BR-SIGNALING KINASE1 (BSK1) is a substrate of the brassinosteroid receptor BRI1. This work shows that BSK1 physically associates with FLS2, and a mutation in BSK1 leads to enhanced susceptibility to multiple pathogens as well as defects in the FLS2-mediated reactive oxygen burst, demonstrating that BSK1 plays an important role in plant innate immunity.
Abstract
Pathogen-associated molecular pattern (PAMP)-trigged immunity (PTI) is the first defensive line of plant innate immunity and is mediated by pattern recognition receptors. Here, we show that a mutation in BR-SIGNALING KINASE1 (BSK1), a substrate of the brassinosteroid (BR) receptor BRASSINOSTEROID INSENSITIVE1, suppressed the powdery mildew resistance caused by a mutation in ENHANCED DISEASE RESISTANCE2, which negatively regulates powdery mildew resistance and programmed cell death, in Arabidopsis thaliana. A loss-of-function bsk1 mutant displayed enhanced susceptibility to virulent and avirulent pathogens, including Golovinomyces cichoracearum, Pseudomonas syringae, and Hyaloperonospora arabidopsidis. The bsk1 mutant also accumulated lower levels of salicylic acid upon infection with G. cichoracearum and P. syringae. BSK1 belongs to a receptor-like cytoplasmic kinase family and displays kinase activity in vitro; this kinase activity is required for its function. BSK1 physically associates with the PAMP receptor FLAGELLIN SENSING2 and is required for a subset of flg22-induced responses, including the reactive oxygen burst, but not for mitogen-activated protein kinase activation. Our data demonstrate that BSK1 is involved in positive regulation of PTI. Together with previous findings, our work indicates that BSK1 represents a key component directly involved in both BR signaling and plant immunity.
INTRODUCTION
Plants have evolved two layers of disease resistance to defend themselves against pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). The first layer of defense is triggered by recognition of pathogen-associated molecular patterns (PAMPs) by transmembrane pattern recognition receptors; this defense is often referred to as PAMP-triggered immunity (PTI) or basal defense. One of the best-studied PAMPs of plant pathogens is flagellin, the protein subunit of the bacterial flagellum (Gómez-Gómez and Boller, 2000; Hayashi et al., 2001). Flagellin is highly conserved among bacterial pathogens and is recognized as a PAMP by many plant and mammalian innate immunity receptors (Felix et al., 1999; Gómez-Gómez and Boller, 2000; Hayashi et al., 2001). In Arabidopsis thaliana, a conserved 22–amino acid peptide in the N terminus of flagellin (flg22) is specifically recognized by the Leucine-rich repeat receptor-like kinase (LRR-RLK) FLAGELLIN SENSING2 (FLS2), and flg22 is sufficient for binding to FLS2 and activation of PTI (Gómez-Gómez and Boller, 2000; Zipfel et al., 2004). FLS2 is present in FLS2-FLS2 complexes in the presence or absence of flg22 (Sun et al., 2012), and ligand perception by FLS2 involves its LRR domains (Mueller et al., 2012). Upon flagellin perception, FLS2 rapidly forms a complex with another LRR-RLK, BRASSINOSTEROID INSENSITIVE1 ASSOCIATED KINASE1 (BAK1) (Chinchilla et al., 2007; Schulze et al., 2010), a positive regulator of brassinosteroid (BR) signaling (Li et al., 2002; Nam and Li, 2002). The activation of PTI leads to the induction of basal defense responses, which include callose deposition, production of reactive oxygen species (ROS), transcriptional induction of a large suite of defense-related genes, and activation of a mitogen-activated protein kinase (MAPK) cascade (Bent and Mackey, 2007). Interestingly, several components involved in FLS2-mediated immune responses, such as BAK1 and BAK1-LIKE1 (BKK1), also play important roles in BR signaling (Li et al., 2002; Nam and Li, 2002; Karlova et al., 2006; He et al., 2007a; Roux et al., 2011), suggesting a mechanistic connection between BR signaling and PTI signaling. BR is a steroid hormone that modulates growth and development in plants. Disruption of BR signaling leads to pleiotropic defects, such as dwarfism and male sterility (Li et al., 1996). Recently, it was shown that activation of BR signaling inhibits PTI mediated by FLS2, and the inhibition of FLS2 by BR signaling is downstream or independent of the FLS2-BAK1 complex (Albrecht et al., 2012). Furthermore, overexpression of BRI1 or a gain-of-function mutation of BRI1 results in inhibition of BAK1-mediated PTI and leads to enhanced susceptibility to the oomycete pathogen Hyaloperonospora arabidopsidis (Belkhadir et al., 2012). In addition, upon flg22 perception, FLS2 also physically associates with BKK1 (Roux et al., 2011), an LRR-RLK that interacts with the BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) as a positive regulator of BR signaling (Karlova et al., 2006; He et al., 2007a). Although BR signaling influences PTI, it is not clear how this interaction between growth and immune signaling occurs.
Powdery mildew fungi are obligate pathogens that infect a large number of plant species, including the model plant Arabidopsis (Schulze-Lefert and Vogel, 2000; Micali et al., 2008). Arabidopsis ecotype Columbia-0 (Col-0) plants are susceptible to the powdery mildew pathogen Golovinomyces cichoracerum, supporting the production of a large number of spores after infection (Adam and Somerville, 1996). Genetic screens led to identification of Arabidopsis genes whose modifications affect powdery mildew resistance. For instance, mutations in ENHANCED DISEASE RESISTANCE1 (EDR1), EDR2, and EDR3 lead to increased resistance to G. cichoracerum, and the resistance in these mutants is accompanied by mildew-induced necrotic cell death (Frye and Innes, 1998; Tang et al., 2005, 2006; Vorwerk et al., 2007). EDR2 encodes a protein consisting of a pleckstrin homology (PH) domain, a StAR transfer domain, and a plant-specific domain of unknown function, DUF1336 (Tang et al., 2005; Vorwerk et al., 2007). The PH and StAR transfer domains have been implicated in lipid binding, suggesting that lipid signaling may be involved in resistance. The PH domain of EDR2 binds to phosphatidylinositol-4-phosphate in vitro, and the binding is essential for EDR2 function (Vorwerk et al., 2007). EDR2 localizes to endoplasmic reticulum, plasma membrane, and endosomes (Vorwerk et al., 2007). The edr2-mediated powdery mildew resistance depends on salicylic acid (SA) signaling, and mutations in SALICYLIC ACID INDUCTION DEFICIENT2 (SID2), NON-EXPRESSOR OF PR1 (NPR1), and AGD2-LIKE DEFENSE RESPONSE PROTEIN1 suppress edr2-associated resistance (Tang et al., 2005; Vorwerk et al., 2007; Nie et al., 2011). In addition, a gain-of-function mutation in SIGNAL RESPONSIVE1 (SR1) and a mutation in RPN1a, a subunit of the 26S proteasome, also suppress edr2-mediated resistance. Both SR1 and RPN1a contribute to SA accumulation upon pathogen infection (Nie et al., 2012; Yao et al., 2012). However, how EDR2 affects SA signaling is not well understood.
To identify new components in the EDR2 signaling pathway, we screened for edr2 suppressor mutants in Arabidopsis (Nie et al., 2011). In this screen, we identified a mutation in a receptor-like cytoplasmic kinase, BR-SIGNALING KINASE1 (BSK1), previously shown by proteomic analyses to be a substrate of the BR receptor BRI1 (Tang et al., 2008). Here, we show that the bsk1 mutation fully suppressed edr2-mediated resistance. The bsk1 single mutant displayed enhanced susceptibility to a variety of pathogens. In addition, BSK1 physically associates with FLS2, and the flg22-induced ROS burst is compromised in the bsk1 mutant. These data indicate that BSK1 is a positive regulator of defense responses and has a significant role in PTI. Our work extends the observation that BR signaling and immune signaling share some components.
