S-acylation or palmitoylation, catalyzed by protein S-acyl transferases (PATs), regulates protein targeting and function. This work shows that Arabidopsis PAT10 regulates pleiotropic developmental processes and stress responses. Putative PAT10 substrates include several calcineurin B-like proteins, whose tonoplast association depends on the function of PAT10.
Abstract
Protein S-acylation, commonly known as palmitoylation, is a reversible posttranslational modification that catalyzes the addition of a saturated lipid group, often palmitate, to the sulfhydryl group of a Cys. Palmitoylation regulates enzyme activity, protein stability, subcellular localization, and intracellular sorting. Many plant proteins are palmitoylated. However, little is known about protein S-acyl transferases (PATs), which catalyze palmitoylation. Here, we report that the tonoplast-localized PAT10 is critical for development and salt tolerance in Arabidopsis thaliana. PAT10 loss of function resulted in pleiotropic growth defects, including smaller leaves, dwarfism, and sterility. In addition, pat10 mutants are hypersensitive to salt stresses. We further show that PAT10 regulates the tonoplast localization of several calcineurin B–like proteins (CBLs), including CBL2, CBL3, and CBL6, whose membrane association also depends on palmitoylation. Introducing a C192S mutation within the highly conserved catalytic motif of PAT10 failed to complement pat10 mutants, indicating that PAT10 functions through protein palmitoylation. We propose that PAT10-mediated palmitoylation is critical for vacuolar function by regulating membrane association or the activities of tonoplast proteins.
INTRODUCTION
S-acylation is a eukaryotically conserved, reversible posttranslational modification catalyzing the addition of a saturated acyl group to the sulfhydryl group of a Cys to form a thioester (Baekkeskov and Kanaani, 2009; Greaves and Chamberlain, 2011). The 16-carbon palmitate is the most common fatty acid added through S-acylation in animals (Greaves and Chamberlain, 2011), although other lipids with different chain lengths, such as stearate, are often added in plants (Sorek et al., 2007; Batistic et al., 2008). Historically and adopted hereafter, palmitoylation is used to refer to S-acylation. Palmitoylation regulates membrane association of soluble proteins, partitioning of transmembrane proteins into lipid rafts, protein activity, protein trafficking into different membrane compartments, as well as protein stability and turnover (Hemsley and Grierson, 2008; Baekkeskov and Kanaani, 2009; Greaves and Chamberlain, 2011). In yeast and animals, proteins subject to palmitoylation, such as transmembrane receptors, small GTPases, and Ca2+ sensors, usually play critical roles in cell signaling. Furthermore, proteomic studies in yeast and animal cells showed that a large number of proteins are palmitoylated (Roth et al., 2006; Kang et al., 2008), implying that palmitoylation may play more important roles in eukaryotic cells than currently appreciated.
Despite a long period of doubt on whether palmitoylation necessitates specific enzymes, there are three types of protein S-acyl transferases currently known to catalyze palmitoylation, including membrane-bound O-acyltransferases, longin domain proteins and Asp-His-His-Cys motif Cys-rich domain (DHHC-CRD) protein S-acyl transferases (PATs) (Greaves and Chamberlain, 2011), among which the DHHC-CRD–type PATs are the most well studied. PATs are present in all eukaryotic genomes as relatively large families, including seven in yeast, 23 in human, and 24 in Arabidopsis thaliana (Hemsley et al., 2005; Greaves and Chamberlain, 2011; Batistic, 2012). PATs typically contain four to six transmembrane domains and the highly conserved DHHC catalytic domain (Hemsley et al., 2005; Hemsley and Grierson, 2008; Greaves and Chamberlain, 2011; Batistic, 2012). Each eukaryotic genome additionally encodes one to two PATs with ankyrin repeat domains (Hemsley et al., 2005; Hemsley and Grierson, 2008; Greaves and Chamberlain, 2011; Batistic, 2012).
An increasing number of plant proteins are found to be regulated by palmitoylation. Palmitoylation regulates membrane anchoring of small GTPase ARA6 (Ueda et al., 2001), protein phosphatases (Gagne and Clark, 2010), heterotrimeric G protein subunits (Zeng et al., 2007), and a few calcium sensors (Martín and Busconi, 2000; Batistic and Kudla, 2009; Batistič et al., 2012). Palmitoylation also regulates microdomain partitioning and possibly subcellular trafficking of Rho of plants (ROP GTPases) (Sorek et al., 2007, 2010). A recent proteomic analysis using Arabidopsis root cell culture identified over 500 proteins to be putatively S-acylated (Hemsley et al., 2013), thus establishing palmitoylation as an important posttranslational modification in plants. Palmitoylated proteins play diverse roles in plant cells, including development (Gagne and Clark, 2010), vesicle trafficking (Ueda et al., 2001), cell morphogenesis (Sorek et al., 2010), and abiotic stress responses (Batistič et al., 2012).
Despite the importance of palmitoylation in plants (Hemsley and Grierson, 2008), progress in understanding the enzymes catalyzing protein palmitoylation has been slow. Arabidopsis TIP GROWTH DEFECTIVE1 (TIP1) is the only plant PAT with characterized function to date. TIP1 loss of function resulted in defective growth of pollen tubes and root hairs (Hemsley et al., 2005). In contrast with TIP1, which encodes an Arabidopsis PAT with the ankyrin domains, functional analysis of other PATs by reverse genetics was considered problematic due to potential functional redundancy (Hemsley and Grierson, 2008) implied by their similar domain organization and overlapping expression patterns (Zimmermann et al., 2004). However, PATs encode transmembrane proteins whose functional specificity is not only determined by their expression patterns but also by their residential membrane compartments (Greaves and Chamberlain, 2011). Indeed, global analysis of yeast and mammalian PATs showed that PATs of a given species could localize at various membrane compartments, including endoplasmic reticulum, Golgi apparatus, plasma membrane, secretory vesicles, and vacuole (Ohno et al., 2006). A recent study using transient expression assays in tobacco (Nicotiana tabacum) showed that Arabidopsis PATs were also localized at different membrane compartments (Batistic, 2012).
We report here functional analysis of PAT10 in Arabidopsis. PAT10 is constitutively expressed. Functional loss of PAT10 resulted in pleiotropic developmental defects, including reduced vegetative and reproductive growth, sporophytic and gametophytic male defects, and compromised ability of pistils to support pollen tube growth. Both cell expansion and cell division were reduced in pat10. Using stable transgenic plants for fluorescent colabeling, we showed that PAT10 localizes at vacuolar membrane (i.e., the tonoplast) but not at Golgi or any post-Golgi vesicles. In addition to the developmental defects, mutants of PAT10 were hypersensitive to salt stresses. We further identified a subfamily of calcineurin B–like proteins (CBLs) as putative substrates of PAT10. Localization of CBL2, CBL3, and CBL6 at the tonoplast depended on functional PAT10. Our results demonstrate that PAT10-mediated protein palmitoylation at the tonoplast is critical for development and salt tolerance in Arabidopsis.
RESULTS
Characterization of PAT10 Loss-of-Function Mutants
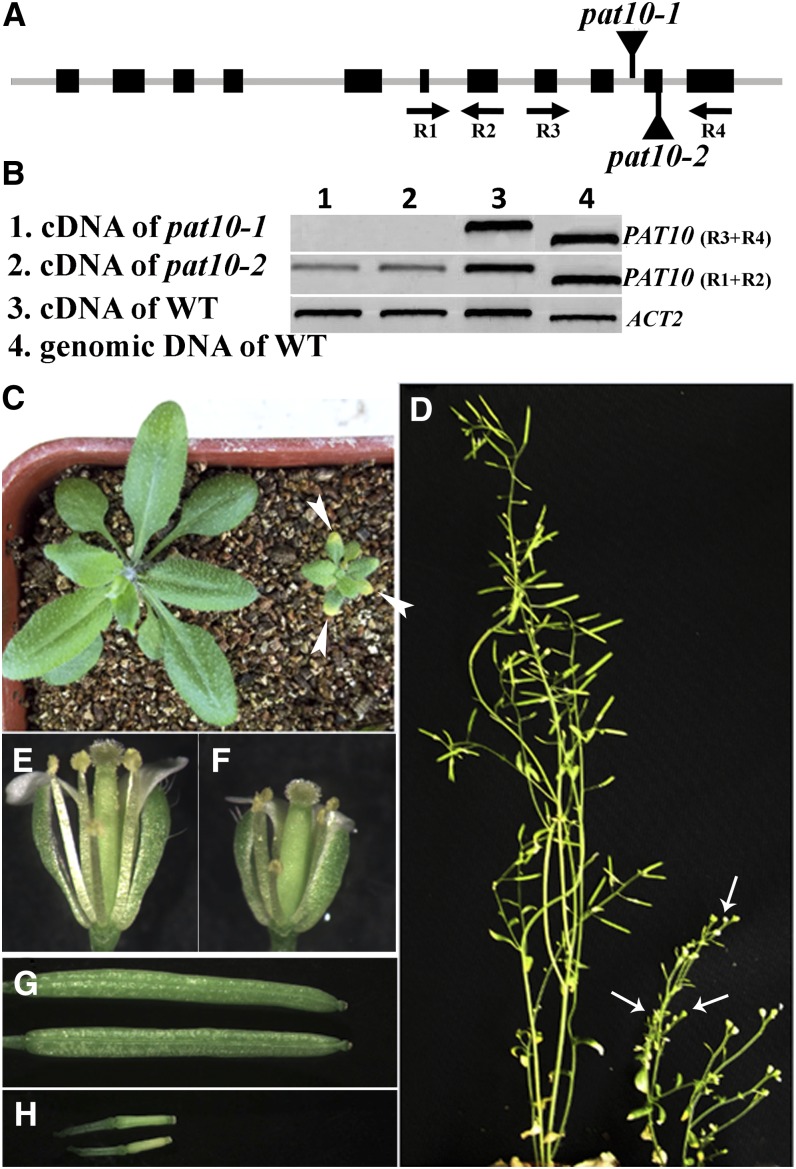
We used a reverse genetic approach to understand the function of Arabidopsis PATs and identified two T-DNA insertion lines for PAT10. T-DNA was inserted at the 10th intron or the 10th exon of PAT10 genomic sequence for pat10-1 (SALK_024964) and pat10-2 (WiscDsLox289_292E10), respectively. By PCR analysis, we confirmed that the genomic sequences upstream and downstream of the insertion sites were intact in mutants. To find out whether the T-DNA insertions disrupted PAT10 expression, we performed RT-PCR analysis on RNAs isolated from seedlings of the wild type and pat10 mutants. RT-PCRs with a primer pair that amplified upstream sequences of T-DNA insertions detected PAT10 transcript in both the wild type and pat10. However, no transcript was detected in pat10 mutants when primer pairs flanking the T-DNA insertion sites were used (Figures 1A and 1B), suggesting that pat10 mutants were not able to produce full-length PAT10. Experiments using both pat10-1 and pat10-2 produced identical results. Therefore, results reported hereafter were those obtained with pat10-1 unless noted.
