This study reports on an anthranilate N-methyltransferase, MT1, required for synthesis of antimicrobial triterpenoid defense compounds (avenacins) in oat and, through the characterization of MT1 and the products of other genes within the avenacin metabolic gene cluster, presents a model of the subcellular organization of triterpenoid biosynthesis.
Abstract
Operon-like gene clusters are an emerging phenomenon in the field of plant natural products. The genes encoding some of the best-characterized plant secondary metabolite biosynthetic pathways are scattered across plant genomes. However, an increasing number of gene clusters encoding the synthesis of diverse natural products have recently been reported in plant genomes. These clusters have arisen through the neo-functionalization and relocation of existing genes within the genome, and not by horizontal gene transfer from microbes. The reasons for clustering are not yet clear, although this form of gene organization is likely to facilitate co-inheritance and co-regulation. Oats (Avena spp) synthesize antimicrobial triterpenoids (avenacins) that provide protection against disease. The synthesis of these compounds is encoded by a gene cluster. Here we show that a module of three adjacent genes within the wider biosynthetic gene cluster is required for avenacin acylation. Through the characterization of these genes and their encoded proteins we present a model of the subcellular organization of triterpenoid biosynthesis.
INTRODUCTION
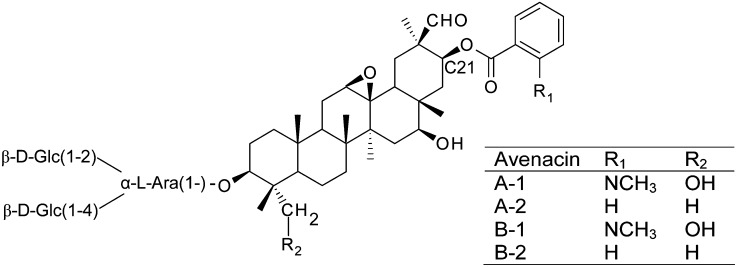
Plants synthesize a diverse range of specialized metabolites (also commonly referred to as natural products), many of which have important roles in protection against disease, herbivory, and/or abiotic stress (Dixon, 2001; Osbourn and Lanzotti 2009). Oats (Avena spp) produce antimicrobial triterpene glycosides (avenacins) in their roots that protect against soil-borne pathogens (Hostettmann and Marston, 1995; Papadopoulou et al., 1999; Osbourn et al., 2011). Although a diverse array of structurally varied triterpene glycosides is found in dicots, the ability to synthesize glycosylated triterpenes is rare in the cereals and grasses. Avenacins exhibit some unusual structural features that are not shared by other plant triterpene glycosides. Most notably, they are acylated at the carbon-21 (C-21) position with either N-methyl anthranilate or benzoate (Figure 1). A recent review of structure-function relationships in triterpenoids indicates that C-21 acylation is important in determining the biological activity of some triterpenes (Podolak et al., 2010). Understanding the mechanisms underlying this type of modification has potential to provide powerful biotechnological tools for the modification of triterpene structure and function in different biological systems.
Figure 1.
Structures of Oat Root Avenacins.
The C-21 acyl group consists of either N-methyl anthranilate or benzoate, depending on the R1 group. The major avenacin found in oat roots is avenacin A-1, which is acylated with N-methyl anthranilate (Crombie et al., 1984).
Acylation orchestrates the amalgamation of complex chemical groups with different biochemical origins into single compounds and so offers great versatility in modulating the form and function of plant natural products (D’Auria, 2006). Plant acyltransferase enzymes have been identified that catalyze the transfer of a wide range of acyl groups onto a diverse set of acceptor substrates belonging to different classes of natural product. These enzymes belong to two unrelated families: the BAHD acyltransferases, which use CoA-thioesters as the acyl donor (Yang et al., 1997; Dudareva et al., 1998; Fujiwara et al., 1998; St-Pierre et al., 1998; Walker et al., 2002; Luo et al., 2007, 2009; Grienenberger et al., 2009); and the serine carboxypeptidase-like (SCPL) acyltransferases, which use O-Glc esters as acyl donors. The SCPL-acyltransferases belong to the serine carboxypeptidase family but have acquired different functions to these ancestral proteases (Lehfeldt et al., 2000; Li and Steffens, 2000; Shirley et al., 2001; Fraser et al., 2007; Weier et al., 2008; Shirley and Chapple, 2003; Milkowski and Strack, 2004; Milkowski et al., 2004; Baumert et al., 2005; Stehle et al., 2008; Mugford et al., 2009; Mugford and Osbourn, 2010; Mugford and Milkowski, 2012). Based on the available evidence, it seems likely that the two different families of acyltransferases are also distinguished by their subcellular localizations. The Arabidopsis thaliana sinapoyl-malate SCPL-acyltransferase SMT has been localized to the vacuole (Hause et al., 2002), and other members of this family are likewise predicted to be targeted to the vacuole in Arabidopsis and other plants (Fraser et al., 2005). Conversely, members of the BAHD family have either been shown to be or are predicted to be cytosolic (Fujiwara et al., 1998; D’Auria, 2006; Yu et al., 2008).
We previously established a collection of avenacin-deficient mutants of the diploid oat species Avena strigosa. The major avenacin, A-1, is esterified with N- methyl anthranilate and fluoresces strongly under ultraviolet illumination. Avenacin-deficient mutants of A. strigosa were isolated following sodium azide mutagenesis, using a simple screen for loss of root fluorescence. These avenacin-deficient mutants were found to have increased susceptibility to infection by soil-borne pathogens (Papadopoulou et al., 1999). This mutant collection has proved to be a valuable resource for investigating triterpenoid biosynthesis, and we cloned genes encoding several steps of the avenacin biosynthetic pathway (Haralampidis et al., 2001; Qi et al., 2004, 2006; Mugford et al., 2009; Owatworakit et al., 2012). An important discovery has been that the loci for the synthesis of avenacins are clustered. Although genes encoding plant metabolic pathways are generally regarded as randomly distributed in the genome, a growing number of plant natural product biosynthetic pathways have been identified that are encoded by operon-like gene clusters in plants (Frey et al., 1997; Gierl and Frey, 2001; Qi et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Field and Osbourn, 2008; Swaminathan et al., 2009; Chu et al., 2011; Falara et al., 2011; Field et al., 2011; Takos et al., 2011; Kliebenstein and Osbourn, 2012; Winzer et al., 2012).
We recently showed that an SCPL acyltransferase, SCPL1, is responsible for the acylation of avenacins in A. strigosa (Mugford et al., 2009). SCPL1 is encoded by Sad7, which forms part of the avenacin gene cluster. This enzyme catalyzes the transfer of N-methyl anthranilate from an O-Glc ester onto triterpenoid backbones to produce avenacins A-1 and B-1. It is also required for the formation of the two benzoylated forms of avenacin, A-2 and B-2 (Figure 1). In Arabidopsis and Brassica species, the glucosylated acyl donor substrates of SCPL acyltransferases are synthesized by family 1 glucosyltransferases (Lim et al., 2001; Baumert et al., 2005). We recently identified a family 1 glycosyltransferase (UGT74H5) in A. strigosa that catalyzes the glucosylation of N-methyl anthranilate to N-methyl anthranilate-O-Glc, the acyl donor substrate for SCPL1. UGT74H5 is encoded by the A. strigosa Sad10 gene, which lies within the avenacin gene cluster, adjacent to the Sad7 (SCPL1) gene (Owatworakit et al., 2012).
Here, we characterize a gene encoding an anthranilate N-methyltransferase (MT1, encoded by Sad9) and show that this enzyme acts together with the UGT74H5 glucosyltransferase and the SCPL1 acyltransferase in the final steps of the synthesis of avenacin A-1. These three genes are adjacent within the wider avenacin biosynthetic gene cluster and so represent an acylation module. We present evidence that acylation of avenacins is important for the potent biological activity of these molecules. This three-gene cassette may therefore be a useful resource for the heterologous modification of triterpenes with altered biological activity. By combining biochemical and genetic approaches with investigation of the subcellular localization of the enzymes of avenacin biosynthesis, we further establish a model for the spatial organization of triterpenoid synthesis.
