This work shows that DELLA proteins physically interact with a member of the BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI) protein family and bind to GA-regulated gene promoters to repress GA-induced responses like seed germination, juvenile-to-adult phase transition, and flowering.
Abstract
DELLA proteins, consisting of GA INSENSITIVE, REPRESSOR OF GA1-3, RGA-LIKE1 (RGL1), RGL2, and RGL3, are central repressors of gibberellin (GA) responses, but their molecular functions are not fully understood. We isolated four DELLA-interacting RING domain proteins, previously designated as BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI), BOI-RELATED GENE1 (BRG1), BRG2, and BRG3 (collectively referred to as BOIs). Single mutants of each BOI gene failed to significantly alter GA responses, but the boi quadruple mutant (boiQ) showed a higher seed germination frequency in the presence of paclobutrazol, precocious juvenile-to-adult phase transition, and early flowering, all of which are consistent with enhanced GA signaling. By contrast, BOI overexpression lines displayed phenotypes consistent with reduced GA signaling. Analysis of a gai-1 boiQ pentuple mutant further indicated that the GAI protein requires BOIs to inhibit a subset of GA responses. At the molecular level, BOIs did not significantly alter the stability of a DELLA protein. Instead, BOI and DELLA proteins are targeted to the promoters of a subset of GA-responsive genes and repress their expression. Taken together, our results indicate that the DELLA and BOI proteins inhibit GA responses by interacting with each other, binding to the same promoters of GA-responsive genes, and repressing these genes.
INTRODUCTION
Gibberellic acid (GA) is a plant hormone that regulates various transition processes during plant development, including seed germination, the juvenile-to-adult phase transition, and flowering (Sun and Gubler, 2004). These GA-regulated processes are exemplified by the phenotypes seen in the GA-deficient mutant, ga1, which harbors a mutation in ent-copalyl diphosphate synthase (Sun and Kamiya, 1994). Ent-copalyl diphosphate synthase is a GA biosynthetic enzyme responsible for converting geranylgeranyl pyrophosphate to copalyl pyrophosphate, which is the first committed step in GA biosynthesis. The ga1 mutant shows defects in various developmental transition processes; it does not germinate unless GA is provided exogenously, and it shows delays in both the juvenile-to-adult phase transition and flowering (Koornneef and van der Veen, 1980; Dill and Sun, 2001; Tyler et al., 2004). In addition, the ga1 mutant displays other pleiotropic mutant phenotypes, including dwarfism, male sterility, increased chlorophyll accumulation, and altered responses to environmental stresses (e.g., cold temperature and osmotic stress) (Cheng et al., 2004; Magome et al., 2004; Thomas and Sun, 2004; Tyler et al., 2004; Yu et al., 2004; Achard et al., 2008). These ga1-associated phenotypes are shared either partially or wholly by various other GA signaling mutants (Koornneef et al., 1985; Talon et al., 1990).
Genetic analyses have identified three key GA signaling components in both rice (Oryza sativa) and Arabidopsis thaliana: the GA receptors (GA INSENSITIVE DWARF1 [GID1] in rice and GID1a, b, and c in Arabidopsis); a group of repressor proteins called the DELLAs (SLENDER-RICE1 [SLR1] in rice and GA INSENSITIVE [GAI], REPRESSOR OF GA1-3 [RGA], RGA-LIKE1 [RGL1], RGL2, and RGL3 in Arabidopsis); and an F-box protein (GID2 in rice and SLEEPY1 [SLY1] in Arabidopsis) (Koornneef et al., 1985; Peng et al., 1997; Silverstone et al., 1997, 1998; Steber et al., 1998; Dill and Sun, 2001; Ikeda et al., 2001; Peng and Harberd, 2002; McGinnis et al., 2003; Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). Among these three components, the DELLAs repress all GA responses, while GID1 and SLY1 (as an SCFSLY1 complex) activate GA responses by binding and ubiquitylating the DELLAs, leading to their degradation in the presence of GA (Peng et al., 1997; Silverstone et al., 1998; Ikeda et al., 2001; Lee et al., 2002; Wen and Chang, 2002; Cheng et al., 2004; Dill et al., 2004; Gomi et al., 2004; Tyler et al., 2004; Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006; Piskurewicz and Lopez-Molina, 2009). The DELLAs are a subfamily of the GRAS (GAI, RGA, SCARECROW [SCR]) protein family, which comprises the DELLAs, SCR, SHR, and SCR-like proteins (SCLs) (Pysh et al., 1999; Bolle, 2004). GRAS family members act in the nucleus as either transcriptional corepressors or transcription factors. The DELLAs differ from other GRAS family members in that they possess N-terminal DELLA and VHYNP motifs, which are needed to interact with GID1 in the presence of GA (Peng et al., 1997; Silverstone et al., 1998; Griffiths et al., 2006; Willige et al., 2007). The C-terminal domains of the DELLAs also contain additional motifs (LHRI-VHIID-LHRII-PFYRE-SAW) that participate in dimerization and other protein–protein interactions (Dill et al., 2004; Bai et al., 2012; Gallego-Bartolomé et al., 2012). In the presence of GA, GID1 binds to the DELLAs, SCFSLY1 recognizes the DELLA-GA-GID1 complexes and ubiquitylates the DELLAs, and then the ubiquitylated DELLAs are degraded by the 26S proteasome (Sun and Gubler, 2004; Thomas and Sun, 2004).
Despite their central position in GA signaling, the molecular functions of the DELLAs have not yet been clearly defined. Protein–protein interactions have been suggested to play key roles in DELLA function. For example, DELLAs have been shown to regulate gene expression by binding to various transcription factors, including the PHYTOCHROME-INTERACTING FACTORs (PIFs), ALCATRAZ, and MYC2 (de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Gallego-Bartolomé et al., 2010; Hong et al., 2012). In the case of the PIFs, the DELLAs have been shown to interact with PIFs and inhibit their binding to target promoters (de Lucas et al., 2008; Feng et al., 2008). Other characterized protein–protein interactions include the DELLA–JAZ9 interaction, whereby JASMONATE-ZIM-DOMAIN PROTEIN9 (JAZ9) prevents DELLAs from binding and inhibiting PIFs (Yang et al., 2012); the DELLA–JAZ1 interaction, whereby DELLAs prevent JAZ1 from binding and inhibiting MYC2 (Hou et al., 2010); and the DELLA–SCL3 interaction, whereby DELLAs bind and inhibit SCL3 (Zhang et al., 2011). In addition to these protein–protein interactions, DELLAs have also been reported to target promoters and regulate the expression levels of genes, such as SCL3 and XERICO, which is a RING domain protein capable of promoting abscisic acid synthesis (Ko et al., 2006; Zentella et al., 2007). However, it is not clear how DELLAs target such promoters and regulate their gene expression.
Beyond the three key components of GA signaling discussed above (GID1, DELLAs, and SCFSLY1), a few additional components have been identified as being related with the function of DELLAs. For example, SPINDLY (SPY) is an O-linked N-acetylglucosamine transferase that, similar to the DELLAs, can repress GA responses (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Shimada et al., 2006; Silverstone et al., 2007). Genetic studies have shown that a spy loss-of-function mutation suppresses the rga-Δ17 mutation (a dominant-active mutation of RGA), suggesting that the function of SPY is required for that of the DELLAs (Silverstone et al., 2007). In the context of cytokinin signaling, SPY was shown to transfer N-acetyglucosamine (GlcNAc) to TCP (TB1, CYC, PCF) transcription factors, thereby activating/repressing them and enhancing cytokinin signaling (Steiner et al., 2012). Analogously, SPY has been suggested to transfer GlcNAc to DELLAs to repress GA responses. Another GA signaling component, rice EARLY FLOWERING1 (EL1), encodes casein kinase I, which represses GA responses through SLR1 (a rice DELLA protein) (Dai and Xue, 2010). Phosphorylation of SLR1 by EL1 stabilizes SLR1 and enhances its activity. However, we do not yet know how GlcNAc or phosphorylation enhances DELLA activity.
RING domain proteins regulate various plant developmental processes (Hotton and Callis, 2008). Among the 469 RING proteins in Arabidopsis, the RING domains can be classified into eight different groups based on their zinc-coordinating amino acid sequences (Stone et al., 2005). RING domains are known to participate in protein–protein interactions and protein ubiquitylation. The majority of RING proteins are predicted to function as ubiquitin E3 ligases, either as single subunit E3s wherein the RING protein provides both the substrate binding site and an E2 binding site, or as multisubunit E3s wherein the RING protein provides an E2 binding site and other components provide the substrate binding sites (Stone et al., 2005; Mazzucotelli et al., 2006; Hotton and Callis, 2008). The multisubunit E3s include the Skp1-Cullin-F-box (SCF)-type E3 ligase, in which a RING protein (RBX1) binds to cullin and recruits ubiquitin E2-conjugating enzyme to the complex, while the F-box protein provides the substrate binding site. RBX1-containing SCF-type multisubunit E3 ligases have been shown to play central roles in hormone signaling, such as the involvement of SCFTIR1/AFB1-3, SCFEBF1/2, SCFSLY1, SCFCOI1, and SCFMAX2 in auxin, ethylene, GA, jasmonate, and strigolactone signaling, respectively (Santner and Estelle, 2010; Kelley and Estelle, 2012). They mediate hormone signaling by ubiquitylating repressor proteins (leading to their degradation) in the presence of a cognate hormone (AUX/indole-3-acetic acid-auxin, DELLAs-GA, and JAZs-jasmonic acid [JA]) (Gray et al., 2001; McGinnis et al., 2003; Sasaki et al., 2003; Griffiths et al., 2006; Chini et al., 2007; Thines et al., 2007) or by removing positive transcription factors in the absence of a cognate hormone (EIN3-ethylene) (Guo and Ecker, 2003; Potuschak et al., 2003). The target of SCFMAX2 has not yet been identified (Stirnberg et al., 2002). Cul3-BTBETO1/EOL1/2 and CUL3-BTBNPR3/4, which target type-2 ACSs (ACS4, 5, and 9) and NPR1, in ethylene and salicylate signaling, respectively, are multisubunit RING E3 ligases that possess a broad complex/tramtrack/bric-a-brac (BTB) protein as the substrate binding protein (Wang et al., 2004; Christians et al., 2009; Fu et al., 2012). Many single subunit RING E3 ligases have also been characterized. For example, CONSTITUTIVE PHOTOMORPHOGENIC1 is an E3 ligase that ubiquitylates positive light signaling components, such as LONG HYPOCOTYL5, LONG HYPOCOTYL IN FAR-RED1, LONG AFTER FAR-RED LIGHT1, phytochrome A, and cryptochrome 2, in concert with the RING proteins SUPPRESSOR OF PHYA-105 (SPA1 to SPA4) in the dark (Osterlund et al., 2000; Shalitin et al., 2002; Seo et al., 2003, 2004; Laubinger et al., 2004; Jang et al., 2005; Yang et al., 2005; Lau and Deng, 2012). Numerous single subunit RING E3 ligases have been implicated in plant stress responses, such as the DRIP1/2-ubiquitylating DREB2A, the Rma1H1-ubiquitylating PIP2;1, and the RGLG2-ubiquitylating ETHYLENE RESPONSE FACTOR53 in drought responses (Qin et al., 2008; Lee et al., 2009; Cheng et al., 2012).
Here, we report that four Arabidopsis RING proteins that we found to have been previously identified as BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI), BOI-RELATED GENE1 (BRG1), BRG2, and BRG3, interact with DELLAs and repress a subset of GA responses, including seed germination, the juvenile-to-adult phase transition, and flowering. The RING domain of the BOI is necessary for its function, but the BOIs do not significantly change the stability of DELLAs. Instead, the BOI is targeted to the same promoters as the DELLAs, suggesting that these RING proteins and DELLAs form a complex, bind to a subset of GA-responsive genes, and regulate their expression.
