Pectic polysaccharides with a low degree of methylesterification form strong gels and facilitate plant cell adhesion, but their biosynthesis and modification are poorly understood. This study identifies an Arabidopsis transmembrane protein that positively regulates the degree of pectin methylesterification in seed mucilage through a mechanism mediated by ubiquitin.
Abstract
Pectins are complex polysaccharides that form the gel matrix of the primary cell wall and are abundant in the middle lamella that holds plant cells together. Their degree of methylesterification (DM) impacts wall strength and cell adhesion since unesterified pectin regions can cross-link via Ca2+ ions to form stronger gels. Here, we characterize flying saucer1 (fly1), a novel Arabidopsis thaliana seed coat mutant, which displays primary wall detachment, reduced mucilage extrusion, and increased mucilage adherence. These defects appear to result from a lower DM in mucilage and are enhanced by the addition of Ca2+ or completely rescued using alkaline Ca2+ chelators. FLY1 encodes a transmembrane protein with a RING-H2 domain that has in vitro E3 ubiquitin ligase activity. FLY1 is orthologous to TRANSMEMBRANE UBIQUITIN LIGASE1, a Golgi-localized E3 ligase involved in the quality control of membrane proteins in yeast. However, FLY1–yellow fluorescent protein (YFP) fusions are localized in punctae that are predominantly distinct from the Golgi and the trans-Golgi network/early endosome in the seed coat epidermis. Wortmannin treatment, which induces the fusion of late endosomes in plants, resulted in enlarged FLY1-YFP bodies. We propose that FLY1 regulates the DM of pectin in mucilage, potentially by recycling pectin methylesterase enzymes in the endomembrane system of seed coat epidermal cells.
INTRODUCTION
Pectin is one of the most complex natural polymers and forms the dynamic gel matrix of primary cell walls in plants (Mohnen, 2008). Pectins, particularly homogalacturonan (HG) with a low degree of methylesterification (DM), are also abundant in the middle lamella, where they mediate cell–cell adhesion (Lord and Mollet, 2002; Wolf et al., 2009a). HG consists of unbranched chains of (1→4)-α-d-galacturonic acid (GalA), and rhamnogalacturonan-II has the same backbone but is a highly substituted polymer. By contrast, the rhamnogalacturonan-I (RG-I) backbone is made of alternating GalA and (1→2)-α-l-rhamnose (Rha) units that have a variable number of branches containing mainly Ara and Gal. These GalA-rich polysaccharides are proposed to form a high molecular weight complex via multiple covalent bonds (Harholt et al., 2010).
Pectin biosynthesis occurs in the Golgi and is predicted to require at least 67 different glycosyltransferases, methyltransferases, and acetyltransferases, although only a small number of enzymes are currently known and well characterized (Mohnen, 2008; Harholt et al., 2010). Analysis of GAUT1, an HG (1→4)-α-galacturonosyltransferase (Sterling et al., 2006), has recently led to the identification of a complex for HG biosynthesis with GAUT1 and GAUT7 at its core, and 12 associated proteins, including two putative pectin methyltransferases (PMTs; Atmodjo et al., 2011). Newly synthesized HG is thought to be highly methylesterified by PMTs in the Golgi, prior to being secreted to the cell wall (Zhang and Staehelin, 1992; Willats et al., 2001c; Pelloux et al., 2007). Pectin methylesterases (PMEs) in the apoplast can demethylesterify HG in a linear (blockwise) fashion, resulting in more carboxyl groups that can form bonds with Ca2+ ions (Moustacas et al., 1991; Goldberg et al., 1996; Wolf et al., 2009a). Linear demethylesterification is known to strengthen pectin gels, to maintain cell–cell adhesion, and to limit cell elongation (Derbyshire et al., 2007). By contrast, certain PMEs can remove methylester groups in a nonlinear manner to render HG more susceptible for degradation by polygalacturonases that facilitate cell separation (Micheli, 2001; Pelloux et al., 2007). Plant cells can also control the pectin DM by secreting PME inhibitor proteins to block the active sites of PMEs in the cell wall (Jolie et al., 2010). The analysis of novel mutants that specifically affect pectin biosynthesis or modification is necessary to increase our understanding of this complex polysaccharide (Wolf et al., 2009a).
In recent years, the Arabidopsis thaliana seed coat epidermis has been successfully employed as a model system for the synthesis, secretion, and modification of cell wall components, particularly pectin (Haughn and Western, 2012). The hallmark of this cell layer is the production of three distinct cell wall structures: an outer primary wall, a pectinaceous mucilage pocket, and a cellulose-rich columella (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000, Mendu et al., 2011). Between 5 and 8 days post anthesis (DPA), large amounts of pectin are secreted to the apoplastic space at the junction of the outer tangential and radial primary walls, forming a donut-shaped pocket of mucilage around a cytoplasmic column (Western et al., 2000; Young et al., 2008). The epidermal cells then synthesize a volcano-shaped secondary wall (9 to 11 DPA), which protrudes through the center of the mucilage pocket and connects to the primary wall. Hydration of mature seeds triggers the rapid expansion of mucilage, releasing a nonadherent layer that easily detaches from the seed and an adherent layer that can only be removed with strong acid or base treatment (Western et al., 2000; Macquet et al., 2007a). During mucilage extrusion from wild-type seeds, the outer tangential primary wall detaches from the radial wall but remains attached to the columella (Western et al., 2000, 2001; Dean et al., 2007; Macquet et al., 2007b).
Analysis of mucilage mutants has led to the identification of several genes that are involved in the biosynthesis of cell wall components. Mucilage is composed primarily of unbranched RG-I, with small quantities of HG, cellulose, and xyloglucan found in the inner adherent layer (Western et al., 2000; Macquet et al., 2007a; Young et al., 2008). Mutants that lack enzymes required for pectin biosynthesis (mum4, Usadel et al., 2004; Western et al., 2004; gaut11, Caffall et al., 2009) or for pectin modification (mum2, Dean et al., 2007; Macquet et al., 2007b; sbt1.7, Rautengarten et al., 2008; bxl1, Arsovski et al., 2009; pmei6, Saez-Aguayo et al., 2013) display reduced mucilage production and/or release. MUCILAGE-MODIFIED4 (MUM4/RHM2) is a UDP-l-rhamnose synthase required for an early step in RG-I biosynthesis (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007). BXL1 encodes an α-l-arabinofuranosidase that removes arabinan residues located on the side chains of RG-I (Arsovski et al., 2009), and MUM2 encodes a β-d-galactosidase (BGAL6) that removes RG-I galactose side chain residues (Dean et al., 2007; Macquet et al., 2007b). Very little is known about HG synthesis in the Arabidopsis seed coat aside from the role of GAUT11, an α-1,4-d-galacturonosyltransferase (Caffall et al., 2009), but there is increasing evidence that establishing the correct DM of HG is essential for mucilage extrusion. Although their PME targets are currently unknown, SBT1.7, an extracellular subtilisin-like protease (Hamilton et al., 2003; Rautengarten et al., 2008), and PMEI6 (Saez-Aguayo et al., 2013) were shown to promote mucilage release by inhibiting PME activity in seed coat epidermal cells.
In addition, analysis of cesa5/mum3 and sos5 mutants revealed that cellulose and arabinogalactan proteins are involved in mucilage adherence to the seed (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011). Despite these discoveries, large gaps remain in the current knowledge of cell wall biogenesis in the seed coat.