RESULTS
Three Genes Encoding a Methyltransferase, Glucosyltransferase, and an Acyltransferase Are Adjacent within the Avenacin Gene Cluster
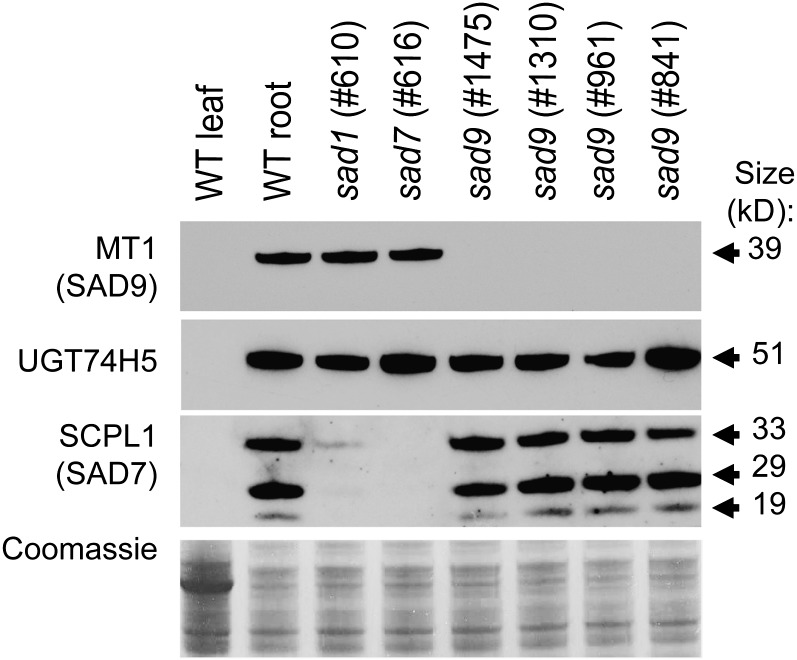
Previously, we identified and sequenced a BAC contig spanning four genes within the avenacin gene cluster: Sad1, 2, 7, and 10 (Qi et al., 2006; Mugford et al., 2009; Owatworakit et al., 2012). Further extension of this BAC contig identified a gene predicted to encode an S-adenosyl-methionine–dependent methyltransferase (MT1), which lies 45 kb from the UGT74H5 (Sad10) gene (Figure 2A). Avenacins A-1 and B-1 are both acylated with N-methyl anthranilate, implicating anthranilate methylation in the avenacin biosynthetic pathway. Phylogenetic analysis shows that MT1 lies within a clade of methyltransferases that consists mainly of O-methyltransferase enzymes with roles in secondary metabolism (Figure 2B; see Supplemental Data Set 1 online), although two characterized members of this clade (Limonium latifolium β-alanine-N-methyltransferase and Ruta graveolens anthranilate-N-methyltransferase) are known to have N-methyltransferase activity (Liscombe and Facchini, 2007; Rohde et al., 2008), consistent with a possible role for MT1 as an N-methyltransferase. Avenacins are synthesized in the epidermal cells of the root tip, and the expression of genes that encode the previously characterized steps in the biosynthetic pathway is restricted to these cells (Qi et al., 2006; Wegel et al., 2009; Mugford et al., 2009). Quantitative PCR analysis revealed that the MT1 transcript is most abundant in whole root and root tip samples, low in the upper root, and not detectable in young leaves (Figure 2C). Immunoblot analysis detected MT1 in extracts from the roots but not the leaves of wild-type A. strigosa seedlings, as is the case for other avenacin biosynthetic enzymes (Figure 3, lanes 1 and 2). Furthermore, immunostaining of root tip tissue sections with specific antisera raised against As-MT1 indicates that the protein is restricted to the epidermal cells of the root tip (Figure 2D). A similar pattern of staining is also observed using antisera raised against the As-SCPL1 protein. Signals were not observed for sections stained with the corresponding preimmune sera (see Supplemental Figure 1 online). Collectively, these results are consistent with a role for As-MT1 in avenacin biosynthesis.
Figure 2.
A Gene Encoding a Predicted Methyltransferase Located within the Avenacin Biosynthetic Gene Cluster Is Implicated in Avenacin Synthesis.
(A) BAC contig showing the avenacin gene cluster and the location of the Sad9 gene, which is predicted to encode a methyltransferase. Gene names are indicated above and encoded proteins below.
(B) Neighbor-joining phylogeny of functionally characterized plant natural product S-adenosyl-Met–dependent methyltransferase enzymes. N-methyltransferase enzymes are indicated with asterisks. The phylogeny includes the sequences used by Liscombe and Facchini, 2007; the MT1 protein (indicated by an arrow); the most similar sequences to As-MT1 encoded by the genomes of Arabidopsis (At4g35160), rice (Oryza sativa; Os9g17560), Brachypodium distachyon (Bd2g19830), and sorghum (Sorghum bicolor; Sb07g024270); and the R. graveolens anthranilate methyltransferase (Rg ANMT; Rohde et al., 2008). Details of these other sequences are provided in Methods, and the full protein sequence alignment can be found in Supplemental Data Set 1 online. Bootstrap values (percentage of 1000 replicates) are shown for key branches. Bar indicates 0.05 substitutions per site.
(C) Sad9 (AsMT1) transcript abundance in young leaves, whole roots, root tips (0.5 mm), and roots minus tips, measured by quantitative PCR (transcript abundance expressed relative to elongation factor 1-α); values are means (n = 3 ± se).
(D) Immunostaining of root tip sections from 3-d-old wild-type A. strigosa seedlings. Sections were stained with (from left to right) anti-AsMT1 antisera, anti-AsSCPL1 antisera, or 4′,6-diamidino-2-phenylindole (DAPI). A bright-field image and an overlay of the MT1, SCPL1, and DAPI images are also shown. Controls probed with preimmune sera are shown in Supplemental Figure 1 online.
Figure 3.
Detection of MT1, UGT74H5, and SCPL1 in Oat Roots.
Immunoblot analysis of soluble protein extracts from 3-d-old A. strigosa seedlings, probed with antisera raised against MT1, UGT74H5, or SCPL1. The bottom panel shows Coomassie blue staining of a replicate gel. Protein extracts were prepared from leaves and roots of 3-d-old A. strigosa wild-type (WT) seedlings and from roots of previously characterized sad1 (mutant line #610) and sad7 (#616) mutants (Papadopoulou et al., 1999; Mugford et al., 2009) and sad9 mutants (#1475, 1310, 961, and 841) (this article). Bands corresponding to the predicted size of the MT1 and UGT74H5 proteins were detected. SCPL1 is posttranslationally cleaved into a large and a small subunit corresponding to the bands detected at 29 and 19 kD and a processing intermediate (33 kD) (Mugford et al., 2009).
MT1 Is Required for N-Methylation of Avenacin A-1 and Corresponds to Sad9, a Locus Previously Defined by Mutation
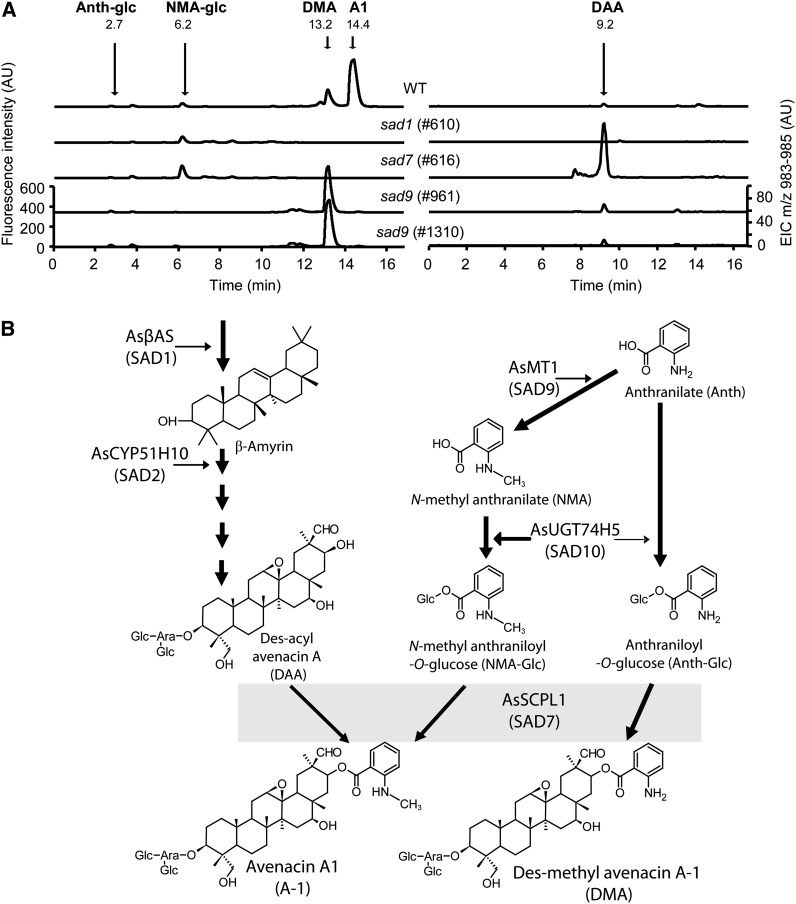
We previously isolated a collection of avenacin-deficient mutants of A. strigosa that were identified from a screen for loss of UV fluorescence in the root tip (Papadopoulou et al., 1999; Qi et al., 2006). Liquid chromatography–mass spectrometry (LC-MS) analysis of this mutant collection identified four independent mutants that lacked avenacin A-1 but accumulated a related compound that is acylated with anthranilate rather than N-methyl anthranilate (des-methyl avenacin A-1 [DMA]) (Figure 4A; see Supplemental Figure 2 online). These four mutant lines (#1475, #1310, #961, and #841) were shown to be allelic. The roots of early avenacin pathway mutants (sad1 and sad2) (Papadopoulou et al., 1999; Qi et al., 2004, 2006; Qin et al., 2010) have little or no fluorescence, while those of sad7 have weak blue autofluorescence due to the accumulation of the acyl donor N-methyl anthraniloyl-O-Glc (Mugford et al., 2009). By contrast, roots of sad9 mutants have a weak purple fluorescence under UV illumination. The fluorescence of extracts from wild-type roots peaks at 440 nm compared with 410 nm from the sad9 mutant. This is consistent with the fluorescence of N-methyl anthranilate (blue) compared with that of anthranilate (purple). LC-MS analysis revealed that DMA is found not only in sad9 mutants, but also in wild-type A. strigosa roots (Figure 4). sad9 mutants lack N-methyl anthraniloyl-O-Glc but accumulate a fluorescent compound with mass spectrum and fragmentation pattern consistent with anthraniloyl-O-glucose (Figure 4A; see Supplemental Figure 2 online). Quantification of acylated avenacins in a selection of mutants showed that DMA accumulates in the sad9 mutants at lower levels (109.0 ± 29.9 pmol/mg fresh weight in #961; 189.2 ± 26.5 pmol/mg fresh weight in #1310) than is found for avenacin A-1 in the wild type (365.3 ± 8.9 pmol/mg fresh weight) (values are means of four replicate samples ± se). In addition, the sad9 mutants accumulate des-acyl avenacin, which is also found in the sad7 mutants (Figure 4), and the nonfluorescent avenacins A-2 and B-2, which are acylated with benzoate and are also present at similar levels in the wild type (see Supplemental Table 1 online).