RESULTS
bsk1-1 Suppresses the edr2 Phenotypes of Powdery Mildew Resistance, Ethylene-Induced Senescence, and Upregulation of Defense Gene Induction
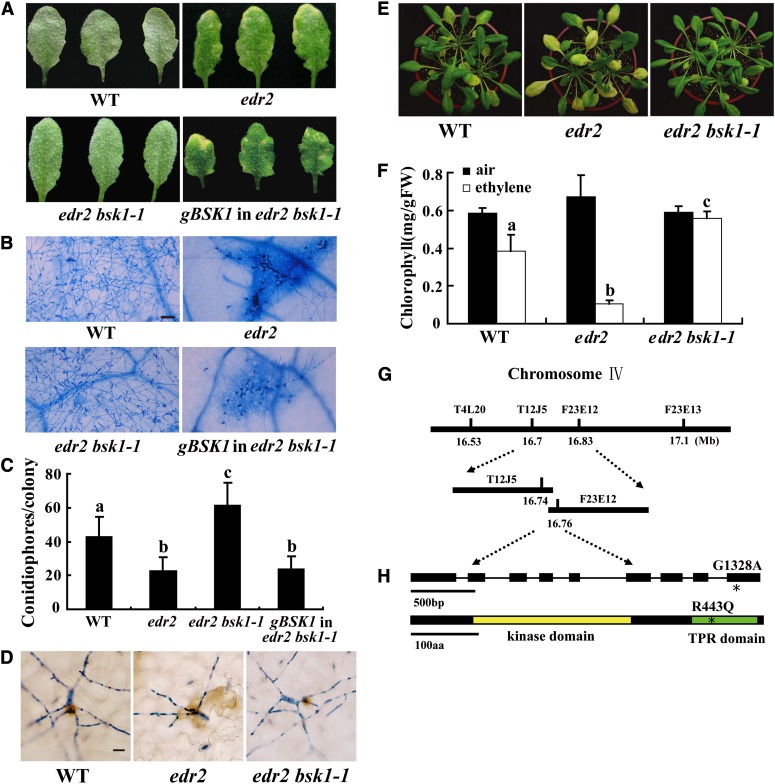
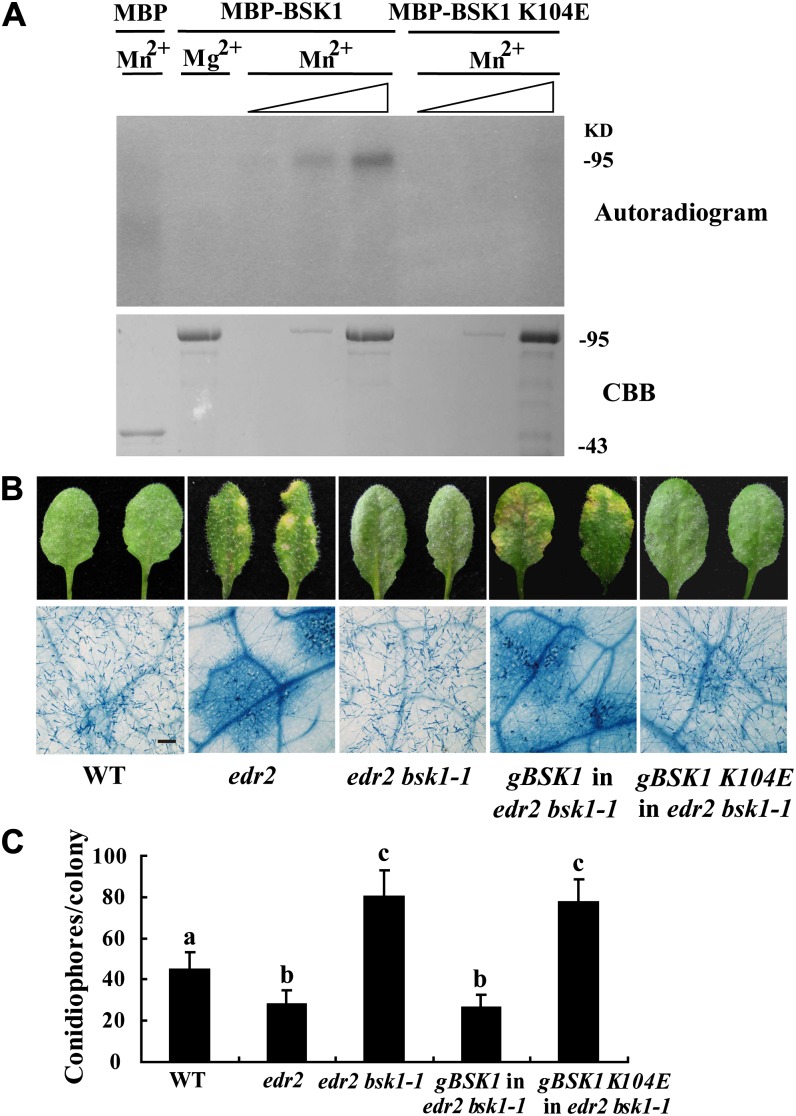
The Arabidopsis edr2 mutant displays enhanced disease resistance to the virulent powdery mildew strain G. cichoracearum UCSC1 (Tang et al., 2005; Vorwerk et al., 2007). To identify components involved in edr2-mediated resistance, we conducted a mutant screen for edr2 suppressors (Nie et al., 2011). In this screen, we identified a mutant that we named bsk1-1 based on subsequent characterization. Upon infection with G. cichoracearum, at 8 d after infection (DAI), the edr2 bsk1-1 mutant displayed a large number of conidia and lacked visible mildew-induced lesions. This is in contrast with edr2 leaves, which produced very few conidia and showed prominent cell death (Figures 1A and 1B). To further assess the edr2 bsk1-1 phenotype, we quantified fungal growth by counting the number of conidiophores (asexual reproductive structures) per colony in the wild type, edr2, and edr2 bsk1-1. The edr2 mutants showed significantly fewer conidiophores than the wild-type plants after infection with G. cichoracearum at 5 DAI; however, the number of conidiophores in the edr2 bsk1-1 plants was significantly more than in edr2 and wild-type plants (Figure 1C), indicating that the bsk1-1 mutation fully suppressed edr2-mediated resistance to powdery mildew.
Figure 1.
Suppression of the edr2-Mediated Phenotypes by bsk1-1.
(A) Four-week-old Arabidopsis plants were infected with G. cichoracearum. The plants were photographed at 8 DAI. The wild-type plants were susceptible, and a large number of spores were produced on the leaves. By contrast, the edr2 mutant was resistant, displaying massive necrotic cell death and very few spores. The edr2 bsk1-1 plants displayed a wild-type-like susceptible phenotype, and no visible necrotic lesions were present on the leaves. A genomic clone containing the BSK1 gene complemented the bsk1-1 mutation. WT, the wild type.
(B) Fungal growth and cell death on 4-week-old plants at 8 DAI with G. cichoracearum. Leaves were stained with trypan blue to show fungal structures and dead cells. A large number of spores were produced in the wild type and edr2 bsk1-1, while extensive mesophyll cell death occurred in edr2. Bar = 50 μm
(C) Quantification of fungal growth in plants at 5 DAI by counting the number of conidiophores per colony. Results were from one experiment, and the bars represent mean and sd (n > 30). Statistically significant differences are indicated by lowercase letters (P < 0.05; one-way analysis of variance [ANOVA]). The experiments were repeated three times with similar results.
(D) Infected leaves were stained with 3,3′-diamino benzidine-HCl and trypan blue sequentially at 2 DAI to visualize H2O2 (brown staining) and fungal structures (blue staining). Bar = 20 μm
(E) and (F) The bsk1-1 mutation suppressed enhanced ethylene-induced senescence in edr2. Four-week-old plants were exposed to 100 μL L−1 ethylene for 3 d, and increased chlorosis was observed in edr2 mutants after ethylene treatment. The edr2 bsk1-1 plants showed a wild-type-like phenotype (E). The chlorophyll content in leaves from the ethylene treated plants (F). Bars represent sd of values obtained from five plants. Lowercase letters represent significant difference from the wild type (P < 0.01, one-way ANOVA). Three independent experiments were performed with similar results. FW, fresh weight.
(G) The bsk1-1 mutation was identified by standard map-based cloning. Markers and BAC clones are indicated.
(H) Structure of the BSK1 gene. The asterisk indicates the bsk1-1 mutation. Exons are indicated by black boxes and introns by black lines. A nucleotide change (G1328A) was identified in the BSK1 gene and leads to an amino acid substitution (R443Q) in the BSK1 protein. The BSK1 protein contains a kinase domain and a TPR domain. The bsk1-1 point mutation (asterisk) is in the TPR domain. aa, amino acids.
Previously, it was shown that edr2 accumulates more hydrogen peroxide (H2O2) upon infection with G. cichoracearum (Vorwerk et al., 2007), and H2O2 accumulation appears to occur at the site of fungal infection (Yao et al., 2012). To examine whether the bsk1-1 mutation affects H2O2 accumulation in edr2, we assessed H2O2 accumulation in wild-type, edr2, and edr2 bsk1-1 plants after infection with G. cichoracearum at 2 DAI. Consistent with previous findings, edr2 accumulated more H2O2 than the wild type at 2 DAI; however, this high level of H2O2 accumulation was suppressed by the bsk1-1 mutation (Figure 1D). In addition to the enhanced resistance to powdery mildew, the edr2 mutant displays increased ethylene-induced senescence phenotypes, indicating possible crosstalk between defense responses and senescence (Tang et al., 2005; Nie et al., 2012). To investigate whether bsk1-1 affects edr2-mediated ethylene-induced senescence, we treated edr2 bsk1-1 with ethylene for 3 d and assessed the leaf senescence. The enhanced ethylene-induced senescence phenotype in edr2 was also suppressed by the bsk1-1 mutation (Figures 1E and 1F).
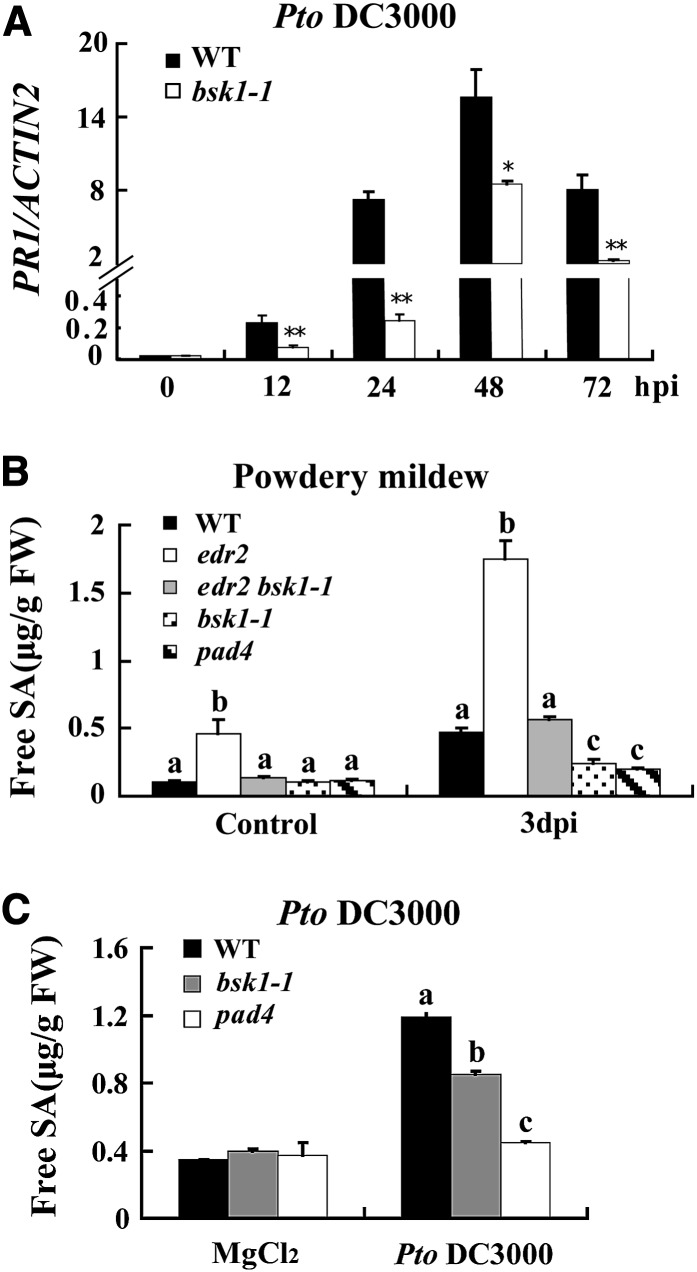
To gain insight into the molecular basis that underlies the suppression of edr2 phenotypes by bsk1-1, we examined the transcript levels of several defense-related genes in edr2 bsk1-1 plants at different time points after G. cichoracearum infection. Previously, it was shown that edr2-mediated resistance requires intact SA signaling; therefore, we selected several marker genes for the SA pathway, including PATHOGENESIS-RELATED GENE1 (PR1), PR2, FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1), PHYTOALEXIN DEFICIENT4 (PAD4), and SID2, which are all widely used marker genes and are upregulated when defense responses are activated (Jirage et al., 1999; Mishina and Zeier, 2006). All those defense-related genes were induced more strongly in edr2 than in wild-type plants at 3 DAI, and in edr2 bsk1-1, the levels of transcript of those genes were significantly lower than in edr2 (see Supplemental Figures 1A to 1E online), indicating that bsk1-1 suppressed upregulation of defense-related genes in edr2 during G. cichoracearum infection. In the bsk1-1 single mutant, the levels of transcript of those genes were similar to that in the wild type, only the PR2 transcript levels accumulated at significantly lower levels than in the wild type at 3 DAI upon G. cichoracearum infection (see Supplemental Figure 1B online).