Here, we characterize a seed coat mutant, flying saucer1 (fly1), which has more compact mucilage capsules and detached outer tangential primary walls when hydrated in water. The fly1 defects apparently result from a lower pectin DM in mucilage and can be intensified by adding Ca2+ ions or completely rescued by alkaline cation chelators. We show that FLY1 is a transmembrane RING E3 ubiquitin ligase, which controls the DM of pectin in seed mucilage.
RESULTS
fly1-1 Seeds Release Discs Upon Hydration in Water
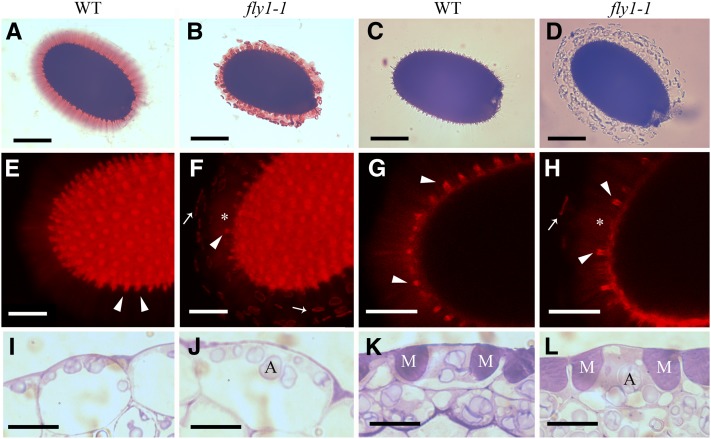
The fly1-1 mutant line was isolated by screening for seed mucilage defects in a Columbia-2 (Col-2) population mutagenized with ethyl methanesulfonate (EMS). Wild-type extruded mucilage is not homogeneous but consists of an outer, nonadherent layer that is easily removed by shaking in water and an inner, adherent layer, which remains attached to the seed even after prolonged shaking (see Supplemental Figure 1 online; Western et al., 2000; Macquet et al., 2007a). Mature wild-type seeds, shaken in water and stained with Ruthenium Red (RR), a pectin dye (Sterling, 1970), are surrounded by a pink gel-like capsule (Figure 1A). In contrast with the wild type, hydrated fly1-1 seeds appear to release smaller mucilage halos, surrounded by a large number of darkly stained small discs (Figure 1B). The difference between wild-type and fly1-1 mucilage halos is enhanced when dry seeds are hydrated directly in RR (see Supplemental Figures 1C and 1D online). The fly1-1 discs appear at the periphery of the adherent mucilage layer and are not detached from the seed after 24 h of shaking with an orbital rotator (see Supplemental Figures 1E and 1F online). This suggests that the discs are strongly bound to the adherent mucilage and may result from reduced mucilage extrusion in fly1-1 compared with the wild type. Although mucilage must be stained to be clearly observed (Figure 1C), the fly1-1 discs are also visible without staining, suggesting that they cannot be composed solely of mucilage (Figure 1D).
Figure 1.
fly1-1 Seeds Display Primary Wall Detachment and Reduced Mucilage Extrusion.
(A) and (B) Mature seeds shaken in water for 2 h and stained with RR. The mucilage capsule of fly1-1 is smaller than the wild type (WT) and is surrounded by disc-like structures.
(C) and (D) Mature seeds shaken in water for 2 h. The fly1-1 discs, unlike mucilage, can be clearly seen without staining.
(E) to (H) Mature seeds shaken in water for 2 h and stained with S4B.
(E) and (F) S4B signal from multiple optical slices.
(G) and (H) Optical slices through the middle of seeds. Arrowheads indicate S4B-labeled primary cell walls attached to columellae. The loss of primary wall attachment (asterisk) correlates with the position of fly1-1 discs (arrow).
(I) to (L) The development of fly1-1 seed coat epidermal cells is indistinguishable from the wild type at 4 DPA ([I] and [J]) and 7 DPA ([K] and [L]). M, mucilage pockets. A, amyloplasts.
Bars = 200 µm in (A) to (D), 100 µm in (E) to (H), and 15 µm in (I) to (L).
Primary Cell Walls Detach From fly1-1 Seed Coat Epidermal Cells
Mature seed coat epidermal cells contain three morphologically and chemically distinct cell walls (Western et al., 2000; Macquet et al., 2007a; Young et al., 2008). The fly1-1 discs resemble the polygonal shape of the outer tangential primary walls, which are normally attached to the columellae of hydrated seeds (Western et al., 2004; Dean et al., 2007; Stork et al., 2010). We therefore examined the position of the outer primary walls in wild-type and fly1-1 seeds shaken in water. All wild-type seed epidermal cells have primary walls attached to their columellae, but many fly1-1 cells display columellae with no attached walls (see Supplemental Figures 1G and 1H online). We further investigated the composition of the fly1-1 discs using Pontamine Fast Scarlet 4B (S4B), which preferentially stains cellulose microfibrils (Anderson et al., 2010). All wild-type seed coat epidermal cells have S4B-stained primary wall fragments attached to their columellae (Figures 1E and 1G), consistent with the observations made using unstained seeds in water. Although wild type and fly1-1 have similar levels of diffuse S4B labeling in the inner mucilage layers, fly1-1 seeds display many S4B-stained discs, which are at the outer edge of the mucilage halo and are positioned above columellae that lack S4B-labeled primary wall attachment (Figures 1F and 1H).
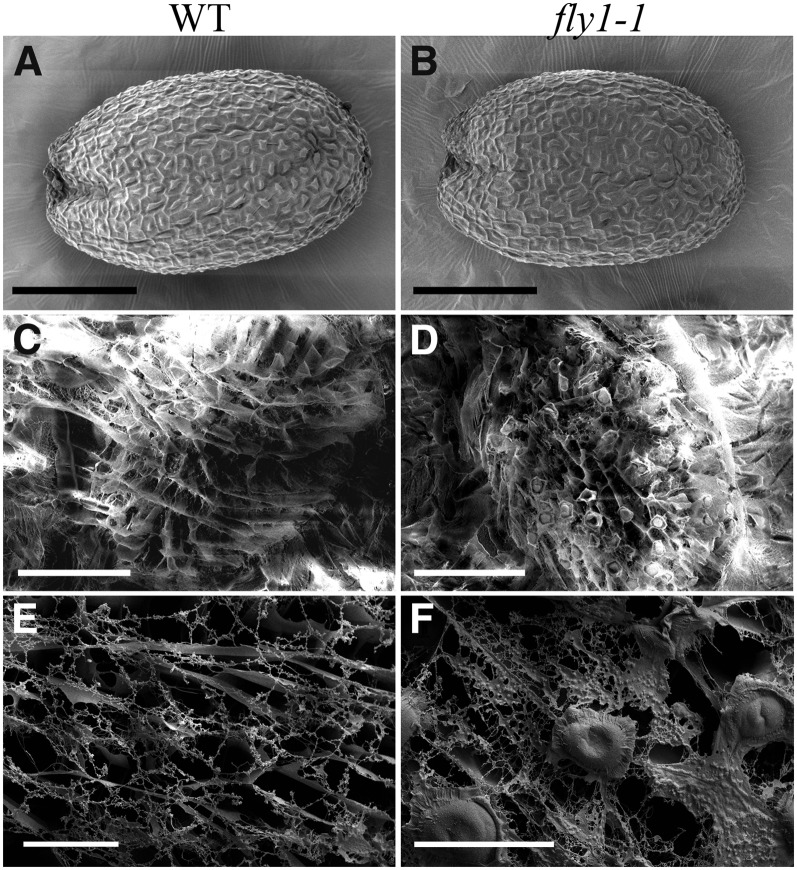
The surface morphology of wild-type and fly1-1 dry seeds was examined by scanning electron microscopy (SEM). Wild-type and fly1-1 seed epidermal cells displayed outer tangential cell walls and columellae with similar shapes and sizes (Figures 2A and 2B). To investigate the structure of extruded mucilage, seeds hydrated in water were immediately frozen in liquid nitrogen and examined with cryo-scanning electron microscopy (cryoSEM). Both wild-type and fly1-1 hydrated seeds display intricate mesh-like networks of mucilage (Figures 2C to 2F). However, only the mutant seeds show electron-dense discs on top of the extruded mucilage matrix (Figures 2D and 2F), consistent with position of RR-stained discs at the edge of the fly1-1 mucilage halo (Figure 1B). Since the outer primary cell walls viewed with SEM (Figure 2B) and the fly1-1 discs observed with cryoSEM (Figures 2D and 2F) or RR staining (Figure 1B) resemble polygons, we measured their diameters (>30 samples per technique) and compared them using Student’s t tests. The diameter of fly1-1 outer tangential primary walls observed with SEM (32.95 ± 0.69 µm; mean ± se) is similar to that of RR-stained discs (32.98 ± 0.88 µm; P = 0.98). The cryoSEM discs (30.88 ± 0.70 µm) are similar to RR-stained discs (P = 0.066, >0.05) but significantly smaller than walls in SEM micrographs (P = 0.038, <0.05).
