This work identified a PPR-DYW subgroup protein EMP5 that is required for the C-to-U editing of multiple transcripts in maize mitochondria. The editing at the rpl16-458 site is considered particularly critical to the mitochondrial functions and, hence, to seed development. Interestingly, a deletion of the DYW domain does not abolish the editing, but only reduces the editing efficiency.
Abstract
In flowering plants, RNA editing is a posttranscriptional mechanism that converts specific cytidines to uridines in both mitochondrial and plastidial transcripts, altering the information encoded by these genes. Here, we report the molecular characterization of the empty pericarp5 (emp5) mutants in maize (Zea mays). Null mutation of Emp5 results in abortion of embryo and endosperm development at early stages. Emp5 encodes a mitochondrion-targeted DYW subgroup pentatricopeptide repeat (PPR) protein. Analysis of the mitochondrial transcripts revealed that loss of the EMP5 function abolishes the C-to-U editing of ribosomal protein L16 at the rpl16-458 site (100% edited in the wild type), decreases the editing at nine sites in NADH dehydrogenase9 (nad9), cytochrome c oxidase3 (cox3), and ribosomal protein S12 (rps12), and surprisingly increases the editing at five sites of ATP synthase F0 subunit a (atp6), apocytochrome b (cob), nad1, and rpl16. Mutant EMP5-4 lacking the E+ and DYW domains still retains the substrate specificity and editing function, only at reduced efficiency. This suggests that the E+ and DYW domains of EMP5 are not essential to the EMP5 editing function but are necessary for efficiency. Analysis of the ortholog in rice (Oryza sativa) indicates that rice EMP5 has a conserved function in C-to-U editing of the rice mitochondrial rpl16-458 site. EMP5 knockdown expression in transgenics resulted in slow growth and defective seeds. These results demonstrate that Emp5 encodes a PPR-DYW protein that is required for the editing of multiple transcripts in mitochondria, and the editing events, particularly the C-to-U editing at the rpl16-458 site, are critical to the mitochondrial functions and, hence, to seed development in maize.
INTRODUCTION
RNA editing is a posttranscriptional mechanism that alters RNA sequences via insertion, deletion, and conversion of nucleotides, resulting in changes in the genetic information encoded by the DNA. In flowering plants, most RNA editing events are conversions of cytidines (C) to uridines (U), and all the reported C-to-U editing events occur in either mitochondria or plastids (Chateigner-Boutin and Small, 2010). C-to-U editing was first reported in mitochondrial mRNAs by three independent groups (Covello and Gray, 1989; Gualberto et al., 1989; Hiesel et al., 1989). The plastid genomes contain ∼120 genes and fewer than 50 editing sites in the transcripts. By contrast, the mitochondrial genomes contain ∼60 genes, and ∼300 to 500 editing sites are predicted in the transcripts (Unseld et al., 1997; Giegé and Brennicke 1999; Notsu et al., 2002), implicating that RNA editing plays a major role in the expression of the mitochondrial genome. RNA editing is important for posttranscriptional regulation and in some cases critical to the functions of the encoded proteins. For example, editing can restore a conserved amino acid codon (Tillich et al., 2005), create an initiation or stop codon (Kotera et al., 2005), or remove a stop codon that leads to a functional larger protein (Grewe et al., 2009). Therefore, deficiency in editing may result in a compromised or complete loss of function for the encoded protein, leading to severe consequence in plant growth and development. Indeed, defects in mitochondrial RNA editing cause severe phenotypes, such as seed development defects, growth and development retardation (Kim et al., 2009; Sung et al., 2010; Hammani et al., 2011; Sosso et al., 2012; Yuan and Liu, 2012). Editing can also occur in introns and untranslated regions, potentially playing a role in regulating transcript stability (Börner et al., 1995; Chateigner-Boutin and Small, 2010).
Although RNA editing is important to the posttranscriptional regulation of organelle gene expression (Shikanai, 2006; Grennan, 2011), little is known about the mechanisms of C-to-U RNA editing in plants until 2005. The first insight came with the identification of CHLORORESPIRATORY REDUCTION4 (CRR4), a pentatricopeptide repeat (PPR) protein that is required for the C-to-U editing of a single site in the chloroplast transcript ndhD in maize (Zea mays) (Kotera et al., 2005). Since then, PPR proteins have been recognized as the trans-acting factors responsible for RNA editing in plastids and mitochondria in plants (Fujii and Small, 2011). All of the PPR proteins responsible for C- to-U editing are localized in either mitochondria or chloroplasts. Domain structure analysis indicated that they belong to E or DYW subclasses of the PPR protein family (Okuda et al., 2007; Schmitz-Linneweber and Small, 2008; Fujii and Small, 2011). The DYW domain shows a significant similarity to deaminase, raising the possibility of being a catalytic domain to convert C to U (Salone et al., 2007). Although this notion is not supported by the finding that truncated proteins lacking the DYW motifs can completely restore the RNA editing in vivo (Okuda et al., 2009), a recently identified protein DYW1, which contains a highly conserved DYW domain but no identifiable PPR motifs, can function in trans with the E subclass PPR protein CRR4 in the ndhD-1 site editing (Boussardon et al., 2012). Recently, a new family of proteins essential for organelle RNA editing has been identified, the multiple organellar RNA editing factor (MORF), including RNA-EDITING FACTOR INTERACTING PROTEIN 1 (RIP1, =MORF8) (Bentolila et al., 2012; Takenaka et al., 2012). Loss-of-function mutations in MORFs alter editing efficiency at multiple sites, distinguishing them from PPR proteins typically affecting one or a few sites. RIP1 is localized in both chloroplasts and mitochondria, affecting numerous editing sites in the two organelles. MORF2 or MORF9 are required for almost all sites of RNA editing in the chloroplast, and mutations of MORF1 and MORF3 affect numerous editing sites in mitochondria. These MORF proteins can interact selectively with PPR proteins to establish a complex editosome, and they can also form hetero- and homodimers in RNA editing (Bentolila et al., 2012; Takenaka et al., 2012).
Increasing evidence indicates that PPR genes play an important role in embryo and endosperm development in flowering plants. In Arabidopsis thaliana, GLUTAMINE-RICH PROTEIN23 (GRP23) is shown to be essential for early embryo development (Ding et al., 2006). GRP23, an exceptional PPR protein that is localized in the nucleus, exerts its function by interacting with RNA Polymerase II Subunit III. Mutation of this gene leads to embryo arrest at 16-cell dermatogen stage (Ding et al., 2006). Similarly, mutation of Arabidopsis PPR2, a gene probably responsible for translation process in chloroplasts, causes defects in cell proliferation during embryogenesis (Lu et al., 2011). A systematic analysis of Arabidopsis embryo-defective mutants (emb) revealed that 17 PPR genes among 250 Emb genes were essential to embryogenesis (Tzafrir et al., 2004; Cushing et al., 2005). In rice (Oryza sativa), OPAQUE AND GROWTH RETARDATION1 (OGR1) is found to be important for seed development (Kim et al., 2009). OGR1 is a PPR-DYW protein responsible for the RNA editing of mitochondrial transcripts nad2, nad4, cox2, cox3, and ABC transporter subunit C (ccmC). In maize, three mitochondrial PPR proteins are shown to be essential for seed development. EMPTY PERICARP4 (EMP4), a PPR protein that is required for the correct expression of a small group of mitochondrial genes, is essential for early embryo and endosperm development (Gutiérrez-Marcos et al., 2007). Recently, PPR2263, which affects kernel size by affecting both the embryo and endosperm development, was reported to be required for nad5 and cob transcript editing in maize mitochondria (Sosso et al., 2012). PPR protein MPPR6 is reported to be directly involved in the 5′ maturation and translation initiation of rps3 mRNA in mitochondria. Mutation of MPPR6 affects both embryo and endosperm development in maize (Manavski et al., 2012).
In this study, we report the molecular characterization of a nuclear gene Emp5, which affects the embryo and endosperm development in maize. Emp5 encodes a mitochondrion-targeted DYW subgroup PPR protein. Functional analysis indicates that EMP5 is responsible for the editing of 10 sites in four genes (rpl16, nad9, cox3, and rps12), of which the rpl16-458 site editing is completely abolished due to the emp5 mutation. Molecular analysis of the emp5-4 allele, which carried a Mu insertion disrupting the E+ and DYW domains, provides genetic evidence that E+ and DYW domains are not essential for the EMP5 editing function. Further analysis of the Emp5 ortholog in rice by RNA interference (RNAi) transgenics confirmed that the editing function, and its role in affecting seed development, is conserved between the two species.
RESULTS
Phenotypic and Genetic Characterization of emp5-1
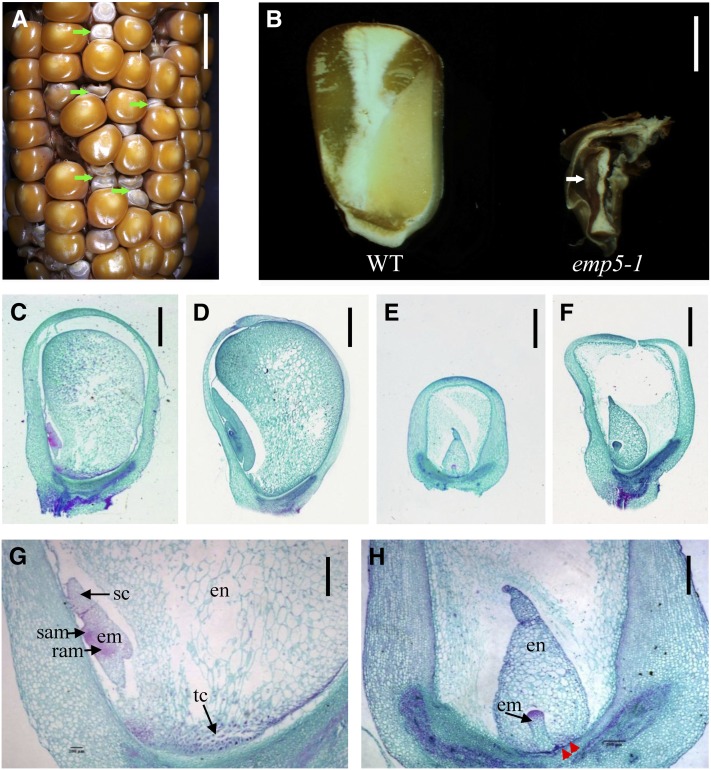
The emp5-1 mutant was isolated from the UniformMu population where the active Mu lines were introgressed into the inbred W22 genetic background (McCarty et al., 2005). When this mutant was isolated, six backcrosses to W22 had been performed. The isolated emp5-1 allele was backcrossed to W22 twice afterward to reduce Mu copy numbers. Therefore, the mutant was considered in nearly isogenic W22 background (99.6%). The selfed progeny of the emp5-1 heterozygotes segregated emp kernels in a 3:1 ratio (WT:emp, 455:167, P > 0.95), indicating that Emp5 is a monogenic, recessive, and nuclear gene. In contrast with the wild type, the mutant emp5-1 kernels at maturity typically were small, containing a white pericarp that was often wrinkled (Figure 1A). During sectioning of the kernels, some residual tissues could be found, but not recognizable embryo or endosperm structures (Figure 1B). This result indicated that the emp5 mutation blocks the development of both the embryo and the endosperm. The emp5-1 allele is an embryo-lethal mutation that is maintained in heterozygotes.
