Phosphorylation of Arabidopsis ETHYLENE RESPONSE FACTOR6 (ERF6) by MPK3/MPK6 results in the accumulation of ERF6 protein in vivo, which positively regulates defense gene expression and plant resistance to the necrotrophic fungal pathogen Botrytis cinerea.
Abstract
Arabidopsis thaliana MPK3 and MPK6, two mitogen-activated protein kinases (MAPKs or MPKs), play critical roles in plant disease resistance by regulating multiple defense responses. Previously, we characterized the regulation of phytoalexin biosynthesis by Arabidopsis MPK3/MPK6 cascade and its downstream WRKY33 transcription factor. Here, we report another substrate of MPK3/MPK6, ETHYLENE RESPONSE FACTOR6 (ERF6), in regulating Arabidopsis defense gene expression and resistance to the necrotrophic fungal pathogen Botrytis cinerea. Phosphorylation of ERF6 by MPK3/MPK6 in either the gain-of-function transgenic plants or in response to B. cinerea infection increases ERF6 protein stability in vivo. Phospho-mimicking ERF6 is able to constitutively activate defense-related genes, especially those related to fungal resistance, including PDF1.1 and PDF1.2, and confers enhanced resistance to B. cinerea. By contrast, expression of ERF6-EAR, in which ERF6 was fused to the ERF-associated amphiphilic repression (EAR) motif, strongly suppresses B. cinerea–induced defense gene expression, leading to hypersusceptibility of the ERF6-EAR transgenic plants to B. cinerea. Different from ERF1, the regulation and function of ERF6 in defensin gene activation is independent of ethylene. Based on these data, we conclude that ERF6, another substrate of MPK3 and MPK6, plays important roles downstream of the MPK3/MPK6 cascade in regulating plant defense against fungal pathogens.
INTRODUCTION
Plants have sophisticated surveillance systems to sense invading pathogens through the recognition of pathogen/microbe-associated molecular patterns or pathogen-derived effectors. After the sensing step, signals generated at the receptors/sensors are converted to a wide range of defense responses through various signal transduction pathways (reviewed in Ausubel, 2005; Glazebrook, 2005; Jones and Dangl, 2006; Boller and Felix, 2009; Dodds and Rathjen, 2010; Nishimura and Dangl, 2010; Spoel and Dong, 2012). Mitogen-activated protein kinase (MAPK) cascades are important signaling modules in this process (reviewed in Zhang and Klessig, 2001; Ichimura et al., 2002; Pedley and Martin, 2005; Zhang, 2008; Pitzschke et al., 2009; Andreasson and Ellis, 2010; Rodriguez et al., 2010; Tena et al., 2011). Arabidopsis thaliana has three pathogen-responsive MAPKs: MPK3, MPK4, and MPK6. MPK3 and MPK6, which show a high level of functional redundancy, are downstream of MKK4 and MKK5, two redundant MAPK kinases (Asai et al., 2002; Ren et al., 2002; Wang et al., 2007; Ren et al., 2008). MPK4 forms the other independent pathogen-responsive MAPK cascade with its upstream MKK1 and MKK2 (two redundant MAPK kinases) and MEKK1 (a MAPK kinase kinase) (Petersen et al., 2000; Suarez-Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008).
In a MAPK cascade, the MAPK kinase kinases receive signals from the sensors/receptors and activate downstream MAPK kinases, which then activate the bottom tier MAPKs through phosphorylation. The outputs of a MAPK cascade are determined by the phosphorylation of MAPK substrates, which can be enzymes, transcription factors, and proteins with other biochemical functions (Zhang, 2008; Tena et al., 2011). Increasing evidence indicates that MPK3 and MPK6 act as positive regulators of defense responses, whereas MPK4 plays both positive and negative roles in regulating plant defense (Ren et al., 2002, 2008; Liu and Zhang, 2004; Gao et al., 2008; Zhang et al., 2012). Identification of the first plant MAPK substrate revealed that MPK3/MPK6 positively regulate ethylene production through phosphorylation and stabilization of the rate-limiting 1-aminocyclopropane-1-carboxylic acid synthase (ACS) isoforms, ACS2 and ACS6 (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). Recently, we demonstrated that MPK3/MPK6 play a positive role in regulating the biosynthesis of camalexin by phosphorylating the WRKY33 transcription factor, which promotes the expression of camalexin biosynthetic genes (Ren et al., 2008; Mao et al., 2011). The MPK3/MPK6 cascade has also been implicated in many other defense responses, including defense gene activation, reactive oxygen species (ROS) generation, hypersensitive response–like cell death, stomatal closure, and disease resistance (Ren et al., 2002; Kim and Zhang, 2004; Liu et al., 2007; Asai et al., 2008; Gudesblat et al., 2009; Galletti et al., 2011). However, the molecular mechanisms underlying the functions of MPK3/MPK6 remain largely unclear due to the lack of information about their substrates.
Ethylene response factors (ERFs) constitute the largest family of transcription factors in Arabidopsis with more than 120 members (Nakano et al., 2006). Many ERFs have been implicated in plant defense responses (Oñate-Sánchez and Singh, 2002; Gutterson and Reuber, 2004; McGrath et al., 2005). Several members of the Arabidopsis ERF family, including ERF1 (At3g23240), At-ERF1 (At4g17500), ERF2 (At5g47220), ERF14 (At1g04370), and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59, At1g06160), are induced at transcriptional level in response to pathogen infection (Oñate-Sánchez and Singh, 2002; Oñate-Sánchez et al., 2007; Pré et al., 2008). ERF1 and ORA59 are activated synergistically by ethylene and jasmonic acid (JA), suggesting that they might function as the integrating points of JA and ethylene signaling pathways (Lorenzo et al., 2003; Pré et al., 2008). Constitutive overexpression of ERF1, ERF2, or ORA59 activates the expression of several defense-related genes, including PDF1.2 and ChiB, and was shown to confer resistance to a range of pathogens (Berrocal-Lobo et al., 2002; Brown et al., 2003; McGrath et al., 2005; Pré et al., 2008). Although most ERFs identified so far are transcription activators, several ERF-associated amphiphilic repression (EAR) motif-containing ERFs, including Arabidopsis ERF3 (At1g50640) and ERF4 (At3g15210), were shown to function as transcriptional repressors (Ohta et al., 2001; Hiratsu et al., 2003).
Recently, several ERF transcription factors have been shown to be the substrates of pathogen-responsive MAPKs (Cheong et al., 2003; Bethke et al., 2009). Arabidopsis ERF104 was identified to be a MPK6 substrate that plays important roles in plant resistance to a nonadapted bacterial pathogen (Bethke et al., 2009). In rice (Oryza sativa), ETHYLENE RESPONSE ELEMENT BINDING PROTEIN1 (EREBP1) was reported to be phosphorylated by the BLAST- AND WOUNDING-INDUCED MAP KINASE1 (BWMK1). (Cheong et al., 2003). However, whether other members of the large ERF family can be phosphorylated by MAPKs and the function of MAPK-mediated phosphorylation of ERFs in plant defense remain to be defined. In this report, we identify ERF6, an ERF transcription factor, as a substrate of both MPK3 and MPK6 in Arabidopsis. The two clustered Ser-Pro sites in the C-terminal region of ERF6 are phosphorylated by MPK3/MPK6 in vitro and in vivo. MPK3/MPK6-mediated phosphorylation of ERF6 increases its protein stability in vivo. Gain-of-function expression of a phospho-mimicking mutant of ERF6 and loss-of-function expression of a chimeric ERF6-EAR repressor demonstrate that ERF6 plays important roles downstream of MPK3/MPK6 in regulating plant defense in response to fungal pathogen, including defensin gene activation and fungal resistance.
RESULTS
In Vitro Phosphorylation of ERF6 by MPK3 and MPK6
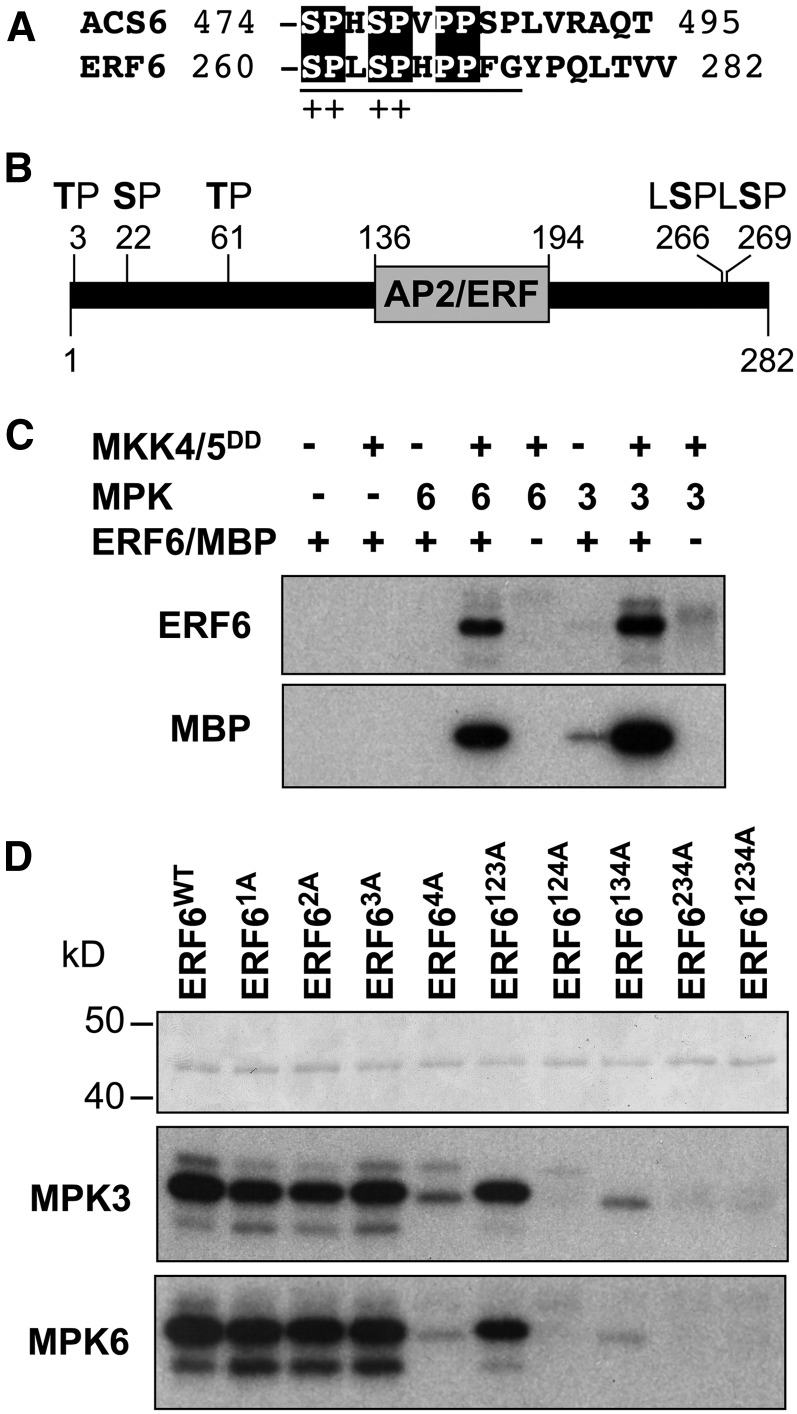
After the identification of the pair of plant MAPK substrates, ACS2 and ACS6, two isoforms of the Arabidopsis ACS family (Liu and Zhang, 2004), we searched the database for proteins with homology to the C-terminal 16–amino acid region of ACS6 that contains the MPK3/MPK6 phosphorylation sites. AAL38331, annotated as an unknown protein at the time, was identified (Figure 1A). This protein is identical to ERF6 (At4g17490). To determine whether MPK3/MPK6 can phosphorylate ERF6, we prepared His6-tagged recombinant ERF6 protein. As a result of either proteolytic degradation, premature termination, or both, the purified recombinant ERF6 preparation contained many truncated ERF6 proteins. As a result, we generated a double-tagged ERF6 with an N-terminal His6 tag and a C-terminal FLAG tag. Sequential purification using His-tag column and anti-FLAG antibody affinity gel yielded almost homogeneous ERF6 recombinant protein to be used in the phosphorylation assays. As shown in Figure 1C, activated recombinant MPK3 and MPK6 strongly phosphorylated ERF6. Without activation by MKK4/MKK5, MPK3 and MPK6 showed little kinase activity toward ERF6. Control reactions using myelin basic protein as a substrate confirmed the normal activation of MPK3 and MPK6 after incubation with the constitutively active MKK4/MKK5.