Figure 1.
Mutations at PAT10 Caused Pleiotropic Developmental Defects.
(A) Schematic illustration of T-DNA insertions within the PAT10 genomic region.
(B) RT-PCR analysis showing that pat10 mutants are devoid of full-length PAT10 transcripts. WT, the wild type.
(C) A representative image of the wild type (left) and pat10 (right) at 20 DAG. Arrowheads indicate necrotic leaf tips in the mutant plant.
(D) A representative image of the wild type (left) and pat10 (right) at 50 DAG. Arrows point to nonelongated siliques.
(E) A representative image of wild-type open flowers.
(F) A representative image of pat10 open flowers.
(G) Fully elongated siliques in the wild type.
(H) Nonelongated siliques in pat10.
Homozygous pat10 mutants displayed pleiotropic developmental defects compared with the wild type under soil growth condition. pat10 was substantially smaller and developed more slowly. As a result, growth differences became more pronounced over time. Rosette leaves of pat10 were much smaller than those of the wild type at 20 d after germination (DAG) (Figure 1C), although leaf shape was normal (Figure 1C). The size reduction was due to reduced cell number as well as reduced cell expansion (see Supplemental Figure 1 online). In addition, pat10 showed early senescence on the leaf tips when growing in soil at 20 DAG (Figure 1C). During floral transition, pat10 showed no correlation between flowering time and rosette leaf number such that they flowered temporally later but had fewer rosette leaves than the wild type (see Supplemental Figure 2 online), likely a consequence of slow development. Inflorescence stems were much shorter for pat10 than for the wild type (Figure 1D). Mutant flowers (Figure 1F) were also smaller than those of the wild type (Figure 1E). Although stamen filaments of pat10 were shorter than the wild type, pistils of pat10 mutants were also shorter (Figures 1E and 1F), allowing reaching of stigma surface by pollen. pat10 growing in soil were completely sterile (Figure 1D) without elongating siliques (Figure 1H).
Sporophytic and Gametophytic Male Defects in pat10
Although we were able to obtain homozygous pat10 mutants from self-pollinated heterozygous pat10, the ratio of homozygous to heterozygous mutants was much lower than expected (Table 1). This suggested that the loss of PAT10 led to gametophytic defects. To find out which gametophyte was defective due to PAT10 loss of function, we performed reciprocal crosses between heterozygous pat10 mutants and the wild type. Results showed that pollen of pat10 did not transmit as well as that of the wild type, while female gametophytes were not affected (Table 1). These results suggested that the defective male gametophytes were caused by PAT10 loss of function.
Table 1. Loss of Function of PAT10 Resulted in Defective Male Transmission.
| Experiment Parent | F1 Segregation | ||
|---|---|---|---|
| Female × Male | Genotype | Expected Ratio | Observed |
| 1. pat10-1+/− × wild type | PAT10+/+: +/− | 1:1 | 90:98a |
| 2. Wild type × pat10-1+/− | PAT10+/+: +/− | 1:1 | 121:7b |
| 3. pat10-2+/− × wild type | PAT10+/+: +/− | 1:1 | 96:71a |
| 4. Wild type × pat10-2+/− | PAT10+/+: +/− | 1:1 | 160:55b |
| 5. pat10-1+/− × pat10-1+/− | PAT10+/+:+/−:−/− | 1:2:1 | 115:208:50c |
| 6. pat10-2+/− × pat10-2+/− | R:Sd | 3:1 | 507:228e |
Not significantly different from the segregation ratio 1:1 (Student’s t test, P > 0.05).
Significantly different from the segregation ratio 1:1 (Student’s t test, P < 0.01).
Significantly different from the segregation ratio 1:2:1 (Student’s t test, P < 0.01).
R for Basta resistant and S for Basta sensitive. The mutant copy of pat10-2 is resistant to Basta salt; therefore, Basta resistance was calculated here.
Significantly different from the segregation ratio 3:1 (Student’s t test, P < 0.01).
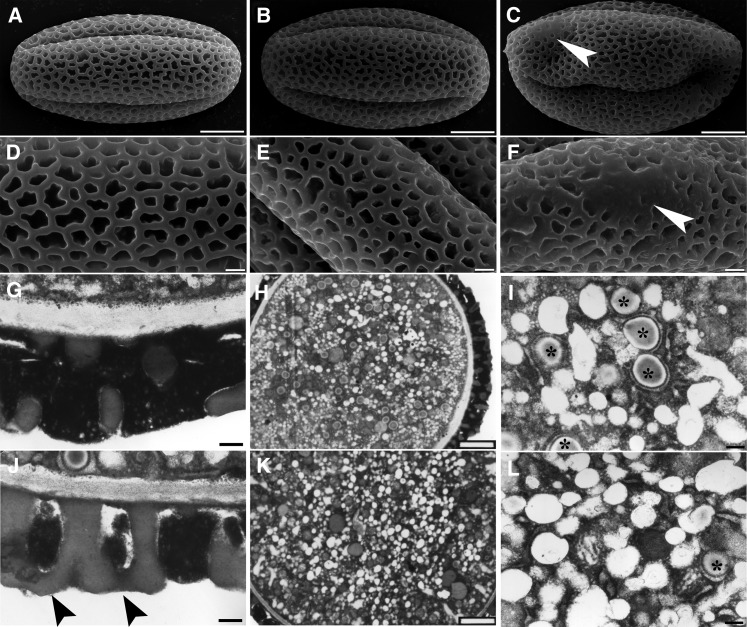
To find out at which stage male gametophytes of pat10 was defective, we next analyzed pollen viability using 4',6-diamidino-2-phenylindole and Alexander’s stain. No differences were observed regarding pollen viability between the wild type and pat10 (see Supplemental Figure 3 online), suggesting that PAT10 did not regulate pollen viability. However, scanning electron microscopy (SEM) showed that the deposition of the pollen coat was abnormal in homozygous pat10 mutants (Figures 2C and 2F), even though no difference was observed in the pollen coats of heterozygous pat10 plants (Figures 2B and 2E) versus in the wild type (Figures 2A and 2D). Because deposition of the pollen coat depends on the sporophytic tapetum, we concluded that PAT10 loss of function also caused sporophytic male defects. Transmission electron microscopy (TEM) further confirmed the pollen coat defect of pat10 (Figure 2J), whereas pat10 pollen contained fewer lipid bodies (Figure 2L) than the wild type (Figure 2I).
Figure 2.
Mutations at PAT10 Caused Pollen Developmental Defects.
(A) to (C) Scanning electron micrographs (SEM) of mature pollen from the wild type (A), heterozygous pat10 mutants (B), and homozygous pat10 mutants (C). Arrowhead indicates defective pollen coat.
(D) to (F) Close-ups of SEM images shown in (A) to (C), respectively. Arrowhead indicates defective pollen coat.
(G) and (J) Transmission electron micrograph (TEM) of mature pollen from the wild type (G) and pat10 (J) showing pollen coat structure. Arrowheads indicate defective pollen coats.
(H) and (K) TEM section of mature pollen from the wild type (H) and pat10 (K).
(I) and (L) Close-ups of TEM shown in (H) and (K), respectively. Asterisks indicate lipid bodies.
Bars = 5 µm in (A) to (C), 1 µm in (D) to (F), 2 µm in (H) and (K), and 200 nm in (G), (I), (J), and (L).
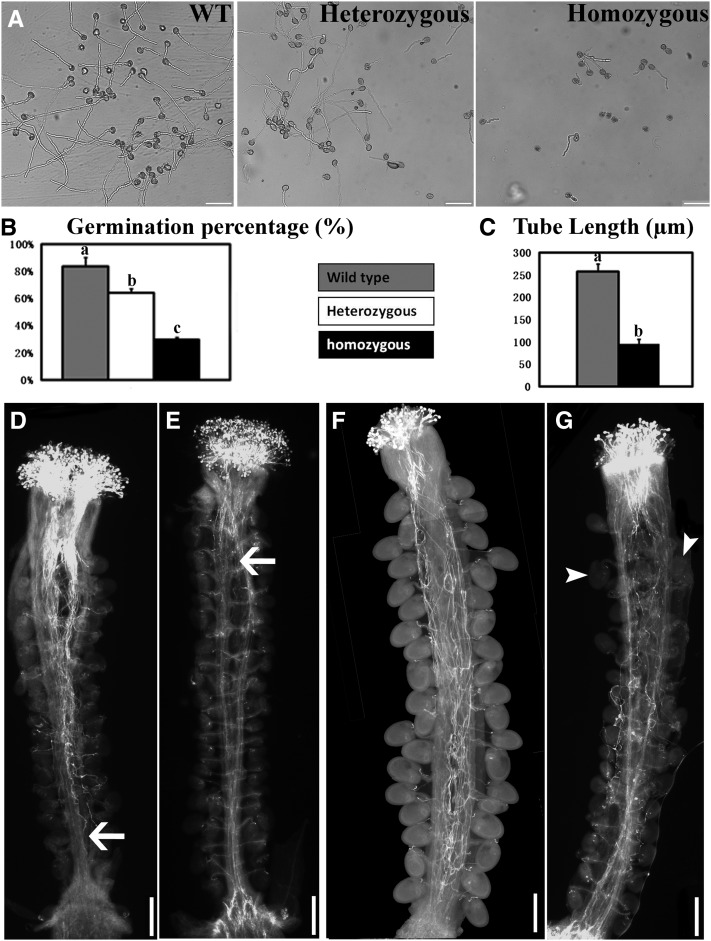
We further analyzed in vitro pollen germination and growth. A low percentage of pat10 pollen germinated (Figures 3A and 3B). Pollen germination of heterozygous pat10 was also significantly reduced compared with the wild type (Figures 3A and 3B), suggesting that mutations at PAT10 not only caused sporophytic male defects but also affected gametophytic male function. Mutant pollen tubes ceased growth early, resulting in short tubes (Figures 3A and 3C).
Figure 3.