Figure 2.
Analysis of Dry and Hydrated Seeds by Scanning Electron Microscopy and Cryo-Scanning Electron Microscopy.
(A) and (B) The surface morphology of dry mature fly1-1 seeds viewed with scanning electron microscopy is indistinguishable from the wild type (WT).
(C) to (F) Cryo-scanning electron microscopy of mucilage extruded from mature seeds hydrated in water. Wild-type seeds showed an irregular mucilage matrix before (C) and after sputter coating (E). fly1-1 seeds have discs on top of the mucilage matrix before (D) and after sputter coating (F).
Bars = 200 µm in (A) to (D) and 50 µm in (E) and (F).
Analysis of hydrated seeds prior to and after S4B staining revealed that fly1-1 discs contain cellulose and are positioned above columellae lacking attached primary walls. The fly1-1 discs in light and cryoSEM micrographs were similar in size to the primary walls of the seed coat epidermis visualized with SEM. Overall, the results in this section strongly support the hypothesis that the outer tangential primary cell walls of many fly1-1 seed coat epidermal cells detach from columellae upon mucilage extrusion in water.
fly1 Seeds Have Normal Development and Monosaccharide Composition
To determine if the fly1-1 mucilage extrusion defects result from abnormal seed coat differentiation, fixed sections of developing seeds were analyzed using light microscopy. The development of fly1-1 seed epidermal cells is indistinguishable from the wild type and mucilage biosynthesis appears unaffected (Figures 1I to 1L), unlike mutants that synthesize less pectin, such as mum4 (Western et al., 2004). Primary wall attachment to the columella appears identical to the wild type in live fly1-1 seed coat epidermal cells at 12 DPA (see Supplemental Figures 1I and 1J online). The unaltered development of fly1-1 seed coat cells is consistent with the normal surface morphology of dry mature seeds (Figures 2A and 2B).
The monosaccharide composition of wild-type and fly1-1 whole seeds was determined using high-performance anion-exchange chromatography (HPAEC). The alcohol-insoluble residues (AIRs) prepared from fly1-1 and wild-type seeds contained similar monosaccharide levels (Table 1), consistent with seed coat mutants that primarily affect pectin modification rather than pectin biosynthesis (Dean et al., 2007; Arsovski et al., 2009). Overall, the fly1-1 seed development and the whole seed HPAEC results suggest that FLY1 is not required for the biosynthesis of a major cell wall polysaccharide.
Table 1. Monosaccharide Composition of Sequentially Extracted Mucilage and Whole Seeds.
| Mucilage Extraction 1 |
Mucilage Extraction 2 |
Whole Seeds |
||||
|---|---|---|---|---|---|---|
| Sugar | Wild Type | fly1-1 | Wild Type | fly1-1 | Wild Type | fly1-1 |
| Fuc | Trace | Trace | Trace | Trace | 4.03 ± 0.20 | 3.98 ± 0.17 |
| Ara | 1.03 ± 0.80 | 0.67 ± 0.12 | 0.35 ± 0.01 | 0.35 ± 0.05 | 182.30 ± 5.75 | 170.40 ± 5.40 |
| Rha | 46.92 ± 1.18 | 26.60 ± 2.72 | 12.27 ± 1.66 | 19.69 ± 1.83 | 192.38 ± 8.52 | 174.88 ± 5.49 |
| Gal | 2.35 ± 0.16 | 1.14 ± 0.15 | 0.64 ± 0.10 | 1.06 ± 0.09 | 110.58 ± 3.98 | 100.48 ± 3.19 |
| Glc | 1.20 ± 0.18 | 0.77 ± 0.07 | 0.80 ± 0.19 | 0.93 ± 0.11 | 178.62 ± 13.04 | 179.46 ± 6.17 |
| Xyl | 2.76 ± 0.05 | 1.96 ± 0.32 | 0.63 ± 0.12 | 1.50 ± 0.12 | 48.51 ± 1.26 | 48.43 ± 2.94 |
| GalA | 58.67 ± 0.60 | 33.70 ± 3.86 | 13.74 ± 2.38 | 26.40 ± 2.26 | 312.89 ± 14.34 | 313.02 ± 7.37 |
Extraction 1 (gently shaking in water for 1 h) contains mucilage without discs, while extraction 2 (an additional 2 h of vigorously shaking in water) includes both mucilage and discs (see Supplemental Figure 2 online). Values are the mean ± se of four (mucilage) or three samples (whole seed) and represent nmol sugar per milligram seed used for mucilage extraction or milligram alcohol-insoluble residue hydrolyzed (whole seed). Results were verified using two additional biological replicates.
fly1 Mucilage Is More Adherent Than Wild-Type Mucilage
We investigated if FLY1 affects mucilage extrusion and adhesion by analyzing the monosaccharide compositions of sequential mucilage extractions from wild-type and fly1-1 seeds. Although fly1-1 discs are not detached from the seed by gentle shaking, vigorous agitation using a vortex mixer can remove the majority of polygonal discs and the adherent mucilage (see Supplemental Figure 2 online). Therefore, we extracted mucilage without discs by gently shaking seeds in water for 1 h, followed by mucilage including discs by vigorously shaking in water for an additional 2 h.
HPAEC monosaccharide analysis of the first mucilage extraction showed a strong reduction of Rha and GalA in fly1-1 mucilage relative to the wild type, but the subsequent fly1-1 mucilage extraction contained almost twice as much Rha and GalA compared with the wild type (Table 1). All other monosaccharides detected are minor mucilage components, but showed similar trends to Rha and GalA.
In addition to the sequential extractions, total mucilage amounts were extracted from water-hydrated seeds by adding NaOH to a final concentration 0.2 M and vortex mixing for 1 h. No RR-stained mucilage remained around wild-type and fly1-1 seeds (see Supplemental Figures 2K and 2L online). Wild-type and fly1-1 mucilage extracted by NaOH have similar monosaccharide compositions (see Supplemental Figure 2M online).
