SUMMARY
The DSS (dextran sulfate sodium) model of colitis is a mouse model of inflammatory bowel disease. Microscopic symptoms include loss of crypt cells from the gut lining and infiltration of inflammatory cells into the colon. An experienced pathologist requires several hours per study to score histological changes in selected regions of the mouse gut. In order to increase the efficiency of scoring, Definiens Developer software was used to devise an entirely automated method to quantify histological changes in the whole H&E slide. When the algorithm was applied to slides from historical drug-discovery studies, automated scores classified 88% of drug candidates in the same way as pathologists’ scores. In addition, another automated image analysis method was developed to quantify colon-infiltrating macrophages, neutrophils, B cells and T cells in immunohistochemical stains of serial sections of the H&E slides. The timing of neutrophil and macrophage infiltration had the highest correlation to pathological changes, whereas T and B cell infiltration occurred later. Thus, automated image analysis enables quantitative comparisons between tissue morphology changes and cell-infiltration dynamics.
INTRODUCTION
Inflammatory bowel disease (IBD) is a disorder of the digestive tract and affects more than 1 million people in the United States alone (Abraham and Cho, 2009). In ulcerative colitis (UC), a type of IBD, inflammation occurs primarily in the mucosa of the large intestines, leading to debilitating conditions including diarrhea, rectal bleeding and weight loss.
Although both genetic and non-genetic factors are associated with the disease, it is thought that UC is largely caused by an inappropriate inflammatory response by the host to intestinal microbes penetrating through a damaged epithelial barrier (Xavier and Podolsky, 2007). The enormous variety of gut flora contributes to the heterogeneity of the disease. This complexity might explain why currently available therapies for UC have only a 50% chance of positive outcome for patients (Pastorelli et al., 2009).
In recent years, an ever growing number of drugs have been tested to treat UC, based on various strategies to regulate the immune response, including steroids, immunomodulators, and antibodies against inflammatory cytokines, with variable success (Pastorelli et al., 2009). To speed up drug discovery, new candidate drugs need to be efficiently screened in appropriate model systems that have clinical relevance.
Mouse (Mus musculus) models are invaluable for this purpose because they are one of the least expensive systems in which mammalian host-microbe relationships can be studied. In the dextran sulfate sodium (DSS) model of colitis, DSS polymers are dissolved in the drinking water of wild-type mice for 5 days (Wirtz et al., 2007). The resulting chemical damage to the mucus layer protecting the epithelial tissue of the colon allows microbe entry into the gut, causing acute inflammation (Johansson et al., 2010). Macroscopically, weight loss, diarrhea and shortening of the colon are observed (Diaz-Granados et al., 2000; Yan et al., 2009). Microscopic observation shows a progressive loss of intestinal crypt cells predominantly near the rectum and in the distal regions of the colon. Concomitantly, there is an increase in innate immune cells (neutrophils, macrophages, dendritic cells and natural killer T cells) and lymphocytes (B cells and T cells) in the intestinal mucosa (Yan et al., 2009; Hall et al., 2011). The inflammatory response is thought to cause further erosion and ulceration of crypts. These microscopic observations, as well as macroscopic signs, recapitulate aspects of the human disease.
Putative therapeutics can be found by searching for molecular agents that effectively reduce pathological scores in colitic mice. These scores are calculated by experienced pathologists based on microscopic observations, through examining hematoxylin and eosin (H&E)-stained slides of the gut (Wirtz et al., 2007). Because manual scoring is time-consuming and requires a specialized pathologist, it easily becomes a bottleneck in the drug-discovery process.
In order to reduce the reliance on manual assessment of microscopic findings, we set out to develop a fully automated method to score DSS-induced colitis in mice by using image analysis software, Definiens. The ‘Developer’ module of Definiens allows the development of algorithms that interpret H&E slide images using an ‘object-oriented’ method (Baatz et al., 2009). A ruleset (algorithm) was designed using Developer, which automatically evaluates the extent of crypt loss in an H&E colon slide.
RESOURCE IMPACT.
Background
Colitis is a highly debilitating disease that affects over a million people in the United States alone. Half of these patients do not respond well to available therapies, highlighting a serious, unmet medical need. Mouse models of colitis are routinely used to screen for potential therapeutics, but the screening time required for a pathologist to score their colon slides manually is a major bottleneck in the drug-discovery process. There is a possibility of automating the process by digital image analysis; however, this requires analysis of hematoxylin and eosin (H&E) slides, which are generally far more difficult to analyze than immunohistochemistry (IHC) slides owing to their signal complexity and variability. Consequently, few attempts have been made to analyze H&E slides thus far, even though it is one of the most common stains used by pathologists for a wide variety of applications.
Results
The authors propose a new automated image analysis method to quantify responses to treatment, using the dextran sulfate sodium (DSS) mouse model of inflammatory bowel disease. This method quantifies responses on the basis of histological changes in the mouse tissue, which are scored on whole H&E slides. The authors demonstrate that the method is sufficiently predictive and robust to support its use in a drug-screening setting. Furthermore, the image-analysis method can be combined with more traditional IHC quantification techniques to gain further insight into the molecular mechanism of action of a particular therapeutic candidate.
Implications and future directions
In addition to its potential for application in research involving the DSS mouse model of colitis, this work is likely to encourage more research into applying image analysis techniques to quantify tissue morphology changes on H&E slides. Considering the similarities in tissue structure between mice and humans, it is conceivable that the method presented here will, in the future, be used to analyze colon tissue from human biopsies, which could help to accelerate clinical diagnoses of colitis.
Another method commonly used to study colitis is immunohistochemistry (IHC). Because immunomodulation is a primary target for potential therapeutics, it is informative to examine colon slides labeled with immune cell markers. Quantification of stain distribution is challenging to the human eye, but can be performed efficiently using Definiens (Diaz et al., 2012).
The methods presented here can decrease the labor required by pathologists and speed up the identification of potential therapeutic targets for patients suffering from UC.
RESULTS
Evaluation of historical studies
In order to decrease the time required for microscopic scoring of colon pathology in the DSS-induced model of colitis, an automated algorithm was developed using specialized image analysis software, Definiens, and applied to whole scanned and digitized H&E slides of mouse large intestines. For an automated algorithm to be useful, it must be significantly faster and less labor intensive than a pathologist’s manual scoring system, and allow the identification of candidate therapeutics in drug-discovery studies in a way that is comparable to using manual scores. To test this, H&E colon slides from nine historical studies with a total of 544 colon samples, for which manual scores had already been obtained, were analyzed.