PAT10 Mutations Resulted in Defective Pollen Germination and Tube Growth.
(A) In vitro germination of pollen from the wild type (WT), heterozygous pat10 mutants (heterozygous), and homozygous pat10 mutants (homozygous).
(B) In vitro germination percentage of the wild type, heterozygous pat10 mutants, and homozygous pat10 mutants. Germination percentage was calculated from three independent experiments, including around 400 pollen grains in each experiment. For each experiment, samples from three genetic backgrounds were placed side by side on the same slide to reduce system variations. Results are given as means ± se. Samples with different letters (a, b, and c) are significantly different from each other by Fisher’s LSD method.
(C) Tube length of in vitro grown pollen from the wild type and pat10. Data were collected from three independent experiments, each including 80 to 100 pollen tubes. For each experiment, samples from the two genetic backgrounds were placed side by side on the same slides to reduce system variations. Results are given as means ± se. Samples with different letters (a and b) are significantly different from each other by Fisher’s LSD method.
(D) and (E) Aniline blue staining of emasculated wild-type pistils hand-pollinated with wild-type pollen (D) or with pat10 pollen (E) at 9 HAP. Arrows point to the growth fronts of pollen tubes.
(F) and (G) Aniline blue staining of emasculated wild-type pistils hand-pollinated with wild-type pollen (F) or pat10 pollen (G) at 48 HAP. Arrowheads point to ovules fertilized by pat10 pollen.
Bars = 100 µm in (A) and 200 µm in (D) to (G).
To find out whether PAT10 regulated pollen tube growth in vivo, we performed aniline blue staining on emasculated wild-type pistils hand-pollinated with pollen either from the wild type or pat10. At 9 h after pollination (HAP) when most wild-type pollen tubes reached to the bottom of a pistil (Figure 3D), pollen tubes of pat10 just emerged from the style (Figure 3E). At 48 HAP, all ovules in pistils pollinated by the wild type were fertilized and developing (Figure 3F). By contrast, few ovules were targeted by pat10 pollen tubes at 48 HAP despite the fact that the mutant pollen tubes had reached the bottom of the pistils (Figure 3G). These results suggested that polar and directional growth of pollen tubes was compromised by PAT10 loss of function.
Sporophytic Female Defects by PAT10 Loss of Function
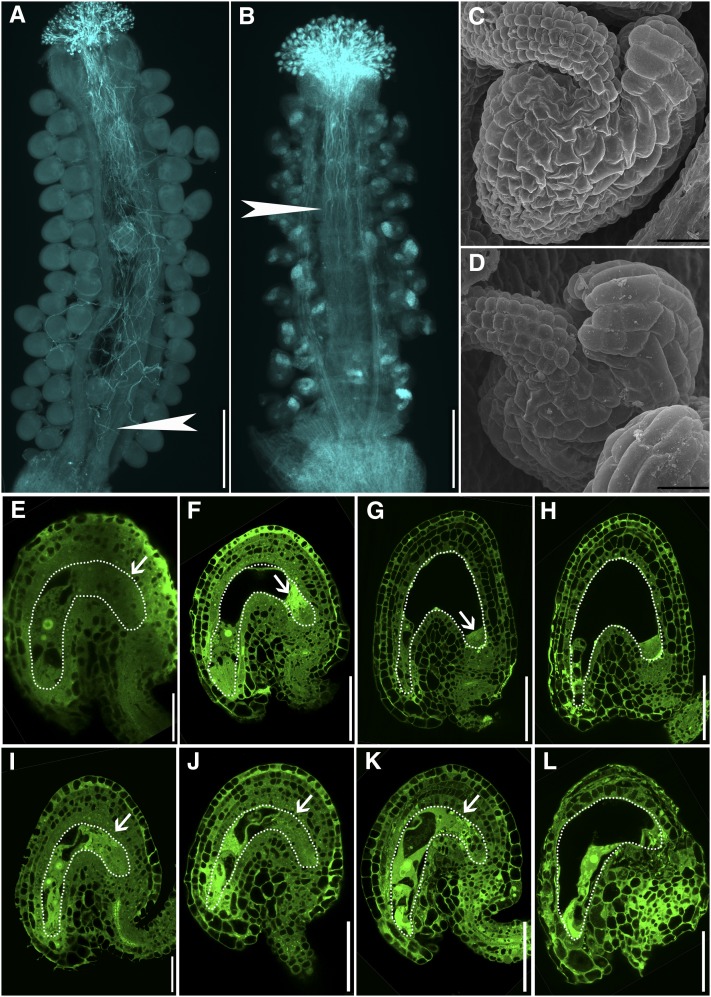
Our reciprocal analysis using the wild type and heterozygous pat10 mutants indicated that female gametophytes were not affected by PAT10 loss of function. However, reciprocal crosses using wild-type pollen and pat10 pistils failed to produce seeds. To find out whether PAT10 loss of function caused sporophytic female defects, we performed aniline blue staining on pat10 pistils pollinated with wild-type pollen. At 24 HAP, when wild-type pollen tubes reached to the bottom of wild-type pistils (Figure 4A), they hardly reached to the middle of pat10 pistils (Figure 4B), suggesting that pat10 pistils could not support pollen tube growth. SEM analysis of mature ovules did not reveal morphological differences between the wild type (Figure 4C) and pat10 (Figure 4D) regarding ovule integuments. We further performed optical sections on unfertilized mature ovules as well as ovules immediately after fertilization. Interestingly, embryo sacs of pat10 were found to be disorganized at the chalazal end (Figure 4I) compared with the ordered organization in the wild type (Figure 4E). In contrast with normal embryo development in the wild type after fertilization (Figures 4F to 4H), embryo sacs of pat10 gradually degenerated likely due to fertilization failure (Figures 4J to 4L). No such embryo sac defects were observed in ovules from heterozygous pat10 mutants (see Supplemental Figure 4 online), which correlated with the normal transmission of mutant female gametophytes during reciprocal crosses (Table 1).
Figure 4.
Female Reproductive Organs Are Defective in pat10.
(A) and (B) Aniline blue staining of emasculated wild-type pistils (A) and pat10 pistils (B) hand-pollinated with wild-type pollen at 24 HAP. Arrows indicate the growth fronts of pollen tubes.
(C) and (D) Scanning electron microscopy of a mature ovule from the wild type (C) and pat10 (D).
(E) to (H) Optical sections of wild-type ovules from unfertilized mature pistils (E) and from pistils at 9 HAP (F), 24 HAP (G), and 48 HAP (H). Dotted lines highlight the embryo sacs. Arrows indicate the chalazal end.
(I) to (L) Optical sections of pat10 ovules from unfertilized mature pistils (I) and from pistils at 9 HAP (J), 24 HAP (K), and 48 HAP (L). Dotted lines highlight the embryo sacs. Arrows indicate the chalazal end.
Bars = 200 µm in (A) and (B), 20 µm in (C) and (D), 25 µm in (E) and (I), and 50 µm in (F) to (H) and (J) to (L).
Constitutive Expression of PAT10
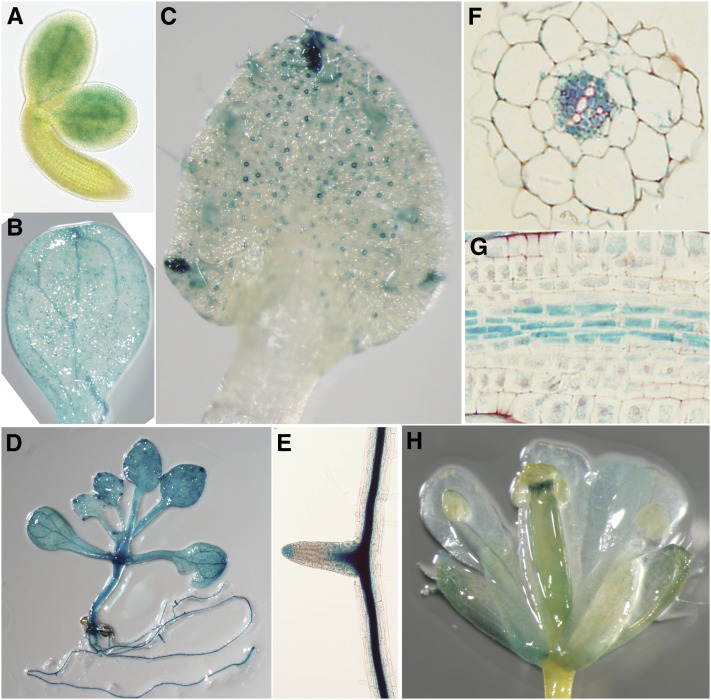
The pleiotropic developmental defects by PAT10 loss of function suggested a constitutive expression pattern. Indeed, PAT10 was previously found to be expressed in all tissues via microarray analysis (Zimmermann et al., 2004). In addition, we analyzed the expression pattern of PAT10 using ProPAT10:GUS (for β-glucuronidase) reporter lines by histochemical analysis. GUS signals were detected in mature embryos, cotyledons, and whole seedlings (Figures 5A, 5B, and 5D). In mature leaves, the GUS signal was highest at hydathodes as well as in guard cells (Figure 5C). PAT10 was strongly expressed at the sites of lateral root initiation and at root tips (Figure 5E). Dissection of GUS signals in vascular bundles showed that PAT10 was expressed strongly in phloem but not in xylem (Figures 5F and 5G).
Figure 5.
Constitutive Expression of PAT10 by ProPAT10:GUS Reporter Analysis.
(A) A mature embryo freshly dissected from rehydrated mature seeds.
(B) A cotyledon showing GUS signals at vascular tissues.
(C) A true leaf showing strong GUS signals at hydathodes, trichomes, and guard cells.
(D) A seedling at 15 DAG showing constitutive GUS signals at both aerial parts and in roots.
(E) A lateral root.
(F) A transverse section of ProPAT10:GUS transgenic roots showing strong GUS signal at the phloem but not at the xylem.
(G) A longitudinal section of ProPAT10:GUS transgenic roots showing strong GUS signal at the vascular bundles.
(H) An open flower of a ProPAT10:GUS transgenic plant showing strong GUS signal at the style and stomata of floral organs.
