14-3-3 proteins function in many cellular processes. Here, we show that 14-3-3s regulate the stability of proteins involved in ethylene biosynthesis. 14-3-3 directly interacts with and regulates the turnover of ACC synthase, a key ethylene biosynthesis enzyme, and the ETO1/EOLs E3 ubiquitin ligases, which regulate ACS protein turnover, thus playing a key role in regulating ethylene biosynthesis.
Abstract
14-3-3 proteins are a family of conserved phospho-specific binding proteins involved in diverse physiological processes. Plants have large 14-3-3 gene families, and many binding partners have been identified, though relatively few functions have been defined. Here, we demonstrate that 14-3-3 proteins interact with multiple 1-aminocyclopropane-1-carboxylate synthase (ACS) isoforms in Arabidopsis thaliana. ACS catalyzes the generally rate-limiting step in the biosynthesis of the phytohormone ethylene. This interaction increases the stability of the ACS proteins. 14-3-3s also interact with the ETHYLENE-OVERPRODUCER1 (ETO1)/ETO1-LIKE (EOLs), a group of three functionally redundant proteins that are components of a CULLIN-3 E3 ubiquitin ligase that target a subset of the ACS proteins for rapid degradation by the 26S proteasome. In contrast with ACS, the interaction with 14-3-3 destabilizes the ETO1/EOLs. The level of the ETO1/EOLs in vivo plays a role in mediating ACS protein turnover, with increased levels leading to a decrease in ACS protein levels. These studies demonstrate that regulation of ethylene biosynthesis occurs by a mechanism in which 14-3-3 proteins act through a direct interaction and stabilization of ACS and through decreasing the abundance of the ubiquitin ligases that target a subset of ACS proteins for degradation.
INTRODUCTION
The controlled degradation of proteins by the ubiquitin-26S proteasome system is an important regulatory mechanism in eukaryotic cells. In plants, the gene families encoding the elements of the ubiquitin-26S proteasome system are greatly expanded relative to their animal counterparts (Vierstra, 2009), suggesting a more prominent role for ubiquitin-based protein degradation in plant cells. The ubiquitin-26S proteasome has been linked to diverse functions in plants, including hormone signaling, photomorphogenesis, and defense responses (Yi and Deng, 2005; Stone and Callis, 2007; Vierstra, 2009; Santner and Estelle, 2010).
The function of the phytohormone ethylene is regulated by the ubiquitin-26S proteasome system at the level of both signaling and biosynthesis. In ethylene signaling, the abundance of ETHYLENE-INSENSITIVE3, a positive regulator of the pathway, is regulated by an SCF E3 ligase containing two F-box substrate specificity factors, EIN3 BINDING F-BOX PROTEIN1 (EBF1) and EBF2 (Potuschak et al., 2003; Guo and Ecker, 2003; Gagne et al., 2004; An et al., 2010). In ethylene biosynthesis, a subset of 1-aminocyclopropane-1-carboxylate synthase (ACS) proteins, which catalyze the generally rate-limiting step in ethylene biosynthesis, are specifically targeted by ETHYLENE OVER-PRODUCER1 (ETO1) for ubiquitination and rapid degradation by the 26S proteasome. ETO1 is a Broad complex/Tramtrack/Bric-a-brac (BTB) domain substrate adaptor for CULLIN-3 E3 (CUL3) ubiquitin ligase, which targets a C-terminal sequence specific to type-2 ACS proteins called the TOE (for Target of ETO1) domain (Vogel et al., 1998a; Chae et al., 2003; Wang et al., 2004; Yoshida et al., 2005, 2006; Christians et al., 2009). Arabidopsis thaliana contains two ETO1 paralogs, ETO1-like (EOL1) and EOL2, that act redundantly with ETO1, at least in etiolated seedlings, to target type-2 ACS proteins for rapid degradation (Christians et al., 2009). Substrate adaptors, such as the ETO1/EOLs, regulate the accessibility of substrate proteins to the CUL3 ligase for ubiquitination, and their availability is important for the ubiquitination and, thus, stability of the cognate substrates.
ACS enzymes can be classified into three types based on the presence or absence of putative phosphorylation sites located on their C termini (Chae and Kieber, 2005). Type-1 ACS proteins contain target sites for both mitogen-activated protein kinase and calcium-dependent protein kinase phosphorylation. Type-2 ACS proteins have only a putative calcium-dependent protein kinase target site, and type-3 ACS proteins have a short C-terminal domain with no recognized phosphorylation sites (Chae and Kieber, 2005). The stability of type-1 ACS proteins is dependent on their phosphorylation status; phosphorylation by the pathogen-regulated mitogen-activated protein kinase MPK6 leads to increased accumulation of these ACS proteins and, hence, increased ethylene production (Liu and Zhang, 2004; Joo et al., 2008). While nonphosphorylated type-1 ACS proteins are rapidly degraded by the 26S proteasome (Joo et al., 2008), the corresponding E3 ligase has not been identified. The stability of type-2 ACS proteins, which are targeted for rapid degradation by the ETO1/EOLs, is specifically increased by the phytohormones cytokinin and brassinosteroids (Vogel et al., 1998a; Hansen et al., 2009). The lack of a regulatory C-terminal domain on type-3 ACS proteins suggests that they may be more stable than other ACS proteins (Chae and Kieber, 2005).
14-3-3s are a family of highly conserved regulatory proteins involved in diverse physiological processes by phosphorylation-dependent protein–protein interactions (Dougherty and Morrison, 2004; Darling et al., 2005; Oecking and Jaspert, 2009; Freeman and Morrison, 2011). There are 13 functional 14-3-3 genes in Arabidopsis, and they have been shown to act in multiple physiological processes (Chevalier et al., 2009; Oecking and Jaspert, 2009; Denison et al., 2011), including brassinosteroid signaling (Gampala et al., 2007; Ryu et al., 2007), the regulation of flowering time (Mayfield et al., 2007; Purwestri et al., 2009), the regulation of activity of nitrate reductase (Huber et al., 2002), light signaling (Mayfield et al., 2007), and nutrient acquisition (Xu et al., 2012; Yang et al., 2013). In animals, 14-3-3s have a well-established role in regulating the turnover of a variety of proteins (Dougherty and Morrison, 2004), though there are relatively few reports of such a function in plants (Weiner and Kaiser, 1999; Nakashima et al., 2007; Oh et al., 2010). For example, in response to DNA damage, mammalian 14-3-3σ positively regulates the stability of p53, a short-lived-tumor suppressor protein. 14-3-3σ also promotes self-ubiquitination and degradation of the p53-targeting E3 ligases MURINE DOUBLE MINUTE2 (MDM2) and CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), leading to the stabilization of p53 (Fang et al., 2000; Lee and Lozano, 2006; Yang et al., 2007; Su et al., 2011). Several ACS isoforms, as well as both EOL1 and EOL2, were identified as 14-3-3 interacting proteins by proteomic profiling of purified complexes from Arabidopsis (Chang et al., 2009).
Here, we report that 14-3-3 regulates ACS protein turnover. We show that 14-3-3 protein positively regulates type-2 ACS protein stability by both increasing the turnover of the ETO1/EOL BTB E3 ligases that target type-2 ACS proteins and by an ETO1/EOL-independent mechanism. We demonstrate that the level of the ETO1/EOL proteins influences the level of ethylene biosynthesis by regulating the stability of the type-2 ACS proteins. Together, our results suggest that 14-3-3 regulates ACS protein stability as well as the abundance of the E3 ligases that target type-2 ACS proteins for degradation by the 26S proteasome system.
RESULTS
14-3-3 Interacts with ACS in Vivo
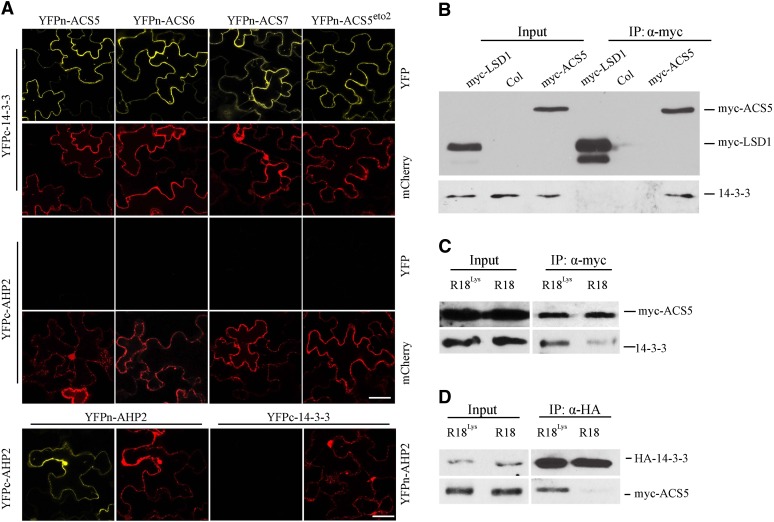
We examined the interaction between ACS and 14-3-3 using a bimolecular fluorescence complementation (BiFC) assay (Figure 1). The 14-3-3ω isoform interacted in this assay with ACS5, ACS6, and ACS7 (Figure 1A), which represent a type-2, type-1, and type-3 ACS, respectively. A strong fluorescent signal was observed in the cytoplasm of tobacco (Nicotiana benthamiana) epidermal cells expressing both YFPn-ACS (YFP, yellow fluorescent protein) and YFPc-14-3-3ω fusion proteins but not with the nonrelevant AHP2 fusions, which serve as negative controls. Disruption of the C-terminal TOE domain of a type-2 ACS (ACS5eto2), which is required for the interaction with ETO1/EOLs, did not affect the interaction with 14-3-3 (Figure 1A). This is consistent with the observation that 14-3-3ω interacted with all three classes of ACS proteins, two of which lack the TOE domain (type-1 and type-3) (Chang et al., 2009). The interaction between 14-3-3 and ACS proteins was confirmed in vivo using a coimmunoprecipitation assay from Arabidopsis transgenic seedlings expressing myc-tagged ACS5 protein (Figure 1B) (Chae et al., 2003). We examined the interaction of other isoforms of 14-3-3 with ACS5 using the BiFC assay in transiently transfected tobacco epidermal cells (see Supplemental Figure 1 online). All four isoforms of 14-3-3 tested (14-3-3ι, 14-3-3ο, 14-3-3κ, and 14-3-3ϕ) interacted with ACS5, indicating that, at least in this assay, there was no specificity in the interaction between 14-3-3s and ACS proteins. Consistent with this, previous results suggested that 14-3-3 isoforms are often at least partially functionally redundant (Roberts and de Bruxelles, 2002; Paul et al., 2012).