Speed of scoring
All studies had been scored manually by one of two experienced pathologists (Table 1). The manual scoring process required up to 5 minutes per slide. The automated algorithm required an average computing time of just under 15 minutes per slide per processor but, using 28 central processing units (CPUs), the entire sample set (544 animals) was processed in ∼4.5 hours, effectively taking 30 seconds per slide. No manual intervention was necessary, except loading the slides (∼4 seconds per slide, or ∼36 minutes for 544 samples) and initiating the computation, which took a minute or less per study.
Table 1.
Study numbers and dose of IL-22 Fc used as a +ve control

Score normalization
The studies were designed to address specific hypotheses regarding the effects of molecular agents on colitis scores in mice administered DSS. All studies contained three types of control groups: animals given water without DSS (Ø control), mice given DSS alone or DSS combined with an inert antibody (−ve control) and mice administered a molecule known to reduce colitis severity, IL-22 Fc (Sugimoto et al., 2008), at various doses (+ve control). In addition, each study contained groups of animals given DSS and several molecular agents, or a single agent at varying doses. Because there was large variation of raw scores given to equivalent control groups in different studies (Fig. 1), raw scores were normalized relative to the mean scores of the Ø and −ve control groups in each study.
Fig. 1.
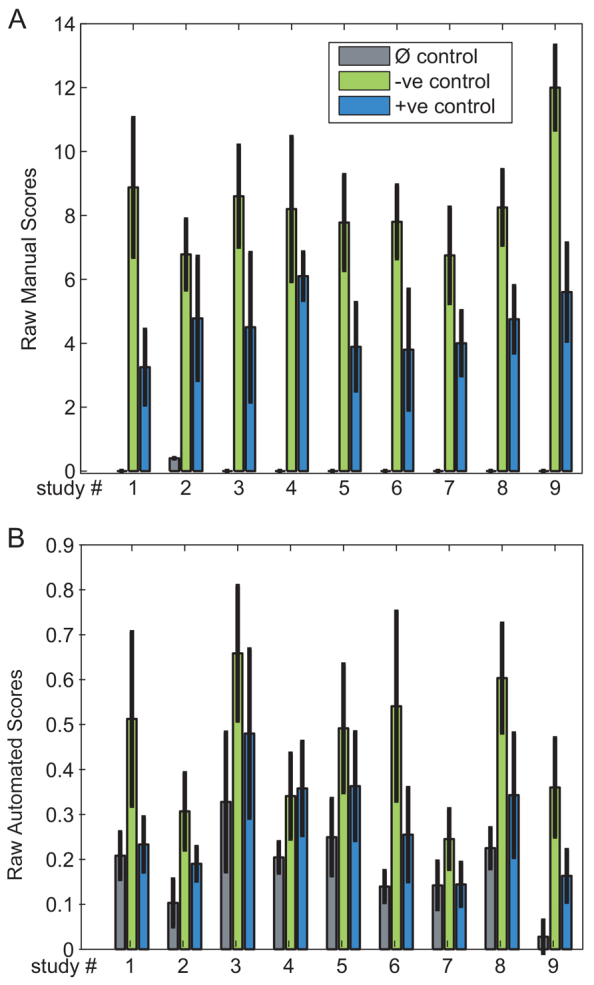
Evaluation of raw scores of control groups in historical studies. The mean and standard deviations of the manual (A) and automated (B) raw scores for equivalent (Ø, −ve and +ve) control groups in each study. The bars indicate the mean score within the group, whereas the error bars indicate 1 s.d. The mean score across all studies for the Ø control groups divided by the mean score of the animals in each corresponding −ve control group is 0.005±0.016 (manual) and 0.430±0.098 (automated). The corresponding values for the −ve control group are 1±0.186 (manual) and 1±0.178 (automated). Finally, within the +ve control group, the corresponding values are 0.506±0.088 (manual) and 0.584±0.160 (automated). The relatively large s.d. of mean scores from equivalent control groups suggest that the raw scores from different studies need to be normalized in order to be comparable.
Role of effect size and P-values in drug target identification
For correct identification of efficacious drug candidates, the automated method needs to be able to identify treatment groups that had scores that were statistically different from those in the −ve control group. However, it might not be able to do so at all effect sizes. Consider, for example, the dose titration study #7 (Fig. 2): in this particular study, IL-22 Fc, which in other studies was used as a +ve control at only one dose, was instead given at doses titrated from 30 to 0.05 μg in combination with DSS. Both automated and manual normalized scores identified a statistically significant difference from the −ve control at the 30 μg and 6 μg dose (Table 2). At 1.25 μg, both automated and manual scores failed to reach statistical significance but, at 0.25 μg, the manual score identified a statistically significant difference not found by the automated method. However, the effect size of the 0.25 μg dose level is small: less than 20%. Because statistical difference was not observed at the 1.25 μg level by either manual or automated scores, this might or might not be a ‘true’ result. In our experience in general, it is unlikely that an effect size of <20% is a biologically meaningful result. The drug candidate identification process thus depends on both effect size (difference between the means of the −ve control group and test group) and statistically significant differences based also on variance (the result of a Student’s t-test between the −ve control group and test group).
Fig. 2.
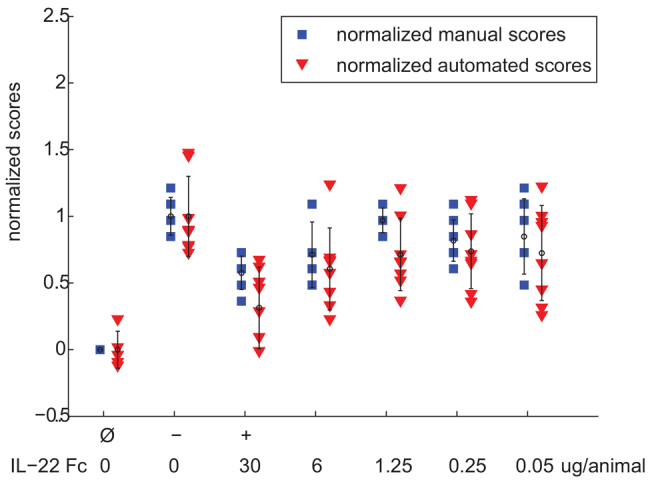
Results for study #7. Each marker indicates a single animal, and is color-coded by whether it was a normalized manual score (blue square) or normalized automated score (red triangle). The markers are plotted for all animals in each experimental test group. Means are indicated by small circles, and error bars correspond to 1 s.d. The dose of IL-22 Fc for each titration group is also indicated.
Table 2.