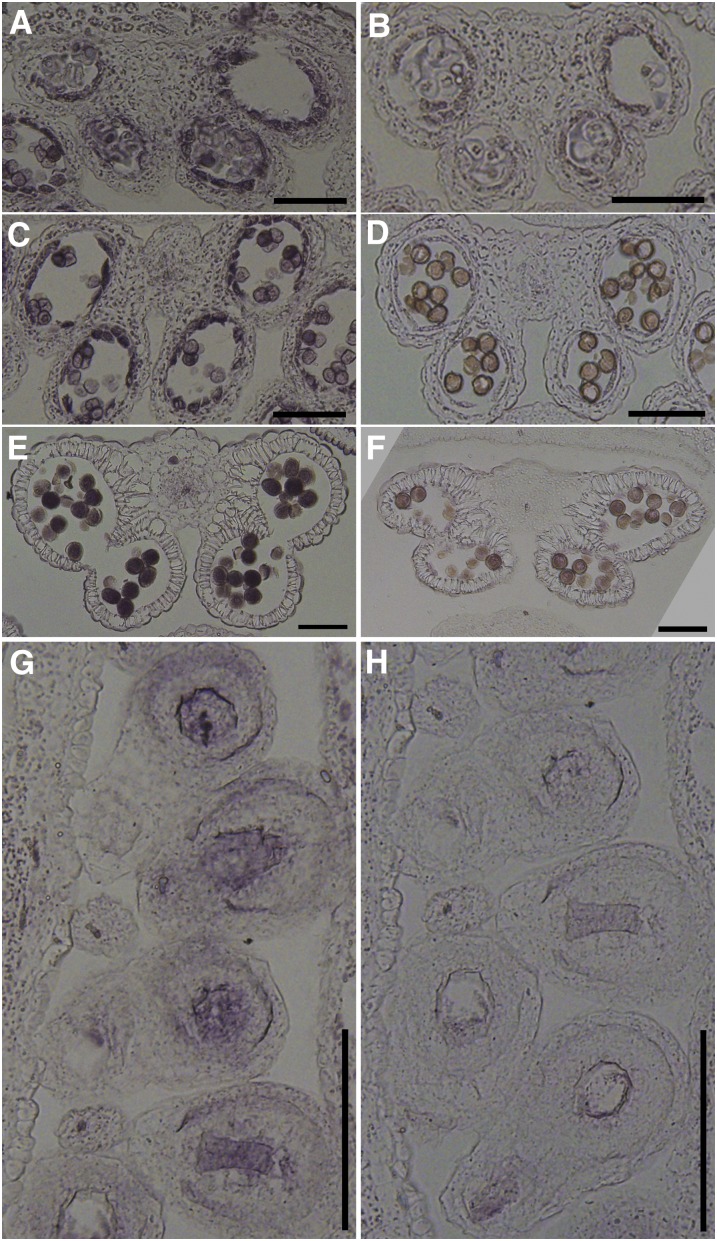
It is intriguing that the GUS signal was not detected at most reproductive tissues (Figure 5H), whereas PAT10 loss of function resulted in developmental defects. We reasoned that the promoter sequences used to generate ProPAT10:GUS reporter lines might not contain all of the information required for expression. Therefore, we next performed RNA in situ hybridization to find out whether PAT10 was expressed in reproductive tissues. Indeed, PAT10 was strongly expressed in the tapetal layer of developing anthers starting from anther developmental stage 6 (Figures 6A and 6B). With tapetal degeneration, PAT10 expression gradually increased in developing microspores (Figures 6C and 6D) and in mature pollen (Figures 6E and 6F). PAT10 was also expressed at the embryo sacs and surrounding cells in mature pistils (Figures 6G and 6H). In addition, no signal was detected in reproductive organs when identical RNA in situ hybridization was performed in pat10 (see Supplemental Figure 5 online), indicating that the probe was specific for PAT10 without cross-reacting with the other 23 PATs (Hemsley and Grierson, 2008; Batistic, 2012).
Figure 6.
Expression of PAT10 in Tapetum, Pollen, and Developing Ovules by RNA in Situ Hybridization Analysis.
(A) and (B) A stage 6 wild-type anther labeled by the antisense probe (A) or the sense probe (B). The tapetal layer shows strong signal.
(C) and (D) A stage 9 wild-type anther labeled by the antisense probe (C) or the sense probe (D). Strong signal is not only detected at the tapetal layer but also in developing pollen.
(E) and (F) A stage 12 wild-type anther labeled by the antisense probe (E) or the sense probe (F). The tapetal layer is fully degenerated. Strong signals are present in mature pollen.
(G) and (H) Longitudinal sections of unfertilized mature pistils from wild-type plants labeled by the antisense probe (G) or the sense probe (H). Strong signals are present at the embryo sacs and its surrounding cells.
Bars = 100 µm.
PAT10 but Not PAT10C192S Complemented pat10
To confirm that the pleiotropic developmental defects were caused by PAT10 loss of function, we performed a complementation study in which we introduced a PAT10 genomic fragment green fluorescent protein (GFP) translational fusion (PAT10-GFP) into pat10. Exogenous PAT10 expression rescued all defects of the pat10 mutants (see Supplemental Figure 6 online), confirming the null mutant identity of pat10. Since the GFP translational fusion did not interfere with the function of PAT10, we therefore used it to study the subcellular localization of PAT10 by fluorescence microscopy. Unlike the ProPAT10:GUS reporter lines, which showed no signal in most reproductive tissues, the GFP signal was detected in pollen (see Supplemental Figure 7 online), confirming that PAT10 expression in reproductive tissues or cells requires intronic sequences.
Because animal PATs can have functions independent of their enzymatic activities (Greaves and Chamberlain, 2011), we wondered whether Arabidopsis PAT10 regulated development through its S-acyl transferase activity. Therefore, we introduced a point mutation at PAT10 so that the conserved DHHC motif was changed to DHHS (PAT10C192S), as such a mutation was previously shown to abolish PAT activity (Hemsley et al., 2005; Baekkeskov and Kanaani, 2009; Greaves and Chamberlain, 2011). The exogenous PAT10C192S-GFP did not rescue pat10 (see Supplemental Figure 8 online), suggesting that PAT10 functions through its palmitate transferase activity.
PAT10 Is Localized at the Tonoplast
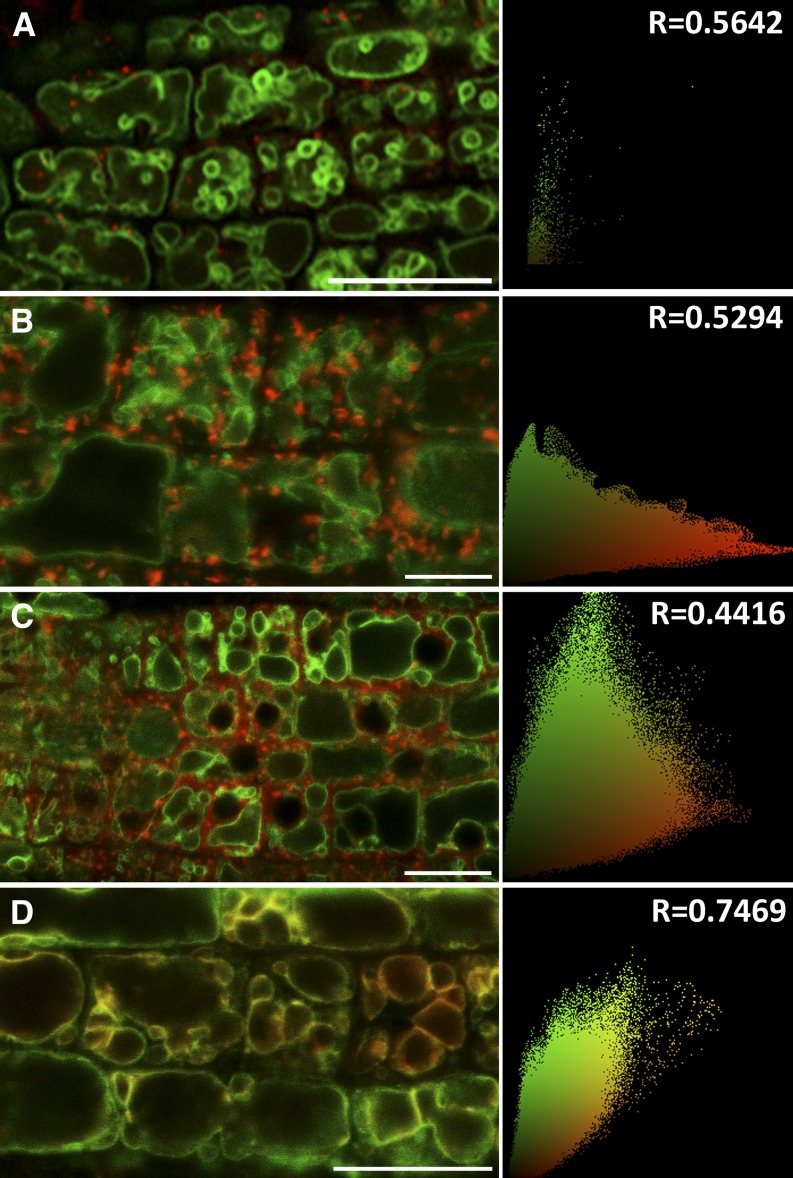
The subcellular localization of Arabidopsis PATs was recently performed via transient expression in Nicotiana benthamiana leaves (Batistic, 2012). When PAT10 was transiently expressed in tobacco epidermal cells, this protein mostly colocalized with a Golgi marker but reached the tonoplast several days later (Batistic, 2012). We introduced the Wave lines expressing red fluorescent protein (RFP)-labeled endomembrane markers (Geldner et al., 2009) into the complemented plants (PAT10-GFP;pat10-1) for analysis via confocal fluorescence microscopy. PAT10 did not colocalize with the trans-Golgi network/early endosome (TGN/EE) marker (Figure 7A) nor the Golgi marker (Figure 7B) or the prevacuolar compartment marker (Figure 7C). Instead, PAT10 showed significantly colocalization with the tonoplast marker WAVE9R (Figure 7D) (Geldner et al., 2009).
Figure 7.
PAT10 Localizes at Tonoplast.
Confocal fluorescence micrographs of transgenic Arabidopsis roots coexpressing PAT10-GFP (green) and fluorescent markers (red) for the TGN/EE (A), Golgi apparatus (B), the prevacuolar compartments (C), and the tonoplast (D). Pearson’s correlation coefficient (R) is shown at the right side of each micrograph. Bars = 10 µm.
We also performed an uptake study in the complemented lines expressing PAT10-GFP in pat10 using the endocytic dye FM4-64 (Lam et al., 2007; Geldner et al., 2009; Bassil et al., 2011a). After 3 h of FM4-64 uptake, the GFP signal completely merged with FM4-64 at the tonoplast (see Supplemental Figure 9 online). Addition of Brefeldin A (BFA), a fungal toxin that interferes with vesicle trafficking at the Golgi and the TGN/EE (Lam et al., 2009), caused aggregation of FM4-64–positive vesicles but not PAT10-labeled compartments.