Figure 1.
14-3-3 Interacts with All Three Classes of ACC Synthases.
(A) BiFC of 14-3-3ω and various ACS isoforms in transiently transformed N. benthamiana. A plasmid expressing a mitochondria, monomeric cherry (mCherry) fluorescent protein was used as a transformation marker. YFPc-AHP2 and YFPn-AHP2 were used as negative control, which were shown to be expressed by their ability to dimerize and produce a BiFC signal. The YFP signal was observed using confocal microscopy. Bars = 50 µm.
(B) Coimmunoprecipitation of 14-3-3 and ACS5. Protein extracts from light-grown seedlings expressing myc-ACS5 and myc-LSD 1 were immunoprecipitated with an anti-myc antibody and the immunoprecipitated proteins were analyzed by protein gel blotting using an anti-myc or anti-14-3-3 antibody. The myc-LSD1 was used as a negative control.
(C) R18 peptide disrupts the interaction of ACS5 and 14-3-3 proteins in seedlings. Protein extracts from Arabidopsis seedlings expressing myc-ACS5 treated with 20 µg/mL R18 or R18Lys for 3 h were immunoprecipitated with an anti-myc antibody, and the immunoprecipitated proteins were analyzed by protein gel blotting using an anti-myc or anti-14-3-3 antibody.
(D) R18 peptide disrupts the interaction of ACS5 and 14-3-3 proteins in protoplasts. Protein extracts from protoplasts expressing HA-14-3-3ω and myc-ACS5 proteins that were treated for 3 h with 10 µg/mL R18 or R18Lys were immunoprecipitated with an anti-HA antibody, and the immunoprecipitated proteins were analyzed by protein gel blotting using an anti-myc or anti-HA antibody.
The R18 peptide is a strong competitive inhibitor of 14-3-3 client protein interactions (Wang et al., 1999). It has a high affinity for different 14-3-3 isoforms, which enables the peptide to disrupt a wide array of 14-3-3-interactions. In plants, R18 has been shown to alter 14-3-3 function in Arabidopsis leaf disks (Paul et al., 2005). The ability of R18 and a nonfunctional form of the peptide (R18Lys) (Masters and Fu, 2001) to disrupt the interaction between ACS5 and 14-3-3 was examined (Figures 1C and 1D; see Supplemental Figure 2 online). We first examined the effect of R18 on the BiFC interaction between ACS5 and 14-3-3 (see Supplemental Figure 2 online). There were two distinctive classes of fluorescence observed in the BiFC assays of ACS5 and 14-3-3ω in Arabidopsis protoplasts. Approximately 90 percent of the transformed protoplasts (marked with a mitochondria, monomeric Cherry cotransformation reporter) generated strong, punctate fluorescence when transformed with plasmids expressing YFPc-ACS5 and YFPn-14-3-3ω, while the rest showed very faint, weak signals distributed throughout the cytoplasm (see Supplemental Figure 2A online). The percentage of the protoplasts showing strong fluorescence was greatly reduced following incubation with R18 peptide (see Supplemental Figure 2B online). By contrast, the mutated R18Lys peptide had little or no effect in this assay. We next examined the effect of R18 on the in planta interaction of ACS5 and 14-3-3. Treatment of seedlings with R18 peptide, but not with an R18Lys control, resulted in a decrease in the amount of 14-3-3 that coimmunoprecipitated with myc-ACS5 (Figure 1C). Likewise, treatment of protoplasts expressing myc-ACS5 and HA-14-3-3ω with R18 peptide resulted in a decrease in the level of myc-ACS5 that coimmunoprecipitated with HA-14-3-3ω protein (Figure 1D). Together, these results indicate that ACS and 14-3-3 proteins exist as a complex and that this interaction can be effectively disrupted in vivo using the R18 peptide.
14-3-3 Is a Positive Regulator of ACS5 Protein Stability
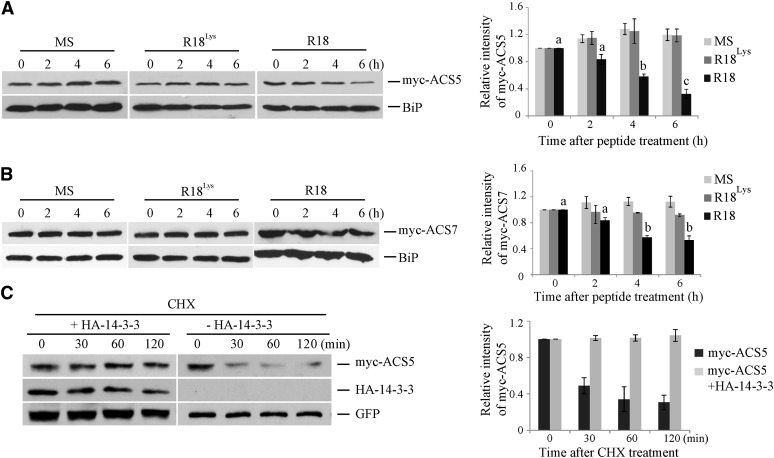
In order to determine whether the interaction with 14-3-3 affects ACS protein stability, the levels of a myc epitope–tagged ACS5 protein were examined following treatment with either R18 or R18Lys peptide. The steady state level of myc-ACS5 protein was reduced approximately threefold in etiolated transgenic seedlings 6 h after treatment with R18 peptide (Figure 2A). By contrast, ACS5 protein levels were comparable in seedlings treated with either R18Lys or a Murashige and Skoog (MS) medium control. The decrease in myc-ACS5 protein levels in R18-treated seedlings was not the result of a decrease in the myc-ACS5 transcript levels (see Supplemental Figure 3 and Supplemental Methods 1 online), and thus likely reflects changes in protein stability. Similar to ACS5, myc-ACS7 (a type-3 ACS) protein levels were reduced in the presence of R18 peptide in etiolated seedlings (Figure 2B). While the decrease in ACS7 levels was less than that observed for ACS5, it was very reproducible and significant (P < 0.01 at 4 and 6 h after R18 treatment) and may reflect the relatively long half-life of ACS7 protein. We next studied whether the decrease in ACS5 protein levels in R18-treated seedlings is posttranscriptionally regulated through ubiquitin/proteasome activity. The proteasome inhibitor MG132 abrogated the decrease in myc-ACS5 in response to R18 treatment in etiolated seedlings (see Supplemental Figure 4 online), suggesting that the destabilization caused by disruption of the 14-3-3/ACS5 interaction is proteasome dependent.
Figure 2.
14-3-3 Positively Regulates ACS Protein Stability.
(A) and (B) R18 peptide results in a decrease in the steady state level of myc-ACS5 (A) or mycACS7 (B) proteins. Seedlings expressing myc-ACS5 or myc-ACS7 were treated with 10 µg/mL R18, R18Lys, or an MS medium control for the indicated times. Total protein extracts from these seedlings were then analyzed by immunoblotting using an anti-myc antibody or an anti-BiP antibody as a loading control. On the right is shown a quantification of the protein levels from three independent biological replicates per treatment using ImageJ software. The myc-ACS5 or myc-ACS7 bands were normalized to the BiP control, and these values were than normalize to the time zero, which was set to 1. Different letters indicate significant differences at P < 0.01 (analysis of variance, Tukey HSD post-hoc test); error bars = se.
(C) The half-life of ACS5 protein is increased by 14-3-3. Mesophyll protoplasts were transformed with a plasmid expressing myc-ACS5 with or without a plasmid expressing 14-3-3ω. At time 0, cycloheximide (CHX) was added (250 µM Cf) and total proteins extracted at the indicated times and then analyzed by immunoblotting using the indicated antibodies. The myc-ACS5 signal was normalized to the GFP signal, and these values expressed relative to the time 0 value, which was set to 1. A quantification of the protein levels from three independent biological replicates is shown on the right. Values are presented as the mean ± se.
A transient expression system in Arabidopsis protoplasts was used to determine if the interaction with 14-3-3 protein increases the half-life of ACS5. Coexpression of HA-14-3-3ω increased the half-life of myc-tagged ACS5 protein in this system (Figure 2C). The level of myc-ACS5 protein decreased approximately threefold after 1 h of inhibition of de novo protein synthesis by cycloheximide treatment. By contrast, when a 14-3-3ω expression plasmid was cotransformed with the ACS5 expression plasmid, there was almost no decrease in myc-ACS5 protein levels even after 2 h of cycloheximide treatment. Together, these results indicate that 14-3-3 positively regulates ACS protein stability via the 26S proteasome–dependent pathway.