Results from study #7
Identification of treatment candidates using the automated algorithm
Applying the reasoning above, we determined how many test groups that were identified as being effective at reducing colitic scores using normalized manual scores would be found using normalized automated scores. ‘Hits’ were defined as those test groups for which mean scores obtained at least a 20% effect size and a Student’s t-test P-value <0.05 compared with −ve controls. Out of 50 test groups, 13 were found to be a hit by both automated and manual methods (true positives), 0 were found to be a hit only by the automated method (false positive), 6 were found to be a hit only by the manual method (false negative), and 31 were found to not be hits by either method (true negatives). The concordance between predictions from the automated scoring and manual scoring was (true positive + true negative) / (total tests) = 44/50 (88%). The fact that the automated method had no false positives implies that it is effective as a first-pass screen to identify efficacious drug candidates: if it detects a hit, it is highly likely to be valid. However, if the automated algorithm determines a treatment group not be a hit, there is a 16% chance [(false negative) / (false negative + true negative) = 6/37] that the manual scores would have picked up the candidate.
Internal scoring consistency
To investigate the internal consistency of each method, four serial sections of colon from study #1 (Table 1) were prepared and stained separately, and scored by a pathologist and the automated algorithm. The internal correlation of a pathologist’s raw scores for this study was 0.929±0.067, whereas that for the automated algorithm was 0.903±0.040.
Meta-analysis
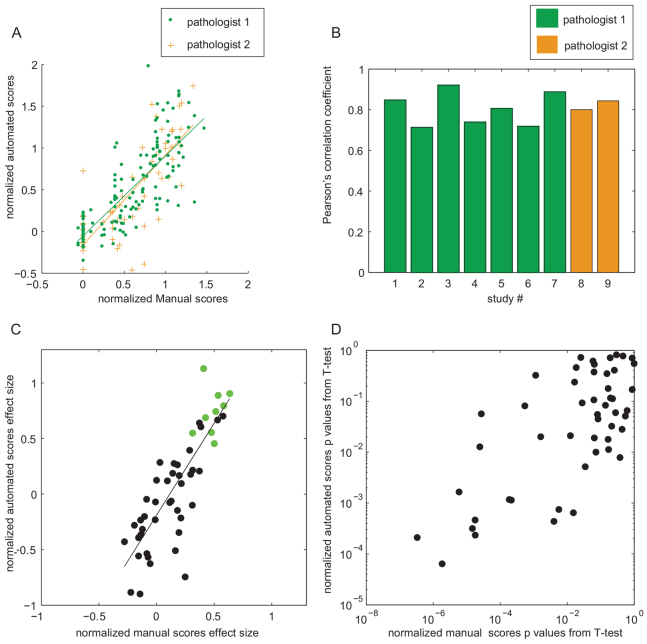
In order to better understand the general differences between the automated and manual scoring methods, normalized scores were pooled from multiple studies. Automated scores in all studies from the three control groups were linearly related to manual scores of both pathologists (Fig. 3A), with similar linear fits. Normalization is crucial because one pathologist assigned systematically higher absolute scores than the other (data not shown), but this discrepancy disappears with normalization. The Pearson’s correlation coefficient between the pooled manual and automated scores was 0.779. Correlation coefficients were also calculated within each study (Fig. 3B) and were similar.
Fig. 3.
Comparisons between normalized manual and automated scores. (A) Scatter plot of normalized manual versus automated scores from the three control groups, across all studies. Two linear fits corresponding to the two pathologists who scored the studies are shown in green and in orange. Pathologist 1: fit: y=0.960x −0.053; Pearson’s correlation: 0.788. Pathologist 2: fit: y=1.075x − 0.159, Pearson’s correlation: 0.766. If the linear fit is applied to the combined manual scores from both pathologists, the fit equation is y=0.99x − 0.080; Pearson’s correlation: 0.779. (B) Individual Pearson’s correlation coefficients within studies, color-coded for those studies scored by different pathologists. (C) Scaled effect sizes for corresponding normalized automated and manual scores in animal groups from all studies: the difference between the −ve control group and test groups are shown as black circles, and the difference between the −ve control group and the +ve control group are shown as green circles. The line indicates a linear fit: y=1.656x − 0.189; Pearson’s correlation: 0.830. (D) A log-log graph of P-values for Student’s t-tests between test animal groups and −ve control groups in all studies.
We next compared the effect sizes of treatment groups computed from all drug studies, using the normalized manual and automated scores. The effect sizes were linearly related, with a correlation coefficient of 0.830 (Fig. 3C). Interestingly, the automated scoring had a tendency to exaggerate the effect size.
The statistical differences between the groups were also compared, by calculating P-values from a Student’s t-test between the −ve control and test groups from all drug treatment studies. The P-values obtained using normalized manual and automated scores appeared broadly related (Fig. 3D). However, the P-values were generally higher for the automated scores compared with the manual scores.
Correlating tissue morphology change with inflammatory cell infiltration
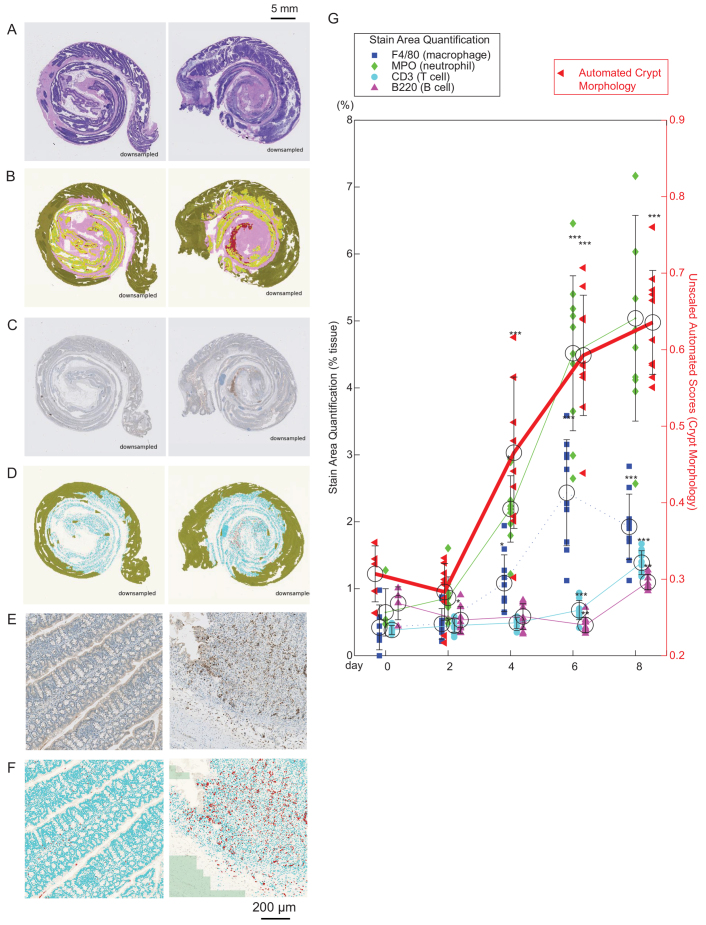
The analysis so far indicated that the automated scoring method, although slightly less sensitive than the manual method, is able to capture pathological tissue morphology change in a consistent manner. Because most treatment candidates that are used in colitis drug discovery are immunomodulatory, it is informative to understand their effect on the dynamics of immune cell infiltration as well as on tissue morphology. Whereas tissue morphology is best evaluated in H&E slides, infiltration of immune cells into the gut is effectively visualized using IHC staining. Fine quantification of stain is difficult for a pathologist to perform by eye, but relatively simple using Definiens. To demonstrate the ability to compare tissue morphology change and inflammatory cell infiltration easily using automation, a timecourse study was performed where animals were given 3% DSS and euthanized after 0, 2, 4, 6 or 8 days. H&E slides were prepared and raw automated scores were calculated. Serial sections of the H&E slides were immunostained with F4/80 (macrophage marker), CD3 (T-cell marker), MPO (neutrophil marker) and B220 (B-cell marker). The area containing cells stained with each marker, relative to total tissue area, was quantified (see Materials and Methods, Fig. 4A–F and supplementary material Fig. S1). The stain quantification took ∼30 seconds per slide per processor.