RESULTS
DELLA Proteins Interact with Four Closely Related RING Domain Proteins, the BOIs
To gain insight into how DELLA proteins work, we used yeast two-hybrid screening to search for DELLA-interacting proteins. We identified an Arabidopsis RING domain protein, At4g19700, as a DELLA-interacting protein. A BLAST search showed that the Arabidopsis genome possesses three closely related homologs (>6e−39): At5g45100, At1g79110, and At3g12920. These four RING domain proteins have HCa-type RING motifs with three amino acids between the 4th and 5th residues of zinc-coordinating residues (see Supplemental Figure 1A online) (Stone et al., 2005). Previously, At4g19700 was shown to interact with BOTRYTIS SUSCEPTIBLE1 (BOS1) protein; thus, the protein was named BOS1 INTERACTOR (BOI), and the three related proteins were named BOI-RELATED GENE1 (BRG1; At5g45100), BRG2 (At1g79110), and BRG3 (At3g12920) (Luo et al., 2010). In our phylogenetic analysis, BOI and the BRGs clustered together with four closely related rice RING domain proteins rather than with other Arabidopsis RING domain proteins (see Supplemental Figure 1B and Supplemental Data Set 1 online), suggesting that these proteins might have similar functions in plants. For succinctness, we will hereafter use “BOIs” to collectively refer to BOI and the BRGs.
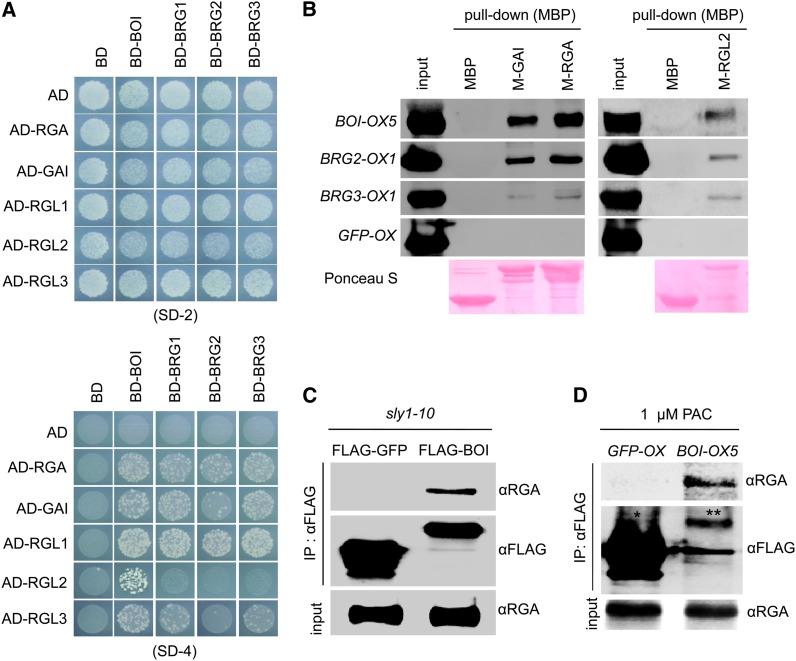
We used yeast two-hybrid and immunoprecipitation assays to systematically determine if the DELLAs could interact with the BOIs. Full-length DELLAs are capable of activating transcription when fused to the DNA binding domain of GAL4 in yeast, so we individually fused five DELLAs (GAI, RGA, RGL1, RGL2, and RGL3) to the GAL4 activation domain and the four BOIs to the DNA binding domain of GAL4. The presence of an interaction was judged by the growth of transformed yeast in selection medium lacking adenine, His, Leu, and Trp. For the most part, a yeast strain grew well in the selection media when it possessed one DELLA and one BOI (Figure 1A). There were two exceptions: For RGL2, yeast grew with BOI but not with the BRGs; and for RGL3, yeast grew well with all of the BOIs except BRG2. However, because yeast two-hybrid assays sometimes fail to identify true interactions, it is unclear whether these exceptions indicate a lack of protein–protein interaction, the presence of a very weak interaction, or an anomaly associated with the yeast two-hybrid assay.
Figure 1.
BOI Proteins Interact with DELLA Proteins.
(A) Yeast two-hybrid data showing interactions between the four BOIs (BOI, BRG1, BRG2, and BRG3) and five DELLAs (GAI, RGA, RGL1, RGL2, and RGL3). SD-2, minimal medium lacking Trp and Leu; SD-4, minimal medium lacking Trp, Leu, His, and adenine.
(B) MBP pull-down assay showing the interaction between plant-expressed FLAG-tagged BOIs and recombinant MBP-DELLAs. Extracts of 7-d-old light-grown stable transgenic seedlings expressing FLAG-tagged BOI, BRG2, BRG3, or GFP (BOI-OX5, BRG2-OX1, BRG3-OX1, or GFP-OX) were mixed with MBP-tagged GAI, RGA, RGL2, or MBP alone and precipitated with MBP resin. Precipitated BOIs were detected with an anti-FLAG antibody. Ponceau S shows the amount of MBP-fused protein stained by Ponceau S dye.
(C) Coimmunoprecipitation assay showing the interaction between FLAG-BOI and endogenous RGA in the sly1-10 mutant. FLAG-tagged BOI or FLAG-tagged GFP was transiently expressed in 10-d-old light-grown sly1-10 mutant by Agrobacterium tumefaciens infiltration and precipitated with an anti-FLAG antibody. Precipitated RGA was detected with an anti-RGA antibody (see Supplemental Figure 2 online). Input shows the amount of RGA detected by anti-RGA antibody.
(D) Coimmunoprecipitation assay showing the interaction between stably expressed FLAG-tagged BOI and endogenous RGA. Stable transgenic seedlings expressing either FLAG-tagged BOI (BOI-OX5) or FLAG-tagged GFP (GFP-OX) were grown on the medium containing 1 µM PAC under the white light condition for 10 d. After precipitating with an anti-FLAG antibody, coimmunoprecipitated endogenous RGA was detected with an anti-RGA antibody. *Immunoprecipitated GFP-FLAG; **immunoprecipitated BOI-FLAG. GFP-FLAG signal is much stronger than BOI-FLAG due to much higher expression level of GFP-FLAG.
[See online article for color version of this figure.]
We further probed the interaction by pull-down assays using recombinant DELLA proteins (GAI, RGA, and RGL2) and extracts from plants expressing three of the BOIs (BOI, BRG2, and BRG3). For the assay, we mixed maltose binding protein (MBP)–tagged recombinant DELLAs with extracts of 7-d-old light-grown stable transgenic Arabidopsis plants expressing FLAG-tagged BOIs or green fluorescent protein (GFP) (BOI-OX5, BRG2-OX1, BRG3-OX1, or GFP-OX) and tested if the DELLAs could precipitate the BOIs. We were not able to generate transgenic lines expressing BRG1, so all subsequent experiments were performed with BOI, BRG2, and BRG3. We found that GAI and RGA precipitated BOI, BRG2, and BRG3, but not GFP. In contrast with results obtained in yeast two-hybrid assays, RGL2 also precipitated BOIs but not GFP, indicating that RGL2 also interacts with BOIs (Figure 1B). FLAG tag has shown not to bind to DELLA proteins (Ariizumi et al., 2008). MBP alone did not pull down any of the BOIs, supporting our contention that DELLAs directly interact with BOIs.
An interaction was also observed between BOI and endogenous RGA in the sly1-10 mutant, which accumulates DELLA proteins due to its inability to ubiquitylate them in response to GA (McGinnis et al., 2003; Dill et al., 2004; Strader et al., 2004). Here, we transiently expressed FLAG-tagged BOI in 10-d-old light-grown sly1-10 mutant and tested if the expressed BOI could bind and precipitate endogenous RGA. As shown in Figure 1C, FLAG-tagged BOI precipitated endogenous RGA, whereas FLAG-tagged GFP did not. We also probed for interactions using a stable transgenic line expressing FLAG-tagged BOI or FLAG-tagged GFP in a wild-type background. For this assay, we grew plants for 10 d in white light conditions in the presence of 1 µM paclobutrazol (PAC), an inhibitor of GA biosynthesis, and determined if BOI bound and precipitated endogenous RGA. Consistent with an interaction between these proteins, FLAG-tagged BOI precipitated endogenous RGA, whereas FLAG-tagged GFP did not (Figure 1D). These results confirm that BOI interacts with endogenous RGA in vivo, suggesting that BOIs might regulate GA signaling by interacting with DELLA proteins.
The BOIs Inhibit a Subset of DELLA-Mediated GA Responses
To investigate the role of BOIs in GA signaling in Arabidopsis, we generated overexpression lines of BOI, BRG2, and BRG3 using the 35S promoter (see Supplemental Figure 3A online). We also isolated boi and brg single mutants that had T-DNAs in their exons and did not express full-length BOI mRNAs, suggesting that they are null alleles (see Supplemental Figures 3B and 3C online). Since BOIs are likely to function redundantly, we further crossed the single mutants to generate a boi quadruple mutant (boi-2 brg1-1 brg2-1 brg3-3; designated boiQ). We then characterized the boiQ and BOI-overexpressing lines and compared them with a DELLA pentuple mutant (dellaP). The dellaP mutant was generated by crossing five della mutants in a Columbia-0 (Col-0) background (rga-28, an introgressed gai-t6, rgl1-SK62, rgl2-SK54, and rgl3-3). The dellaP displayed constitutive GA responses even in the absence of GA treatment.
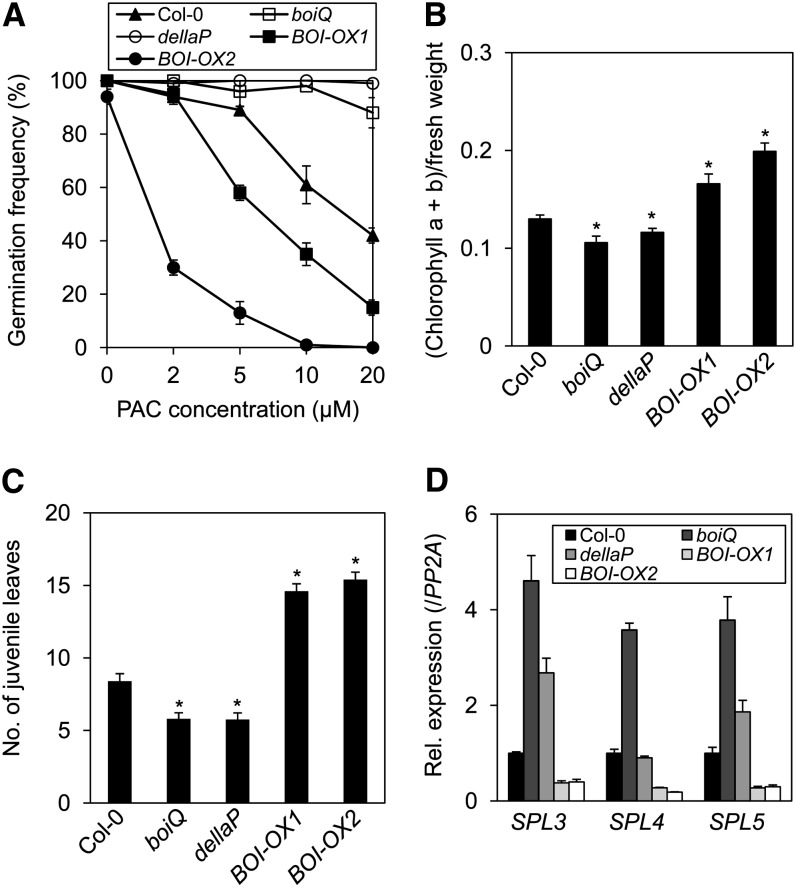
We first determined whether BOIs regulate GA responses during seed germination. Germination frequency was determined under white light in the presence of increasing concentrations of PAC. Consistent with the inhibitory roles of DELLA proteins in GA signaling during seed germination (Lee et al., 2002; Tyler et al., 2004; Cao et al., 2005; Piskurewicz and Lopez-Molina, 2009; Piskurewicz et al., 2009; Gallego-Bartolomé et al., 2010), only about half of the wild-type seeds germinated in the presence of 20 μM PAC, whereas 100% of the dellaP mutant seeds germinated (Figure 2A). Single mutations in the BOI and BRG genes did not significantly affect germination, but the boiQ quadruple mutant showed ∼90% germination in the presence of 20 μM PAC, suggesting that the BOIs inhibit GA responses during seed germination (Figure 2A; see Supplemental Figure 3D online). Consistent with this inhibitory role, all of the BOI overexpression lines germinated less than wild-type seeds in the presence of PAC (Figure 2A; see Supplemental Figure 3E online). Taken together, these results indicate that BOIs redundantly inhibit GA responses during seed germination.