Figure 1.
MPK3 and MPK6 Phosphorylate ERF6 on the C-Terminal Motif That Shares Homology with the C Terminus of ACS6.
(A) Alignment of the C termini of ACS6 and ERF6. Plus symbols indicate the conserved MAPK phosphorylation sites.
(B) Diagram of ERF6 protein with the putative MAPK phosphorylation sites marked.
(C) Phosphorylation of ERF6 recombinant protein by activated MPK3 and MPK6. Activated MPK3 and MPK6 were used to phosphorylate purified double-tagged ERF6 (top panel). Side-by-side control reactions using myelin basic protein (MBP) as a substrate validated the activation of MPK3 and MPK6 by the constitutively active MKK4 and MKK5 (bottom panel).
(D) Identification of the MPK3 and MPK6 phosphorylation sites in ERF6 based on in vitro phosphorylation assay. Single and high-order Ser/Thr-to-Ala mutant ERF6 proteins were purified and normalized to the same concentration as validated by Coomassie blue staining (top panel). The ability of ERF6 and its mutants to be phosphorylated by MPK3 (middle panel) and MPK6 (bottom panel) was determined by in vitro phosphorylation assays.
In addition to Ser-266 and Ser-269 at the C terminus of ERF6 that share similarity to the MPK3/MPK6 phosphorylation sites in ACS2 and ACS6, there are three additional potential MAPK phosphorylation sites with Ser/Thr residues followed by a Pro residue, including Thr-3, Ser-22, and Thr-61 (Figure 1B). To map the MPK3/MPK6 phosphorylation sites in ERF6, we generated a serial of Ser/Thr to Ala mutants. For simplicity, we designated these putative phosphorylation sites with numbers, Thr-3 as 1, Ser-22 as 2, Thr-61 as 3, and Ser-266/Ser-269 together as 4. Because of the close proximity between Ser-266 and Ser-269 residues, we mutated both residues at the same time. Double-tagged recombinant proteins with various combinations of mutations were purified (Figure 1D, top panel), and their ability to serve as MPK3/MPK6 substrates was determined by in vitro phosphorylation assays. As shown in Figure 1D (middle and bottom panels), mutation of Thr-3 (ERF61A), Ser-22 (ERF62A), and Thr-61 (ERF63A) one at a time, or all three at once (ERF6123A), did not affect their phosphorylation by MPK3 or MPK6. By contrast, phosphorylation of the S266A/S269A mutant (ERF64A) protein by MPK3 and MPK6 was greatly reduced, demonstrating that these two Ser residues are the major MPK3/MPK6 phosphorylation sites in vitro. Additional higher order mutants containing the 4A mutation (ERF6124A, ERF6134A, ERF6234A, and ERF61234A) revealed that the weak phosphorylation of ERF64A was on the Ser-22 residue since ERF6134A, but not ERF6124A, ERF6234A, or ERF61234A, can be weakly phosphorylated by MPK3 and MPK6.
Phosphorylation of ERF6 by MPK3 and MPK6 in Vivo in Response to Botrytis cinerea Infection
To determine if ERF6 can be phosphorylated by MPK3/MPK6 in vivo, we transformed ERF6 and its Ser/Thr to Ala mutants into the conditional gain-of-function GVG-Nt-MEK2DD transgenic Arabidopsis (abbreviated as DD) (Yang et al., 2001; Ren et al., 2002). Constitutive 35S promoter was used to drive ERF6 transgenes, and a 4myc tag was added to the N terminus of ERF6 for protein detection. Transgenic lines with ERF6 expression were screened from T1 populations by immunoblot analysis using anti-myc antibody. T2 lines with a single transgene insertion and similar levels of transgene expression were used for experiments.
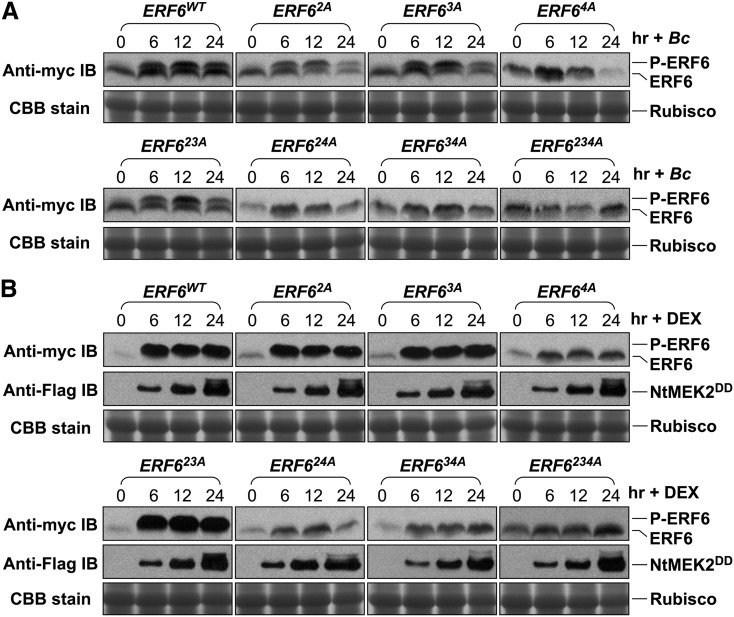
We first investigated the in vivo phosphorylation of ERF6 protein in response to pathogen infection. Twelve-day-old ERF6WT/DD, ERF62A/DD, ERF63A/DD, ERF64A/DD, ERF623A/DD, ERF624A/DD, ERF634A/DD, and ERF6234A/DD transgenic seedlings were inoculated with B. cinerea spores, and samples were collected at different times after inoculation. In the absence of the dexamethasone (DEX) inducer, DD transgene was not expressed, and the effect of B. cinerea infection can be determined. We observed an upshift of myc-tagged wild-type ERF6 protein (Figure 2A, first panel), suggesting a modification of ERF6WT protein, possibly by phosphorylation, in response to B. cinerea. The use of Phos-Tag reagent confirmed that the upshift of ERF6 band after B. cinerea inoculation was a result of phosphorylation modification (see Supplemental Figure 1 and Supplemental Methods 1 online). Furthermore, mutation of Ser-266/Ser-269 residues (ERF64A) either alone or in combination with other putative MAPK phosphorylation sites (ERF624A, ERF634A, and ERF6234A) abolished the upshift of ERF6 protein. By contrast, mutants with Ser-266/Ser-269 residues unchanged (ERF62A, ERF63A, and ERF623A) had the normal band upshift as the wild-type ERF6 protein (Figure 2A). These results indicate that (1) the upshifted bands are indeed a result of phosphorylation, and (2) S266 and S269 residues of ERF6, two MPK3/MPK6 phosphorylation sites identified based on in vitro phosphorylation assay, were phosphorylated in vivo in response to B. cinerea infection.
Figure 2.
Phosphorylation of ERF6 on Ser-266 and Ser-269 Residues by MPK3 and MPK6 in Vivo.
(A) Twelve-day-old 35S:ERF6WT, 35S:ERF62A, 35S:ERF63A, 35S:ERF64A, 35S:ERF623A, 35S:ERF624A, 35S:ERF634A, and 35S:ERF6234A transgenic seedlings in DD background were inoculated with B. cinerea spores, and samples were collected at different times after inoculation. ERF6 and its mutant proteins were detected by immunoblot (IB) analysis using anti-myc antibody (top panels). Equal loading of proteins was confirmed by Coomassie brilliant blue (CBB) stained gels (bottom panels). Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
(B) The same set of transgenic seedlings as in (A) was treated with DEX (1 µM final concentration), and samples were collected at different times. Protein extracts were prepared and analyzed by immunoblot analysis using anti-myc antibody (top panels). Induction of DD protein by DEX treatment was determined by immunoblots using anti-Flag antibody (middle panels). Equal loading of proteins was confirmed by Coomassie blue–stained gels (bottom panels).
To confirm that ERF6 can be phosphorylated by MPK3/MPK6 in vivo, we also analyzed the phosphorylation status of ERF6 in the ERF6WT/DD, ERF62A/DD, ERF63A/DD, ERF64A/DD, ERF623A/DD, ERF624A/DD, ERF634A/DD, and ERF6234A/DD transgenic plants treated with DEX, which induces the expression of constitutively active Nt-MEK2DD and activates the downstream MPK3/MPK6 (Ren et al., 2002, 2008; Mao et al., 2011). As shown in Figure 2B, within 6 h after DEX treatment, the majority of the ERF6WT, ERF62A, ERF63A, and ERF623A proteins were phosphorylated, as indicated by the upshift of the protein bands. By contrast, no such upshift was observed in ERF64A/DD, ERF624A/DD, ERF634A/DD, and ERF6234A/DD plants after DEX treatment. In all the transgenic lines, the induction of DD expression after DEX treatment was similar (Figure 2B, middle panels), eliminating the potential of variable DD transgene induction in different lines, which would change the band-shift patterns as well. The mobility upshift of ERF6 proteins induced by gain-of-function MPK3/MPK6 activation is dependent on the same Ser residues as that induced by B. cinerea infection (Figures 2A and 2B), indicating that ERF6 is phosphorylated in vivo by MPK3/MPK6 on the Ser-266 and Ser-269 residues in response to B. cinerea infection.
MPK3/MPK6-Mediated Phosphorylation Stabilizes ERF6 Protein in Vivo
We noticed that, within 6 h after DEX treatment, ERF6 proteins accumulated to much higher levels in ERF6WT/DD, ERF62A/DD, ERF63A/DD, and ERF623A/DD plants despite the expression of ERF6 transgenes was driven by the constitutive 35S promoter (Figure 2B). The accumulated ERF6 proteins were in the phosphorylated form, as indicated by their slower mobility in the gel. Mutation of Ser-266/Ser-269, two MPK3/MPK6 phosphorylation sites, abolished the accumulation of ERF6 proteins as seen in ERF64A/DD, ERF624A/DD, ERF634A/DD, and ERF6234A/DD plants. These results indicate that phosphorylation of ERF6 by MPK3/MPK6 could increase the stability of ERF6 protein. Although to a lesser extent than that induced in the gain-of-function DD plants after DEX treatment, B. cinerea infection also resulted in the increase in total and phosphorylated ERF6 proteins in ERF6WT, ERF62A, ERF63A, and ERF623A plants (Figure 2A). Due to the progressive nature of pathogen infection, only a fraction of the cells are responding to the pathogen with MAPK activation at a certain time. By contrast, in DD plants, application of DEX can activate MPK3/MPK6 in all cells synchronously, thus leading to higher levels of ERF6 accumulation.