Fig. 4.
Correlating tissue morphology change with inflammatory cell infiltration. (A–F) Example images from an animal given no DSS (left panels) and an animal taken down on day 6, after 5 days of 3% DSS administration (right panels). (A) Example H&E slide. (B) Pseudo-colored automated analysis result of A, where non-analyzed area (green), muscle (pink), healthy crypt area (yellow) and pathological area (red) are indicated. (C) Original image of colon stained with marker (here MPO). (D) Automated analysis based on C, where non-analyzed area (green), tissue area (blue) and positively stained cells (in this case neutrophils; red) are shown (see also supplementary material Fig. S1). (E,F) Examples of high-magnification regions within C and D, respectively. (G) Timecourse analysis of the area of positive IHC staining (axis and labels on left) over relevant tissue area, with an overlay of a graph of H&E automated morphology scores (axis and labels on right). Note that the automated H&E morphology scores are not normalized, because the experiment did not contain a +ve control group as in the drug-treatment studies. Dunnett’s test was performed for each group against day 0. *P<0.05 compared with control; **P<0.01; ***P<0.001.
A statistically significant increase in F4/80 and MPO staining was found concomitant with crypt loss, closely correlating to the automated scores (Fig. 4G). Note that raw automated scores were used because this was not a drug treatment study and no −ve control group was run. Late in the disease (day 8), a gradual increase in CD3 (T cells) and B220 (B cells) was also observed. Spearman’s correlations between individual automated crypt scores and the proportion of stained area for each marker was 0.84 for MPO stain (neutrophil), 0.74 for F4/80 (macrophage), 0.70 for CD3 (T cells) and 0.23 for B220 (B cells). The results show that the crypt morphology change was most linearly correlated to the change in number of neutrophils, followed by macrophages and T-cell infiltration. It was least correlated to B-cell infiltration.
DISCUSSION
In order to increase drug-discovery efficiency, it is desirable to screen a large number of candidates in the shortest possible time and at minimal cost (Paul et al., 2010). We thus developed a method to automatically analyze H&E-stained slides of colons from mice with DSS-induced colitis to speed up drug candidate identification and save pathologists’ time.
Speed
The automated quantification method allowed analysis of 544 slides in 4.5 hours, which could be run overnight. By using a larger number of processors running Definiens in parallel, the analysis is scalable to any number of slides.
Use for drug discovery
The automated algorithm classified 88% of tested agents in the same way as manual scores, as effective or not effective. Importantly, all the discrepancies were false negatives – the automated algorithm failed to identify some potential candidates. The fact that no false positives were identified indicates that the algorithm is suitable for fast screening because, if a target is identified, it is likely to be worth the effort to validate it. The automated method is not, however, appropriate for evaluation of studies in which subtle effects need to be measured, for example to examine fine differences in efficacy between two compounds.
Robustness
For an automated algorithm to be useful, it needs to be robust to routine variation during sample preparation. The algorithm was therefore designed to rely less on variables that are easily affected by H&E sample preparation, such as color, and relied mostly on ‘contextual’ features. As examples of this approach, a hierarchical classification algorithm was used to establish regions of interest in a low-magnification image and, at high resolution, healthy crypts were identified by geometric and neighborhood features. In fact, this strategy is similar to the human visual interpretation of images as composed of ‘objects’, rather than classical computer-based image analysis, which treats images as arrays of pixels.
Owing to the robustness of the algorithm, nine out of ten examined studies were assessed as appropriate quality for image analysis, and all studies had a correlation coefficient between manual and automated scores greater than 0.7. Furthermore, the automated scores had an internal consistency of over 0.9, very similar to that of the pathologist. The high correlation argues that the algorithm is highly robust to sample preparation differences.
Correlation to pathologists
By performing meta-analysis, we also demonstrate that, across all studies, the normalized scores from the automated algorithm correlates well with scores from both pathologists. The pathologists cannot be compared directly to each other, because they scored different studies. But the fact that both of their scores correlate well to the automated scores suggests that the algorithm is assessing general features that are accepted signs of pathology by multiple pathologists.
Quantification of immune cell infiltration
Although the predominant H&E morphology observed in DSS colitis is ulceration and crypt-cell loss, another hallmark of colitis is infiltration of immune cells into the gut, which is most effectively visualized using IHC staining. In order to study the relationship between crypt loss and immune-cell infiltration, serial sections of slides used for automated crypt analysis were stained for neutrophils, macrophages, T cells and B cells. Stain intensity was automatically quantified. Whereas H&E morphological assessment was optimized to be independent of stain intensity, stain intensity is the primary readout of IHC quantification and, therefore, is by nature more sensitive to sample preparation. Any procedural changes in staining the slides will give inconsistent values; therefore, all slides in a study should be ideally stained at the same time. If sample preparation is consistent, chromogenic IHC on tissues can be used in a highly effective quantitative manner (Taylor and Levenson, 2006; Walker, 2006).