Figure 2.
BOIs Inhibit GA Responses during Seed Germination and Vegetative Phase Transition.
(A) Increased seed germination in the boiQ and dellaP mutants in the presence of PAC. Nonimbibed seeds were germinated under white light in the presence of various concentrations of PAC. boiQ, BOI quadruple mutant (boi-2 brg1-1 brg2-1 brg3-3); dellaP, DELLA pentuple mutant (rga-28 gai-t6 rgl1-SK62 rgl2-SK54 rgl3-3); BOI-OX1 and -OX2, two independent BOI-overexpressing lines (sd, n = 3).
(B) Decreased chlorophyll accumulation in the boiQ and dellaP mutants. Chlorophyll was measured by spectroscopy using the 5th and 6th leaves of 20-d-old LD-grown plants, and the values were normalized with respect to the fresh leaf weight (sd, n = 6; *P < 0.05, Student’s t test).
(C) Accelerated juvenile-to-adult phase transition in the boiQ and dellaP mutants. The juvenile leaves were counted in 1-month-old LD-grown plants (sd, n = 10; *P < 0.05, Student’s t test).
(D) Increased expression of SPL mRNAs in the boiQ and dellaP mutants. Seventeen-day-old LD-grown plants were sampled for expression analysis at the 12th hour of light. SPL mRNA expression levels are indicated relative to those of PP2A and wild-type plants (([SPLx]/[PP2Ax])/([SPLCol-0]/[PP2ACol-0])) (sd, n = 3 biological replicates).
Chlorophyll accumulation has been shown to be inhibited by GA (Cheminant et al., 2011). Here, we report that BOIs also inhibit chlorophyll accumulation, which was determined by extracting and measuring chlorophyll from the 5th or 6th leaves of 20-d-old long-day (LD)-grown plants. Consistent with the role of GA, the dellaP and boiQ mutants had paler leaves and mildly but significantly lower chlorophyll levels compared with the wild type. By contrast, the BOI-OXs had higher chlorophyll levels (Figure 2B). Similar to the BOI-OXs, the other BRG-OX lines also had slightly but significantly higher chlorophyll accumulations (see Supplemental Figure 3F online). These results indicate that both the BOIs and the DELLAs promote chlorophyll accumulation.
The BOIs and DELLAs also both inhibited the juvenile-to-adult phase transition, as determined by the appearance of abaxial trichomes. When grown under the LD condition, wild-type plants developed abaxial trichomes on approximately the 9th leaf (Figure 2C). By contrast, dellaP mutant developed abaxial trichomes earlier, on the 6th leaf, supporting the notion that GA promotes this phase transition. The boiQ mutant also developed abaxial trichomes earlier, on the 5th or 6th leaf, while the BOI-OXs developed abaxial trichomes later, on the 14th leaf of BOI-OX1 and the 15th leaf of BOI-OX2. Similarly, the BRG-OXs also developed abaxial trichomes later than wild-type plants (see Supplemental Figure 3G online).
This phase transition is known to be promoted by a set of SQUAMOSA PROMOTER BINDING PROTEIN (SPL) transcription factors (Huijser and Schmid, 2011). To determine if the BOIs and DELLAs regulate the transition by altering SPL expression, we grew Arabidopsis under the LD condition and determined the expression levels of SPL3, -4, and -5. The boiQ mutant expressed higher levels of all three SPL mRNAs compared with the wild type, whereas the BOI-OXs expressed lower levels (Figure 2D). Overexpression of the other BRGs also decreased the levels of all three SPL mRNAs (see Supplemental Figure 3H online). Similar to boiQ, the dellaP mutant expressed higher levels of SPL3 and SPL5 mRNAs compared with the wild type; however, dellaP plants expressed the SPL4 mRNA at a level similar to that of wild-type plants. The increased levels of SPL mRNA in the dellaP mutant is consistent with previous reports showing a decrease in the levels of SPL mRNA in a transgenic line expressing a GA catabolic gene or in the triple gid1a-c mutant (Galvão et al., 2012; Porri et al., 2012). Taken together, our results suggest that BOIs and DELLAs both inhibit the juvenile-to-adult phase transition by repressing SPL expression but are not identical in their regulation of SPL4. A previous study reported that RGA also interacts with SPL3 and SPL9 at the protein level (Yu et al., 2012); thus, our results further suggest that DELLAs regulate SPLs at both the transcriptional and posttranslational levels.
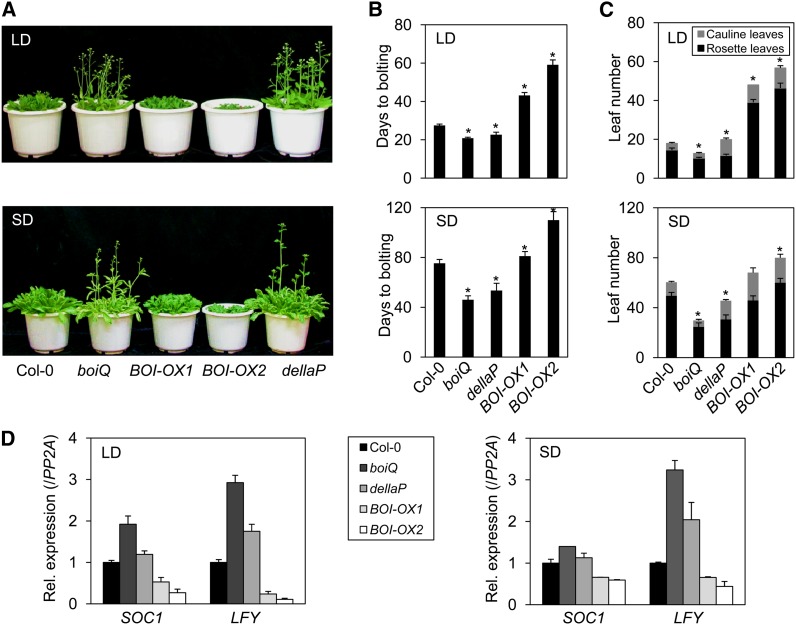
The BOIs and DELLAs inhibited flowering under both LD and short-day (SD) conditions (Figures 3A to 3C). Under the LD condition, wild-type plants bolted at 27 d after sowing and showed 15 rosette leaves. By contrast, the boiQ mutant bolted at 20 d with 10 rosette leaves, and the BOI-OXs flowered much later (43 and 59 d with 39 and 46 rosette leaves, respectively) (Figures 3B and 3C). Similar to boiQ, the dellaP mutant also flowered early, at 22 d with 11 rosette leaves. The boiQ and dellaP plants also flowered earlier than the wild type under the SD condition. Wild-type plants flowered at 75 d after sowing and showed 50 rosette leaves. By contrast, the boiQ mutant flowered at 46 d with 25 rosette leaves, whereas the BOI-OX2 flowered at 110 d or later after developing more than 60 rosette leaves (Figures 3B and 3C). The dellaP mutant also flowered earlier, at 53 d with 30 rosette leaves. Overexpression of the other BOIs also delayed flowering under the LD condition (see Supplemental Figure 4 online). Taken together, these results indicate that the BOIs and DELLAs inhibit flowering under both LD and SD conditions.
Figure 3.
BOIs and DELLAs Inhibit Flowering under Both LD and SD Conditions.
(A) Early flowering of boiQ and dellaP mutants under the LD and SD conditions. Pictures were taken at 4 weeks for LD-grown plants and at 8 weeks for SD-grown plants.
(B) Quantification of bolting days under the LD and SD conditions (sd, n = 10; *P < 0.05, Student’s t test).
(C) Quantification of rosette and cauline leaves at bolting in LD- and SD-grown plants (sd, n = 10; *P < 0.05, Student’s t test).
(D) Expression of SOC1 and LFY mRNAs in boiQ and dellaP mutants grown under the LD and SD conditions. Seventeen-day-old LD-grown and 5-week-old SD-grown plants were sampled for expression analysis at the 12th hour and 8th hour of light, respectively. The mRNA levels of SOC1 or LFY are indicated relative to PP2A expression and wild-type values (([LFYx]/[PP2Ax])/([LFYCol-0]/[PP2ACol-0])) (sd, n = 3 biological replicates).
[See online article for color version of this figure.]
GA is known to promote flowering by activating the expressions of LEAFY (LFY) and SUPPRESSOR OF CONSTANS1 (SOC1) (Blazquez et al., 1998; Moon et al., 2003). Thus, we examined the mRNA expression levels of LFY and SOC1 in samples taken at the 12th hour and 8th hour after light exposure under the LD and SD conditions, respectively. The boiQ mutant expressed higher levels of LFY mRNA compared with the wild type under both conditions, whereas the BOI-OXs expressed lower levels of LFY under these conditions (Figure 3D). The SOC1 mRNA was expressed at a higher level in boiQ and lower levels in the BOI-OXs. The dellaP mutant expressed a higher level of LFY mRNA compared with the wild type under both LD and SD conditions, whereas the SOC1 mRNA was relatively unchanged (Figure 3D). These results indicate that both BOIs and DELLAs inhibit flowering by repressing the expression of LFY mRNA. However, the boiQ and dellaP mutants were not identical in their regulations of SOC1 mRNA expression, indicating that the BOIs and DELLAs inhibit flowering both redundantly and independently.