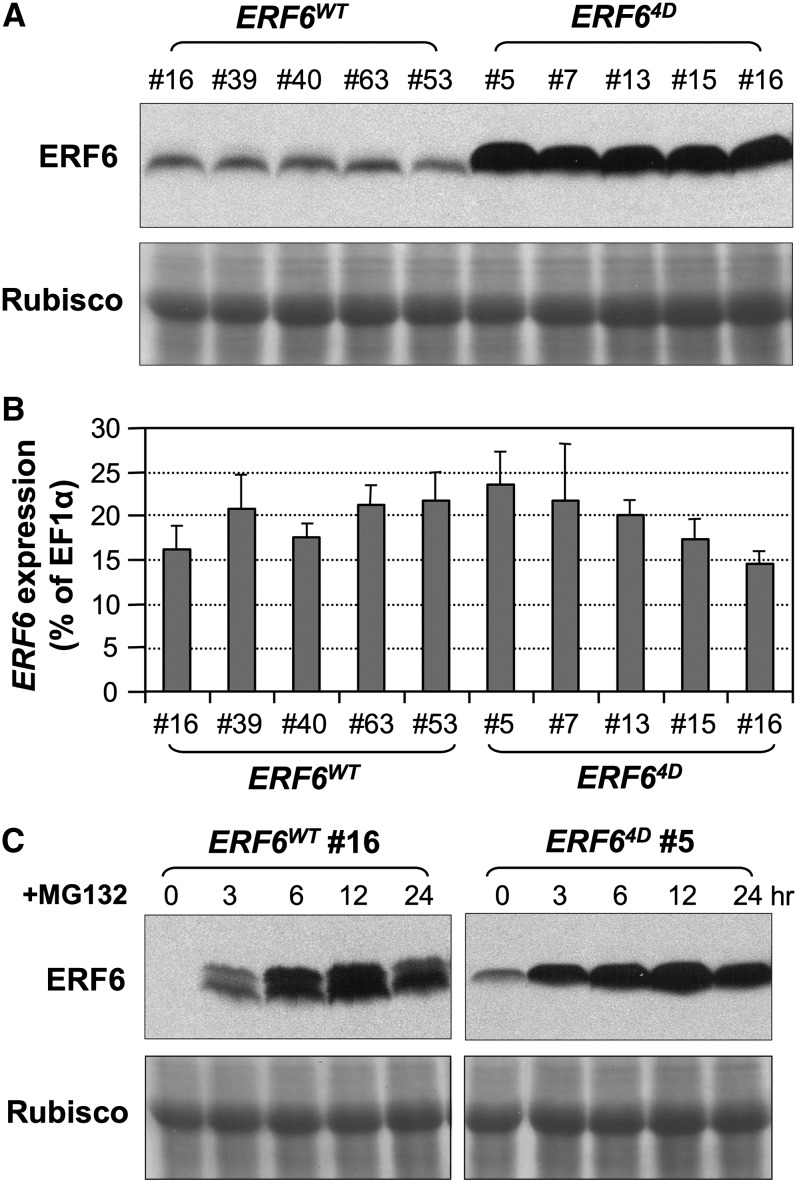
To further demonstrate the regulation of ERF6 stability by phosphorylation, we generated transgenic plants that express the 4myc-tagged phospho-mimicking ERF64D, ERF6 with Ser-266/Ser-269 mutated to Asp, under the control of 35S promoter in Columbia-0 (Col-0). With the negative charge on Asp, ERF64D may mimic the phosphorylated form of ERF6 and lead to the constitutive accumulation of ERF6 in 35S:ERF64D transgenic plants. Consistent with this speculation, 35S:ERF64D plants accumulated much higher levels of ERF6 protein than 35S:ERF6WT plants (Figure 3A), although the transcript levels of ERF6 transgenes were comparable in 35S:ERF64D and 35S:ERF6WT lines (Figure 3B).
Figure 3.
Stability Regulation of ERF6 Protein in Vivo by Phosphorylation.
(A) and (B) Differential accumulation of ERF6WT and phospho-mimicking ERF64D proteins in lines with similar levels of transgene expression. Twelve-day-old 35S:ERF6WT and 35S:ERF64D transgenic seedlings (five independent lines each) were collected for protein and RNA preparations. Levels of ERF6 protein were determined by immunoblots using anti-myc antibody (A), and transgene expression at mRNA level was determined by real-time qPCR (B). Error bars indicate se (n = 3). Primers used for qPCR detect both endogenous ERF6 and transgene transcripts. Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
(C) Degradation of ERF6 protein by the proteasome pathway. Twelve-day-old 35S:ERF6WT and 35S:ERF64D transgenic seedlings were treated with MG132, a 26S proteasome inhibitor, and samples were collected at different times. Levels of ERF6 protein were determined by immunoblots using anti-myc antibody. Equal loading of proteins was confirmed by Coomassie blue–stained gels (bottom panels).
We then treated 35S:ERF6WT and 35S:ERF64D transgenic seedlings with MG132, a specific inhibitor of the 26S proteasome, to determine whether the ubiquitin-proteasome pathway is involved in the degradation of ERF6. As shown in Figure 3C, MG132 treatment greatly increased the levels of ERF6WT and ERF64D proteins in a time-dependent manner, indicating that both ERF6WT and ERF64D could be degraded by the proteasome-dependent pathway. However, the levels of ERF64D without MG132 treatment were much higher than that of ERF6WT (Figures 3A and 3C), suggesting that phosphorylation of ERF6 attenuates the degradation of ERF6, thus leading to an increase in ERF6 protein stability. The accumulation of doublet bands in 35S:ERF6WT plants is likely to be a result of the accumulation of both phosphorylated and unphosphorylated ERF6 due to the inhibition of the 26S proteasome degradation pathway. The presence of the phosphorylated ERF6 form could be a result of the handling of seedlings during inhibitor treatment, which is known to activate MPK3/MPK6.
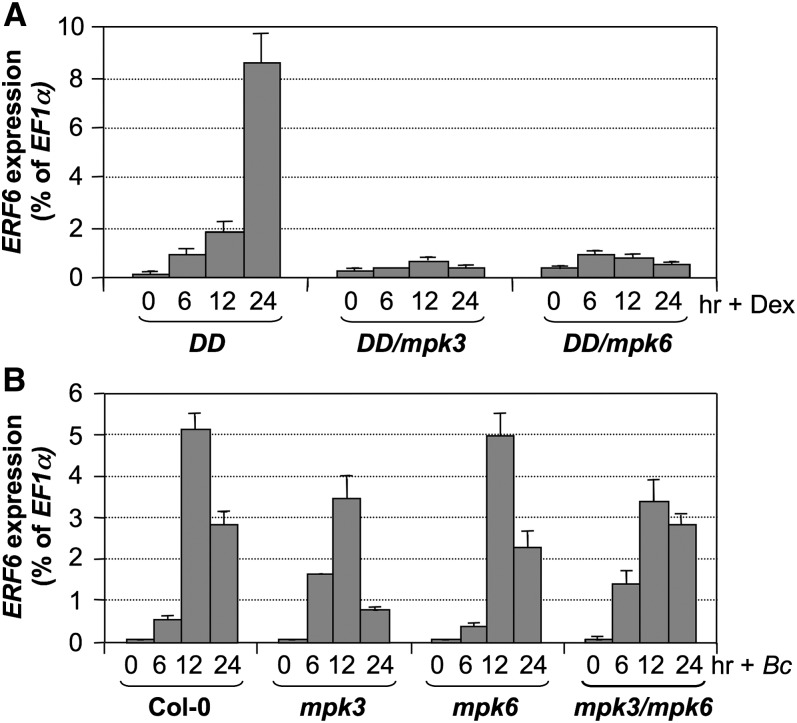
Induction of ERF6 Gene Expression by Pathogen Infection and Gain-of-Function Activation of MPK3/MPK6
Previously, we observed that MPK3/MPK6 regulate the expression of their substrate genes, such as in the cases of ACS2/ACS6 and WRKY33 (Mao et al., 2011; Li et al., 2012). As a result, we examined the expression of ERF6 in the gain-of-function DD plants after DEX treatment. As shown in Figure 4A, DEX treatment of DD plants strongly induced the expression of ERF6 gene, and the induction was dependent on the endogenous MPK3 and MPK6. Mutation of either MPK3 or MPK6 compromised the ERF6 gene activation in DD plants after the DEX treatment. The induction of DD expression and the activation of MPK3/MPK6 in DD, DD/mpk3, and DD/mpk6 plants have been validated in our previous report (Mao et al., 2011), and the same reverse transcription sample sets were used for this study. These results indicate that the MPK3/MPK6 cascade is involved in ERF6 gene activation.
Figure 4.
Activation of ERF6 Gene Expression during B. cinerea Infection Involves Both MPK3/MPK6-Dependent and -Independent Pathways.
(A) Induction of ERF6 expression in the conditional gain-of-function DD plants. Twelve-day-old DD, DD/mpk3, and DD/mpk6 seedlings were treated with 1 µM DEX. Samples were collected at indicated times. Expression of ERF6 was quantified by real-time PCR and calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
(B) Induction of ERF6 expression in response to B. cinerea infection. Twelve-day-old wild-type (Col-0), mpk3, mpk6, and rescued mpk3 mpk6 double mutant seedlings were inoculated with B. cinerea spores. Samples were collected at indicated times. Expression levels of ERF6 were quantified by real-time PCR and were calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
To provide loss-of-function evidence, we further examined ERF6 expression in B. cinerea–infected wild-type plants, single mpk3 or mpk6 mutants, and a rescued mpk3 mpk6 double mutant (Wang et al., 2007; Ren et al., 2008). As shown in Figure 4B, infection of wild-type plants by B. cinerea also strongly induced ERF6 expression. The activation of ERF6 was slightly reduced in mpk3 single mutant and the rescued mpk3 mpk6 double mutant, but not the mpk6 single mutant (Figure 4B). These data suggest that, although the gain-of-function activation of MPK3/MPK6 is sufficient to activate ERF6 expression, the MPK3/MPK6 cascade is not the only signaling pathway that can activate ERF6 gene expression in response to B. cinerea infection.
Expression of Phospho-Mimicking ERF64D and Dominant-Negative ERF6-EAR Result in Opposite Plant Growth Phenotypes
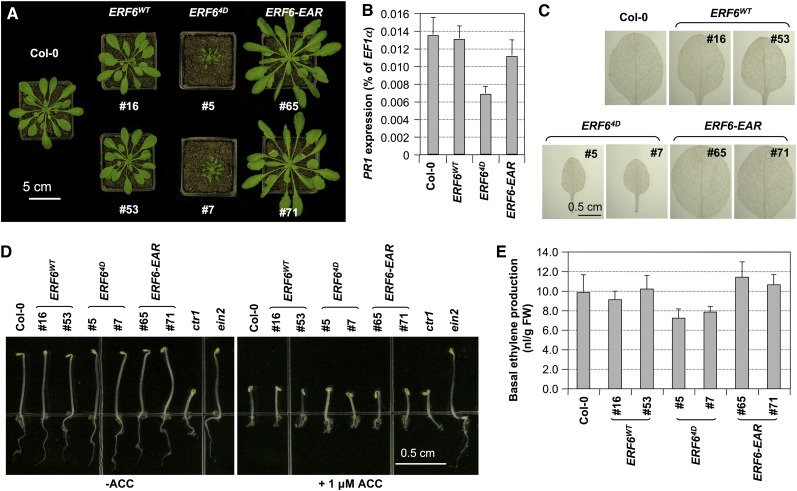
In the process of generating transgenic plants for functional analysis, we noticed that 35S:ERF64D plants exhibited a severe dwarf phenotype. They were normal at the seedling stage, but after 2 weeks, their growth was retarded. By 7 weeks under short-day growth conditions, the rosettes of 35S:ERF64D plants were much smaller than those of the wild type, despite having a similar number of leaves (Figure 5A). By contrast, 35S:ERF6WT plants were almost the same size as the wild type (Figure 5A), suggesting that mimicking phosphorylation of ERF6 is important for the dwarf phenotype of the 35S:ERF64D plants.
Figure 5.
Morphological Phenotypes of the Loss- and Gain-of-Function ERF6 Transgenic Plants Are Not Associated with Constitutive PR1 Expression, ROS Generation, or Alteration in Ethylene Signaling/Production.
(A) Expression of the phospho-mimicking ERF64D and dominant-negative ERF6-EAR confer opposite morphological phenotypes. Images of 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR, along with Col-0 control plants grown under short-day conditions, were taken at 7 weeks.
(B) PR1 gene expression in Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants was quantified by real-time qPCR. PR1 transcript levels were calculated as percentages of the EF1α transcript. Error bars indicate sd (n = 3).
(C) ROS accumulation in Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants was detected by DAB staining.
(D) Normal triple responses of etiolated 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings. ACC, 1-aminocyclopropane-1-carboxylic acid.
(E) Normal basal level production of ethylene in 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings. FW, fresh weight.
[See online article for color version of this figure.]
To gain loss-of-function evidence to support the role of ERF6, we identified a T-DNA insertion mutant of ERF6 from the ABRC seed collection (SALK_087356). The erf6 mutant plants did not show any visible phenotype, possibly due to the presence of three other closely related ERF homologs, including ERF5 (At5g47230), ERF104 (At5g61600), and ERF105 (At5g51190) in the Arabidopsis genome (Nakano et al., 2006). Similar to ERF6, these three homologs also contain conserved MAPK phosphorylation sites at their C termini (see Supplemental Figure 2 online), suggesting a similar mode of regulation and function. We acquired T-DNA insertion mutants of these three ERF genes and generated erf6 erf5, erf6 erf104, and erf6 erf105 double mutants. Attempt to generate high-order mutants by genetic cross failed. ERF5, ERF104, and ERF105 are all located at the lower arm of chromosome 5. We could not identify any progenies with crossovers between erf5 and erf104, erf5 and erf105, or erf104 and erf105. Physically, ERF5 and ERF104 are more than 5.5 million base pairs apart. However, we failed to identify a single crossover after genotyping more than 1000 F2 progenies.