IHC analysis showed that neutrophils and macrophages increased concomitantly with crypt loss, with a latent increase in B and T cells. The correlation coefficients were highest between crypt morphology scores and staining of neutrophils and macrophages. This shows that crypt loss was more closely associated with infiltration of innate immune cells. The results do not imply causation – whether neutrophils drive damage to crypts, or vice versa – but implicate a role of neutrophils in an early stage of the disease. Our results are consistent with immune-cell infiltration quantification using flow cytometry in DSS-induced colitis (Hall et al., 2011), but ours is a less laborious and faster method.
By applying IHC analysis to drug-treatment studies, it would be possible to investigate the role of a drug candidate not only on tissue morphology, but also on immune-cell infiltration, in a relatively easy, but quantitative, manner.
Understanding errors in the automated algorithm
When the relationship between effect sizes was examined using normalized automated and manual scores, there was a tendency for the automated scores to ‘overshoot’, leading to an exaggerated effect size. This shows that, at the upper range of the effect size, the algorithm has no problem recognizing molecular agents as candidates. However, higher P-values in the automated algorithm compared with manual scores shows that the intra-group variance is higher in the automated method, and so subtle differences with small effect sizes would be harder to detect. These results demonstrate that, for future drug-discovery studies, it would be optimal to administer drug candidates at the maximal feasible dose. The larger the effect size, the higher the chance that the candidate would be detected by the automated algorithm.
It is unclear what exactly causes the greater ‘noise’ in the automated algorithm compared with analysis by pathologists, but one possibility is that pathologists are better at assimilating a larger number of morphological cues in the tissue features, and assessing their relative importance in determining pathology severity. This effect is particularly evident in the scoring of Ø control animals, where pathologists scored every animal as 0, in all but one study. This demonstrates the pathologists’ phenomenal ability to correctly recognize tissue that is within ‘normal range’, a skill that is largely acquired through experience. Such less tangible aspects of pathology scoring is a challenge to translate into artificial intelligence.
Recommendations
Here are recommendations for using the automated algorithms for screening for drug candidates:
Include Ø, −ve and +ve controls in all studies. The +ve control should be a therapeutic agent known to be effective. In our hands, IL-22 Fc was found to be a particularly suitable positive control owing to its reproducible and robust therapeutic effect. IL-22 Fc (Ota et al., 2011) binds to the IL-22 receptor, which is restricted to cells of epithelial origins, and is crucial for wound healing, restoration of goblet cells and mucosal repair (Sugimoto et al., 2008), as well as for mucosal defense (Zheng et al., 2008).
Administer the maximal feasible doses of test agents to enhance target identification.
After slide preparation, briefly quality control slides for gross defects.
Run automated H&E morphology analysis on the number of computer processors appropriate for the size of the drug screen.
Normalize all scores using the Ø control and −ve control, and ensure that the +ve control exhibits at least 20% effect size and statistically significant difference compared with the −ve control. If the +ve control does not meet these criteria, it is advised to score the study manually.
Identify other treatment groups that show a >20% effect size and statistical difference compared with the −ve control, and validate these ‘hits’ manually.
If desired, perform IHC on serial sections with immune cell markers, run the IHC image analysis algorithm and compare these results with the H&E morphology analysis.
Comparison to other image analysis techniques
Digital image analysis studies, especially applied to H&E slides, are relatively rare in the literature. Unlike quantitative IHC, where intensities of individual image pixels can simply be added to obtain overall stain intensity, H&E-stained slides contain complex tissue structures that are distinguished by morphology rather than stain intensity. Definiens Developer is especially suitable as image analysis algorithm design software because it is capable of measuring both pixel-based intensity and analyzing properties of images as objects.
Definiens has been used in the past to analyze colon cancer (Persohn et al., 2007), but this was done by counting BrdU-positive cells in IHC, which is similar to our method to analyze lymphocyte infiltration count. Some other studies have used textural analysis to automatically identify colon tumors in H&E slides (Hamilton et al., 1997; Esgiar et al., 2002). These papers assessed tumors by examining textural properties (i.e. distribution of pixels) in randomly selected areas of the colon, and not by identifying ‘crypt-like’ objects as we have done. Unlike tumor identification, colonic tissue with DSS-induced pathological crypt loss is often proximal to healthy crypts and, therefore, gross textural analysis alone was insufficient to categorize the extent of crypt loss. However, we utilize an extension of this approach, because a part of the criteria to identify objects as crypts is standard deviation of pixel intensities within the object area, a concept related to textural analysis.
Applications to genetically engineered mice
The DSS model can be readily used to evaluate genetically altered mice, because it is effective in a variety of mouse strains and does not require crossing to strains that are genetically susceptible to colitis. One useful application is in wound-healing research, where extensive epithelial loss and ulceration associated with DSS makes it an attractive model (Cox et al., 2012; Zhang et al., 2012). The automated methods described here could be applied to genetically engineered mice in such studies, so long as appropriate controls are included.
Conclusions
Overall, we have used Definiens Developer to analyze images in a similar way to a pathologist’s eye. The methods described here are highly applicable to many other morphology-based tissue scoring techniques that pathologists perform routinely on slides stained with H&E, for which few other effective automated analysis methods exist.
MATERIALS AND METHODS
Animals
All procedures were carried out with Institutional Animal Care and Use Committee (IACUC) approval in accordance with the institution’s ethical guidelines.
Colitis was induced in C57BL/6 mice (Jackson Laboratory) by administration of 3% DSS in drinking water ad libitum for 5 consecutive days, similarly as described previously (Wirtz et al., 2007). Animals were euthanized on day 8. In drug-treatment studies, IL-22 Fc (PRO312045) (Ota et al., 2011) or the mouse anti-Ragweed isotype-matched control antibody was administered to mice in relevant experimental groups at various doses ranging from 0.05 to 200 μg/100 μl intraperitoneally at days −1, 1, 4 and 6. In the time-course study, no antibodies were administered.
H&E slide preparation and manual scoring
Colons were prepared as a ‘Swiss roll’ (Wirtz et al., 2007) and fixed in formalin. Tissues were embedded into paraffin blocks and 5-μm sections were prepared. Slides were stained with H&E and scored by one of two experienced pathologists in four anatomical regions of the colon: the proximal colon (PC), middle colon (MC), distal colon (DC) and rectum (R). Each region was given a raw score based on crypt epithelial cell loss with consideration of the extent of inflammatory cell infiltrate, on a scale from 0 (healthy) to 5 (severe diffuse colitis characterized by complete loss of colonic epithelial cells). The raw scores from each region were summed to give a total raw colitis-severity score for each animal, which ranged from 0 (least severe) to 20 (most severe). Thus, the score is a composite readout of severity as well as extensiveness of epithelial cell degeneration and inflammation, and is a simplified form of the Dieleman scoring scheme (Dieleman et al., 1998). Studies were ‘mock-blinded’, where the pathologist chose not to look at treatment groups, although this information was accessible to the pathologist.