bsk1-1 is a recessive mutant, as the edr2/edr2 bsk1-1/BSK1 plants displayed edr2-like powdery mildew resistance phenotypes. To identify the bsk1-1 mutation, we performed a standard map-based cloning approach (Figure 1G) and identified a mutation (G→A) in At4g35230, which caused an amino acid change (R443Q) in the predicted open reading frame (Figure 1H). To confirm that At4g35230 is the responsible gene, we made a genomic clone construct for At4g35230 and transformed edr2 bsk1-1 with the construct. We inoculated multiple T1 plants with G. cichoracearum and found that the transgenic plants were all resistant to powdery mildew, a phenotype indistinguishable from the edr2 single mutant. Therefore, the At4g35230 genomic clone complemented the bsk1-1 phenotype (Figures 1A to 1C), indicating that the mutation identified in At4g35230 causes the bsk1-1 phenotype. At4g35230 was previously designated BR-SIGNALING KINASE1 (BSK1) (Tang et al., 2008). BSK1 encodes a receptor-like cytoplasmic kinase (RLCK) belonging to the RLCK-XII phylogenetic clade, which comprises 12 proteins in Arabidopsis (Shiu et al., 2004; Tang et al., 2008). The BSK1 protein consists of an N-terminal kinase domain and a C-terminal tetratricopeptide repeat (TPR) domain. The TPR domain is thought to be involved in protein–protein interactions (Blatch and Lässle, 1999). The bsk1-1 mutation (R443Q) is in the TPR domain and the Arg residue at position 443 in BSK1 is strictly conserved in the Arabidopsis BSK1 protein family (see Supplemental Figure 2 online). In addition, knockdown of BSK1 by RNA interference (RNAi) also suppressed edr2-mediated powdery mildew resistance, indicating that suppression of edr2 phenotypes in edr2 bsk1-1 was caused by loss of BSK1 function (see Supplemental Figure 3 online). The mutation in several BSK1-like genes, including BSK2, BSK3, BSK6, BSK8, and BSK12, did not suppress edr2-mediated powdery mildew resistance (see Supplemental Figure 2 online).
bsk1-1 Suppresses edr1, mlo2, and pmr4-Mediated Powdery Mildew Resistance Phenotypes
To further investigate the role of BSK1 in powdery mildew resistance, we crossed bsk1-1 with several well-characterized resistant mutants, including edr1, mildew resistance locus O2 (mlo2), powdery mildew resistant4 (pmr4), and autophagy-related2 (atg2), which all show SA-dependent powdery mildew resistance, except for mlo2 (Frye et al., 2001; Nishimura et al., 2003; Tang et al., 2005; Consonni et al., 2006; Wang et al., 2011). The edr1 bsk1-1, mLo2 bsk1-1, and pmr4 bsk1-1 plants were susceptible to G. cichoracearum, showing a large number of spores, indicating that bsk1-1 suppressed the edr1, mlo2, and pmr4-mediated powdery mildew resistance phenotypes (see Supplemental Figures 4A to 4C online). Previously, mlo2 was shown to display an early senescence phenotype, and this phenotype is SA dependent but jasmonic acid and ethylene independent (Consonni et al., 2006). The mlo2-mediated senescence was suppressed in the mlo2 bsk1-1 double mutant, indicating that this phenotype also requires BSK1 function (see Supplemental Figure 5 online). By contrast, the atg2 bsk1-1 plants displayed mildew-induced cell death phenotypes similar to atg2, indicating that bsk1-1 could not suppress the disease resistance and mildew-induced cell death phenotypes of atg2 (see Supplemental Figure 6 online).
The bsk1-1 Mutant Displays Enhanced Susceptibility to G. cichoracearum, Pto DC3000, and H. arabidopsidis
To further explore the role of BSK1 in defense responses, we first challenged the bsk1-1 single mutant with G. cichoracearum, followed by quantification of conidiophore formation. The bsk1-1 mutant was more susceptible and supported significantly more spores than wild-type plants (see Supplemental Figures 4A to 4C online), indicating that BSK1 is required for basal resistance to G. cichoracearum.
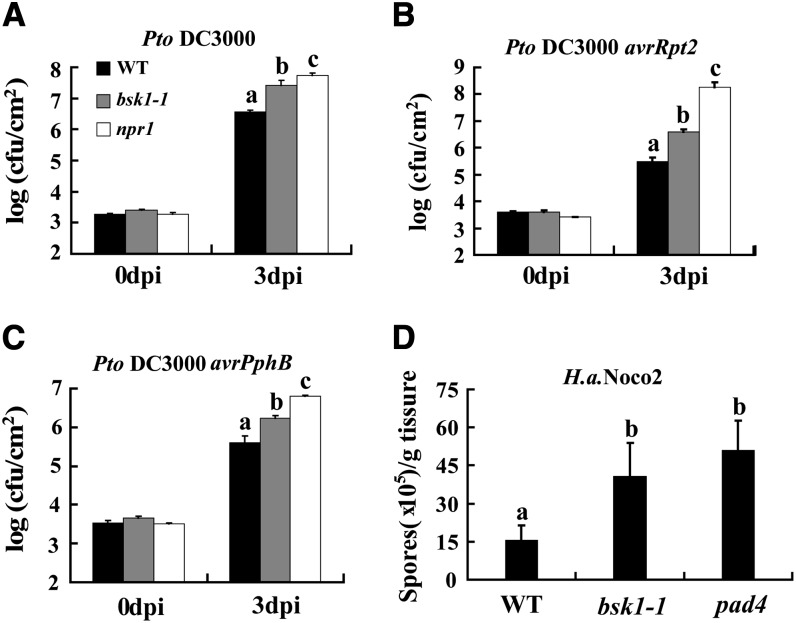
To investigate whether BSK1 plays a role in resistance to other pathogens, we infected bsk1-1 with the virulent bacterium Pseudomonas syringae pv tomato (Pto) DC3000 and the avirulent Pto DC3000 strains that carry the effector genes avrRpt2 or avrPphB. The effectors encoded by avrRpt2 and avrPphB are recognized by the coiled-coil-NBS-LRR resistance proteins RESISTANCE TO PSEUDOMONAS SYRINGAE2 (RPS2) or RPS5, respectively. We found that the bsk1-1 mutant was more susceptible to the virulent and avirulent strains of Pto DC3000, as it supports significantly more growth of all the bacterial strains than the wild type 3 DAI (Figures 2A to 2C). In addition, bsk1-1 accumulated lower PR1 transcript levels than the wild type during infection with Pto DC3000 (Figure 3A). Similarly, the BSK1-RNAi plants also displayed enhanced susceptibility to Pto DC3000 (see Supplemental Figure 7 online). These data indicate that BSK1 plays an important role in resistance to both virulent and avirulent Pto DC3000 strains.
Figure 2.
Responses of the bsk1-1 Mutant to Pathogens.
(A) to (C) Four-week-old plants were infected with Pto DC3000 (A), Pto DC3000 avrRpt2 (B), and Pto DC3000 avrPphB (C). The plants were inoculated with bacterial suspensions at OD600 = 0.0005. The number of bacteria was counted at 4 h after infection and 3 DAI. cfu, colony-forming units; WT, the wild type.
(D) Two-week-old plants were infected with H. a. Noco 2. The number of sporangiophores was counted at 7 DAI.
Bars represent sd of values obtained from three independent samples. Lowercase letters represent significant differences from the wild type (P < 0.05, one-way ANOVA). At least three independent experiments were performed with similar results.
Figure 3.
SA Levels Are Affected by the bsk1-1 Mutation.
Four-week-old plants were infected with G. cichoracearum or Pto DC3000.
(A) Accumulation of PR1 transcripts was examined by quantitative real-time PCR at various time points after inoculation with Pto DC3000 (OD600 = 0.001). ACTIN2 was used as an internal control. Bars represent mean and sd from three independent experiments. Statistically significant differences are indicated by one asterisk (P < 0.05, Student’s t test) or two asterisks (P < 0.01, Student’s t test). hpi, h after infection; WT, the wild type.
(B) Free SA levels were measured in the uninfected and infected (3 DAI) leaves after inoculation with G. cichoracearum. FW, fresh weight.
(C) Free SA levels were measured in the 10 mM MgCl2 treated or Pto DC3000 (OD600 = 0.001) infected (2 DAI) leaves.
(B) and (C) Bars represent mean and sd from three independent biological experiments. Lowercase letters indicate statistically significant differences (P < 0.01, one-way ANOVA).
To further investigate the role of BSK1 in defense responses, we infected bsk1-1 with H. arabidopsidis (H. a.) Noco2, a virulent oomycete pathogen that causes downy mildew disease in Arabidopsis. The bsk1-1 mutant was more susceptible than wild-type plants, supporting the growth of more H. a. Noco2 sporangiophores than the wild type (Figure 2D; see Supplemental Figure 8 online), indicating that the disease resistance to downy mildew also requires BSK1. Taken together, these data demonstrate that BSK1 plays an important role in resistance to a variety of pathogens.
bsk1-1 Accumulates Lower Levels of SA after Pathogen Infection
The bsk1-1 suppressed edr1, edr2, and pmr4-mediated resistance and showed enhanced susceptibility to several biotrophic pathogens, suggesting that bsk1-1 may have defects in SA accumulation. To investigate whether bsk1-1 affects SA production, we measured the SA levels in bsk1-1 before and after G. cichoracearum infection. We used pad4, an enhanced susceptibility mutant that accumulates lower levels of SA upon pathogen infection (Zhou et al., 1998) as a susceptible control. As shown in Figure 3B, edr2 accumulated much higher levels of SA than the wild type upon G. cichoracearum infection; however, bsk1-1 suppressed higher accumulation of SA levels in edr2. Also, SA levels were lower in the bsk1-1 mutant than the wild type at 3 DAI with G. cichoracearum. Similarly, the accumulation of SA in bsk1-1 was significantly lower than in the wild type after Pto DC3000 infection (Figure 3C). This observation indicates that the enhanced susceptibility to G. cichoracearum and Pto DC3000 may, at least partially, be caused by lower SA production in the bsk1-1 mutant. This may also partially explain why bsk1-1 suppresses edr2 resistance, as previously it was shown that the powdery mildew resistant phenotype in edr2 was suppressed by the sid2 and npr1 mutations, which cause defects in SA accumulation and signaling, respectively (Tang et al., 2005; Vorwerk et al., 2007).