14-3-3 Interacts Directly with and Promotes Destabilization of ETO1/EOLs
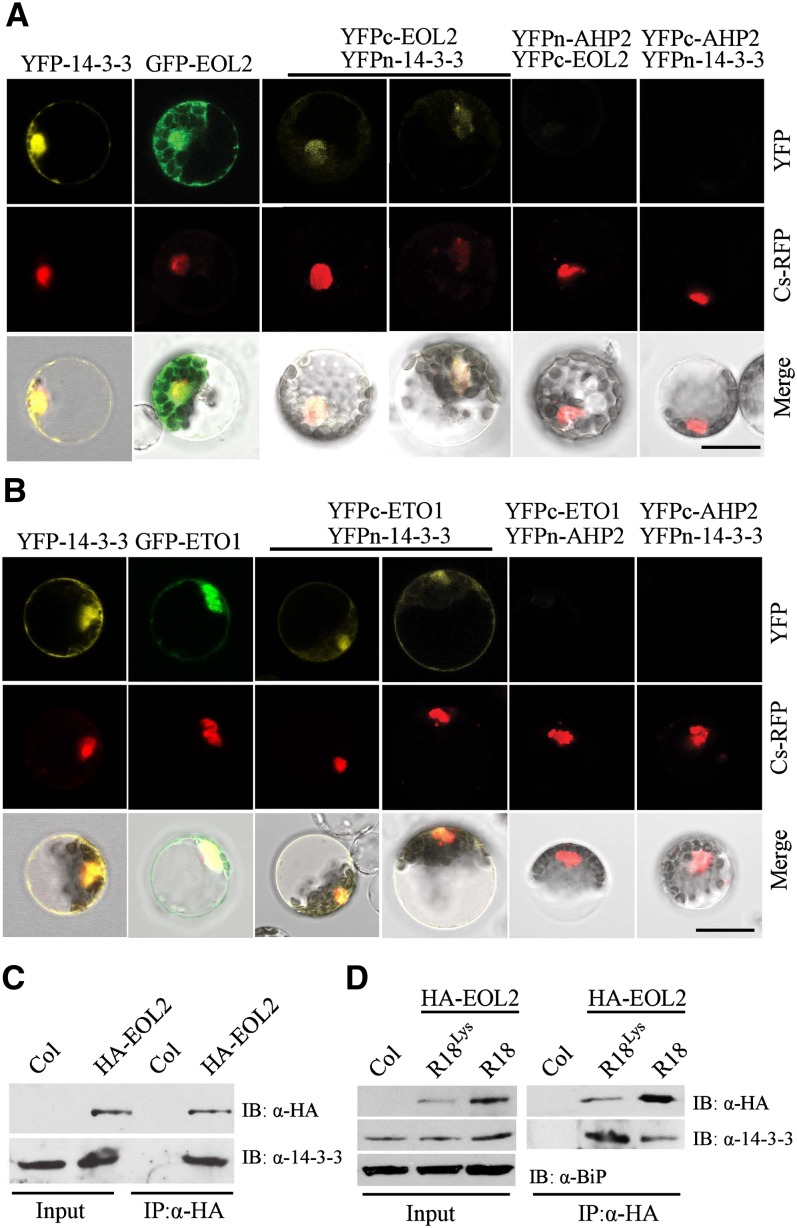
In addition to the ACS protein, both EOL1 and EOL2 were identified in 14-3-3ω complexes using a tandem affinity purification approach (Chang et al., 2009). As previously demonstrated in etiolated seedlings (Christians et al., 2009), ETO1 acts partially redundantly with EOL1 and EOL2 to negatively regulate ethylene biosynthesis in light-grown seedlings (see Supplemental Figure 5 online). We hypothesized that similar to the function of 14-3-3σ in its interactions with both p53 and its cognate E3 ligases in mammalian cells (Fang et al., 2000; Lee and Lozano, 2006; Yang et al., 2007; Su et al., 2011), 14-3-3ω may downregulate the stability of the ETO1/EOL E3 ligase components in Arabidopsis. To test this, we first confirmed the previously reported interaction of 14-3-3 and EOLs (Chang et al., 2009) using a BiFC approach (Figures 3A and 3B). Consistent with the prior report, we observed a positive interaction between 14-3-3ω and both ETO1 and EOL2. Surprisingly, the BiFC fluorescence observed in these interactions was localized not only in the cytoplasm, but also in the nucleus, colocalizing with a nuclear red fluorescent protein marker (Cs-RFP) (Asai et al., 2002). We thus examined the localization of EOL2 and ETO1 green fluorescent protein (GFP) fusions and 14-3-3ω YFP fusion protein. All three proteins were localized in both the nucleus and cytoplasm (Figures 3A and 3B), consistent with the BiFC results. Similar to the 14-3-3/ACS5 interaction, the ETO1/EOLs interacted with multiple different isoforms of 14-3-3 (14-3-3ο, 14-3-3κ, 14-3-3ι, and 14-3-3ϕ) (see Supplemental Figure 6 online), suggesting that, at least with the BiFC assay, there is no specificity in the interactions of 14-3-3s with either ACS or ETO1/EOLs. Interaction of EOL2 and 14-3-3 was confirmed in vivo with a coimmunoprecipitation assay using Arabidopsis HA epitope–tagged EOL2 transgenic seedlings (EOL2-OX) (Figure 3C), and this interaction was disrupted by R18 peptide (Figure 3D). Together, these results confirmed that EOL2 exists as a complex with 14-3-3 protein and its interaction is disrupted by R18 peptide.
Figure 3.
14-3-3 Interacts with ETO1/EOLs in Vivo.
(A) and (B) BiFC analysis of 14-3-3ω interacting with ETO1 (A) or EOL2 (B). The left-most panel shows a full-length YFP fused to 14-3-3ω. The second panel shows a full-length GFP fused to EOL2 (A) or ETO1 (B). The remaining panels show the result of cotransformation with the indicated BiFC plasmids. A Cs-RFP plasmid was used as a marker for the nucleus and transformation control. The top row shows the YFP or GFP signal, the second row the RFP signal, and the lowest row a merge of the GFP/YFP, RFP, and differential interference contrast images. Bars = 20 µm.
(C) Coimmunoprecipitation of EOL2 and 14-3-3 proteins. Protein extracts from Arabidopsis seedlings expressing HA-EOL2 were immunoprecipitated with an anti-HA antibody, and the immunoprecipitated proteins were analyzed by immunoblotting using an anti-HA or an anti-14-3-3 antibody. Col, Columbia.
(D) R18 peptide disrupts the interaction between EOL2 and 14-3-3 proteins. Arabidopsis seedlings expressing HA-EOL2 were treated with 20 µg/mL R18 or R18Lys peptide, total protein extracts were immunoprecipitated (IP) with an anti-HA antibody, and the immunoprecipitated proteins were analyzed by immunoblotting (IB) using an anti-HA or anti-14-3-3 antibody.
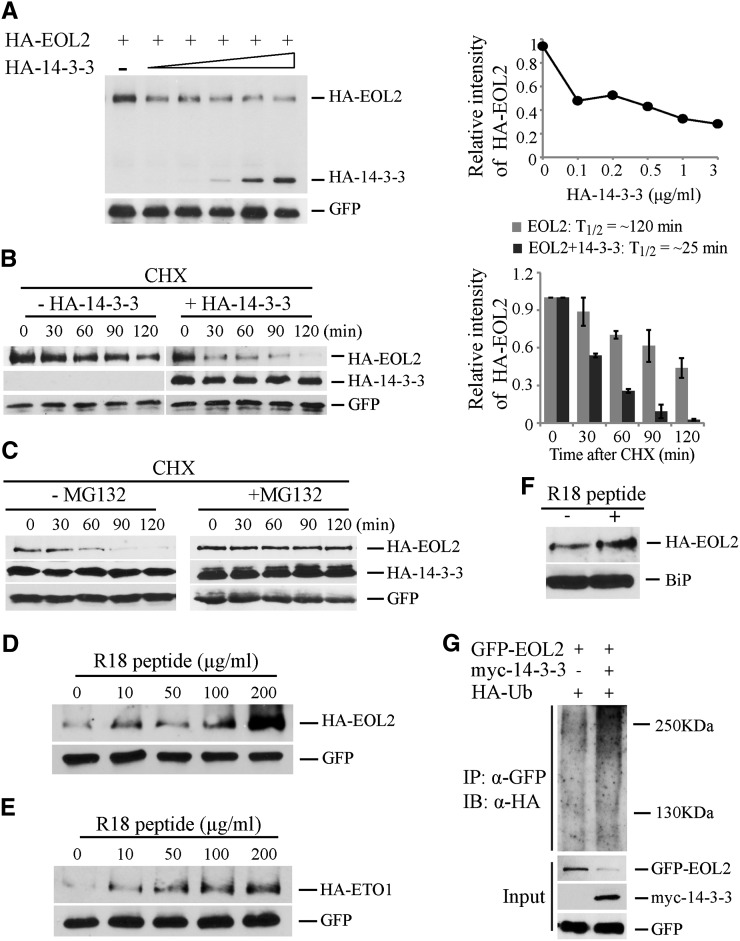
The effects of 14-3-3 on the steady state level of EOL2 protein was examined using transiently transformed Arabidopsis protoplasts. A fixed quantity of plasmid designed to express HA-EOL2 and increasing amounts of plasmid designed to express HA-14-3-3ω were cotransformed along with a compensating amount of empty vector plasmid (Figure 4A). The steady state level of HA-EOL2 protein decreased as the expression of HA-14-3-3ω increased (Figure 4A). The half-life of HA-EOL2 was greatly shortened in the presence of HA-14-3-3ω (∼25 min) compared with that of HA-EOL2 without added 14-3-3 (∼120 min) (Figure 4B). MG132 blocked the effect of 14-3-3 expression on HA-EOL2 turnover, suggesting that 14-3-3–mediated destabilization of EOL2 is dependent on ubiquitin/proteasome activity (Figure 4C). To test this, we examined the effect of 14-3-3 expression on the ubiquitination of EOL2 using an HA-tagged ubiquitin in mesophyll protoplasts (Figure 4G). As expected, coexpression with myc-14-3-3 reduced the steady state level of GFP-EOL2. Furthermore, there was substantially more of the polyubiquitinated form GFP-EOL2 when coexpressed with myc-14-3-3, suggesting that 14-3-3 promotes the ubiquitination and subsequent destabilization of EOL2 and therefore increases ACS stability. Furthermore, disruption of the ETO1/EOL2-14-3-3 interaction by application of R18 peptide resulted in increased stability of HA-EOL2 and HA-ETO1 proteins in a dosage-dependent manner (Figures 4D and 4E). In vivo experiment using Arabidopsis seedlings expressing HA-EOL2 protein also confirmed that R18 treatment enhances the steady state levels of EOL2 protein (Figure 4F). Because R18 treatment increased the steady state levels of HA-EOL2 protein and inhibited the interaction of EOL2 and 14-3-3, we conclude that 14-3-3 acts a negative regulator of EOL2 protein stability.