Despite having whole seed and total mucilage monosaccharide compositions similar to the wild type, fly1-1 seeds display a reduced mucilage extractability in water. The fly1-1 mucilage capsule is more adherent than the wild type since vigorous shaking in water partially compensates for the reduced monosaccharide levels in the first fly1-1 water extraction.
The fly1 Mucilage Defects Are Calcium and pH Dependent
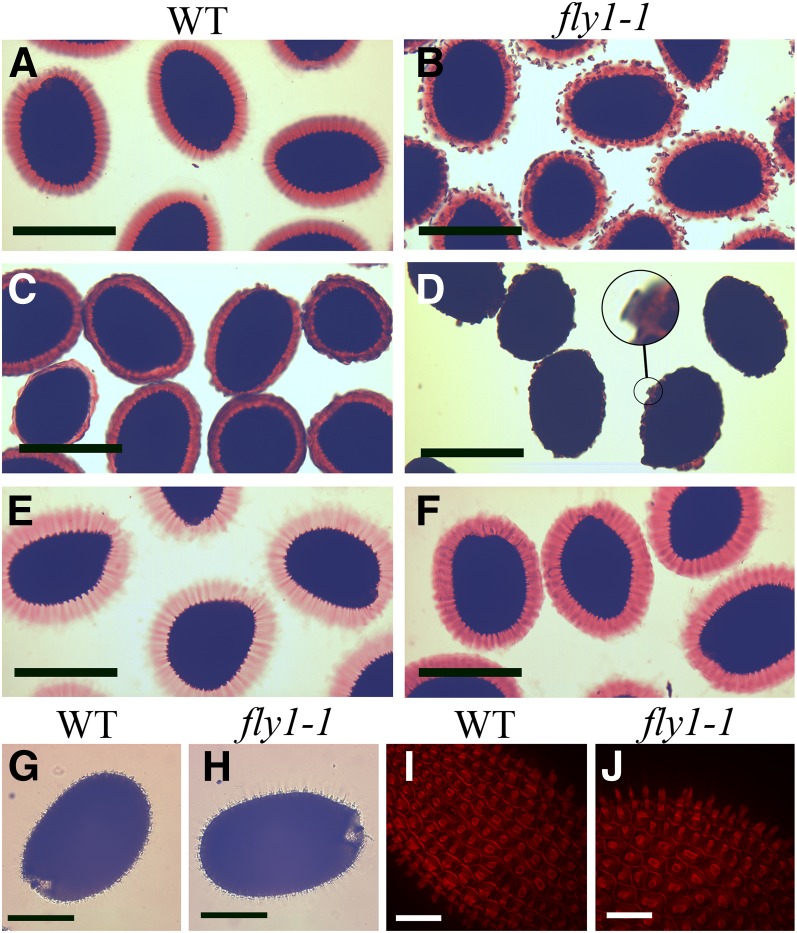
Similar to the wild type, fly1-1 seeds release a mucilage halo within 10 s of hydration in water (see Supplemental Figure 3 online). However, fly1-1 discs continue to be released after more than 60 s (see Supplemental Figure 3D online), suggesting that some fly1-1 cells have delayed mucilage release. Compared with the wild type, fly1-1 seeds have a more compact and adherent mucilage halo (Figures 3A and 3B, Table 1). To identify the cause of reduced mucilage expansion in the fly1-1 mutant, we examined the effects of hydrating seeds in a CaCl2 solution. Ca2+ ions are required for the formation of cross-links between unesterified HG blocks and can therefore strengthen the pectin gel matrix (Jarvis, 1984). Interestingly, hydration of seeds directly in 50 mM CaCl2 almost completely impairs mucilage extrusion from the fly1-1 mutant but not from wild-type seeds, which show only a small reduction in mucilage halo size (Figures 3C and 3D). This suggests that pectins in fly1-1 mucilage can form more calcium cross-links than in the wild type. Although only a few CaCl2-treated fly1-1 seed epidermal cells release mucilage, all of these appear to have discs atop their compact mucilage columns (Figure 3D, inset).
Figure 3.
Effects of Ca2+ and EDTA on fly1-1 Mucilage Extrusion.
(A) to (F) Mature seeds shaken for 45 min in water ([A] and [B]), 50 mM CaCl2 ([C] and [D]), or 50 mM EDTA ([E] and [F]) and then stained with RR. Only a few CaCl2-treated fly1-1 cells release mucilage, but these appear to have discs atop compact mucilage columns ([D], inset). EDTA-treated wild-type (WT) and fly1-1 seeds have equally large mucilage capsules.
(G) and (H) Unstained wild-type and fly1-1 seeds shaken in EDTA have primary walls attached to all columellae and do not display discs.
(I) and (J) S4B staining confirms that the columellae of EDTA-hydrated seeds have primary cell walls attached to them. Note the absence of fly1-1 discs ([H] and [J]).
Bars = 200 µm in (A) to (F), 150 µm in (G) and (H), and 75 µm in (I) and (J).
In stark contrast with the addition of Ca2+ ions, cation chelators such as EDTA can disrupt cross-links between unesterified HG chains and facilitate seed mucilage extrusion (Rautengarten et al., 2008; Arsovski et al., 2009). Hydration of mature seeds in 50 mM EDTA pH 8.0 completely rescued the fly1-1 mucilage extrusion defect, resulting in equally large wild-type and fly1-1 mucilage capsules without any visible discs (Figures 3E and 3F). RR-stained discs were not observed in EDTA-hydrated fly1-1 seeds (Figure 3F), indicating that EDTA may also rescue the primary wall detachment phenotype of this mutant. Surprisingly, unstained fly1-1 seeds hydrated directly in EDTA look identical to wild-type seeds and have primary cell wall fragments attached to all columellae (Figures 3G and 3H). S4B staining confirmed that the primary walls stay attached to columellae in fly1-1 seeds hydrated with EDTA (Figures 3I and 3J).
Since pectin is stabilized at acidic pH and can be degraded at alkaline pH (Sriamornsak, 2003), we investigated if the suppression of the fly1-1 mucilage defects is also pH dependent. The EDTA pH 6.0 treatment largely rescued the fly1-1 defects similar to EDTA pH 8.0 (see Supplemental Figures 4A to 4C online). Treatment with 50 mM HEPES buffer, which does not bind Ca2+, partially rescued the fly1-1 defects at pH 8.0 but not at pH 6.0 (see Supplemental Figures 4D to 4F online). Seeds treated with both EDTA and HEPES resembled the EDTA-treated seeds (see Supplemental Figures 4G to 4I online). Removal of Ca2+ by EDTA is largely sufficient to rescue the fly1-1 mucilage defects, but an alkaline pH also contributes to rescue the mutant phenotype.