IHC staining procedure
Four serial sections of 5-μm thickness were prepared from paraffin blocks, each mounted on separate slides.
Detection of macrophages, T cells and B cells
All steps were carried out using the DAKO Autostainer. Slides were incubated in Target retrieval solution (DAKO S1700). Macrophages were detected with rat anti-F4/80 monoclonal antibody (Serotec MCAP497), B cells with anti-CD45r (B220) monoclonal antibody (Pharmingen 557390) and T cells with anti-CD3 monoclonal antibody (NeoMarkers RM-9107-S). Stains were visualized using the VECTASTAIN Elite ABC Kit (Vector Labs PK-6101). Pierce Metal Enhanced DAB (Thermo Scientific PI-34065) was used for chromogenic detection, and nuclear counterstain was performed using Mayer’s hematoxylin.
Detection of neutrophils
All steps were carried out using the Ventana Discovery XT Autostainer. Samples were pre-treated with Cell Conditioner CC1 (Ventana 950-124), and the anti-myeloperoxidase (MPO; NeoMarkers RB-373-A) polyclonal antibody was used as a primary, and visualized using OmniMap anti-Rb HRP (Ventana 760–4311). The ChromoMap DAB kit (Ventana 760-159) was used for chromogenic detection, and nuclear counterstain was performed using Hematoxylin II (Ventana 790–2208).
Digitization of sections, image analysis software and IT infrastructure
Slides were scanned using a Hamamatsu Nanozoomer 2.0 HT digital slide scanner running NDP Scan software, with an Olympus UPlan SApo 0.75 NA 20× objective lens. All slides were only scanned in the area where specimen tissue was present. Resulting images (0.46 μm/pixel resolution) were saved in the NDPI format, a type of JPEG, with 20% default compression, on an Isilon NAS drive. Image analysis was performed with custom-written rulesets for Definiens Developer software (Munich, AG), and run on up to 28 CPUs at a time, on two nodes each containing 16 Intel Xeon based logical CPUs, with 96 GB of RAM, and over 220 GB of free space. The Ethernet network speed was 10 Gbit/second.
Historical study selection
H&E colon slides from ten historical studies, for which manual scores had already been obtained, were scanned. Each study included 6 to 11 (median of 8) experimental groups, each group containing 4 to 10 (median of 7) animals. Each study was visually inspected for gross sample preparation problems. One study was excluded owing to serious artifacts that made scoring challenging even for pathologists. The nine included studies, with a total of 544 samples, contained artifacts that would be expected during routine laboratory practice.
Morphological image analysis on H&E sections
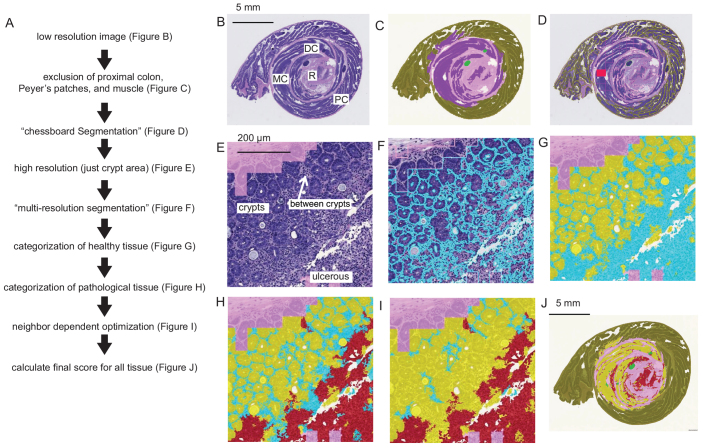
An algorithm (available on request) was designed to differentiate relevant areas of the colon for assessing colitis at low resolution, and then at high resolution, to calculate the relative proportions of healthy and pathological areas in H&E slides of mouse colon (Fig. 5A). Images are analyzed using the RGB (red, green and blue) spectra. Definiens denotes pixel intensities within each spectrum from 0 (darkest) to 255 (brightest). The term ‘brightness’ without specifying a single spectrum refers to the mean pixel intensities of red, green and blue spectra.
Fig. 5.
Method for morphological image analysis on H&E sections. (A) The automated image analysis workflow. (B) The H&E image is scaled to 1% of the original size. Each region of the colon rectum (R), distal colon (DC), middle colon (MC) and proximal colon (PC) are labeled. (C) The image is subdivided by multi-resolution segmentation algorithm (Definiens). The procedure divides the image into discrete units by grouping pixels that have common morphological features. These units are false colored in this figure by the categories assigned to different regions of the H&E slide (note the coloring is for aiding understanding only, no color filters are applied to the original image). The hematoxylin stain is used to separate the darkly stained units, shown in purple (intestinal region where, in healthy animals, crypts would be present) from the units shown in pink (muscle). Peyer’s patches, shown in light green, are excluded. Only units near the geometric center of the detected tissue area are analyzed (a circular region within 4.55 mm), and the region outside of this area. (D) A ‘chessboard segmentation’, where the image is conceptually divided into equally sized square tiles except for the boundaries of tissues where they are truncated, is used to analyze individual areas in the colon. Classifications of the tissue units are shown here in the Definiens ‘outline’ view, and an example of an analyzed region is shown in red. (E) The individual tiles are analyzed at high magnification (10×). The false-colored pink shade indicates area that was already categorized at low resolution as ‘muscle’. This region is pixelated in appearance due to the down-sampling (analysis at low magnification) processes in C. The relevant analysis area is shown here without any false coloring. Within this region, healthy crypts, regions between crypts and regions were ulcers are visible are labeled. (F) The first step is a multi-resolution segmentation in the relevant region. Cyan outlines show cohesive pixels that were grouped into units. (G) A combination of pixel features within the units were used to categorize some as crypts (false colored in yellow), and others as still undefined (false colored cyan). (H) Within the undefined area, units with pixel features characteristic of pathology were classified as pathological (false colored red). (I) The uncategorized region was further categorized depending on their proximity to neighboring regions that were already categorized (compare with H). (J) The entire classified tissue area is used to compute the final score (see also supplementary material Fig. S3).