Figure 4.
14-3-3 Destabilizes EOL2 Protein Stability.
(A) 14-3-3 destabilizes EOL2 protein. Protein extracts from protoplasts transformed with plasmid expressing HA-EOL2, and increasing amounts of plasmid expressing HA-14-3-3ω were analyzed by immunoblotting with an anti-HA or anti-GFP antibody, which was used as a transformation control. The right shows a quantification of the immunoblots. The HA-EOL2 signal was normalized to the GFP signal, and these values expressed relative to the no HA-14-3-3ω control, which was set to 1.
(B) 14-3-3 decreases the half-life of EOL2 protein. Protoplasts were transformed with a plasmid expressing HA-EOL2 with or without a plasmid expressing 14-3-3ω. At time 0, cycloheximide was added (250 µM Cf), and total proteins were extracted at the indicated times and then analyzed by immunoblotting using the indicated antibodies. Right: The HA-EOL2 signals were normalized to the matching GFP signals and these values expressed relative to the time 0 value, which was set to 1. A quantification of the protein levels from three independent biological replicates using ImageJ software is shown on right. Values are presented as the mean ± se.
(C) 14-3-3–mediated destabilization of EOL2 protein is ubiquitin/proteasome dependent. Protoplasts expressing HA-EOL2 and HA-14-3-3ω were incubated with or without MG132 for the indicated times, and total proteins were extracted at the indicated times and then analyzed by immunoblotting using the indicated antibodies.
(D) and (E) R18 peptide promotes accumulation of EOL2 (D) and ETO1 (E) proteins. The indicated concentrations of R18 peptide were added to the protoplasts transformed with plasmids expressing either HA-EOL2 or HA-ETO1. The blots were probed with anti-HA and anti-GFP antibodies.
(F) R18 peptide promotes accumulation of EOL2 protein in planta. Arabidopsis seedlings expressing HA-EOL2 were treated with 20 µg/mL R18 peptide or a mock control. Total protein was extracted and analyzed by immunoblotting using an anti-HA antibody or anti-BiP antibody as a loading control.
(G) 14-3-3 promotes the ubiquitination of EOL2 protein. Protoplasts expressing GFP-EOL2 and HA-ubiquitin in the presence or absence of a myc-14-3-3ω expression plasmids as indicated were incubated with 50 µM MG132 for 6 h. The protein extracts were immunoprecipitated (IP) with an anti-GFP antibody and then analyzed by immunoblotting (IB) using an anti-HA or anti-GFP antibody. The GFP signal is used as a transformation control.
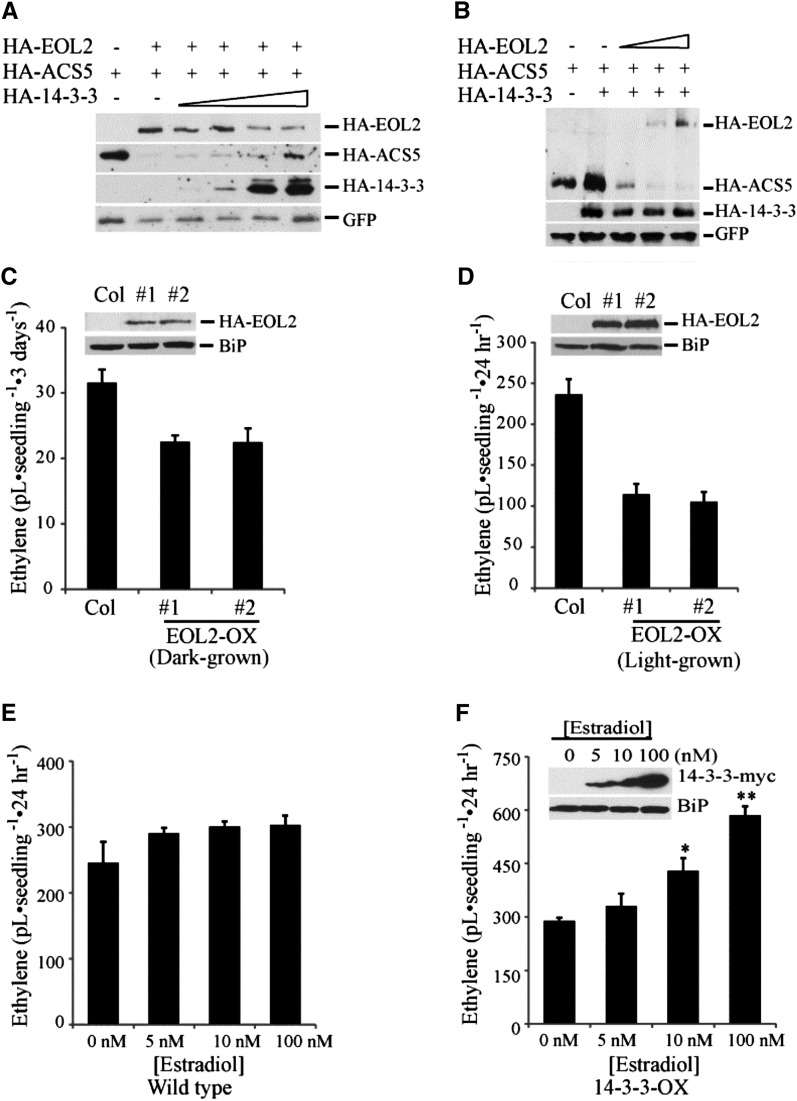
We next examined the interaction of ACS5, EOL2, and 14-3-3 proteins when plasmids expressing all proteins were introduced into Arabidopsis protoplasts simultaneously (Figure 5). Fixed amounts of HA-EOL2 and HA-ACS5 plasmid were cotransformed with increasing amounts of HA-14-3-3ω plasmid. The steady state level of HA-EOL2 and HA-ACS5 proteins displayed opposite responses to increasing HA-14-3-3ω expression (Figure 5A; see Supplemental Figure 7 online). As the level of 14-3-3 increased, the level of EOL2 declined and that of ACS5 increased, consistent with the results when these proteins were examined individually. Together, these results suggest that 14-3-3 has reciprocal effects on the stability of ACS and ETO1/EOL proteins.
Figure 5.
ACS Protein Stability Is Regulated by the Level of ETO1/EOL and 14-3-3 Proteins.
(A) The steady state level of HA-EOL2 and HA-ACS5 proteins with increasing HA-14-3-3ω expression. Protoplasts were transformed with plasmids expressing HA-EOL2, HA-ACS5, and increasing levels of HA-14-3-3ω. An empty vector was added as appropriate to ensure equal amounts of plasmid DNA was added to each reaction. Total proteins were analyzed by immunoblotting using an anti-HA antibody or an anti-GFP antibody. The GFP signal is used as a transformation control.
(B) Increased expression of EOL2 protein promotes degradation of ACS5 protein. Protoplasts were transformed with plasmids expressing HA-14-3-3ω, HA-ACS5, and increasing levels of HA-EOL2. An empty vector was added as appropriate to ensure equal amounts of plasmid DNA was added to each reaction. Total proteins were analyzed by immunoblotting using an anti-HA antibody or an anti-GFP antibody.
(C) and (D) Ethylene production from two independent transgenic lines overexpressing EOL2 (EOL2-OX) as etiolated (C) or light-grown (D) seedlings. The inset shows the expression of the HA-EOL2 proteins in the lines. mean ± se; n = 3. Col, Columbia.