Figure 1.
Mutant emp5-1 Kernels Abort Early in Seed Development.
(A) The ear segregates 3:1 for wild-type and emp5-1 mutant kernels (arrows).
(B) Dissection of mature wild-type (WT; left) and emp5-1 (right) kernels.
(C) to (H) Developmental comparisons of wild-type and emp5-1 kernels at 8 and 13 DAP. Wild-type kernels at 8 DAP ([C] and [G]) and 13 DAP (D); emp5-1 kernels at 8 DAP ([E] and [H]) and 13 DAP (F). en, endosperm; em, embryo; ram, root apical meristem; sam, shoot apical meristem; sc, scutellum; tc, transfer cells. Red arrows point to positions where transfer cells were not clearly formed in the mutant.
Bars = 1 cm in (A), 2.5 mm in (B), 1 mm in (C) to (F), and 500 µm in (G) and (H).
To examine the developmental arrest in emp5-1, we analyzed the seed development process by light microscopy. Maize embryo development is characterized by three stages: transition, coleoptilar, and late embryogenesis. Endosperm development includes coenocytic, cellularization, differentiation, and maturation stages (Olsen, 2001). A close comparison was made by analyzing both the wild type and the emp5-1 mutant in the same segregating ear. The emp5-1 mutant kernel can be clearly distinguished from the wild-type siblings as early as 8 d after pollination (DAP), with characteristics of a small size and translucent appearance resulted from arrested embryo and endosperm development. The identification was confirmed by PCR genotyping. Dissection of the emp5-1 mutant seeds at 8 and 13 DAP revealed that the mutant embryogenesis was arrested at the transition stage, characterized by the establishment of radial asymmetry that was introduced by the formation of an internal wedge-shape meristematic region in the upper part of the embryo. The endosperm development was blocked at the differentiation stage, characterized by the formation of distinct starchy endosperm and aleurone layer (Figure 1E, 1F, and 1H). By contrast, the wild-type embryo at 8 DAP already differentiated a scutellum and shoot apical meristem, and the endosperm was much larger than the mutant (Figures 1C and 1G).
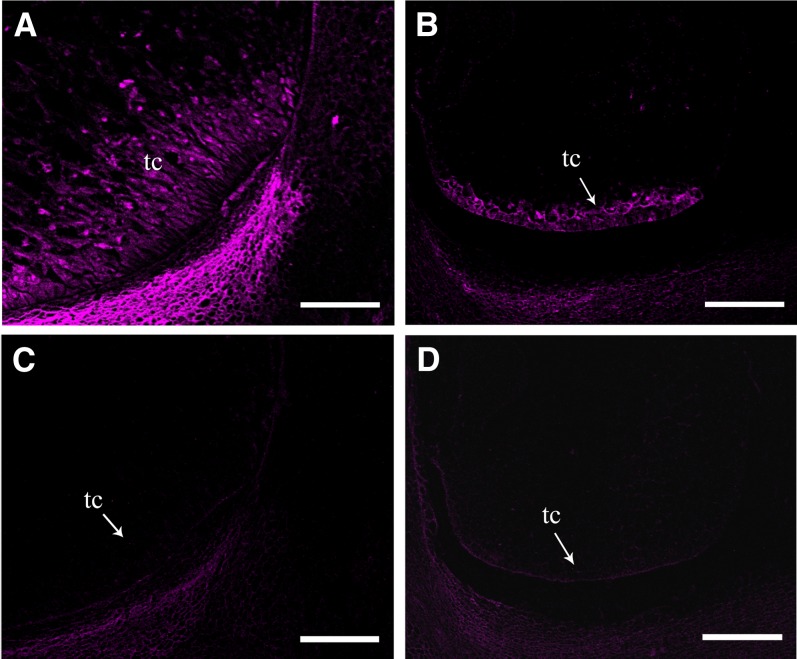
Besides the dramatic difference in embryo and endosperm, we noticed that emp5-1 appeared lacking basal transfer cells. Basal transfer cell layer is responsible for the uptake of solutes critical to seed development (Pate and Gunning, 1972). To investigate the development of basal transfer cell layer in the emp5-1 mutant seeds, we performed the immunohistochemistry analysis using BETL-2 antibody. BETL-2 is a basal endosperm transfer layer–specific protein expressed in early and mid-term endosperm development (Hueros et al., 1999). Therefore, it is a marker of basal transfer cell formation. The wild-type kernels at 13 DAP differentiated more than three layers of transfer cells, and the BETL-2 protein was expressed in the whole basal transfer cell layer region (Figure 2A). By contrast, the emp5-1 mutant kernels from the same ear only formed a single layer of transfer cells, and BETL-2 was detected in these cells (Figure 2B). During subsequent microscopy analyses, we did not observe the formation of multiple cell layers of transfer cells in the mutant. This result indicated that the basal transfer cell development in the emp5-1 kernels is arrested.
Figure 2.
The Basal Transfer Cell Development in the emp5-1 Kernels Is Arrested.
Confocal fluorescent microscopy visualization of BETL-2 by immunofluorescence in 13-DAP wild-type ([A] and [C]) and emp5-1 mutant ([B] and [D]) kernels. To visualize the basal endosperm transfer layer, specific antibody BETL-2 was used ([A] and [B]), and no BETL-2 antibody PBS buffer was used as control ([C] and [D]). tc, transfer cell. Bars = 200 µm.
Cloning of Emp5
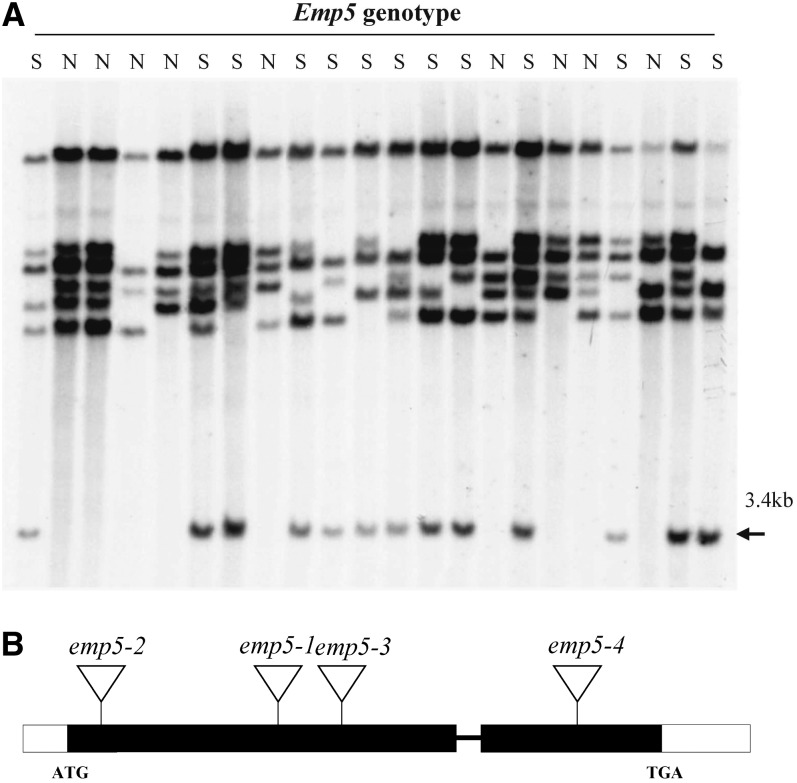
Because the emp5-1 allele was potentially tagged by Mu transposons, we performed DNA gel blot analysis to identify whether a Mu insertion was linked to the mutation. A segregating F2 population was created by self-crossing a heterozygous emp5-1/Emp5 plant. Genomic DNA was isolated from the individual F2 plants, and the genotype was determined by selfing the plant and checking for emp5-1 mutant segregation. Because homozygous emp5-1 is not viable, only heterozygotes (segregating) or the wild type (nonsegregating) were available for this analysis. The blots were hybridized with several Mu elements, including Mu1/Mu2, Mu3, Mu4, Mu8, MuDR, and Mu13 (Tan et al., 2011), and only the hybridization with a Mu1/Mu2-specific probe identified a 3.4-kb HindIII fragment that cosegregated with the emp5 mutation (i.e., the presence of this fragment in emp5-1 heterozygote and its absence in the wild type) (Figure 3A). No recombination was detected in the initial 22 F2 individuals tested. Increasing the population to 120 individuals still did not produce any recombination, suggesting a tight linkage between this Mu insertion and the emp5-1 mutation. Because the Mu1/Mu2 probe hybridizes to both Mu1 and Mu2, we could not tell which one was inserted in this fragment at this stage.
Figure 3.
Emp5 Gene Cloning.
(A) DNA gel blot cosegregation analysis of an emp5-1 segregation population using the internal sequence of Mu1 as a probe. The Mu2 transposon-tagged 3.4-kb HindIII fragment (arrowhead) cosegregated with the emp5-1 mutation. N, nonsegregating (the wild type); S, segregating (heterozygous).
(B) Gene structure of Emp5 and locations of the Mu insertions in four independent alleles. Exons are closed boxes and introns are lines. Mu insertion sites of emp5 alleles were confirmed by sequencing and marked by triangles.
The Mu1 or Mu2 flanking sequence in the 3.4-kb HindIII fragment was amplified by inverse PCR using Mu TIR primers. The size of the product was 1.7 kb with TIR sequences on both ends and a HindIII site in the middle. A 9-bp target site duplication was found flanking the Mu insertion. This suggests that the Mu element is likely a Mu2 (1.7 kb), not a Mu1 (1.4 kb). BLAST search of the National Center for Biotechnology Information GenBank with the 1.7-kb Mu2 flanking sequence identified an expression mRNA in maize (accession number EU956937). This gene is located on chromosome 3. This gene appears to be a single-copy gene based on the analysis of B73 whole genomic sequence draft AGPv2 (Schnable et al., 2009). Using two gene-specific primers designed according to this clone, the full genomic sequence of this Emp5 candidate gene was cloned from W22. And the cDNA was amplified by RT-PCR from leaf RNAs.