Low resolution
Scanned images of ‘Swiss roll’ preparations of mouse colon slides (H&E) are first analyzed at low resolution (0.4×). The image is subdivided into ‘objects’ using the Definiens built-in ‘multi-resolution segmentation’ algorithm (Baatz and Schäpe, 2000). The segmentation method divides the image areas into smaller areas that have similar features, in a hierarchical fashion. This procedure separates the majority of the tissue into two distinct areas (Fig. 5B): the muscle, which is highly eosinophilic (pinkish hue), and the gut lining, where healthy animals would have intestinal crypts, which are basophilic (purplish hue). Only the basophilic area is selected for further analysis (Fig. 5C). Peyer’s patches are normally occurring lymphoid nodules in the intestines, containing a large number of B and T cells. These areas are selected by their dark basophilic staining and removed from the analysis area. Without removal, the lymphoid regions would be mis-categorized as a tissue area showing signs of colitis-induced inflammation. During manual assessment, the pathologist categorizes four sub-regions of the colon: rectum, distal colon, middle colon and proximal colon (Fig. 5B), and select a ‘representative’ region from each area, which is examined at a higher magnification. The automated algorithm was not designed to explicitly distinguish between these sub-regions; however, the relative distance from the center of the Swiss roll is used to confine analysis within the inner area of the colon (area within a 4.55-mm radius from the center of the roll), mostly excluding the proximal region (Fig. 5C). This is because DSS-induced colitis predominantly affects the rectal, middle and distal regions of the colon.
High resolution
The entire colon tissue within the selected region (within 4.55 mm from the center) is too large to analyze at a high resolution at once owing to limited computational capacity. Therefore, this region is tiled in a chessboard pattern, using the Definiens algorithm ‘chessboard segmentation’, and each tile (approximate size of 0.8×0.8 mm) is analyzed at higher magnification (Fig. 5D).
At 10× magnification, only relevant areas (within 4.55 mm from the center, excluding muscle and Payer’s patches) are analyzed (Fig. 5E). Healthy crypts appear as relatively basophilic epithelial and goblet cells, neatly arranged in round or oblong rings. When pathological changes (such as ulcers and erosions) are present, crypts appear severely disrupted or are completely absent, so that the entire area appears less basophilic. Typically, a large number of round nuclei, smaller than those of epithelial and goblet cells, are found in affected areas, indicating inflammation.
The automated algorithm assesses the loss of crypts as follows: the regions where crypts should be present in healthy animals are first subdivided into multiple smaller areas, of sizes corresponding to average crypt diameter (50–100 μm), using the Definiens ‘multi-resolution segmentation’ algorithm (Fig. 5F). The effect is that cohesive structures, such as crypts, are identified and appear as relatively round structures. Pathological regions that appear relatively homogenous are subdivided into areas of random shapes (compare Fig. 5E and 5F).
The classification strategy of these areas relies on a likelihood score, which depends on a geometrical combination of morphological and textural features characteristic of healthy crypts or pathological tissue, namely the overall shape (crypts are ellipsoid, pathological tissue is amorphous), staining intensity (crypts tend to stain more darkly) and standard deviation of pixels within a region, using Definiens ‘evaluation classes’. The standard deviation of pixel intensities is higher when a large number of small dark objects are present, as is the case when many inflammatory nuclei are concentrated in a region of tissue. Areas that have a combination of shape, staining and standard deviation metrics, indicating a high likelihood of belonging to crypts, are classified as healthy (Fig. 5G, yellow), and areas with characteristics of ulcers or erosions are classified as pathological (Fig. 5H, red). Some areas with indeterminate features are left uncategorized (Fig. 5H, cyan). These are often areas between crypts where some immune cell nuclei are visible even when the intestinal tissue is perfectly healthy (Fig. 5E) so that areas have properties of both healthy and pathological tissues. To improve the classification, after defining areas that are comprised of healthy crypts, the probability score for a unit to be classified as pathological is decreased if it is adjacent to an area that is already defined as healthy. In this way, narrow strips of tissue that contain some infiltrates are more likely to be classified as healthy (Fig. 5I), which is consistent with the interpretation by a pathologist.
Causes of misclassification
Several artifacts due to sample preparation affected the automated analysis algorithm. These included:
unraveling or distortion of the ‘Swiss roll’, resulting in a mis-categorization of intestinal regions
incomplete sections or tissue dropout due to sectioning defects
inclusion of non-intestinal tissue in the slide, such as the perianal glands, which can appear similar to ‘healthy crypts’
uneven sectioning of the tissue, causing irregular staining in various regions.
Illustrative images of poor sample preparation are shown in supplementary material Fig. S2, whereas Fig. 5B shows an example of a good preparation.
Automated scoring method
The areas categorized as either healthy or pathological were summed together for all tiles (Fig. 5J and supplementary material Fig. S3) to compute a total raw score as follows: Score=2 × Ap/(Ap+Ah), where Ap=total area scored as pathological, and Ah=total area scored as healthy. In theory, the raw score is within 0 (all healthy) and 2 (all pathological), but most raw scores were under 1. The factor of 2 in the equation was introduced so that most raw scores varied between 0 and 1.
Staining area quantification on IHC sections
IHC slides were analyzed using the RGB spectra, as described in the H&E image analysis section. Images are analyzed (available on request) initially at low resolution (0.2×), and ‘bright’ areas are excluded as background. The muscle area is not separated from crypts for analysis but, as with the H&E-stained slides, only the region within a 4.55-mm radius from the center is used for analysis. The very dark areas (areas with brightness less than 70% of mean tissue brightness) are excluded because they probably correspond to lymph nodes. On the rest of the tissue area, the Definiens algorithm ‘chessboard segmentation’ strategy is applied to analyze tissue regions at high resolution (10×) tile by tile. At high magnification, stained areas of the tissue are isolated by an intensity threshold only on the blue spectrum, using the Definiens algorithms ‘auto-threshold’ followed by ‘multi-threshold segmentation’, which divides the image into dark-blue and bright-blue regions after determining an optimal intensity threshold. Of the objects selected as ‘dark blue’, only regions with sizes greater than 2 μm2 and under 104 μm2 are selected, because tiny specks are unlikely to be intact nuclei, and larger dark stains are likely to be artifacts, such as non-specifically bound antibody. The density of cells is calculated as follows: Score=Ts/Ta, where Ts=total area occupied by the stained areas of the tissue, and Ta=total area occupied by analyzed tissue, in the entire slide.
Analysis methods
Statistical methods
All data analysis was performed using MATLAB’s (MathWorks) Statistics Toolbox.
Calculation of internal consistency of scoring
Consistency for a particular scoring method was computed by applying either the manual or automated scoring method to read the same study four times. Instead of scoring the exact same slides, four serial sections were prepared for each and every animal in a study, and scored independently (supplementary material Fig. S4). Then, the Pearson’s correlation coefficients between four independent sets of raw scores were computed. With four sets of slides, this constitutes 4×3/2=6 comparisons. The root mean square (RMS) and root mean square error (RMSE) of all six correlation coefficients was computed to estimate the consistency of either the manual or automated method for this study.