The fly1 Mucilage Has More Unesterified HG Than the Wild Type
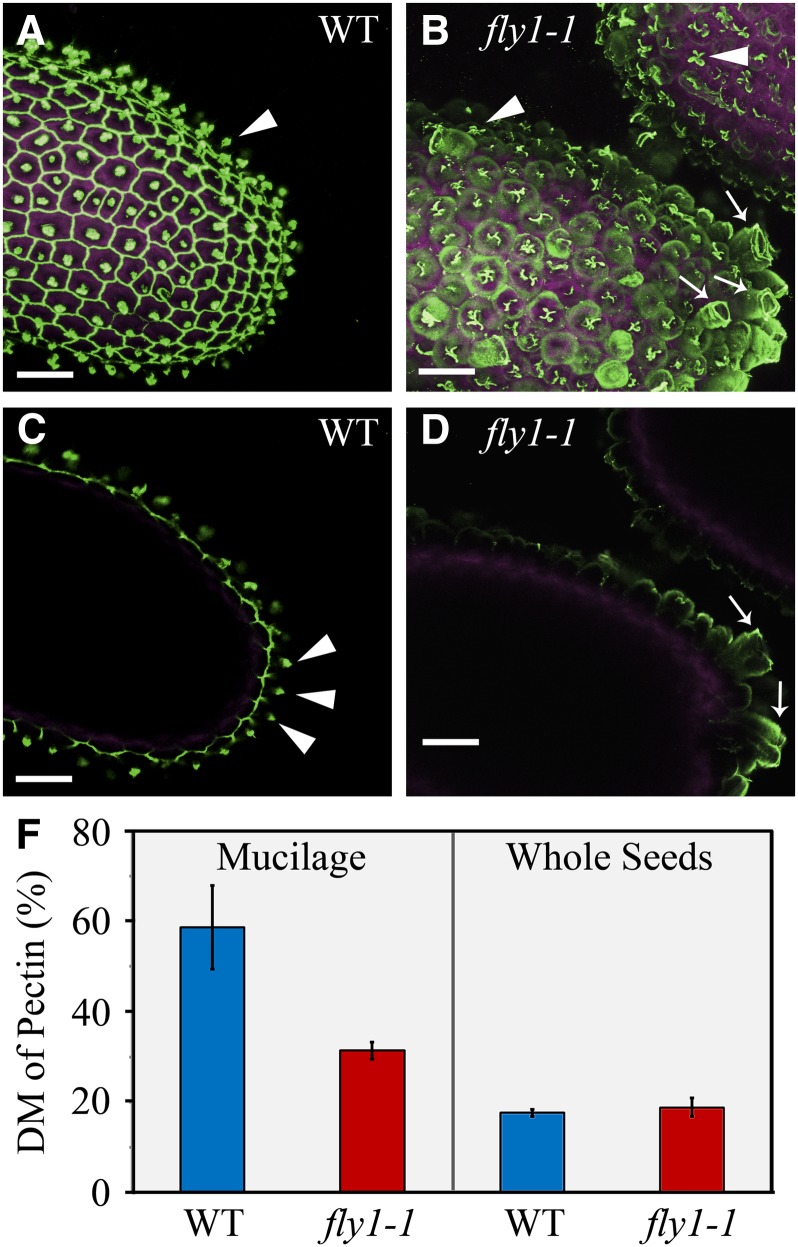
Since the fly1-1 defects are sensitive to the presence of Ca2+ ions, fly1-1 mucilage likely contains more unesterified GalA than wild-type mucilage. Immunolabeling of mature seeds with three anti-HG antibodies was conducted to determine if fly1-1 mucilage displays an altered pattern of methylesterification. The same image acquisition settings were used within each immunolabeling experiment. 2F4, an antibody that specifically binds unesterified blocks of HG cross-linked by Ca2+ ions (Liners et al., 1989), only labeled primary cell wall material in wild-type seeds (Figure 4), including the outer tangential walls attached to columellae and the radial walls. Interestingly, fly1-1 seeds displayed 2F4 signal in both the primary cell wall and in the extruded mucilage. The abundance of 2F4 signal in fly1-1 mucilage, along its absence in wild-type mucilage, is consistent with increased amounts of unesterified HG in the mutant.
Figure 4.
Immunolabeling and Biochemical Analysis of Pectin DM in Seeds and Mucilage.
(A) to (D) 2F4 immunolabeling (green) of unesterified HG and seed intrinsic fluorescence (magenta).
(A) and (B) Maximum intensity signals from multiple optical stacks.
(C) and (D) Optical slices through the middle of seeds. In the wild type (WT), 2F4 labels attached primary walls (arrowheads). For fly1-1, 2F4 labels both the detached primary walls (arrows) and the underlying mucilage. Bars = 50 µm.
(F) Biochemical determination of pectin DM in mucilage and whole seeds. Wild-type and fly1-1 whole seeds have a similar DM, but fly1-1 mucilage has a significantly lower DM than wild-type mucilage. Values represent the mean ± se of four technical replicates. Two additional biological replicates showed the same trend.
Two additional antibodies with broader specificities were also used to characterize the seed mucilage: JIM7, which binds to heavily (35 to 81%) methylesterified HG (Knox et al., 1990), and JIM5, which binds partially (up to 40%) methylesterified HG (Vandenbosch et al., 1989). JIM5 and JIM7 recognize partially overlapping domains of seed mucilage and both labeled larger regions of fly1-1 mucilage compared with the wild type (see Supplemental Figure 5 online). The results of all three antibodies are consistent with fly1-1 mucilage containing more unesterified HG than the wild type.
To exclude the possibility that increased immunolabeling of fly1-1 mucilage results from higher epitope accessibility rather than larger amounts of unesterified HG, the DM of mucilage and whole seeds was determined using biochemical assays. The DM was calculated as a percentage molar ratio of methanol released after cell wall saponification (Lionetti et al., 2007) to uronic acids measured using the m-hydroxydiphenyl assay (van den Hoogen et al., 1998). Mucilage was extracted by vigorously shaking seeds in 50 mM EDTA for 1 h (see Supplemental Figures 2I and 2J online). Whereas wild-type and fly1-1 whole seeds contain pectin with a similar DM (Student’s t test, P = 0.60, >0.05), fly1-1 mucilage has a significantly lower DM than wild-type mucilage (Student’s t test, P = 0.02, <0.05; Figure 4F). The lower DM in fly1-1 mucilage corroborates the increased 2F4 immunolabeling of unesterified HG (Figures 4B and 4D) and accounts for the strong effects of CaCl2 and EDTA treatments on fly1-1 mucilage defects (Figure 3). Since the altered DM appears to be the primary defect of fly1-1 mucilage, the detached discs may result from increased binding of mucilage to the primary wall. Along with the normal DM and monosaccharide composition in fly1-1 whole seeds (Figure 4F, Table 1), these results suggest that FLY1 is required to establish the correct DM in seed mucilage.