We then used the EAR suppressor domain identified in tobacco (Nicotiana tabacum) ERF3 (Ohta et al., 2001) to convert ERF6 to a dominant suppressor to gain loss-of-function data. This approach has been successfully used in several studies (Hiratsu et al., 2003; Mitsuda et al., 2011). The EAR suppressor domain was introduced into the C terminus of ERF6 before the stop codon in 35S:ERF6WT, and the resulting construct was named 35S:ERF6-EAR. We found that the expression of ERF6-EAR resulted in an opposite effect on plant growth in comparison with the expression of ERF64D. As shown in Figure 5A, 35S:ERF6-EAR transgenic plants were much bigger than wild-type plants.
The dwarf phenotype of 35S:ERF64D plants is similar to that of ctr1 mutant, which has constitutive activation of ethylene responses (Kieber et al., 1993). As a result, we examined the triple response of 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings. As shown in Figure 5D, all three genotypes had the wild-type etiolated seedling phenotype. In addition, all of them could respond to 1-aminocyclopropane-1-carboxylic acid treatment and displayed a typical triple response the same way as wild-type seedlings. In addition, 35S:ERF64D plants did not overproduce ethylene (Figure 5E). These results demonstrate that the dwarf phenotype of 35S:ERF64D plants is not a result of constitutive ethylene responses or ethylene overproduction.
The dwarf phenotype of 35S:ERF64D plants is also reminiscent of the mpk4 mutant, which shows constitutive activation of salicylic acid (SA)–mediated defense pathways (Petersen et al., 2000). We examined the PR1 gene expression in 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants and found no accumulation of PR1 transcripts (Figure 5B). Furthermore, no accumulation of ROS was detected in 35S:ERF64D plants as determined by 3,3′-diaminobenzidine (DAB) staining (Figure 5C), which is again different from that of mpk4 mutant (Ichimura et al., 2006; Nakagami et al., 2006). These results suggest that the dwarf phenotype of 35S:ERF64D plants is not associated with constitutive activation of SA signaling pathway, which distinguishes it from mpk4 mutant.
Expression of Phospho-Mimicking ERF64D and Dominant-Negative ERF6-EAR Results in Opposite Phenotype in Plant Resistance to Fungal Pathogen
To identify molecular markers associated with the phenotypes of loss- and gain-of-function ERF6 transgenic plants, we profiled gene expression in Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants by Illumina RNA sequencing. 35S:ERF64D plants showed 245 up- and 75 downregulated genes (at least twofold changes, P value ≤ 0.001) (see Supplemental Figure 3 and Supplemental Data Set 1 online). A large number of the upregulated genes in 35S:ERF64D plants are defense-related genes involved in plant resistance against fungal pathogens, such as plant defensins (see Supplemental Data Set 1 online). Most genes that showed higher expression in 35S:ERF64D plants were repressed in 35S:ERF6-EAR plants, but 35S:ERF6WT plants only showed elevated expression of a few genes and the levels of induction were much lower in comparison to those in 35S:ERF64D plants. These results further support that the phosphorylation of ERF6 is important for its function.
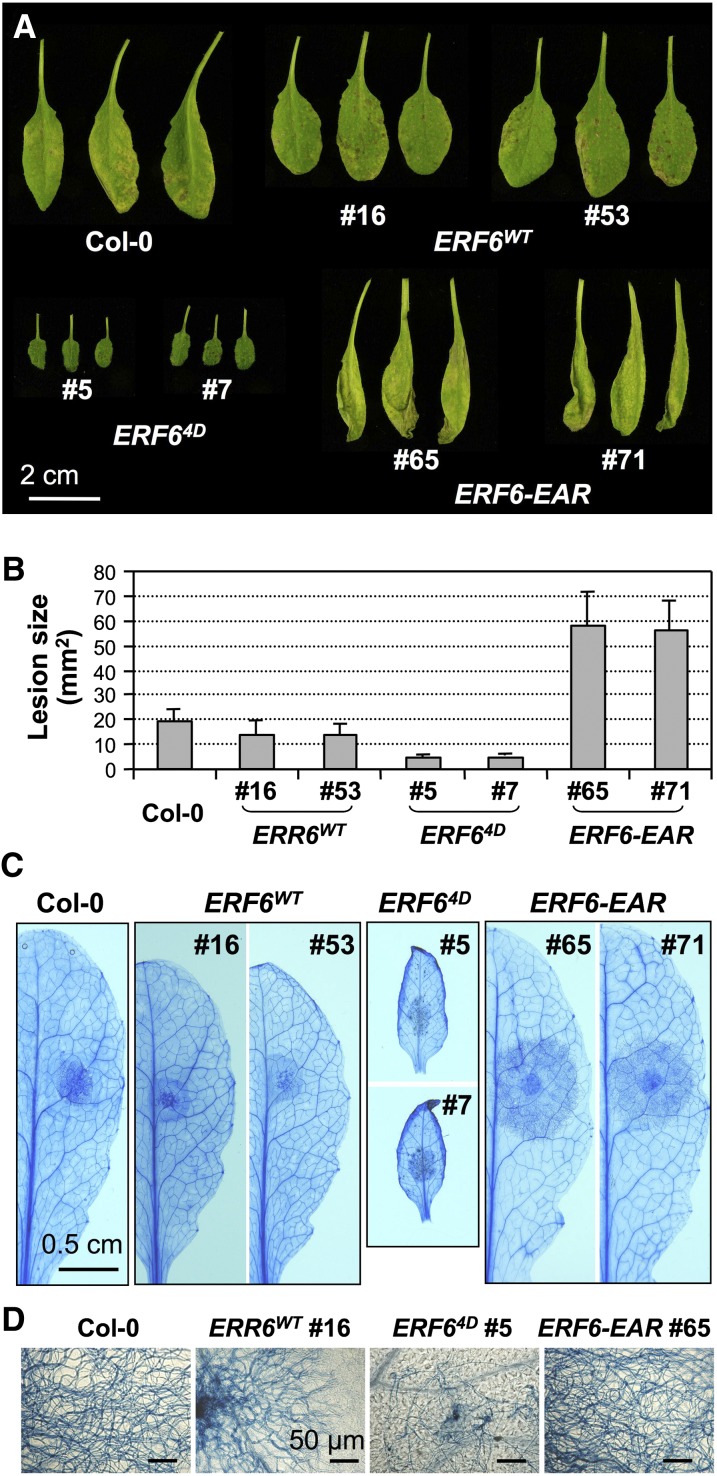
The high-level induction of fungal-related defense genes in 35S:ERF64D plants prompted us to test the resistance of 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants against B. cinerea. When spray-inoculated, B. cinerea caused much more damage to 35S:ERF6-EAR plants than to the wild-type control plants, suggesting that the suppression of ERF6 function increased plant susceptibility to B. cinerea (Figure 6A; see Supplemental Figure 4 online). By contrast, 35S:ERF64D plants showed higher resistance with no obvious necrosis, and 35S:ERF6WT plants were slightly more resistant to B. cinerea than the wild-type control plants. To better quantify plant resistance to B. cinerea, we used drop inoculation, in which 5-μL droplets of spore suspension were placed on fully expanded leaves. After 3 d, the lesion sizes were measured. As shown in Figure 6B, compared with wild-type plants, 35S:ERF6-EAR plants exhibited much larger lesions, while 35S:ERF64D plants showed significantly smaller lesions. The lesion size of 35S:ERF6WT plants was a little smaller than that of wild-type plants. These results are in agreement with those obtained using spray inoculation (Figure 6A).
Figure 6.
Expression of Phospho-Mimicking ERF64D Confers Enhanced Resistance to B. cinerea, Whereas Expression of a Dominant-Negative ERF6-EAR Results in an Opposite Phenotype.
(A) Six-week-old Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants grown under short-day conditions were spray inoculated with B. cinerea spore suspension (1 × 105 spores/mL). Leaf images were taken at 3 d after inoculation.
(B) Fully expanded leaves from Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants were drop inoculated with B. cinerea spore suspension (5 μL per droplet, and 1 × 105 spores/mL). Lesion size was measured 3 d after inoculation. Error bars indicate sd (n = 40 to 50).
(C) Visualization of hyphae growth in drop-inoculated leaves by lactophenol-trypan blue staining.
(D) The absence of hyphal growth in 35S:ERF64D plants. The lactophenol-trypan blue stained leaves shown in (C) were examined under a microscope.
To visualize the growth of B. cinerea in plant tissues, we stained the hyphae by lactophenol-trypan blue staining. As shown in Figure 6C, the growth of B. cinerea in ERF6-EAR plants was very extensive, spread far beyond the inoculated areas. By contrast, no or little hyphal growth in plant tissues was visible in ERF64D plants. Observation under a microscope revealed no invasive growth of B. cinerea hyphae in ERF64D plant tissues at all, and the hyphal growth stopped soon after spore germination (Figure 6D). The hyphal growth in ERF6WT was slightly inhibited, consistent with the visual phenotype on these plants after B. cinerea spore inoculation.
Opposing Effects of ERF64D and ERF6-EAR Transgenes on Plant Resistance to B. cinerea Correlate with the Expression of Antifungal Defensin Genes
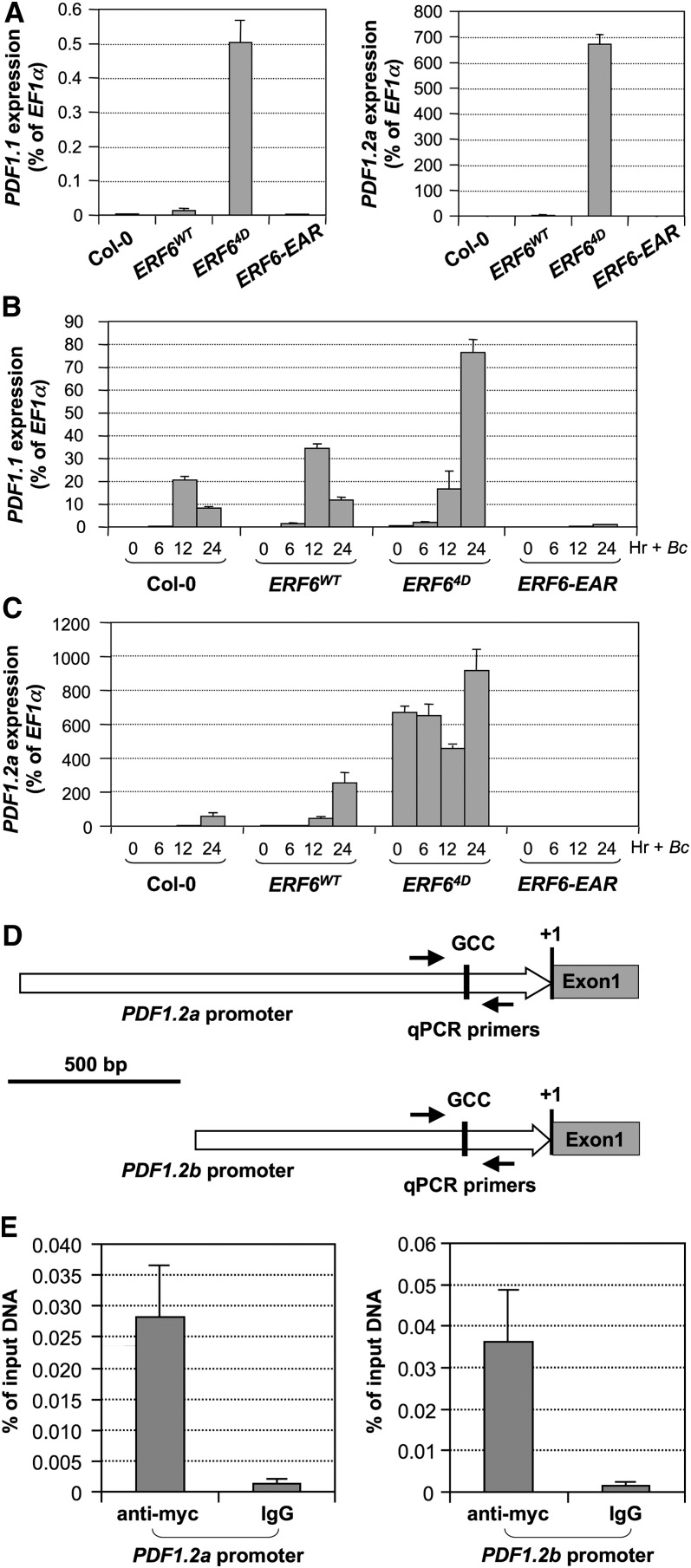
Plant defensins play an important role in resistance against necrotrophic fungal pathogens (Terras et al., 1995; Penninckx et al., 1996; Gao et al., 2000; Lay and Anderson, 2005). Gene expression profiling of the gain-of-function 35S:ERF64D plants revealed high basal-level expression of several defensin genes, including PDF1.1 and PDF1.2a (see Supplemental Data Set 1 online). This was further confirmed by real-time quantitative RT-PCR analysis (Figure 7A). To investigate the role of ERF6 in regulating defensin gene expression in response to B. cinerea infection, we also quantified the induction of PDF1.1 and PDF1.2a gene expression at different times after B. cinerea spore inoculation. As shown in Figure 7B, PDF1.1 expression in 35S:ERF64D plants was further elevated after inoculation with B. cinerea. Different from PDF1.1, the basal-level expression of PDF1.2a in 35S:ERF64D had already reached a level close to the maximum after B. cinerea infection (Figure 7C). As a result, B. cinerea inoculation failed to elevate PDF1.2a expression further. Induction of both PDF1.1 and PDF1.2a in 35S:ERF6WT plants was higher than that in Col-0, but much lower than that in the 35S:ERF64D plants, which is consistent with the slightly higher resistance of 35S:ERF6WT plants to B. cinerea (Figure 6). In contrast with the gain-of-function 35S:ERF64D transgenic plants, the basal expression levels of PDF1.1 and PDF1.2a were much lower in 35S:ERF6-EAR transgenic plants. Furthermore, the induction of PDF1.1 and PDF1.2 in 35S:ERF6-EAR plants by B. cinerea infection was almost completely abolished (Figures 7A and 8B, left panels).