Figure 4.
sad9 Mutants Are Blocked in the Methylation of Avenacin Acyl-Groups.
(A) Analysis of wild-type and mutant (sad1, sad7, and sad9) root extracts by fluorescence LC-MS. Fluorescence detection shows the absence of avenacin A-1 (A1) in all mutants and the accumulation of DMA in sad9 mutants. N-methyl anthraniloyl-O-Glc (NMA-glc) is not detectable in root extracts from sad9 mutants, but anthraniloyl-O-Glc (Anth-glc) is detected at low levels. Similar metabolite profiles were observed in the other sad9 mutants analyzed (see Supplemental Figure 2 online). Confirmation of the identity of these compounds by mass spectrometry is presented in Supplemental Figure 2 online. LC-MS shows the accumulation of des-acyl avenacin A (DAA; extracted ion chromatogram for mass-to-charge ratio = 983 to 985) in the sad7 mutant, as previously shown by Mugford et al. (2009). AU, arbitrary units.
(B) Proposed pathway for acylation of avenacin A-1. N-methyl anthranilate (NMA) is synthesized by the methylation of anthranilate (Anth) by MT1. The glucosyltransferase UGT74H5 glucosylates N-methyl anthranilate to give the activated acyl donor substrate N-methyl anthraniloyl-O-Glc (NMA-Glc). UGT74H5 has a clear preference for N-methyl anthranilate but also has weak activity toward anthranilate to give anthraniloyl-O-Glc (Anth-Glc) (Owatworakit et al., 2012). In wild-type A. strigosa, the SCPL1 acyltransferase uses NMA-Glc as the acyl donor to give avenacin A-1 (A1). In sad9 mutants (lacking MT1), the triterpenoid backbone is acylated with anthranilate instead of N-methyl anthranilate to give DMA.
The failure of sad9 mutants to methylate avenacin and the presence of MT1 (encoding a predicted methyltransferase) in the avenacin gene cluster suggests that Sad9 and MT1 may be synonymous. DNA sequence analysis of MT1 in the sad9 mutants revealed nonsynonymous point mutations in all four alleles (#841, C998T and A333V; #961, C89T and S30F; #1310, G978A and W326Stop; #1475, C235T and R79W) that alter single amino acids or in the case of #1310 introduce a stop codon, indicating that Sad9 is likely to encode MT1. These data suggest a pathway for acylation of avenacin as shown in Figure 4B that involves N-methylation of anthranilate by MT1 (SAD9), glucosylation of N-methyl anthranilate by UGT74H5 (SAD10), and the transfer of the acyl group to the triterpene backbone by SCPL1 (SAD7). In the absence of MT1, SCPL1 is apparently still able to acylate avenacins using anthraniloyl-O-Glc as the acyl donor instead of N-methyl anthraniloyl-O-Glc.
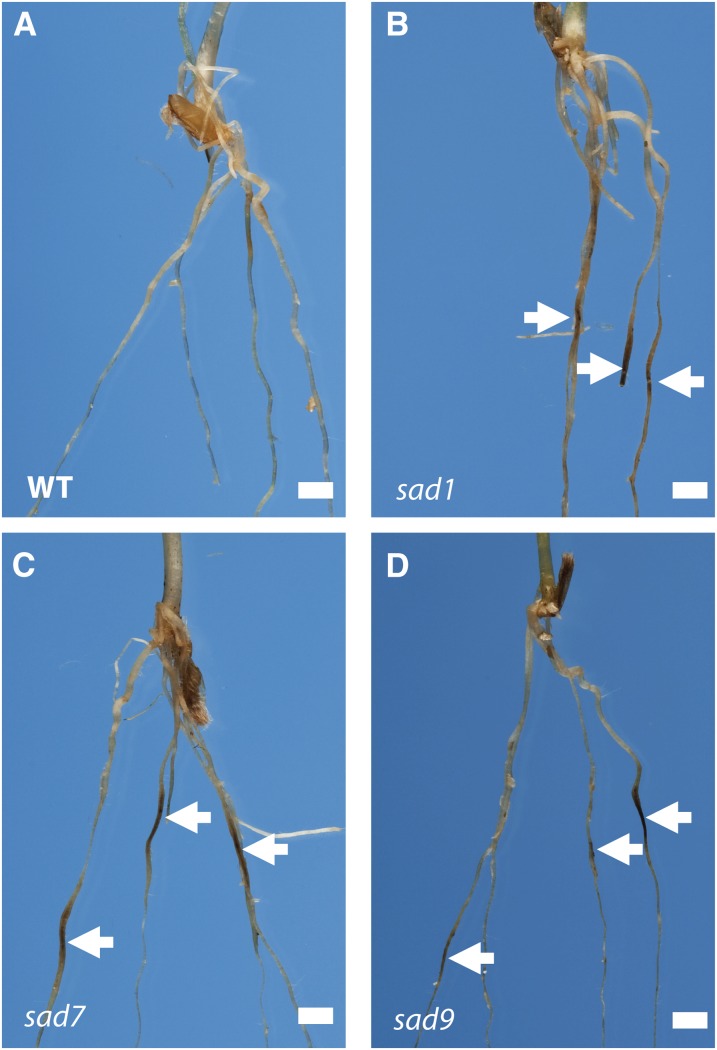
Previously we have shown that sad1, sad2, and sad7 mutants of A. strigosa have increased susceptibility to soil-borne fungal pathogens (Papadopoulou et al., 1999; Qi et al., 2004, 2006; Mugford et al., 2009). Pathogenicity tests with Gaeumannomyces graminis var tritici, the causal agent of take-all disease of cereals, revealed that, like sad1, sad2, and sad7 mutants, sad9 mutants are more susceptible to infection than wild-type seedlings (Figure 5). The increased susceptibility of sad7 and sad9 mutants to disease may be because correct acylation of avenacins is required for antifungal activity or, alternatively, because the unacylated/incorrectly acylated avenacins accumulate at lower levels in the mutants. We measured the total concentrations of avenacins and des-acyl avenacins in wild-type and sad7 mutant oat roots and found that the levels of unacylated avenacins in the sad7 mutant roots were comparable to the levels of acylated avenacins in the wild type (see Supplemental Figure 3 online), indicating that the acyl group is likely to be important in protecting against disease. Furthermore, bioassays with extracts from wild-type and sad7 mutant root tips separated by thin layer chromatography (TLC) show that while avenacin A-1 in the wild-type sample strongly inhibits the growth of the indicator fungus Colletotrichum orbiculare, the presence of similar amounts of desacyl avenacin A in the sad7 mutant extract has no such inhibitory effect (see Supplemental Figure 4 online). Thus, the acyl group is important for biological activity. The presence in the sad9 mutants of benzoylated forms of avenacin in addition to DMA (see Supplemental Table 1 online) complicates efforts to establish the importance of the methyl group for biological activity. However, a thin layer chromatography bioassay with extracts from sad9 mutant roots clearly shows zones of C. orbiculare growth inhibition associated with both of the major forms of avenacin present in this mutant, desmethyl avenacin A-1 and avenacin A-2, indicating that desmethyl avenacin A-1 does possess antifungal activity (see Supplemental Figure 4 online). It is therefore likely that the increased susceptibility of sad9 mutants to fungal infection is due to the overall lower concentrations of acylated avenacins relative to the wild type (see Supplemental Table 1 online).
Figure 5.
sad9 Mutants Have Enhanced Disease Susceptibility.
Seedlings were inoculated with the take-all fungus (G. graminis var tritici strain T2) and scored for disease symptoms 3 weeks later as described by Papadopoulou et al. (1999). Dark-brown/black lesions on the roots (arrowed) are typical symptoms of infection. The wild type (WT) (A), sad1 mutant #610 (B), sad7 mutant #616 (C), and sad9 mutant #1475 (D). Images are representative of six to 10 biological replicates across two independent experiments. Bars = 0.25 cm.