(E) and (F) Ethylene production from the wild type (E) or a transgenic line expressing 14-3-3ω-myc from an estradiol-inducible promoter (F). Seedlings were grown in the light in the presence of the indicated level of estradiol for 12 d and then capped for 24 h and the level of ethylene accumulated measured. The inset shows the expression of 14-3-3ω-myc in response to different concentration of estradiol. Significance was determined using analysis of variance and Tukey’s HSD post-hoc test. *P < 0.05 and **P < 0.01. Error bars indicate se; n = 3.
Levels of EOL2 and 14-3-3 Proteins Affect ACS Stability
As our results indicated that 14-3-3s regulated the stability of the ETO1/EOL proteins, we examined if altered levels of the ETO1/EOLs affected ACS protein levels and, hence, ethylene biosynthesis in planta. Consistent with this, there is a quantitative effect of disruption of the six copies of the ETO1/EOL1/EOL2 genes on ethylene production from etiolated (Christians et al., 2009) and light-grown (see Supplemental Figure 5 online) Arabidopsis seedlings. In order to test if elevated ETO1/EOLs can have the opposite effect from the loss-of-function mutations, we used Arabidopsis protoplasts and transgenic plants overexpressing EOL2. In Arabidopsis protoplasts, increased EOL2 protein led to a decrease in ACS5 protein levels (Figure 5B), suggesting that the expression level of ETO1/EOL proteins could act as a regulatory input affecting ACS protein stability. Consistent with this, transgenic seedlings overexpressing HA-EOL2 protein (EOL2-OX) exhibited reduced ethylene production compared with wild-type seedlings in both dark- and light-grown seedlings (Figures 5C and 5D). This is consistent with results in tomato (Solanum lycopersicum), in which overexpression of ETO1 suppressed auxin-induced ethylene biosynthesis (Yoshida et al., 2005), and in Arabidopsis, in which overexpression of ETO1 reduced cytokinin-induced ethylene biosynthesis and suppressed the etiolated seedling phenotype of an ACS5 overexpressing transgenic line (Wang et al., 2004). Consistent with 14-3-3 acting to stabilize ACS proteins, induced overexpression of 14-3-3ω from an estradiol-inducible promoter (14-3-3-OX) enhanced ethylene production in light-grown Arabidopsis seedlings (Figures 5E and 5F). Together, these results support the hypothesis that the levels of both ETO1/EOL2 and 14-3-3 regulate the stability of ACS proteins.
14-3-3 Increases Type-2 ACS Stability Partially Independently of the ETO1/EOLs
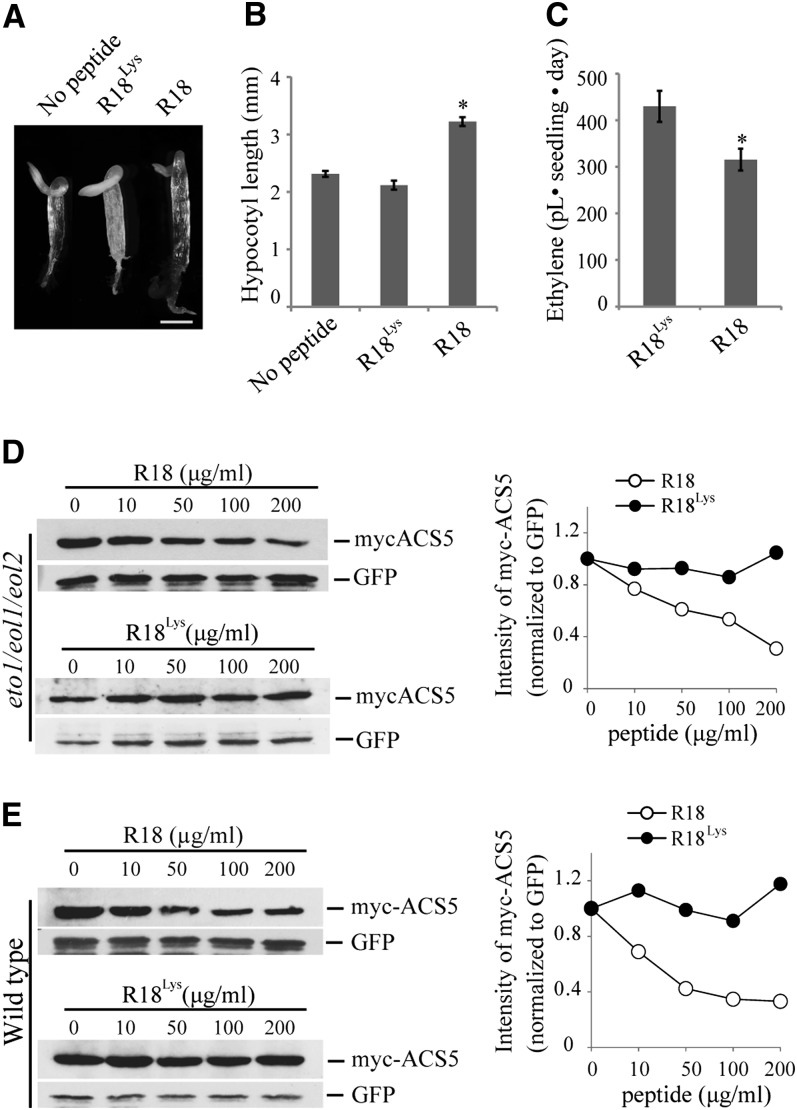
We examined whether the effect of 14-3-3 on ACS can be attributed solely to its alteration of ETO1/EOL levels or if there is also an ETO1/EOL-independent effect of 14-3-3 on ACS stability. To address this, we examined the effect of disrupting 14-3-3 interactions via R18 treatment on ethylene function in the eto1-13 eol1-1 eol2-2 triple mutant, in which all three ETO1/EOL1/EOL2 genes harbored T-DNA insertions (Gingerich et al., 2005). The three T-DNA insertions in the eto1-13, eol1-1, and eol2-2 mutants all occur within exons and eliminate full-length transcript (Gingerich et al., 2005) and are thus likely null alleles. As shown previously, eto1/eol1/eol2 etiolated seedlings display a strong triple response (Figure 6A) (Christians et al., 2009). The R18 peptide, but not the inactive R18Lys peptide, caused a decrease in the triple response in this triple mutant, as evidenced by the increased hypocotyl length (P < 0.001; Figures 6A and 6B). R18 did not affect the hypocotyl length of wild-type seedlings grown in the presence of ACC (see Supplemental Figure 8 online), suggesting the effects of R18 in eto1/eol1/eol2 etiolated seedlings were the result of an effect on ACS function. Consistent with this, the eto1/eol1/eol2 mutant produced less ethylene in the presence of R18 peptide compared with seedlings grown in the presence of the R18Lys peptide (P < 0.02; Figure 6C). Finally, we examined the steady state level of myc-ACS5 protein in transiently transformed eto1/eol1/eol2 mutant protoplasts treated with increasing concentrations of R18 or R18Lys peptide. R18, but not R18Lys, treatment caused a dose-dependent reduction of ACS5 protein levels in eto1/eol1/eol2 (Figure 6D), similar to the effect in wild-type protoplasts (Figure 6E), though somewhat reduced in magnitude. Together, these results suggest that 14-3-3 affects ACS5 protein stability in a manner that is partially independent of its effect on the ETO1/EOLs.
Figure 6.
14-3-3 Stabilizes ACS Proteins via Downregulation of an ETO1/EOL E3 Ligase-Independent Pathway.
(A) Triple response phenotype of etiolated eto1/eol1/eol2 seedlings grown in the presence of no peptide or 200 µg/mL R18 or R18Lys peptide. Bar = 1 mm.
(B) Hypocotyl lengths of 3-d-old etiolated eto1/eol1/eol2 seedlings grown in the presence of no peptide or 200 µg/mL R18 or R18Lys peptide. n ≥ 18; mean ± se; *P < 0.001 (Student’s t test).
(C) Ethylene production from etiolated eto1/eol1/eol2 seedlings grown in the presence or absence of R18 or R18Lys peptide. Mean ± se; n = 3; *P < 0.02 (Student’s t test).
(D) and (E) R18 peptide induces destabilization of myc-ACS5 proteins in eto1/eol1/eol2 background. eto1/eol1/eol2 (D) or wild-type (E) protoplasts were cotransfected with a plasmid expressing myc-ACS5 and incubated with the indicated concentrations of R18 or R18Lys peptide. Total proteins were analyzed by immunoblotting using an anti-myc or anti-GFP antibody. Right: The myc-ACS5 signals were normalized to the matching GFP signals, and these values expressed relative to the 0 concentration of R18, which was set to 1.
DISCUSSION
In mammalian cells, 14-3-3σ interacts with p53 and its cognate E3 ligases COP1 and MDM2; 14-3-3σ stabilizes p53 by downregulation of MDM2 and COP1 protein stability (Yang et al., 2007; Su et al., 2011). Here, we demonstrate that a similar mechanism acts in Arabidopsis to regulate the stability of ACS proteins, which catalyzed a key step in ethylene biosynthesis. 14-3-3s increase the stability of ACS proteins, both by destabilizing the cognate E3 ligase components ETO1/EOLs and through an ETO1/EOL-independent mechanism. Furthermore, in animals, in addition to the MDM2 and COP1, there are several cases in which the stability of E3 ligase components is regulated, generally by ubiquitination and subsequent degradation by the 26S proteasome. For example, the F-box protein S-phase kinase-associated protein 2 (Skp2) is polyubiquitinated and hence targeted for degradation by anaphase-promoting complex in the G1 phase of the cell cycle (Wei et al., 2004). Likewise, the F-box protein Atrogin-1 is polyubiquitinated and degraded in a p38 mitogen-activated protein kinase–dependent manner (Li et al., 2011). In Arabidopsis, 14-3-3s were recently shown to interact with the F-box protein FBS1 and hypothesized to enhance their autoubiquitination activity (Sepúlveda-García and Rocha-Sosa, 2012). The gene families encoding E3 ligase components have greatly expanded in plants, with a likely increase in associated functions. The regulation of the ETO1/EOL protein stability represents an added level of complexity in the plant ubiquitin–26S proteasome system.