Membrane Localization Is Required for BSK1 Function
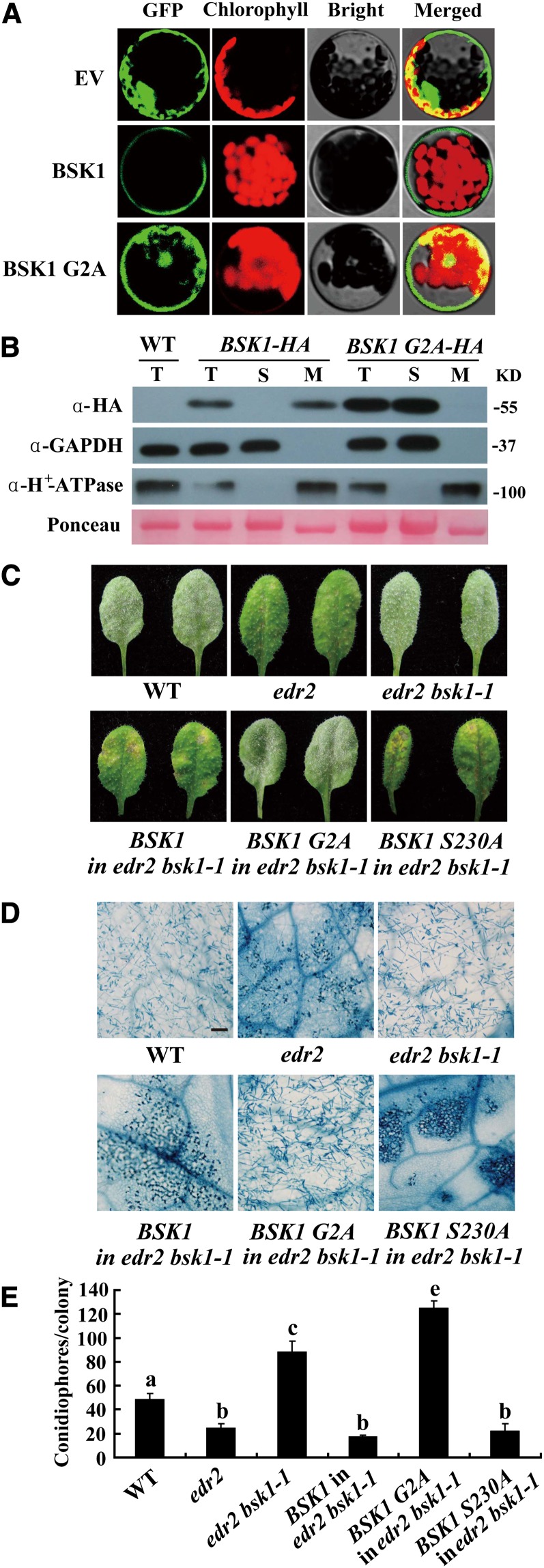
Previously, it was shown that BSK1 localizes to the plasma membrane (Tang et al., 2008). The BSK1 protein contains a myristoylation site at its N terminus, which is a potential membrane localization signal (Thompson and Okuyama, 2000). To examine whether the myristoylation site is critical for BSK1 membrane localization, we created a BSK1 mutant with a disrupted myristoylation site (G2A) and fused it to green fluorescent protein (GFP). We then transiently transformed Arabidopsis protoplasts with the construct. The BSK1 G2A-GFP fusion protein did not exclusively localize to the cell periphery (Figure 4A). To further investigate the role of the myristoylation site in BSK1, we generated constructs to express BSK1-HA or the G2A mutant form of BSK1-HA (BSK1 G2A-HA) under the control of the BSK1 promoter and stably transformed edr2 bsk1-1 plants with the construct. Both the BSK1-HA and BSK1 G2A-HA transgenic plants expressed fusion proteins of the correct sizes, as detected by immunoblot analysis (see Supplemental Figure 9 online). We then performed immunoblots with total protein and soluble and membrane fractions from 4-week-old transgenic plants. Consistent with the confocal microscopy observations, BSK1-HA was detected in the membrane fraction, but not in the soluble fraction; by contrast, BSK1 G2A-HA was detected only in the soluble fraction (Figure 4B). We infected the transgenic plants with G. cichoracearum and examined whether BSK1-HA and BSK1 G2A-HA restored the edr2-like resistant phenotype in the edr2 bsk1-1 background; as shown in Figures 4C to 4E, transgenic edr2 bsk1-1 plants carrying BSK1-HA were indistinguishable from edr2 mutants in powdery mildew resistance, indicating that BSK1-HA is functional. However, by contrast, BSK1 G2A-HA was unable to complement the bsk1-1 phenotype (Figures 4C to 4E), strongly suggesting that membrane localization is essential for BSK1 function.
Figure 4.
BSK1-GFP Localizes to the Plasma Membrane, and Disruption of the Predicted Myristoylation Site Compromises BSK1 Function.
(A) BSK1-GFP was transformed into Arabidopsis protoplasts, and the GFP signal was detected by confocal microscopy. BSK1-GFP localizes to cell periphery, and disruption of the myristoylation site of BSK1 (G2A) compromised BSK1 cell periphery localization. EV, empty vector.
(B) Subcellular fractionation and immunoblot assays. Total protein was extracted from 4-week-old plants. Total (T), soluble (S), and membrane (M) fractions of protein from BSK1-HA or BSK1 G2A-HA transgenic plants were loaded on SDS-PAGE gels and subjected to immunoblotting with anti-HA antibody. GAPDH and H+-ATPase were used as soluble marker or plasma membrane marker, respectively. Molecular masses of protein markers are shown on the right. Ponceau S staining of ribulose-1,5-bis-phosphate carboxylase/oxygenase is shown as a loading control. The experiments were repeated three times with similar results. WT, the wild type.
(C) to (E) The BSK1 G2A-HA clone was unable to restore edr2 bsk1-1 to the edr2 phenotype. Four-week-old plants were infected with G. cichoracearum.
(C) Representative leaves were removed and photographed at 8 DAI. Leaves of two independent transgenic lines for each construct are shown. More than 10 independent transgenic lines expressing the fusion proteins of the correct sizes were examined for each construct. All the transgenic lines examined showed consistent phenotypes.
(D) The infected leaves at 8 DAI were stained with trypan blue. Bar = 100 μm.
(E) The number of conidiophores per colony was counted at 5 DAI. Bars represent mean and sd (n > 30). Lowercase letters represent statistically significant differences (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
BSK1 Has Protein Kinase Activity in Vitro, and the Kinase Activity Is Required for BSK1 Function
Previously, Tang et al. (2008) showed that amino acid Ser-230 in BSK1 is the major phosphorylation site for the BR receptor BRI1. To examine whether the phosphorylation of this amino acid is important for BSK1 function in disease resistance, we generated a mutant form of BSK1-HA that contained an S230A substitution and transformed edr2 bsk1-1 with BSK1 S230A. Interestingly, this construct was able to fully restore the edr2 resistant phenotypes in the edr2 bsk1-1 background (Figures 4C to 4E), indicating that the phosphorylation of this amino acid is not essential for BSK1 function in suppression of the edr2-mediated defense responses.
To examine whether BSK1 has protein kinase activity, we produced BSK1 in Escherichia coli and performed in vitro kinase assays. BSK1 could autophosphorylate in vitro, and its kinase activity required Mn2+, instead of Mg2+, as a divalent cation cofactor (Figure 5A). To test whether the kinase activity is required for the BSK1 function, we generated and transformed edr2 bsk1-1 with a kinase-deficient form of BSK1 (K104E). We infected 4-week-old transgenic plants with G. cichoracearum. The kinase-deficient form of BSK1 failed to restore the edr2 resistant phenotypes in the edr2 bsk1-1 background (Figures 5B and 5C), indicating that kinase activity is required for BSK1 function.
Figure 5.
BSK1 Has Kinase Activity in Vitro, and the Kinase Activity Is Required for Defense Function.
(A) BSK1 can autophosphorylate in vitro. Recombinant maltose binding–BSK1 fusion protein (MBP-BSK1) or MBP-BSK1 (K104E) was incubated in a kinase assay buffer containing [γ-32P]ATP and 10 mM MnCl2 or 10 mM MgCl2 as indicated, then separated by SDS-PAGE and detected by autoradiography. Kinase activity was assessed with a dose–response assay. First lane, 1 μg MBP; second lane, 2 μg MBP-BSK1; the next six lanes, 20 ng, 200 ng, or 2 μg of MBP-BSK1 or MBP-BSK1(K104E). Increasing amounts of protein are denoted with a triangle. Top panel, autoradiogram; bottom panel, Coomassie blue (CBB) staining. The molecular weight markers are indicated. The experiment was repeated three times with similar results.
(B) BSK1 (K104E), the kinase-dead mutant form of BSK1, cannot complement the bsk1-1 mutation. The BSK1 or BSK1(K104E) genomic clone was transformed into edr2 bsk1-1 mutants, and the transgenic plants were infected with G. cichoracearum. Representative leaves were removed and photographed at 8 DAI (top panel). The infected leaves at 8 DAI were stained with trypan blue to visualize the fungal structures and mesophyll cell death in the leaves. WT, the wild type. Bar = 100 μm.
(C) Quantification of fungal growth by counting the number of conidiophores per colony at 5 DAI. Bars represent mean and sd (n > 30). Lowercase letters indicate statistically significant differences (P < 0.01, one-way ANOVA). Experiments were repeated three times with similar results.
Previously, BSK1 was shown to be involved in BR signaling as a substrate for the BR receptor BRI1 (Tang et al., 2008). Also, the BR signaling component BAK1 was shown to play a role in basal defense; therefore, we investigated whether BAK1 and BSK1 share similar functions in plant immunity. However, the edr2 bak1-4 double mutant displayed edr2-like powdery mildew resistance phenotypes, indicating that edr2-activated disease resistance does not require BAK1 (see Supplemental Figure 10 online). Previously, it was shown that BRI1 SUPPRESSOR1 (BSU1), an important player in BR signaling, interacts with BSK1 in vivo (Kim et al., 2009). We also tested whether BSU1 and BSU-LIKE1 (BSL1) are required for edr2-mediated disease resistance. Neither bsu1 nor bsl1 mutations affected edr2-mediated powdery mildew resistance (see Supplemental Figure 10 online), indicating that edr2-activated defense signaling is independent of BSU1 and BSL1 function.
The bsk1-1 mutants did not show obvious growth defects, but bsk1-1 leaves were slightly narrower than wild-type leaves, and this phenotype was complemented by a genomic BSK1 clone (see Supplemental Figure 11 online). The narrow leaf phenotype in bsk1-1 could be an indication of activation of BR signaling. To assess the BR responses in the bsk1-1 mutant, we treated the bsk1-1 seedlings with different concentrations of brassinolide (BL) for 7 d and measured the root lengths. The wild-type seedlings displayed typical BL responses, with much shorter roots than those of the bri1-9 mutant, which was consistent with previous observations (Noguchi et al., 1999), while the bsk1-1 seedlings and BSK1 RNAi lines were very similar to the wild type, displaying inhibited root growth (see Supplemental Figure 12A online). In addition, to test the activity of BL signaling in the bsk1-1 mutant, we also performed hypocotyl elongation assays on etiolated seedlings in the presence or absence of brassinazole (BRZ), a BR biosynthesis inhibitor (Asami et al., 2000). The bsk3-1 and bri1-9 mutants were more sensitive to BRZ than the wild type and showed lower relative hypocotyl lengths (Tang et al., 2008); by contrast, the bsk1-1 mutant and BSK1 RNAi line were indistinguishable from the wild type in response to BRZ (see Supplemental Figure 12B online). In addition, the transcript accumulation of two BL-responsive genes, SAUR-AC and DWF4 in the bsk1-1 and BSK1 RNAi plants, was very similar to that of the wild type in the presence or absence of 100 nM BL (see Supplemental Figures 12C and 12D online). These data indicate that bsk1 displays wild-type phenotypes in response to BL treatment.