MT1 was detected in protein preparations from the roots of wild-type A. strigosa seedlings and in sad1 and sad7 mutant lines. However, it was not detected in the four sad9 mutants (Figure 3). The sad9 mutants were isolated from a forward screen for loss-of-function mutations. Mutant #1310 has a premature stop codon, while, based on comparison with the structure of the Medicago truncatula isoflavone O-methyltransferase (the most similar sequence to As-MT1 for which a crystal structure is available; National Center for Biotechnology Information accession number 2QYO_A), the amino acid substitutions in the other three mutants are all likely to disrupt α-helices integral to the structure of the protein. All four sad9 mutants had wild-type levels of the SCPL1 (SAD7) and UGT74H5 (SAD10) proteins. Interestingly, the levels of SCPL1 (SAD7) protein were substantially reduced in sad1 mutants, which are blocked in the first committed step in avenacin biosynthesis (Qi et al., 2004) (shown for sad1 mutant #610 in Figure 3). Examination of the levels of SCPL1 (SAD7) protein in a wider range of early pathway mutants (Qi et al., 2004, 2006; Qin et al., 2010) revealed that the SCPL1 protein level is reduced or abolished in all sad1 and sad2 mutants, while the levels of the MT1 and UGT74H5 proteins were not substantially altered in these mutants (see Supplemental Figure 5 online). The levels of the Sad7 transcript were not significantly altered in any of the mutants (see Supplemental Figure 5 online), suggesting that SCPL1 may be posttranscriptionally regulated. A scenario consistent with these data is that the triterpene backbone is required for the accumulation of SCPL1 protein, since mutants that are unable to synthesize this backbone fail to accumulate SCPL1.
MT1 Encodes an Anthranilate N-Methyltransferase
MT1 was expressed in Escherichia coli with a C-terminal 6XHis epitope tag and purified by metal affinity chromatography. Accumulation of des-methyl avenacin and anthraniloyl-O-Glc in the sad9 mutants indicates that the likely substrate for this enzyme in planta is either anthranilate or anthraniloyl-O-Glc. We found no detectable activity of the recombinant protein against anthraniloyl-O-Glc at a range of substrate concentrations (0.5 to 5 mM), while under the same conditions, MT1 effectively catalyzed the formation of N-methyl anthranilate from anthranilate. Kinetic analysis of the MT1 enzyme in vitro showed that the enzyme has a strong affinity for anthranilic acid (Km = 5.51 ± 0.11 µM), with a pH optimum of 8.0, and a high catalytic efficiency (kcat/Km = 5.55 ± 0.16 nM−1 s−1), which compare favorably with the reported kinetic properties of other plant methyl transferases that are active toward small phenolic compounds (Wang and Pichersky, 1999; Gang et al., 2002; Rohde et al., 2008; Jonczyk et al., 2008). Thus, As-MT1 catalyzes the first committed step in the synthesis of the N-methyl anthraniloyl acyl donor that is used by the As-SCPL1 acyltransferase, namely, methylation of the primary shikimate pathway intermediate anthranilate. This is consistent with the proposed pathway in Figure 4B and with the catalytic properties of As-UGT74H5, which has a strong affinity for N-methyl anthranilate compared with anthranilate (Owatworakit et al., 2012).
Coexpression of As-MT1 and As-UGT74H5 in Nicotiana benthamiana Leaves Results in the Accumulation of N-Methyl Anthraniloyl-O-Glc
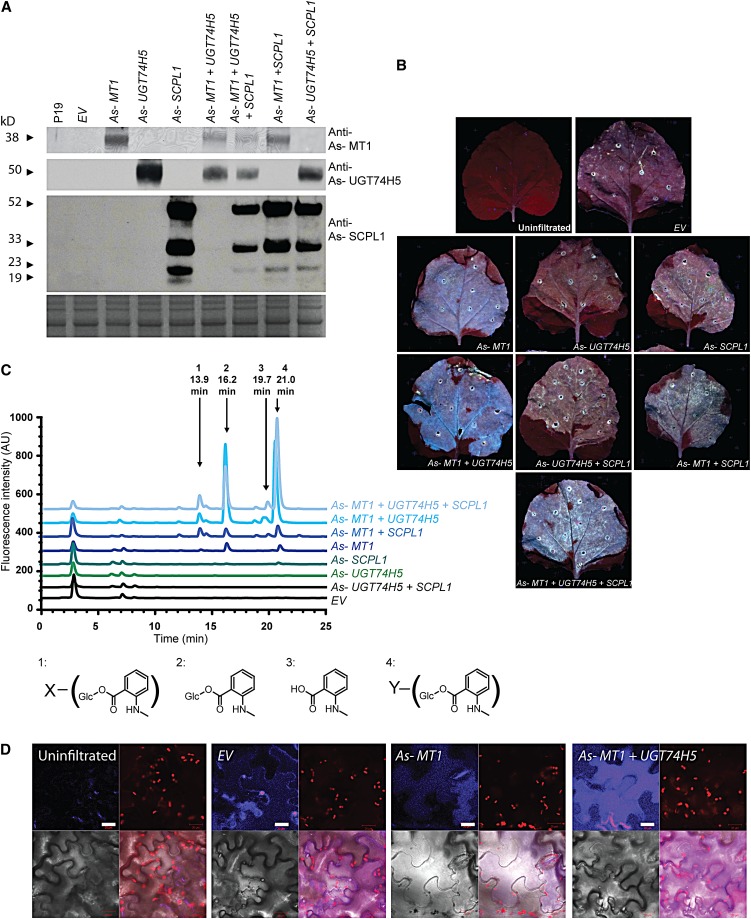
The above evidence coupled with the previous characterization of As-SCPL and As-UGT74H5 (Mugford et al., 2009; Owatworakit et al., 2012) implicates As-MT1, As-UGT74H5, and As-SCPL1 in the synthesis and transfer of the N-methyl anthraniloyl acyl group onto the triterpene backbone. To further test the roles of these enzymes, we transiently coexpressed the As-MT1, As-UGT74H5, and As-SCPL1 cDNAs in N. benthamiana leaves using a cowpea mosaic virus (CPMV)–derived expression system (Sainsbury et al., 2008; Mugford et al., 2009) (Figure 6). Immunoblot analysis confirmed the expression of the three proteins, expressed either alone or in combination (Figure 6A). Interestingly, expression of MT1 alone was sufficient to cause the leaves to fluoresce blue under UV illumination, reminiscent of the fluorescence of oat root tips (Figure 6B), and this fluorescence was enhanced when MT1 and UGT74H5 were expressed in combination. HPLC analysis of methanolic extracts of the leaf material revealed that the expression of MT1 and UGT74H5 together results in the accumulation of N-methyl anthraniloyl-O-Glc (39.8 ± 6.5 nmol/mg fresh weight) (Figure 6C; see Supplemental Table 2 online). Expression of UGT74H5 alone did not lead to the production of detectable levels of any fluorescent compounds. Expression of the SCPL1 acyltransferase together with MT1 and UGT74H5 did not lead to accumulation of any additional detectable fluorescent compounds, indicating that SCPL1 is unlikely to be able to acylate endogenous compounds in N. benthamiana, which is perhaps not surprising since there are no reports of triterpenoid saponins or related compounds in this species. Further analysis by LC-MS failed to identify any other compounds that accumulated in an SCPL1-dependent manner. The expression of MT1 alone led to the accumulation of low levels of N-methyl anthranilate and N-methyl anthraniloyl-O-Glc, suggesting that N. benthamiana possesses enzymes capable of glucosylating N-methyl anthranilate at low levels, presumably as a side reaction of an endogenous glucosyltransferase. Also, two other fluorescent compounds were found to accumulate in leaves expressing MT1 and UGT74H5 together (and at lower levels in leaves expressing MT1 alone). These were identified by LC-MS as derivatives of N-methyl anthraniloyl-O-Glc, apparently modified by the addition of a hexose or malate residue, respectively (Figure 6C; see Supplemental Table 2 and Supplemental Figure 6 online) and are likely to have arisen as a result of endogenous N. benthamiana enzyme activities acting on N-methyl anthraniloyl-O-Glc. Confocal microscopy of N. benthamiana leaves suggests that the fluorescent material generated by coexpression of MT1 and UGT74H5 is located in the vacuole (Figure 6D).
Figure 6.
Coexpression of As-MT1 and As-UGT74H5 in Tobacco Leaves Results in the Accumulation and Sequestration of N-Methyl Anthraniloyl-O-Glc.
(A) Immunoblot analysis of soluble protein extracts of N. benthamiana leaves probed with antisera raised against MT1, UGT74H5, and SCPL1. The bottom panel shows a replicate gel stained with Coomassie blue. Lanes were loaded with protein extracted from leaves 6 d after infiltration with Agrobacterium tumefaciens strains containing (from left to right) the P19 silencing suppressor alone (P19) or in combination with the CPMV empty vector (EV); vectors containing the coding sequences of MT1, UGT74H5, or SCPL1 alone or coinfiltrated in combination as indicated.
(B) Leaves of N. benthamiana photographed under UV illumination 6 d after infiltration with Agrobacterium strains.
