Arabidopsis BRANCHED1 is required for branch suppression in response to shade. Transcriptional profiling of shade-treated wild-type and brc1 axillary buds revealed a group of ABA response genes and a network of cell cycle– and ribosome-related genes whose mRNA levels are dependent on BRC1 function. These genes may play a key role in the growth-to-dormancy transition in buds.
Abstract
Plants interpret a decrease in the red to far-red light ratio (R:FR) as a sign of impending shading by neighboring vegetation. This triggers a set of developmental responses known as shade avoidance syndrome. One of these responses is reduced branching through suppression of axillary bud outgrowth. The Arabidopsis thaliana gene BRANCHED1 (BRC1), expressed in axillary buds, is required for branch suppression in response to shade. Unlike wild-type plants, brc1 mutants develop several branches after a shade treatment. BRC1 transcription is positively regulated 4 h after exposure to low R:FR. Consistently, BRC1 is negatively regulated by phytochrome B. Transcriptional profiling of wild-type and brc1 buds of plants treated with simulated shade has revealed groups of genes whose mRNA levels are dependent on BRC1, among them a set of upregulated abscisic acid response genes and two networks of cell cycle– and ribosome-related downregulated genes. The downregulated genes have promoters enriched in TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) binding sites, suggesting that they could be transcriptionally regulated by TCP factors. Some of these genes respond to BRC1 in seedlings and buds, supporting their close relationship with BRC1 activity. This response may allow the rapid adaptation of plants to fluctuations in the ratio of R:FR light.
INTRODUCTION
Plants obtain their energy from light by converting photons into chemical energy through photosynthesis. To optimize this process, they have evolved mechanisms to maximize light harvesting and avoid shade from other plants. Even before they are completely shaded, plants can perceive decreases in the ratio of red to far-red light (R:FR) due to absorption of red (R) light by the photosynthetic pigments of neighboring plants. A low R:FR triggers a series of developmental responses collectively known as the shade avoidance syndrome (SAS) (Casal, 2012). These responses include upward movement of leaves and promotion of elongation of stem-like organs (including hypocotyl and petioles) at the expense of leaf expansion. This generates tall plants with erect leaves less likely to become shaded by neighbors. Long-term exposure to low R:FR also leads to early flowering, an escape mechanism that shortens generation time (Halliday et al., 1994). In addition, a common SAS response of adult plants is the suppression of shoot branching. In a wide variety of species ranging from conifers and grasses to eudicots, plants grown in low R:FR, at high density or under plant canopies, develop fewer lateral branches (Smith and Jordan, 1994; Branka Tucić, 2005; Aguilar- Kearney et al., 2007; Martínez et al., 2007; Finlayson et al., 2010).
Considerable work has been performed to understand the genetics of the SAS in Arabidopsis thaliana seedlings. A family of five photoreceptors (phytochromes) detect changes in the R:FR, of which phytochrome B (phyB) seems to be the main receptor responsible for the initial detection of these changes (Ballaré, 1999). Phytochromes act as dimers and exist in two photoconvertible forms: Pr and Pfr, with the Pr:Pfr ratio reflecting the R:FR of the environment (Quail, 2002). Upon photoconversion into active Pfr, part of the cytoplasmic phytochrome pool goes into the nucleus, where it regulates gene expression by interacting with several PHYTOCHROME INTERACTING FACTOR (PIF) and PIF3-LIKE (PIL) proteins, which belong to the basic helix-loop-helix (bHLH) transcription factors. Through their direct interaction with PIF/PIL proteins, phytochromes regulate the transcription of light-responsive G-box-containing genes (Li et al., 2011). These global transcriptional responses have been studied (Devlin et al., 2003; Salter et al., 2003; Sessa et al., 2005; Tao et al., 2008; Hornitschek et al., 2012). In addition, several hormone signaling pathways (brassinosteroid, auxin, ethylene, cytokinin [CK], and gibberellins) have been involved in the SAS in seedlings (Stamm and Kumar, 2010). By contrast, in spite of its great ecological and economic impact, little is known about the mechanisms underlying the suppression of shoot branching in adult plants in response to shade.
Candidates to play a role in this process are genes that regulate shoot branching in white (W) light, whose activity could be modulated by changes in light quality. The control of lateral shoot growth is coordinated by a conserved network of genes that regulate the synthesis and signaling of the hormones auxin, strigolactone (SL), and CK. Auxin and SL, synthesized in the shoot apex and root, respectively, prevent branching, while CK, synthesized in the root and stem, promotes branching (Domagalska and Leyser, 2011). Also, class II TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) transcription factors Teosinte branched1 (tb1)-like, in monocots, and BRANCHED1 (BRC1)-like, in dicots, act locally, inside the axillary buds, to cause growth arrest (Doebley et al., 1997; Aguilar-Martínez et al., 2007; Finlayson, 2007; Martín-Trillo et al., 2011).
Recent studies have proposed that tb1- and BRC1-like genes are indeed involved in the shade-induced response of branch suppression. In Sorghum bicolor, phyB seems to negatively regulate Sb-TB1 mRNA levels in response to light signals (Kebrom et al., 2006). In Arabidopsis, BRC1 is upregulated in axillary buds of plants grown at high density and is required for complete branch suppression in these conditions (Aguilar-Martínez et al., 2007). Moreover, it has been proposed that BRC1 and the closely related BRC2 could differentially contribute to the response of branch suppression and act through divergent pathways in Arabidopsis plants grown under constitutive shade (Finlayson et al., 2010). However, studies on the shade control of shoot branching in Arabidopsis have analyzed, so far, plants grown from seed germination through to flowering under low R:FR light. These long-term treatments not only affect axillary bud development (initiated in adult plants) but also seedling development and flowering time, which in turn affects rosette leaf and axillary bud number. Moreover, shade induces photosynthetic acclimation (or long-term response; Walters, 2005; Dietzel and Pfannschmidt, 2008) involving changes in gene expression and metabolism (Roig-Villanova et al., 2007). All these effects complicate the comparison with plants constitutively grown in W. To compensate for variations in flowering time, standardization procedures have been devised (Finlayson et al., 2010), but the contribution of other factors to the branching phenotypes is still unclear.
In this work, we investigated the SAS of newly formed axillary buds in adult plants grown in W and transiently exposed to low R:FR. This approach not only minimized phenotypic differences between plants before the treatment, but also allowed the study of the rapid response of buds to transient changes in light quality, a phenomenon so far completely unknown. We observed that, in response to a treatment of low R:FR after flowering, axillary buds become arrested. BRC1 seems to play an important role in this response, while BRC2 may play a partially redundant one. The global transcriptomic response of wild-type axillary buds is strongly dependent on BRC1, and the gene categories affected support a central role for BRC1 in causing axillary bud arrest. A comparison of genes responding to shade-triggered bud arrest and decapitation-triggered bud activation helped us identify a list of bud dormancy and bud activation genes tightly associated with bud status irrespective of the stimulus involved. This may help us understand the genetic mechanisms controlling the reversible transition of growth to dormancy in Arabidopsis axillary buds.
RESULTS
BRC1 Plays a Role in Branch Suppression during the SAS
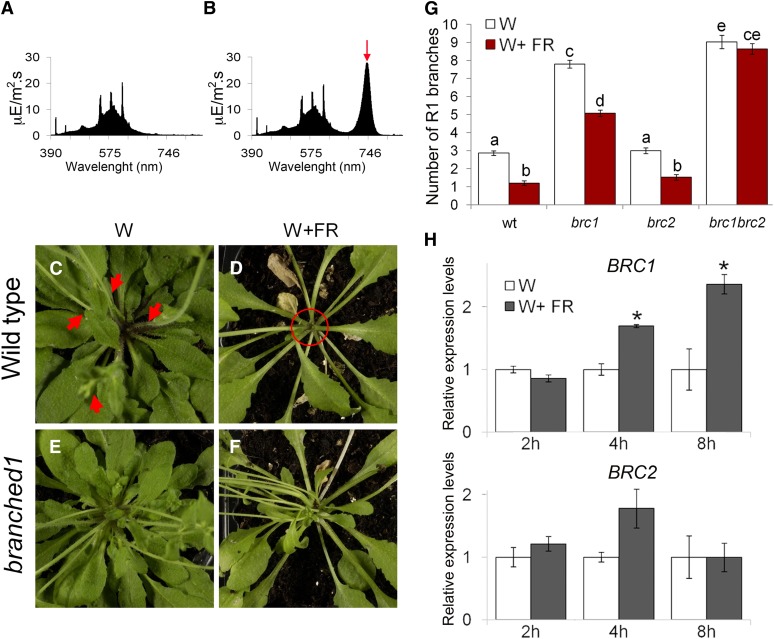
To investigate whether BRC1 and BRC2 have a relevant role during SAS, we grew wild-type plants in long days under W (R:FR = 11.7; Figure 1A) until flowering, when axillary meristems begin to initiate in long days (Hempel and Feldman, 1994; Grbić and Bleecker, 2000; Long and Barton, 2000). We transferred half of the plants to a growth chamber with identical PAR but where W was supplemented with far-red light (FR), simulating a canopy shade (W+FR, R:FR = 0.2; Figure 1B). We started the W+FR treatment after flowering to avoid early flowering of plants treated with W+FR, which would reduce rosette leaf and axillary bud number relative to W-treated plants. Two weeks later, we counted primary rosette branches (RI) and found that wild-type plants grown in W+FR had 3 times fewer branches than plants grown in W (Figures 1C, 1D, and 1G), indicating that exposure of plants with young buds to a low R:FR ratio promotes bud arrest in Arabidopsis. By contrast, the brc1-2 mutants were partially insensitive to this condition: W+FR-treated brc1-2 plants still had 5 times more branches than W+FR-treated wild-type plants. Moreover, their response to low R:FR was reduced compared with the wild-type response: brc1-2 plants grown in W+FR had only 1.5 times fewer branches than plants grown in W (Figures 1E to 1G). Other SA responses, such as hyponasty and stem and petiole elongation, were indistinguishable between the wild-type and mutant plants (Figures 1D and 1F). The branching patterns of brc2-1 mutants were like those of wild-type plants both in W and W+FR (Figure 1G). However, the phenotype of the double mutants was insensitive to the W+FR treatment, revealing a certain degree of genetic redundancy between BRC1 and BRC2 in these conditions.
Figure 1.