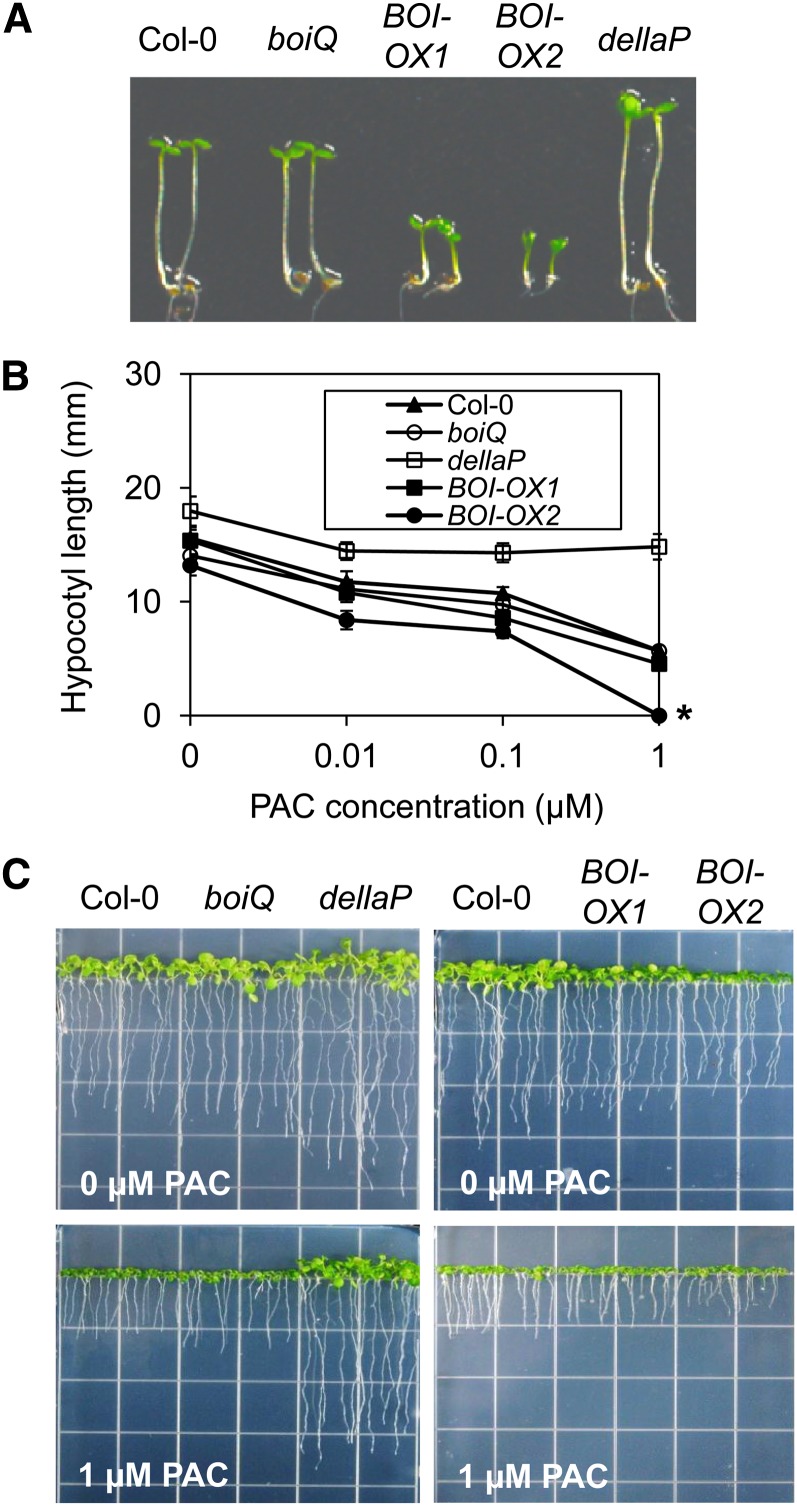
We also determined whether the BOIs regulate hypocotyl and root elongation. When seedlings were grown under red light, the dellaP mutant had longer hypocotyls than wild-type plants; the boiQ mutant had hypocotyl lengths similar to those of the wild type; and the hypocotyls of the BOI-OXs were shorter than those of the wild type (Figure 4A). This indicates that although BOI overexpression could repress hypocotyl elongation under red light, the tested BOI mutations were not sufficient to cause hypocotyl elongation. To examine this effect further, we determined hypocotyl length in the presence of various concentrations of PAC. Seedlings were grown for 6 d in the dark on Murashige and Skoog (MS) media containing low concentrations of PAC. In wild-type plants, the hypocotyl length decreased from 16 to 6 mm when the concentration of PAC was increased from 0 to 1 μM. The hypocotyl lengths of both boiQ and the BOI-OXs also decreased in a manner similar to that of the wild type, whereas the hypocotyl length of dellaP remained relatively unchanged (Figure 4B). Similar to our findings for hypocotyl length, the dellaP mutant had longer root lengths than the wild type in the absence of PAC, the boiQ mutant had root lengths similar to those of the wild type, and the BOI-OXs had shorter root lengths than the wild type. When plants were grown in the presence of 1 µM PAC, the root lengths of wild-type plants were reduced by 58%, those of the dellaP mutant were not significantly altered, and those of boiQ and the two BOI-OXs were reduced by 56, 58, and 74%, respectively (Figure 4C). The further reduction in the root length of BOI-OX2 in the PAC-containing media is likely to reflect the slower germination kinetics of these seeds. Taken together, these results indicate that BOIs are not necessary for hypocotyl and root elongation, but their overexpression could reduce elongation independent of the DELLAs.
Figure 4.
BOIs Do Not Regulate DELLA-Mediated Hypocotyl and Root Elongation.
(A) Hypocotyl elongation of boiQ, BOI-OXs, and dellaP grown under continuous red light (20 μmol m−2 s−1) for 4 d.
(B) Hypocotyl elongation of boiQ, BOI-OXs, and dellaP grown in the dark in the presence of PAC for 6 d. Asterisk indicates that BOI-OX2 did not germinate in the presence of 1 μM PAC.
(C) Root elongation of boiQ, BOI-OXs, and dellaP mutants grown under the LD condition in the absence or presence of 1 μM PAC for 10 d. Notice that while BOI-OX2 did not germinate in the dark in the presence of 1 μM PAC, it successfully germinated under white light.
[See online article for color version of this figure.]
DELLA Requires BOIs to Regulate a Subset of GA Responses
To further investigate the functional relationship between BOIs and DELLAs, we crossed the gai-1 mutant, a dominant gain-of-function mutant of GAI, with boiQ to generate a gai-1 boiQ pentuple mutant. Various GA responses were determined in the pentuple mutant, and these results were compared with those from the gai-1 single mutant.
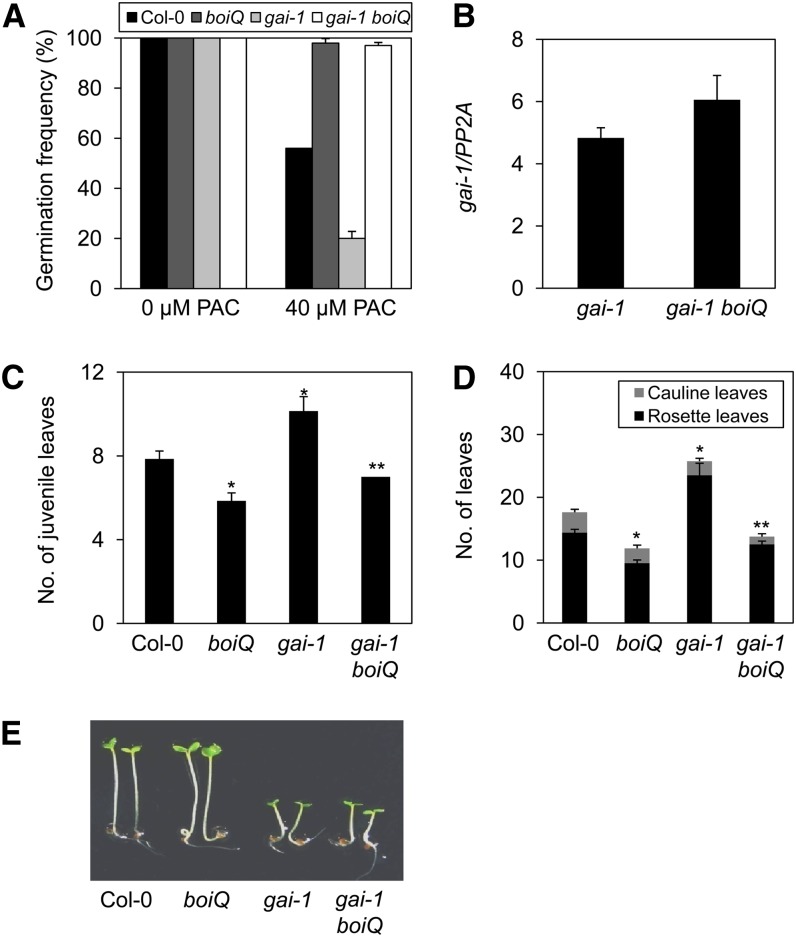
We first determined the germination frequency of cold-imbibed seeds in the presence of PAC. All seeds germinated well in the absence of PAC. In the presence of PAC, 56% of wild-type seeds germinated, almost 100% of the boiQ mutant seeds germinated, only 20% of the gai-1 mutant germinated, and almost 100% of the gai-1 boiQ pentuple mutants germinated (Figure 5A). Thus, the boiQ mutation appears to be epistatic to the gai-1 mutation. The rescue of germination by the boiQ mutation was not caused by an alteration in gai-1 mRNA expression, as both gai-1 single and gai-1 boiQ pentuple mutants expressed similar levels of the gai-1 mRNA (Figure 5B). These results indicate that gai-1 requires BOIs to inhibit seed germination.
Figure 5.
GAI Requires BOIs to Inhibit a Subset of GA Responses.
(A) The boiQ mutation suppresses the low germination frequency phenotype of gai-1 mutant. Imbibed seeds were germinated under white light with or without 40 μM PAC (sd, n = 3).
(B) Similar expression levels of gai-1 mRNA were seen in the gai-1 single and gai-1 boiQ pentuple mutants (sd, n = 3 biological replicates).
(C)The delayed juvenile-to-adult phase transition of gai-1 mutant is suppressed by the boiQ mutation. Plants were grown under the LD condition and transition was determined by the appearance of abaxial trichomes (sd, n = 10; *significant difference from the wild-type; **significant difference from the gai-1 mutant; P < 0.05, Student’s t test).
(D) Suppression of the late flowering of gai-1 mutant by the boiQ mutation. Flowering time was determined by counting rosette leaves at bolting (sd, n = 10; *significant difference from the wild type; **significant difference from the gai-1 mutant; P < 0.05, Student’s t test).
(E) The shortened hypocotyl phenotype of gai-1 mutant is not suppressed by the boiQ mutation. Seedlings were grown under continuous red light (20 μmol m−2 s−1) for 4 d.
[See online article for color version of this figure.]
We next determined the timing of the juvenile-to-adult phase transition by determining the appearance of abaxial trichomes. Wild-type plants developed abaxial trichomes on the 8th leaf under the LD condition, whereas boiQ plants developed theirs on the 6th leaf. Under the same conditions, the gai-1 mutants developed abaxial trichomes on the 10th leaf, while the gai-1 boiQ pentuple mutant developed abaxial trichomes on the 7th leaf (Figure 5C). The gai-1 also required BOIs to inhibit flowering. Under the LD condition, wild-type plants flowered after developing 14 rosette leaves, whereas boiQ mutants flowered after developing around 10 rosette leaves. Under the same conditions, the gai-1 mutants flowered after developing 23 rosette leaves, whereas the gai-1 boiQ pentuple mutants flowered much earlier, after developing only ∼12 rosette leaves (Figure 5D). However, gai-1 did not require BOIs to exhibit decreased hypocotyl elongation. When grown in red light, gai-1 plants had much shorter hypocotyls than wild-type plants, boiQ plants had a hypocotyl length similar to that of the wild type, and the gai-1 boiQ pentuple mutant had a hypocotyl length similar to that of gai-1 plants (Figure 5E), indicating that the BOIs are not required for the shortened hypocotyl phenotype of the gai-1 mutant. Taken together, our results indicate that GAI requires BOIs to inhibit GA responses such as seed germination, phase transition, and flowering, but not hypocotyl elongation.