(C) HPLC analysis with fluorescence detection (excitation at 353 nm; emission at 441 nm) of methanolic extracts from N. benthamiana leaves. Peaks 2 and 3 are N-methyl anthraniloyl-O-Glc and N-methyl anthranilic acid, respectively. Peaks 1 and 4 were found to be derivatives of N-methyl anthraniloyl-O-Glc, which were further modified. Accurate mass determination and fragmentation by MS2 provides putative identification of the additional group on compound 1 (X) as a hexose and on compound 2 (Y) as malate (see Supplemental Figure 6 online). AU, arbitrary units.
(D) Confocal microscopy of lower epidermal cells of leaves (from left to right) uninfiltrated or infiltrated with Agrobacterium strains bearing the CPMV empty vector (EV), the CPMV vector carrying the MT1 (Sad9) coding sequence, or MT1 and UGT74H5. Clockwise from top left for each set of four panels: UV fluorescence, chlorophyll autofluorescence, overlay, and transmitted light. Blue fluorescence is detectable across the central body of cells coexpressing MT1 and UGT74H5 (and more weakly, also from cells expressing MT1 alone), consistent with vacuolar compartmentalization of the fluorescent metabolites. The epidermal cells are highly vacuolated; the chlorophyll autofluorescence shows how the chloroplasts are restricted to the periphery of the cells by the enlarged vacuole. Conversely, weak blue fluorescence in the empty vector control leaves is mainly associated with the cell wall; this may be due to a response to Agrobacterium infiltration. Bars = 20 μm.
Subcellular Localization of the Components of the Avenacin Acylation Pathway
Avenacins accumulate in the vacuoles of the epidermal cells of oat root tips (Mylona et al., 2008). However, the primary shikimate pathway leading to the synthesis of the precursors of the acyl groups is expected to be cytosolic or, more likely, plastidial (Radwanski and Last, 1995; Schmid and Amrhein, 1995). Immunogold labeling of epidermal cells in ultrathin sections of 3-d-old oat root tips was performed using antisera raised against SCPL1, UGT74H5, or MT1 (Figure 7; see Supplemental Table 3 online). The density of gold particles associated with different cellular compartments was determined across replicate samples probed with either specific antisera or the corresponding preimmune sera (see Supplemental Table 3 online). The MT1 antisera principally labeled the cytosol in wild-type oat root epidermal cells. Signal was not detected in the control sad9 mutant line #961 or in samples probed with the corresponding preimmune sera (Figures 7A and 7B; see Supplemental Table 3 online). Sections taken from N. benthamiana leaves expressing MT1 protein were also probed with the anti-AsMT1 antisera (Figures 7C and 7D; see Supplemental Table 3 online). These were found to show strong labeling mainly in the cytosol, whereas empty vector control samples and samples probed with the preimmune sera did not give significant signals (see Supplemental Table 3 online).
Figure 7.
Subcellular Localization of As-MT1, As-UGT74H5, and As-SCPL1.
(A) to (D) Transmission electron microscopy images from immunogold labeling of ultrathin sections probed with antisera raised against MT1.
(A) and (B) Root tip epidermal cells of 3-d-old wild-type (WT) (A) and sad9 (#961) mutant (B) oat seedlings.
(C) and (D) Sections from N. benthamiana leaves expressing the CPMV-AsMT1 construct (C) or the CPMV-EV control (D).
(E) to (G) Sections probed with antisera raised against UGT74H5.
(E) Wild-type oat root tip epidermis.
(F) and (G) N. benthamiana leaves expressing CPMV-AsUGT74H5 (F) or CPMV-EV (G).
(H) and (I) Sections probed with antisera raised against SCPL1. Wild-type (H) or sad7 (#616) (I) mutant oat root tip epidermis.
Gold particle labeling is visible as black dots. Selected subcellular compartments are labeled as follows: C, cytoplasm; V, vacuole; P, plastid; M, mitochondria; and W, cell wall. Images are representative of 10 to 20 biological replicates, and quantification of gold particle densities in different compartments is presented in Supplemental Table 2 online. Bars = 0.5 μm.
Antisera raised against UGT74H5 labeled the cytoplasm and also the vacuole in wild-type oat root samples (Figure 7E) but not in samples probed with the corresponding preimmune sera (see Supplemental Table 3 online) (mutants lacking the UGT74H5 protein are not available). It should be noted that two close homologs of UGT74H5 are also expressed in oat roots and cross-react with the anti-AsUGT74H5 antisera (Owatworakit et al., 2012). To determine the localization of UGT74H5 in the absence of these other proteins, sections of N. benthamiana leaves expressing UGT74H5 were probed with the anti-AsUGT74H5 antisera. Strong labeling of the cytosol but not the vacuole was observed (Figures 7F and 7G; see Supplemental Table 3 online). This labeling was not seen in empty vector controls or in samples probed with the corresponding preimmune sera (see Supplemental Table 3 online). This suggests that UGT74H5 may also be restricted to the cytosol in oat roots and that the signal obtained from the vacuole in oat roots might be attributable to one of the close homologs of UGT74H5.
The SCPL1 antiserum specifically labeled the vacuole in wild-type oat roots. No labeling was seen in sad7 mutants, which lack SCPL1, or in samples probed with preimmune serum (Figures 7H and 7I; see Supplemental Table 3 online). Taken together, our localization experiments suggest a model for the organization of the pathway as shown in Figure 8.
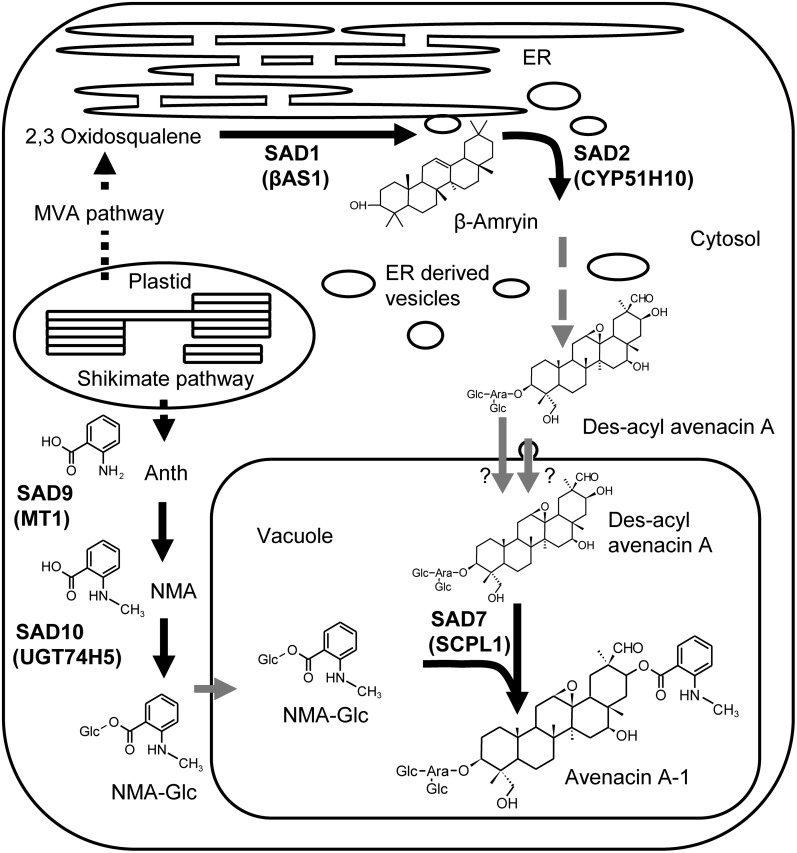
Figure 8.
Model of the Subcellular Organization of Avenacin A-1 Biosynthesis in Oat.
The acyl group of avenacin A-1 originates from the shikimate pathway in the plastid. Anthranilate (Anth) (which fluoresces purple under UV illumination) is methylated by MT1 (encoded by Sad9) to produce N-methyl anthranilate (NMA) (which fluoresces blue) and then glucosylated by UGT74H5 to give N-methyl anthraniloyl-O-glucose (NMA-Glc). NMA-Glc is transported into the vacuole by an unknown mechanism, where it serves as the acyl donor substrate for SCPL1 (SAD7). The triterpene glycoside is synthesized from the cytoplasmic mevalonate (MVA) pathway products, and the early steps are catalyzed by β-amyrin synthase (SAD1/AsβAS1) and CYP51H10 (SAD2). These steps are likely to be associated with the endoplasmic reticulum (ER) membrane (Wegel et al., 2009). Des-acyl avenacin A is then formed by a series of as yet uncharacterized oxidation and glycosylation steps. The triterpene glycoside is transported into the vacuole, either by a vacuolar membrane transporter or possibly through vesicular trafficking from the endoplasmic reticulum to the vacuole.
DISCUSSION
We have shown that three consecutive genes within the avenacin gene cluster are involved in the acylation of avenacins at the C-21 position and that the failure to correctly acylate avenacins results in enhanced disease susceptibility. This is in line with the finding that C-21 acylation is a common feature of many of the most cytotoxic triterpenoid glycosides, suggesting that it is a key contributor to the biological activity of these compounds (Podolak et al., 2010). The identification of a cassette of genes for triterpene acylation is thus of practical significance and opens up opportunities for triterpene modification using metabolic engineering approaches.