Effect of a Simulated Shade on Lateral Shoot Elongation in Arabidopsis Wild-Type, brc1, and brc2 Mutant Plants and Response of BRC1 and BRC2 to W+FR.
(A) W spectrum used to grow plants presented in (C), (E), and (G).
(B) W+FR spectrum used to grow plants in (D), (F), and (G).
(C) and (D) Close-up of wild-type Arabidopsis rosettes grown continuously in W (C) or in W until flowering and then for 2 weeks in W+FR (D). Red arrows indicate lateral shoots. In W+FR, axillary buds are arrested (red circle).
(E) and (F) Close-up of brc1 rosettes grown in W (E) or W+FR (F). Buds of brc1 mutants are unresponsive to W+FR, although other shade responses (stem and petiole elongation) are normal.
(G) Branching phenotypes of the wild type (wt) and brc1 and brc2 mutants grown in W or W+FR for 3 weeks after flowering (n = 37 to 40).
(H) and Transcript levels of BRC1 (H) and BRC2 analyzed by qPCR, in buds of W+FR-treated plants, relative to levels in W-treated plants.
Error bars are se of three biological replicates. Different letters in (G) denote significant differences (one-way ANOVA, P < 0.05) among means. Asterisks in (H) are significant differences (Student’s t test, P < 0.05) between control and treated plants.
[See online article for color version of this figure.]
Transcriptional Response of BRC1 and BRC2 to Changes in Light Quality
We then studied the short-term transcriptional response of BRC1 and BRC2 to the simulated shade treatment that triggered bud arrest in wild-type plants (W+FR; Figure 1B). For that, we grew wild-type plants in W until bolting (when rosette leaves had small vegetative buds; Aguilar-Martínez et al., 2007) and then transferred half of them to W+FR. We compared the mRNA levels of buds treated for 2, 4, and 8 h with W or W+FR and observed that, after 4 h, BRC1 mRNA levels were higher in W+FR-treated plants (Figure 1H). These results indicated that BRC1 expression was rapidly promoted in low R:FR. By contrast, BRC2 did not show significantly different transcription levels in these conditions (Figure 1H).
As many genes responding to changes in light quality have daily oscillating mRNA levels (Finlayson et al., 1998; Yamashino et al., 2003) and some SAS responses are coupled to the circadian clock (Salter et al., 2003; Alabadí and Blázquez, 2009; Sellaro et al., 2012), we investigated whether BRC1 and BRC2 were regulated in a circadian-dependent manner. We grew wild-type plants in a 12-h-day/12-h-night photoperiod for 3 weeks and then transferred them to constant light for 2 d, during which we studied BRC1 and BRC2 transcript levels (see Supplemental Figure 1 online). We confirmed that BRC1 and BRC2 expression levels changed during the day: they were highest in the afternoon, around 4 h after the peak of CCA1 expression (Mizoguchi et al., 2002), and lowest at the beginning of the night. This suggested that, like for other TCP factors (Pruneda-Paz et al., 2009; Giraud et al., 2010), BRC1 and BRC2 expression had a daily oscillation and that a circadian control of these genes could be gating the response of bud arrest. These oscillations were therefore carefully taken into account for all the expression experiments performed in this work.
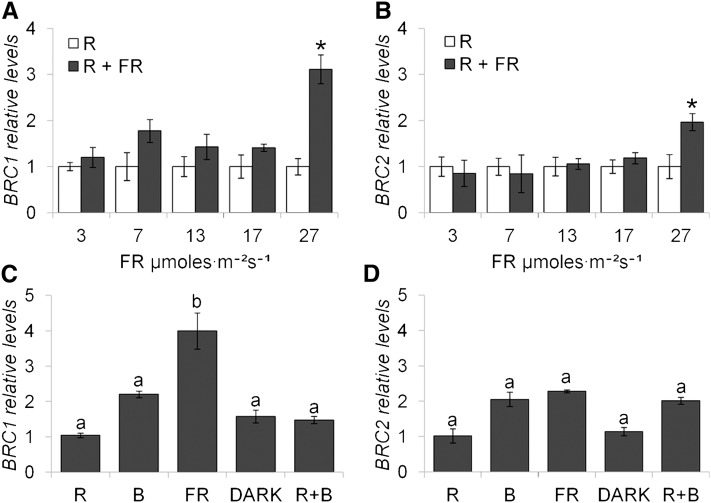
We then studied the sensitivity of BRC1 and BRC2 to changes in the R:FR ratio in more detail. We treated bolting plants with constant amounts of R light alone or supplemented with increasing amounts of FR (R:FR = 9.8 to 1.2) for 8 h. In the R:FR = 1.2 treatment, BRC1 expression was upregulated 3 times relative to the expression in pure R-treated plants (Figure 2A) but did not respond to other R+FR treatments. BRC2 responded mildly to the R:FR = 1.2 treatment (Figure 2B). To compare the sensitivity of these genes to different light wavelengths, we studied their transcriptional response to monochromatic light by treating bolting plants with pure R, blue (B), R+B, FR light, or darkness for 8 h. BRC1 mRNA levels were 2 to 4 times higher in FR-treated plants than in plants treated with any other type of light or with darkness (Figure 2C). By contrast, BRC2 responses to R, B, and FR light or darkness were not significantly different (Figure 2D).
Figure 2.
Response of BRC1 and BRC2 to Changes in Light Quality.
BRC1 (A) and BRC2 (B) mRNA levels after 8 h of exposure to R and increasing amounts of FR relative to levels in plants treated with pure R light. BRC1 (C) and BRC2 (D) mRNA levels after 8 h exposure to pure R, B, FR, R+B, or darkness relative to levels in R treatment. Error bars are se of three biological replicates. Asterisks are significant differences (Student’s t test, P < 0.05) between control and treated plants. Letters denote significant differences (one-way ANOVA, P < 0.05) among means.
In summary, BRC1 is required for bud arrest in response to low R:FR. Moreover, BRC1 expression is upregulated early in response to increasing amounts of FR, even when R:FR > 1 and strongly responds to pure FR. On the other hand, BRC2 expression does not change after short treatments of simulated shade and shows only a mild response to changes in the R:FR ratio when R:FR > 1. Moreover, brc2 mutants and brc1 brc2 mutant phenotypes suggest that BRC2 could play a role partially redundant with BRC1 in this response. Based on these results that support a more relevant role of BRC1 in the shade-induced response of bud suppression, we focused on this gene for subsequent studies.
phyB Mediates BRC1 Upregulation in Low R:FR
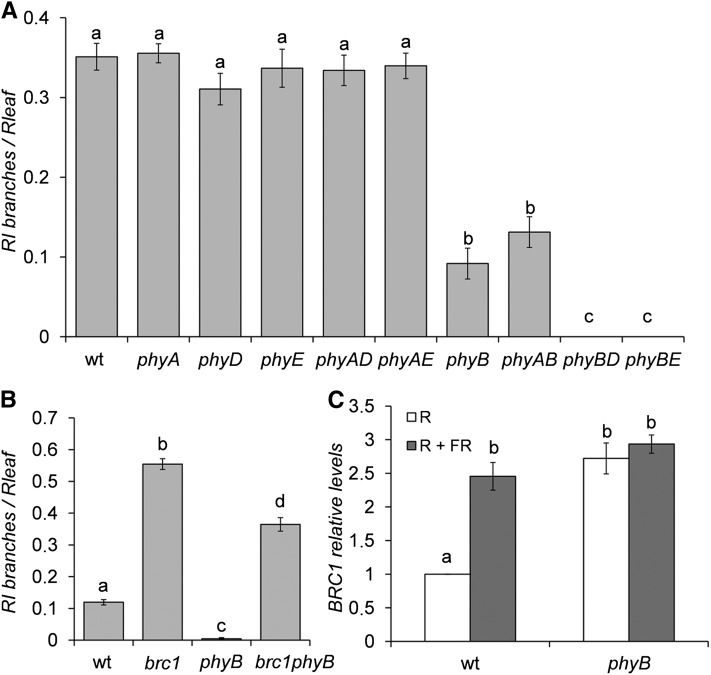
Of the five Arabidopsis phytochromes, phyB has been reported to play the most relevant role in the SAS regulation (Franklin and Whitelam, 2005). In R, phyB suppresses SAS. In low R:FR, phyB is inactivated and SAS is triggered. In agreement, phyB mutants display a constitutive SAS phenotype of stem and petiole elongation and early flowering. To study whether phyB or the other phytochrome (phy) mutants had increased branch suppression, we compared their branching patterns with those of wild-type plants in W. To reduce a potential bias due to variations in rosette leaf number (phy mutants are early flowering), we grew plants at 19°C, a condition that attenuates this early flowering phenotype. In addition, we divided RI number by the number of rosette leaves in each genotype to normalize the measurements. phyB mutants had fewer branches than wild-type Landsberg erecta (Ler) plants. Moreover, all the combinations of phyB with the other phy mutants also had fewer branches than wild-type plants (Figure 3A). In addition, phyB phyD and phyB phyE had an enhanced branch suppression phenotype compared with phyB single mutants, indicating some partial redundancy with phyB. To test whether BRC1 was required for branch suppression in phyB mutants, we generated phyB-9 brc1-2 double mutants in the Columbia background. We found that brc1-2 completely suppressed the phyB phenotype of reduced branching (Figure 3B), while it did not suppress other phyB phenotypes, such as the long hypocotyl of seedlings (see Supplemental Figure 2 online). Then, we measured BRC1 mRNA abundance in phyB-9 mutants treated with either R or R+FR (R:FR = 9.8 and 1.1, respectively) and found that, in R, phyB mutants had more BRC1 transcripts than wild-type plants. In R+FR, phyB and wild-type plants had similarly high mRNA levels comparable to those of phyB mutants in R (Figure 3C).
Figure 3.
Role of Phytochromes in Branch Suppression during Shade Avoidance.
(A) Shoot branching phenotype of phytochrome mutants. Number of RI relative to the number of rosette leaves is represented (n = 20 to 22). phyAD, phyA phyD; phyAE, phyA phyE; phyAB, phyA phyB; phyBD, phyB phyD; phyBE, phyB phyE. wt, the wild type.
(B) Shoot branching phenotype of wild-type, phyB, brc1, and phyB brc1 mutant plants (n = 72 to 76).
(C) BRC1 mRNA levels in axillary buds of wild-type and phyB mutant plants after 8 h of exposure to R or R+FR, analyzed by qPCR. Levels are relative to those of R-treated wild-type plants. Error bars are se of three biological replicates. Different letters denote significant differences (one-way ANOVA, P < 0.05) among means.