To confirm that the cloned gene is the bona fide causative gene for the emp5-1 phenotype, we isolated additional Mu insertional alleles from the Pioneer Hi-Bred International Trait Utility System for Corn (TUSC) population using Emp5 gene-specific primers in combination with Mu-TIR primers (Bensen et al., 1995). Three independent Mu insertions in the Emp5 candidate gene were identified, named emp5-2, emp5-3, and emp5-4 (Figure 3B). The insertion sites were confirmed by sequencing the PCR products that were amplified with TIR8 primers and Emp5-specific primers. The selfed progeny of heterozygote emp5-2 and emp5-3 produced empty pericarp kernels segregation at a ratio of 1:3 (emp:WT), but that of the emp5-4 produced all normal kernels. Crosses between emp5-1 heterozygotes with heterozygotes for emp5-2 and emp5-3 alleles produced ears segregating empty pericarp kernels at a 1:3 ratio (emp:WT), whereas crosses between emp5-1 and emp5-4 produced all wild-type kernels. Genotyping using gene-specific primers indicated that homozygous emp5-4 seeds were viable and contained a normal embryo and endosperm. Later analysis indicated that the emp5-4 mutation was leaky and only partially abolished the Emp5 function. Because three independent alleles carried Mu insertions in the Emp5 gene conditioned the empty pericarp phenotype, we concluded that the Emp5 locus was cloned.
Emp5 Encodes a Mitochondrion-Targeted PPR-DYW Subclass Protein
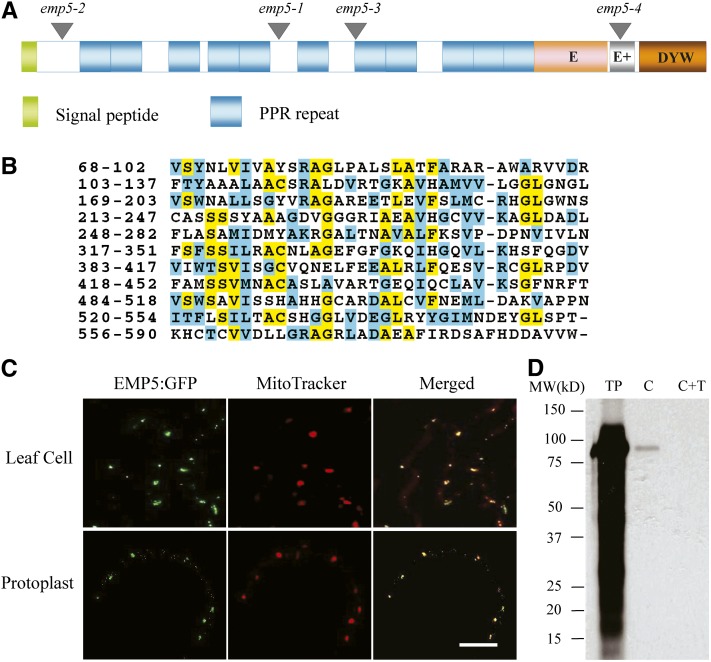
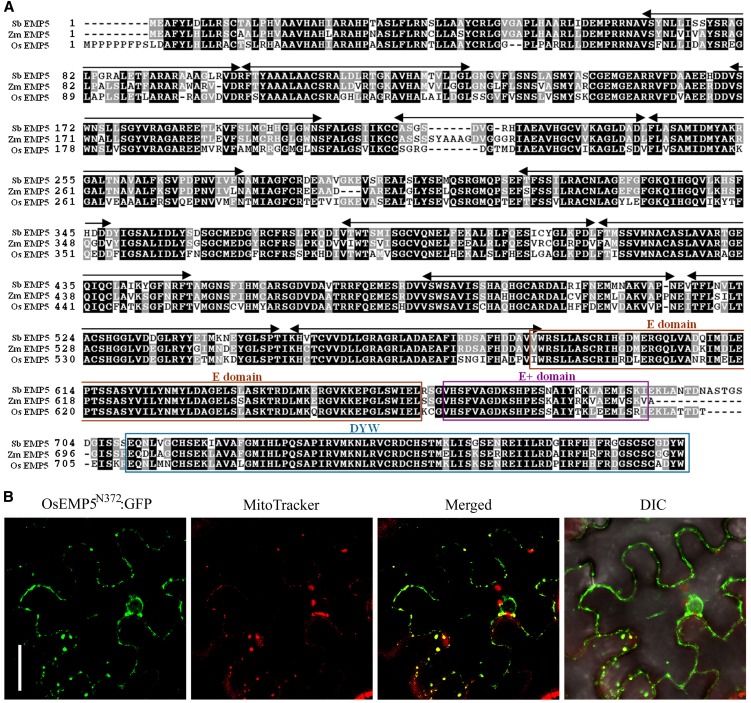
Sequence analysis indicated that the Emp5 gene consists of two exons, and in emp5-1, a Mu2 was inserted in the first exon (Figure 3B).This Emp5 gene encodes a 776–amino acid protein. Motif prediction analysis by algorithm TPRpred (http://tprpred.tuebingen.mpg.de/tprpred) revealed that EMP5 contains 11 PPR motifs, classifying this protein as a member of the PPR protein family (Figures 4A and 4B). The C-terminal region between residues 590 and 776 shows a strong similarity to the consensus sequences of the E, E+, and DYW domains (Lurin et al., 2004), indicating that EMP5 is a DYW subclass PPR protein. Analysis of the rice and sorghum (Sorghum bicolor) genome identified an ortholog in each genome, named Os EMP5 and Sb EMP5, respectively. EMP5 showed clear divergence between monocots and dicots. Among the closely related proteins, it shares a high degree of similarity with sorghum (91%), rice (84%), and barley (Hordeum vulgare) (83%) but a low degree of similarity with grape (Vitis vinifera) (69%) and Arabidopsis (64%). Functions of these proteins are not known yet.
Figure 4.
EMP5 Is a Mitochondrion-Localized PPR-DYW Protein.
(A) The EMP5 protein contains 11 PPR motifs and E, E+, and DYW motifs at the C terminus. Locations of four Mu insertion alleles are marked with triangles.
(B) Alignment of 11 PPR motifs found in EMP5 proteins. Identical residues are shaded in yellow and similar residues in light blue.
(C) A EMP5N469-GFP fusion protein that carried the N terminus 469 amino acids fused with GFP was expressed in transgenic Arabidopsis leaves. The leaf samples and protoplasts were used for GFP signal detection by confocal microscopy. Fluorescent signals from EMP5N469-GFP are green and MitoTracker stained mitochondria are red. Bar = 10 µm.
(D) Chloroplast import of full-length EMP5 protein into pea chloroplasts. C, chloroplasts after imported with 3H-EMP5; C+T, 3H-EMP5 imported chloroplasts treated with thermolysin to remove surface adhered proteins; TP, translated precursor EMP5 labeled with 3H-Leu.
Analysis of EMP5 with the TargetP algorithm predicted a putative mitochondrial localization, but with marginal confidence (http://www.cbs.dtu.dk/services/TargetP/). To experimentally determine the subcellular localization of EMP5, we fused full-length EMP5 with green fluorescent protein (GFP) and transformed Arabidopsis. Ten lines of transgenic Arabidopsis were generated, and among these transgenics, GFP signals were either absent or low. We suspected that the length of the EMP5-GFP fusion may be the cause for this problem. So, the N-terminal 469 amino acids of EMP5 that contains eight PPR repeats was fused with GFP, resulting in fusion construct EMP5N469-GFP. Fourteen lines of transgenic Arabidopsis were produced and analyzed. Confocal laser scanning microscopy analysis of the transgenic Arabidopsis leaf samples and protoplasts showed in vivo colocalization of the GFP signal of EMP5N469-GFP with MitoTracker Red in mitochondria (Figure 4C). To independently test the possibility of chloroplast localization, we also performed pea (Pisum sativum) chloroplast import assays in which the EMP5 protein was labeled with 3H-Leu and tested its capability in importing to live chloroplasts. The result showed that EMP5 was not imported into the chloroplasts as protease thermolysin treatment of the imported chloroplasts eliminated all the radioactive 3H-EMP5 protein (Figure 4D). Thermolysin protease treatment degrades proteins unprotected by the chloroplast envelopes. Together, these results indicated that EMP5 is a PPR-DYW protein localized in maize mitochondria.
Expression of Emp5
BLAST analysis of EMP5 identified an EST from maize cDNA library of mix tissues, indicating that Emp5 may be expressed in multiple tissues. Analysis of Os Emp5, the ortholog in rice, also identified two ESTs derived from rice panicle and callus. However, the Emp5 mRNA could not be detected by conventional RNA gel blot analysis, suggesting that it may be expressed at low levels. Similarly, the maize mitochondrial PPR gene Emp4 was also reported having a low expression level that could not be detected by RNA gel blot analysis (Gutiérrez-Marcos et al., 2007). The Emp5 expression could be detected by RT-PCR, and the results confirmed that Emp5 was expressed in all vegetative and reproductive tissues tested (Figure 5). Relative high mRNA expression was in stem, leaf, root, and ear and weak expression in tassel and kernels at developmental stages. This suggests that the EMP5 function may not be limited to embryo and endosperm development. Rather, it may have functions in other vegetative tissues during plant growth and development.
Figure 5.
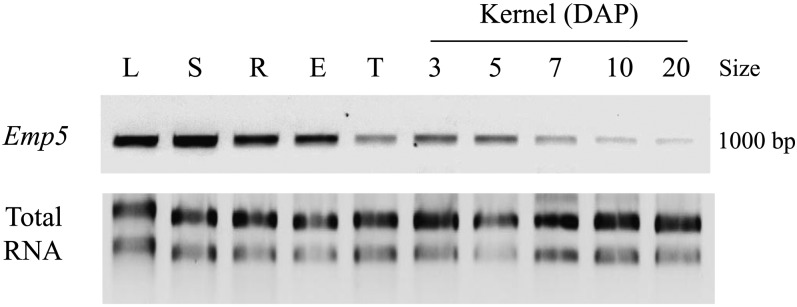
RT-PCR Analysis Indicates Expression of Emp5 in Multiple Organs and during Seed Development in Maize.