The biochemical characterization and localization data presented here suggest a model for the subcellular organization of the triterpene pathway as shown in Figure 8. The shikimate pathway intermediate anthranilate is methylated and subsequently glucosylated in the cytosol, producing the activated acyl donor N-methyl anthraniloyl-O-glucose. This compound is then transported into the vacuole where it serves as a substrate for SCPL1. Evidence for the transport of N-methyl anthraniloyl-O-Glc comes from sad7 mutants, which accumulate this compound, where it can be seen that the fluorescence originates from the vacuole (see Supplemental Figure 7 online). The capacity to transport the acyl donor substrate into the vacuole is also present in N. benthamiana (Figure 6D). The identity of the transporter(s) involved remains to be established. Previous reports of the localization of members of the S-adenosyl Met-dependent methyltransferase and family 1 glucosyltransferase families indicate that most of these enzymes are in the cytosol (Yazaki et al., 1995; Ruelland et al., 2003; Achnine et al., 2005; Dhaubhadel et al., 2008; Hugueney et al., 2009), although members of both families have been reported in other compartments (Halkier and Møller, 1989; Anhalt and Weissenböck, 1992; Ibrahim, 1992; Jones and Vogt, 2001). The localization of the As-SCPL1 acyltransferase to the vacuole is consistent with predictions for this family (Fraser et al., 2005; Mugford et al., 2009) and with experimental data for one other SCPL acyltransferase, which was found to be vacuolar (Hause et al., 2002).
An interesting aspect of the regulation of the avenacin biosynthetic pathway has emerged as a part of this work. We found that the As-SCPL1 acyltransferase protein, which catalyzes the final step in avenacin biosynthesis, was not detectable in mutants that are blocked in early stages of the pathway (sad1 and sad2 mutants) but was detectable in mutants blocked late in the pathway (sad7 and sad9 mutants; Figure 3; see Supplemental Figure 2 online). As-SCPL1 transcript levels were not altered in any of the mutants tested (see Supplemental Figure 2 online). This suggests that the levels of As-SCPL1 may be subject to posttranscriptional regulation, possibly in response to the presence of an intermediate in the avenacin biosynthetic pathway. This regulatory mechanism seems to be specific to As-SCPL1, since other avenacin biosynthetic enzymes are not affected in the same way. This adds another level of regulation to the tight cotranscriptional regulation that restricts the expression of the avenacin gene cluster to root tip epidermal cells.
The mechanisms by which plant natural product gene clusters form are not understood. It has been speculated that the formation of gene clusters encoding plant natural product biosynthetic pathways is associated with the toxicity of pathway intermediates and is driven by selection to prevent partial pathway inheritance (Takos and Rook, 2012). However, we found that the sad9 mutants do not exhibit any obvious growth defects or other abnormalities that might be expected as a result of the accumulation of a toxic intermediate compound. This is similar in the case of other mutants blocked in this pathway, with the exception of the sad3 and sad4 mutants, which accumulate a toxic form of avenacin that is incompletely glucosylated and that disrupts root development (Mylona et al., 2008). While containment of toxic intermediates may contribute to the selection driving the evolution of plant natural product gene clusters, it seems likely that other factors, such as coinheritance of beneficial combinations of alleles of the different pathway genes and coordinated regulation at the level of chromatin, are also likely to be important (Wegel et al., 2009; Chu et al., 2011).
Previously, we reported that several of the avenacin biosynthetic genes bear signatures of recent and strong selective pressure (Qi et al., 2004, 2006; Mugford et al., 2009; Mugford and Osbourn, 2010), consistent with a change of function since the divergence of these sequences from their relatives in other species. It remains to be established whether the neofunctionalization of these enzymes preceded the clustering of their genes or vice versa. In-depth phylogenetic analysis coupled with comparative genomics for all of these large multigene families will be required to address these questions.
METHODS
Plant Material
The wild-type and mutant Avena strigosa lines used in this study are described by Papadopoulou et al. (1999), Qi et al. (2006), and Mugford et al. (2009). Pathogenicity assays were performed as previously described (Papadopoulou et al., 1999).
BAC Contig Expansion and Expression Analysis
The A. strigosa BAC contig spanning the Sad1, Sad2, Sad7, and UGT74H5 (Sad10) genes has been previously described (Qi et al., 2006; Mugford et al., 2009; Owatworakit et al., 2012). Overlapping BAC clones were identified by screening the BAC library with single/low-copy number probes from the ends of BAC clones and verified by PCR and restriction mapping. New BAC clones were verified by shotgun sequencing.
The predicted full-length coding sequence of the Sad9 gene was amplified by PCR and cloned into the pET24a vector (Novagen) with the addition of sequence encoding a 6XHis epitope tag to the C terminus of the protein. The His-tagged Sad9 protein was expressed in Escherichia coli (BL21) and purified by Ni-affinity chromatography and subsequently by gel filtration chromatography according to standard methods. Purified protein (1 mg) was supplied to Biogenes for the production of antisera in rats. Immunoblot analysis was performed as described by Mugford et al., (2009), and the anti-Sad9 antisera was used at a dilution of 1:500.
RNA was extracted and cDNA synthesized as described previously (Qi et al., 2006).
Quantitative real-time PCR was performed using a Bio-Rad CFX96 real-time system C1000 thermal cycler using SYBR Green JumpStart Taq ReadyMix for quantitative PCR (Sigma-Aldrich). Oligonucleotide sequences used were Ef1α, 5′-TCCCCATCTCTGGATTTGAG-3′ and 5′-TCTCTTGGGCTCGTTGATCT-3′; As-SCPL1, 5′-GCTTCACCGTCGAGTATTCC-3′ and 5′-GATCCATCTTCGGACCATGT-3′; and As-MT1, 5′-ACCCCACGACAACATCAAG-3′ and 5′-CGCGGTTCTACTCAAGTGGT-3′.
Phylogenetic Methods
Protein sequence alignment was performed using Muscle 3.6 (Edgar 2004), and the neighbor joining phylogeny was constructed using Mega 4.0 (Tamura et al., 2007). The full sequence alignment used to reconstruct the phylogeny can be found in Supplemental Data Set 1 online. Accession numbers for sequences included in the phylogeny are listed below. The abbreviations used are as follows: CNMT, (S)-coclaurine N-methyltransferase; 4OMT, (S)-3-hydroxy-N-methylcoclaurine 4-O-methyltransferase; 6OMT, (S)-norcoclaurine 6-O-methyltransferase; 7OMT, (R,S)-reticuline 7-O-methyltransferase; BANMT, β-Ala N-methyltransferase; BOMT, benzoic acid carboxyl methyltransferase; CaOMT, catechol O-methyltransferase; ChOMT, chavicol O-methyltransferase; COMT, caffeic acid O-methyltransferase; CoOMT, columbamine O-methyltransferase; DMXNMT, dimethylxanthine N-methyltransferase; EOMT, (iso)eugenol O-methyltransferase; FOMT, flavonoid 8-O-methyltransferase; MXNMT, 7-methylxanthine N-methyltransferase; PEANMT, phosphoethanolamine N-methyltransferase; PNMT, putrescine N-methyltransferase; SAMT, salicylic acid carboxyl methyltransferase; SOMT, scoulerine 9-O-methyltransferase; and XNMT, xanthosine methyltransferase.
sad9 Mutant Identification and Metabolite Analysis
Previously uncharacterized avenacin-deficient A. strigosa mutants (Papadopoulou et al., 1999; Qi et al., 2006) were screened for single nucleotide polymorphisms in the Sad9 gene using the Surveyor mutation detection kit (Transgenomic) according to the manufacturer’s protocol and candidates verified by sequencing. Metabolites were extracted from root tips of 5-d-old seedlings as described previously (Mugford et al., 2009). Fluorescent compounds were analyzed by LC-MS, coupled with fluorescence detection (Mugford et al., 2009). Glc esters of anthranilate and N-methyl anthranilate were quantified by HPLC (also described in Mugford et al., 2009).
Biochemical Characterization of As-MT1
Activity assays were performed using 4 μg of purified recombinant As-MT1 protein in 50 mM Tris-HCl, pH 8.0, and 2.5 mM S-adenosyl-Met, in a volume of 200 μL at 30°C. Reactions were started by the addition of the methyl acceptor substrate and stopped after 7 min by the addition of 10 μL of trichloro-acetic acid (TCA). For the determination of kinetic parameters, activity was tested at anthranilate concentrations ranging from 0.05 to 7.5 mM and anthraniloyl-O-Glc from 0.5 to 5 mM, each tested in triplicate. Concentrations of methylated product were determined by HPLC using a Dionex UltiMate 3000 HPLC with a Luna C18 160 × 4, 60-mm column (Phenomenex) running a gradient of acetonitrile against 0.1% trifluoroacetic acid in water from 10 to 90% over 22 min. The flow rate was constant at 0.5 mL/min, the injection volume was 60 μL, and the detection wavelength used was 320 nm. Data were plotted as a Lineweaver-Burk plot, and Michaelis-Menten kinetic parameters were determined.