These results suggest that phyB negatively regulates BRC1 mRNA levels in R and that variations in R:FR during the SAS could affect BRC1 expression through the phyB pathway.
Genome-Wide Transcriptional Profiles of Axillary Buds after a Low R:FR Treatment
To gain further insight into the role of BRC1 in bud suppression in low R:FR, we analyzed the global expression profiles of buds of wild-type and brc1 mutant plants exposed either to W or to W+FR for 8 h, long before phenotypic differences are detectable between W- and W+FR-treated buds. We obtained mRNAs from axillary buds and hybridized sextuplicate arrays representing over 26,000 annotated genes and microRNAs of Arabidopsis.
First, we confirmed, in the wild type, the BRC1 upregulation after the W+FR treatment (1.76-fold increase, FDRLiMMA = 0.04 [FDR, false discovery rate]) and the lack of significant response of BRC2 (FDRLiMMA = 0.3) (see Supplemental Data Set 1 online). We also confirmed that in brc1 plants, BRC1 mRNA was undetectable. Second, we observed that in wild-type and brc1 samples, genes responding to low R:FR treatments in seedlings (ATHB-2, PIL1, PIL2, HFR1, and IAA29; Carabelli et al., 1993; Salter et al., 2003; Sessa et al., 2005) were upregulated, indicating that the W+FR treatment had been effective (see Supplemental Data Set 1 online). Third, we confirmed that marker genes for axillary bud dormancy, such as DORMANCY-ASSOCIATED PROTEIN (DRM1) and DORMANCY/AUXIN-ASSOCIATED PROTEIN (Stafstrom et al., 1998; Tatematsu et al., 2005), were upregulated in wild-type but not in brc1 plants (see Supplemental Data Set 1 online), indicating that buds were becoming dormant in wild-type but not in brc1 plants in response to low R:FR.
We selected a robust group of differentially expressed genes with significant changes of expression (FDRLiMMA < 0.05; PvalLiMMA < 0.0005) for further analysis. In the wild type, 362 genes changed after the W+FR treatment, of which 262 were upregulated and 100 downregulated. In brc1 mutants, 94 genes were altered after the treatment, of which 73 were upregulated and 21 downregulated (see Supplemental Data Set 1 and Supplemental Figures 3A and 3B online). These changes were confirmed by quantitative real-time PCR (qPCR) for six upregulated and five downregulated genes (see Supplemental Figures 4A and 4B online). The qPCR and microarray data showed a very high average Pearson correlation coefficient (0.93 and 0.89 for the wild type and brc1 mutants, respectively; see Supplemental Figures 4C and 4D online), confirming the high reliability of the array data. However, the array results strongly underestimated the magnitude of expression level differences, emphasizing the relevance of changes found significant in our microarray analysis.
We first analyzed the global response of wild-type and brc1 mutant plants using the Gene Ontology (GO) Classification Superviewer (University of Toronto; http://bar.utoronto.ca). In the wild type, several gene categories were overrepresented among genes responding to W+FR. Among the upregulated genes, overrepresented Biological Processes were Development, Signal transduction, Transcription, Response to abiotic and biotic stimulus, and Stress (see Supplemental Figure 5A online). Among the downregulated genes were Development and Cell organization and biogenesis (see Supplemental Figure 5C online). In brc1 mutants, the categories Development (for up- and downregulated genes) and Cell organization and biogenesis (for downregulated genes) were no longer overrepresented (arrows in Supplemental Figures 5B and 5D online). We hypothesized that genes whose expression changed in the wild type but not in brc1 mutants could directly or indirectly depend on BRC1 function and termed them BRC1-dependent genes (see Supplemental Figures 3A and 3B and Supplemental Data Set 1 online). They represented 84% of the genes responding in the wild type, confirming the relevance of BRC1 function for the axillary bud response to low R:FR. Conversely, genes responding to low R:FR both in the wild type and brc1 mutants could be related to a general SAS independent of BRC1. We focused on the list of BRC1-dependent genes for further analysis.
Upregulated BRC1-Dependent Genes: Hormone Signaling
A more detailed characterization of the BRC1-dependent genes using MapMan (Thimm et al., 2004) indicated that upregulated genes belonged to the categories Cell wall remodeling, Lipid metabolism, Transcription (including homeodomain, bHLH, and AP2/EREB-like, MYB, bZIP, and CO-like protein coding genes), Development, Amino acid and Protein degradation (RING and F-box proteins), Mitochondrial electron transport, Drought and Salt stress, Metabolite transport, and Hormone metabolism and response (see Supplemental Data Set 1 online). However, FatiGO analyses (Al-Shahrour et al., 2004) indicated that the most significantly enriched terms were those related to hormone responses (see Supplemental Data Set 1 online).
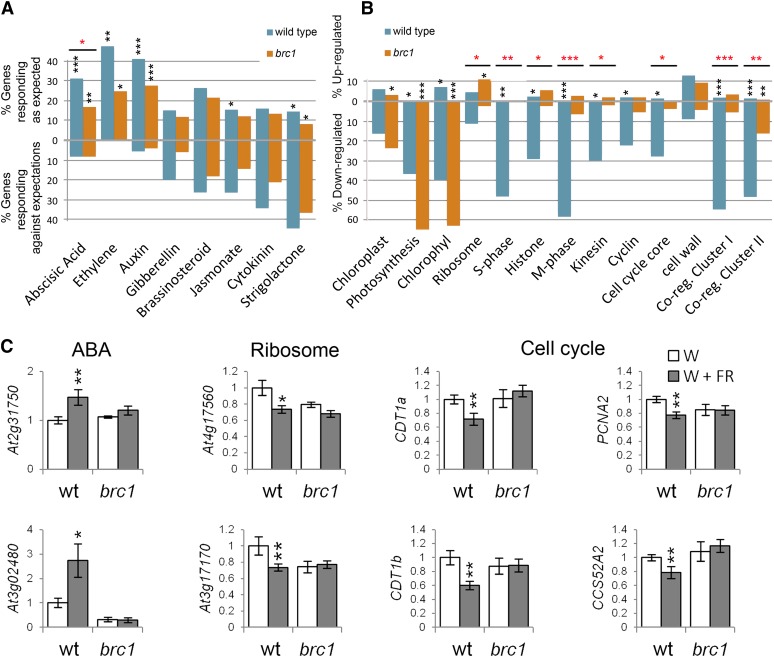
As hormone signaling plays a key role during the SAS and in the control of bud outgrowth, we investigated further the significance of this overrepresentation. We examined in our arrays the expression of a large group of genes defined as hormone specific markers by Nemhauser et al. (2006) (see Supplemental Data Set 2 online) and tested whether they behaved as if a particular hormone pathway was active. The high number of genes analyzed (between 57 and 777 genes per category) allowed us to test the statistical significance of their global response using a test of proportions (Wilson, 1927). In wild-type buds, a significant proportion of abscisic acid (ABA), auxin, and ethylene marker genes behaved like in tissues with high levels of these hormones (Figure 4A; see Supplemental Data Set 3 online). In brc1 mutants, the ABA pathway showed a significantly reduced response (P value = 1.49⋅10−5) relative to the wild-type response (Figure 4A; see Supplemental Data Set 3 online). We analyzed by qPCR the expression levels of two ABA markers and confirmed that they were significantly upregulated in the wild type but not in brc1 mutants (Figure 4C). In wild-type plants, we also detected a slightly reduced jasmonic acid (JA) signaling, in agreement with the low JA sensitivity observed in Arabidopsis plants grown in low R:FR (Cerrudo et al., 2012). Although a list of SL-specific marker genes is not available, we studied the response of the genes described by Mashiguchi et al. (2009) as genes responding to SL in seedlings (see Supplemental Data Set 2 online). Their expression did not support a strong SL signaling in buds (Figure 4A; see Supplemental Data Set 3 online).
Figure 4.
Transcriptomic Response of Wild-Type and brc1 Mutant Axillary Buds to Low R:FR.
(A) Global response of hormone marker genes in wild-type and brc1 buds treated with W+FR. Top, percentage of hormone marker genes responding as expected after a hormone treatment (Nemhauser et al., 2006; Mashiguchi et al., 2009). Bottom, percentage of hormone marker genes responding opposite to expected after hormone treatment.
(B) Global responses of gene categories. Black asterisks indicate statistically significant differences with the expected distribution in nontreated tissue (A) and statistically significant differences with the response of a random gene collection (B). Red asterisks indicate statistically significant differences between wild-type and brc1 responses ([A] and [B]). *P < 5⋅10−3; **P < 5⋅10−6, and ***P < 5⋅10−9. NABA = 777, NEth = 57, NAuxin = 259, NGA = 121, NBr = 61, NJA = 749, NCk = 76, NSL = 69, NChpl = 68, NPh = 41, NChl = 43, NRib = 413, NS-ph = 50, NHis = 93, NM-ph = 79, NKin = 57, NCyc = 113, NCell-cyc-core = 83, Ncell-wall = 656, NCR-ClusterI = 187, and NCR-ClusterII = 168.
(C) Relative mRNA levels of representative genes of the categories ABA, ribosome, and cell cycle analyzed by qPCR. Error bars are se. Asterisks represent significant differences (Student’s t test, *P < 0.1 and **P < 0.05) between W- and W+FR-treated plants. wt, the wild type.
[See online article for color version of this figure.]
These results showed that, after a shade treatment, buds exhibit a general increase in auxin, ethylene, and ABA signaling and a reduction in JA signaling. BRC1 could play a role in the maintenance of ABA signaling.