Primers Emp5-F2 and Emp5-R2 were used in RT-PCR. E, ear; L, leaf; S, stem; R, root; T, tassel. Kernel at 3, 5, 7, 10, and 20 DAP.
EMP5 Is Required for Mitochondrial RNA Editing
Up to now, DYW1, MORF, and PPR proteins are the only identified plant RNA editing trans-factors, and most of the PPR proteins belong to DYW subclass (Fujii and Small, 2011). Since EMP5 is a typical mitochondria-targeted DYW subclass PPR protein, it is highly likely that EMP5 functions in RNA editing in maize mitochondria.
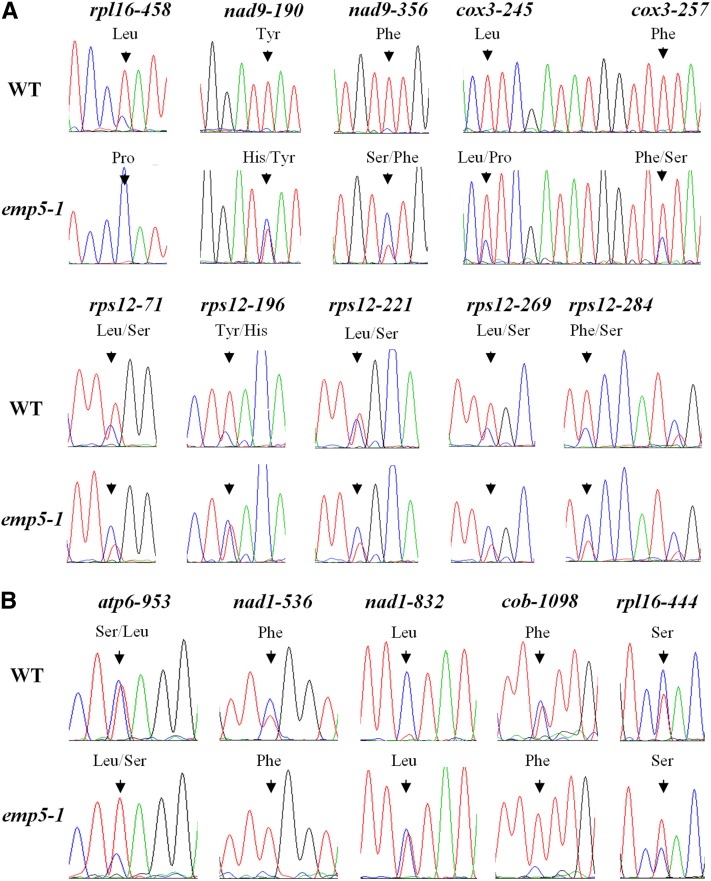
A direct comparison of the mitochondrial transcripts between the emp5 mutant and the wild type was performed by amplifying the transcripts and analyzing their sequences. Based on the maize mitochondrial genome (Clifton et al., 2004), 35 sets of primers were designed to cover the predicted 35 mitochondrial protein-coding genes (see Supplemental Table 1 online). These protein-coding mitochondrial genes include 22 genes of the electron transport chain, 11 ribosomal proteins genes, a maturase gene (mat-r), and a transporter gene (mttB). The reference allele emp5-1 was chosen for its near-isogenic W22 genetic background. To eliminate possible contamination of mitochondrial DNA (most mitochondrial genes are intronless), RNAs were treated with RNase-free DNase and confirmed to be DNA free by PCR amplification without reverse transcription. The RNA was isolated from the 13-DAP kernels as the mutant was very distinct from the wild type. The pericarp was carefully removed to prevent contamination by the heterozygous maternal tissues. RT-PCR was performed using proofreading DNA polymerase Phusion (New England Biolabs). The PCR products were purified from the gel and sequenced without cloning into a vector. This allows detection of both edited and unedited sites in one sequencing reaction and also eliminates cloning bias and random DNA polymerase errors. We amplified the mitochondrial genes in the emp5-1, and the sequences indicated that the W22 contains a NB-type mitochondrial genome (Clifton et al., 2004). As shown in Figure 6A, the C-to-U editing of rpl16-458 was completely abolished in the emp5-1 allele, whereas it was completely edited in the wild type. The unedited sequence codes for a Pro, whereas the edited sequence codes for a Leu. Therefore, the lack of C-to-U editing in rpl16-458 resulted in an amino acid change from Leu to Pro in RPL16 protein in the emp5-1 mutant. In addition, 100% C-to-U editing was found at nad9-190, nad9-356, cox3-245, and cox3-257 sites in the wild type, whereas it was dramatically diminished in emp5-1, especially for nad9-190 and nad9-356 sites (Figure 6A). Editing at five sites of the rps12 transcript (rps12-71, rps12-196, rps12-221, rps12-269, and rps12-284) was also reduced in the emp5-1 mutant (Figure 6A). All this editing lead to a change of the encoded amino acid as indicated in Figure 6A.
Figure 6.
RNA Editing Defects of Mitochondria Genes in the emp5-1 Mutant.
(A) RNA editing of 10 sites in four mitochondrial gene transcripts decreased in emp5-1. WT, the wild type.
(B) RNA editing of five sites in four mitochondrial gene transcripts increased in emp5-1. Sequence chromatogram of PCR-amplified wild-type cDNA or emp5-1 cDNA of the editing sites is shown. The position of RNA editing represents the name of transcripts and the edited C position. The amino acid changes are indicated at the top, and the prevalent amino acids are put in front of the less prevalent ones. Arrowheads indicate the editing sites.
Interestingly, we also found that the editing of several sites was increased in the emp5-1 mutant. Editing of rpl16-444, atp6-953, nad1-536, nad1-832, and cob-1098 was dramatically increased in the emp5-1 mutant, comparing with weaker editing of these sites in the wild type (Figure 6B). However, this editing, except atp6-953, does not change the encoded amino acids. Increased editing was also reported in the required for efficiency of mitochondrial editing1 (reme1) mutants encoding a PPR protein and the rip1 T-DNA insertional mutants (Bentolila et al., 2010, 2012). However, the underlying mechanism is not clear.
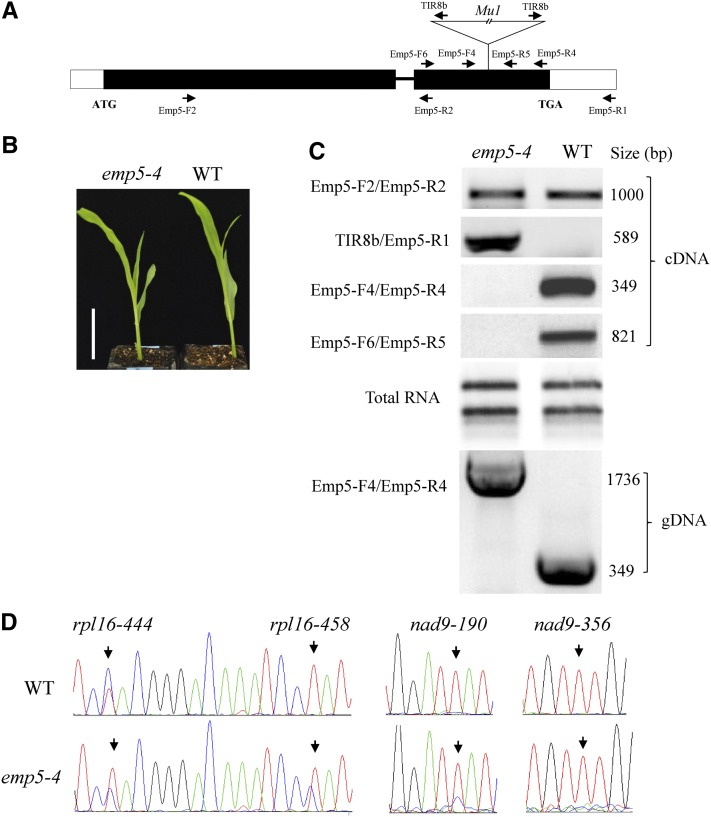
Molecular Characterization of the emp5-4 Allele
As described previously, homozygous emp5-4 produced viable seeds. Genotyping with primers Emp5-F4 and Emp5-R4 and sequencing of the amplicon identified that the insertion is a 1.4-kb Mu1 element in the middle of the E+ motif, thereby disrupting the E+ and DYW domain (Figure 7A). However, homozygous emp5-4 seedlings and adult plants were macroscopically indistinguishable from the wild type (Figure7B). In contrast with the use of transgenes, this allele provided excellent genetic material for studying the effect of disrupted E+ and DYW domains on the function of EMP5. First, we examined the expression using different Emp5 primers anchored on different regions of the emp5-4 allele (Figure 7A). As indicated in Figure 7C, RT-PCR results of the seedling leaf transcripts indicated that the region 5′ of the Mu1 insertion in emp5-4 was expressed at roughly the similar level as the wild type. The region including Mu1 insertion was only expressed in emp5-4, as indicated in the RT-PCR using Mu primer TIR8 and Emp5-R1 in RT-PCR (Figure 7C). However, PCR with primers Emp5-F6/Emp5-R5 as well as Emp5-F4/Emp5-R4 across the Mu1 insertion did not produce any products in emp5-4 (Figure 7C). To address whether this was caused by a difficulty in PCR amplification across the Mu1 element, we used wild-type and homozygous emp5-4 genomic DNA as template. PCR amplification with primers Emp5-F4 and Emp5-R4 reliably produced a 1.7-kb fragment in emp5-4 (predicted 1736 bp) and a 350-bp fragment in the wild type (predicted 349 bp). Sequencing confirmed that the 1.7-kb amplicon contains the Mu1 insertion in the emp5-4 allele (Figure 7C), indicating the primer set worked well in amplifying across the Mu1 insertion. We did not detect any other alternatively spliced transcripts as some Mu inserted alleles showed splicing of the element. The spliced transcripts should be favored in amplification because of a shorter size. RT-PCR analysis indicated that the emp5-4 allele does not produce a detectable level of Emp5 transcripts that is likely to be translated into a wild-type protein, although the gene including the Mu1 element can produce two transcripts: One contains the 5′ region of the Emp5 and ends in the Mu1 insertion, and the second starts from somewhere inside the Mu1 and ends probably where wild-type Emp5 ends.
Figure 7.
Insertion of a Mu1 Element in the E+ Domain in the emp5-4 Allele Reduces Editing Efficiency.