Cloning of the FLY1 Gene and Expression Analysis
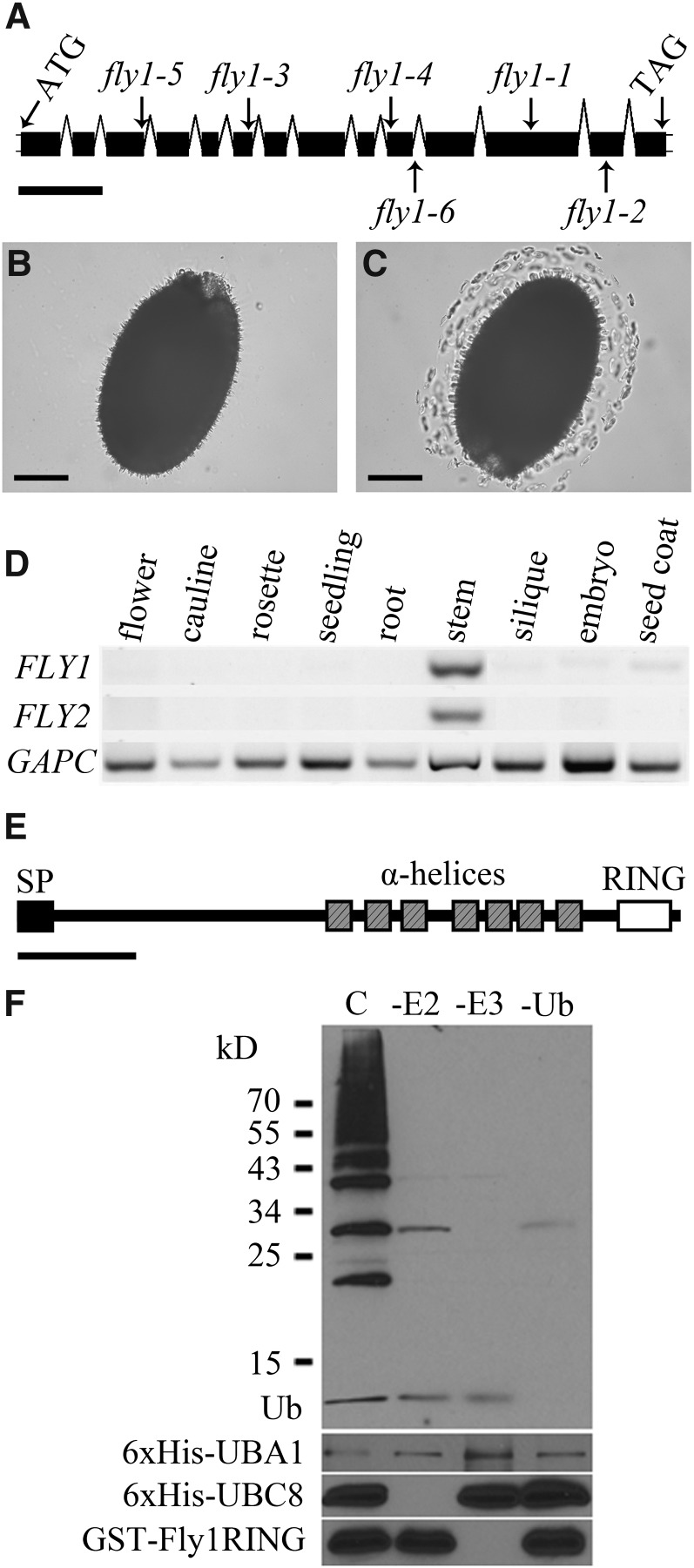
The fly1-1 phenotype was shown to segregate as a single recessive nuclear mutation (3 wild type:1 fly1; χ2 = 0.333; P > 0.5; n = 169). The position of the fly1-1 mutation was mapped to a 180-kb region containing 60 genes near the end of chromosome IV. We screened nearly 100 independent T-DNA insertions in 58 of the 60 candidate genes for mucilage defects. Five lines, fly1-2 (SALK_067290), fly1-3 (SALK_144822), fly1-4 (SALK_139156), fly1-5 (SALK_000015), and fly1-6 (SALK_062423), all contained mutations in At4g28370 (Figure 5A) and produced seeds that release compact mucilage halos and discs (see Supplemental Figure 6 online). PCR-based segregation analysis using these five independent lines indicated that the T-DNA insertions were responsible for the fly1 phenotype and were consistent with recessive loss-of-function mutations.
Figure 5.
Characterization of the FLY1 Gene and Its Encoded Protein.
(A) FLY1 gene structure and mutations. T-DNA insertions (fly1-2 to fly1-6) are indicated with arrows. Boxes and connecting lines represent exons and introns.
(B) and (C) Genomic complementation of fly1-1 with At4g28370.
(B) fly1-1 plants transformed with FLY1pro:FLY1-YFP produce seeds that do not release discs.
(C) fly1-1 plants transformed with an empty pGreenII0229 vector produced seeds that still released discs.
(D) RT-PCR analysis of FLY1 and FLY2 transcripts in Col-2 tissues. Embryo and seed coat RNA was isolated from seeds at 7 DPA. GAPC was used as a loading control.
(E) Predicted architecture of the FLY1 protein. FLY1 has a signal peptide (SP), seven α-helices (Schwacke et al., 2003), and a RING-H2 domain. Bars = 300 bp in (A), 150 µm in (B) and (C), and 100 amino acids in (E).
(F) The FLY1 RING domain has E3 ubiquitin ligase activity in vitro. Complete (C) ubiquitination assays contained E1 enzyme 6xHis-At-UBA1, E2 enzyme 6xHis-At-UBC8, E3 enzyme GST-Fly1RING (only includes the soluble RING domain), and Ubiquitin (Ub). Top: Immunoblot using anti-Ub. The ladder and smear represent poly-Ub species of the enzymes present in the reaction. No poly-Ub is detected when omitting 6xHis-At-UBC8 (-E2 lane), GST-Fly1RING (-E3 lane), or ubiquitin (-Ub lane) from the assay. The asterisk marks
anti-Ub cross-reacting with the E3 enzyme. Center: anti-His to detect E1 and E2 enzymes. Bottom: anti-GST to detect GST-Fly1RING.
The At4g28370 gene encodes a previously uncharacterized protein belonging to the Really Interesting New Gene (RING) finger superfamily (TAIR 10; Lamesch et al., 2012). Sequencing of At4g28370 in the fly1-1 background revealed a G-to-A transition, consistent with EMS mutation, at 3903 bp that changes the amino acid Trp (residue 460) to a stop codon. This early stop codon is predicted to truncate the peptide encoded by the FLY1 gene, resulting in the loss of the conserved RING domain.
The fly1-2 and fly1-3 alleles were characterized in greater detail and were shown to be indistinguishable from fly1-1 with cryoSEM, S4B staining, and anti-HG immunolabeling (data not shown). Complementation tests between fly1-1, fly1-2, and fly1-3 failed to rescue the mucilage defects, consistent with mutations occurring in the same gene. In addition, we performed molecular complementation of fly1-1 with a FLY1pro:FLY1-YFP (Yellow Fluorescent Protein) construct, containing wild-type At4g28370 genomic DNA (from 1 kb upstream of the ATG site to, but not including, the stop codon) fused to citrine YFP in the pAD vector (DeBono, 2012). Multiple independent fly1-1 lines transformed with FLY1pro:FLY1-YFP produced seeds that resemble the wild type (Figure 5B), but fly1-1 plants transformed with empty pAD vector produced seeds that still released many discs (Figure 5C).
Consistent with the seed mucilage defects of the fly1 mutant alleles, the Electronic Fluorescent Pictograph browser (eFP; Winter et al., 2007) revealed very high FLY1 transcript levels in seeds relative to other Arabidopsis organs and preferential expression in the seed coat compared with the endosperm and embryo (Schmid et al., 2005; Le et al., 2010). Interestingly, FLY1 has one paralog in Arabidopsis (FLY2, At2g20650) and at least one ortholog in fungi, protists, green algae, mosses, and other vascular plant species with a sequenced genome (Ostlund et al., 2010; Goodstein et al., 2012; Van Bel et al., 2012). FLY2 encodes a protein with 84.5% amino acid identity to FLY1 (Huang and Miller, 1991). Although FLY2 transcripts are less abundant than FLY1 in developing seeds (Zimmermann et al., 2004; Winter et al., 2007; Le et al., 2010), FLY2 shows preferential expression in the seed coat compared with other seed tissues. To investigate if FLY2 is also involved in mucilage biosynthesis, we isolated homozygous fly2-1 (SALK_140887), fly2-2 (SALK_023653), fly2-3 (SALK_074297), fly2-4 (SAIL_515_C07), and fly2-5 (WiscDsLox412F02) T-DNA lines (see Supplemental Figure 7 online). Hydrated fly2 seeds display wild-type RR staining (see Supplemental Figure 6 online), S4B staining, and 2F4, JIM5, and JIM7 immunolabeling (data not shown).