Previous studies have revealed specific posttranscriptional regulatory inputs into ACS proteins: Type-1 ACS proteins are phosphorylated by MPK6 in response to stress and pathogen interaction (Liu and Zhang, 2004; Joo et al., 2008); cytokinin and brassinosteroid regulate the stability of type-2 ACS proteins (Vogel et al., 1998b; Hansen et al., 2009). In contrast with these regulatory inputs, 14-3-3 likely acts on all three classes of ACS proteins as they directly interact with all the ACS proteins tested and they alter the stability of both ACS5 (type-2) and ACS7 (type-3). Thus, the regulation by 14-3-3 may represent a mechanism to increase the stability of all ACS proteins.
The only demonstrated targets for the ETO1/EOLs are ACS proteins, which are in the cytoplasm. Furthermore, there is no canonical nuclear localization signal in either ETO1 or EOL2. Thus, it was somewhat surprising that GFP-EOL2 and GFP-ETO1 were localized to both the nucleus and the cytoplasm and that the interaction with 14-3-3 also occurred at both of these cellular locations, suggesting that there may be other substrates of these E3 ligases in addition to the type-2 ACS proteins. Alternatively, the localization of ETO1/EOL2 proteins may play a role in regulating their ubiquitination, similar to the regulation of COP1, whose localization is affected by interaction with 14-3-3σ (Su et al., 2010). It is possible that overexpression of the ETO1/EOL proteins by the cauliflower mosaic virus 35S promoter could lead to their mislocalization, though the large size of the GFP-ETO1/EOL2 fusion protein precludes free diffusion into nucleus.
Our results from the half-life experiment using the proteasome inhibitor MG132 (Figures 4B and 4C) and ubiquitination assay (Figure 4G) suggest that 14-3-3 promotes the degradation of ETO1/EOLs, likely via the 26S proteasome pathway, and therefore positively regulates the ACS protein stability. Several studies have suggested that F-box proteins, such as mammalian SKP2 and β-TrCP1/2 and yeast Cdc4p, Grr1p, and Met30p, undergo ubiquitination and degradation in the absence of substrate via an autocatalytic process regulated by substrate availability and COP9 signalosome-dependent deneddylation of the CUL1 SCF complex (Galan and Peter, 1999; Wee et al., 2005; Ho et al., 2008). It is possible that the 14-3-3 regulates the availability of the ACS substrates for the ETO1/EOL E3 ligase, thus regulating the turnover of both sets of proteins. Alternatively, the interaction with dimeric 14-3-3 proteins may cause the ETO1/EOLs to dimerize, thereby promoting self-ubiquitination and subsequent degradation. Finally, the interaction with 14-3-3s could enhance the interaction with distinct E3 ligases, leading to the ubiquitination and subsequent degradation of the ETO1/EOLs.
While 14-3-3 regulates the stability of the ETO1/EOLs, several lines of evidence suggest that they also regulate ACS stability independently of these proteins. First, 14-3-3 interacted with the ACS5eto2 protein, a type-2 ACS that lacks the C-terminal TOE domain, which is required for the ETO1/EOL binding. Secondly, 14-3-3 interacts with and stabilizes ACS7, a type-3 ACS, whose stability is not regulated by ETO1/EOLs. Finally, 14-3-3 increases ACS stability in an eto1 eol1 eol2 triple mutant. Thus, there is at least one other system acting to degrade type 2 ACS proteins in addition to the ETO1/EOLs and the 14-3-3 proteins antagonize this second degradation pathway. This is consistent with the observation that cytokinin and brassinosteroid increase type-2 ACS function partly through a TOE-independent mechanism (Hansen et al., 2009). Furthermore, the RING-type E3 ligase XBAT32 has recently been identified that regulates ACS7 (type-3) and ACS4 (type-2) protein stability (Lyzenga et al., 2012). As type-3 ACS proteins lack a C-terminal TOE sequence, XBAT32 could represent this TOE-independent mechanism. One possible mechanism for the ETO1/EOL-independent stabilization of ACS protein by 14-3-3 is that binding of 14-3-3 to ACS blocks access by other E3 ligases, such as XBAT32.
14-3-3s interact with both ACS and ETO1/EOL proteins, and prior studies indicate that ACS interacts directly with the ETO1/EOLs (Christians et al., 2009), suggesting that these three proteins may exist as a complex in the cell. However, we cannot rule out the possibility that ACS, ETO1/EOLs, and 14-3-3 proteins are present in independent complexes, perhaps populated with distinct 14-3-3 isoforms.
14-3-3 likely binds to ACS through noncanonical binding sites as neither the mode I [Arg-Ser-X-(pSer)-X-Pro] nor mode II [Arg-X-X-X-(pSer)-X-Pro] (Tzivion and Avruch, 2002) canonical binding sites are found in the ACS proteins. Furthermore, the interactions between 14-3-3s and their client proteins are often dependent on the phosphorylation status of the target proteins, suggesting that the target ACS and ETO1/EOL proteins may be phosphorylated. In the case of the ACS proteins, this would likely occur within the catalytic domain, as our data are most consistent with that being the 14-3-3 interaction domain.
Ethylene biosynthesis is regulated by multiple endogenous and exogenous factors, including light, fruit ripening, other phytohormones, and pathogen and stress responses (Yang and Hoffman, 1984; Mattoo and Suttle, 1991; Abeles et al., 1992; Argueso et al., 2007), and some of these inputs may act in part through 14-3-3s and/or regulation of ETO1/EOL levels. While high redundancy among 14-3-3 isoforms can present a major obstacle to study the function of individual isoforms, a defined subset display defects in specific processes, such as the regulation of stomatal opening, flowering time, and phytochrome signaling in Arabidopsis (Mayfield et al., 2007, 2012; Folta et al., 2008; Purwestri et al., 2009; Tseng et al., 2012). The identification of the specific 14-3-3 isoforms that play roles in ethylene biosynthesis is an important question for the future. Furthermore, defining the 14-3-3 interacting domain and phosphorylation site on the ACS and ETO1/EOLs will bring detailed mechanistic insight into the 14-3-3–mediated ethylene biosynthesis.
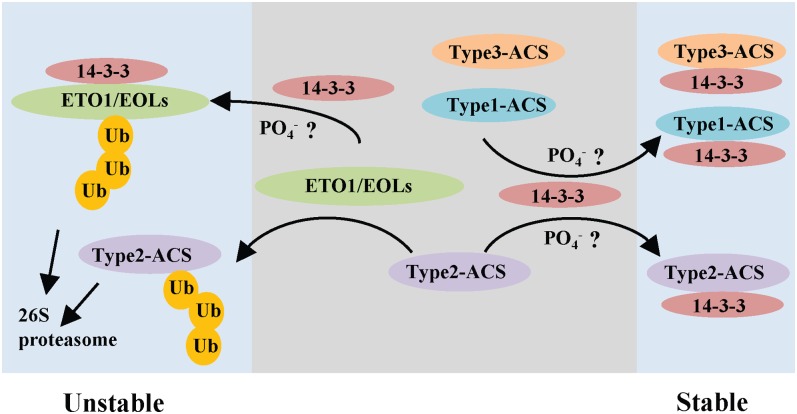
A tentative model for regulation of ACS protein stability by 14-3-3 is shown in Figure 7. The stability of ACS proteins is negatively regulated by their corresponding E3 ligases, keeping ethylene levels low. Upon exposure to various inducers of ethylene biosynthesis, we postulate that 14-3-3 interacts with the ETO1/EOLs and likely promotes their ubiquitination and degradation, leading to increased stability of type-2 ACS proteins. Phosphorylation is likely involved in the interactions between 14-3-3 and its substrates, though this has yet to be demonstrated in the ACS/ETO1/EOL interactions. In addition to the effect on ETO1/EOLs, 14-3-3 acts positively on all three classes of ACS proteins independently of the ETO1/EOLs, again likely through a phosphorylation-dependent pathway.
Figure 7.
Model for the Regulation of ACS Stability.
The ETO1/EOL E3 ligases target the type-2 ACS proteins for degradation by the 26S proteasome, and 14-3-3 increases degradation of the ETO1/EOLs via promotion of the ubiquitination of ETO1/EOLs, thus stabilizing these ACS proteins. 14-3-3 also directly interacts with all three classes of ACS proteins, likely through a phosphorylation-dependent mechanism, to increase their stability. See text for additional details.
[See online article for color version of this figure.]