Since PAT10C192S-GFP did not rescue pat10, we next wanted to find out if the C192S mutation altered its subcellular localization, thus failing to complement. An FM4-64 uptake study was performed in lines expressing PAT10C192S-GFP. Similar to PAT10-GFP, the majority of PAT10C192S-GFP localized at the tonoplast together with FM4-64 after 3 h uptake (see Supplemental Figure 10 online). However, in contrast with PAT10 (see Supplemental Figure 9 online), additional punctate vesicles were detected for PAT10C192S (see Supplemental Figure 10 online). Such punctate localization was also detected in pollen tubes expressing PAT10C192S but not in pollen tubes expressing PAT10 (see Supplemental Figure 7 online). Upon BFA treatment, these GFP-labeled vesicles aggregated to regions surrounding the FM4-64–positive BFA compartments (se Supplemental Figure 10 online), indicating that these vesicles were Golgi apparatus (Lam et al., 2009).
PAT10 Loss of Function Caused Hypersensitivity to Salt Stresses
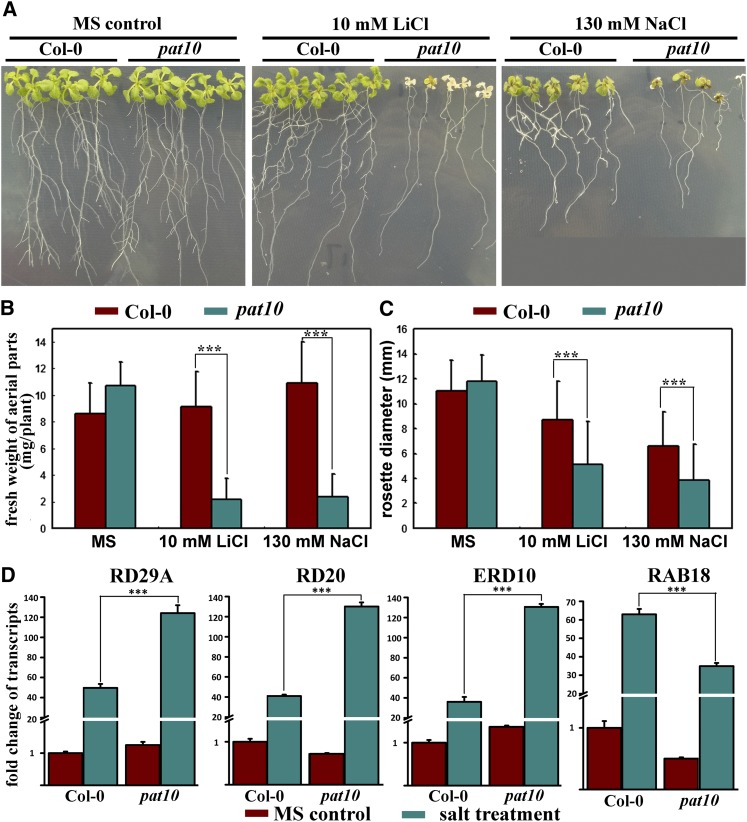
The tonoplast localization of PAT10 prompted us to test the responses of pat10 to salt and osmotic stresses because vacuoles are critical organelles for plant responses to these abiotic stresses (Apse et al., 2003; Cheng et al., 2005; Kim et al., 2007; Bassil et al., 2011b; Kim and Bassham, 2011; Barragán et al., 2012). We applied various stresses to pat10, including high concentrations of NaCl, LiCl, KCl, CsCl, CaCl2, mannitol, and abscisic acid. The pat10 mutants were hypersensitive to NaCl and LiCl, as pat10 grew poorly under high NaCl and LiCl stresses (Figures 8A to 8C).
Figure 8.
pat10 Mutants Are Hypersensitive to Salt Stresses.
(A) Seedlings growing either on MS medium, on MS medium supplemented with 10 mM LiCl, or on MS medium supplemented with 130 mM NaCl. Col-0, Columbia-0.
(B) and (C) Fresh weight (B) and rosette diameter (C) of seedlings (aerial parts only) grown on either MS medium, MS medium supplemented with 10 mM LiCl, or MS medium supplemented with 130 mM NaCl. All seedlings were grown on MS for 4 d before being transferred to MS medium or MS supplemented with salts for an additional 14 d. Data were collected from three independent experiments in which at least 30 seedlings were included in each experiment. Results are given as means ± se. Asterisks indicate significant differences (Student’s t test, P < 0.01).
(D) Induction of salt responsive genes RD29A, RD20, ERD10, and RAB18 in the wild type or pat10. Asterisks indicate significant differences (Student’s t test, P < 0.01).
Next, we tested the expression of salt-responsive genes, such as RESPONSIVE TO DESSICATION 29A (RD29A), RD20, EARLY RESPONSIVE TO DESSICATION10 (ERD10), and RESPONSIVE TO ABA18 (RAB18), whose expression was greatly induced by abiotic stresses (Magnan et al., 2008; Ryu et al., 2010; Bassil et al., 2011a). Real-time quantitative RT-PCR (qRT-PCR) analysis showed that salt stresses significantly induced higher expression of RD29A, RD20, and ERD10 in pat10 than in the wild type (Figure 8D). Although the absolute increase of RAB18 was slightly lower in pat10 than in the wild type, the induction ratio of RAB18 in pat10 was comparable to that of the wild type (Figure 8D). However, PAT10 itself was not induced by salt stresses (Zimmermann et al., 2004).
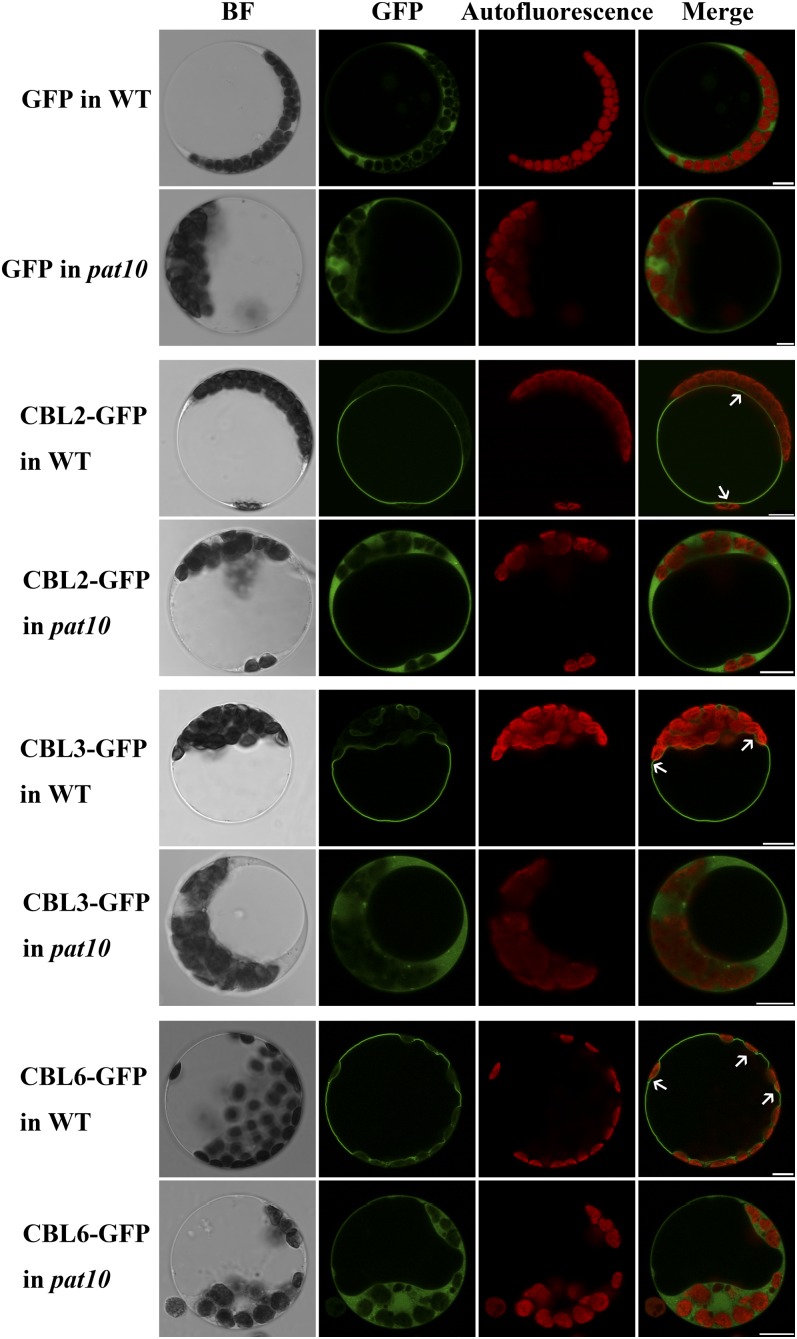
Palmitoylation-Regulated Tonoplast Localization of CBL2/3/6 Depended on PAT10
The hypersensitivity of pat10 to salt stresses gave us a hint about candidate substrates of PAT10. Arabidopsis CBLs play critical roles in abiotic stress responses (Batistic and Kudla, 2009; Luan, 2009). Tonoplast localization of CBL2 is regulated by palmitoylation (Batistic et al., 2008; Batistic and Kudla, 2009; Batistič et al., 2012). In addition, the Cys residues for palmitoylation were conserved among several tonoplast-localized CBLs, such as CBL3 and CBL6, indicating that they are likely modified by palmitoylation. We therefore hypothesized that the tonoplast-localized, palmitoylated CBLs are substrates of PAT10.
To find out whether the palmitoylation-regulated tonoplast localization of CBL2 relies on PAT10, we tested the subcellular localization of GFP-fused CBL2 in wild-type or pat10 protoplasts. CBL2-GFP expressed in the wild-type protoplasts showed tonoplast localization (Figure 9). However, CBL2-GFP expressed in pat10 was relocated to the cytoplasm (Figure 9), similar to soluble GFP (Figure 9). Because the Cys residues proven to be the palmitoylation sites in CBL2 (Batistič et al., 2012) were also present in CBL3 and CBL6, we suspected that their tonoplast localization was also regulated by PAT10. Indeed, both CBL3 and CBL6 were localized at the tonoplast in wild-type protoplasts, but they became cytosolic in pat10 protoplasts (Figure 9). Treatment with 2-bromopalmitate (2-BP), a specific inhibitor of protein palmitoylation (Hemsley et al., 2005; Batistič et al., 2012), caused relocalization of CBL2/3/6 from the tonoplast to cytoplasm in wild-type protoplasts (see Supplemental Figure 11 online), confirming their palmitoylation-regulated tonoplast localization. Taken together, these results suggested that CBL2/3/6 are putative substrates of PAT10.
