDNA microarray technology. Life Sci. 76, 2559–2568 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Moore M. W. (2012). Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis. Model. Mech. 5, 289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr A. E., Lerch J. P., Spring S., Kabani N., Henkelman R. M. (2008). High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage 42, 60–69 [DOI] [PubMed] [Google Scholar]
- Geyer S. H., Mohun T. J., Weninger W. J. (2009). Visualizing vertebrate embryos with episcopic 3D imaging techniques. ScientificWorldJournal 9, 1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Lee I., Marcotte E. M., Hurles M. E. (2010). Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Markowetz F., Unwin R. D., Leek J. T., Airoldi E. M., MacArthur B. D., Lachmann A., Rozov R., Ma’ayan A., Boyer L. A., et al. (2009). Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J. K., Montgomery S. B., et al. (2012). A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Choi S. (2011). Gene expression analysis of Gα13−/− knockout mouse embryos reveals perturbations in Gα13 signaling related to angiogenesis and hypoxia. Genomics Inform. 9, 161–172 [Google Scholar]
- Rodriguez-Zas S. L., Schellander K., Lewin H. A. (2008). Biological interpretations of transcriptomic profiles in mammalian oocytes and embryos. Reproduction 135, 129–139 [DOI] [PubMed] [Google Scholar]
- Spring S., Lerch J. P., Henkelman R. M. (2007). Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage 35, 1424–1433 [DOI] [PubMed] [Google Scholar]
- Spring S., Lerch J. P., Wetzel M. K., Evans A. C., Henkelman R. M. (2010). Cerebral asymmetries in 12-week-old C57Bl/6J mice measured by magnetic resonance imaging. Neuroimage 50, 409–415 [DOI] [PubMed] [Google Scholar]
- Tsutsumi T., Kuwabara H., Arai T., Xiao Y., Decaprio J. A. (2008). Disruption of the Fbxw8 gene results in pre- and postnatal growth retardation in mice. Mol. Cell. Biol. 28, 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyadi M., Baghdadi L., Lerch J. P., Bhattacharya S., Schneider J. E., Henkelman R. M., Sled J. G. (2010). Mouse embryonic phenotyping by morphometric analysis of MR images. Physiol. Genomics 42A, 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gu W., Hurles M. E., Lupski J. R. (2009). Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Schneider J. E., Portnoy S., Bhattacharya S., Henkelman R. M. (2010). Comparative SNR for high-throughput mouse embryo MR microscopy. Magn. Reson. Med. 63, 1703–1707 [DOI] [PubMed] [Google Scholar]