Score normalization
All drug-treatment studies contained a control group given water without DSS (Ø control), and mice given DSS alone or DSS combined with an inert antibody (−ve control).
The raw scores (Fig. 1) were normalized by setting the mean score of the Ø control group as 0, and the mean score of the −ve control group as 1, by the following formula: ScoreNorm=[Scoreorig − (Scoremean(Ø))]/(Scoremean(−)−Scoremean(Ø)), where ScoreNorm= normalized individual score, Scoreorig=original individual raw score, Scoremean(Ø)=mean raw score of all animals in the Ø control group, and Scoremean(−)=mean raw score of all animals in −ve control group.
Supplementary Material
Acknowledgments
The authors thank Thomas Bengtsson for statistical advice and critical evaluation of the manuscript, Wenjun Ouyang for help regarding use of IL-22Fc as a positive control, and the Genentech pathology core labs for all slide preparation.
Footnotes
COMPETING INTERESTS
All authors are employed full-time by Genentech Inc., which provided all funding. All authors hold stock options, bonds and a pension plan with Genentech.
AUTHOR CONTRIBUTIONS
C.K. performed all image analysis, prepared all figures, and drafted the manuscript. S.J., J.L. and X.W. carried out experiments. J.D., L.D. and J.B. conceived and guided experiments. C.K. and S.G. performed statistical analysis. All authors were involved in writing the paper and had final approval of the submitted and published versions.
FUNDING
This work was funded by Genentech, Inc.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.011759/-/DC1
REFERENCES
- Abraham C., Cho J. H. (2009). Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baatz M., Zimmermann J., Blackmore C. G. (2009). Automated analysis and detailed quantification of biomedical images using Definiens Cognition Network Technology. Comb. Chem. High Throughput Screen. 12, 908–916 [DOI] [PubMed] [Google Scholar]
- Baatz M., Schape A. (2000). Multiresolution Segmentation: an optimization approach for high quality multi-scale image segmentation. In Angewandte Geographische Informationsverarbeitung XII. Beiträge zum AGIT-Symposium Salzburg 2000 (ed. Strobl J.). Karlsruhe: Herbert Wichmann Verlag [Google Scholar]
- Cox J. H., Kljavin N. M., Ota N., Leonard J., Roose-Girma M., Diehl L., Ouyang W., Ghilardi N. (2012). Opposing consequences of IL-23 signaling mediated by innate and adaptive cells in chemically induced colitis in mice. Mucosal Immunol. 5, 99–109 [DOI] [PubMed] [Google Scholar]
- Diaz D., Allamneni K., Tarrant J. M., Lewin-Koh S. C., Pai R., Dhawan P., Cain G. R., Kozlowski C., Hiraragi H., La N., et al. (2012). Phosphorous dysregulation induced by MEK small molecule inhibitors in the rat involves blockade of FGF-23 signaling in the kidney. Toxicol. Sci. 125, 187–195 [DOI] [PubMed] [Google Scholar]
- Diaz-Granados N., Howe K., Lu J., McKay D. M. (2000). Dextran sulfate sodium-induced colonic histopathology, but not altered epithelial ion transport, is reduced by inhibition of phosphodiesterase activity. Am. J. Pathol. 156, 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieleman L. A., Palmen M. J., Akol H., Bloemena E., Peña A. S., Meuwissen S. G., Van Rees E. P. (1998). Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin. Exp. Immunol. 114, 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esgiar A. N., Naguib R. N., Sharif B. S., Bennett M. K., Murray A. (2002). Fractal analysis in the detection of colonic cancer images. IEEE Trans. Inf. Technol. Biomed. 6, 54–58 [DOI] [PubMed] [Google Scholar]
- Hall L. J., Faivre E., Quinlan A., Shanahan F., Nally K., Melgar S. (2011). Induction and activation of adaptive immune populations during acute and chronic phases of a murine model of experimental colitis. Dig. Dis. Sci. 56, 79–89 [DOI] [PubMed] [Google Scholar]
- Hamilton P. W., Bartels P. H., Thompson D., Anderson N. H., Montironi R., Sloan J. M. (1997). Automated location of dysplastic fields in colorectal histology using image texture analysis. J. Pathol. 182, 68–75 [DOI] [PubMed] [Google Scholar]
- Johansson M. E., Gustafsson J. K., Sjöberg K. E., Petersson J., Holm L., Sjövall H., Hansson G. C. (2010). Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE 5, e12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota N., Wong K., Valdez P. A., Zheng Y., Crellin N. K., Diehl L., Ouyang W. (2011). IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat. Immunol. 12, 941–948 [DOI] [PubMed] [Google Scholar]
- Pastorelli L., Pizarro T. T., Cominelli F., Vecchi M. (2009). Emerging drugs for the treatment of ulcerative colitis. Expert Opin. Emerg. Drugs 14, 505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. M., Mytelka D. S., Dunwiddie C. T., Persinger C. C., Munos B. H., Lindborg S. R., Schacht A. L. (2010). How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203–214 [DOI] [PubMed] [Google Scholar]
- Persohn E., Seewald W., Bauer J., Schreiber J. (2007). Cell proliferation measurement in cecum and colon of rats using scanned images and fully automated image analysis: validation of method. Exp. Toxicol. Pathol. 58, 411–418 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A. K., Blumberg R. S., Xavier R. J., Mizoguchi A. (2008). IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. R., Levenson R. M. (2006). Quantification of immunohistochemistry – issues concerning methods, utility and semiquantitative assessment II. Histopathology 49, 411–424 [DOI] [PubMed] [Google Scholar]
- Walker R. A. (2006). Quantification of immunohistochemistry – issues concerning methods, utility and semiquantitative assessment I. Histopathology 49, 406–410 [DOI] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., Neurath M. F. (2007). Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546 [DOI] [PubMed] [Google Scholar]
- Xavier R. J., Podolsky D. K. (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 [DOI] [PubMed] [Google Scholar]
- Yan Y., Kolachala V., Dalmasso G., Nguyen H., Laroui H., Sitaraman S. V., Merlin D. (2009). Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS ONE 4, e6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Dubé P. E., Washington M. K., Yan F., Polk D. B. (2012). ErbB2 and ErbB3 regulate recovery from dextran sulfate sodium-induced colitis by promoting mouse colon epithelial cell survival. Lab. Invest. 92, 437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Valdez P. A., Danilenko D. M., Hu Y., Sa S. M., Gong Q., Abbas A. R., Modrusan Z., Ghilardi N., de Sauvage F. J., et al. (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.