Figure 9.
Localization of CBLs at the Tonoplast Depends on Functional PAT10.
Representative images of each treatment (i.e., exogenous expression) are shown. In total, 60 to 80 protoplasts from three independent experiments were visualized for each treatment. BF, bright-field; WT, the wild type. Bars = 10 µm.
To exclude the possibility that loss of CBL2/3/6 tonoplast localization was caused by a general reduction of cellular palmitoylation owing to PAT10 loss of function, we introduced CBL9 and ARA6 into pat10 protoplasts. The plasma membrane–localized CBL9 (Batistic et al., 2010) and the endosome-localized ARA6 (Ueda et al., 2001) both contain sequences for palmitoylation and regulate plant responses to abiotic stresses (Pandey et al., 2004; Ebine et al., 2011). The plasma membrane localization of CBL9 was abolished by 2-BP treatment (see Supplemental Figure 11 online), suggesting that its membrane association depended on palmitoylation. However, membrane association of both CBL9 and ARA6 was retained in pat10 (see Supplemental Figure 10 online), indicating that PAT10 loss of function did not reduce protein palmitoylation globally. The membrane localization of ARA7 (a marker for TGN/EE) (Ueda et al., 2001) as well as that of ERD2 (a marker for Golgi apparatus) (Cheung et al., 2002) were not affected by PAT10 loss of function (Supplemental Figure 12 online), suggesting that the integrity of the endomembrane system was intact.
DISCUSSION
Arabidopsis PAT10 Regulates Vacuolar Function through Protein Palmitoylation
Plant vacuoles are acidic organelles essential for nutrient remobilization, maintenance of turgor pressure, sequestration of toxic compounds, ions and secondary metabolites, and accumulation of storage proteins (Maeshima, 2001; Mimura et al., 2003; Hamaji et al., 2009; Agee et al., 2010; Krebs et al., 2010; Barragán et al., 2012). Vacuoles occupy ∼80 to 90% of the total cell volume and provide cells with the ability to withstand salt and osmotic stresses (Mimura et al., 2003; Hamaji et al., 2009; Barragán et al., 2012). Vacuole function is performed mainly by tonoplast proteins (Maeshima, 2001), including vacuolar H+-ATPase (Dettmer et al., 2006; Krebs et al., 2010), vacuolar PPase, Ca2+-ATPases (Pittman, 2011), Ca2+/H+ antiporters (Cheng et al., 2005), K+ and Na+/H+ antiporters (NHXs) (Bassil et al., 2011b; Barragán et al., 2012), tonoplast intrinsic proteins, and CBLs (Kim et al., 2007; Batistic et al., 2010). Functional loss of these tonoplast proteins caused pleiotropic developmental defects and hypersensitivity to abiotic stresses (Apse et al., 2003; Cheng et al., 2005; Batistic et al., 2010; Krebs et al., 2010; Bassil et al., 2011a, 2011b; Barragán et al., 2012), suggesting defective vacuolar function.
In this study, PAT10 was shown to localize at vacuolar membrane. Loss of function of PAT10 resulted in pleiotropic developmental defects, reduced cell expansion, and hypersensitivity to salt stresses, similar to those caused by the functional loss of tonoplast proteins (Cheng et al., 2005; Krebs et al., 2010; Bassil et al., 2011b; Barragán et al., 2012). pat10 also displayed a spectrum of reproductive defects, including sporophytic male and female defects as well as compromised growth of pollen tubes (Figures 2 to 4). The highly dynamic tubular vacuoles of pollen tubes (Hicks et al., 2004) may play critical roles in pollen germination and tube growth because disrupting vacuolar protein sorting often resulted in reduced pollen germination and poor male gametophytic transmission (Wang et al., 2010). Little is known about vacuole function in reproductive tissues or cells. However, it is clear that male and female sporophytic cells undergo extensive secretion for the maturation of pollen (tapetal cells) or to support pollen tube growth (the transmitting tract), whereas genetically or pharmacologically interfering with vacuole function can disrupt both endocytic and secretory trafficking (Dettmer et al., 2006; Rojo and Denecke, 2008). Another possibility is that ion homeostasis regulated by vacuoles, as indicated by studies of tonoplast proteins (Cheng et al., 2005; Bassil et al., 2011b; Barragán et al., 2012; Tang et al., 2012), was disrupted in pat10, leading to the pleiotropic reproductive defects. It was shown that several animal PATs could act as cation transporters (Goytain et al., 2008; Hines et al., 2010); we therefore could not exclude the possibility that PAT10 plays similar roles directly rather than through its substrates.
Not all DHHC-type PATs identified so far show palmitate transferase activity (Ohno et al., 2012). However, the C192S mutation could not rescue pat10 mutants, despite the fact that the majority of the mutant protein remained at the tonoplast (see Supplemental Figure 8 online), suggesting that PAT10 functions through its PAT activity. Therefore, we propose that PAT10 catalyzes the palmitoylation of proteins peripheral or integral to the tonoplast and thereby mediates vacuole function. Indeed, a similar regulatory paradigm occurred early during evolution. Pfa3, one of the seven DHHCs in yeast, catalyzes palmitoylation of Vac8, whose association with vacuolar membrane is critical for vacuolar trafficking and fusion (Hou et al., 2005; Smotrys et al., 2005).
Substrates of Arabidopsis PAT10
Our study suggests that tonoplast-localized CBL2/3/6 are putative substrates of PAT10. First, tonoplast-localized CBL2 is modified by palmitoylation, and mutating its potential palmitoylation sites rendered the protein cytosolic (Batistič et al., 2012). The Cys residues for palmitoylation were conserved for CBL3 and CBL6. Second, localization of CBL2 and CBL6 at the tonoplast did not rely on vesicle trafficking (Bottanelli et al., 2011; Batistič et al., 2012), suggesting that their palmitoylation was conducted by tonoplast-localized PATs. Third, we showed here that tonoplast localization of CBL2/3/6 was abolished by 2-BP treatment as well as in pat10. Because transient expression of PAT10 resulted in mistargeting of PAT10 to the Golgi apparatus (Batistic, 2012), we could not test the effects of exogenous PAT10 on the localization of CBLs in pat10 nor the in planta interaction between PAT10 and CBL2/3/6.
More proteins are likely subjected to PAT10-mediated palmitoylation. First, pat10 displayed pleiotropic developmental defects in the absence of abiotic stresses, which were not observed in loss-of-function mutants of CBLs (Batistic et al., 2010), although cbl2 cbl3 double mutants showed pleiotropic defects, including leaf tip necrosis and reproductive defects (Tang et al., 2012). Second, although the Arabidopsis genome encodes other 21 PATs sharing the same domain organization with PAT10 (Hemsley et al., 2005; Hemsley and Grierson, 2008; Batistic, 2012), PAT10 is one of the only two PATs detected at the tonoplast (Batistic, 2012), suggesting low functional redundancy.
Global analysis of protein palmitoylation in yeast (Roth et al., 2006), in rat neuron (Kang et al., 2008), and in Arabidopsis (Hemsley et al., 2013) indicated that a large number of proteins are modified by palmitoylation, although this has yet to be shown experimentally. Because of the lack of consensus sequence for protein palmitoylation, it is impossible to perform an in silico prediction to evaluate the likely substrates of palmitoylation. Based on the subcellular localization and phenotypes of loss-of-function mutants, we consider some vacuolar transporters likely to be the substrates of PAT10, such as Arabidopsis NHX1 and NHX2 (Apse et al., 2003; Bassil et al., 2011a, 2011b; Barragán et al., 2012). Double mutants of NHX1 and NHX2 showed a spectrum of developmental and cellular defects (Bassil et al., 2011b) similar to those of pat10.
Palmitoylation may regulate the activities of transmembrane proteins, protein sorting, or protein–protein interaction. Unlike peripheral membrane proteins that will become cytosolic in the absence of their corresponding PATs, transmembrane proteins are membrane localized irrespective of palmitoylation. Perhaps for these reasons, only the receptor-like kinase FLAGELLIN SENSITIVE2 was experimentally shown to be regulated by palmitoylation, although many other receptor-like kinases as well as transporters were identified as putatively palmitoylated (Hemsley et al., 2013). Future technological improvements should help lead to the identification of transmembrane substrates of PAT10 at the tonoplast.
Subcellular Targeting of PAT10
PATs are multispan transmembrane proteins whose catalytic activities occur at or close to the membranes in which they reside (Hou et al., 2009). Therefore, there must be stringent mechanisms regulating their subcellular targeting to their final destination compartments. Although PATs of a given species can localize at diverse membrane compartments (Ohno et al., 2006; Greaves and Chamberlain, 2011; Batistic, 2012), little is known about the targeting determinants. Mutation at a palmitoyltransferase conserved C terminus (PaCCT) motif of yeast Pfa3 resulted in its functional loss and mistargeting to the vacuolar lumen (González Montoro et al., 2009). However, the PaCCT motif is not present in Arabidopsis PAT10.