BOIs and DELLAs Are Targeted to the Same Promoters
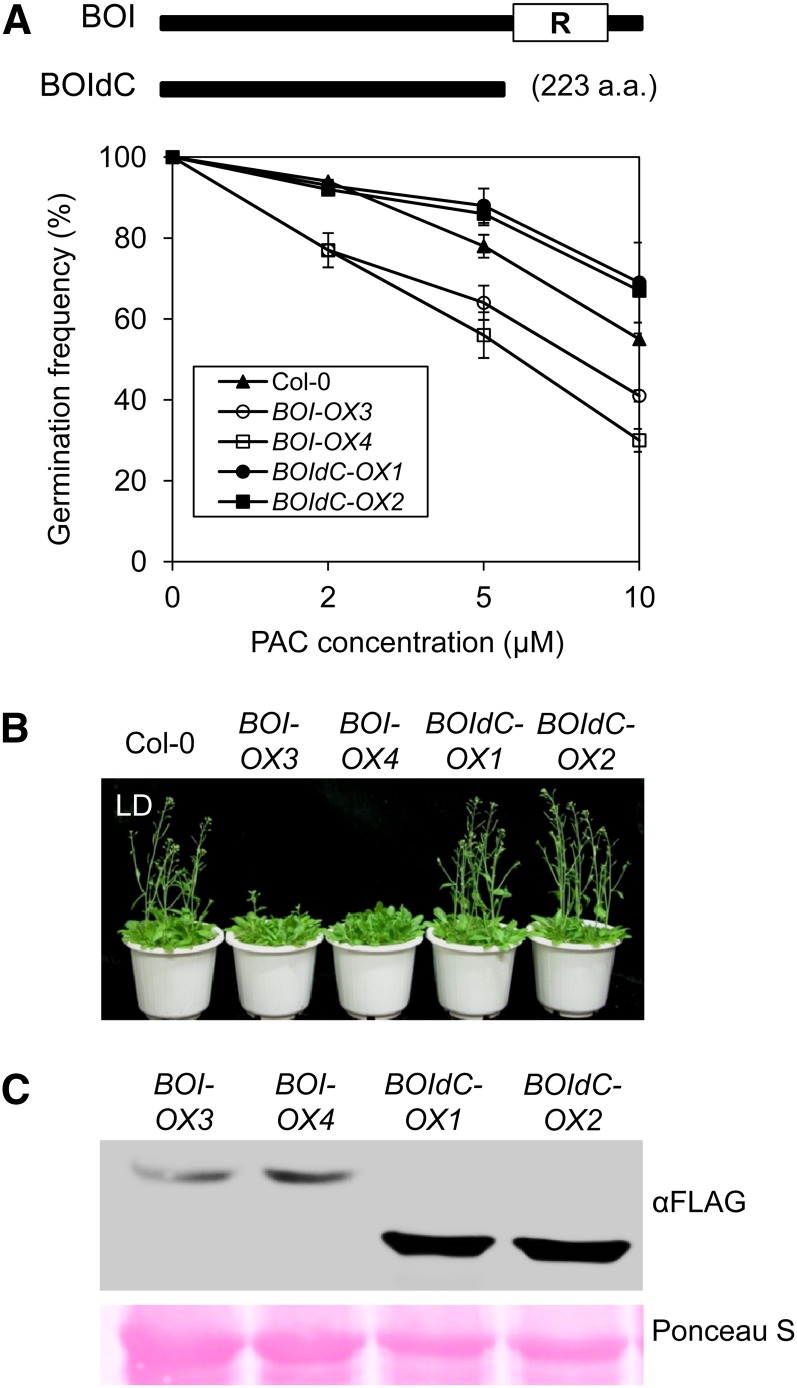
Since the BOIs are RING domain proteins that reportedly possess ubiquitin E3 ligase activity, we tested whether the RING motif of the BOI protein was necessary to inhibit GA responses (Luo et al., 2010). We generated transgenic plants expressing FLAG-tagged BOI lacking the RING motif (BOIdC-OX1 and BOIdC-OX2) and determined the germination and flowering phenotypes (Figure 6A). We also generated transgenic lines expressing FLAG-tagged full-length BOI (BOI-OX3 and BOI-OX4). BOI-OX3 and -OX4 germinated less than wild-type seeds in the presence of PAC, whereas the BOIdC-OXs germinated at similar or slightly higher rates than the wild type (Figure 6A). Similarly, the BOIdC-OXs did not show any alteration in flowering time, whereas BOI-OX3 and -OX4 delayed flowering under the LD condition (Figure 6B). The inability of the RINGless BOI to function was not due to decreased expression, as the BOIdC protein was expressed at a much higher level than the full-length BOI protein (Figure 6C). Taken together, these results indicate that the RING motif is necessary for BOI to inhibit a subset of GA responses.
Figure 6.
The RING Motif of BOI Is Necessary to Inhibit GA Responses.
(A) Inhibition of germination by overexpression of BOI but not BOIdC. Nonimbibed seeds were germinated under white light in the presence of PAC (sd, n = 3). Top panel is a diagram showing schematics of full-length BOI and BOI lacking the RING motif (BOIdC). a.a., amino acids.
(B) Late flowering by BOI- but not BOIdC-overexpressing plants. Plants were grown under the LD condition and photographed at 5 weeks.
(C) Comparison of BOI and BOIdC protein levels in the BOI-OX and BOIdC-OX lines.
[See online article for color version of this figure.]
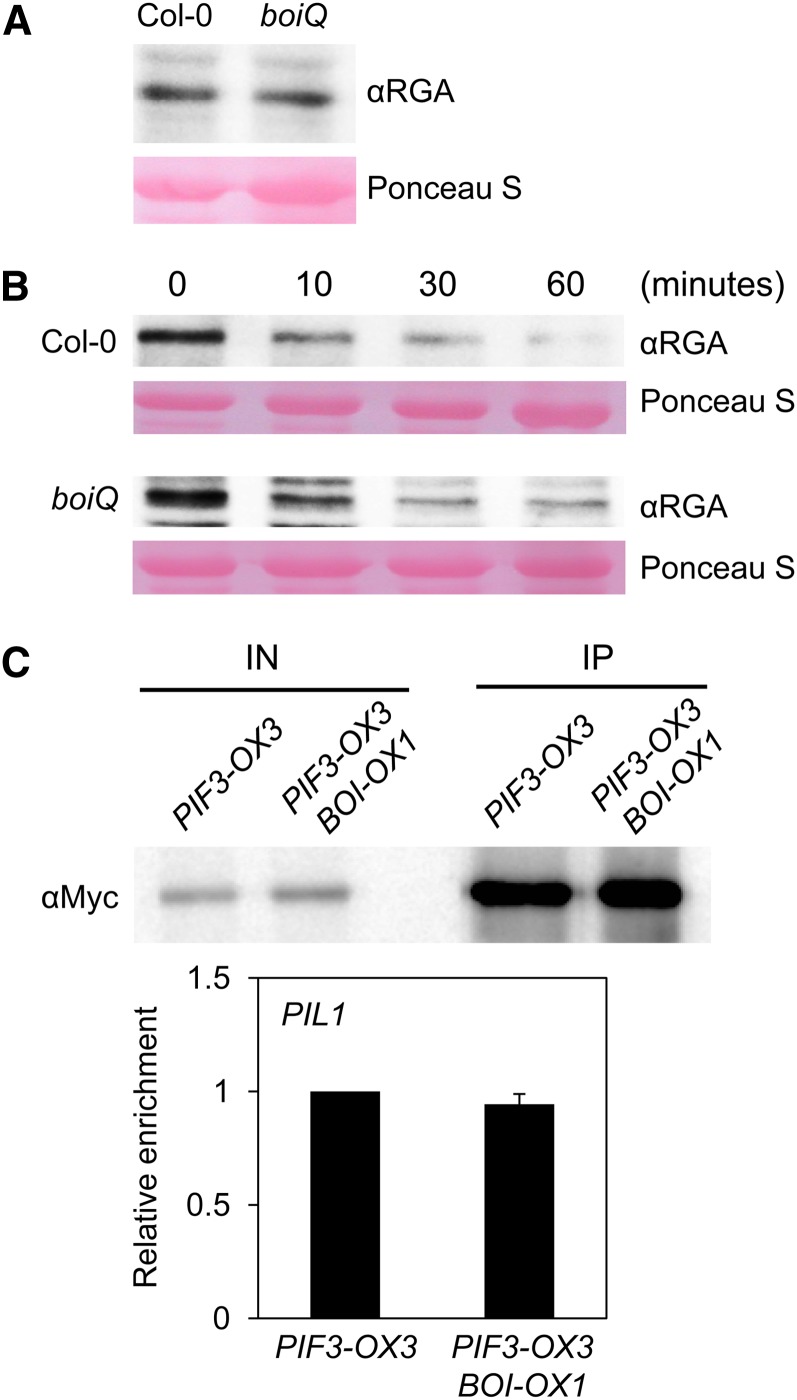
Since the RING domain of BOI appears to be necessary for its function, it is feasible that BOIs ubiquitylate and promote degradation of their interacting proteins. Since both BOIs and DELLAs inhibit GA responses, it did not seem likely that BOIs destabilize DELLAs. Nevertheless, we tested endogenous RGA protein levels in wild-type and boiQ mutant seedlings. The initial RGA levels were similar between wild-type and boiQ mutant seedlings, and when exogenous GA was applied, RGA was degraded rapidly in wild-type and boiQ seedlings, showing similar kinetics (Figures 7A and 7B). These results indicate that BOIs do not drastically alter the stability of RGA in plants, further suggesting that BOIs repress GA responses via other means.
Figure 7.
BOI Does Not Regulate the Stability of RGA or the DNA Binding of PIF3.
(A) Comparable levels of endogenous RGA protein were found in wild-type and boiQ plants.
(B) Comparable degradation kinetics of RGA in response to GA were observed in wild-type and boiQ plants.
(C) PIF3-ChIP data showing comparable enrichment of PIF3 at its target promoter irrespective of BOI overexpression. The top panel shows similar PIF3 protein levels in PIF3-OX3 and PIF3-OX3 BOI-OX1 plants before (IN) and after (IP) ChIP. The bottom panel shows the relative enrichment of PIF3 at the PIL1 promoter, as assessed by PIF3-ChIP (sd, n = 3).
[See online article for color version of this figure.]
Previous reports indicated that DELLAs interact with PIFs and inhibit their binding to target DNA (de Lucas et al., 2008; Feng et al., 2008; Gallego-Bartolomé et al., 2010). Thus, we examined whether BOI enhances DELLA to inhibit binding of PIF3 to its target promoters, using transgenic plants expressing either PIF3-Myc alone (PIF3-OX3) or PIF3-Myc together with BOI (PIF3-OX3 BOI-OX1). The PIF3-OX3 BOI-OX1 was generated by crossing PIF3-OX3 and BOI-OX1 and selecting double homozygous lines expressing levels of PIF3-Myc and His-BOI similar to those of the parental lines. PIF3-ChIP (chromatin immunoprecipitation) analyses showed that a representative target promoter (the PIL1 promoter) was enriched comparably in PIF3-OX3 and PIF3-OX3 BOI-OX1 lines (Figure 7C). These results indicate that BOI overexpression does not enhance the ability of DELLAs to inhibit the binding of DNA by PIF3.
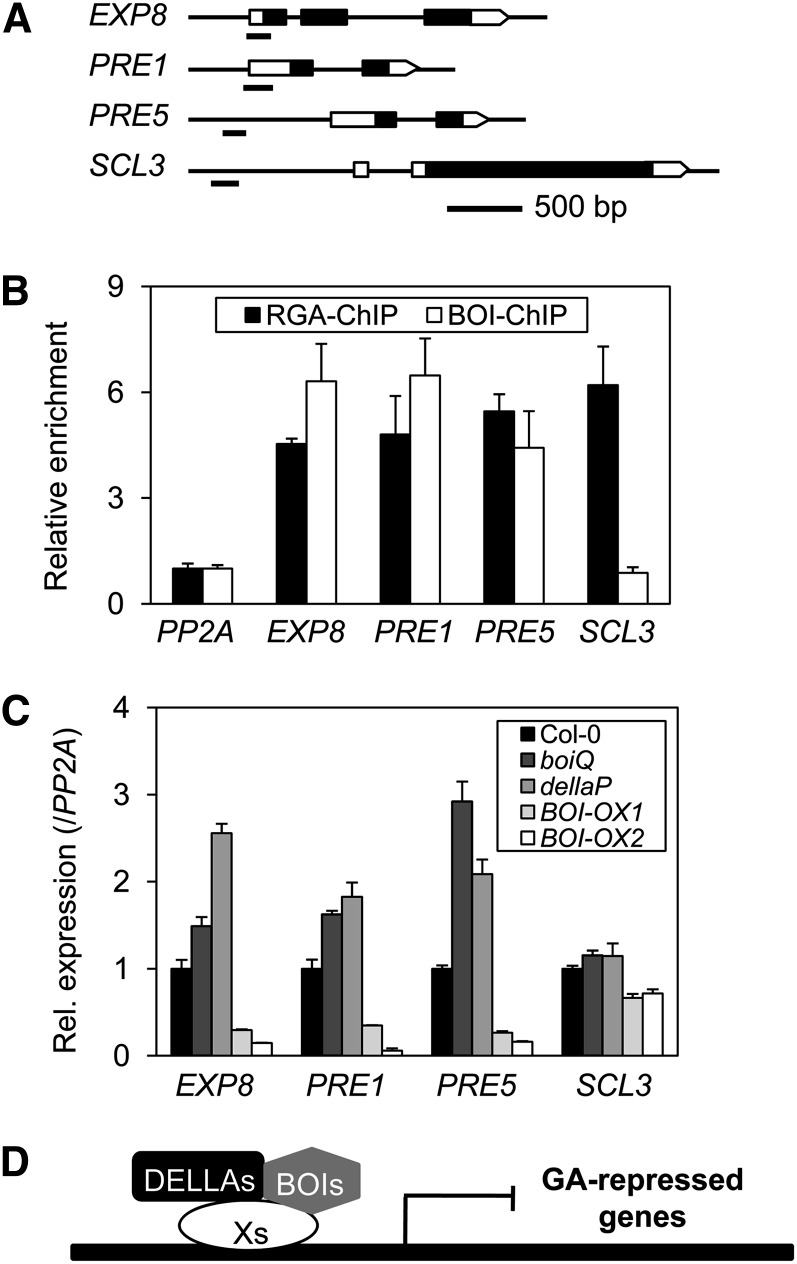
DELLAs are also known to target and regulate various gene promoters (Zentella et al., 2007; Gallego-Bartolomé et al., 2011; Stamm et al., 2012). Since BOIs and DELLAs interact, we hypothesized that the two protein families could target and regulate the same regulatory regions of genes. We thus examined if the two proteins were targeted to the regulatory regions of various GA-responsive genes (EXP8, PRE1, PRE5, and SCL3). These GA-responsive genes were chosen because they were shown to be direct target genes of DELLAs (Zentella et al., 2007; Gallego-Bartolomé et al., 2011; Stamm et al., 2012). We used ChIP analysis to determine if RGA and BOI were targeted to these promoters. Consistent with a previous report, RGA was targeted to the promoter of SCL3, as shown by strong enrichment of a specific SCL3 promoter fragment by anti-RGA antibody (RGA-ChIP) in the sly1-10 mutant (Figures 8A and 8B). Similarly, RGA-ChIP also strongly enriched specific promoter fragments of EXP8, PRE1, and PRE5, whereas it did not enrich thePP2A promoter, indicating that RGA is targeted to theSCL3, EXP8, PRE1, and PRE5 promoters. Similar ChIP assays using anti-BOI (BOI-ChIP) also strongly enriched specific fragments of EXP8, PRE1, and PRE5, indicating that BOI is also targeted to these promoters. Unlike the RGA-ChIP, however, BOI-ChIP did not enrich the SCL3 promoter (Figure 8B). Thus, our results indicate that RGA and BOI are targeted to the regulatory regions of shared but distinct gene sets.