CPMV-Based Coexpression
The coding sequences of the As-MT1, As-SCPL1, and As-UGT74H5 cDNAs were amplified by PCR and cloned into the modified pM81S2NT plasmid (Cañizares et al., 2006) to flank the sequences by the 5′ and 3′ untranslated regions of CPMV RNA-2 prior to transfer to pBINPLUS. Expression from the cassette was under control of the cauliflower mosaic virus 35S promoter. Constructs were transformed into Agrobacterium tumefaciens LBA4404 and infiltrated into Nicotiana benthamiana leaves as described previously (Cañizares et al., 2006; Mugford et al., 2009). For coexpression experiments, Agrobacterium cultures carrying each of the three constructs were mixed prior to infiltration or were diluted in infiltration medium for expression alone. Leaves were harvested after 6 d and snap-frozen in liquid nitrogen or photographed under UV illumination. Metabolite extracts and immunoblot analysis was performed as described previously (Mugford et al., 2009). Accurate mass identification of unknown metabolites was performed by LC-MS. Compounds were separated on a 50 × 2-mm 3-µ Luna C18(2) column (Phenomenex) on a Surveyor HPLC system (Thermo Finnigan) using a gradient of methanol versus 0.1% formic acid in water from 10 to 80% over 20 min. The column outflow was connected directly to an Orbitrap mass spectrometer electrospray spray chamber (Thermo Finnigan) using spray chamber conditions of 250°C capillary temperature, 32 units sheath gas, 6 units aux gas, 4.3-kV spray voltage, and tube lens voltage of 40 V. The Orbitrap was set up to collect full mass spectra at a resolution of 60,000 from mass-to-charge ratio 160 to 1500 and to collect data dependent MS2 at a resolution of 15,000. MS2 was collected by Fourier Transform Mass Spectrometry (FTMS). MS2 was performed by collision-induced dissociation at 35% collision energy and with an isolation width of 2.0.
Confocal microscopy was performed on leaf sections from infiltrated plants and on whole oat roots using a Zeiss LSM510 microscope, with illumination from a violet diode laser at 405 nm.
Immunostaining
For immunofluorescence staining, roots were harvested from 5-d-old seedlings, fixed in 2% formaldehyde, and sectioned on a vibratome (50 µm). Sections were digested for 1 h with 1% driselase, 0.5% cellulose, and 0.025% pectolyase (Duchefa), preincubated in 3% BSA in tris-buffered saline (TBS) for 30 min, and then incubated for 1 h with the antisera (or preimmune sera). Anti-AsMT1 antisera was used at 1:200 dilution and anti-AsSCPL1 antisera at 1:1000 dilution. Incubations were performed overnight at 4°C, followed by washing in TBS. Sections were then incubated in a 1:200 dilution of fluorescent dye–conjugated secondary antibodies (Life Technologies; goat anti-rabbit-alexa488 for As-SCPL1 and goat anti-rat-alexa456 for As-MT1) in 3% BSA in TBS for 1 h, washed in TBS, stained in 1 µg/mL 4′,6-diamidino-2-phenylindole for 5 min, and imaged using a Nikon Eclipse e600 microscope.
Tissue fixation for immunogold labeling was performed as by Simpson et al. (2009).
Primary antibodies were used at the following dilutions: 1/100 for anti-AsMT1; 1/200 for anti-AsUGT74H5 and 1/500 for anti-AsSCPL1, with 1/50 goat anti-rabbit (for As-UGT74H5 and As-SCPL1) or goat anti-rat (for As-MT1) secondary antibody conjugated to 10-nm gold particles (BioCell, Agar Scientific). The grids were viewed in a Tecnai 20 transmission electron microscope (FEI) at 200 kV, and digital TIFF images were taken using an AMT XR60B digital camera (Deben).
Accession Numbers
The sequence of the Sad9 gene can be found in GenBank under accession number JQ071450. Sequences included in the phylogenetic tree in Figure 2 are as follows: Thalictrum flavum 6OMT, AAU20765; Coptis japonica 6OMT, BAB08004; Papaver somniferum 6OMT, AAQ01669; P. somniferum 4OMT, AAP45313; T. flavum 4OMT, AAU20768; C. japonica 4OMT, BAB08005; C. japonica CoOMT, BAC22084; A. strigosa MT1, JQ071450; Mentha x piperita FOMT, AY337459; Ocimum basilicum EOMT, AAL30424; O. basilicum ChOMT, AAL30423; P. somniferum 7OMT, AAQ01668; Limonium latifolium BANMT, AAP03058; T. flavum SOMT, AAU20770; C. japonica SOMT, BAA06192; Ammi majus COMT, AAR24095; R. graveolens ANMT, DQ884932; P. somniferum CaOMT, AAQ01670; O. basilicum COMT1, AAD38189; O. basilicum COMT2, AF154918; Clarkia breweri EOMT, AAC01533; C. breweri COMT, O23760; Atropa belladonna PNMT, BAA82264; Datura stramonium PNMT, CAE47481; Solanum lycopersicum PEANMT, AAG59894; Coffea arabica DMXNMT1, BAC75663; C. arabica MXNMT, BAB39216; C. arabica XNMT, BAB39215; Camellia sinensis DMXNMT, BAB12278; Antirrhinum majus BOMT, AAF98284; C. breweri SAMT, AAF00108; Mesorhizobium loti CFAPS, BAB53730; Arabidopsis At4g33110, AAM65762; Arabidopsis At4g33120, ORF NP 195038; P. somniferum CNMT, AAP45316; T. flavum CNMT, AAU20766; and C. japonica CNMT, BAB71802.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Wild-Type Oat Root Sections Probed with Preimmune Sera.
Supplemental Figure 2. Identification of Des-Methyl Avenacin A-1 and Anthraniloyl-O-Glc by LC-MS.
Supplemental Figure 3. 1H-NMR Spectra of the Avenacin C30-Aldehyde Region in Oat Root Extracts.
Supplemental Figure 4. Acylation of Avenacins Is Important for Biological Activity.
Supplemental Figure 5. As-SCPL1 Expression Is Reduced or Abolished in Oat Mutants Blocked in the Early Steps in the Synthesis of the Avenacin Triterpene.
Supplemental Figure 6. LC-MS Analysis of Fluorescent Compounds from N. benthamiana Plants Coexpressing As-MT1 and As-UGT74H5.
Supplemental Figure 7. Fluorescent Compounds Accumulate in the Vacuole of sad7 Mutants.
Supplemental Table 1. Quantification of Avenacins and Des-Methyl Avenacin A-1 in Wild-Type and Mutant Oat Root Tips.
Supplemental Table 2. Quantification of Fluorescent Compounds in N. benthamiana Leaves Coexpressing Avenacin Biosynthetic Enzymes.
Supplemental Table 3. Subcellular Localization of the As-MT1, As-UGT74H5, and As-SCPL1 Proteins.
Supplemental Data Set 1. Protein Sequence Alignment Used in Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Kim Findlay and Sue Bunnewell for immunogold labeling and transmission electron microscopy, Grant Calder for confocal microscopy, and Ian Colquhoun for NMR. This project was funded by the Biotechnology and Biological Sciences Research Council, UK (Grant BB/E009912/1 to S.T.M.), by a joint Engineering and Physical Sciences Research Council/National Science Foundation award to A.O. and S.J.R. as part of the Syntegron Consortium (EP/H019154/1), and by the University of Glasgow work placement program (S.H.). R.M., L.H., G.P.L., and A.O. are supported by the Biotechnology and Biological Sciences Research Council Institute Strategic Program Grant “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004561/1) and the John Innes Foundation.
AUTHOR CONTRIBUTIONS
S.T.M, T.L., R.M., X.Q., S.B., T.T., L.H., and S.H. performed the research. S.T.M., X.Q., G.P.L, S.J.R., and A.O. designed the research. S.T.M. and A.O. wrote the article.