Overrepresented Motifs in the Promoter of BRC1-Dependent Upregulated Genes
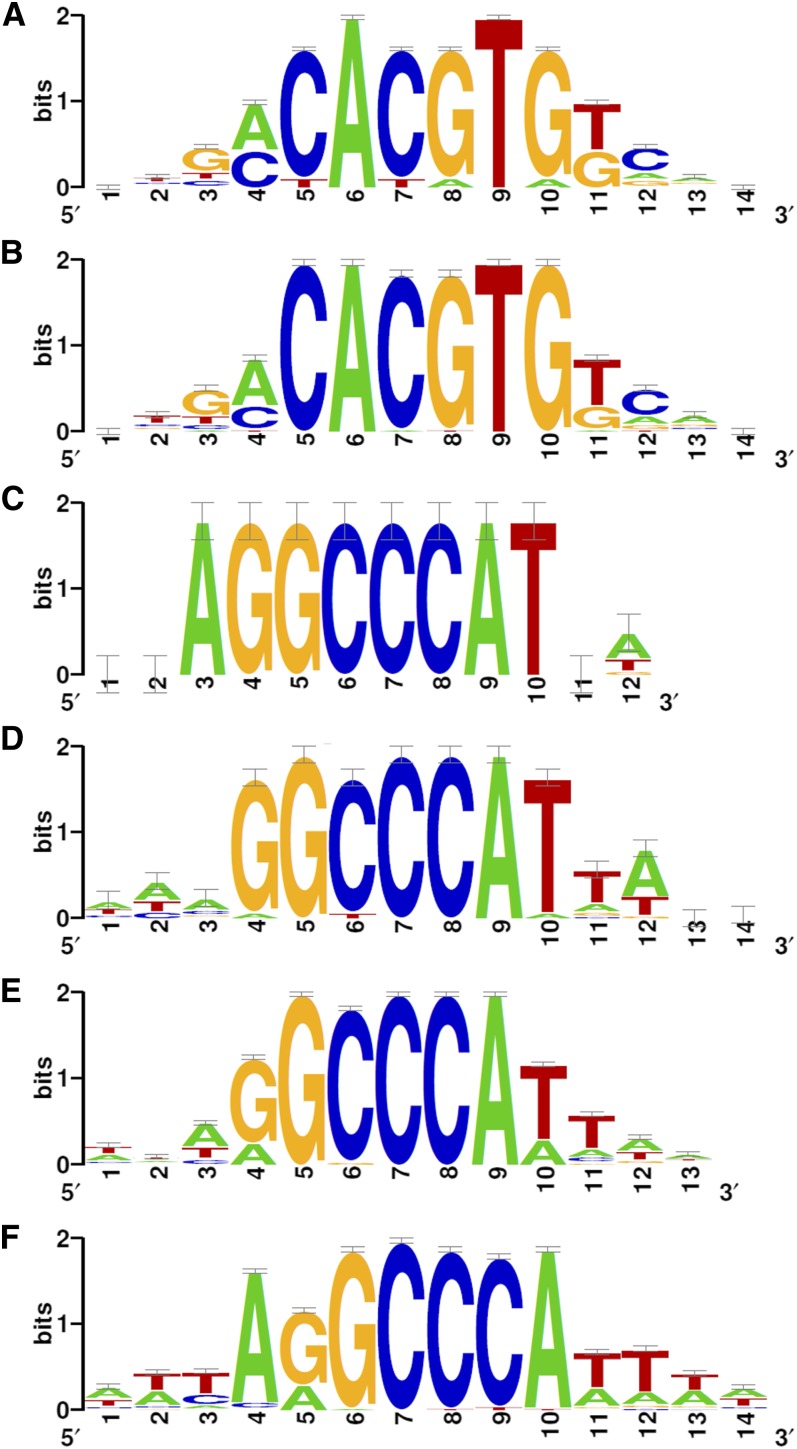
To identify putative transcriptional regulatory elements associated with the upregulation of BRC1-dependent genes, we searched for enriched 6-bp motifs within the 500-bp upstream region of their predicted transcription start sites using Motif discovery (van Helden et al., 1998). We found nine overrepresented motifs, the most frequent of which was CACGTG, found 3 times more often than expected at random (see Supplemental Data Set 4 online). The nine motifs were assembled into frequency matrices and rendered the consensus motif aCACGTGt (Figure 5A). This motif contained the ABA-responsive element (ACGT) associated with ABA and drought responses (Simpson et al., 2003), the E-box core element (CANNTG; Toledo-Ortiz et al., 2003), and the G-box (CACGTG) recognized by bHLH and bZIP proteins and overrepresented in gene promoters responding to stress and light (Menkens et al., 1995; Chattopadhyay et al., 1998; Martínez-García et al., 2000). These results are consistent with our finding that ABA signaling is predominant in axillary buds after a W+FR treatment.
Figure 5.
Consensus Sequences of Overrepresented Motifs in Promoters of BRC1-Dependent Genes.
Logo representing the frequency matrix of the consensus motif in BRC1-dependent upregulated genes (A), bud dormancy genes (B), BRC1-dependent downregulated genes (C), bud activation genes (D), and genes coregulated with clusters I (E) and II (F).
[See online article for color version of this figure.]
Downregulated BRC1-Dependent Genes: Protein Synthesis and Cell Division
MapMan analysis of BRC1-dependent downregulated genes indicated that these genes belonged to categories related to chloroplast function and chlorophyll synthesis (PORA, PORB, CHLM, and Mg2+ chelatases), amino acid and protein synthesis (ribosomal proteins), chromatin structure (HISTONE H4, HISTONE 2B, and HISTONE B9), and cell cycle division and organization (CYCLIN-A3;2, KINESIN-12B, and FtsH) (see Supplemental Data Set 1 online). To test whether our filtered gene list reflected a more global trend, we studied the general behavior of genes containing annotations related to these categories (see Supplemental Data Set 2 online) as described above. Chlorophyll- and photosynthesis-related genes were strongly downregulated both in wild-type and brc1 samples (Figure 4B; see Supplemental Data Set 3 online). By contrast, genes involved in the cell cycle and cell division were strongly downregulated in the wild type but had a significantly reduced response in brc1 mutants (Figure 4B; see Supplemental Data Set 3 online). Finally, genes containing ribosome-related annotations were slightly downregulated in the wild type but were significantly upregulated in brc1 mutants. We analyzed by qPCR the expression of four genes involved in the control of the cell cycle and two ribosomal genes and confirmed that they were significantly downregulated in the wild type but not in brc1 mutants (Figure 4C).
Among the BRC1-dependent downregulated genes, three overlapping 6-bp motifs were found to be overrepresented (see Supplemental Data Set 4 online). These motifs were assembled into frequency matrices and rendered the consensus motif AGGCCCAT (Figure 5C; see Supplemental Data Set 4 online). This motif resembled the Up1 motif, which is overrepresented in genes upregulated in axillary buds after decapitation (GGCCCAWW; Tatematsu et al., 2005), the binding site of class I TCP proteins (GGNCCCAC; Kosugi and Ohashi, 2002), the site IIa of the rice (Oryza sativa) PCNA gene promoter bound by TCP factors PCF1 and PCF2 (AGGTGGGCCCGT; Kosugi and Ohashi, 1997), and the GCCCR motif recognized by AtTCP20 (Li et al., 2005).
These results indicate that after a low R:FR treatment, axillary buds undergo a general transcriptional repression of genes involved in photosynthesis, cell division, and protein synthesis. While the control of photosynthesis-related genes seems mostly independent of BRC1, the expression of a large number of ribosomal genes and genes involved in cell cycle and cell division is strongly influenced by BRC1 activity. Moreover, the enrichment of their promoters in TCP motifs could indicate that their transcription is regulated by TCP transcription factors.
A Group of BRC1-Dependent Genes Is Closely Related to Bud Activity
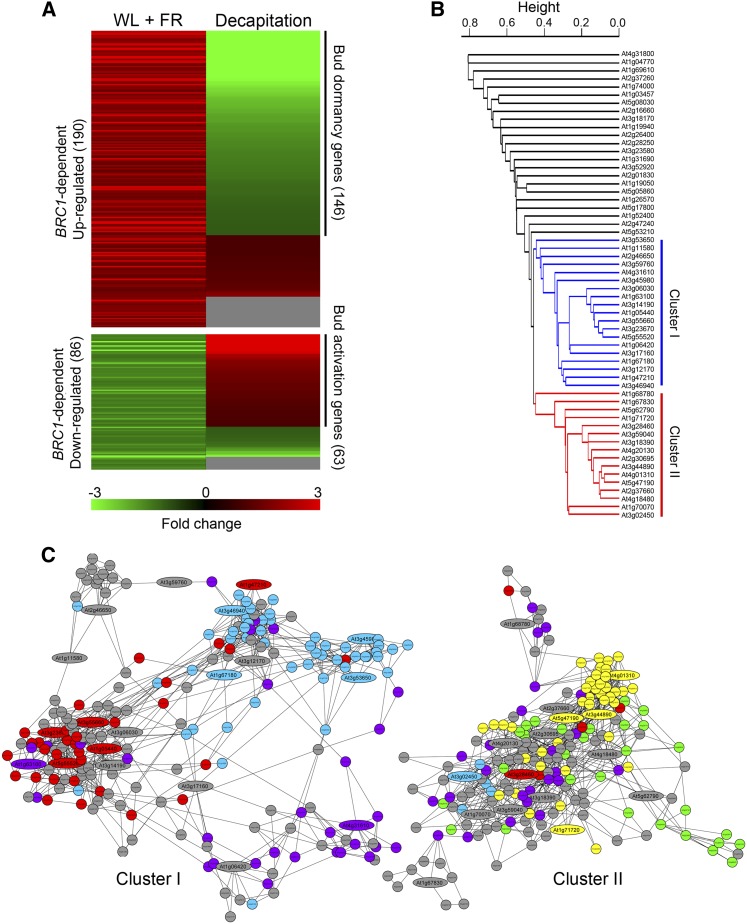
A total of 276 of the 306 BRC1-dependent genes were represented in the Affymetrix chips (190 upregulated and 86 downregulated genes); therefore, their response in other experiments could be analyzed using Genevestigator (Zimmermann et al., 2004). The behavior of these genes showed an excellent negative correlation with the transcriptional profiling of dormant versus active axillary buds (from either intact or decapitated plants, respectively) performed by Tatematsu et al. (2005). A significant proportion of the BRC1-dependent genes had opposite responses in low R:FR and after decapitation, conditions causing bud arrest and bud activation, respectively: 77% of genes upregulated in shade (146) were downregulated after decapitation, and 73% of genes downregulated in shade (63) were upregulated after decapitation (Figure 6A; see Supplemental Figures 3C and 3D and Supplemental Data Set 1 online). During decapitation-induced bud activation, BRC1 is rapidly downregulated (Aguilar-Martínez et al., 2007). Therefore, these 209 BRC1-dependent genes were closely associated with axillary bud activity and BRC1 function regardless of the stimulus involved (either light quality or decapitation) and could play a central role in the dormancy-to-growth transition in buds. We termed them bud dormancy and bud activation genes, respectively.