35S-driven transient expression of PAT10 showed a strong signal at the Golgi apparatus (Batistic, 2012), but such results could not be confirmed by in vivo approaches. We showed that the translational fusion of GFP at the C terminus of PAT10 functionally rescued pat10 (see Supplemental Figure 6 online), thus allowing the analysis of its subcellular localization by fluorescence microscopy. Although transmembrane proteins are usually synthesized at the endoplasmic reticulum and delivered to their final destinations via vesicular trafficking, pharmacological treatments, together with fluorescent colabeling, showed that no PAT10 signal was detected at either the TGN/EE or the Golgi (see Supplemental Figure 9 online) upon BFA treatment, suggesting that PAT10 is transported to the vacuole through a route independent of the Golgi. We noticed that the C192S mutation resulted in partial translocation of PAT10 to the Golgi (see Supplemental Figure 10 online). It is known that the Cys residue within the DHHC motif is not only critical for PAT activity but is also itself a palmitoylation site (Greaves and Chamberlain, 2011). Therefore, the mislocalization caused by the C192S mutation could be due to interfered trafficking of PAT10, as PAT10C192S is not completely retained at the tonoplast. Because the functionality of PATs is determined by their membrane localization, future efforts should be dedicated to verify the results obtained from a transient heterogeneous study (Batistic, 2012) using various in vivo approaches.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana WAVE lines expressing RFP fusions for the TGN (WAVE2R), Golgi (WAVE22R), prevacuolar compartment (WAVE3R), and the tonoplast (WAVE9R) (Geldner et al., 2009) were obtained from the European Arabidopsis Stock Center (http://Arabidopsis.info). The T-DNA insertion lines SALK_024964 (pat10-1) and WiscDsLox289_292E10 (pat10-2) were obtained from the ABRC (http://www.Arabidopsis.org). The Columbia-0 ecotype was used as the wild type. Arabidopsis plants were grown in a 4:1:1 mix of Fafard 4P:perlite:vermiculite under an 18-h-light/6-h-dark cycle at 21°C. For growth on plates, surface-sterilized Arabidopsis seeds were placed on half-strength Murashige and Skoog (MS) basal medium with vitamins (Phytotechlab) except where noted. Plates were kept at 4°C in darkness for 4 d before being transferred to a growth chamber with a 16-h-light/8-h-dark cycle at 21°C. Stable Arabidopsis transformations were produced using the floral dipping method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS medium supplemented with 30 µg/mL Basta salt (Sigma-Aldrich). Measurement of epidermal cell sizes in mature leaves was performed as described (Ohto et al., 2005).
PCR, RT-PCR, and qRT-PCR
The segregation ratios in the reciprocal analysis were determined by PCR using the following primers: G1/G2 for PAT10, G3/G2 for pat10-1, G1/G4 for PAT10, and G5/G4 for pat10-2.
Total RNAs were isolated using a Qiagen RNeasy plant mini kit according to the manufacturer’s instructions. Oligo(dT)-primed cDNAs were synthesized using Superscript III reverse transcriptase with on-column DNase digestion (Invitrogen). Primers used in RT-PCR to characterize pat10 were R1/R2/R3/R4. Primers used in RT-PCR to verify complementation were R5/R6 for the endogenous PAT10 and R5/R7 for the exogenous PAT10. Primers to amplify ACTIN2 were as described (Zhang and McCormick, 2007).
The qRT-PCR analysis of salt-induced gene expression was performed with the Bio-Rad CFX96 real-time system using SYBR Green real-time PCR master mix (Toyobo). Each 20-μL reaction system contained 2 μL SYBR Green real-time PCR master mix, 2 μL cDNA, and 2 μL primers (5 µM). GAPDH and TUBLIN2 were used as internal controls (Ryu et al., 2010). Primers used for RD29A, RD20, ERD20, RAB18, GAPDH, and TUBLIN2 were as described (Ryu et al., 2010; Jiang et al., 2011). The cycling conditions were as followed: 95°C for 2 min and 42 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 15 s. Fluorescence data were collected during the 72°C step and analyzed with the Bio-Rad CFX Manager. RNA extractions were performed for three biological replicates for each treatment, and three technical replicates were performed for each sample. The data presented in Figure 8 are means of the three biological replicates for which three technical replicates were averaged. All primers used in this study are listed in Supplemental Table 1 online.
Plasmid Construction
All constructs were generated using Gateway technology (Invitrogen). The whole genomic fragment of PAT10 was cloned with primers PAT10gF/PAT10gR from genomic DNA. The C192S mutation was generated with primers PAT10gmF/PAT10gmR from the PAT10g entry vector using a Phusion site-directed mutagenesis kit according to the manufacturer’s instructions (Finnzyme). The 1242-bp sequence upstream of the start codon of PAT10 was used as its promoter (ProPAT10). ProPAT10 was cloned with primers ProF/ProR from genomic DNA. The entry vectors for CBL2, CBL3, CBL6, CBL9, and ERD2 were generated from a mixed Arabidopsis cDNA library using the following primer pairs: CBL2-F/R for CBL2, CBL3-F/R for CBL3, CBL6-F/R for CBL6, CBL9-F/R for CBL9, and ERD2-F/R for ERD2. All entry clones were generated in the pENTR/D/TOPO vector (Invitrogen).
The destination vector used to generate ProPAT10:GUS were described previously (Curtis and Grossniklaus, 2003). The destination vector for PAT10-GFP and PAT10C192S-GFP was modified from a previously described vector (Zhang and McCormick, 2007) by removing the ProLAT52 sequence. The expression vector for ARA6 and ARA7 was described (Zhang et al., 2010). Expression vectors were generated by LR reactions using LR Clonase II (Invitrogen). Expression vectors for mbSUS were generated by in vivo homologous recombination in yeast as described (Obrdlik et al., 2004).
All PCR amplifications used Phusion hot start high-fidelity DNA polymerase with the annealing temperature and extension times recommended by the manufacturer (Finnzyme). All entry vectors were sequenced, and sequences were analyzed using Vector NTI (Invitrogen). The Bioneer PCR purification kit and Bioneer Spin miniprep kit were used for PCR product recovery and plasmid DNA extraction, respectively.
GUS Histochemistry
Roots of ProPAT10:GUS transgenic plants were fixed, cleared, and embedded in paraffin (Sigma-Aldrich). Fifteen embedded root tissues from three individual transgenic lines were analyzed. To show the outlines of cells, sectioned samples on slides were stained with 100 mg/L ruthenium red (Sigma-Aldrich) for 3 to 5 min before being visualized and photographed with an Olympus BX51 microscope equipped with a charge-coupled device camera.
RNA in Situ Hybridization
RNA in situ hybridization was performed as previously described (Li et al., 2010). Briefly, open flowers and young pistils were fixed in formaldehyde solution (formalin:acetic acid:ethanol:H2O=1:2:10:7) overnight at 4°C, embedded in Paraplast (Sigma-Aldrich) after dehydration, and sectioned at 6-µm thickness. A 216-bp fragment specific for PAT10 was amplified from the coding region of PAT10 with primers ProbeF/ProbeR. The sense probe and the antisense probe were cloned into pMD19 T-simple vector (Takara), modified in vitro with digoxigenin-UTP by SP6 or T7 RNA polymerases (Roche), respectively. Sections were hybridized with 1.5 ng/μL probes at 42°C overnight in a hybridization solution that contained 50% formamide. Hybridization signals were detected using antidigoxigenin antibody (Anti-Digoxigenin-Ap Fab fragments; Roche). The samples were observed using an Olympus BX51 microscope.
Pollen Analysis
To detect pollen viability, mature anthers were soaked overnight in Alexander stain (Johnson-Brousseau and McCormick, 2004) and observed with a BX51 microscope (Olympus). Pollen from open flowers was stained with 1 mg/mL (w/v) 4',6-diamidino-2-phenylindole (Sigma-Aldrich) for 30 min and observed with an Axio Observer D1 microscope (Zeiss). Pollen germination in vitro and aniline blue staining of pollen tubes growing in vivo were performed as described (Zhang et al., 2009). For pollen SEM, mature pollen coated with gold palladium was observed under a JSM-6610LV scanning electron microscope (JEOL). For pollen TEM, mature flowers were fixed in 4% (v/v) glutaraldehyde and 1% (w/v) osmic acid (Sigma-Aldrich). After washing with a phosphate buffer, the specimens were dehydrated in an ethanol series and embedded in Epon 812 resin (SPI-CHEM). Ultrathin sections were stained with acetate uranium and lead citrate. The specimens were observed using a JEM-1200EX transmission electron microscope (JEOL).
Pharmacological Treatment
For double labeling experiments with FM4-64, roots of 4- to 5-DAG seedlings were dipped in liquid MS media supplemented with 4 µM FM4-64 for 5 min (except for the 1-min assay). The roots were then taken out, washed three times with liquid MS medium without FM4-64, and visualized for FM4-64 intake at the designated times. For BFA treatment, roots were treated first with FM4-64 for 5 min, then washed and incubated with MS medium supplemented with 50 µM BFA for 50 min and visualized for the formation of BFA compartments. For treatment of protoplasts with 2-BP, 2-BP at the final concentration of 10 µM was added into cultural medium immediately after polyethylene glycol–mediated transformation. Samples were examined between 16 and 24 h after treatment.
Salt Treatment
For salt treatment, wild-type and pat10 seedlings at 4 DAG on MS media were transferred to fresh MS media or to MS media supplemented with either 10 mM LiCl or 130 mM NaCl for additional 14 d. Plates were photographed with a BX51 microscope (Olympus) equipped with a digital camera. Rosette diameter was measured using ImageJ (http://rsbweb.nih.gov/ij/). For each treatment, a representative image from three independent experiments was shown. For the measurement of rosette diameter and fresh weight, each treatment was repeated three times, and each experiment included at least 30 plants from each genetic background.
For salt induction of gene expression, wild-type and pat10 seedlings growing vertically on MS medium for 7 d were transferred to 3M filter paper presoaked with either MS medium or MS medium supplemented with 300 mM NaCl. After 6 h of incubation, whole seedlings were used for RNA extraction.
Expression in Arabidopsis Protoplasts
Preparation of Arabidopsis protoplasts was performed according to a previous report (Wu et al., 2009). Transformation of Arabidopsis protoplasts was performed according to standard protocols (Yoo et al., 2007). Three independent experiments for each treatment or combination were conducted to ensure that the results were consistent.
Fluorescent and Confocal Microscopy
In vitro growing pollen from PAT10-GFP or PAT10C192S-GFP transgenic plants was captured by epifluorescence using Axio Observer D1 equipped with a charge-coupled device camera (Zeiss). Other images were captured by an inverted laser scanning confocal microscope (LSM780; Zeiss) with a Plan-Neofluar ×40/1.3 oil differential interference contrast objective or ×63/1.45 oil differential interference contrast objective. GFP-RFP double-labeled materials were captured alternately using line switching with the multitrack function (488 nm for GFP and 545 nm for RFP). Fluorescence was detected using a 505- to 550-nm band-pass filter for GFP and a 575- to 650-nm band-pass filter for RFP. Postacquisition image processing was performed with LSM image processing software (Zeiss). Pearson coefficients analysis was performed as described (French et al., 2008).