(A) Gene structure of Emp5 and location of the Mu1 insertion in the emp5-4 allele. The positions of primers used in RT-PCR analysis are shown as arrows.
(B) Seedling phenotype comparison between the homozygous emp5-4 and the wild type (WT). Bar = 5 cm.
(C) RT-PCR analysis of the transcripts in the emp5-4 mutant and wild-type seedling leaves. Primer combinations are shown in the left. PCR amplification with Emp5-F4 and Emp5-R4 primers on genomic DNA was used as a control.
(D) Sequence chromatograms showing the C-to-U editing of rpl16 and nad9 transcripts in the emp5-4 mutant and the wild type. The RNA editing site is described as the name of transcripts and the edited position from translation start codon. Arrows indicate the editing sites.
To determine the impact of this mutation on editing, we amplified and sequenced the mitochondrial transcripts using the same strategy as in emp5-1. The templates were the seedling RNAs from the wild type and the mutants from the same ear. The genotype was confirmed by PCR analysis. The results revealed that the emp5-4 homozygotes showed similar levels of editing in most transcripts targeted by EMP5 in comparison to the wild type. These include nad9-356, cox3-245, cox3-257, cob-1098, atp6-953, nad1-536, and nad1-832 and five editing sites in rps12 (rps12-71, rps12-196, rps12-221, rps12-269, and rps12-284). However, the rpl16-458 editing was diminished but clearly detectable in the emp5-4 allele. This editing converts a Pro to a Leu residue, which is edited completely in the wild type, but the editing was completely abolished in emp5-1. Similarly, editing of nad9-190 site was increased in emp5-4 in comparison to emp5-1. This site is also edited completely in the wild type, which converts a His to a Tyr residue (Figures 6A and 7D). Even in the same transcript, the other EMP5-targeted site (nad9-356) showed the same editing level as in the wild type. Similar to emp5-1, the editing of rpl16-444 was also increased in emp5-4, but this editing does not change in coded amino acid (Figure 7D). These results indicated that the mutation in emp5-4 does not affect the editing of most EMP5 targeted sites but does decrease the editing in rpl16-458 and nad9-190.
Functional Analysis of the Rice Emp5 Gene
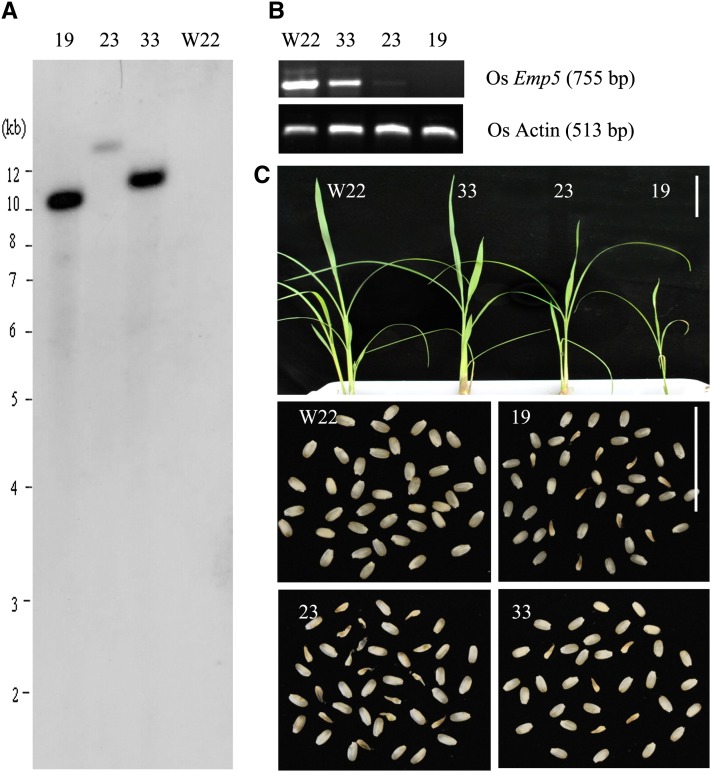
Rice EMP5 protein is a putative ortholog of EMP5 in maize (Zm EMP5), sharing a high degree of similarity (84%) with EMP5 and containing 11 PPR repeats in the N terminus and E/E+/DYW domains in the C terminus (Figure 8A). To address whether Os EMP5 has a conserved function similar to the Zm EMP5, we characterized EMP5 in rice. First, we determined the subcellular localization of rice EMP5 by expressing OsEMP5N372:GFP fusion in tobacco (Nicotiana tabacum) leaf epidermal cells. Confocal laser scanning microscopy analysis revealed that the GFP signal of OsEMP5N372:GFP was colocalized with MitoTracker Red in mitochondria (Figure 8B), confirming that EMP5 is targeted to the mitochondrion. To study the function of EMP5, Os Emp5 RNAi transgenic rice was created. It is likely that severe knockdown transgenic lines would be lethal considering the essential role of EMP5 in maize, therefore only weak knockdown lines were likely generated. About 20 lines of transgenic rice were obtained and three representational lines were used for further molecular analysis. DNA gel blot analysis of the three Os Emp5 RNAi transgenic lines (lines 19, 23, and 33), using the hygromycin phosphotransferase (hpt) gene as a probe, confirmed that they were independent transgenic lines, each carrying one copy of transgene (Figure 9A). RT-PCR analysis revealed the Emp5 expression level was significantly decreased in lines 19 and 23 and slightly decreased in line 33 compared to the wild type (Figure 9B). The transgenic plants grew much slower than the wild type at seedling stage. But after the seedling stage, the plants gradually recovered and grew to normal adult plants (Figure 9C). The T1 progeny of all these three transgenic lines segregated defective seeds as different ratio (Figure 9C). The ratio of defective seeds in line 19 is 24.7% (199:807), line 23 is 22.3% (93:417), and line 33 is 9% (53:584). The severity of seed phenotype was roughly consistent with the suppressed Emp5 expression level. This result indicated that similar to Emp5 in maize, rice Emp5 is essential to seed development in rice.
Figure 8.
Os EMP5 Is a Mitochondrion-Localized PPR-DYW Protein.
(A) Alignment of maize Zm EMP5 protein with rice ortholog Os EMP5 and sorghum ortholog Sb EMP5 protein.
(B) OsEMP5N372:GFP fusion protein was transient expressed in tobacco epidermal cells. Fluorescence signals from OsEMP5N372:GFP (green) and MitoTracker stained mitochondria (red) were detected by confocal microscopy. DIC, differential interference contrast. Bar = 10 µm.
Figure 9.
RNAi Knockdown Analysis of Os Emp5 Expression in Transgenic Rice.
(A) DNA gel blot analysis of independent Os Emp5 RNAi transgenic lines (lines 19, 23, and 33) using the hpt gene as a probe.
(B) RT-PCR analysis of endogenous Emp5 expression in three transgenic lines. Os Actin (X15865.1) was used as control. Primers Osactin-F, Osactin-R, OsEmp5-F, and OsEmp5-R (see Supplemental Table 1 online) were used in RT-PCR.
(C) Phenotypes of Os Emp5 RNAi transgenic plants at the seedling stage and in T1 progeny segregating defective seeds. Bars = 5 cm.
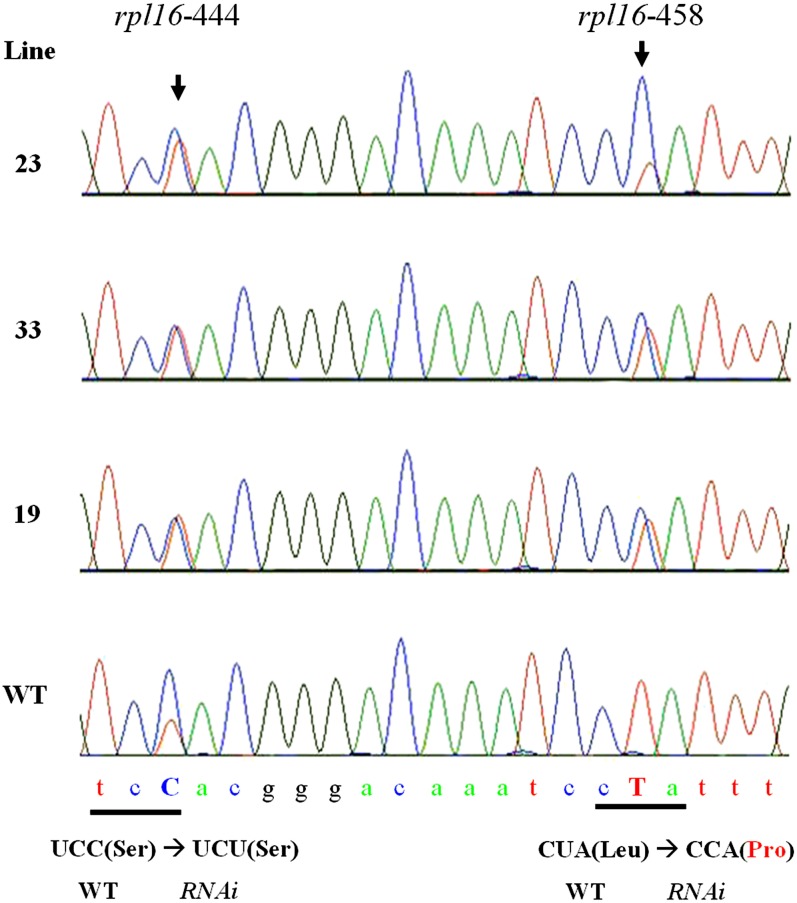
To reveal the effect on mitochondrial RNA editing in these Os Emp5-RNAi transgenic lines, mitochondrial rpl16 transcripts were amplified and sequenced in the three Os Emp5 RNAi transgenic lines and wild-type plants to compare the RNA editing difference. This analysis revealed that <50% rpl16-458 site in all three transgenic lines was edited, compared to 100% editing in the wild type (Figure 10). For rpl16-444 site, the editing level was increased in the three transgenic lines, which is similar to the maize emp5-1 allele (Figure 10). This result confirmed that similar to maize EMP5, Os EMP5 is required for the editing of rpl16-458 in rice mitochondria.
Figure 10.
Os EMP5 Is Required for rpl16 Editing in Rice Mitochondria.
Sequence chromatograms of the rice rpl16 cDNA amplified by RT-PCR in wild-type (WT) and Os Emp5 RNAi transgenic lines showed the editing sites. Arrowheads indicate the editing sites. The amino acid change is indicated at the bottom.