14-3-3 proteins have been implicated in the regulation of other plant signaling systems, including other phytohormones (abscisic acid, gibberellin, and brassinosteroids), light, plant defense, and biotic stress (Denison et al., 2011). The regulation of protein stability by 14-3-3 may represent a widespread mechanism in regulating signaling/biosynthetic components in plants. Furthermore, the regulation of the stability of E3 ligase components in plants may play an important role in regulating the function of these abundant elements.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds (Columbia ecotype) were surface sterilized and plated on 1× MS/1% Suc/0.8% agar medium. The plates were stratified for 3 d at 4°C in the dark and then incubated in light for 3 to 5 h at 22°C. Seedlings were incubated at 22°C in the dark or constant light as indicated. Nicotiana benthamiana plants were grown in soil under 16 h light/8 h dark at 20°C under white fluorescent light.
Arabidopsis transgenic lines used were dexamethasone-inducible myc-tagged ACS5 (Chae et al., 2003) and ACS7 and estradiol-inducible myc-tagged 14-3-3 and 35S promoter–driven HA-tagged EOL2. p35S:HA-EOL2 and p35S:14-3-3-myc constructs were introduced into wild-type Columbia to generate EOL2-OX and 14-3-3-OX. Transgenic T1 seedlings were selected on MS agar plates supplemented either 30 µg/mL hygromycin or 10 µg/mL Basta. Transgenic expression was confirmed in hygromycin-resistant or Basta-resistant T2 seedlings by immunoblotting using an anti-myc antibody or anti-HA antibody (Roche). Homozygous T3 lines of 14-3-3-OX and T2 lines of EOL2-OX were used in this study.
Plasmid Constructs
The open reading frames of 14-3-3ω, 14-3-3ι, 14-3-3ο, 14-3-3φ, and 14-3-3κ were amplified from cDNA (see Supplemental Tables 1 and 2 online) and cloned into pENTR/D-TOPO vector (Invitrogen) to generate entry clones. Along with theses entry clones, entry clones of ACS5, ACS5eto2, ACS6, ACS7, AHP2, ETO1, and EOL2 clones were sequence verified and then moved into pCL112 and pCL113 (BiFC) or pEarleyGate binary (protein expression) Gateway-compatible destination vectors (see Supplemental Table 3 online) via LR reactions (Invitrogen). pMDC7-GW-myc was constructed by inserting a 6X myc cassette into the pMDC7 cassette B vector (Curtis and Grossniklaus, 2003). The pTA7002-DEX-GW vector was constructed by inserting a gateway cassette into pTA7002 (Aoyama and Chua, 1997).
BiFC Using N. benthamiana
Agrobacterium tumefaciens–mediated infiltration was performed as previously described (Yang et al., 2000). Agrobacterium strain GV3102 was cultured in Luria-Bertani broth supplemented with rifampicin and gentamycin. To enhance transgene expression, we coinfiltrated Agrobacterium carrying the p19 suppressor of gene silencing (Voinnet et al., 2003). For infiltration, 50 mL of Agrobacterium culture was centrifuged, washed, and suspended in a solution containing 10 mM MES, 10 mM MgCl2, and 200 μM acetosyringone. Three-week-old N. benthamiana plants were infiltrated and incubated for 3 d at room temperature and subsequently analyzed at day 3 after infiltration.
R18 and R18Lys Peptide Treatment in Arabidopsis Seedlings
For R18-mediated changes in steady state levels of ACS5 and ACS7 protein experiments, either myc-tagged ACS5 or myc-tagged ACS7 transgenic seedlings were grown on MS plates with dexamethasone in the dark for 3 d. The seedlings were harvested on day 3 and subsequently washed with liquid MS medium for 60 min to remove residual dexamethasone, following incubation with R18 peptide (AnaSpec) (10 µg/mL) or R18Lys (University of North Carolina High-Throughput Peptide Synthesis Facility) (10 µg/mL) for different time periods. The seedlings were then harvested at different time points and the total proteins were extracted with 2× SDS sample buffer and subjected to immunoblotting. For coimmunoprecipitation from R18 or R18Lys-treated Arabidopsis seedlings, Arabidopsis seedlings were grown for 10 d in the light. The seedlings were then carefully removed from the plate and subsequently washed with MS medium for 60 min, followed by further incubation with R18 and R18Lys control peptides for indicted time. The seedlings were then harvested and total proteins were extracted with 2× SDS sample buffer and subjected to immunoblotting. Anti-myc, anti-HA, and anti-GFP antibodies (Roche), anti-14-3-3 antibody (Santa-Cruz), and anti-BiP antibody (Enzo-Life Science) were used for immunoblotting assay.
Arabidopsis Protoplast Isolation and Polyethylene Glycol–Mediated Transformation
Ten-day-old light-grown Arabidopsis seedlings were transferred to soil and grown under a 12-h-light/12-h-dark regime at 22°C. Protoplast were isolated and transfected as previously described (Yoo et al., 2007). For transfection, 5 × 105 of protoplasts were incubated with appropriate amounts of plasmid DNA in 20% polyethylene glycol (Fluka) for 5 min. Transfected protoplasts were then washed twice with W5 solution (Yoo et al., 2007) and incubated for 12 to 16 h in the dark for BiFC assay, protein expression, or determination of turnover rates. For protein turnover assay, protoplasts were transfected with appropriate plasmids using polyethylene glycol–mediated method and incubated for 16 h, followed by further incubation with 250 µM cycloheximide (Sigma-Aldrich). Protoplasts were then harvested at different time points for immunoblotting analysis. For the turnover assay including MG132 (Calbiochem), after 16 h of incubation, the protoplasts were preincubated with 50 µM MG132 before adding cycloheximide.
Confocal Microscopy Analysis
Images for intact tobacco and Arabidopsis protoplasts containing GFP, BiFC-YFP, mitochondria monomeric Cherry, and Cs-RFP constructs were captured and analyzed using confocal scanning laser microscopy with a Zeiss LSM 510 META scanning confocal microscope. The following excitation lines and emission ranges were used: GFP (excited using a 488-nm line, detected between 505 and 530 nm); YFP (excited using a 514-nm line, detected between 530 and 560 nm); mitochondria monomeric Cherry and Cs-RFP (excited using a 560-nm line, detected between 566 and 612 nm).
Coimmunoprecipitation Assay
Transgenic seedlings expressing myc-ACS5 protein and myc-LSD1 (Coll et al., 2010) were grown on MS medium containing 1× MS salts (Research Products International), 0.05% MES buffer, 1% Suc, and 10 nM dexamethasone (Sigma-Aldrich), pH 5.7. Total protein extracts were isolated and suspended in coimmunoprecipitation buffer (100 mM sodium phosphate, pH 7.0, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM NaVO3, 5 mM NaF, 1× protease inhibitor cocktail [Roche], and 1 mM PMSF). Magnetic anti-myc-beads (Miltenyi Biotech) were added to total protein extracts suspended in coimmunoprecipitation buffer and further incubated for 60 min on ice with gentle shaking. The total protein suspension containing the anti-myc beads was applied to a magnetic column, washed with three times with wash buffer (100 mM sodium phosphate, pH 7.0, 250 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM NaVO3, 5 mM NaF, 1× protease inhibitor cocktail, and 1 mM PMSF), and eluted with boiled 2× SDS buffer followed by immunoblotting.
Measurements of Ethylene Production
Ethylene measurements were performed as previously described (Hansen et al., 2009). Surface-sterilized seeds were germinated in 22-mL gas chromatography vials containing 3 mL of MS with 1% Suc. Following 4 d of stratification at 4°C, the seeds were exposed to white light for 3 to 5 h. For ethylene measurement of dark-grown seedlings, the vials were capped after stratification and incubated at 22°C for 3 d in the dark. For light-grown seedlings, the vials were incubated at 22°C for 12 d after stratification, capped at day 12, and incubated for additional 24 h in the light. For analysis of the effects of R18 peptide on ethylene biosynthesis of eto1-13/eol1-1/eol2-2 (Christians et al., 2009) seedlings, the seeds were grown in 22-mL gas chromatography vials containing 2 mL of MS with either 200 µg/mL R18 or R18Lys peptide for 3 d in the dark. After growth at 22°C, the accumulated ethylene was measured by gas chromatography. All genotypes and treatments were measured from at least three vials each.
Measurements of Protein Half-Lives
Cycloheximide (250 µM) was added to the suspended protoplasts harboring the appropriate plasmids and the protoplasts then harvested at various time points, followed by immunoblot analysis. For the half-life experiment including MG132 treatment, 50 µM MG132 was added 3 h before cycloheximide addition.
Quantification of the Signals from Immunoblotting Analysis
Image J software (http://rsb.info.nih.gov/ij/) was used for a quantification of the bands intensity from immunoblots. Intensity of the bands was normalized to BiP or GFP control band intensity, and these values normalized to the 0 time points or no treatment, which set to a value of 1.
Ubiquitination Assay
Arabidopsis protoplasts were cotransfected with 35S:GFP-EOL2 and 35S:HA-ubiquitin with or without 35S:myc-14-3-3 plasmids. An empty vector plasmid was cotransformed in order to normalize the level of 35S plasmid in all transformations. After 16 h of incubation, 50 µM MG132 was added into the protoplasts and further incubated for 6 h. The protoplasts were then harvested and used for coimmunoprecipitation assay. Total proteins were extracted in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM PMSF, and 1× protease inhibitor cocktail) and incubated with α-GFP magnetic beads (Miltenyi Biotech) for 2 h at 4°C, followed by washing with lysis buffer three times. The precipitate was then eluted with boiled 2× SDS sample buffer and used for further analysis with immunoblotting.