Glossary
- SCPL
Ser carboxypeptidase-like
- LC-MS
liquid chromatography–mass spectrometry
- DMA
des-methyl avenacin A-1
- MS2
tandem mass spectrometry
- CPMV
cowpea mosaic virus
References
- Achnine L., Huhman D.V., Farag M.A., Sumner L.W., Blount J.W., Dixon R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41: 875–887 [DOI] [PubMed] [Google Scholar]
- Anhalt S., Weissenböck G. (1992). Subcellular localization of luteolin glucuronides and related enzymes in rye mesophyll. Planta 187: 83–88 [DOI] [PubMed] [Google Scholar]
- Baumert A., Milkowski C., Schmidt J., Nimtz M., Wray V., Strack D. (2005). Formation of a complex pattern of sinapate esters in Brassica napus seeds, catalysed by enzymes of a serine carboxypeptidase-like acyltransferase family. Phytochemistry 66: 1334–1345 [DOI] [PubMed] [Google Scholar]
- Cañizares M.C., Liu L., Perrin Y., Tsakiris E., Lomonossoff G.P. (2006). A bipartite system for the constitutive and inducible expression of high levels of foreign proteins in plants. Plant Biotechnol. J. 4: 183–193 [DOI] [PubMed] [Google Scholar]
- Chu H.Y., Wegel E., Osbourn A. (2011). From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 66: 66–79 [DOI] [PubMed] [Google Scholar]
- Crombie L., Crombie W.M.L., Whiting D.A. (1984). Structure of the four avenacins, oat root resistance factors to take-all disease. J. Chem. Soc. Chem. Commun. 4: 246–248 [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S., Farhangkhoee M., Chapman R. (2008). Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 59: 981–994 [DOI] [PubMed] [Google Scholar]
- Dixon R.A. (2001). Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Dudareva N., D’Auria J.C., Nam K.H., Raguso R.A., Pichersky E. (1998). Acetyl-CoA:benzylalcohol acetyltransferase—An enzyme involved in floral scent production in Clarkia breweri. Plant J. 14: 297–304 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V., Akhtar T.A., Nguyen T.T., Spyropoulou E.A., Bleeker P.M., Schauvinhold I., Matsuba Y., Bonini M.E., Schilmiller A.L., Last R.L., Schuurink R.C., Pichersky E. (2011). The tomato terpene synthase gene family. Plant Physiol. 157: 770–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Fiston-Lavier A.-S., Kemen A., Geisler K., Quesneville H., Osbourn A.E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Fraser C.M., Rider L.W., Chapple C. (2005). An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol. 138: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M., Thompson M.G., Shirley A.M., Ralph J., Schoenherr J.A., Sinlapadech T., Hall M.C., Chapple C. (2007). Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Tanaka Y., Yonekura-Sakakibara K., Fukuchi-Mizutani M., Nakao M., Fukui Y., Yamaguchi M., Ashikari T., Kusumi T. (1998). cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J. 16: 421–431 [DOI] [PubMed] [Google Scholar]
- Gang D.R., Lavid N., Zubieta C., Chen F., Beuerle T., Lewinsohn E., Noel J.P., Pichersky E. (2002). Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Frey M. (2001). Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213: 493–498 [DOI] [PubMed] [Google Scholar]
- Grienenberger E., Besseau S., Geoffroy P., Debayle D., Heintz D., Lapierre C., Pollet B., Heitz T., Legrand M.A.F. (2009). A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J. 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Møller B.L. (1989). Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol. 90: 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralampidis K., Bryan G., Qi X., Papadopoulou K., Bakht S., Melton R., Osbourn A. (2001). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. USA 98: 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B., Meyer K., Viitanen P.V., Chapple C., Strack D. (2002). Immunolocalization of 1-O-sinapoylglucose:malate sinapoyltransferase in Arabidopsis thaliana. Planta 215: 26–32 [DOI] [PubMed] [Google Scholar]
- Hostettmann, K., and Marston, A. (1995). Saponins: Chemistry and Pharmacology of Natural Products. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Hugueney P., Provenzano S., Verriès C., Ferrandino A., Meudec E., Batelli G., Merdinoglu D., Cheynier V., Schubert A., Ageorges A. (2009). A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 150: 2057–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, R.K. (1992). Immunolocalization of flavonoid conjugates and their enzymes. In Phenolic Metabolism in Plants, H.A. Stafford and R.K. Ibrahim, eds (New York: Plenum Press), pp. 25–61. [Google Scholar]
- Jonczyk R., Schmidt H., Osterrieder A., Fiesselmann A., Schullehner K., Haslbeck M., Sicker D., Hofmann D., Yalpani N., Simmons C., Frey M., Gierl A. (2008). Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol. 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Vogt T. (2001). Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 213: 164–174 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Osbourn A. (2012). Making new molecules - Evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 15: 415–423 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C., Shirley A.M., Meyer K., Ruegger M.O., Cusumano J.C., Viitanen P.V., Strack D., Chapple C. (2000). Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.X., Steffens J.C. (2000). An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc. Natl. Acad. Sci. USA 97: 6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.K., Li Y., Parr A., Jackson R., Ashford D.A., Bowles D.J. (2001). Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Liscombe D.K., Facchini P.J. (2007). Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282: 14741–14751 [DOI] [PubMed] [Google Scholar]
- Luo J., Fuell C., Parr A., Hill L., Bailey P., Elliott K., Fairhurst S.A., Martin C., Michael A.J. (2009). A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Milkowski C., Baumert A., Schmidt D., Nehlin L., Strack D. (2004). Molecular regulation of sinapate ester metabolism in Brassica napus: Expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J. 38: 80–92 [DOI] [PubMed] [Google Scholar]
- Milkowski C., Strack D. (2004). Serine carboxypeptidase-like acyltransferases. Phytochemistry 65: 517–524 [DOI] [PubMed] [Google Scholar]
- Mugford S.T., Milkowski C. (2012). Serine carboxypeptidase-like acyltransferases from plants. Methods Enzymol. 516: 279–297 [DOI] [PubMed] [Google Scholar]
- Mugford S.T., Osbourn A. (2010). Evolution of serine carboxypeptidase-like acyltransferases in the monocots. Plant Signal. Behav. 5: 193–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford S.T., et al. (2009). A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P., Owatworakit A., Papadopoulou K., Jenner H., Qin B., Findlay K., Hill L., Qi X., Bakht S., Melton R., Osbourn A. (2008). Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A., Goss R.J.M., Field R.A. (2011). The saponins: Polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 28: 1261–1268 [DOI] [PubMed] [Google Scholar]
- Osbourn, A.E., and Lanzotti, V. (2009). Plant-Derived Natural Products. (New York: Springer). [Google Scholar]
- Owatworakit, A., et al. (2012). Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. J. Biol. Chem. 288: 3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K., Melton R.E., Leggett M., Daniels M.J., Osbourn A.E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 96: 12923–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolak I., Galanty A., Sobolewska D. (2010). Saponins as cytotoxic agents: A review. Phytochem. Rev. 9: 425–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bakht S., Qin B., Leggett M., Hemmings A., Mellon F., Eagles J., Werck-Reichhart D., Schaller H., Lesot A., Melton R., Osbourn A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 103: 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B., Eagles J., Mellon F.A., Mylona P., Peña-Rodriguez L.M., Osbourn A.E. (2010). High throughput screening of mutants of oat that are defective in triterpene synthesis. Phytochemistry 71: 1245–1252 [DOI] [PubMed] [Google Scholar]
- Radwanski E.R., Last R.L. (1995). Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 7: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde B., Hans J., Martens S., Baumert A., Hunziker P., Matern U. (2008). Anthranilate N-methyltransferase, a branch-point enzyme of acridone biosynthesis. Plant J. 53: 541–553 [DOI] [PubMed] [Google Scholar]
- Ruelland E., Campalans A., Selman-Housein G., Puigdomènech J., and Rigaul, J. (2003). Cellular and subcellular localization of the lignin biosynthetic enzymes caffeic acid-O-methyltransferase, cinnamyl alcohol dehydrogenase and cinnamoyl-coenzyme A reductase in two monocots, sugarcane and maize. Physiol. Plant. 117: 93–99 [Google Scholar]
- Sainsbury F., Lavoie P.-O., D’Aoust M.-A., Vézina L.-P., Lomonossoff G.P. (2008). Expression of multiple proteins using full-length and deleted versions of cowpea mosaic virus RNA-2. Plant Biotechnol. J. 6: 82–92 [DOI] [PubMed] [Google Scholar]
- Schmid J., Amrhein N. (1995). Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39: 737–749 [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Shirley A.M., Chapple C. (2003). Biochemical characterization of sinapoylglucose:choline sinapoyltransferase, a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. J. Biol. Chem. 278: 19870–19877 [DOI] [PubMed] [Google Scholar]
- Shirley A.M., McMichael C.M., Chapple C. (2001). The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J. 28: 83–94 [DOI] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle F., Stubbs M.T., Strack D., Milkowski C. (2008). Heterologous expression of a serine carboxypeptidase-like acyltransferase and characterization of the kinetic mechanism. FEBS J. 275: 775–787 [DOI] [PubMed] [Google Scholar]
- St-Pierre B., Laflamme P., Alarco A.M., De Luca V. (1998). The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 14: 703–713 [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Morrone D., Wang Q., Fulton D.B., Peters R.J. (2009). CYP76M7 is an ent-cassadiene C11alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A.M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M.S., Olsen C.E., Sato S., Tabata S., Jørgensen K., Møller B.L., Rook F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68: 273–286 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Rook F. (2012). Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 17: 383–388 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Walker K., Long R., Croteau R. (2002). The final acylation step in taxol biosynthesis: Cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc. Natl. Acad. Sci. USA 99: 9166–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pichersky E. (1999). Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch. Biochem. Biophys. 368: 172–180 [DOI] [PubMed] [Google Scholar]
- Wilderman P.R., Xu M., Jin Y., Coates R.M., Peters R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E., Koumproglou R., Shaw P., Osbourn A. (2009). Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier D., Mittasch J., Strack D., Milkowski C. (2008). The genes BnSCT1 and BnSCT2 from Brassica napus encoding the final enzyme of sinapine biosynthesis: Molecular characterization and suppression. Planta 227: 375–385 [DOI] [PubMed] [Google Scholar]
- Winzer T., et al. (2012). A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Yang Q., Reinhard K., Schiltz E., Matern U. (1997). Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol. Biol. 35: 777–789 [DOI] [PubMed] [Google Scholar]
- Yazaki K., Inushima K., Kataoka M., Tabata M. (1995). Intracellular localization of UDPG:p-hydroxybenzoate glucosyltransferase and its reaction product in Lithospermum cell cultures. Phytochemistry 38: 1127–1130 [Google Scholar]
- Yu X.-H., Chen M.-H., Liu C.-J. (2008). Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J. 55: 382–396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.