Figure 8.
BOI and RGA Target the Same Gene Promoters.
(A) A diagram showing the genomic structures of four GA-responsive genes and the ChIP amplicons used. Untranslated regions are indicated by empty rectangles, exons are indicated by filled rectangles, and the ChIP amplicons are indicated by short underlines.
(B) Targeting of RGA and BOI to the GA-responsive gene promoters examined by ChIP. The PP2A gene promoter was used as a nonbinding control. ChIP values are indicated relative to that of PP2A (sd, n = 3 biological replicates).
(C) Expression of target gene mRNAs in wild-type, boiQ, dellaP, and BOI-OX plants. Seven-day-old LD-grown seedlings were sampled for expression analysis (sd, n = 3 biological replicates).
(D) A promoter targeting model for the actions of DELLA and BOI. The model proposes that DELLAs and BOIs are targeted to gene promoters by interacting with yet unidentified DNA binding transcription factors (Xs). Once at the target promoters, they regulate gene expression to inhibit GA responses.
We further examined whether BOI also represses these direct target genes of DELLA. We determined mRNA expression levels in 7-d-old LD-grown seedlings and found that the levels of EXP8, PRE1, and PRE5 were strongly elevated in the dellaP mutant compared with the wild type, whereas the mRNA level of SCL3 was comparable to that of the wild type under our experimental conditions (Figure 8C). In the boiQ mutant, the mRNA level of SCL3 was unchanged but the others were elevated either moderately (EXP8 and PRE1) or strongly (PRE5), indicating that BOI represses the expression of its target genes. Consistent with its repressor function, the mRNA levels of EXP8, PRE1, and PRE5 (but not SCL3) were strongly repressed in the BOI-OXs. Taken together, these results indicate that DELLA and BOI are targeted to a subset of GA-responsive gene promoters and repress their expression levels. As shown in the case of SCL3, however, DELLAs and BOIs may also be targeted separately to the regulatory regions of other genes.
DISCUSSION
DELLA proteins are central repressors of GA responses. However, despite their central position in GA signaling, the molecular roles of the DELLA proteins are still being actively examined. In this article, we show that DELLAs interact with four RING domain proteins that were previously identified as BOI and its homologs, BRG1, BRG2, and BRG3 (Luo et al., 2010). Similar to the DELLAs, the BOIs repress various GA responses, including GA-mediated seed germination, the juvenile-to-adult phase transition, and flowering. The repressive function of the BOIs distinguishes them from other DELLA-interacting proteins, in that both DELLAs and BOIs act to repress GA responses, whereas the DELLAs and their other interacting proteins (BZR1, JAZs, MYC2, PIF3, PIF4, and SCL3) act antagonistically (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Zhang et al., 2011; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Hong et al., 2012; Yang et al., 2012). Our genetic analyses of the gai-1 and boi mutants indicated that DELLAs require BOIs to fully repress GA responses. This requirement is not due to the stabilization of DELLAs by BOIs or the enhanced inhibition of PIFs by DELLAs in the presence of BOIs. Rather, we herein show that DELLAs and BOIs target and regulate both shared and distinct sets of GA-responsive genes. Taken together, our results indicate that DELLAs and BOIs form complexes, bind to target gene promoters, regulate the expression levels of these target genes, and thus repress GA responses. It should be noted, however, that our data do not completely exclude the possibility that BOIs might repress some GA responses not only through interactions with DELLAs but also through indirect mechanisms.
The molecular mechanism through which BOIs and DELLAs repress gene expression is not yet fully understood. A previous study showed that BOI ubiquitylates BOS1, an R2R3MYB transcription factor, and targets it for degradation (Luo et al., 2010). We found that a truncated BOI lacking the RING domain is unable to repress GA responses, indicating that the RING domain of BOI, which is essential for ubiquitin E3 ligase activity, is also essential for the repression of GA responses. BOIs do not seem to ubiquitylate DELLAs, as the absence of BOIs did not affect the molecular weights or stabilities of DELLAs. Instead, we postulate that the BOI-DELLA complex may repress gene expression by ubiquitylating other transcription factors or components of the transcriptional machinery. Alternatively, since RING domains also serve as interacting domains for other proteins in addition to ubiquitin E3 ligase (Houvras et al., 2000), BOIs may be necessary to recruit other proteins to the BOI-DELLA complex to repress gene expression. Further investigations will be needed to clarify how BOIs regulate gene expression at the molecular level.
BOIs and DELLAs Work Both Dependently and Independently
Although we found that the BOIs repress a subset of GA responses, including seed germination, the juvenile-to-adult phase transition, and flowering, not all GA responses are repressed by BOIs. Two notable examples are GA-mediated hypocotyl elongation and root elongation, which we found to be repressed by DELLAs but not by BOIs. Consistent with this result, although BOIs were required to repress a subset of GA-responsive genes in the gai-1 mutant, not all gai-1 phenotypes were suppressed by the boiQ mutations. Among the gai-1 mutant phenotypes, low germination frequency, delayed juvenile-to-adult phase transition, and late flowering were suppressed by boiQ, whereas other phenotypes (e.g., short hypocotyl length) were not. This partial suppression implies that the DELLAs repress GA responses through both BOI-dependent and -independent pathways. However, it should be noted that the BOI-independent action of DELLAs does not necessarily mean that the DELLAs do not need RING domain proteins to repress BOI-independent GA responses. As shown in Supplemental Figure 1 online, Arabidopsis possesses seven other BOI-related RING domain proteins when BLAST E values lower than e−10 are employed. While BOIs form a tight cluster that is phylogenetically distinct from these other BOI-related RING domain proteins, the latter may substitute for the BOIs in repressing BOI-independent GA responses.
BOIs also possess DELLA-independent actions. For example, the boiQ and dellaP mutants both flowered early compared with wild-type plants under the SD condition, as assessed by days to bolting and number of rosette leaves (Figures 3B and 3C). However, in terms of the number of cauline leaves, the two mutants displayed different phenotypes. The boiQ mutant developed five cauline leaves before developing flowers, while wild-type plants developed 11 cauline leaves, and the dellaP mutant developed 15 cauline leaves. This indicates that (unlike the DELLAs), the BOIs can inhibit the conversion of vegetative meristems to reproductive meristems. DELLA-independent BOI actions were also seen in the context of gene expression. Among the tested genes, the mRNA levels of FT and SPL4 were specifically increased in the boiQ mutant but not in the dellaP mutant, whereas those of LFY, EXP8, PRE1, and PRE5 were increased in both the boiQ and dellaP mutants. BOI and RGA also showed both shared and distinct promoter targeting abilities, with BOI targeting the EXP8, PRE1, and PRE5 promoters, but not the SCL3 promoter, whereas RGA targeted all four promoters. Taken together, these results suggest that the BOIs and DELLAs work both dependently and independently.
Two Modes of DELLA Action: The Interfering Model and the Targeting Model
Previous studies have shown that DELLAs function partly by interacting and interfering with transcription factors, including PIFs (PIF1, -3, and -4, which are basic helix-loop-helix (bHLH) transcription factors responsible for repressing light responses), ALCATRAZ (ALC; a bHLH transcription factor that separates valve cells from the replum during fruit dehiscence), SPATULA (SPT; a bHLH transcription factor that regulates fruit development and seed germination), MYC2 (a bHLH transcription factor that mediates JA responses), and BZR1 (a bHLH transcription factor that mediates BR responses) (de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Gallego-Bartolomé et al., 2010, 2012; Josse et al., 2011; Bai et al., 2012; Hong et al., 2012). Functionally, DELLAs interact with BZR1, PIF3, and PIF4, inhibiting their binding to target promoters (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012; Gallego-Bartolomé et al., 2012). It is not yet known if the DELLAs inhibit the DNA binding of PIF1, ALC, SPT, and MYC2. DELLAs have also been shown to interact with other transcriptional regulators, including JAZ1 and JAZ9 (two JA coreceptors that repress JA signaling), and SCL3 (a positive GA signaling component) (Hou et al., 2010; Zhang et al., 2011; Yang et al., 2012). The interactions between DELLAs and JAZ1 or JAZ9 were proposed to be functionally opposites; it is believed that the interaction of DELLAs with JAZ1 inhibits the binding of JAZ1 to MYC2, whereas the interaction of DELLAs with JAZ9 inhibits the binding of DELLAs to PIFs (Hou et al., 2010; Yang et al., 2012). Meanwhile, DELLAs and SCL3 inhibit each other (Zhang et al., 2011). Overall, these studies have indicated that the DELLAs regulate gene expression by interacting and interfering with other regulators of transcription.
By contrast, our results support the idea that DELLAs directly regulate gene expression by targeting promoters. This promoter-targeting model for DELLA action was originally suggested by a previous report demonstrating that the DELLAs are targeted to the promoters of various GA-responsive genes, including GID1a, GID1b, MYB (At3g11280), bHLH137, WRKY27, SCL3, LBD40, and XERICO (Zentella et al., 2007). The modulation of rice DELLA protein (SLR1) action by the fusion of transcription activation motif (VP16) or transcription repression motif (SRDX) also support the notion that DELLAs may act as a direct transcriptional regulator rather than simply dissociating other DNA binding transcription factors from promoters (Hirano et al., 2012). Our data further show that RGA and BOI are targeted to the promoters of EXP8, PRE1, and PRE5. Our results are consistent with a previous report showing that PRE1 and PRE5 were direct target genes of gai-1, as demonstrated by transgenic plants expressing gai-1 fused to glucocorticoid receptor (Gallego-Bartolomé et al., 2011). EXP8, PRE1, and PRE5 are also GA-responsive genes, indicating that DELLAs bind to the regulatory regions of some GA-responsive genes, thereby regulating their expressions. However, the directionality of this DELLA-mediated regulation appears to differ among genes. For example, DELLAs repress the expression of EXP8, PRE1, and PRE5 but activate the expressions of GID1a, GID1b, MYB (At3g11280), bHLH137, WRKY27, SCL3, LBD40, and XERICO (Zentella et al., 2007; Gallego-Bartolomé et al., 2011; Stamm et al., 2012). This suggests that DELLAs can act as transcriptional activators or repressors depending on the target gene. Notably, the expression levels of SCL3 did not differ significantly in wild-type versus dellaP plants under our experimental conditions, perhaps indicating that some GA-responsive genes are influenced by environmental conditions.