Figure 6.
Bud Dormancy and Bud Activation Genes.
(A) Heat map illustrating the expression profiling of BRC1-dependent genes in axillary buds after a W+FR treatment and after decapitation (Tatematsu et al., 2005). Green, downregulation; red, upregulation; gray, no data.
(B) Hierarchical clustering representation of bud activation genes based on degree of coregulation. Cluster I, highlighted in blue, corresponds to genes highly coregulated with S-phase and cytokinesis genes (blue and red circles, respectively, in [C]). Cluster II, highlighted in red, corresponds to genes coregulated with ribosomal genes (yellow circles in [C]).
(C) Network of genes most coregulated with clusters I and II. Bud activation genes are indicated as ellipses with Arabidopsis Genome Initiative numbers.
The 146 bud dormancy genes (Figure 6A; see Supplemental Figure 3C and Supplemental Data Set 1 online) could be directly involved in the promotion of axillary bud arrest. Consistently, this group included MAX2, known to play a key role in this process (Stirnberg et al., 2002), and DRM1 and DORMANCY/AUXIN-ASSOCIATED PROTEIN, genes whose expression is tightly correlated with bud dormancy (Tatematsu et al., 2005). It also included a significant number of genes related to ABA, ethylene, auxin, and gibberellin signaling (ABF3, GFB3, NTR2.1, EBF2, WES1, and GID1C), transcription (ATHB1, ATHB21, ATHB40, and ATHB53), and protein degradation (RHA1B, EBF2, and MAX2). A search for overrepresented 6-bp motifs in their gene promoters confirmed an enrichment in CACGTG motifs (Figure 5B; see Supplemental Data Set 4 online), which could indicate that ABA signaling plays an important role in the transition to bud dormancy independently of the stimulus involved. On the other hand, the 63 bud activation genes (Figure 6A; see Supplemental Figure 3D and Supplemental Data Set 1 online) could be involved in promoting axillary bud growth. This group included many genes classified as unknown and others with largely unrelated annotations, making it unclear whether they played a coordinated role in promoting bud activation. As their gene promoters exhibited a significant enrichment (occ_E = 2.8⋅10−4) in the consensus TCP binding site GGCCCAT (Figure 5D; see Supplemental Data Set 4 online), we analyzed this gene list further to study their putative connection with BRC1.
Global Analysis of Bud Activation Genes
To know more about the function of the bud activation genes, we investigated whether they belonged to larger coregulation networks related to known biological functions. For this, we studied their degree of coregulation in other transcriptomic experiments using Hcluster and NetworkDrawer (Obayashi et al., 2007). We found two groups of 19 and 16 bud activation genes (Clusters I and II, respectively) strongly coregulated in 1388 microarray experiments (Figure 6B; see Supplemental Data Set 1 online). Genes of Cluster I (which included genes related to cell cycle and cell division, [i.e., CYCA3;2, HISTONE H2B, HISTONE B9, HISTONE H4, and KINESIN12B]) were coregulated with a larger network of genes whose GO annotations were enriched in the terms DNA replication, DNA repair, chromatin structure, cell cycle, and cytokinesis (Figure 6C; see Supplemental Data Sets 1 and 2 online). Cluster II (which comprised ribosomal protein-encoding genes (i.e., RPL5, RPL9, and RPL19) was coregulated with a network of genes mainly involved in ribosomal assembly, chloroplast protein synthesis, and photosynthesis (Figure 6C; see Supplemental Data Sets 1 and 2 online). Seventy-nine percent of the genes in this network were predicted to be targeted to chloroplasts according to TargetP (Emanuelsson et al., 2007) or WoLF PSORT (Horton et al., 2007; see Supplemental Data Set 1 online). By contrast, the proportion of putative chloroplast genes in Cluster I network was 22% (see Supplemental Data Set 1 online).
We tested whether genes in these two networks were downregulated (although perhaps below our stringent significance threshold, FDR < 0.05) in our experiment. We found that 54 and 48% of the genes coregulated with Clusters I and II, respectively, were also downregulated after the low R:FR treatment. This response was drastically reduced in brc1 mutants (Figure 4B; see Supplemental Data Sets 2 and 3 online). Motif analyses revealed a significant enrichment of the sequence GGCCCA in their promoters (Figures 5E and 5F; see Supplemental Data Set 4 online).
In summary, two gene networks coregulated with a group of bud activation genes and likely involved in the control of cell division (in particular, S-phase and M-phase) and protein synthesis, respectively, could play an important role during the reversible bud growth-to-dormancy transition. These networks could be globally regulated at the transcriptional level by TCP factors and their expression is strongly influenced by the presence of BRC1.
Overexpression of BRC1 Causes Downregulation of Some BRC1-Dependent Genes
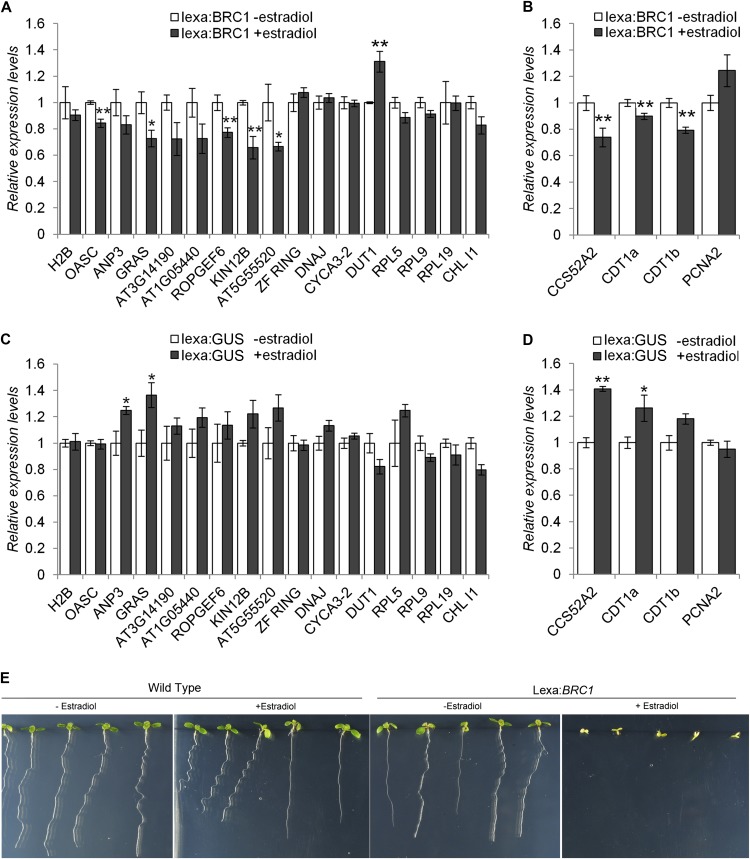
To further test the relationships between BRC1 and bud activation genes, in particular genes from Clusters I and II, we generated BRC1 estradiol-inducible lines and studied their response after BRC1 induction in seedlings. We analyzed the mRNA levels of several bud activation genes 7 h after estradiol application, when BRC1 was highly transcribed (see Supplemental Figure 6A online). In spite of the slight upregulation caused by the estradiol treatment alone (control lines, Figures 7C and 7D), lines expressing BRC1 showed downregulation of nine out of 13 genes of cluster I, and the downregulation was highly significant for five of them (Figure 7A). By contrast, the four ribosome genes tested from cluster II did not respond to BRC1 in seedlings. We also studied the expression of four additional cell cycle genes (PCNA2, CCS52A2, CDT1a, and CDT1b) that changed significantly in our experiment (Figure 4C). Three of them responded negatively to BRC1, including CDT1a and CDT1b, which were previously described as being negatively regulated by class II At-TCP24 (Figures 7B and 7D; Masuda et al., 2008). PCNA2, a potential class I TCP target (Kosugi and Ohashi, 1997; Li et al., 2005), did not respond to BRC1. We tested the phenotype of plants expressing BRC1 after estradiol induction and confirmed that the treatment caused a fast and generalized growth arrest in the root and shoot apical meristems and in leaf primordia (Figure 7E).
Figure 7.
BRC1 Negatively Regulates Bud Activation Genes and Cell Cycle Genes in Seedlings.
(A) and (C) mRNA levels, quantified by qPCR, of bud activation genes in 7-d-old seedlings carrying an estradiol-inducible construct lexa:BRC1 (A) or lexa:GUS (C), 7 h after the beginning of treatment (mock or 10 μM estradiol).
(B) and (D) mRNA levels of selected cell division genes using the same mRNA samples as in (A) and (C), respectively. Error bars are se. Asterisks are significant differences between mock and estradiol-treated plants (Student’s t test, *P < 0.1 and **P < 0.05).
(E) Phenotype of 7-d-old wild-type and lexa:BRC1 seedlings treated with either mock or 10 μM estradiol after germination.
[See online article for color version of this figure.]
Then, we used this line to test the response to BRC1 in axillary buds. We grew plants in vitro until bolting and then added 20 μM estradiol to the medium. Seven hours later, we studied the mRNA levels of bud activation genes. Most genes (except seven) were strongly downregulated in the control lines and could not be analyzed (see Supplemental Figure 7A online). However, the seven remaining genes were downregulated after BRC1 induction (see Supplemental Figure 7B online). Five of these genes had not shown a response to BRC1 in seedlings (H2A, ZF-RING, DNAJ, DUT1, and CYCA3;2).
Altogether, these results indicate that at least some of the bud activation genes could be responding to BRC1, regardless of the developmental stage and light conditions, and that a key role of BRC1 during bud arrest would be to trigger a moderate but generalized downregulation of genes related to cell replication and cytokinesis.
DISCUSSION
In most plant species, one of the responses of the SAS of adult plants is the suppression of axillary bud outgrowth. However, little is known about the genetic mechanisms acting in buds to cause growth arrest in low R:FR. We studied this response in adult plants grown in W and exposed transiently to a shade simulated with W+FR. This allowed us to minimize phenotypic differences between treated and nontreated plants prior to axillary bud formation and to identify early responses of buds. We confirmed that exposure to low R:FR after flowering causes branch suppression and that BRC1 is required to prevent bud outgrowth in these conditions. We monitored the transcriptional changes taking place in axillary buds and found that BRC1 (but not BRC2) mRNA accumulated shortly after exposure to shade. These results are in contrast with those of Finlayson et al. (2010), who reported that BRC1 expression was unaffected in plants constitutively grown in low R:FR, while BRC2 expression was upregulated in these conditions. These contrasting results could indicate that different genetic pathways act in response to either long-term or transient treatments of low R:FR.