BSK1 Forms a Protein Complex with FLS2 in Nicotiana benthamiana and Arabidopsis
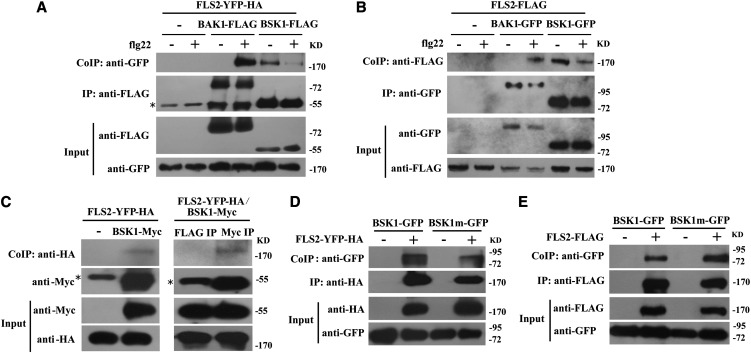
Previously, BSK1 was identified as a possible component of an RPS2 protein complex, and the PAMP receptor FLS2 has also been shown to physically associate with RPS2 (Qi et al., 2011). To investigate whether BSK1 forms a protein complex with FLS2, we performed coimmunoprecipitation (Co-IP) assays by transiently expressing BSK1-FLAG and FLS2-YFP-HA in N. benthamiana leaves. We expressed FLS2-YFP-HA alone as a negative control. BSK1 was immunoprecipitated using anti-FLAG antibody, and in the precipitate, FLS2 was detected with anti-GFP antibody only from the leaves that coexpressed both BSK1-FLAG and FLS2-YFP-HA, not from the negative control leaves that only expressed FLS2-YFP-HA (Figure 6A). These observations indicate that BSK1 and FLS2 form a protein complex in N. benthamiana. Previously, it was shown that FLS2 and BAK1 form a complex in a ligand-dependent manner (Chinchilla et al., 2007). To validate the N. benthamiana system, we expressed both BAK1-FLAG and FLS2-YFP-HA in N. benthamiana leaves as a control; consistent with previous findings, BAK1 only associated with FLS2 upon flg22 treatment. This observation indicated that ectopic expression in the N. benthamiana system did not alter the behavior of the FLS2 and BAK1 proteins. However, the association of BSK1 and FLS2 occurs without flg22 treatment (Figure 6A), and it appears that there is less interaction in the presence of flg22 (Figure 6A). This observation was reproducible in four independent experiments.
Figure 6.
BSK1 Forms a Protein Complex with FLS2.
(A) Co-IP of BSK1 and FLS2 from N. benthamiana transiently expressing BSK1-FLAG and FLS2-YFP-HA before (−) or 10 min after (+) elicitation with 1 μM flg22, as indicated. Total protein was extracted and subjected to immunoprecipitation of BSK1 protein by FLAG antibody, followed by immunoblot analysis with anti-GFP antibody. BAK1-FLAG was used an internal control. Asterisk indicates the heavy chain of IgG (∼55 kD).
(B) Co-IP of BSK1 and FLS2 from Arabidopsis protoplasts transiently expressing BSK1-GFP and FLS2-FLAG before (−) or 10 min after (+) elicitation with 1 μM flg22 as indicated. FLS2-FLAG alone was used as a negative control. The BSK1 protein was immunoprecipitated by GFP antibody, followed by immunoblot analysis with anti-FLAG antibody. BAK1-GFP was used as an internal control.
(C) Co-IP of BSK1 and FLS2 from transgenic Arabidopsis plants. Total protein was extracted from 3-week-old plants expressing both BSK1-Myc and FLS2-YFP-HA. Plants expressing FLS2-YFP-HA alone (left panel) or an immunoprecipitation with unrelated antibody (right panel) were used as negative controls. The BSK1 protein was immunoprecipitated by anti-Myc antibody, and the presence of FLS2-YFP-HA protein was detected by immunoblot analysis with anti-HA antibody.
(D) and (E) The bsk1-1 mutation did not affect BSK1 and FLS2 association in N. benthamiana or Arabidopsis protoplasts. Co-IP of BSK1 and FLS2 from N. benthamiana transiently expressing BSK1m-GFP (carrying the bsk1-1 mutation) and FLS2-YFP-HA (D) or Arabidopsis protoplasts transiently expressing BSK1m-GFP and FLS2-FLAG (E). BSK1-GFP or BSK1m-GFP alone was used as a negative control. Total protein was subjected to immunoprecipitation with anti-HA (D) or anti-FLAG (E) antibody, followed by immunoblot analysis using anti-GFP antibody.
These experiments were repeated four times with similar results.
To investigate the interaction of BSK1 and FLS2 in Arabidopsis, we performed a Co-IP assay by expressing BSK1-GFP and FLS2-FLAG in Arabidopsis protoplasts. We expressed FLS2-FLAG alone as a negative control. We extracted total protein from the protoplasts, immunoprecipitated BSK1 protein with GFP antibody, and then examined whether FLS2-FLAG coimmunoprecipitated with BSK1. FLS2 was detected with anti-FLAG antibody only from the protoplasts that coexpressed both BSK1-GFP and FLS2-FLAG, not from the negative control (Figure 6B), indicating that BSK1 and FLS2 form a protein complex in Arabidopsis. Similarly, we also expressed both BAK1-GFP and FLS2-FLAG in Arabidopsis protoplasts as controls and found that BAK1 only associates with FLS2 upon flg22 elicitation (Figure 6B), validating the protoplast system.
To further confirm the physical association of BSK1 and FLS2, we performed Co-IP assays using stably transformed transgenic plants that express both BSK1pro:BSK1-Myc and 35Spro:FLS2-YFP-HA. Transgenic plants that express only 35Spro:FLS2-YFP-HA were used as a negative control. BSK1-Myc restored the phenotype of bsk1-1 mutants (see Supplemental Figures 13A and 13B online), and the 35Spro:FLS2-YFP-HA clone was able to complement the fls2 phenotype (see Supplemental Figure 13C online), indicating that the BSK1-Myc and FLS2-YFP-HA proteins are functional. We extracted total protein from the transgenic plants and immunoprecipitated the BSK1 protein with Myc antibody or with unrelated FLAG antibody as a negative control. We then examined whether FLS2 was in the precipitate by immunoblotting with HA antibody. The FLS2-YFP-HA protein was detected only in the sample immunoprecipitated with Myc antibody, not in the negative controls (Figure 6C). This observation indicated that BSK1-Myc physically associates with FLS2-YFP-HA in Arabidopsis.
BSK1 is a receptor-like cytoplasmic protein kinase with an N-terminal kinase domain and a C-terminal TPR domain. The TPR motif was originally identified in yeast as a protein–protein interaction domain (Hirano et al., 1990; Sikorski et al., 1990). Many TPR-containing proteins serve as scaffolds for the assembly of multiprotein complexes, and the TPR motif facilitates specific interactions between partners (Blatch and Lässle, 1999; Smith, 2004). The bsk1-1 mutation causes a substitution of a strictly conserved residue (Arg-443) in the TPR motif (see Supplemental Figure 2 online). This mutation may disrupt BSK1 function by affecting interactions between BSK1 and its partner(s). To investigate whether this mutation affects the association of BSK1 and FLS2, we examined whether the mutated version of BSK1 forms a complex with FLS2. As shown in Figures 6D and 6E, the mutated version of BSK1 forms a complex with FLS2 in both N. benthamiana and Arabidopsis, which was similar to the wild-type BSK1 protein, indicating that the bsk1-1 mutation did not affect the association of BSK1 and FLS2. This observation suggests that the bsk1-1 phenotype is not caused by the defects of BSK1(R443) in association with FLS2.