The dual abilities of DELLAs to regulate transcription both positively and negatively do not favor either the targeting model or the interfering model. In terms of the targeting model, many transcription factors have been shown to regulate their directly targeted genes both positively and negatively depending on the target gene. Dorsal, which is a Rel family transcription factor responsible for regulating dorso-ventral axis formation in Drosophila melanogaster, can activate or repress transcription depending on the presence of a neighboring ventral activation region or a ventral repression region (Belvin and Anderson, 1996). Furthermore, a recent genome-wide determination of transcription factor binding sites coupled with expression analysis further showed that dual roles for a transcription factor are the norm rather than the exception (Oh et al., 2009; Hornitschek et al., 2012; Oh et al., 2012). This is consistent with the ability of DELLAs to target and regulate various promoters either positively or negatively depending on the promoter context. However, the dual abilities of the DELLAs could be also explained by the interfering model. For example, DELLAs regulate the expression of PIF-targeted genes by dissociating PIFs from target promoters. Although PIFs mainly act as transcription activators, previous studies have shown that they can also repress some direct target genes (de Lucas et al., 2008; Oh et al., 2009, 2012; Hornitschek et al., 2012). Thus, in the interfering model, the dissociation of PIFs by DELLAs would result in transcriptional activation or repression depending on the target gene. At this point, we do not know what conditions cause DELLAs to act as transcriptional activators or repressors. The promoter context and/or the presence of different neighboring transcriptional regulators may determine the activities of the DELLAs themselves or of the DELLA-interacting transcriptional regulators. Alternatively, specific modifications could cause switching between the opposing activity states of the DELLAs and/or DELLA-dissociable transcription regulators.
It is not yet clear how the two different action modes of DELLA operate simultaneously in plants, but we offer a few different possibilities. First, DELLAs may interact differently with different transcriptional regulators, either inhibiting the function of DELLA-interacting transcriptional regulators or targeting DELLAs to a promoter together with their interacting transcriptional regulators. Second, DELLAs may exist in two different forms generated either by covalent modification (e.g., phosphorylation) or via the formation of complexes with different components, with one form interfering with the functions of DELLA-interacting transcriptional regulators while the other collaborates with them. Alternatively, the two action modes of DELLA may not occur simultaneously in the same cell, but rather may be seen in different cells or tissues. Future work on identifying and characterizing DELLA-interacting transcriptional regulators could help clarify these possibilities.
BOIs May Link Various Signals to That of GA
BOIs may link both endogenous and environmental signals to GA signaling through dynamic changes in expression. Among the plant hormones, ethylene and salicylate have been shown to induce BOI mRNA expression, whereas GA and methyl jasmonate have been shown to repress BOI mRNA expression. Regarding environmental factors, biotic factors such as Botrytis cinerea and Pseudomonas syringae pv tomato DC3000 and abiotic factors such as drought, NaCl, and methylviolagen have been shown to induce BOI mRNA expression (Luo et al., 2010). An analysis of the heat map profile of the BAR site (Toufighi et al., 2005) indicates that BOIs are broadly expressed, with relatively higher expression levels in cauline and aging leaves, flower, and seeds (see Supplemental Figure 5A online). Experimental analyses further corroborate the broad expression of BOIs, with slightly higher expression levels in seeds (see Supplemental Figure 5B online). Altered expression of BOIs is likely to modify the strength of GA signaling. Given that GA signaling has been shown to regulate plant responses to these endogenous and environmental factors, it would be interesting to determine the possible roles of BOIs in these plant responses.
Finally, previously reported microarray data and a genome-wide binding site analysis of PIF1 (also known as PIL5) suggested that the BOIs are direct target genes of PIF1 in the regulation of seed germination (Oh et al., 2009). Microarray data showed that red light strongly represses the expression levels of BOI and BRG3 in imbibed seeds, and we further confirmed this with our RT-PCR analysis (see Supplemental Figure 6A online). BOI and BRG3 were repressed in pif1 mutant seeds irrespective of the light condition, suggesting that red light–dependent repression of BOI and BRG3 is mediated by PIF1. These expression patterns resemble those of many direct target genes of PIF1. Indeed, a previously reported genome-wide binding site analysis of PIF1 indicated that BOI and BRG3 are direct target genes of PIF1, and we further confirmed this finding with our ChIP-PCR analysis (see Supplemental Figures 6B and 6C online). These findings suggest that PIF1 represses GA responses not only by directly binding to the promoters of GAI and RGA and increasing their expression levels, but also by directly binding the promoters of BOI and BRG3 and increasing their expression levels in imbibed seeds. Similar analyses indicated that other PIFs also bind to the promoters of BOIs (all BOIs were bound by PIF4, while the BRG3 promoter was bound by PIF5) and regulate their expression levels (Hornitschek et al., 2012; Oh et al., 2012). However, since PIFs and DELLAs play antagonistic roles in seedling development, further investigation will be needed to evaluate the involvement of BOIs downstream of PIFs in seedling development.
METHODS
Plants Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a growth room at 22 to 24°C, with a 16-h-light/8-h-dark cycle for the LD condition or an 8-h-light/16-h-dark cycle for the SD condition. T-DNA insertion mutants of brg1-1, brg2-1, and brg3-3 in a Col-0 background were obtained from the Salk Institute and the Nottingham Arabidopsis Stock Centre (SALK_010178, SAIL_95_F06, and GABI_661B07, respectively) (Alonso et al., 2003). The boi-2 mutant allele from RIKEN (RATM11-5576-1_G) in the No-0 background was isolated and introgressed into the Col-0 by backcrossing four times (Ito et al., 2002). The boi quadruple mutant (boiQ) was generated by crossing, followed by selection of introgressed boi-2, brg1-1, brg2-1, and brg3-3 mutants. To generate the 35S:His-BOI (for the BOI-OX1 and BOI-OX2 lines) and 35S:BOI-FLAG transgenic lines (for the BOI-OX5 line), the BOI gene was amplified from Arabidopsis cDNA, cloned into the pBARH8 and pbFLAG2 binary vectors, respectively, and subsequently transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998). To generate 35S:FLAG-BOI (for the BOI-OX3 and BOI-OX4 lines), 35S:FLAG-BRG2 (for the BRG2-OX1 line), 35S:FLAG-BRG3 (for the BRG3-OX1 line), and 35S:FLAG-BOIdC (223 amino acids) transgenic plants, the respective inserts were cloned into the phNF binary vector for Arabidopsis transformation.
The sly1-10 mutant in Landsberg erecta was introgressed into Col-0 by backcrossing three times before use. The gai-1 and gai-t6 mutants were backcrossed to Col-0 six times (Oh et al., 2007). The rga-28 (SALK_089146), rgl1-SK62 (SALK_136162), rgl2-SK54 (SALK_027654), and rgl3-3 (CS16355) lines were generated in the Col-0 background at the Salk Institute (Alonso et al., 2003; Tyler et al., 2004). The della pentuple mutant (dellaP) was generated by crossing rga-28, rgl1-SK62, rgl2-SK54, rgl3-3, and introgressed gai-t6. All of the oligomers used in cloning and testing T-DNA insertions are listed in Supplemental Table 1 online, and vectors are described below and in Supplemental Table 2 online.
Construction of Plasmids
The multicloning sites (MCSs) of binary vectors (pCAMBIA; http://www.cambia.org/daisy/cambia/585), Escherichia coli expression vectors (pET and pMAL; Invitrogen and NEB), and yeast two-hybrid vectors (pGADT7 and pGBKT7; Clontech) were modified to accommodate the need to minimize unnecessary PCR amplification and sequencing. Two oligomers were designed to have common serial recognition sites for restriction enzymes (XbaI-XmaI-AvrII-BamHI-SalI) and to maintain the correct reading frame of genetic codes. Adapter DNA fragments specific for the MCS of each vector were prepared by denaturing oligomers at 94°C for 1 min and then annealing by gradually cooling to 20°C using a stepwise protocol with 1-min intermittent incubations; prepared double-stranded adapter DNAs were kept at 4°C until use. Original vectors from each company were digested, purified, and ligated with the respective adapters to create pCAMBIAM (phNIL), pETM, pMALM, pGADM, and pGBKM (see Supplemental Table 1 online for each adapter sequence). pBARH8 and pbFLAG2 binary vectors were modified from pCB301 by adapter ligation by adding an 8XHis-tag at the N terminus and a 3XFLAG-tag at the C terminus, respectively. phNF was generated by ligating SpeI-SmaI digests of phNIL and 3XFLAG amplified from pbFLAG2.
Depending on the abundance of the mRNA of interest (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi), different tissues were used as sources of template cDNA. PCR oligomers were designed to have restriction enzyme sites compatible for binary vectors, yeast vectors, and recombinant protein expression vectors. Forward oligomers were made to contain a start codon (ATG) following the restriction site, and reverse oligomers were designed to not have stop codons. Stop codons in all three different reading frames are present downstream of the MCS in all vectors. BOIs and DELLAs amplified with Pfu DNA polymerase (Solgent) were cloned and confirmed to be free of mutation by automatic sequencing. Sequences and restriction sites are described in Supplemental Table 1 online. Supplemental Table 2 online lists all of the vectors and related information.
Phenotypic Characterization
Germination assays were performed with batch seeds harvested and dried for at least 2 months to remove dormancy. Briefly, sterilized seeds were plated on 0.5× MS medium (Duchefa) with different concentrations of PAC. After 6 d of incubation under continuous white light at 22°C, the germination frequency was determined by counting the seeds that showed emerging radicles. For the gai-1 and gai-1 boiQ mutants, Col-0, boiQ, gai-1, and gai-1 boiQ seeds were dried for 1 month, stratified at 4°C for 4 d, and then placed under constant light for 6 d.
To measure hypocotyl and root elongation on PAC, seeds were sown on 0.5× MS plus 1% Suc plates supplemented with different concentrations of PAC. Hypocotyl experiments were performed similarly to that previously described (Cowling and Harberd, 1999) with some minor modifications. After a 4-d stratification, the plates were illuminated for 6 h and then incubated in the dark for an additional 6 d. For the root tests, seedlings were vertically grown under the LD condition for 10 d as previously described for the white light condition (Zhang et al., 2011). For measurement of hypocotyl length, seedlings were stratified and then grown on 0.5× MS medium under continuous red light (20 μmol m−2 s−1) for 4 d as previously described (Oh et al., 2004). Hypocotyls from at least 15 seedlings were measured using Scion Image software (http://scion-image.software.informer.com).
Chlorophyll levels were measured from the 5th or 6th leaves of LD-grown plants. The extraction method was similar to those previously described (Kim et al., 2011; Paik et al., 2012) with some modifications. Leaves were submerged in 1 mL of 95% ethanol and kept at 4°C in the dark for at least 10 h. The sums of the absorbance at 664.2 and 648.6 nm divided by the leaf fresh weight were used as the indexes for chlorophyll levels.
Vegetative-phase transition and flowering time were measured similarly to the previous description (Dill and Sun, 2001). For the analysis of vegetative-phase transition, the number of juvenile leaves lacking an abaxial trichome was counted in 1-month-old LD-grown plants. Flowering time was determined by counting the numbers of rosette leaves and noting the number of growth days when plants had 1-cm bolts.
Quantification of mRNA Expression
For expression analysis, total mRNA was extracted using the Spectrum plant total RNA kit (Sigma-Aldrich) and then converted to cDNA using MMLV-RTase (Promega) according to the manufacturer’s protocol. Quantitative PCR was performed using an iCycler IQ5 (Bio-Rad) real-time PCR system using SYBR Green, and the relative transcript level of each gene, as compared with that of PP2A, was determined by the delta cycle threshold method (Livak and Schmittgen, 2001). Gene expression patterns were confirmed using three biological replicates. All of the oligomers used for quantitative PCR are summarized in Supplemental Table 1 online.
Protein Analysis and Antibody Production
To determine protein levels, 7-d-old light-grown seedlings were harvested and pulverized with a TissueLyser (Qiagen) in liquid nitrogen. Total proteins were extracted from the resulting seedling powders using denaturing buffer (100 mM Na2HPO4, 8 M urea, and 10 mM Tris-Cl, pH 8.0) as previously described (Oh et al., 2007). Supernatants were collected by centrifugation at 4°C for 10 min, mixed with 5× SDS sample buffer, and boiled for 5 min. Equal amounts of total proteins were separated by SDS-PAGE and subjected to immunoblotting.