Optical sectioning of ovules was performed as described (Shi et al., 2005). Briefly, pistils were fixed in 4% glutaraldehyde (in 12.5 mM phosphate buffer, pH 6.9) for at least 20 min with vacuum and overnight at room temperature. After fixation, the tissues were dehydrated through a conventional ethanol series and cleared in 2:1 (v/v) benzyl benzoate:benzyl alcohol for at least 1 h. The pistils were then dissected, mounted with immersion oil, and observed with a Leica TCS SP5II laser scanning microscope (Leica) with a 488-nm argon laser and an LP 500 filter.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (www.Arabidopsis.org) under the following accession numbers: PAT10 (At3g51390), CBL2 (AT5G55990), CBL3 (AT4G26570), CBL6 (AT4G16350), CBL9 (AT5G47100), ARA6 (At3g54840), ARA7 (AT4G19640), ERD2 (AT1G29330), RD29A (AT5G52310), RD20 (AT2G33380), ERD10 (AT1G20450), RAB18 (AT5G66400), GAPDH (AT3G04120), and TUBLIN2 (AT5G62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cell Size and Number Were Both Reduced in pat10.
Supplemental Figure 2. PAT10 Mutations Caused Temporally Delayed but Developmentally Early Floral Transition.
Supplemental Figure 3. Pollen Viability Was Not Affected by PAT10 Mutations.
Supplemental Figure 4. Embryo Sac Was Normal in Ovules from Heterozygous pat10 Mutants.
Supplemental Figure 5. The Probe Used in RNA in Situ Hybridization Was Specific for PAT10.
Supplemental Figure 6. PAT10-GFP Complemented pat10.
Supplemental Figure 7. Subcellular Localization of PAT10 and PAT10C192S in Transgenic Arabidopsis Pollen Tubes Growing in Vitro.
Supplemental Figure 8. PAT10C192S-GFP Did Not Complement pat10.
Supplemental Figure 9. PAT10 Localizes at the Tonoplast.
Supplemental Figure 10. PAT10C192S Localizes at the Tonoplast as well as BFA-Sensitive Vesicles.
Supplemental Figure 11. Treatment with 2-Bromopalmitate Abolished the Membrane Association of CBL2, CBL3, CBL6, and CBL9 in Wild-Type Protoplasts.
Supplemental Figure 12. Membrane Association of CBL9, ARA6, ARA7, and ERD2 Did Not Rely on Functional PAT10.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC and European Arabidopsis Stock Center for the plant materials described in this article. We thank Xian Sheng Zhang for giving us access to the microscope facilities of his laboratory. This research was supported by Major Research Plan (2013CB945102) from the Ministry of Science and Technology of China and by a grant from the National Science Foundation of China (NSFC) (31261160490). Y.Z.’s laboratory is supported by the Tai-Shan Scholar program from Shandong Provincial Government. L.J. is supported by the Research Grants Council of Hong Kong and NSFC/RGC (N_CUHK406/12).
AUTHOR CONTRIBUTIONS
L.-Z.Z., S.L., Q.-N.F., Y.-L.Z. (SDAU), X.Z., Y.-L. Z. (CUHK), and H.W. performed the experiments. L.J. provided materials and helped with microscopy. Y.Z. and S.L. designed the experiments and wrote the article.
Glossary
- PAT
protein S-acyl transferase
- DAG
days after germination
- TEM
transmission electron microscopy
- HAP
hours after pollination
- GUS
β-glucuronidase
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- TGN
trans-Golgi network
- EE
early endosome
- BFA
Brefeldin A
- qRT-PCR
quantitative RT-PCR
- 2-BP
2-bromopalmitate
- MS
Murashige and Skoog
References
- Agee A.E., et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152: 120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse M.P., Sottosanto J.B., Blumwald E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36: 229–239 [DOI] [PubMed] [Google Scholar]
- Baekkeskov S., Kanaani J. (2009). Palmitoylation cycles and regulation of protein function (Review). Mol. Membr. Biol. 26: 42–54 [DOI] [PubMed] [Google Scholar]
- Barragán V., Leidi E.O., Andrés Z., Rubio L., De Luca A., Fernández J.A., Cubero B., Pardo J.M. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E., Ohto M.A., Esumi T., Tajima H., Zhu Z., Cagnac O., Belmonte M., Peleg Z., Yamaguchi T., Blumwald E. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23: 224–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E., Tajima H., Liang Y.C., Ohto M.A., Ushijima K., Nakano R., Esumi T., Coku A., Belmonte M., Blumwald E. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23: 3482–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O. (2012). Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol. 160: 1597–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Kudla J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793: 985–992 [DOI] [PubMed] [Google Scholar]
- Batistič O., Rehers M., Akerman A., Schlücking K., Steinhorst L., Yalovsky S., Kudla J. (2012). S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Bottanelli F., Foresti O., Hanton S., Denecke J. (2011). Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell 23: 3007–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Shigaki T., Lachmansingh J., LeClere S., Lahner B., Salt D.E., Hirschi K.D. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Chen C.Y., Glaven R.H., de Graaf B.H.J., Vidali L., Hepler P.K., Wu H.M. (2002). Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859 [DOI] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Gagne J.M., Clark S.E. (2010). The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22: 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Montoro A., Quiroga R., Maccioni H.J., Valdez Taubas J. (2009). A novel motif at the C-terminus of palmitoyltransferases is essential for Swf1 and Pfa3 function in vivo. Biochem. J. 419: 301–308 [DOI] [PubMed] [Google Scholar]
- Goytain A., Hines R.M., Quamme G.A. (2008). Huntingtin-interacting proteins, HIP14 and HIP14L, mediate dual functions, palmitoyl acyltransferase and Mg2+ transport. J. Biol. Chem. 283: 33365–33374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J., Chamberlain L.H. (2011). DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem. Sci. 36: 245–253 [DOI] [PubMed] [Google Scholar]
- Hamaji K., et al. (2009). Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 50: 2023–2033 [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Grierson C.S. (2008). Multiple roles for protein palmitoylation in plants. Trends Plant Sci. 13: 295–302 [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Kemp A.C., Grierson C.S. (2005). The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17: 2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A., Weimar T., Lilley K.S., Dupree P., Grierson C.S. (2013). A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197: 805–814 [DOI] [PubMed] [Google Scholar]
- Hicks G.R., Rojo E., Hong S., Carter D.G., Raikhel N.V. (2004). Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol. 134: 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines R.M., Kang R., Goytain A., Quamme G.A. (2010). Golgi-specific DHHC zinc finger protein GODZ mediates membrane Ca2+ transport. J. Biol. Chem. 285: 4621–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., John Peter A.T., Meiringer C., Subramanian K., Ungermann C. (2009). Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic 10: 1061–1073 [DOI] [PubMed] [Google Scholar]
- Hou H., Subramanian K., LaGrassa T.J., Markgraf D., Dietrich L.E., Urban J., Decker N., Ungermann C. (2005). The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. USA 102: 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Zhang X.F., Wang X.F., Zhang D.P. (2011). Arabidopsis 3-ketoacyl-CoA thiolase-2 (KAT2), an enzyme of fatty acid β-oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau S.A., McCormick S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Kang R., et al. (2008). Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456: 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-G., Waadt R., Cheong Y.H., Pandey G.K., Dominguez-Solis J.R., Schültke S., Lee S.C., Kudla J., Luan S. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Bassham D.C. (2011). TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol. 156: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M., Beyhl D., Görlich E., Al-Rasheid K.A., Marten I., Stierhof Y.D., Hedrich R., Schumacher K. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Cai Y., Tse Y.C., Wang J., Law A.H., Pimpl P., Chan H.Y., Xia J., Jiang L. (2009). BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J. 60: 865–881 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.G., Su Y.H., Zhao X.Y., Li W., Gao X.Q., Zhang X.S. (2010). Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 450: 109–120 [DOI] [PubMed] [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Maeshima M. (2001). TONOPLAST TRANSPORTERS: Organization and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 469–497 [DOI] [PubMed] [Google Scholar]
- Magnan F., Ranty B., Charpenteau M., Sotta B., Galaud J.P., Aldon D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Martín M.L., Busconi L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24: 429–435 [DOI] [PubMed] [Google Scholar]
- Mimura T., Kura-Hotta M., Tsujimura T., Ohnishi M., Miura M., Okazaki Y., Mimura M., Maeshima M., Washitani-Nemoto S. (2003). Rapid increase of vacuolar volume in response to salt stress. Planta 216: 397–402 [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y., Kihara A., Sano T., Igarashi Y. (2006). Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761: 474–483 [DOI] [PubMed] [Google Scholar]
- Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., Kihara A. (2012). Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23: 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.K., Cheong Y.H., Kim K.N., Grant J.J., Li L., Hung W., D’Angelo C., Weinl S., Kudla J., Luan S. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman J.K. (2011). Vacuolar Ca(2+) uptake. Cell Calcium 50: 139–146 [DOI] [PubMed] [Google Scholar]
- Rojo E., Denecke J. (2008). What is moving in the secretory pathway of plants? Plant Physiol. 147: 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A.F., Wan J., Bailey A.O., Sun B., Kuchar J.A., Green W.N., Phinney B.S., Yates J.R., III, Davis N.G. (2006). Global analysis of protein palmitoylation in yeast. Cell 125: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M.Y., Cho S.K., Kim W.T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys J.E., Schoenfish M.J., Stutz M.A., Linder M.E. (2005). The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J. Cell Biol. 170: 1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N., Poraty L., Sternberg H., Bar E., Lewinsohn E., Yalovsky S. (2007). Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell. Biol. 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N., Segev O., Gutman O., Bar E., Richter S., Poraty L., Hirsch J.A., Henis Y.I., Lewinsohn E., Jürgens G., Yalovsky S. (2010). An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr. Biol. 20: 914–920 [DOI] [PubMed] [Google Scholar]
- Tang R.-J., Liu H., Yang Y., Yang L., Gao X.-S., Garcia V.J., Luan S., Zhang H.-X. (2012). Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 22: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tse Y.C., Law A.H., Sun S.S., Sun Y.B., Xu Z.F., Hillmer S., Robinson D.G., Jiang L. (2010). Vacuolar sorting receptors (VSRs) and secretory carrier membrane proteins (SCAMPs) are essential for pollen tube growth. Plant J. 61: 826–838 [DOI] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeng Q., Wang X., Running M.P. (2007). Dual lipid modification of Arabidopsis Ggamma-subunits is required for efficient plasma membrane targeting. Plant Physiol. 143: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., He J., Lee D., McCormick S. (2010). Interdependence of endomembrane trafficking and actin dynamics during polarized growth of Arabidopsis pollen tubes. Plant Physiol. 152: 2200–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., He J., McCormick S. (2009). Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes. Plant J. 58: 474–484 [DOI] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.