Figure 7.
Opposing Effects of ERF64D and ERF6-EAR Transgenes on Defensin Gene Induction, Which Are Associated with Their Sensitivities to B. cinerea Infection.
(A) Elevated basal level expression of defensin genes in 35S:ERF64D plants. Twelve-day-old Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings were collected without any treatment. The expression of PDF1.1 (left) and PDF1.2a (right) genes was quantified by real-time qPCR.
(B) and (C) Opposing effects of ERF64D and ERF6-EAR transgenes on defensin gene induction. Twelve-day-old Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings were inoculated with B. cinerea spore, and samples were collected at indicated times. The expression of PDF1.1 (B) and PDF1.2a (C) genes was quantified by real-time qPCR and presented as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
(D) Diagrams showing the GCC box in the promoters of PDF1.2a and PDF1.2b genes.
(E) ERF6 binds to the promoters of PDF1.2a and PDF1.2b genes in vivo. ChIP-qPCR analysis was performed using 35S:ERF64D plants. Input chromatin was isolated from 12-d-old seedlings. Epitope-tagged ERF64D-chromatin complex was immunoprecipitated with an anti-myc antibody. A control reaction was processed side-by-side using mouse IgG. ChIP- and input-DNA samples were quantified by real-time qPCR using primers specific to the promoters of PDF1.2a (left) and PDF1.2b (right) genes. The ChIP results are presented as a percentage of input DNA. Error bars indicate sd (n = 3).
Figure 8.
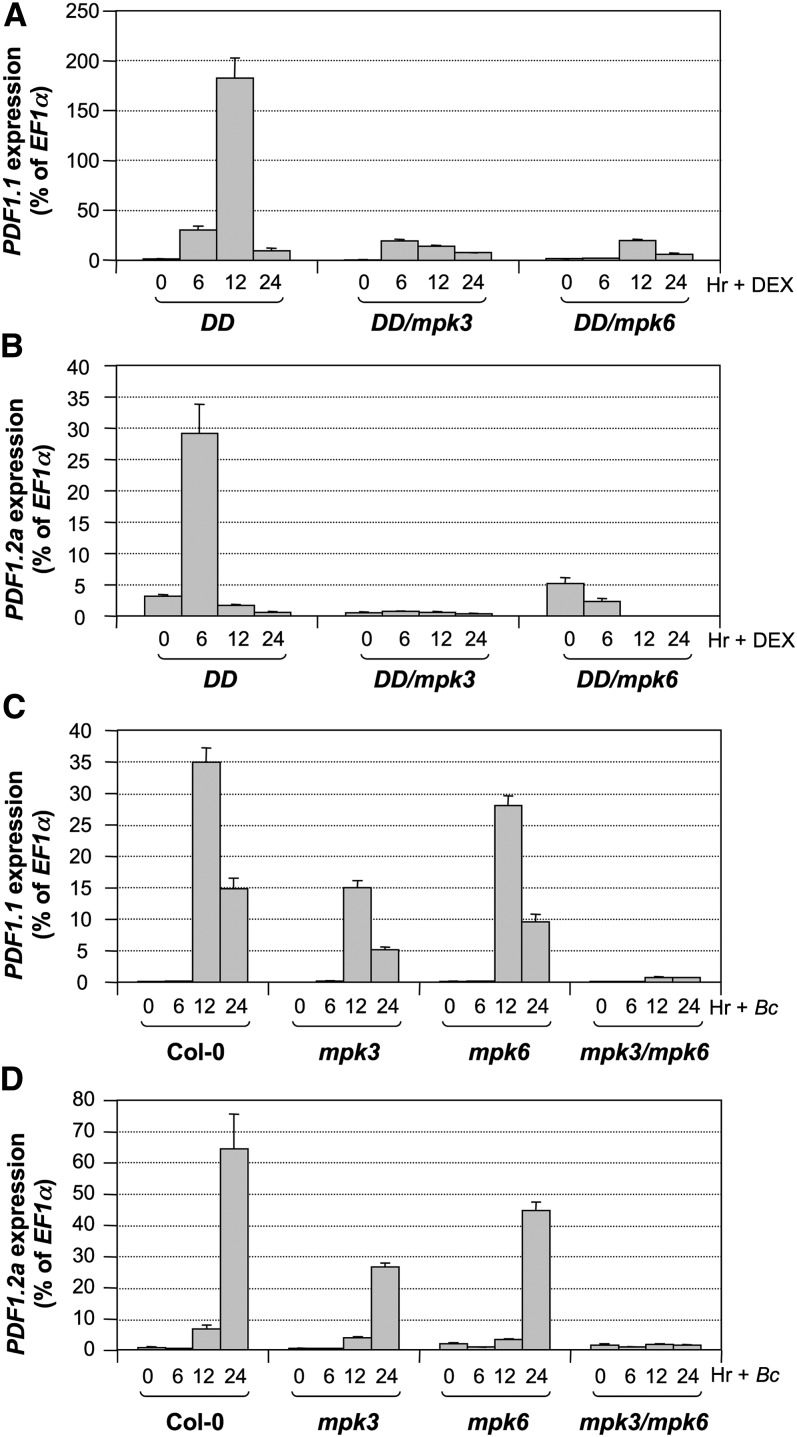
MPK3/MPK6-Dependent Activation of PDF1.1 and PDF1.2a Expression in Response to B. cinerea Infection.
(A) and (B) Induction of PDF1.1 (A) and PDF1.2a (B) expression in DD, DD/mpk3, and DD/mpk6 seedlings after DEX treatment.
(C) and (D) Induction of PDF1.1 (C) and PDF1.2a (D) expression in Col-0, mpk3, mpk6, and mpk3 mpk6 seedlings infected by B. cinerea. Gene expression was quantified by real-time PCR and calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
Quantification of additional defensin genes, including PDF1.2b, PDF1.2c, and PDF1.3, and other fungal resistance-related genes, ChiB, PR5, and HEL, revealed a similar pattern of regulation mediated by ERF6 (see Supplemental Figure 5 online). Therefore, both gain- and loss-of-function data suggest that ERF6 is an important regulator of plant defensin gene expression in response to pathogen infection. It is likely that the higher susceptibility of 35S:ERF6-EAR plants to B. cinerea is a result of the suppression of defensin gene activation. We also quantified the expression of endogenous ERF6 gene and ERF6WT, ERF64D, and ERF6-EAR transgenes (see Supplemental Figure 6 online). The expression of the 35S promoter–driven transgenes was about twice of that of the endogenous ERF6 in Col-0 after B. cinerea infection.
To determine whether ERF6 directly interacts with the promoters of defensin genes, we analyzed the promoters of PDF1.1 and PDF1.2 and performed chromatin immunoprecipitation (ChIP) assays. The PDF1.1 promoter does not contain a GCC-box (De Coninck et al., 2010), excluding it as a direct target of ERF6 transcription factor. By contrast, PDF1.2a and PDF1.2b promoters have GCC-box elements (Figure 7D). We performed ChIP–quantitative PCR (qPCR) assays to determine whether they are direct targets of ERF6. As shown in Figure 7E, the GCC-box–containing promoter regions of PDF1.2a and PDF1.2b were greatly enriched with an anti-myc antibody that immunoprecipitates the 4myc-tagged ERF64D protein. By contrast, IgG control failed to pull down the promoter regions of PDF1.2a and PDF1.2b. This result demonstrated that ERF64D directly binds to the promoters of PDF1.2a and PDF1.2b in vivo. By contrast, PDF1.1 gene should be an indirect target of ERF6 with an additional transcription factor(s) involved. The conclusion that PDF1.1 and PDF1.2a are differentially regulated by ERF6 is also consistent with their differential activation patterns in ERF64D plants after B. cinerea infection (Figures 7B and 7C).
Role of MPK3/MPK6 in ERF6-Mediated Defensin Gene Activation
To establish a link between MPK3/MPK6 and defensin gene activation, we examined the expression of PDF1.1 and PDF1.2a in the DD transgenic Arabidopsis plants after DEX treatment. As shown in Figures 8A and 8B, the expression of both genes was highly induced. In mpk3 or mpk6 mutant background, the induction was partially compromised, suggesting that the gain-of-function DD transgene functions through the endogenous MPK3/MPK6. We also examined the expression of PDF1.1 and PDF1.2a in mpk3 single, mpk6 single, and the rescued mpk3 mpk6 double mutants after B. cinerea inoculation. If the phosphorylation of ERF6 by MPK3/MPK6 is essential to the activation of downstream defense genes, we expect that the activation of these genes would be abolished in the loss of function of mpk3 mpk6 plants. As shown in Figures 8C and 8D, B. cinerea–induced PDF1.1 and PDF1.2a gene activation was partially inhibited in mpk3 and mpk6 single mutants and was completely abolished in the rescued mpk3 mpk6 double mutant, providing the loss-of-function evidence to support the role of MPK3/MPK6 in defensin gene activation.
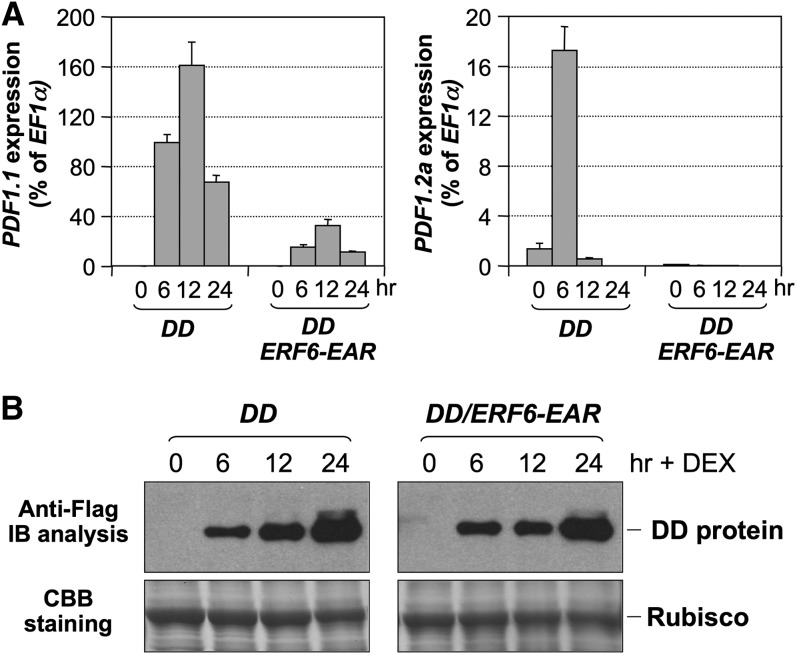
We also generated DD/ERF6-EAR double transgenic plants to characterize the MPK3/MPK6-induced defensin gene activation in the loss-of-function 35S:ERF6-EAR background. As shown in Figure 9A, the induction of both PDF1.1 and PDF1.2a in DD plants after DEX treatment was largely suppressed by ERF6-EAR transgene. Immunoblot analysis using anti-Flag antibody revealed a similar DD protein induction in DD and DD/ERF6-EAR seedlings after DEX treatment (Figure 9B), excluding the possibility of DD gene silencing in DD/ERF6-EAR seedlings, which could also compromise downstream gene activation. Together, these results demonstrate that ERF6 functions downstream of MPK3/MPK6 in mediating the activation of Arabidopsis defensin genes.