For GA3 treatments, 7-d-old light-grown seedlings on plates were submerged in 100 µM GA3 solution and harvested at different time points. Total proteins were extracted, and immunoblotting with a rabbit anti-RGA antibody was used to detect endogenous RGA levels. The anti-RGA antibody was raised against the peptide, LysArgAspHisHisGlnPheGlnGlyArgLeuSerAsnHisGly, and the antiserum was subjected to affinity purification (Abfrontier). The specificity and sensitivity of the antibody were tested using Col-0 and rga-28. To generate the anti-BOI antibody, recombinant BOI-his protein was produced in E. coli BL21 (DE3) RIL using pET29a (Invitrogen), and the protein was affinity purified under denaturing conditions and injected into rabbits (Abfrontier).
In Vitro and in Vivo Binding Assays
For our in vitro binding assays, recombinant MBP-RGA and MBP-GAI proteins were produced in E. coli BL21 (DE3) RIL using the pMAL-c2X vector and purified on an amylose resin (NEB) according to the manufacturer’s instructions. FLAG-tagged BOIs were extracted from 7-d-old light-grown BOI-overexpressing seedlings using a nondenaturing lysis buffer containing 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.05% Nonidet P-40, 50 μM MG132, and 1× protease inhibitor cocktail (Roche). Lysis buffer (1 mL) was mixed with 1 mL of seedling powder, and the lysates were further homogenized by being passed through a QiaShredder column (Qiagen). After centrifugation at 4°C for 10 min, equal amounts of supernatants were incubated with MBP-RGA, MBP-GAI, or MBP-bound resin at 4°C for 2 h, followed by four washes with lysis buffer lacking MG132 and the protease inhibitor. FLAG-tagged BOIs bound to MBP-RGA and MBP-GAI were visualized by immunoblotting with a rabbit anti-FLAG antibody (Sigma-Aldrich).
For our in vivo coimmunoprecipitation assay involving FLAG-BOI and endogenous RGA, Agrobacterium tumefaciens infiltration was used to transiently express 35S:FLAG-BOI and 35S:FLAG-GFP (in the binary vector, phNF) in 10-d-old light-grown sly1-10 mutants in a Landsberg erecta background. The protocol was similar to those previously described (Marion et al., 2008; Li et al., 2009) with some modifications. Briefly, Agrobacterium harboring FLAG-BOI and FLAG-GFP binary vectors were suspended in 5% Suc solution and infiltrated into sly1-10 seedlings on a plate using a vacuum method (2 min of vacuum, break, and 2 min of vacuum). Infiltrated seedlings were incubated in a growth chamber for 3 d after three washes with autoclaved water and harvested for coimmunoprecipitation experiments. The sly1-10 seedlings expressing FLAG-BOI and FLAG-GFP were ground in liquid nitrogen, and total proteins were extracted using the nondenaturing lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 2 mM DTT, 0.05% Nonidet P-40, 50 μM MG132, and 1× protease inhibitor cocktail). The obtained supernatants were incubated with anti-FLAG conjugated resin (Sigma) for 2 h at 4°C, washed four times with lysis buffer lacking MG132, boiled in SDS sample buffer, and then subjected to immunoblotting. Anti-FLAG and anti-RGA antibodies were used to detect FLAG-BOI, GFP-FLAG, and endogenous RGA.
For in vivo coimmunoprecipitation assay using the BOI-OX5 and GFP-OX lines, transgenic seedlings were grown on the medium containing 1 µM PAC under white light for 10 d, and the coimmunoprecipitation assay was performed as described above.
Yeast Two-Hybrid Screening
Each of the BOI genes was cloned into pGBKM, a derivative of pGBKT7 bait vector, and each of the DELLA genes was cloned into pGADM, a derivative of pGADT7 prey vector, using PCR-based methods and oligomers summarized in Supplemental Table 1 online. Yeast (strain AH109) was simultaneously cotransformed with various combinations of bait and prey vectors according to the manufacturer’s instructions (Clontech). Briefly, transformed yeast cells were collected and spotted onto synthetic dropout medium lacking Trp and Leu (SD-2), which selects for yeast harboring both pGBKM and pGADM vectors, or onto synthetic dropout medium lacking Trp, Leu, adenine, and His (SD-4), which selects for yeast harboring pGBKM and pGADM encoding an interacting pair of proteins. 3-Amino-1,2,4-triazole was not used in these assays.
ChIP
For the ChIP analyses, sly1-10 in the Col-0 background and BOI-OX1 were grown for 7 d under the LD condition, and PIF3-OX3 and PIF3-OX3 BOI-OX1 were grown for 7 d in the dark. ChIP was performed as previously described (Oh et al., 2007; Park et al., 2011). Briefly, formaldehyde-cross-linked seedlings were ground under liquid nitrogen, and 1 mL of powder was dissolved in 1 mL of ChIP lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1× protease inhibitor cocktail, and 40 μM MG132) and sonicated to obtain DNA fragments of ∼500 bp. After centrifugation, the chromatin was mixed with the appropriate antibody and the mixture was incubated for 5 h. Then, 20 μL of salmon sperm DNA-coated protein A agarose resin (Millipore) was added, and the sample was incubated for an additional 3 h. The resin was sequentially washed with low-salt wash buffer, high-salt wash buffer, lithium chloride wash buffer, and TE buffer. The immunoprecipitated DNA was eluted from the resin in 200 μL of elution buffer (20 mM Tris-Cl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 1% SDS, and 50 mg/mL proteinase K) at 65°C overnight, and the residual DNA was further eluted in 50 μL of elution buffer. After purification with a PCR purification kit (Solgent), the eluted DNA samples were subjected to quantitative PCR.
All of the oligomers used for quantification of the ChIP DNA were designed manually due to problems with low GC content, particularly in the promoter regions. To test the amplification specificities and efficiencies of the generated oligomers, we confirmed the amplification of single DNA bands and used parallel amplification curves to minimize the bias from the delta cycle threshold method. The oligomers used are listed in Supplemental Table 1 online.
In Silico Analyses of DNA and Protein Sequences
DNA sequences were manipulated using the pDRAW32 program for oligomer design, prediction of restriction enzyme sites, in silico cloning, and gene fusion (http://www.acaclone.com). ClustalW and MEGA5 were used to align and generate a neighbor joining tree among the BOI homologs (http://www.megasoftware.net) (Tamura et al., 2011). For heat map analyses, normalized mRNA data were obtained from the BAR expression browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi) and visualized using a BAR heat mapper tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi) (Toufighi et al., 2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BOI (At4g19700), BRG1 (At5g45100), BRG2 (At1g79110), BRG3 (At3g12920), SPL3 (At2g33810), SPL4 (At1g53160), SPL5 (At3g15270), SOC1 (At2g45660), LFY (At5g61850), RGA (At2g01570), GAI (At1g14920), EXP8 (At2g40610), PRE1 (At5g39860), PRE5 (At3g28857), SCL3 (At1g50420), and PP2A (At1g13320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of BOI and BRGs.
Supplemental Figure 2. The Specificity of RGA Antibody.
Supplemental Figure 3. Redundant Roles of BOI Family Members.
Supplemental Figure 4. Regulation of Flowering by BRGs.
Supplemental Figure 5. Expression Patterns of BOI and BRGs.
Supplemental Figure 6. Regulation of BOI and BRGs by PIF1.
Supplemental Table 1. Primer List.
Supplemental Table 2. Vector List.
Supplemental Data Set 1. Sequence Alignment for NJ Tree.
Supplementary Material
Acknowledgments
sly1-10 in the Landsberg erecta background was a kind gift from Philip N. Benfey (Duke University). Camille M. Steber (Washington State University) and Tai-ping Sun (Duke University) supported us by providing sly1-2 and pRG51. We thank The Arabidopsis Information Resource and the Nottingham Arabidopsis Stock Centre for providing information and mutant seeds. This work was supported in part by grants from the National Research Foundation of Korea (2012R1A2A1A01003133, 2011- 0031955, and 2011-0031350) and the Rural Development Administration (SSAC-PJ009580) to G.C.
AUTHOR CONTRIBUTIONS
J.P., K.T.N., J.-S.J., and G.C. designed the research. J.P., K.T.N., and E.P performed all experiments. J.P., K.T.N., and G.C. wrote the article.
Glossary
- GA
gibberellic acid
- GlcNAc
N-acetyglucosamine
- GFP
green fluorescent protein
- PAC
paclobutrazol
- Col-0
Columbia-0
- LD
long-day
- SD
short-day
- MS
Murashige and Skoog
- ChIP
chromatin immunoprecipitation
- bHLH
basic helix-loop-helix
- JA
jasmonic acid
- MCS
multicloning site
References
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Murase K., Sun T.P., Steber C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T.A., Lawrenson T., Sablowski R., Østergaard L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin M.P., Anderson K.V. (1996). A conserved signaling pathway: The Drosophila toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 12: 393–416 [DOI] [PubMed] [Google Scholar]
- Blazquez M.A., Green R., Nilsson O., Sussman M.R., Weigel D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cheng M.C., Hsieh E.J., Chen J.H., Chen H.Y., Lin T.P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Christians M.J., Gingerich D.J., Hansen M., Binder B.M., Kieber J.J., Vierstra R.D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cowling R.J., Harberd N.P. (1999). Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J. Exp. Bot. 50: 1351–1357 [Google Scholar]
- Dai C., Xue H.W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., Dong X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Alabadí D., Blázquez M.A. (2011). DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Galvão V.C., Horrer D., Küttner F., Schmid M. (2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., Matsuoka M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634 [DOI] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hirano K., Kouketu E., Katoh H., Aya K., Ueguchi-Tanaka M., Matsuoka M. (2012). The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 71: 443–453 [DOI] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hotton S.K., Callis J. (2008). Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59: 467–489 [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Houvras Y., Benezra M., Zhang H., Manfredi J.J., Weber B.L., Licht J.D. (2000). BRCA1 physically and functionally interacts with ATF1. J. Biol. Chem. 275: 36230–36237 [DOI] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Motohashi R., Kuromori T., Mizukado S., Sakurai T., Kanahara H., Seki M., Shinozaki K. (2002). A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 129: 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Binkowski K.A., Olszewski N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Olszewski N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse E.M., Gan Y., Bou-Torrent J., Stewart K.L., Gilday A.D., Jeffree C.E., Vaistij F.E., Martínez-García J.F., Nagy F., Graham I.A., Halliday K.J. (2011). A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.R., Estelle M. (2012). Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Murphy A.S., Baek D., Lee S.W., Yun D.J., Bressan R.A., Narasimhan M.L. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 62: 3981–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Yang S.H., Han K.H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Elgersma A., Hanhart C.J., van Loenen-Martinet E.P., van Rijn L., Zeevaart J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65: 33–39 [Google Scholar]
- Koornneef M., van der Veen J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo H., Laluk K., Lai Z., Veronese P., Song F., Mengiste T. (2010). The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154: 1766–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37: 720–729 [DOI] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Nakajima M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Paik I., Yang S., Choi G. (2012). Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. USA 109: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23: 1404–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Harberd N.P. (2002). The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 5: 376–381 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Lopez-Molina L. (2009). The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal. Behav. 4: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., Sazuka T., Ashikari M., Matsuoka M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Mak P.Y., Martínez E.C., Sun T.P. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Tseng T.S., Swain S.M., Dill A., Jeong S.Y., Olszewski N.E., Sun T.P. (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P., Ravindran P., Mohanty B., Tan E.L., Yu H., Kumar P.P. (2012). Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 12: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C.M., Cooney S.E., McCourt P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Ritchie S., Soule J.D., McGinnis K.M., Steber C.M. (2004). Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sun T.P., Kamiya Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M., Koornneef M., Zeevaart J.A. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87: 7983–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Sun T.P. (2004). Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol. 135: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wang K.L., Yoshida H., Lurin C., Ecker J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wen C.K., Chang C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E.M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Galvão V.C., Zhang Y.C., Horrer D., Zhang T.Q., Hao Y.H., Feng Y.Q., Wang S., Schmid M., Wang J.W. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.