bsk1-1 Displayed Defects in flg22-Induced ROS Burst
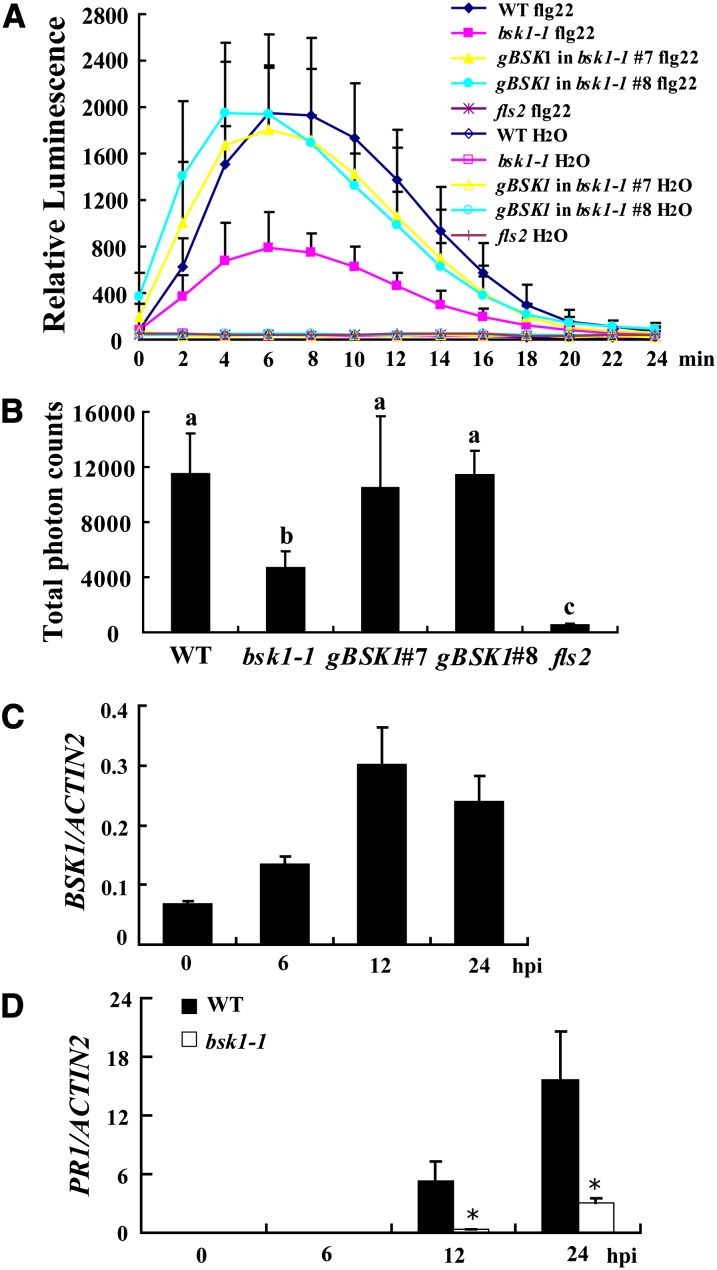
Since BSK1 physically associates with FLS2, we hypothesized that BSK1 may be involved in some flg22-induced responses mediated by FLS2. To test this hypothesis, we treated wild-type and bsk1-1 leaves with 100 nM flg22 and measured the ROS burst, a marker for early basal defense responses. The fls2 mutant was insensitive to flg22; upon flg22 treatment, ROS accumulated rapidly in the wild type, but not in the fls2 mutant. By contrast, the accumulation of ROS in bsk1-1 was partially impaired, indicating that bsk1-1 had defects in the flg22-induced ROS burst (Figures 7A and 7B). The defects of bsk1-1 in flg22-trigged ROS accumulation were complemented by a genomic BSK1 clone (Figures 7A and 7B). Similarly, the BSK1 RNAi plants also showed bsk1-1 like defects in ROS accumulation upon flg22 treatment (see Supplemental Figures 14A and 14B online). When treated with higher concentrations of flg22, bsk1-1 also showed similar defects in ROS accumulation (see Supplemental Figures 14C and 14D online). To examine whether bsk1-1 has a general defect in ROS accumulation, we examined ROS accumulation in bsk1-1 in response to elf18 (derived from bacterial PAMP elongation factor Tu), which is specifically recognized by EF-TU RECEPTOR (EFR) in Arabidopsis (Zipfel et al., 2006). Consistent with previous findings, the efr-1 mutant was insensitive to elf18, but bsk1-1 rapidly accumulated large amounts of ROS upon elf18 treatment, which was very similar to the wild type (see Supplemental Figures 15A and 15B online). Although the effect of bsk1-1 mutation on elf18 responses needs to be further characterized, this observation suggests that bsk1-1 does not have a substantial defect in ROS accumulation triggered by elf18. Consistent with the role of BSK1 in flg22-induced responses, BSK1 transcript accumulation increased following flg22 treatment (Figure 7C). The level of flg22-induced PR1 expression was much lower in bsk1-1 than in the wild type (Figure 7D). By contrast, PR1 accumulation induced by elf18 was similar between bsk1-1 and the wild type (see Supplemental Figure 15C online). To further investigate the role of BSK1 in flg22-induced responses, we also examined the effects of bsk1-1 on the activation of MAPKs by flg22. In the wild type, MPK3, MPK4, and MPK6 were rapidly activated, and the activation of MPK3, MPK4, and MPK6 was abolished in the fls2 mutant, which is consistent with previous findings (Schwessinger et al., 2011). However, MPK3, MPK4, and MPK6 were strongly activated in bsk1-1, which was similar to the wild type (see Supplemental Figure 16A online). Similarly, under our experimental conditions, the induction of three flg22-induced genes, At1g51890, At2g17740, and At2g19190 (He et al., 2006), was not significantly affected in bsk1-1 (see Supplemental Figures 16B to 16D online). To characterize the late responses of bsk1-1 to flg22, we treated bsk1-1 and wild-type seedlings with flg22 for 10 d and found that flg22 inhibited seedling growth in the wild type and bsk1-1, but not in the fls2 mutant (see Supplemental Figure 16E online). Taken together, these data suggest that the bsk1-1 mutant only compromises a subset of FLS2-mediated responses. However, we cannot exclude the possibility that the reason we did not observe effects of bsk1-1 was due to a relatively high dose of flg22 (100 nM) used in the experiments. Such potential quantitative effects of flg22 treatment on bsk1-1–related phenotypes should be investigated in the future.
Figure 7.
bsk1-1 Displayed Defects in flg22-Induced ROS Burst.
(A) Leaves of the wild type, bsk1-1, fls2, and two bsk1-1 complementation lines were treated with 100 nM flg22 and incubated with luminol and horseradish peroxidase to detect ROS. Luminescence was recorded at different time points as indicated. Error bars represent sd of data derived from replicate samples (n = 12). WT, the wild type.
(B) Total photon counts during 30 min of treatment are presented to indicate the ROS production. Bars represent sd (n = 12). Statistically significant differences were indicated with lowercase letters (P < 0.01, one-way ANOVA).
(C) Accumulation of BSK1 transcript in response to flg22. The wild-type seedlings were treated with 100 nM flg22 at different time points. hpi, h after infection.
(D) The transcript accumulation of PR1 was examined by quantitative real-time PCR at various time points after treatment with 100 nM flg22. ACTIN2 was used as an internal control. Bars represent mean and sd from three independent experiments. Statistically significant difference is indicated by an asterisk (P < 0.05, Student’s t test).
The experiments were repeated three times with similar results.
DISCUSSION
BSK1 Plays an Important Role in Defense Responses
The edr2 mutant displays enhanced powdery mildew resistance and mildew-induced cell death, and edr2-mediated resistance is dependent on the SA pathway (Tang et al., 2005; Vorwerk et al., 2007). To dissect the molecular mechanisms underlying edr2-mediated resistance, we screened edr2 suppressors and identified the bsk1-1 mutation, which fully suppressed powdery mildew resistance in edr2, indicating that BSK1 is required for edr2-mediated resistance, or BSK1 and EDR2 function in parallel and their additive effect is observed as phenotypes.
The bsk1-1 single mutant displays enhanced susceptibility to a number of pathogens, including bacteria, fungi, and oomycete pathogens, indicating that BSK1 is a positive regulator of plant defense responses. In addition to EDR2, several genes in Arabidopsis have been shown to play negative roles in disease resistance to powdery mildew, such as EDR1, PMR4, MLO2, and ATG2 (Frye et al., 2001; Nishimura et al., 2003; Consonni et al., 2006; Vorwerk et al., 2007). Previously, it has been shown that PMR4, EDR1, and MLO2 define different genetic pathways in plant immunity (Frye et al., 2001; Nishimura et al., 2003; Consonni et al., 2006; Wawrzynska et al., 2010). The observation that bsk1-1 suppressed all edr1, mlo2, and pmr4-mediated powdery mildew resistance phenotypes indicates that BSK1 may play a general but important role in plant defense responses. Although it is not clear how BSK1 functions in plant immunity, the bsk1-1 mutant accumulates lower levels of SA, which partially explains the effects of bsk1-1 in edr2. Although BSK1 was identified from an edr2 suppressor screen, the relationship between BSK1 and EDR2 is not clear. It is worth noting that both EDR2 and BSK1 are membrane proteins; however, whether EDR2 and BSK1 interact directly or indirectly remains to be addressed.
BSK1 has previously been identified by a quantitative proteomic approach as one of the substrates for the BR receptor kinase BRI1 (Tang et al., 2008). BRI1 interacts with BSK1 in vivo, and BR treatment reduces the BRI1 and BSK1 interaction. In addition, BRI1 phosphorylates BSK1, and the Ser-230 of BSK1 is the major site for BRI1 phosphorylation. Furthermore, overexpression of BSK1 suppresses dwarf phenotypes in the bri1-5 or det2-1 mutants. Those observations indicate that BSK1 is the substrate of BRI1 kinase and is involved in BR signal transduction (Tang et al., 2008). In addition to BSK1, some BSK1 homologs, such as BSK2, BSK3, and BSK5, are also involved in BR signaling (Tang et al., 2008). When BR activates BRI1, BSKs are phosphorylated and dissociate from the BRI1 complex, which leads to activation of downstream signaling. These data indicate that BSK1 is an important regulator in BR signaling. It is worth noting that bsk1-1 did not show obvious defects in BR signaling based on several assays, which may be because bsk1-1 is a point mutation, and the function of BSK1 is only partially impaired by bsk1-1. Another possibility is that other BSK1-like proteins have redundant roles in BR signaling, as BSK1 belongs to a small RLCK protein family. It would be very interesting to define how BSK1 modulates both BR signaling and immune response. The bsk1-1 mutant identified in this study provides a valuable tool to dissect the function of BSK1 in both BR signaling and plant innate immunity.
RLCKs and Plant Innate Immunity
BSK1 belongs to the RLCK subfamily XII, which consists of 12 members, and several proteins in this family are involved in BR signaling (Tang et al., 2008). Although Qi et al. (2011) identified BSK1 and BSK8 as candidate components of RPS2 complexes in a proteomic study, to date, no genetic evidence has shown that any proteins in this family regulate plant disease resistance. To extend our genetic analyses, we tested whether several BSK1-like genes in this family are involved in plant defense responses by infecting T-DNA insertion mutant lines with Pto DC3000. However, unlike the bsk1-1 mutant, those T-DNA insertion lines all displayed wild-type-like phenotypes. Also, the disruption of these genes did not suppress edr2-mediated resistance, indicating that these genes may have different roles than BSK1. Consistent with this notion, the expression patterns of BSK1-like genes are different. For instance, BSK12 is exclusively expressed in pollen, compared with much higher expression of BSK1 in the other parts of plants (Liu and Adams, 2010). As the bsk1-1 mutation affects an amino acid that is highly conserved in the Arabidopsis BSK1 protein family (see Supplemental Figure 2 online), it would be worthwhile to generate transgenic plants express those RLCKs including the same mutation as the bsk1-1 protein. Phenotyping those plants may provide new insights into the role of these proteins in innate immunity and other cellular processes.
Several RLCKs have been shown to be involved in plant innate immunity, including Arabidopsis AVRPPHB SUSCEPTIBLE1, BOTRYTIS-INDUCED KINASE1 (BIK1), and RPM1-INDUCED PROTEIN KINASE (RIPK) and tomato (Solanum lycopersicum) Pto (Innes, 2011; Feng and Zhou, 2012). For instance, BIK1, originally identified as a positive regulator of resistance to Botrytis cinerea (Veronese et al., 2006), physically associates with and transphosphorylates BAK1 and FLS2 (Lu et al., 2010). Mutation in BIK1 causes defects in FLS2-mediated responses and disease resistance to nonpathogenic bacterial infection (Lu et al., 2010). In addition, BIK1 also plays an important role in immune signaling mediated by other immune receptor kinases, including EFR and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) (Zhang et al., 2010), which recognize bacterial elongation factor EF-Tu and fungal chitin, respectively. A BIK1-related RLCK, RIPK, is involved in RESISTANCE TO P. SYRINGAE PV MACULICOLA1–mediated defense responses by physically associating with RPM1 INTERACTING PROTEIN4 (Liu et al., 2011). Intriguingly, all of these RLCKs have been shown to be targeted by pathogen effectors (Innes, 2011; Feng and Zhou, 2012), indicating that RLCKs are important modules in plant innate immunity.