Figure 9.
Suppression of MPK3/MPK6-Induced Defensin Gene Expression by ERF6-EAR Transgene.
(A) Induction of PDF1.1 (left panel) and PDF1.2a (right panel) expression in DD and DD/ERF6-EAR seedlings after DEX treatment. Gene expression was quantified by real-time PCR and calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
(B) Normal DD protein induction in DD/ERF6-EAR seedlings after DEX treatment. DD protein levels were determined by immunoblot (IB) analysis using anti-Flag antibody (top panels). Equal protein loading was verified by Coomassie Brilliant Blue (CBB) staining (bottom panels). Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Ethylene-Independent Regulation and Function of ERF6 in Fungal Disease Resistance
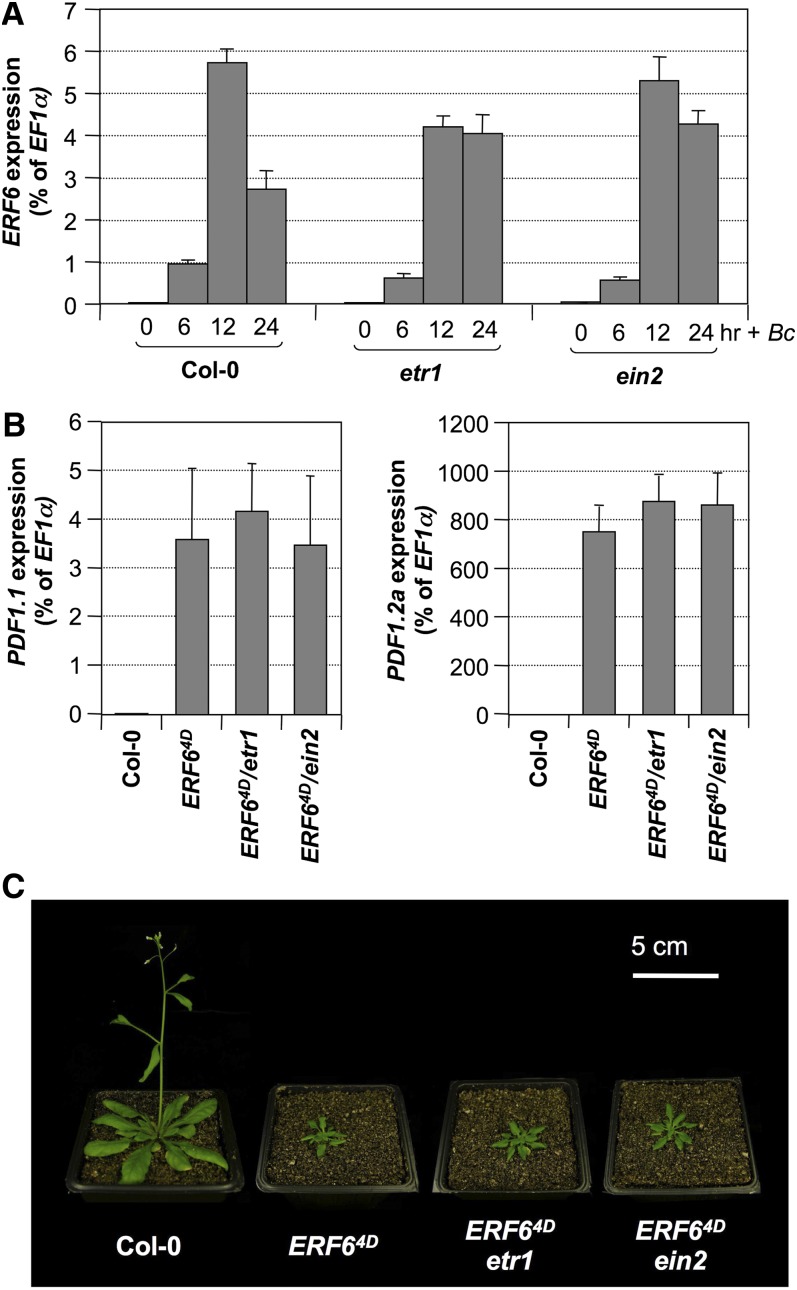
Plants infected by B. cinerea or DD plants after DEX treatment produce high levels of ethylene (Liu and Zhang, 2004; Han et al., 2010). To determine whether the activation of ERF6 gene expression seen in Figures 4A and 4B resulted from ethylene induction, we quantified the expression of ERF6 in etr1 and ein2 mutants infected with B. cinerea. As shown in Figure 10A, B. cinerea–induced ERF6 gene expression was not affected in either mutant, suggesting that the pathogen-inducible expression of ERF6 is ethylene independent. This is different from the well-studied ERF1 transcription factor, whose expression is dependent on ethylene signaling (Solano et al., 1998).
Figure 10.
Ethylene-Independent Regulation and Function of ERF6 in Fungal Disease Resistance.
(A) Twelve-day-old Col-0, etr1-1, and ein2 seedlings were inoculated with B. cinerea spores. Samples were collected at indicated times. The expression of ERF6 was quantified by real-time PCR and calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
(B) Ethylene-independent upregulation of PDF1.1 and PDF1.2a gene expression in the gain-of-function 35S:ERF64D seedlings. Double mutants were generated by crossing, and F3 double homozygous plants were used for experiment. The expression of PDF1.1 (left panel) and PDF1.2a (right panel) in 12-d-old 35S:ERF64D, 35S:ERF64D/etr1-1, and 35S:ERF64D/ein2 seedlings were quantified by real-time qPCR and calculated as a percentage of the EF1α transcript. Error bars indicate sd (n = 3).
[See online article for color version of this figure.]
To determine whether PDF1.1 and PDF1.2 gene activation seen in 35S:ERF64D plants is dependent on ethylene sensing/signaling, we crossed 35S:ERF64D transgene into etr1-1 or ein2 background. Expression of defensin genes in F3 double homozygous seedlings was quantified using real-time qPCR. As shown in Figure 10B, the high-level expression of PDF1.1 and PDF1.2a induced by the phospho-mimicking ERF64D was not affected by the mutation of ETR1 or EIN2, two key components in ethylene sensing/signaling, demonstrating that the constitutive high-level expression of defensin genes in 35S:ERF64D is not dependent on ethylene. Together with the normal basal level ethylene production in 35S:ERF64D plants (Figure 5E), we conclude that the constitutive defensin gene expression in 35S:ERF64D plants is independent of ethylene. Furthermore, the dwarf phenotype of 35S:ERF64D plants was not reversed in etr1-1 or ein2 mutant background (Figure 10C), demonstrating that the morphological phenotype of 35S:ERF64D plants is also independent of ethylene signaling.
DISCUSSION
Arabidopsis MPK3 and MPK6 regulate multiple defense responses (reviewed in Pedley and Martin, 2005; Zhang, 2008; Pitzschke et al., 2009; Andreasson and Ellis, 2010; Rodriguez et al., 2010; Tena et al., 2011). Previously, we characterized the regulation of camalexin biosynthesis by Arabidopsis MPK3/MPK6 cascade and its downstream WRKY33 substrate (Ren et al., 2008; Mao et al., 2011). Here, we report that ERF6 is another substrate of MPK3/MPK6. Based on both gain- and loss-of-function data, we further demonstrate that ERF6 is a key component downstream of MPK3/MPK6 cascade in regulating defense gene activation and fungal disease resistance.
Conflicting Reports about the Functions of ERF5/ERF6 in Plant Disease Resistance
There are conflicting reports about the functions of ERF5/ERF6 in plant defense responses in recent publications (Moffat et al., 2012; Son et al., 2012). Son et al. concluded that ERF5 and ERF6 negatively regulate plant defense against the fungal pathogen Alternaria brassicicola and positively regulate plant defense against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Son et al., 2012). To the contrary, Moffat et al. reported that ERF5 and ERF6 play a positive role in plant resistance against B. cinerea, a necrotrophic fungal pathogen with the same lifestyle as A. brassicicola, and a negative role in resistance against P. syringae pv tomato DC3000 (Moffat et al., 2012). In both reports, similar erf5 erf6 double mutants and transgenic lines overexpressing wild-type ERF5 or ERF6 were used as loss- and gain-of-function systems, respectively. In addition to the opposite disease resistance phenotypes, the two groups also described opposite functions of ERF5/ERF6 in modulating SA-regulated PR gene expression. How can the two groups have reached completely opposite conclusions using similar genetic experimental systems?
We did not observe clear phenotypes with the erf5 erf6 double mutant, possibly as a result of the existence of the functionally redundant ERF104 and ERF105. By contrast, the ERF6-EAR, which should be dominant negative over all four homologs, gave clear loss-of-function phenotypes (Figures 6 to 8). The opposite conclusions reached by the two groups may suggest that the phenotypes of erf5 erf6 double mutants and 35S:ERF5 and/or 35S:ERF6 transgenic lines were subtle and influenced by experimental conditions. In our study, only the phospho-mimicking ERF64D transgene resulted in clear gain-of-function phenotypes. By contrast, overexpression of wild-type ERF6 gave no or very weak phenotypes (Figures 6 to 8). In the recent publications, the expression of ERF5/ERF6 was not characterized at the protein level (Moffat et al., 2012; Son et al., 2012), which may also cause the inconsistency in the two studies. It is possible that the elevated transcript levels in the characterized lines were not associated with an increase in protein levels. This is compounded by the lack of in vivo analysis of ERF5/ERF6 at the posttranslational level in the two studies.
ERF6 protein is quite unusual in its properties, which hampered our progress on this project for several years. For a long time, we could not detect the ERF6 or its mutant proteins despite the detection of elevated levels of transcripts. Indeed, we found that a standard protein extraction procedure is unable to extract the ERF6 protein from cells. As a result, we were unable to correlate the phenotype with protein levels. After we solved this problem by extracting ERF6 protein from plant tissues using extraction buffer containing SDS followed by a boiling step, we were able to reliably detect the ERF6 protein using immunoblot analysis. This improvement allowed us to study the regulation of ERF6 at the posttranslational level and relate the phenotypes to the levels of ERF6 protein. The need for including SDS in the extraction buffer and the boiling step also indicates that ERF6 is tightly associated with chromatin.
Dual-Level Regulation of ERF6 at the Transcriptional and Posttranslational Levels by MPK3/MPK6
Previously, we identified ACS2/ACS6 as MPK3/MPK6 substrates. Phosphorylation of ACS2/ACS6 by MPK3/MPK6 leads to the accumulation of ACS proteins by slowing down the proteasome-mediated degradation (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). Here, we found that MPK3/MPK6 regulate ERF6 protein stability similarly. MPK3/MPK6 phosphorylate ERF6 on the C-terminal Ser sites, which are very similar to the phosphorylation sites of ACS6 (Figure 1). ERF6 is a highly unstable protein as indicated by rapid accumulation of the protein after MG132 treatment (Figure 3C). Phosphorylation of ERF6 by MPK3/MPK6 leads to the accumulation of ERF6 protein in vivo (Figure 2). Consistent with this, phospho-mimicking ERF64D mutant protein is much more stable than ERF6 wild-type protein (Figure 3A). All these data suggest that MPK3/MPK6-mediated phosphorylation stabilizes ERF6 in vivo. Furthermore, when 35S:ERF64D seedlings were treated with MG132, we still observed the accumulation of ERF64D protein, suggesting that phosphorylation of ERF6 does not prevent it from degradation completely, but only slows down its degradation by the 26S proteasome pathway. Finally, the strong phenotypes in plants expressing the phospho-mimicking form of ERF6, but not the wild-type ERF6, indicated the necessity of MPK3/MPK6 phosphorylation in the signaling process.