Wild-type axillary buds exposed to low R:FR display, as early as 8 h after the start of treatment, a general upregulation of genes related to auxin, ethylene, and ABA signaling and a global downregulation of genes related to the cell cycle, protein synthesis, and photosynthesis. The enhanced ethylene and auxin signaling and the reduced expression of photosynthesis-related genes is observed both in wild-type and brc1 treated buds, suggesting that it is mostly independent of BRC1. Enhanced ethylene and auxin production has already been described as a response to low R:FR (Finlayson et al., 1998; Hornitschek et al., 2012), and auxin (1-naphthaleneacetic acid, NAA) has been shown to have a negative effect on photosynthetic efficiency and photosystem II activity in Malus (Zhu et al., 2011). The potential raise in auxin signaling could be partly due to the upregulation of YUCCA3 and YUCCA8, encoding flavin monooxygenases that catalyze a rate-limiting step of the main IAA biosynthesis pathway (Mashiguchi et al., 2011). BRC1-independent downregulation of photosynthesis-related genes could cause a drop in sugar levels, a signal associated with bud arrest (Kebrom et al., 2012). This low sugar signaling could be a BRC1-independent mechanism to suppress branching under certain stress conditions.
We mainly focused on the genetic responses that may be directly or indirectly regulated by BRC1. Some of the most prominent BRC1-dependent gene categories are, among the upregulated genes, ABA-related genes and, among the downregulated genes, cell cycle and protein synthesis genes.
ABA Signaling and the Promotion of Bud Dormancy
We detected an increase in the response of ABA marker genes in wild-type buds after a shade treatment. ABA has been classically related to the promotion and maintenance of bud dormancy in many plant species (Phaseolus vulgaris, Lupinus angustifolius, potato [Solanum tuberosum], and Populus): a correlation has been found between ABA content in axillary buds and bud dormancy (Gocal et al., 1991; Emery et al., 1998; Destefano-Beltrán et al., 2006; Ruttink et al., 2007) and, in pea (Pisum sativum) and Populus, many ABA-responsive genes are upregulated in dormant buds (Shimizu-Sato and Mori, 2001; Ruttink et al., 2007). So far, no ABA measurements have been reported in Arabidopsis axillary buds, and only a few ABA-inducible genes have been studied (Finlayson et al., 2010). However, our transcriptomic results support a correlation between ABA levels and bud arrest in Arabidopsis: Wild-type axillary buds (which become dormant after a shade treatment) display a significant increase in the global response of ABA-related genes, while brc1 buds (which continue to grow) show reduced ABA-related responses. The precise role of ABA in the promotion of bud dormancy in Arabidopsis is still unclear. It has been proposed that ABA induces the expression of ICK1, an inhibitor of CDKs at the G1/S-phase transition (Wang et al., 1998), but we have not detected expression changes of this gene in our treated samples. Direct ABA applications support a role of ABA in bud suppression (Chatfield et al., 2000), but the genetic evidence is contradictory: Arabidopsis sextuple mutants for six ABA receptors (PYR1, PYL1, PYL2, PYL4, PYL5, and PYL8), which display dramatically impaired ABA-dependent gene expression (Gonzalez-Guzman et al., 2012), do not have an excess of branching (see Supplemental Figure 8 online) and some ABA-deficient mutants of Arabidopsis have reduced shoot growth (LeNoble et al., 2004). These phenotypes could be explained by developmental plasticity and phenotypic adaptation of the mutants to low levels of ABA or reduced ABA signaling. Therefore, experiments involving transient ABA treatments may be more informative to investigate this question. Nevertheless, the observation that ABA-related responses are significantly reduced (but not abolished) in brc1 mutants could indicate that BRC1 is required for the maintenance of this pathway, for instance, by promoting the transcription of ABA responsive regulators, such as ABF3 and ABI5, BRC1-dependent genes that contain TCP binding sites in their promoters. On the other hand, a correlation has been found in several plant species between increased auxin levels and a raise of ABA in axillary buds (Eliasson, 1975; Tucker, 1976; Knox and Wareing, 1984; Begonia and Aldrich, 1990). Therefore, the detected increment in auxin signaling could also be participating in the promotion of this response.
Bud Dormancy and Downregulation of Cell Cycle– and Ribosome-Related Genes
A group of BRC1-dependent genes, not only downregulated in low R:FR but also upregulated in buds after decapitation, are coexpressed with a network of S-phase- and cytokinesis-related genes. This supports the view that, during the growth-to-dormancy transition in buds, cells become quiescent at checkpoints before S- and M-phase (Devitt and Stafstrom, 1995; Shimizu and Mori, 1998; Kebrom et al., 2010). Genes in these coregulation networks have promoters with significant enrichment in TCP binding sites, indicating that they could be regulated, at least in part, by TCP factors. Interestingly, the TCP gene At-TCP4 has been reported to inhibit cell division in budding yeast by blocking G1→S transition (Aggarwal et al., 2011). Some other TCP factors have been proposed to regulate the expression of individual cell cycle genes: At-TCP24 negatively regulates the expression of CDT1a and CDT1b, prereplication control factor genes required for licensing cells into the S-phase (DePamphilis, 2003; Masuda et al., 2008). Class I TCP factors (At-TCP20, TCP15, and PCFs) promote the expression and/or bind in vivo to the promoter of G1/S-phase transition genes (E2FB, CDT1A, and PCNA1; Kosugi and Ohashi, 1997; Trémousaygue et al., 2003; Li et al., 2012), endoreduplication (FZR2, RBR, and CYCA2;3; Li et al., 2012), and G2/M genes (WEE1 and CYCB1;1; Li et al., 2005). Our global profiling analysis suggests a more general regulation of this gene network by TCPs, indicating that these factors may play a key role in regulating cell cycle progression. The strong dependence of this network on BRC1 activity (brc1 mutants have a dramatically reduced response to low R:FR) and the observation that the transcription of several of these genes is downregulated by the sole induction of BRC1 in seedlings or buds indicates that BRC1 could be negatively controlling the expression of some S-phase and cytokinesis genes either directly or by competing with TCP transcriptional activators. Experiments to test the binding of BRC1 to their gene promoters will help us test this proposal.
A second group of BRC1-dependent downregulated genes is coregulated with many ribosome-related genes, in agreement with the observation that dormant buds in pea have low mRNA levels of genes encoding the ribosomal proteins RIBOSOMAL PROTEIN L27 (RPL27) and RPL34 (Stafstrom and Sussex, 1992). Genes in this network also have promoters with a significant enrichment in TCP binding sites, consistent with previous in silico analyses showing that the ribosome-related gene promoters in Arabidopsis have an overrepresentation of TCP binding sites (Trémousaygue et al., 2003; Li et al., 2005). Moreover, At-TCP20 has been shown to bind the promoter of ribosomal genes RIBOSOMAL PROTEIN S27 and RIBOSOMAL PROTEIN L24 (Li et al., 2005). Our results further support that TCP genes are closely involved in the general transcriptional regulation of ribosomal genes in Arabidopsis. However, we have not been able to find a clear response of these genes to BRC1 induction, suggesting that their relation with BRC1 may be mediated by other factors, for instance, other TCP proteins.
Models of Bud Growth Regulation and the Role of BRC1
Little is known about the genetic mechanisms acting in Arabidopsis during the transition from active-to-dormant buds and their relationship with BRC1 activity. Several models have been proposed to explain the control of axillary bud growth, involving systemic signals and local control of gene expression (Beveridge and Kyozuka, 2010; Domagalska and Leyser, 2011; Müller and Leyser, 2011). One model relates bud dormancy to basipetal polar auxin transport in the main stem. Bud growth would require that axillary buds (auxin sources) establish their own polar auxin transport stream to export auxin into the main stem. Dampening of auxin transport in the main stem (caused by SLs) could enhance the competition between buds for their common auxin sink in the stem, leading to bud arrest (Prusinkiewicz et al., 2009). In this context, the increased auxin levels detected in wild-type buds could partly reflect reduced auxin export. Either increased auxin levels or reduced auxin export could be responsible for BRC1 upregulation. An auxin-mediated regulation of BRC1 expression would be in agreement with our observation that BRC1 mRNA levels drop as early as 1 h after decapitation (Aguilar-Martínez et al., 2007).
A second model suggests that auxin and SL signaling cause reduced CK levels in buds by downregulating CK synthesis and/or upregulating CK degradation (Nordström et al., 2004; Dun et al., 2012). Reduced CK levels would directly cause bud arrest. In addition, SL and CK could also act antagonistically by promoting or repressing BRC1-like gene expression, respectively (Mashiguchi et al., 2009; Braun et al., 2012; Dun et al., 2012). In our experiment, neither SL-responsive genes nor CK-related global responses seemed dramatically affected in wild-type buds. This may suggest that, inside the buds, changes in SL and CK signaling are not significant during the promotion of bud dormancy in response to low R:FR. A second, more trivial, explanation could be that SL and CK gene lists (obtained from experiments performed in seedlings; Nemhauser et al., 2006; Mashiguchi et al., 2009) do not accurately reflect the response to these hormones in axillary buds.
BRC1 is an excellent candidate to act as second messenger and trigger or maintain bud dormancy. We identified a collection of genes closely associated both with BRC1 function and bud activity, irrespective of whether the stimulus is apical dominance or changes in light quality. In particular, BRC1 activity is linked to the negative regulation of cell cycle and ribosome gene expression and to the promotion of ABA signaling, all of which are responses accompanying bud dormancy in a wide range of species. Moreover, when ubiquitously expressed in seedlings, BRC1 causes SAM and leaf primordia growth arrest, effects observed in axillary buds expressing BRC1. This supports the view that BRC1 is a local promoter of bud dormancy and probably a target for systemic and local signaling regulating branch outgrowth (Aguilar-Martínez et al., 2007). It is possible that the interplay between auxin canalization dynamics along with the antagonistic relation between CK (which promotes meristem activity and may negatively regulate BRC1 in some conditions; Braun et al., 2012; Dun et al., 2012) and BRC1 could act together to determine the growth status of the buds.