The function of FLY1 and FLY2 may not be limited to the seed coat since RT-PCR analysis detected the highest transcript levels in stems (Figure 5D). According to multiple microarray data sets, both genes are most strongly expressed in stem and root xylem cells (see Supplemental Figure 8A online; Brady et al., 2007; Hruz et al., 2008), suggesting that they may play a role in vasculature development. However, neither fly1 nor fly2 mutants show any obvious defects in leaf, stem, or root morphology. In the GeneCAT database (Mutwil et al., 2008), FLY1 and FLY2 are highly coexpressed with KNOTTED1-LIKE HOMEODOMAIN PROTEIN7 (KNAT7), which encodes a transcription factor shown to regulate cell wall biosynthesis in xylem and seed coat cells (Romano et al., 2012; Bhargava et al., 2013). Although knat7 mutants display an irregular xylem cell phenotype, both fly1 and fly2 xylem cells resembled the wild type (see Supplemental Figure 8B online).
Analysis of the FLY1 Peptide Sequence
FLY1 contains an N-terminal signal peptide that targets the protein to the endomembrane system (Petersen et al., 2011) and is predicted to be cleaved in the endoplasmic reticulum between amino acids 32 and 33. The consensus prediction of the ARAMEMNON plant membrane protein database is that FLY1 has seven transmembrane-spanning α-helices (Figure 5E; Schwacke et al., 2003). The C-terminal end of the FLY1 protein contains a RING finger, which is conserved in all eukaryotes and coordinates two zinc ions (Quevillon et al., 2005; Sigrist et al., 2010; Lamesch et al., 2012). The predicted ARAMEMNON topology of the FLY1 protein indicates that the C-terminal RING domain faces the cytosol. Both FLY1 and FLY2 have a RING domain with a C3H2C3 motif (abbreviated RING-H2; Stone et al., 2005) based on the arrangement of the eight zinc binding Cys and His residues (see Supplemental Figure 9 online). Interestingly, the ortholog of FLY1 (28% amino acid identity) in the yeast Saccharomyces cerevisiae is TRANSMEMBRANE UBIQUITIN LIGASE1 (TUL1), a Golgi-localized E3 ligase that tags misfolded membrane proteins with ubiquitin for transport to endosomes and degradation in the vacuole (Reggiori and Pelham, 2002). TUL1 has a C-terminal RING-H2 domain similar to FLY1 and FLY2 (see Supplemental Figure 9 online), along with an N-terminal signal peptide and seven α-helices (Reggiori and Pelham, 2002).
To determine if FLY1 functions as an E3 ligase, we conducted an in vitro ubiquitination assay (Hardtke et al., 2002). Ubiquitination results from the sequential action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) and can regulate a protein’s stability, activity, or location (Hicke and Dunn, 2003; MacGurn et al., 2012). Since recombinant expression and purification of transmembrane proteins is challenging, we expressed only the soluble C terminus of FLY1 containing the RING domain fused to glutathione S-transferase (GST-Fly1RING). The complete in vitro ubiquitination assay contained ubiquitin and three recombinant Arabidopsis proteins: E1 enzyme 6xHis-UBA1, E2 enzyme 6xHis-UBC8, and E3 enzyme GST-Fly1RING. The polyubiquitin smear, detected by immunoblot analysis using an antiubiquitin antibody in the complete assay (Figure 5F), demonstrates that the FLY1 RING domain is capable of mediating polyubiquitination. Omission of the E2 enzyme, the E3 ligase, or the ubiquitin components abolished formation of polyubiquitinated species as expected (Figure 5F).
The E3 ubiquitin ligase activity of FLY1 and the lower pectin DM of fly1 mucilage suggest that FLY1 may positively regulate PMTs or negatively regulate PMEs. While putative PMTs are membrane bound (Mouille et al., 2007, Held et al., 2011; Miao et al., 2011), most PMEs are soluble or have a signal peptide that is cleaved in the endoplasmic reticulum (Pelloux et al., 2007) and therefore do not have cytosolic Lys residues, which can be ubiquitinated (MacGurn et al., 2012). Thus, we investigated if PMEs coexpressed with FLY1 have membrane domains and putative ubiquitination sites. PME6 (At1g23200) and PME16 (At2g43050) are preferentially expressed in the seed coat and are coexpressed with FLY1 in GeneCAT (Mutwil et al., 2008) and ATTED-II databases (Obayashi et al., 2011). Unlike most PMEs, PME6 and PME16 are predicted to have an internal α-helix near the N terminus and at least one cytosolic Lys residue (Schwacke et al., 2003).
FLY1-YFP Expression and Subcellular Localization
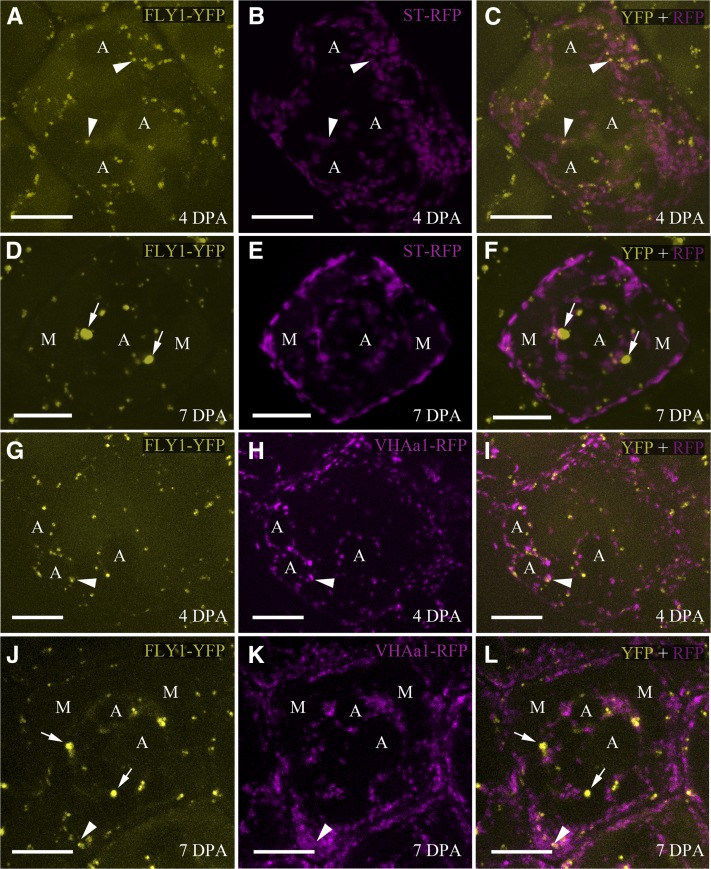
The expression and subcellular localization of FLY1 was analyzed using the previously described FLY1pro:FLY1-YFP construct that complements the fly1-1 mutation (Figure 5B). We used identical image acquisition settings for the different tissues and developmental stages presented within each figure. Multiple independent plants containing the FLY1pro:FLY1-YFP transgene displayed strong YFP fluorescence in the seed coat epidermis (Figure 6; see Supplemental Figure 10 online). FLY1-YFP expression begins at 4 DPA, is highest at 7 DPA, when mucilage synthesis and secretion is at its peak, and decreases by 10 DPA, at the start of columella deposition (see Supplemental Figures 10A to 10F online). No FLY1-YFP signal was observed in dissected embryos at 7 DPA (see Supplemental Figure 10H online) or in tissues more suitable for cell imaging such as leaves and roots.