In addition to the posttranslational regulation, expression of ERF6 gene is highly induced by B. cinerea infection (Figure 4B). Gain-of-function activation of MPK3/MPK6 was sufficient to induce ERF6 expression (Figure 4A), suggesting that the MPK3/MPK6 cascade is involved in pathogen-induced ERF6 expression. However, in the mpk3 mpk6 double mutant, B. cinerea–induced ERF6 induction was only slightly reduced. These results suggest that, in addition to MPK3/MPK6 cascade, other signaling pathways can also activate the expression of ERF6 gene in response to B. cinerea infection. This situation is similar to the activation of WRKY33 gene expression by MPK3/MPK6 in response to B. cinerea (Mao et al., 2011). Based on these data, we conclude that the upregulation of ERF6 at the transcriptional level and the protein stabilization by MPK3/MPK6 phosphorylation are both involved in the regulation of ERF6 function, similar to the dual-level regulation of ACS2/ACS6 by MPK3/MPK6 signaling cascade (Liu and Zhang, 2004; Li et al., 2012).
Regulation of Defensin Gene Expression by Both Ethylene-Dependent and -Independent ERF Transcription Factors
Plant defensins play important roles in resistance against fungal pathogens (Terras et al., 1995; Penninckx et al., 1996; Lay and Anderson, 2005). Constitutive overexpression of several defensins, including PDF1.1 and PDF1.2, confers higher resistance against fungal pathogens (Terras et al., 1995; Gao et al., 2000; Stotz et al., 2009; De Coninck et al., 2010). Regulation of PDF1.2 expression in Arabidopsis in response to pathogens has been well documented (Penninckx et al., 1998; Solano et al., 1998; Lorenzo et al., 2003; Pré et al., 2008). By contrast, PDF1.1 has been considered as a seed-specific defensin (Terras et al., 1993; Penninckx et al., 1996) until a recent study demonstrated that it is also highly induced in vegetative tissues by pathogens, and its overexpression enhances plant resistance against fungal pathogens (De Coninck et al., 2010).
In this article, we demonstrate that ERF6 is an important transcription factor downstream of MPK3/MPK6 in regulating the expression of plant defense genes. Expression of phospho-mimicking ERF64D leads to constitutive high-level expression of defensin genes PDF1.1, PDF1.2a, PDF1.2b, PDF1.2c, and PDF1.3, as well as other defense related genes, such as ChiB, PR5, and HEL (Figure 7; see Supplemental Figure 5 online). However, expression of ERF6WT only showed a weak effect on the expression of these genes, suggesting the importance of MPK3/MPK6-meidated phosphorylation of ERF6 in regulating defensin gene activation. By contrast, expression of ERF6-EAR suppressor strongly suppressed B. cinerea–induced defense gene expression. The regulation of these defense genes could be either direct or indirect with additional unidentified transcription factors downstream of ERF6. ChIP-qPCR demonstrated that ERF64D directly binds to the promoters of PDF1.2a and PDF1.2b in vivo (Figure 7), suggesting that they are the direct target genes of ERF6. However, no GCC-box is found in the promoter of PDF1.1, excluding it as a direct target of ERF6. Nonetheless, both are downstream of MPK3/MPK6 cascade (Figure 8), and the expression of ERF6-EAR suppresses the induction of defensin genes in the conditional gain-of-function DD plants (Figure 9). Taken together, we can conclude that ERF6, an MPK3/MPK6 substrate, plays important roles in regulating plant defense gene expression downstream of pathogen-responsive MPK3/MPK6 cascade.
ERF1, a transcription factor downstream of EIN3 in the ethylene signaling pathway, is also involved in the induction of PDF1.2 (Solano et al., 1998). It is the integration point of ethylene and JA signaling pathways that synergistically regulate PDF1.2 expression. Unlike ERF1, the pathogen-inducible expression of ERF6 is ethylene independent (Figure 10A). The induction of ERF6 gene expression was not affected by the mutation of either ETR1 or EIN2 in response to B. cinerea infection, suggesting that the induction of ERF6 expression is not dependent on ethylene signaling. Different from 35S:ERF1 plants, which show constitutive triple response (Solano et al., 1998), neither 35S:ERF6WT nor 35S:ERF64D plants showed constitutive triple response (Figure 5D). In addition, 35S:ERF64D plants do not overproduce ethylene, suggesting that the high-level induction of defense genes is not a result of the activation of ethylene biosynthesis. Consistent with this finding, the elevated defensin gene expression in 35S:ERF64D plants was not suppressed by ethylene sensing/signaling mutant etr1 and ein2 (Figure 10B). These results further suggest that ERF6 is an ethylene-independent transcription factor downstream of MPK3/MPK6 pathway, which can also activate defensin genes.
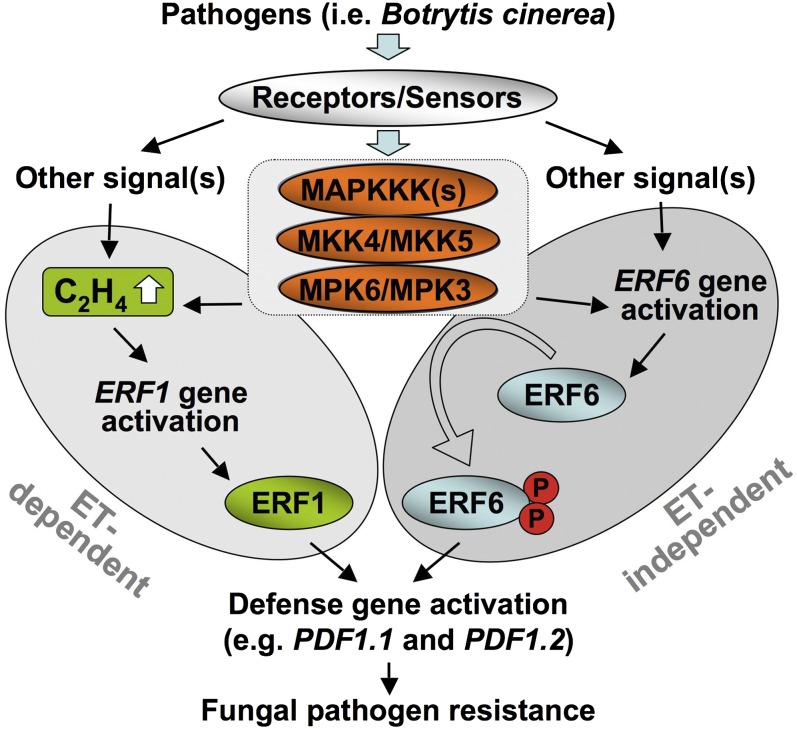
Ectopic overexpression of ERF1 is sufficient to activate downstream PDF1.2 genes (Lorenzo et al., 2003). By contrast, ERF6 is regulated at both transcriptional and posttranslational levels, and the overexpression of wild-type ERF6 is not sufficient to activate downstream defense genes. By contrast, the expression of the phospho-mimicking form of ERF6, ERF64D, constitutively activates plant defense gene expression and fungal resistance. Ethylene production in response to B. cinerea infection involves both ACS gene activation and phosphorylation-induced stabilization of ACS proteins, and MPK3 and MPK6 play critical roles in both processes (Liu and Zhang, 2004; Han et al., 2010; Li et al., 2012). As a result, we can conclude that MPK3/MPK6 cascade regulates the expression of defensin genes through both ethylene-dependent ERF1 and ethylene-independent ERF6 transcription factors, as illustrated in Figure 11, and MPK3/MPK6 cascade regulates both ERF transcription factors via (1) direct phosphorylation of ERF6 and (2) activation of ERF1 gene expression via the induction of ethylene biosynthesis.
Figure 11.
Coordinated Regulation of Plant Defensin Gene Expression and Fungal Resistance by Two ERF Transcription Factors through Ethylene-Dependent and -Independent Mechanisms.
Both ERF1 and ERF6 are involved in regulation of plant defensin gene expression. ERF1 is downstream of ethylene (ET). Overexpression of ERF1 is sufficient to activate downstream PDF1.2 genes. By contrast, ERF6 is regulated at both transcriptional and posttranslational levels. ERF6 gene activation induced by B. cinerea is independent of ethylene pathway, and overexpression of wild-type ERF6 is not sufficient to activate downstream defense genes. By contrast, the expression of the phospho-mimicking form of ERF6, ERF64D, constitutively activates plant defense gene expression and fungal resistance. The MPK3/MPK6 cascade is involved in regulating both ERF transcription factors by (1) direct phosphorylation of ERF6 and (2) activation of ERF1 gene expression via the induction of ethylene biosynthesis. MAPKKK, MAPK kinase kinase.
[See online article for color version of this figure.]
MPK3/MPK6 Regulate Multiple Defense Pathways in Plant Fungal Resistance
MPK3/MPK6 are involved in regulating multiple defense responses triggered by pathogen invasion, including the induction of ethylene biosynthesis, defense gene activation, and camalexin induction (Liu and Zhang, 2004; Ren et al., 2008; Mao et al., 2011; this report), all of which are important to plant fungal resistance. It appears that the multifunctionality of MPK3/MPK6 is a result of their ability to phosphorylate different substrates. A subset of ACS isoforms can be directly phosphorylated by MPK3 and MPK6, which stabilizes the ACS proteins and leads to ethylene induction (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). Phosphorylation of WRKY33 by MPK3/MPK6 enhances its activity in promoting the expression of downstream camalexin biosynthetic genes, which drives the metabolic flow to camalexin production in Arabidopsis challenged by pathogens (Mao et al., 2011). In this article, we found that MPK3/MPK6-mediated phosphorylation stabilizes ERF6, which activates defensin gene expression and confers fungal resistance. Previously, we associated the compromised B. cinerea resistance in MAPK mutants with reduced camalexin induction (Ren et al., 2008). In light of the findings in this article, we have to revise the previous hypothesis and conclude that the compromised B. cinerea resistance in MPK3/MPK6 mutant is likely to be a result of the loss or reduction of multiple defense responses.
Based on the total number of kinases and phosphoproteins, it is predicted that each kinase should have ∼20 to 40 substrates on average (Johnson and Hunter, 2005). Although each substrate can define a specific function downstream of a kinase, forming a functional module, these different modules can crosstalk, which contributes to the complexity of a signaling network. For instance, phosphorylation of WRKY33 by MPK3/MPK6 not only activates the expression of camalexin biosynthetic genes, driving the induction of camalexin biosynthesis in response to pathogen invasion, it is also is involved in the transcriptional activation of ACS2 and ACS6 genes, which contributes to the high-level ethylene induction by fungal pathogen (Li et al., 2012). In a recent survey using tandem metal oxide affinity chromatography followed by tandem mass spectrometry analysis, more than 30 novel putative MPK3/MPK6 substrates were identified (Hoehenwarter et al., 2013). Detailed function analysis of these substrates and their crosstalk will allow us to have a better understanding of MPK3/MPK6 cascade in plant signaling.
METHODS
Plant Materials, Growth Conditions, and Treatments
Arabidopsis thaliana mutants and transgenic lines used in this study are all in Col-0 ecotype background. Steroid-inducible promoter-driven tobacco (Nicotiana tabacum) MEK2DD transgenic Arabidopsis line (DD), mpk3-1, mpk6-2, ctr1-1, etr1-1, and ein2-1 mutants were described previously (Alonso et al., 1999; Kieber et al., 1993; Liu and Zhang, 2004; Ren et al., 2002). T-DNA insertion mutant alleles of erf6 (SALK_087356), erf5 (GABI_681E07), erf104 (SALK_057720), and erf105 (GABI_680C11) were obtained from the ABRC and the European Arabidopsis Stock Centre (Alonso et al., 2003; Rosso et al., 2003). DD/mpk3, DD/mpk6, and rescued mpk3 mpk6 double mutant were previously described (Wang et al., 2007; Ren et al., 2008). 35S:ERF64D/etr1 and 35S:ERF64D/ein2 double mutants were generated by genetic cross, and F3 double homozygous lines were used for experiments.
Arabidopsis seedlings were grown and treated as previously described (Ren et al., 2008; Mao et al., 2011). For the triple response assay, seeds were plated on half-strength Murashige and Skoog medium without or with 1 μM ACC and incubated in a 22°C growth chamber in darkness for 3 d.
At least two independent repetitions were performed for experiments with multiple time points. For single time point experiments, at least three independent repetitions were done.