DISCUSSION
Abortion of emp5-1 Mutant Seed Development Is Caused by Defective Mitochondrial RNA Editing
The emp mutants are defined by a dramatic reduction in embryo and endosperm size, yet possess a normal pericarp (Sheridan and Neuffer, 1980). In maize, three mutants with a similar phenotype have been characterized. Emp2 encodes a repressor of a heat shock response in seeds, mutation of which unleashes a heat shock response causing seed development abortion (Fu et al., 2002). Emp4 encodes a PPR protein that is required for normal level expression of several mitochondrial genes (Gutiérrez-Marcos et al., 2007). Recently, MPPR6, a PPR gene whose mutation causes an empty pericarp phenotype, is required for maturation and translation initiation of rps3 mRNA in mitochondria (Manavski et al., 2012). In this study, we show that Emp5 encodes a DYW subclass PPR protein that functions in the editing of several maize mitochondrial gene transcripts. The cloning of Emp5 is supported by multiple independent insertions in the Emp5 gene that condition a typical emp phenotype and also by the further functional analysis in editing. Such function in mitochondrial editing is conserved in its ortholog Emp5 in rice. The results indicate that the deficiency of editing in these mRNAs compromises the mitochondrial function, causing the embryo and endosperm development to be arrested at transition stage. Since the pericarp tissue is maternal, growth of the pericarp was not expected to be affected in heterozygous Emp5/emp5 plants. Thus, the mutant kernels are appeared as empty pericarp. Considering the critical functions of mitochondria to both the embryo and the endosperm, the emp mutants should be enriched with genes that have key functions in mitochondria. In maize, the emp mutants form a distinct class that will be ideal genetic materials for dissecting the mitochondrial PPR functions.
In the severe allele of emp5-1, the editing of rpl16-458 was completely abolished, and the other nine editing sites in three different transcripts (nad9, cox3, and rps12) were also diminished. rpl16 and rps12 encode ribosomal proteins of the mitochondrial translation machinery. nad9 and cox3 are required for Complex I and Complex IV function in the electron transport chain, respectively. However, a significant portion of correctly edited transcripts of nad9, cox3, and rps12 still exist. It is not clear of the impact on seed development as a result of reduced editing in these three transcripts. A fraction of the correctly edited mRNA may be sufficient for mitochondrial function to complete embryogenesis. This is indicated in other mitochondrial genes, such as apocytochrome b (cob). A complete loss of cob-908 editing severely reduces the growth in maize, whereas a residual level of cob-908 editing is sufficient to assure normal plant growth in Arabidopsis (Sosso et al., 2012). In addition, our editing analysis in maize mitochondrial transcripts showed that partial editing for a particular site is common in wild-type mitochondrial transcripts. This argues a possibility that the reduced editing in the nine sites of three transcripts (nad9, cox3, and rps12) may not be the major cause for the emp phenotype. Instead, the complete loss of a single rpl16-458 editing site may be attributable to the abortion of emp5 mutant seed development.
The rpl16 transcript lacking rpl16-458 editing translates to a protein with a Pro instead of Leu residue at position 153, which may severely compromise the RPL16 function in mitochondrial protein translation. Pro often acts as a structural disruptor in protein secondary structure, such as α-helices and β-sheets. Thus, this change may severely affect the structure and the function of the RPL16 protein. There are several cases where an unedited site coding for Pro causes a severe impact on plant growth and development. The maize mitochondrial cob-908 is normally C-to-U edited by PPR2263 to render a Pro-to-Leu change. The unedited cob-908 in ppr2263 mutants caused defects in seed development and seedling growth (Sosso et al., 2012). In the slg1 mutant of Arabidopsis, RNA editing in a single site nad3-250 in mitochondria is abolished, resulting in a codon for Pro instead of the edited for Ser. This single amino acid mutation caused slow growth and delayed development (Yuan and Liu, 2012). The mutations of OGR1, SLOW GROWTH1 (SLO1), and MITOCHONDRIAL EDITING FACTOR11(MEF11) also leaded to absence of RNA editing of mitochondrial transcripts, which all result in Leu-to-Pro changes in nad4-416, nad4-449, and cox3-422, respectively (Kim et al., 2009; Sung et al., 2010; Verbitskiy et al., 2010). All three mutants exhibited significant defects in plant growth and development. Conceivably, genetic screens will enrich the identification of changes critical to protein functions. These cases demonstrate that the editing for transition from Pro to other amino acids is critical to the function of multiple mitochondrial proteins.
The rpl16 gene has been proved to be essential to mitochondrial gene expression as a key component in the translation machinery. Thus far, a complete deletion of the rpl16 gene in mitochondria has not been isolated, presumably due to its lethality (Newton et al., 1996). In the Arabidopsis maternal distorted leaf mutant, rearrangement in two mitochondrial DNA fragments associated with the rps3-rpl16 polycistron resulted in a deletion of part of the intron and exon b of rps3 that is upstream of rpl16 but without affecting the rpl16 coding region. The mutant showed poor growth, distorted rough leaves, and aborted flowering organs (Sakamoto et al., 1996). A deletion of the 5′ untranslated region sequences, including the promoter for the transcription unit of the rps3-rpl16 polycistron in maize nonchromosomal stripe3 (ncs3) and ncs4 mutants, caused severe stunted and striped leaves and male fertility, respectively (Hunt and Newton, 1991; Newton et al., 1996). The mRNA abundance corresponding to the rps3/rpll6 coding region was specifically reduced in these mutants. Mitochondrial protein synthesis was dramatically reduced in severely affected mutant plants. These results reveal that the RPL16 protein is essential for protein synthesis in mitochondria, which consequently will be crucial for plant development. Therefore, the failed development in both embryo and endosperm in the emp5 mutants is likely due to a loss of rpl16-458 editing.
Increased Editing in the emp5 Mutant
The mechanism by which RNA editing is performed in plastids and mitochondria is not clear. PPR proteins are considered as the specificity determinants that recognize different transcripts (Shikanai, 2006; Chateigner-Boutin and Small, 2010). A single PPR protein, such as EMP5, that is responsible for multiple editing sites in multiple transcripts has been reported in several PPR-DYW proteins, such as MEF1, MEF11, and OGR1 in Arabidopsis and rice (Kim et al., 2009; Zehrmann et al., 2009; Verbitskiy et al., 2010). A considerate amount of RNA was found still correctly edited in the emp5-1 mutant, raising a reasonable possibility that overlapping editing on the same site may be mediated by different PPR proteins. In addition, we found that five editing sites on four mitochondrial transcripts were increased in the emp5-1 mutants, interspersing in atp6, nad1, cob, and rpl16 transcripts (Figure 6B). This result was also confirmed in Emp5 RNAi knockdown transgenic rice where the rpl16-444 editing level is increased in all three transgenic lines compared with the wild type. Intriguingly, the editing in these sites do not change the amino acid coding except in atp6-953 where editing converts Ser to Leu. This phenomenon has also been reported in PPR protein REME1, mutation of which reduces the editing of nad2-558 and orfX-552, but increases the editing extent in at least two sites, matR-1771 and rpl5-92 (Bentolila et al., 2010). It seems that PPR proteins may work as a positive and negative regulator of organelle RNA editing at the same time. One possibility is that overlapping functions by PPR proteins in editing a single site exist in plants such that the deficiency in one PPR leads to a compensational expression of another PPR protein that edits another set of sites, overlapping but not identical. In this case, some sites will be edited more than the wild type as a result of the compensational PPR proteins. Another possibility is that although a Mu2 is inserted into the Emp5 gene, it can still be transcribed and translated into a truncated protein that recognizes different RNA substrates. For the emp5-1 allele, the Mu2 insertion disrupted six of the 11 PPR motifs, allowing five PPR repeats remained in the mutant protein if it can be translated (Figure 4A).
Targeting Sequences of EMP5 Are Not Conserved
Two PPR proteins, CRR4 and RESTORER OF FERTILITY1 (RF1), were reported as possessing the RNA binding activity in vitro without other factors (Okuda et al., 2006; Kazama et al., 2008), leading to a hypothesis that PPR proteins can specifically recognize the cis-element of an editing site. Some RNA editing sites were shown to have conserved sequences upstream of the editing sites (Karcher et al., 2008; Kobayashi et al., 2008; Sosso et al., 2012). However, these conserved sequences were derived by comparing merely two editing sites. In cases where a PPR protein is involved in the editing of multiple sites, such as OGR1, MEF1, MEF11, and CRR22, no conserved sequences in the corresponding region can be identified (Kim et al., 2009; Okuda et al., 2009; Zehrmann et al., 2009; Verbitskiy et al., 2010). In CHLOROPLAST BIOGENESIS19 (CLB19), the sequences surrounding the two editing sites showed little sequence similarity (Chateigner-Boutin et al., 2008). In an attempt to identify conserved sequences for EMP5 edited sites, we aligned the adjacent sequences near the editing sites (region from −40 to +20). We did not find any conserved sequences except most of the −1 base is T (see Supplemental Figures 1 and 2 online). The likely possibility is that PPR proteins recognize a specific secondary structure of the transcripts, not the primary sequence. It is hypothesized that PPR editing factors can only distinguish pyrimidines from purines and, at some positions, must be able to recognize specific bases (Hammani et al., 2009). Recently, Barkan et al. (2012) used computational methods to infer a PPR-RNA recognition mechanism where a combination of position 6 amino acid residues in a PPR motif and the 1′ position of the next PPR motif recognizes one nucleotide in the RNA substrate. As such, the tandem PPR motifs are decoded to a nucleotide sequence that specifies the substrate RNA of the PPR protein. It was validated by recoding a PPR protein to bind novel RNA sequences in vitro. However, we failed to make the connection between EMP5 and rpl16.
The E+ and DYW Motifs of EMP5 Are Not Essential for PPR Protein Function
PPR proteins are classified into P, PPR-E, and PPR-DYW subclasses based on the presence of additional C-terminal motifs (Lurin et al., 2004). The PPR repeats are proposed to recognize the target RNA sequences (Shikanai, 2006). In MEF11, the second PPR repeat was shown to be crucial for the specific editing of cox3-422, nad4-124, and ccb203-344 (Verbitskiy et al., 2010). The DYW domain that showed significant similarity to cytidine deaminases was proposed to have catalytic editing activity (Salone et al., 2007). Indeed, its presence is correlated with presence of RNA editing in plant evolution (Fujii and Small, 2011). Although PPR proteins, such as CRR4, CLB19, MEF9, and SLO1, all lacking a DYW domain, still possess the function of C-to-U RNA editing (Kotera et al., 2005; Chateigner-Boutin et al., 2008; Sung et al., 2010; Takenaka, 2010), a recent report showed that the DYW domain can be supplied in trans to the CRR4 protein and it is essential for the editing (Boussardon et al., 2012). Therefore, the hypothesis that the DYW domain functions as the deaminase enzyme remains. Up to this work, all the PPR proteins involved in editing contain an E domain, but the E domain lacks any obvious catalytic characteristics, suggesting that the E domain is indispensable for editing with unknown function (Shikanai, 2006; Okuda et al., 2007). It is possible that the E domain mediates protein–protein interaction to recruit another protein with deaminase activity, which may include PPR-DYW proteins.