The PTI mediated by FLS2 involves multiple components, including BAK1, BIK1, and RIPK, as well as BSK1. The bsk1-1 mutant displays enhanced susceptibility to multiple pathogens, so it would be reasonable to hypothesize that BSK1 may interact with other RLKs and modulate multiple defense response pathways. Consistent with this notion, BIK1, another RLCK, has been shown to associate with multiple RLKs, including FLS2, EFR, and CERK1, and modulates defense responses activated by different ligands (Lu et al., 2010; Zhang et al., 2010). However, the functions of BIK1 and BSK1 in plant innate immunity appear to be different; for instance, it was previously shown that the bik1 mutant displays enhanced disease resistance to Pto DC3000, but the bsk1 mutant is more susceptible to Pto DC3000. In addition, it is worth noting that the kinase activities of BIK1, FLS2, and BAK1 are not required for BIK1 association with FLS2 or BAK1 (Lu et al., 2010). Further study on the relationship between BSK1 and those well-characterized RLKs involved in plant immunity may shed new light on how BSK1 modulates plant defense responses.
In conclusion, we demonstrated that BSK1 plays critical roles in plant disease resistance and forms a complex with PAMP receptor FLS2. Our findings provide insights into the role of BSK1, the substrate of BR receptor BRI1, in plant innate immunity.
METHODS
Plant Growth Conditions
The edr2 bsk1-1 mutant was identified from an ethyl methanesulfonate–mutagenized edr2 population (Nie et al., 2011). Arabidopsis thaliana plants were grown in the growth room at 20 to 22°C under a 9-h-light/15-h-dark cycle for phenotyping or a 16-h-light/8-h-dark cycle for seed set, as described previously (Nie et al., 2011).
Pathogen Infections and Microscopy
Powdery mildew pathogen Golovinomyces cichoracearum strain UCSC1 was maintained on pad4-1 as described previously (Frye et al., 2001). To achieve an even distribution of conidia, inoculations were performed as described (Wang et al., 2011). To quantify fungal growth and conidiation, the number of conidiophores per colony was counted at 5 DAI as described by Consonni et al. (2006). To visualize fungal hyphae and dead cells, infected leaves were stained with trypan blue as described (Frye and Innes, 1998). H2O2 accumulation was observed by 3,3′-diamino benzidine-HCl staining, and to visualize fungal structures, the infected leaves were further stained with trypan blue (Xiao et al., 2005). Pto DC3000 infection was performed as described (Mengiste et al., 2003). Hyaloperonospora arabidopsidis (H. a.) Noco 2 infection was also performed as described previously (Li et al., 2010).
Ethylene-Induced Senescence Assay
Ethylene-induced senescence assays and measurement of the chlorophyll content were performed as previously described (Frye et al., 2001).
Map-Based Cloning and Complementation
To create the mapping population, we crossed edr2 bsk1-1 (in Col-0 background) with ecotype Landsberg erecta. The F2 plants homozygous for edr2 were identified and inoculated with G. cichoracearum. Initially, we mapped bsk1-1 to chromosome 4 between markers M4I22 and nga1107. We then developed our own molecular markers at intervals between these two markers using Monsanto Col-0 and Landsberg erecta polymorphism data (http://www.Arabidopsis.org/browse/Cereon/index.jsp). We used ∼2500 plants from F3 families that were homozygous for edr2, but segregating for powdery mildew resistance. Ultimately, bsk1-1 was mapped to a 20-kb region between markers T12J5-AflII and F23E12-PstI. All five genes in the genetic interval were amplified and sequenced directly.
To complement the edr2 bsk1-1 mutant, the genomic sequence including 1622 bp upstream of the ATG start codon and 1210 bp downstream of the stop codon of At4g35230 was cloned into binary vector pCambia1300 (http://www.cambia.org.au) for complementation analysis. The derived genomic construct was verified by sequencing and transformed into Agrobacterium tumefaciens strain GV3101. The edr2 bsk1-1 plants were transformed using the floral dip method (Clough and Bent, 1998).
Sequences of BSKs family members were identified by BLAST searches at http://www.Arabidopsis.org/Blast/index.jsp. Multiple sequence alignments were produced using ClustalW with default parameters (http://sci.cnb.uam.es/Services/MolBio/clustalw/). The multiple-alignment file was further analyzed with BoxShade online software (http://www.ch.embnet.org/software/BOX_form.html).
Construction of Double Mutants
The following T-DNA mutants were obtained from the ABRC (www.Arabidopsis.org): Salk_001600 (bsk2-3), Salk_096500C (bsk3-1) (Tang et al., 2008), Salk_063711C (bsk6-1), Salk_077982C (bsk8-1), Salk_051462 (bsk12-1), Salk_079850C (mlo2-7), Salk_030721 (bsu1-1) (Kim et al., 2009), and Salk_151344C (bsl1-2). To generate double mutants, the edr2 bsk1-1 mutant was crossed with edr1, pmr4-1, atg2-2, bak1-4, and the above T-DNA insertion mutants. All the plants mentioned above were in the ecotype Col-0 background.
BSK1-GFP Subcellular Localization
To visualize BSK1 subcellular localization, the 1.6-kb BSK1 promoter region and the full-length BSK1 coding sequence (CDS) without stop codon were amplified from Col-0 genomic DNA or cDNA. The BSK1pro:BSK1 CDS was cloned into the pEGAD vector between the PacI and AgeI multiple cloning sites to remove the 35S promoter from the vector with GFP in frame at the C terminus. Protoplasts isolated from 4-week-old Col-0 plants were transfected with the GFP constructs as described (He et al., 2007b). Twelve hours after transfection, GFP fluorescence was visualized using an Olympus FV500 confocal laser scanning microscope (Olympus) at an excitation wavelength of 488 nm and emission wavelength of 505 to 550 nm.
Quantitative Real-Time RT-PCR
Real-time quantitative PCR was performed as described previously (Wang et al., 2011). Total RNA was isolated using TRIzol reagent (Invitrogen). First-strand cDNA from 2 μg of total RNA was synthesized using M-MLV reverse transcriptase (Promega). Real-time PCR was performed with SYBR Green Premix Extaq (Takara) following the manufacturer’s instructions. The ACTIN2 gene was used as an internal control for normalizing the amount of cDNA.
SA Extraction and Measurement
SA extraction and measurement were performed as previously described (Gou et al., 2009).
Vector Construction and Site-Directed Mutagenesis
The full-length BSK1 CDS without stop codon was amplified by PCR from Col-0 cDNA or bsk1-1 mutant cDNA and inserted into the Gateway vector pDONR207 using a BP Clonase kit (Invitrogen) to create a pDONA207-BSK1 CDS or pDONA207-BSK1 CDSm entry clone. The inserts were next subcloned using an LR Clonase kit (Invitrogen) into the pEarleyGate 103 destination vector with 35S promoter and C-terminal GFP fusion (Earley et al., 2006) or into the pGWB11 vector with C-terminal FLAG fusion (Nakagawa et al., 2007). FLS2 CDS was amplified from Col-0 cDNA and subcloned into pEarleyGate 101 (C-terminal YFP-HA) through Gateway technology. BAK1 CDS was amplified from Col-0 cDNA and subcloned into the pEarleyGate 103 vector. The FLS2pro:FLS2-FLAG and BAK1pro:BAK1-FLAG plasmids were kindly provided by Jian-Min Zhou (Zhang et al., 2010).
To generate a BSK1 RNAi construct, a 418-bp fragment at the 3′-end was amplified by PCR. The PCR product was first cloned into pKANNIBAL (Wesley et al., 2001), resulting in an inverted repeat separated by an intron fragment. The derived construct was then digested with NotI and ligated to pART27 expression vector (Gleave, 1992).
To produce a kinase-dead version of BSK1, a conserved Lys residue at position 104 in the ATP binding site was substituted with a Glu using site-directed mutagenesis (Stratagene). Both the genomic DNA and CDS of BSK1 were mutated. The K104E mutated genomic DNA was cloned into the binary vector pCambia1300 for complementation tests. The wild-type and mutant CDS were digested with BamHI and SalI and inserted into the pMAL-c2G expression vector for kinase activity assays. Similarly, BSK1 (G2A) and BSK1 (S230A) were also generated using site-directed mutagenesis as described above. The mutated fragments were subcloned into the pEGAD vector to create the BSK1pro:BSK1 CDS G2A-GFP vector for GFP observation in protoplasts. In addition, BSK1pro:BSK1 CDS, BSK1pro:BSK1 CDS G2A, and BSK1pro:BSK1 CDS S230A were PCR amplified and inserted into the Gateway vector pDONR207 and then cloned into the pEarleyGate 301 destination vector with C-terminal HA fusion. The BSK1pro:BSK1 CDS was also cloned into pEarleyGate 303 destination vector with C-terminal Myc fusion.
ROS Assay
ROS assay was performed as described previously (Zhang et al., 2010). Each data point consists of at least 12 replicates.
Seedling Growth Inhibition
Arabidopsis seeds were surface sterilized, rinsed in sterile water, and grown vertically on plates containing half-strength Murashige and Skoog (MS) salts plus 0.8% agar. Five-day-old seedlings were transferred into liquid half-strength MS (two seedlings per well of 24-well plate) with or without flg22 and incubated for 10 d. The fresh weight of seedlings was measured, and the relative growth was documented by comparing with the untreated control.
In Vitro Kinase Assays
Expression of the MBP-BSK1 fusion proteins in pMAL-c2G expression vector were induced by 0.3 mM Isopropyl β-D-1-thiogalactopyranoside for 4 h at 28°C. The recombinant proteins were affinity purified using amylose resin (New England Biolabs) according to the manufacturer’s instructions. In vitro kinase assays were performed as described (Tang et al., 2008).
MAPK Activation
MAPK activation was performed as described (Schwessinger et al., 2011) with minor modifications. Fourteen-day-old seedlings in liquid half-strength MS media were treated with 100 nM flg22 for 0, 5, or 15 min. Seedlings were frozen and ground in liquid nitrogen. Proteins were extracted with MAPK extraction buffer. Samples were centrifuged at 12,000g at 4°C for 20 min. Crude proteins were quantified with a BCA protein assay kit (Thermo Scientific) using BSA as a standard. Equal amounts of proteins (200 μg) were subjected to 10% SDS-PAGE and electroblotting. Phosphorylated MAPKs were detected by incubation with anti-p42/44 MAPK antibodies (1:1000; Cell Signaling Technology) in 5% BSA in TBS-Tween overnight, followed by incubation with anti-rabbit-horseradish peroxidase secondary antibodies for 1 h.