Preparation of Recombinant ERF6 Proteins and in Vitro Phosphorylation Assay
Full-length ERF6 cDNA was PCR amplified using the ERF6-F1 and ERF6-B1 primer pair and cloned into pBluescript II KS+ vector. After sequencing verification, the ERF6 cDNA was subcloned into pET28a in frame with the N-terminal His tag. To generate loss-of-phosphorylation ERF6 mutants, Ser/Thr-to-Ala mutations were introduced by QuickChange site-directed mutagenesis (Stratagene) and confirmed by sequencing. The primers used for cloning and mutagenesis are listed in Supplemental Table 1 online. Higher-order ERF6 mutants were generated by successive mutagenesis steps. Double-tagged ERF6 constructs were generated using PCR to introduce the FLAG tag to the C terminus of ERF6 in the pET-28a-ERF6 or its mutant constructs. All the prokaryotic expression constructs were transformed into Escherichia coli strain BL21 (DE3). Recombinant protein expression was induced with 0.25 mM isopropylthio-β-galactoside for 3 h at 25°C. His-tagged proteins were purified using HiTrap nickel columns (GE Healthcare), and His/FLAG double-tagged proteins were sequentially purified using HiTrap columns and anti-FLAG M2 Affinity Gel (Sigma-Aldrich) according to the manufacturer’s instructions.
In vitro phosphorylation assay was performed as previously described (Liu and Zhang, 2004; Han et al., 2010). Briefly, recombinant ERF6 or its mutant proteins were incubated with the activated MPK3 or MPK6 (20:1 substrate enzyme ratio) in the kinase reaction buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, and 1 mM DTT) with 25 µM ATP and [γ-32P]ATP (1 µCi per reaction). The reactions were stopped by the addition of SDS sample buffer after 30 min. Phosphorylated ERF6 was visualized by autoradiography after being resolved in a 10% SDS-PAGE gel.
Generation of ERF6 Binary Constructs and Transgenic Plants
For the generation of plant expression constructs, cDNA fragments of wild-type ERF6 and Ser/Thr-to-Ala mutants were amplified from the corresponding prokaryotic expression constructs using the ERF6-F1 and ERF6-B2 primer pair (see Supplemental Table 1 online), and the PCR products were cloned into a modified pBlueScript II KS+ vector in frame with a N-terminal 4myc epitope tag coding sequence. Phospho-mimicking ERF6 mutant was generated by introducing Ser to Asp mutations using the 4D-F1/B1 primer pair (see Supplemental Table 1 online). The EAR suppressor motif was introduced into the C terminus of ERF6 before the stop codon in 35S:ERF6WT by PCR to generate the 35S:ERF6-EAR construct. All ERF6 cDNA fragments with 4myc epitope tag sequence were moved into the SpeI-XhoI sites of a modified pBI121 vector, which were then transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The floral dipping method was used to transform Arabidopsis (Clough and Bent, 1998). Independent lines with expression of 4myc-tagged ERF6 were identified based on immunoblot analyses. Transformants with a single copy of T-DNA insertion were identified based on the 3:1 segregation of antibiotic resistance in T2 generation. T3 homozygous lines were used for experiments.
Protein Extraction and Immunoblot Analysis
Proteins for immunoblot detection of Flag-tagged DD were extracted as previously described (Zhang and Klessig, 1997). For detection of 4myc-tagged ERF6 proteins, total proteins were extracted using three volumes (v/w) of SDS loading buffer without bromophenol blue dye (Joo et al., 2008). The concentration of protein was determined using the Bio-Rad protein assay kit with BSA as the standard. Immunoblot detection of tagged proteins was performed as previously described (Liu and Zhang, 2004).
qPCR Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). After DNase treatment, 1 µg of total RNA was used for reverse transcription. qPCR analysis was performed using an Optican 2 real-time PCR machine (MJ Research) as previously described (Ren et al., 2008). The levels of gene expression were calculated as percentages of the EF-1α transcript. At least three independent biological replicates were examined with similar results. The primer pairs used for qPCR are listed in Supplemental Table 1 online.
Botrytis cinerea Resistance Assay and Lactophenol-Trypan Blue Staining of Fungal Structures
For B. cinerea resistance assays, seeds were sown in soil and grown under an 8-h-light/16-h-dark cycle in a growth chamber at 22°C and 65% relative humidity for 6 weeks. The plants were then sprayed with B. cinerea spore suspension with a density of 1 × 105 spores/mL (∼1 mL for each plant). Inoculated plants were covered with a transparent plastic dome to maintain high humidity for 24 h, and disease symptoms were scored 3 d after inoculation. To quantify disease resistance, mature rosette leaves were detached and inoculated with 5-μL drops of spore suspension (1 × 105 spores/mL). Inoculated leaves were kept in Petri dishes on wet filter paper. Three days later, the lesion size was measured, and the fungal structures were stained with lactophenol-trypan blue staining as previously described (Han et al., 2010).
Detection of ROS Generation by DAB Staining
In vivo hydrogen peroxide generation in plants was detected by an endogenous peroxidase-dependent in situ histochemical staining procedure using DAB as previously described (Liu et al., 2007).
ChIP-qPCR Analysis
Twelve-day-old 35S:ERF64D seedlings were used for ChIP assay as previously described (Mao et al., 2011). Chromatin was isolated from 0.8 g of frozen tissue and sonicated with a Bioruptor sonicator (15 s on and 15 s off cycles, medium-energy settings) for 6 min. Immunoprecipitation was performed by incubating chromatin with 2 μg of anti-myc antibody (Millipore) or mouse IgG (negative control) for 1 h at 4°C. The protein-chromatin immunocomplexes were captured using Protein G-Dynal magnetic beads (Invitrogen). After Proteinase K digestion, the immunoprecipitated DNA was purified using ChIP DNA clean and concentrator kit (Zymo Research). Immunoprecipitated DNA and input DNA were analyzed by qPCR using primers specific for the promoter regions of PDF1.2a and PDF1.2b (see Supplemental Table 1 online). The ChIP results are presented as percentage of input DNA.
Illumina RNA Sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen) from 12-d-old Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR seedlings. After DNase treatment, total RNA was purified using RNA clean and concentrator kit. RNA sequencing libraries were constructed using Illumina TruSeq RNA library preparation kit and sequenced using the Illumina Genome Analyzer IIx according to the manufacturer’s instructions. Image analysis and base calling were performed using the standard Illumina analysis pipeline. After the dirty raw reads were filtered out, clean reads were mapped to the reference genome and reference gene sequences, respectively. Gene expression levels were calculated using the RPKM method (reads per kb per million reads) (Mortazavi et al., 2008). Genes with differential expression in 35S:ERF64D and the Col-0 seedlings (P-value ≤ 0.001) were identified, and their expression patterns in Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR plants were presented as heat map generated using the R statistical computing package (http://www.r-project.org).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MPK3 (At3g45640), MPK6 (At2g43790), ERF5 (At5g47230), ERF6 (At4g17490) ERF104 (At5g61600), ERF105 (At5g51190), PDF1.1 (At1g75830), PDF1.2a (At5g44420), PDF1.2b (At2g26020), PDF1.2c (At5g44430), PDF1.3 (At2g26010), ChiB (At3g12500), HEL (At3g04720), and PR5 (At1g75040).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phos-Tag Mobility Shift Detection of in Vivo Phosphorylated ERF6 in 35S:ERF6WT Plants after B. cinerea Inoculation.
Supplemental Figure 2. Alignment of the C Termini of ERF6, ERF5, ERF104, and ERF105 with the C Terminus of ACS6 That Contains the MPK3/MPK6 Phosphorylation Sites.
Supplemental Figure 3. Differential Gene Expression in 12-d-Old Col-0, 35S:ERF6WT, 35S:ERF64D, and 35S:ERF6-EAR Seedlings.
Supplemental Figure 4. Expression of Phospho-Mimicking ERF64D Confers Strong Resistance to B. cinerea, Whereas Expression of a Dominant-Negative ERF6-EAR Results in an Opposite Phenotype.
Supplemental Figure 5. Opposing Effects of ERF64D and ERF6-EAR Transgenes on Defense Gene Expression.
Supplemental Figure 6. The Expression Levels of Endogenous ERF6 and Transgenes before and after Botrytis Infection.
Supplemental Table 1. Primers Used in This Study.
Supplemental Methods 1. Methods for Supplemental Figure 1.
Supplemental Data Set 1. Differentially Expressed Stress/Defense-Related Genes in 35S:ERF64D Transgenic Plants.
Supplementary Material
Acknowledgments
This work was supported by Zhejiang University 985 third-stage Grant 118000-193411801 to J.X. and National Science Foundation Grant IOS-0743957 to S.Z. Illumina sequencing was supported by Grant 13321 from the Missouri Life Science Research Board. B.M. was supported by the University of Missouri Life Sciences Undergraduate Research Opportunity Program Summer Internship Fund. We thank the ABRC and the European Arabidopsis Stock Centre for mutant seeds and Dongtao Ren (China Agricultural University) for participating in the screening of ERF double and triple mutants.
AUTHOR CONTRIBUTIONS
X.M., J.X., and S.Z. designed the research. X.M., J.X., Y.H., K.-Y.Y., B.M., Y.L., and S.Z. performed the research. X.M., J.X., and S.Z. analyzed data and wrote the article.
Glossary
- MAPK
mitogen-activated protein kinase
- ROS
reactive oxygen species
- ERF
ethylene response factor
- JA
jasmonic acid
- DEX
dexamethasone
- Col-0
Columbia-0
- SA
salicylic acid
- DAB
3,3′-diaminobenzidine
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
References
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113 [DOI] [PubMed] [Google Scholar]
- Asai S., Ohta K., Yoshioka H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Ausubel F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brown R.L., Kazan K., McGrath K.C., Maclean D.J., Manners J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y.H., et al. (2003). BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Coninck B.M., Sels J., Venmans E., Thys W., Goderis I.J., Carron D., Delauré S.L., Cammue B.P., De Bolle M.F., Mathys J. (2010). Arabidopsis thaliana plant defensin AtPDF1.1 is involved in the plant response to biotic stress. New Phytol. 187: 1075–1088 [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A.G., Hakimi S.M., Mittanck C.A., Wu Y., Woerner B.M., Stark D.M., Shah D.M., Liang J., Rommens C.M. (2000). Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18: 1307–1310 [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gudesblat G.E., Torres P.S., Vojnov A.A. (2009). Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 149: 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N., Reuber T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Han L., Li G.J., Yang K.Y., Mao G., Wang R., Liu Y., Zhang S. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W., Thomas M., Nukarinen E., Egelhofer V., Röhrig H., Weckwerth W., Conrath U., Beckers G.J. (2013). Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell. Proteomics 12: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K., et al. ; MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Johnson S.A., Hunter T. (2005). Kinomics: Methods for deciphering the kinome. Nat. Methods 2: 17–25 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim C.Y., Zhang S. (2004). Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 38: 142–151 [DOI] [PubMed] [Google Scholar]
- Lay F.T., Anderson M.A. (2005). Defensins—Components of the innate immune system in plants. Curr. Protein Pept. Sci. 6: 85–101 [DOI] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ren D., Pike S., Pallardy S., Gassmann W., Zhang S. (2007). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51: 941–954 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K.C., Dombrecht B., Manners J.M., Schenk P.M., Edgar C.I., Maclean D.J., Scheible W.R., Udvardi M.K., Kazan K. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Matsui K., Ikeda M., Nakata M., Oshima Y., Nagatoshi Y., Ohme-Takagi M. (2011). CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol. Biol. 754: 87–105 [DOI] [PubMed] [Google Scholar]
- Moffat C.S., Ingle R.A., Wathugala D.L., Saunders N.J., Knight H., Knight M.R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M.T., Dangl J.L. (2010). Arabidopsis and the plant immune system. Plant J. 61: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Anderson J.P., Young J., Singh K.B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 143: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Singh K.B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Penninckx I.A., Eggermont K., Terras F.R., Thomma B.P., De Samblanx G.W., Buchala A., Métraux J.P., Manners J.M., Broekaert W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Thomma B.P., Buchala A., Métraux J.P., Broekaert W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son G.H., Wan J., Kim H.J., Nguyen X.C., Chung W.S., Hong J.C., Stacey G. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25: 48–60 [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Stotz H.U., Spence B., Wang Y. (2009). A defensin from tomato with dual function in defense and development. Plant Mol. Biol. 71: 131–143 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F.R., et al. (1995). Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell 7: 573–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F.R., Torrekens S., Van Leuven F., Osborn R.W., Vanderleyden J., Cammue B.P., Broekaert W.F. (1993). A new family of basic cysteine-rich plant antifungal proteins from Brassicaceae species. FEBS Lett. 316: 233–240 [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. (2008). Mitogen-activated protein kinase cascades in plant intracellular signaling. In Annual Plant Reviews 33: Intracellular Signaling in Plants, Z. Yang, ed (Oxford, UK: Wiley-Blackwell), pp. 100–136 [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.