Accession Numbers
Sequence data for the genes in this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: 14-3-3ω (AT1G78300), 14-3-3κ (AT5G65430), 14-3-3φ (AT1G35160), 14-3-3ο (AT1G34760), 14-3-3ι (AT1G26480), ACS5 (AT5G65800), ACS6 (AT4G11280), ACS7 (AT4G26200), ETO1 (AT3G51770), EOL2 (AT5G58550), and Ubiquitin3 (AT5G03240.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ACS5 Interacts with Multiple Isoforms of 14-3-3.
Supplemental Figure 2. In Vivo Interaction between ACS5 and 14-3-3ω in Arabidopsis Protoplasts.
Supplemental Figure 3. Real-Time RT-PCR Assay of the myc-ACS5 Transcripts in Arabidopsis Seedlings in Response to R18 Peptide Treatment.
Supplemental Figure 4. R18-Mediated Destabilization of myc-ACS5 Protein Is Dependent on Ubiquitin/26s Proteasome Activity.
Supplemental Figure 5. Ethylene Production of eto1, eol1, and eol2 Mutant Seedlings in Light.
Supplemental Figure 6. ETO1/EOL2 Interacts with Multiple Isoforms of 14-3-3.
Supplemental Figure 7. Quantification of the Steady State Level of HA-EOL2 and HA-ACS5 Proteins with Increasing 14-3-3 Expression.
Supplemental Figure 8. R18 Does Not Affect the Hypocotyl Length of Wild-Type Seedlings Grown in the Presence of ACC.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Table 2. List of Plasmids Used in This Study.
Supplemental Table 3. List of Gateway Constructs Used in This Study.
Supplemental Methods 1. Quantitative RT-PCR Analysis.
Supplementary Material
Acknowledgments
This work was supported by a National Science Foundation grant to J.J.K. (MCB-1021704). We thank members of the Kieber lab for critical reading of the article, and Tony Purdue for helping with the confocal microscopy.
AUTHOR CONTRIBUTIONS
G.M.Y. and J.J.K. designed research, analyzed data, and wrote the article. G.M.Y. performed research.
Glossary
- ACS
1-aminocyclopropane-1-carboxylate synthase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- MS
Murashige and Skoog
- RFP
red fluorescent protein
- GFP
green fluorescent protein
References
- Abeles, F.B., Morgan, P.W., and Saltveit, M.E., Jr. (1992). Ethylene in Plant Biology. (San Diego, CA: Academic Press). [Google Scholar]
- An F., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Argueso C.T., Hansen M., Kieber J.J. (2007). Regulation of ethylene biosynthesis. J. Plant Growth Regul. 26: 92–105 [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Chae H.S., Faure F., Kieber J.J. (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.S., Kieber J.J. (2005). Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10: 291–296 [DOI] [PubMed] [Google Scholar]
- Chang I.-F., Curran A., Woolsey R., Quilici D., Cushman J.C., Mittler R., Harmon A., Harper J.F. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9: 2967–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D., Morris E.R., Walker J.C. (2009). 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60: 67–91 [DOI] [PubMed] [Google Scholar]
- Christians M.J., Gingerich D.J., Hansen M., Binder B.M., Kieber J.J., Vierstra R.D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling D.L., Yingling J., Wynshaw-Boris A. (2005). Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68: 281–315 [DOI] [PubMed] [Google Scholar]
- Denison F.C., Paul A.L., Zupanska A.K., Ferl R.J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22: 720–727 [DOI] [PubMed] [Google Scholar]
- Dougherty M.K., Morrison D.K. (2004). Unlocking the code of 14-3-3. J. Cell Sci. 117: 1875–1884 [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen J.P., Ludwig R.L., Vousden K.H., Weissman A.M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Folta K.M., Paul A.L., Mayfield J.D., Ferl R.J. (2008). 14-3-3 isoforms participate in red light signaling and photoperiodic flowering. Plant Signal. Behav. 3: 304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A.K., Morrison D.K. (2011). 14-3-3 proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 22: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.-M., Peter M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala, S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed]
- Gingerich D.J., Gagne J.M., Salter D.W., Hellmann H., Estelle M., Ma L., Vierstra R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hansen M., Chae H.S., Kieber J.J. (2009). Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 57: 606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M.S., Ou C., Chan Y.R., Chien C.T., Pi H. (2008). The utility F-box for protein destruction. Cell. Mol. Life Sci. 65: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S.C., MacKintosh C., Kaiser W.M. (2002). Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 50: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Lee M.H., Lozano G. (2006). Regulation of the p53-MDM2 pathway by 14-3-3 sigma and other proteins. Semin. Cancer Biol. 16: 225–234 [DOI] [PubMed] [Google Scholar]
- Li J.J., Zhang T.P., Meng Y., Du J., Li H.H. (2011). Stability of F-box protein atrogin-1 is regulated by p38 mitogen-activated protein kinase pathway in cardiac H9c2 cells. Cell. Physiol. Biochem. 27: 463–470 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga W.J., Booth J.K., Stone S.L. (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71: 23–34 [DOI] [PubMed] [Google Scholar]
- Masters S.C., Fu H. (2001). 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 276: 45193–45200 [DOI] [PubMed] [Google Scholar]
- Mattoo, A.K., and Suttle, J.C. (1991). The Plant Hormone Ethylene. (Boca Raton, FL: CRC Press). [Google Scholar]
- Mayfield J.D., Folta K.M., Paul A.-L., Ferl R.J. (2007). The 14-3-3 proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 145: 1692–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J.D., Paul A.L., Ferl R.J. (2012). The 14-3-3 proteins of Arabidopsis regulate root growth and chloroplast development as components of the photosensory system. J. Exp. Bot. 63: 3061–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Hayashi N., Kaneko Y.S., Mori K., Sabban E.L., Nagatsu T., Ota A. (2007). RNAi of 14-3-3eta protein increases intracellular stability of tyrosine hydroxylase. Biochem. Biophys. Res. Commun. 363: 817–821 [DOI] [PubMed] [Google Scholar]
- Oecking C., Jaspert N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12: 760–765 [DOI] [PubMed] [Google Scholar]
- Oh C.S., Pedley K.F., Martin G.B. (2010). Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK alpha. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.L., Denison F.C., Schultz E.R., Zupanska A.K., Ferl R.J. (2012). 14-3-3 phosphoprotein interaction networks - Does isoform diversity present functional interaction specification? Front. Plant Sci. 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.L., Sehnke P.C., Ferl R.J. (2005). Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol. Biol. Cell 16: 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Purwestri Y.A., Ogaki Y., Tamaki S., Tsuji H., Shimamoto K. (2009). The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Roberts M.R., de Bruxelles G.L. (2002). Plant 14-3-3 protein families: Evidence for isoform-specific functions? Biochem. Soc. Trans. 30: 373–378 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda-García E., Rocha-Sosa M. (2012). The Arabidopsis F-box protein AtFBS1 interacts with 14-3-3 proteins. Plant Sci. 195: 36–47 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Callis J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10: 624–632 [DOI] [PubMed] [Google Scholar]
- Su C.H., Zhao R., Velazquez-Torres G., Chen J., Gully C., Yeung S.C., Lee M.H. (2010). Nuclear export regulation of COP1 by 14-3-3σ in response to DNA damage. Mol. Cancer 9: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.H., Zhao R., Zhang F., Qu C., Chen B., Feng Y.H., Phan L., Chen J., Wang H., Wang H., Yeung S.C., Lee M.H. (2011). 14-3-3σ exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 71: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.S., Whippo C., Hangarter R.P., Briggs W.R. (2012). The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24: 1114–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G., Avruch J. (2002). 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277: 3061–3064 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vogel J.P., Schuerman P., Woeste K.W., Brandstatter I., Kieber J.J. (1998b). Isolation and characterization of Arabidopsis mutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics 149: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., Woeste K.E., Theologis A., Kieber J.J. (1998a). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang B., Yang H., Liu Y.C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999). Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38: 12499–12504 [DOI] [PubMed] [Google Scholar]
- Wang K.L.-C., Yoshida H., Lurin C., Ecker J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wee S., Geyer R.K., Toda T., Wolf D.A. (2005). CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7: 387–391 [DOI] [PubMed] [Google Scholar]
- Wei W., Ayad N.G., Wan Y., Zhang G.J., Kirschner M.W., Kaelin W.G.J., Jr (2004). Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428: 194–198 [DOI] [PubMed] [Google Scholar]
- Weiner H., Kaiser W.M. (1999). 14-3-3 proteins control proteolysis of nitrate reductase in spinach leaves. FEBS Lett. 455: 75–78 [DOI] [PubMed] [Google Scholar]
- Xu W., Shi W., Jia L., Liang J., Zhang J. (2012). TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ. 35: 1393–1406 [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Wen Y.Y., Lin Y.I., Pham L., Su C.H., Yang H., Chen J., Lee M.H. (2007). Roles for negative cell regulator 14-3-3sigma in control of MDM2 activities. Oncogene 26: 7355–7362 [DOI] [PubMed] [Google Scholar]
- Yang J.L., Chen W.W., Chen L.Q., Qin C., Jin C.W., Shi Y.Z., Zheng S.J. (2013). The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 197: 815–824 [DOI] [PubMed] [Google Scholar]
- Yang S.F., Hoffman N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35: 155–189 [Google Scholar]
- Yang Y., Li R., Qi M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Nagata M., Saito K., Wang K.L., Ecker J.R. (2005). Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Wang K.L., Chang C.-M., Mori K., Uchida E., Ecker J.R. (2006). The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol. Biol. 62: 427–437 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.