In summary, bud arrest triggered by shade, a response of major adaptive importance for plants, may share local regulators with other stimuli that regulate branching, such as apical dominance. One of these local regulators, BRC1, could promote bud dormancy by a combined negative action on cell proliferation and protein synthesis and by promoting ABA signaling. Understanding these mechanisms that control bud arrest may have a significant impact on crop yield in high-density plantings common in agriculture.
METHODS
Plant Material and Growth Conditions
The wild-type Arabidopsis thaliana used in this study were of the Columbia or Ler ecotypes, unless otherwise indicated. The brc1-2 and brc2-1 mutants were described by Aguilar-Martínez et al. (2007). The phy single and double mutants (Ler background) were provided by G.C. Whitelam (University of Leicester, UK). The phyB-9 mutant (Columbia background) was provided by J.A. Jarillo (Instituto Nacional de Investigaciones Agrarias, Madrid, Spain). The sextuple ABA receptor mutant (aba6) was provided by P.L. Rodriguez (Instituto de Biología Molecular y Celular de Plantas., Valencia, Spain). Wild-type and mutant seeds were sown on commercial soil and vermiculite in a 3:1 proportion. They were stratified in darkness at 4°C for 3 d and grown in a 16/8-h photoperiod at 22°C in W light (PAR, 100 μmol m−2 s−1; R:FR ratio = 11.7) or W supplemented with FR (W+FR, PAR, 100 μmol m−2 s−1, R:FR ratio = 0.2), unless otherwise specified.
Light Sources
W was provided by tubes of white light-emitting diodes (LEDs) T8-160CM-DW (CCT 4000-4500; Prosolda) or cool-white 20-W F20T12/CW tubes (Phillips). Supplemental FR light was provided by lamp tubes carrying FR 735-nm LEDs (L735-03AU; Epitex). Monochromatic light treatment experiments were performed in light-insulated growth chambers with rows of Phillips GreenPower monochromatic LED modules (HF deep-red 660 nm, FR 730 nm, and B 455 nm). All light measurements were monitored with a JAZ spectroradiometer (Ocean Optics).
Phenotypic Analysis
Wild-type and brc1 and brc2 mutant plants were either maintained in W (see above) or transferred to W+FR (see above) after flowering. Two weeks after the main inflorescence became visible, branches (shoots longer than 0.5 cm) were counted. phy (Ler) plants were grown at 19°C to attenuate the early flowering phenotype. phyB and phyB brc1 (Columbia) mutants were grown at 22°C in PAR = 60 μmol m−2 s−1. The number of RI branches/number of rosette leaves was counted.
Expression Analyses
For Figures 1H and 2, plants were grown in W as described above until the bolts were <1 cm long. At this stage, rosette leaves had small vegetative buds of 150 to 400 μm (Aguilar-Martínez et al., 2007). Plants were then kept in W or transferred to W+FR (Figure 1H) or LED light chambers (Figure 2). Samples were collected at 2, 4, and 8 h after the beginning of the treatment (Figure 1H) or 8 h after the beginning of the treatment (Figure 2). Plants in Figures 2A and 2B were exposed to constant amounts of R light (32 μmol m−2 s−1) alone or supplemented with 3, 7, 13, 17, and 27 μmol m−2 s−1 of FR light (R:FR ratio = 9.8, 4.8, 2.4, 1.9, and 1.2, respectively). Plants in Figures 2C and 2D were treated with pure R (32 μmol m−2 s−1), B (15 μmol m−2 s−1), R+B (32+15 μmol m−2 s−1), or FR (5 μmol m−2 s−1) light or darkness for 8 h. Plants for the microarray hybridization were grown in W or W+FR for 8 h as described for Figure 1H. Plants of the experiment in Figure 3C were grown at 22°C and PAR = 60 μmol m−2 s−1 as described for the phenotypic analysis of phyB mutants and then were transferred to LED chambers with either R or R+FR light for 8 h. For circadian expression analyses, wild-type plants were grown in a 12/12-h photoperiod at 22°C in W until the bolts were <1 cm long and then were transferred to constant W. Twenty-four hours later, axillary bud material was collected every 4 h for 4 d. For the BRC1 estradiol induction experiment (Figure 7E), seeds were sown in Murashige and Skoog, 0.7% agar, and 1% Suc and stratified in darkness at 4°C for 3 d. Seven days after germination and growth of the seedlings in 22°C, W and 16/8 photoperiod, 2 mL of 10 µM estradiol prepared from a 20 mM stock (in DMSO) or 0.0005% (v/v) DMSO (mock) were added to each plate. Seedlings were collected 7 h later. For BRC1 induction in buds, plants were grown in Murashige and Skoog until bolting, and then 3 mL of 20 µM estradiol or mock was added to the medium and axillary buds were collected 7 h later.
RNA Preparation and qPCR Analyses
Rosettes leaves with their petioles, main inflorescence, and roots were carefully dissected out, leaving a tissue highly enriched in axillary buds and subtending tissue. Material was harvested from at least eight individuals, and a minimum of three biological replicates per genotype/treatment and was stored in N2(l). For the estradiol induction experiment in seedlings, 20 seedlings were collected per biological replicate. RNA was isolated with the RNeasy plant mini kit (Qiagen). Possible traces of DNA were eliminated with RNase-Free DNase set (Qiagen). Three micrograms of RNA was used to make cDNA with the High-Capacity cDNA archive kit (Applied Biosystems). The qPCR reactions were performed with Power SYBR Green (Applied Biosystems) and the Applied Biosystems 7300 real-time PCR system, according to the manufacturer’s instructions. Three technical replicates were done for each biological replicate. For Figure 4, the qPCR reactions were performed as described by Aguilar-Martínez et al. (2007). Pairs of primers used are described in Supplemental Data Set 5 online. Cycle threshold values were obtained with the 7300 Systems SDS Software Version 1.3 (Applied Biosystems). Three reference genes were used: SAND, PP2A, and PPR (Czechowski et al., 2005). Relative fold expression changes were calculated using qBASE software (Hellemans et al., 2007).
BRC1-Inducible Construct
The coding sequence of BRC1 was cloned in the Gateway vector pMDC7 (Curtis and Grossniklaus, 2003) using an LR reaction according to the manufacturer’s instructions (Invitrogen). Transgenic plants (Columbia) were generated by agroinfiltration using the floral dip method (Clough and Bent, 1998). T3 homozygous lines generated from T1 individuals carrying a single insertion of the transgene were analyzed.
Hybridization and Analysis of Agilent Genome Arrays
Six independent biological samples per genotype and treatment (wild-type W, wild-type W+FR, brc1 W, and brc1 W+FR) were used to hybridize Agilent genome arrays. RNA amplification, labeling, and hybridization were performed basically as described by Adie et al. (2007).
Microarray slides were composed of synthetic 70-mer oligonucleotides from the Operon Arabidopsis Genome Oligo Set Version 3.0 (Qiagen) spotted on aminosilane-coated slides (Telechem) by the University of Arizona. Slides were rehydrated and UV cross-linked according to the supplier’s website: http://ag.arizona.edu/microarray/methods.html. Images from Cy3 and Hyper5 channels were equilibrated and captured with a GenePix 4000B (Axon) and spots quantified using GenPix software (Axon). Background correction and normalization of expression data were performed using LIMMA (Smyth and Speed, 2003; Smyth, 2004). LIMMA is part of Bioconductor, an R language project (Ihaka and Gentleman, 1996). First, the data set was filtered based on the spot quality. A strategy of adaptive background correction was used that avoids exaggerated variability of log ratios for low-intensity spots. For local background correction, the “normexp” method in LIMMA to adjust the local median background was used. The resulting log ratios were print-tip loess normalized for each array (Smyth and Speed, 2003). To have similar distribution across arrays and achieve consistency among arrays, log ratio values were scaled using as scale estimator the median absolute value (Smyth and Speed, 2003). Linear model methods were used for determining differentially expressed genes. Each probe was tested for changes in expression over replicates using an empirical Bayes moderated t-statistic (Smyth, 2004). To control the false discovery rate, P values were corrected using the method of Benjamini and Hochberg (1995). The expected false discovery rate was controlled to be <5 or 10% where specified.
Statistical Tests of Expression Analyses
To test the statistical significance of results, Student’s t test and one-way analysis of variance (ANOVA) with Tukey test for significance were used. Asterisks denote significant differences in Student’s t tests. Different letters denote significant differences in Tukey’s tests.
Statistical Analysis of Response in Gene Lists
Statistical analyses of the response of gene categories were performed by a Wilson approximation to the hypothesis test of equality of two proportions defining binomial distributions (Wilson, 1927) (R function: prop.test) described in Supplemental Methods 1 online.
Promoter Motif Analysis
Five hundred base pair sequences 5′ of the ATG of BRC1-dependent genes were retrieved with Sequence Bulk Download (The Arabidopsis Information Resource, www.Arabidopsis.org). Identification of overrepresented hexamer motifs was performed with Motif discovery (RSAT, www.rsat.ulb.ac.be; van Helden et al., 1998). Oligo analysis was used to find significantly overrepresented motifs. These motifs were assembled into frequency matrices with Pattern assembly and default parameters. Matrices were converted into consensus motifs with Convert-matrix and represented using WebLogo (Crooks et al., 2004).
Global Gene Expression Comparisons
Expression data of the BRC1-dependent genes in the decapitation experiment (Tatematsu et al., 2005) was retrieved when available (276/307 genes) with Genevestigator (www.genevestigator.com). Hierarchical clustering of bud activation genes based on degree of coregulation was performed with Hcluster (ATTED-II; Obayashi et al., 2007). The 187 genes most coregulated with cluster I and 168 coregulated with cluster II were obtained with NetworkDrawer (ATTED-II; Obayashi et al., 2007). Gene network data from NetworkDrawer was represented with Cytoscape (Shannon et al., 2003). Statistical Analysis of GO annotation enrichment was performed using FatiGO (Al-Shahrour et al., 2004).
Accession Numbers
Array data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE27273.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Circadian Expression of BRC1 and BRC2.
Supplemental Figure 2. Seedling Phenotype of brc1, phyB, and phyB brc1 Plants.
Supplemental Figure 3. Venn Diagrams Showing the Distribution of Genes Whose Expression Changes Significantly (FDR < 0.05) in Our Experiment.
Supplemental Figure 4. Validation of Microarray Data by qPCR.
Supplemental Figure 5. Functional Classification of Genes Responding to W+FR.
Supplemental Figure 6. BRC1 mRNA Levels in Estradiol-Inducible Lines.