In the emp5-4 allele, a Mu1 element is inserted in the middle of E+ motif, disrupting the E+ motif and DYW motif. RT-PCR analysis using several sets of primers revealed the presence of two transcripts where one contained the 5′ region of the Emp5 gene and ended inside the Mu1, and the other contained the 3′ region of the Emp5 gene, but no transcript across the Mu1 element existed. We analyzed the two transcripts for open reading frames that would possibly encode proteins with a scenario like the PPR-E protein with a trans-supplied DYW protein. As shown in Supplemental Figure 3 online, the translation of first transcript is predicted to terminate by a stop codon, TAA, only adding two amino acids encoded by the Mu1 TIR sequence. This will produce a truncated EMP5 protein, similar to a PPR-E protein, and if translated, it should be able to target to mitochondria. The second transcript predicts only one open reading frame in frame with the DYW domain. The translation adds five amino acid residues (MAIIS) from the Mu1 sequence to the N terminus of this hypothetical protein. However, the N terminus sequence does not predict a mitochondrion signal peptide by currently available algorisms. Therefore, it is highly unlikely that the protein would be targeted to the mitochondrion, even if it can be translated. Alternatively spliced transcripts were not detected, although we intentionally set the conditions favoring the amplification of potentially spliced emp5-4 transcripts. This leads us to conclude that the insertion in the emp5-4 allele causes a possible deletion of the C-terminal E+ domain and the entire DYW domain. However, a truncated version of the EMP5-4 protein may be produced. Genetic analysis indicated that the emp5-4 allele appeared normal in growth and development. And most of the editing events targeted by EMP5 showed similar editing level comparing to the wild type, except the editing of rpl16-458, which is considered critical showed partial editing. In addition, rpl16-444 and nad9-190 editing efficiency was also changed slightly. The presence of editing in emp5-4 that lacks the E+ and DYW motif indicates that E+ and DYW domains are not essential for EMP5 editing function. This conclusion is consistent with the plastid-located CRR22, CRR28, and ORGANELLE TRANSCRIPT PROCESSING28 (OTP28) and the mitochondrial factors MEF3 and MEF11 (Okuda et al., 2009, 2010; Verbitskiy et al., 2010, 2012; Zehrmann et al., 2011). However, the DYW domain of MEF1 cannot be destroyed without severe effects on its function in editing (Zehrmann et al., 2009). Therefore, it is possible that EMP5 associates with a DYW container partner, which can almost completely complement E+ and DYW motif truncation and carry out editing, although inefficiently. However, for MEF1, there is no such DYW container partner to associate, so the DYW domain cannot be deleted or mutated.
METHODS
Plant Materials
The emp5-1 reference allele was isolated from the UniformMu population by introgressing Mu active lines into the inbred W22 genetic background (McCarty et al., 2005). The wild-type plants were either siblings of the mutant or W22. The emp5-2, emp5-3, and emp5-4 alleles were isolated from the TUSC population (Pioneer Hi-Bred International) by PCR screening with Emp5-specific primers and Mu primers. The maize (Zea mays) plants were grown in the experimental field at the Chinese University of Hong Kong under natural conditions. Rice (Oryza sativa ssp japonica; cv Nipponbare) was used as the plant material for Agrobacterium tumefaciens–mediated rice transformation.
Light Microscopy of Cytological Sections
To enable a precise comparison, wild-type and emp5-1 mutant kernels were harvested from the same ear of a self-pollinated heterozygous plant at 8 and 13 DAP. The kernel was cut along longitudinal axis, and the slice containing the embryo was fixed for 1 d at room temperature in 4% paraformaldehyde. The fixed material was dehydrated in an ethanol gradient series (50, 70, 85, 95, and 100% ethanol). After clearing with xylene and paraffin wax infiltration, the sample was embedded and sectioned at 6- to 10-µm thickness under a Leica 2035 Biocut. The sections were stained with Johansen’s Safranin O and Fast Green and observed with a Nikon ECLIPSE 80i microscope.
Immunohistochemistry Analysis
The sections containing 13 DAP wild-type and emp5-1 seeds were deparaffinized, then air-dried for 15 min, rinsed with PBS, and incubated in 0.2 to 0.3% Triton X-100 in PBS for 15 min. Nonspecific antibody binding was blocked with 3% BSA for 2 h. The tissues were then incubated with the primary antibody (a gift from Gregorio Hueros, Universidad de Alcalá, Spain; 1:400 diluted in 1% BSA and 0.05% Triton X-100 in PBS) at 4°C overnight. The slides were washed three times with PBS, 15 min each to completely remove the primary antibody. Then the slides were incubated with Alexa Fluor-568 anti-rabbit secondary antibodies (Invitrogen) for immunofluorescent detection (diluted 1:500 in PBS, room temperature, 1 h). After washing three times with PBS, the slides were viewed and imaged under an Olympus FluoView FV1000 confocal microscope. Control sections were incubated with PBS without the primary antibody and subsequently processed as described above.
DNA Gel Blot Analysis and Inverse PCR Cloning
For cosegregation analysis, maize genomic DNA was extracted from emp5-1/+ and wild-type seedlings using the urea extraction method as described previously (Tan et al., 2011) and digested by HindIII. DNA gel blot analysis was performed as previously described (Tan et al., 2011). The hybridization probe was the ∼1-kb HinfI fragment of the Mu1 element internal sequence. Because Mu1 and Mu2 share a high degree of sequence identity, this probe hybridizes to both elements.
For inverse PCR cloning of the Mu flanking sequences in the 3.4-kb HindIII fragment, genomic DNA of heterozygous emp5-1 plants was digested by HindIII and separated on 0.7% agarose gel. The 3.4-kb fragment was enriched by cutting a small gel slice around 3.4 kb and the DNA was purified. The DNA was self-ligated at 50 ng/μL concentration overnight at 4°C. Then, the ligated DNA was digested with NotI. Mu1 and Mu2 elements contain a NotI site in the internal sequences; hence, the digestion linearized the ligated circular DNA for efficient amplification. This DNA was used as the template in the inverse PCR amplification of the Mu flanking sequences. Annealing temperature for the first-round PCR is 60°C and Mu-specific degenerated TIR6 primer (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′) was used. The second-round PCR was performed with a 56°C annealing temperature and Mu1-62 primer (5′-CCCTTCCCTCTTCGTCCATAAT-3′). The amplified fragment was cloned into pCR4-TOPO and sequenced.
RNA Extraction and RT-PCR
Approximately 100 mg of fresh tissue was quickly frozen in liquid nitrogen and ground to fine powder with a mortar and pestle. Total RNA was extracted with 1 mL Trizol reagent according to the manufacturer’s instructions (Invitrogen). After isopropanol precipitation, the RNA was resuspended in 40 μL RNase-free water and treated with RNase-free DNase. RT-PCR analyses were performed using the SuperScript III One-Step RT-PCR system according to the manufacturer’s instructions (Invitrogen). RT-PCR for the expression pattern analysis of Emp5 in maize organs was performed with primers Emp5-F2 and Emp5-R2 and 57°C annealing temperature for 30 cycles. For analysis of Emp5 expression in rice transgenic lines, RT-PCR was performed with primers OsEmp5-F and OsEmp5-R and 55°C annealing temperature for 28 cycles. The information of all primers is listed in Supplemental Table 1 online.
Subcellular Localization of EMP5
To generate a translational protein fusion between the EMP5 signal peptide and GFP, full-length Emp5 and Emp5N469 fragment was amplified by PCR from maize inbred line W22 and cloned into pENTR/D-TOPO (Invitrogen), respectively. The fusion was introduced to binary vector pGWB5 (a gift from Tsuyoshi Nakagawa, Shimane University) by Gateway site-specific recombination. The OsEMP5N372:GFP fusion expression construct was constructed similarly. These fusion proteins were placed under the cauliflower mosaic virus 35S promoter for constitutive expression. Then, these constructs were transformed into Agrobacterium strain EHA105. The resulting strains harboring full-length EMP5:GFP and EMP5N469:GFP expression plasmid were used to transform Arabidopsis thaliana Columbia ecotype by the flower dip method (Clough and Bent, 1998). The transgenic Arabidopsis was identified with PCR amplification of the Hpt gene in pGWB5 vector with primer Hpt-F3 and Hpt-R3. The protoplasts were isolated from the transgenic leaves by digesting with an enzyme solution (1.5% cellulose R10, 0.3% pectolyase Y23, 20 mM MES, pH 5.7, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 0.1% BSA). Using established protocols (van Herpen et al., 2010), the EHA105 strain harboring the OsEMP5N372:GFP fusion construct was infiltrated into tobacco (Nicotiana tabacum) leaves to transiently express the fusion protein. The infiltrated tobacco leaves, transgenic Arabidopsis leaf samples, and protoplasts were used for GFP and MitoTracker red signals detection by an Olympus FluoView FV1000 confocal microscope. The working concentration of MitoTracker was 30 nM, and the samples were incubated at 37°C for 30 min.
In vitro chloroplast protein import assay was performed as described previously (Cline, 1986; Martin et al., 2009). Emp5 cDNA was placed under the SP6 promoter in pGem-3Z vector. RNA transcripts of this construct were produced by in vitro transcription with SP6 polymerase (Promega). The protein was translated with a homemade wheat germ translation system in the presence of 3H-Leu (50 mCi, 3000 Ci/mol). Pea (Pisum sativum cv Laxton’s Progress 9 Improved) used for chloroplast isolation were grown as described (Cline, 1986). The chloroplast import was performed as described (Martin et al., 2009).