Co-IP Assay in Nicotiana benthamiana and in Arabidopsis
Agrobacterium GV3101 carrying different plasmids were suspended in infiltration buffer as described (Liu et al., 2010) to OD600 = 0.8. For coinfiltration, equal volumes of agrobacteria carrying different constructs were mixed prior to infiltration so that the concentration of each strain was OD600 = 0.4. Five-week-old N. benthamiana leaves were used for transient expression. Two days later, leaf samples were syringe infiltrated with or without 1 μM flg22 for 10 min before freezing in liquid nitrogen. Leaves were ground with a mortar and pestle in liquid nitrogen and extracted with 2 mL of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 1% [v/v] IGEPAL CA-630 [Sigma-Aldrich], and 1× protease inhibitor cocktail [Sigma-Aldrich])/g tissue powder. The samples were left on ice with gentle shaking for 1 h to solubilize membrane proteins and centrifuged at 16,000g at 4°C for 30 min. Supernatants (1 mL) were incubated with 3 μL α-HA or α-FLAG antibody (Sigma-Aldrich) for 2 h at 4°C with gentle rotation, and then 40 μL of 50% (v/v) protein G agarose beads (Millipore) slurry was added and incubated for another 4 h. Following incubation, the beads were washed four times with PBS containing 0.1% (v/v) IGEPAL CA-630 and once with PBS. After the last centrifugation, the PBS buffer was removed completely. Eighty microliters of PBS and 20 μL 5× SDS-PAGE sample buffer were added, and the beads were boiled for 10 min. The presence of FLS2 or BSK1 protein was detected by α-HA or α-GFP (Roche) immunoblot. For Co-IP experiments in protoplasts, 1 mL of protoplasts was transfected with 200 μg plasmid DNA and incubated for 16 h before being treated with or without 1 μM flg22 for 10 min as described (He et al., 2007b). For Co-IP experiments in transgenic Arabidopsis, transgenic fls2 plants expressing 35Spro:FLS-YFP-HA were crossed with edr2 bsk1-1 transgenic plants expressing BSK1pro:BSK1-Myc. Total protein was extracted from 3-week-old F2 seedlings expressing both the FLS2-YFP-HA and BSK1-Myc proteins or from the fls2 transgenic seedlings expressing 35Spro:FLS-YFP-HA alone. Co-IP assay was performed as described above.
Microsomal and Soluble Protein Fractionation and Immunoblotting
Microsomal and soluble protein fractionations were prepared according to Heidrich et al. (2011) with minor modifications. Briefly, ∼100 mg of 14-d-old seedlings was ground in liquid nitrogen and extracted with cold Suc buffer as described (Belkhadir et al., 2012). Samples were centrifuged at 2000g at 4°C for 15 min to remove nuclei and cell debris. The supernatants were further ultracentrifuged at 100,000g for 1 h at 4°C to separate soluble and microsomal fractions. Immunoblots were performed as described (Pan et al., 2012). GAPDH and H+-ATPase were used as cytosolic marker or plasma membrane marker, detected by anti-GAPDH (Sigma-Aldrich) or anti-H+-ATPase (Agrisera), respectively.
Oligonucleotide Sequences
The primers used in this study are listed in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis BSK1 (At4g35230), FLS2 (At5g46330), EDR2 (At4g19040), EDR1 (At1g08720), PMR4 (At4g03550), MLO2 (At1g11310), BAK1 (At4g33430), BSU1 (AT1G03445), BSL1 (AT4G03080), BRI1 (At4g39400), BIK1 (At2g39660), EFR (At5g20480), PR1 (At2g14610), PR2 (AT3G57260), PAD4 (At3g52430), FMO1 (AT1G19250), SID2 (AT1G74710), and ACTIN2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. bsk1-1 Affects edr2-Mediated Changes in Gene Expression.
Supplemental Figure 2. T-DNA Insertion Mutations of BSK1 Homologs Did Not Suppress edr2-Mediated Powdery Mildew Resistance, and Mutants of BSK1 Homologs Displayed Wild-Type-Like Responses to Pto DC3000.
Supplemental Figure 3. BSK1 RNAi Lines Displayed a Similar Phenotype to bsk1-1.
Supplemental Figure 4. The bsk1-1 Mutation Suppresses edr1, pmr4, and mlo2-Mediated Powdery Mildew Resistance Phenotypes.
Supplemental Figure 5. The bsk1-1 Mutation Partially Suppresses mlo2-Mediated Senescence.
Supplemental Figure 6. The bsk1-1 Mutation Did Not Suppress atg2-Mediated Powdery Mildew Resistance and Cell Death Phenotypes.
Supplemental Figure 7. The BSK1 RNAi Plants Displayed Enhanced Susceptibility to Pto DC3000.
Supplemental Figure 8. The Phenotype of bsk1-1 in Response to H. a. Noco2.
Supplemental Figure 9. Transgenic Plants Expressed Correctly Sized Fusion Proteins.
Supplemental Figure 10. The bak1, bsu1, and bsl1 Mutations Did Not Suppress edr2-Mediated Powdery Mildew Resistance Phenotypes.
Supplemental Figure 11. The bsk1-1 Mutant Displayed a Narrow Leaf Phenotype.
Supplemental Figure 12. bsk1-1 Displayed Wild-Type-Like Responses to BL Treatment.
Supplemental Figure 13. The BSK1-Myc and FLS2-YFP-HA Proteins Are Functional.
Supplemental Figure 14. The BSK1 RNAi Plants Showed Defects in flg22-Induced ROS Burst.
Supplemental Figure 15. The bsk1-1 Plants Showed Wild-Type-Like Responses in elf18-Induced ROS Burst and PR1 Accumulation.
Supplemental Figure 16. bsk1-1 Showed Wild-Type-Like MAP Kinase Activation, Expression of PTI Marker Genes, and Seedling Growth Inhibition in Response to flg22.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC for providing T-DNA insertion lines, Yuelin Zhang for the H. a. Noco 2 strain, Jianming Li for bak1-4 and bri1-9 seeds, Jian-Min Zhou for the fls2 and efr-1 seeds and FLS2pro:FLS2-FLAG and BAK1pro:BAK1-FLAG vectors, Lu Gan for assistance with SA measurements, and Bairu Zhang for assistance with confocal microcopy. This work was supported by grants from National Basic Research Program of China (2011CB100700 and 2009CB118306), the National Transgenic Program of China (2011ZX08009-003) to D.T., the National Natural Science Foundation of China (31071067) to T.Z., and the National Science Foundation (Grants MCB-0918908 and IOS-1121425) to F.K.
AUTHOR CONTRIBUTIONS
D.T. and H.S. initiated the project, designed the experiments, and wrote the article. H.S., Q.S., Y.Q., H.Y., H.N., Y.C., and T.Z. performed research. H.S., Y.Q., F.K., and D.T. analyzed data. Y.Q. and F.K. contributed to the discussion and revised the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- LRR-RLK
Leucine-rich repeat receptor-like kinase
- BR
brassinosteroid
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- Col-0
Columbia-0
- PH
pleckstrin homology
- SA
salicylic acid
- DAI
days after infection
- H2O2
hydrogen peroxide
- RLCK
receptor-like cytoplasmic kinase
- TPR
tetratricopeptide repeat
- Pto
Pseudomonas syringae pv tomato
- GFP
green fluorescent protein
- BL
brassinolide
- BRZ
brassinazole
- Co-IP
coimmunoprecipitation
- CDS
coding sequence
- MS
Murashige and Skoog
- ANOVA
analysis of variance
References
- Adam L., Somerville S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9: 341–356 [DOI] [PubMed] [Google Scholar]
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T., Min Y.K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J.L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Blatch G.L., Lässle M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Consonni C., Humphry M.E., Hartmann H.A., Livaja M., Durner J., Westphal L., Vogel J., Lipka V., Kemmerling B., Schulze-Lefert P., Somerville S.C., Panstruga R. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Feng F., Zhou J.M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15: 469–476 [DOI] [PubMed] [Google Scholar]
- Frye C.A., Innes R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gou M., Su N., Zheng J., Huai J., Wu G., Zhao J., He J., Tang D., Yang S., Wang G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., Aderem A. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410: 1099–1103 [DOI] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., Russell S.D., Li J. (2007a). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17: 1109–1115 [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.C., Martin G.B., Kemmerling B., Nürnberger T., Sheen J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Sheen J. (2007b). The use of protoplasts to study innate immune responses. Methods Mol. Biol. 354: 1–9 [DOI] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J.E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Hirano T., Kinoshita N., Morikawa K., Yanagida M. (1990). Snap helix with knob and hole: Essential repeats in S. pombe nuclear protein nuc2+. Cell 60: 319–328 [DOI] [PubMed] [Google Scholar]
- Innes R.W. (2011). Activation of plant nod-like receptors: How indirect can it be? Cell Host Microbe 9: 87–89 [DOI] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li Y., Li S., Bi D., Cheng Y.T., Li X., Zhang Y. (2010). SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Liu S.L., Adams K.L. (2010). Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol. Biol. Evol. 27: 2817–2828 [DOI] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, C., Göllner, K., Humphry, M., Consonni, C., and Panstruga, R. (2008). The powdery mildew disease of Arabidopsis: A paradigm for the interaction between plants and biotrophic fungi. The Arabidopsis Book 6: e0115, doi/10.1199/tab.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Bittel P., Chinchilla D., Jehle A.K., Albert M., Boller T., Felix G. (2012). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nie H., Wu Y., Yao C., Tang D. (2011). Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J. Genet. Genomics 38: 137–148 [DOI] [PubMed] [Google Scholar]
- Nie H., Zhao C., Wu G., Wu Y., Chen Y., Tang D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158: 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M.T., Stein M., Hou B.H., Vogel J.P., Edwards H., Somerville S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Liu S., Tang D. (2012). HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 69: 831–843 [DOI] [PubMed] [Google Scholar]
- Qi Y., Tsuda K., Glazebrook J., Katagiri F. (2011). Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 12: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Vogel J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5: 343–348 [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Boguski M.S., Goebl M., Hieter P. (1990). A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60: 307–317 [DOI] [PubMed] [Google Scholar]
- Smith D.F. (2004). Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Cao Y., Jansen Labby K., Bittel P., Boller T., Bent A.F. (2012). Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2005). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2006). A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G.A., Jr, Okuyama H. (2000). Lipid-linked proteins of plants. Prog. Lipid Res. 39: 19–39 [DOI] [PubMed] [Google Scholar]
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R.A., Hirt H., Mengiste T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S., Schiff C., Santamaria M., Koh S., Nishimura M., Vogel J., Somerville C., Somerville S. (2007). EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Wawrzynska A., Rodibaugh N.L., Innes R.W. (2010). Synergistic activation of defense responses in Arabidopsis by simultaneous loss of the GSL5 callose synthase and the EDR1 protein kinase. Mol. Plant Microbe Interact. 23: 578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Xiao S., Calis O., Patrick E., Zhang G., Charoenwattana P., Muskett P., Parker J.E., Turner J.G. (2005). The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42: 95–110 [DOI] [PubMed] [Google Scholar]
- Yao C., Wu Y., Nie H., Tang D. (2012). RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J. 71: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.