Supplemental Figure 7. BRC1 Negatively Regulates Bud Activation Genes and Cell Cycle Genes in Axillary Buds.
Supplemental Figure 8. Branching Phenotype of Sextuple ABA Receptor Mutants.
Supplemental Data Set 1. Filtered Microarray Results and Analyses.
Supplemental Data Set 2. Genes Lists Used in This Work.
Supplemental Data Set 3. Global Gene Response Analyses of W+FR Experiment.
Supplemental Data Set 4. Promoter Motif Overrepresentation Analysis.
Supplemental Data Set 5. Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Jaime Martínez, Crisanto Gutierrez, Florian Chevalier, Michael Nicolas, and Desmond Bradley for helpful comments on the article, Garry C. Whitelam, José Antonio Jarillo, and Pedro Rodríguez for seed stocks, and Crisanto. Gutierrez for cell cycle gene primers. The BRC1-inducible line was generated in the TRANSPLANTA consortium. This work was supported by the Ministerio de Educación y Ciencia (BIO2008-00581 and CSD2007-00057) and the Ministerio de Ciencia y Tecnología (BIO2011-25687). E.G.-G. was a predoctoral fellow of Fundación Ramón Areces.
AUTHOR CONTRIBUTIONS
E.G.-G. and C.P.-C. performed research and analyzed data. C.O.S.S. contributed new computational tools and analyzed data. P.C. designed the research, analyzed data, and wrote the article.
Glossary
- R:FR
ratio of red to far-red light
- R
red
- SAS
shade avoidance syndrome
- bHLH
basic helix-loop-helix
- W
white
- SL
strigolactone
- CK
cytokinin
- RI
primary rosette branches
- FR
far-red
- B
blue
- Ler
Landsberg erecta
- FDRLiMMA
false discovery rate
- qPCR
quantitative real-time PCR
- GO
Gene Ontology
- ABA
abscisic acid
- JA
jasmonic acid
- LED
light-emitting diode
- ANOVA
analysis of variance
References
- Adie B.A.T., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal P., Padmanabhan B., Bhat A., Sarvepalli K., Sadhale P.P., Nath U. (2011). The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1→S transition. Biochem. Biophys. Res. Commun. 410: 276–281 [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Blázquez M.A. (2009). Molecular interactions between light and hormone signaling to control plant growth. Plant Mol. Biol. 69: 409–417 [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F., Díaz-Uriarte R., Dopazo J. (2004). FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (1999). Keeping up with the neighbours: Phytochrome sensing and other signalling mechanisms. Trends Plant Sci. 4: 97–102 [DOI] [PubMed] [Google Scholar]
- Begonia G.B., Aldrich R.J. (1990). Changes in endogenous growth regulator levels and branching responses of soybean to light quality altered by velvetleaf (Abutilon theophrasti Medik.). Biotronics 19: 7–18 [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B 57: 289–300 [Google Scholar]
- Beveridge C.A., Kyozuka J. (2010). New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Branka Tucić J.D.D.P. (2005). Phenotypic responses to early signals of neighbour proximity in Picea omorika, a pioneer conifer tree. Basic Appl. Ecol. 7: 443–454 [Google Scholar]
- Braun N., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M., Sessa G., Baima S., Morelli G., Ruberti I. (1993). The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J. 4: 469–479 [DOI] [PubMed] [Google Scholar]
- Casal J.J. (2012). Shade avoidance. The Arabidopsis Book 10: e0157, doi/10.1199/tab.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I., Keller M.M., Cargnel M.D., Demkura P.V., de Wit M., Patitucci M.S., Pierik R., Pieterse C.M., Ballaré C.L. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield S.P., Stirnberg P., Forde B.G., Leyser O. (2000). The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 24: 159–169 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Puente P., Deng X.W., Wei N. (1998). Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 15: 69–77 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L. (2003). The ‘ORC cycle’: A novel pathway for regulating eukaryotic DNA replication. Gene 310: 1–15 [DOI] [PubMed] [Google Scholar]
- Destefano-Beltrán L., Knauber D., Huckle L., Suttle J. (2006). Chemically forced dormancy termination mimics natural dormancy progression in potato tuber meristems by reducing ABA content and modifying expression of genes involved in regulating ABA synthesis and metabolism. J. Exp. Bot. 57: 2879–2886 [DOI] [PubMed] [Google Scholar]
- Devitt M.L., Stafstrom J.P. (1995). Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol. Biol. 29: 255–265 [DOI] [PubMed] [Google Scholar]
- Devlin P.F., Yanovsky M.J., Kay S.A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel L., Pfannschmidt T. (2008). Photosynthetic acclimation to light gradients in plant stands comes out of shade. Plant Signal. Behav. 3: 1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. (2012). Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson L. (1975). Effect of indoleacetic acid on the abscisic acid level in stem tissue. Physiol. Plant. 34: 117–120 [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Emery R.J.N., Longnecker N.E., Atkins C.A. (1998). Branch development in Lupinus angustifolius L. II. Relationship with endogenous ABA, IAA and cytokinins in axillary and main stem buds. J. Exp. Bot. 49: 555–562 [Google Scholar]
- Finlayson S.A. (2007). Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson S.A., Krishnareddy S.R., Kebrom T.H., Casal J.J. (2010). Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S.A., Lee I.-J., Morgan P.W. (1998). Phytochrome B and the regulation of circadian ethylene production in Sorghum. Plant Physiol. 116: 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Whitelam G.C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. (Lond.) 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C.P., Van Aken O., Millar A.H., Murcha M., Whelan J. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal G.F., Pharis R.P., Yeung E.C., Pearce D. (1991). Changes after decapitation in concentrations of indole-3-acetic acid and abscisic acid in the larger axillary bud of Phaseolus vulgaris L. cv tender green. Plant Physiol. 95: 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V., Bleecker A.B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21: 215–223 [DOI] [PubMed] [Google Scholar]
- Halliday K.J., Koornneef M., Whitelam G.C. (1994). Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 104: 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel F.D., Feldman L.J. (1994). Bidirectional inflorescence development in Arabidopsis thaliana - Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35 (Web Server issue): W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Statis. 5: 299–314 [Google Scholar]
- Kearney D., James R., Montagu K., Smith R.G. (2007). The effect of initial planting density on branching characteristics of Eucalyptus pilularis and E. grandis. Australian Forestry Journal 70: 7 [Google Scholar]
- Kebrom T.H., Brutnell T.P., Finlayson S.A. (2010). Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33: 48–58 [DOI] [PubMed] [Google Scholar]
- Kebrom T.H., Burson B.L., Finlayson S.A. (2006). Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T.H., Chandler P.M., Swain S.M., King R.W., Richards R.A., Spielmeyer W. (2012). Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 160: 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.P., Wareing P.F. (1984). Apical dominance in Phaseolus vulgaris L. J. Exp. Bot. 35: 239–244 [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis-elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- LeNoble M.E., Spollen W.G., Sharp R.E. (2004). Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55: 237–245 [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.Y., Li B., Dong A.W. (2012). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Long J., Barton M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218: 341–353 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E.G., Serra F., Marcel F., Rodríguez-Buey M.L., Schmitz G., Theres K., Bendahmane A., Dopazo H., Cubas P. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67: 701–714 [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Sasaki E., Shimada Y., Nagae M., Ueno K., Nakano T., Yoneyama K., Suzuki Y., Asami T. (2009). Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73: 2460–2465 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H.P., Cabral L.M., De Veylder L., Tanurdzic M., de Almeida Engler J., Geelen D., Inzé D., Martienssen R.A., Ferreira P.C.G., Hemerly A.S. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens A.E., Schindler U., Cashmore A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20: 506–510 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Müller D., Leyser O. (2011). Auxin, cytokinin and the control of shoot branching. Ann. Bot. (Lond.) 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nordström A., Tarkowski P., Tarkowska D., Norbaek R., Astot C., Dolezal K., Sandberg G. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 (Database issue): D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P., Crawford S., Smith R.S., Ljung K., Bennett T., Ongaro V., Leyser O. (2009). Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 106: 17431–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T., Arend M., Morreel K., Storme V., Rombauts S., Fromm J., Bhalerao R.P., Boerjan W., Rohde A. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19: 2370–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sellaro R., Pacín M., Casal J.J. (2012). Diurnal dependence of growth responses to shade in Arabidopsis: Role of hormone, clock, and light signaling. Mol. Plant 5: 619–628 [DOI] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Mori H. (1998). Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol. 39: 255–262 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S., Mori H. (2001). Control of outgrowth and dormancy in axillary buds. Plant Physiol. 127: 1405–1413 [PMC free article] [PubMed] [Google Scholar]
- Simpson S.D., Nakashima K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33: 259–270 [DOI] [PubMed] [Google Scholar]
- Smith J.E., Jordan P.W. (1994). Stand density effects on branching in an annual legume (Senna obtusifolia). Ann. Bot. (Lond.) 74: 17–25 [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: No. 1, Article 3. [DOI] [PubMed]
- Smyth G.K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stafstrom J.P., Ripley B.D., Devitt M.L., Drake B. (1998). Dormancy-associated gene expression in pea axillary buds. Cloning and expression of PsDRM1 and PsDRM2. Planta 205: 547–552 [DOI] [PubMed] [Google Scholar]
- Stafstrom J.P., Sussex I.M. (1992). Expression of a ribosomal protein gene in axillary buds of pea seedlings. Plant Physiol. 100: 1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P., Kumar P.P. (2010). The phytohormone signal network regulating elongation growth during shade avoidance. J. Exp. Bot. 61: 2889–2903 [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Ward S., Leyser O., Kamiya Y., Nambara E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trémousaygue D., Garnier L., Bardet C., Dabos P., Hervé C., Lescure B. (2003). Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Tucker D.J. (1976). Effects of far-red light on the hormonal control of side shoot growth in the tomato. Ann. Bot. (Lond.) 40: 1033–1042 [Google Scholar]
- van Helden J., André B., Collado-Vides J. (1998). Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Walters R.G. (2005). Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 56: 435–447 [DOI] [PubMed] [Google Scholar]
- Wang H., Qi Q., Schorr P., Cutler A.J., Crosby W.L., Fowke L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wilson E.B. (1927). Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 22: 209–212 [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. (2003). A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629 [DOI] [PubMed] [Google Scholar]
- Zhu H., Dardick C.D., Beers E.P., Callanhan A.M., Xia R., Yuan R. (2011). Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol. 11: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.