Analysis of Mitochondrial RNA Editing
For RNA editing analysis in the wild type and the emp5-1 allele, total RNAs were isolated from the immature embryos and endosperms by carefully removing the pericarp. For the emp5-4 allele and rice RNAi transgenic lines, total RNAs were isolated from seedling leaves, all using the Trizol reagent according to the instructions of the manufacturer (Invitrogen). The RNA was treated with DNase I (New England Biolabs), and the complete removal of DNA was checked by PCR on genomic DNA. Then, the DNA-free RNAs were reverse transcribed with random hexamers and the high-fidelity reverse transcriptase SuperScript III (Invitrogen). Full sequences of total 35 protein-coded maize mitochondrial genes and rice mitochondrial rpl16 gene were amplified by PCR. The RT-PCR products were sequenced directly. This analysis was done with three biological replicates using seeds of different days after pollination. These primers are listed in Supplemental Table 1 online.
Rice Transformation
For OsEmp5 RNAi vector construction, a 518-bp Emp5 (position: 1362 to 1879 bp from ATG) fragment was PCR amplified with primers OsEmp5-KpnI and OsEmp5-BamHI, and OsEmp5-SpeI and OsEmp5-SacI, respectively (see Supplemental Table 1 online). The PCR products were digested with BamHI and KpnI, SpeI, and SacI, respectively, and ligated into the binary vector pTCK303. The resulting RNAi vector, pTCK303-OsEmp5, was introduced into the wild-type cultivar Nipponbare using Agrobacterium-mediated transformation. Transformation was performed as previously described (Hiei et al., 1994). DNA gel blot analysis of transgenic lines was performed as previously described (Tan et al., 2011), and the hpt gene amplified from vector pTCK303 with primer Hpt-F1 and Hpt-R1 (see Supplemental Table 1 online) was used as a probe.
Accession Numbers
Sequence data for Emp5 genomic DNA, cDNA, alleles emp5-1, emp5-2, emp5-3, and emp5-4 can be found in the GenBank/EMBL database under accession numbers JX308938, JX308939, JX308940, JX308941, JX308942, and JX308943, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of −40 to +20 Sequences of All 15 RNA Editing Sites in Seven Mitochondrial Gene Transcripts Changed in emp5-1.
Supplemental Figure 2. Alignment of −40 to +20 Sequences of 10 RNA Editing Sites in Four Mitochondrial Gene Transcripts Decreased and Increased in emp5-1.
Supplemental Figure 3. The Hypothetical Proteins as Predicted Based on the Two Transcripts Detected in the emp5-4 Mutant Seeds.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Kenneth Cline (University of Florida) for performing pea chloroplast import assay of Emp5, Gregorio Hueros (Universidad de Alcalá, Spain) for the anti-BETL2 antibody, and Tsuyoshi Nakagawa (Shimane University, Japan) for the pGWB vectors. This work was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region to B.-C.T. (Project 473512)
AUTHOR CONTRIBUTIONS
Y.-J.L., Z.-H.X., and B.-C.T. designed and performed the research. R.M. isolated the alleles of emp5-2 to -4. Y.-J.L., R.M., and B.-C.T. analyzed the data. Y.-J.L. and B.-C.T. wrote the article.
Glossary
- PPR
pentatricopeptide repeat
- RNAi
RNA interference
- DAP
days after pollination
- GFP
green fluorescent protein
References
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen R.J., Johal G.S., Crane V.C., Tossberg J.T., Schnable P.S., Meeley R.B., Briggs S.P. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Heller W.P., Sun T., Babina A.M., Friso G., van Wijk K.J., Hanson M.R. (2012). RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA 109: E1453–E1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Knight W., Hanson M. (2010). Natural variation in Arabidopsis leads to the identification of REME1, a pentatricopeptide repeat-DYW protein controlling the editing of mitochondrial transcripts. Plant Physiol. 154: 1966–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner G.V., Mörl M., Wissinger B., Brennicke A., Schmelzer C. (1995). RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol. Gen. Genet. 246: 739–744 [DOI] [PubMed] [Google Scholar]
- Boussardon, C., Salone, V., Avon, A., Berthome, R., Hammani, K., Okuda, K., Shikanai, T., Small, I. and Lurin, C. (2012). Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24: 3684–3694. [DOI] [PMC free article] [PubMed]
- Chateigner-Boutin A.L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L.F., Lurin C., Small I., León P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., Small I. (2010). Plant RNA editing. RNA Biol. 7: 213–219 [DOI] [PubMed] [Google Scholar]
- Clifton S.W., et al. (2004). Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136: 3486–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. (1986). Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J. Biol. Chem. 261: 14804–14810 [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Covello P.S., Gray M.W. (1989). RNA editing in plant mitochondria. Nature 341: 662–666 [DOI] [PubMed] [Google Scholar]
- Cushing D.A., Forsthoefel N.R., Gestaut D.R., Vernon D.M. (2005). Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221: 424–436 [DOI] [PubMed] [Google Scholar]
- Ding Y.H., Liu N.Y., Tang Z.S., Liu J., Yang W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Meeley R., Scanlon M.J. (2002). Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Small I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191: 37–47 [DOI] [PubMed] [Google Scholar]
- Giegé P., Brennicke A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan A.K. (2011). To thy proteins be true: RNA editing in plants. Plant Physiol. 156: 453–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F., Viehoever P., Weisshaar B., Knoop V. (2009). A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 37: 5093–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J.M., Lamattina L., Bonnard G., Weil J.H., Grienenberger J.M. (1989). RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341: 660–662 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Dal Prà M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., Dickinson H.G., Consonni G. (2007). empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., des Francs-Small C.C., Takenaka M., Tanz S.K., Okuda K., Shikanai T., Brennicke A., Small I. (2011). The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 286: 21361–21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Okuda K., Tanz S.K., Chateigner-Boutin A.L., Shikanai T., Small I. (2009). A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. (1989). RNA editing in plant mitochondria. Science 246: 1632–1634 [DOI] [PubMed] [Google Scholar]
- Hueros G., Royo J., Maitz M., Salamini F., Thompson R.D. (1999). Evidence for factors regulating transfer cell-specific expression in maize endosperm. Plant Mol. Biol. 41: 403–414 [DOI] [PubMed] [Google Scholar]
- Hunt M.D., Newton K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D., Kahlau S., Bock R. (2008). Faithful editing of a tomato-specific mRNA editing site in transgenic tobacco chloroplasts. RNA 14: 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T., Nakamura T., Watanabe M., Sugita M., Toriyama K. (2008). Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 55: 619–628 [DOI] [PubMed] [Google Scholar]
- Kim S.R., Yang J.I., Moon S., Ryu C.H., An K., Kim K.M., Yim J., An G. (2009). Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 59: 738–749 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Matsuo M., Sakamoto K., Wakasugi T., Yamada K., Obokata J. (2008). Two RNA editing sites with cis-acting elements of moderate sequence identity are recognized by an identical site-recognition protein in tobacco chloroplasts. Nucleic Acids Res. 36: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Lu Y., Li C., Wang H., Chen H., Berg H., Xia Y. (2011). AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J. 67: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Guyon V., Meurer J., Wienand U., Brettschneider R. (2012). An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell 24: 3087–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.R., Harwood J.H., McCaffery M., Fernandez D.E., Cline K. (2009). Localization and integration of thylakoid protein translocase subunit cpTatC. Plant J. 58: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61 [DOI] [PubMed] [Google Scholar]
- Newton K.J., Mariano J.M., Gibson C.M., Kuzmin E., Gabay-Laughnan S. (1996). Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19: 277–286 [DOI] [PubMed] [Google Scholar]
- Notsu Y., Masood S., Nishikawa T., Kubo N., Akiduki G., Nakazono M., Hirai A., Kadowaki K. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268: 434–445 [DOI] [PubMed] [Google Scholar]
- Okuda K., Chateigner-Boutin A.L., Nakamura T., Delannoy E., Sugita M., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2009). Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Hammani K., Tanz S.K., Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2010). The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 61: 339–349 [DOI] [PubMed] [Google Scholar]
- Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 104: 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281: 37661–37667 [DOI] [PubMed] [Google Scholar]
- Olsen O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 233–267 [DOI] [PubMed] [Google Scholar]
- Pate J.S., Gunning B.E.S. (1972). Transfer cells. Annu. Rev. Plant Physiol. 23: 173–196 [Google Scholar]
- Sakamoto W., Kondo H., Murata M., Motoyoshi F. (1996). Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8: 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. (2007). A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581: 4132–4138 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sheridan W.F., Neuffer M.G. (1980). Defective kernel mutants of maize II. Morphological and embryo culture studies. Genetics 95: 945–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2006). RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 63: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D., Mbelo S., Vernoud V., Gendrot G., Dedieu A., Chambrier P., Dauzat M., Heurtevin L., Guyon V., Takenaka M., Rogowsky P.M. (2012). PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24: 676–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, T.Y., Tseng, C.C., and Hsieh, M.H. (2010). The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63: 499–511 [DOI] [PubMed] [Google Scholar]
- Takenaka M. (2010). MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol. 152: 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. (2012). Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 109: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, B.C., Chen, Z., Shen, Y., Zhang, Y., Lai, J., and Sun, S.S. (2011). Identification of an active new mutator transposable element in maize. G3 (Bethesda) 1: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Funk H.T., Schmitz-Linneweber C., Poltnigg P., Sabater B., Martin M., Maier R.M. (2005). Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43: 708–715 [DOI] [PubMed] [Google Scholar]
- Tzafrir I., Pena-Muralla R., Dickerman A., Berg M., Rogers R., Hutchens S., Sweeney T.C., McElver J., Aux G., Patton D., Meinke D. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M., Marienfeld J.R., Brandt P., Brennicke A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15: 57–61 [DOI] [PubMed] [Google Scholar]
- van Herpen T.W.J.M., Cankar K., Nogueira M., Bosch D., Bouwmeester H.J., Beekwilder J. (2010). Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE 5: e14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitskiy D., Merwe J.A., Zehrmann A., Härtel B., Takenaka M. (2012). The E-class PPR protein MEF3 of Arabidopsis thaliana can also function in mitochondrial RNA editing with an additional DYW domain. Plant Cell Physiol. 53: 358–367 [DOI] [PubMed] [Google Scholar]
- Verbitskiy D., Zehrmann A., van der Merwe J.A., Brennicke A., Takenaka M. (2010). The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J. 61: 446–455 [DOI] [PubMed] [Google Scholar]
- Yuan H., Liu D. (2012). Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 70: 432–444 [DOI] [PubMed] [Google Scholar]
- Zehrmann A., Verbitskiy D., Härtel B., Brennicke A., Takenaka M. (2011). PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol. 8: 67–70 [DOI] [PubMed] [Google Scholar]
- Zehrmann A., Verbitskiy D., van der Merwe J.A., Brennicke A., Takenaka M. (2009). A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.