Figure 6.
Localization of FLY1-YFP, ST-RFP, and VHAa1-RFP in Seed Coat Epidermal Cells.
Maximum intensity signals from multiple stacks. Composite images (right panels) of FLY1-YFP (left panels) and RFP-tagged markers (middle panels). FLY1-YFP is localized in punctae at 4 DPA ([A] and [G]) and in punctae and larger bodies (arrows) at 7 DPA ([D] and [J]). FLY1-YFP punctae are associated with (arrowheads) but predominantly distinct from the Golgi marker ST-RFP ([C] and [F]) and the TGN/EE marker VHAa1-RFP ([I] and [L]). A, amyloplasts. M, mucilage pockets. Bars = 10 µm.
FLY1-YFP fusion proteins primarily localize in a small number of intracellular punctae in seed coat epidermal cells and are not observed in the mucilage pocket or in the primary cell wall (Figure 6). At 7 DPA, FLY1-YFP fluorescence also appears in several larger compartments (Figures 6D and 6J, arrows). Since TUL1 is known to be Golgi localized (Reggiori and Pelham, 2002), we compared the localization of FLY1-YFP with the distribution of the Golgi marker α-2,6-sialyltransferase (ST) tagged with red fluorescent protein (RFP; Batoko et al., 2000; Brandizzi et al., 2002; Lee et al., 2002). ST-RFP labels a large number of intracellular punctae (Figures 6B and 6E), which are in close proximity to FLY1-YFP bodies and partially overlap with a few of them (Figures 6A to 6F, arrowheads). Surprisingly, however, the majority of FLY1-YFP compartments do not colocalize with the Golgi marker ST-RFP at either 4 or 7 DPA (Figures 6C and 6F). We also analyzed the subcellular localization of the trans-Golgi network/early endosome (TGN/EE) marker VHAa1-RFP (Dettmer et al., 2006), which is distinct from the Golgi marker ST-RFP (Viotti et al., 2010). Although a few FLY1-YFP punctae partially colocalize with VHAa1-RFP in seed coat epidermal cells at 4 and 7 DPA (Figures 6G to 6L, arrowheads), the localization of FLY1-YFP is largely distinct from that of the TGN/EE. The fluorescent markers tested were excluded from seed coat mucilage pockets and the amyloplasts (Figure 6).
We were unable to stain seeds expressing FLY1-YFP with the endocytic tracer FM4-64 (Viotti et al., 2010) due to a lack of dye uptake by developing seed coat cells. Seed epidermal cells pretreated with 10% DMSO to increase permeability were also not affected by Brefeldin A, a drug commonly used in secretion and endocytosis studies (Robinson et al., 2008). However, 7 DPA seed coat epidermal cells treated with wortmannin, a drug that induces the fusion of late endosomes/multivesicular bodies/prevacuolar compartments (TLEs/MVBs/PVCs) in plants (Wang et al., 2009), showed a greater number of large FLY1-YFP bodies than control seeds (see Supplemental Figures 11A and 11B online). Wortmannin treatment increased the ratio of large to small FLY1-YFP bodies from 2 to 39% in one biological replicate and from 13 to 28% in a second biological replicate (see Supplemental Figures 11C and 11D online).
DISCUSSION
We identified the FLY1 locus through a forward genetic screen for Arabidopsis mutants with abnormal seed mucilage extrusion. Histological and biochemical analyses revealed that loss-of-function mutations in the FLY1 gene cause two distinct mucilage defects when seeds are hydrated in water. The most obvious phenotype of fly1 mutant seeds is the presence of a large number of disc-like structures at the edge of the extruded mucilage capsule. The discs appear to be mucilage-bound primary cell walls that detach from the columellae of epidermal cells. Close examination of fly1 seeds revealed that mucilage extrusion is also impaired, resulting in a more compact RR-stained capsule that is more difficult to extract than wild-type mucilage. Despite these defects, fly1 and wild-type seeds produce similar amounts of mucilage and display indistinguishable morphology during development. We found that hydration of fly1 seeds in a CaCl2 solution significantly impairs mucilage release, suggesting that fly1 mucilage can form more pectin cross-links mediated by Ca2+ ions than the wild type. Calcium bridges are required to cross-link unesterified HG chains and to maintain the strength of pectin gels (Willats et al., 2001a). EDTA, a cation chelator, completely rescues the fly1 mutant phenotype at pH 8.0, including both the size of the mucilage capsule and the disc formation. Alkaline solutions can degrade pectin gels (Sriamornsak, 2003), but a HEPES pH 8.0 treatment, which does not remove Ca2+ ions, only partially rescued the mutant phenotype. All fly1 seed epidermal cells treated with EDTA pH 8.0 surprisingly had outer tangential primary walls attached to their columellae and no visible discs, suggesting that fly1 primary wall detachment does not result from weaker connections to the columellae. Instead, the force generated by slowly expanding mucilage may detach the primary walls from columellae of fly1 seeds hydrated in water. Therefore, the primary defect in fly1 appears to be increased mucilage cohesion.
Since HG with a low DM represents the only cell wall component that uses Ca2+ ions as its main mechanism for cross-linking, fly1 mucilage was expected to contain more unesterified HG than the wild type. Indeed, seed immunolabeling with three anti-HG antibodies and biochemical assays of the pectin DM demonstrate that fly1 mucilage has a significantly lower DM than wild-type mucilage. An increased number of unesterified HG blocks would strengthen the pectin gel matrix, reduce mucilage expansion, and increase its adherence to the seed, consistent with the fly1 mucilage defects. Increased binding of mucilage to the primary wall combined with reduced extrusion may generate sufficient pressure for mucilage to detach outer primary walls from columellae when fly1 is hydrated in water. The 2F4 immunolabeling supports this model, as unesterified HG was detected in both the fly1 discs and the underlying compact mucilage columns. Although mucilage is mainly composed of unbranched RG-I (Western et al., 2000, 2004; Penfield et al., 2001; Usadel et al., 2004; Dean et al., 2007; Macquet et al., 2007a, 2007b), changes in the DM of HG can have a profound impact on the entire pectin gel matrix if RG-I and HG are covalently bonded as the current models suggest (Harholt et al., 2010). Overall, the analysis of fly1 seeds suggests that FLY1 is required for maintaining a high pectin DM in seed mucilage.
We identified FLY1 as At4g28370 using positional cloning and showed that multiple independent T-DNA alleles have the same phenotype as the fly1-1 EMS line. Molecular complementation of fly1-1 with a FLY1pro:FLY1-YFP transgene rescued the mutant phenotype. The FLY1 gene encodes a previously uncharacterized protein with seven transmembrane spans and a C-terminal RING-H2 domain. RING fingers are the catalytic sites of E3 ubiquitin ligases, which mediate the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to target proteins (Glickman and Ciechanover, 2002). Since FLY1 and FLY2 are putative E3 ligases (Stone et al., 2005) and their yeast ortholog, TUL1, is a known yeast E3 ligase (Reggiori and Pelham, 2002), we tested the ability of the FLY1 RING-H2 domain to transfer ubiquitin to a substrate in vitro (Hardtke et al., 2002). As predicted in silico, the FLY1 RING domain functions as an E3 ligase in the in vitro ubiquitination assay.
We investigated the expression and subcellular localization of FLY1-YFP under the transcriptional control of the native FLY1 promoter. FLY1-YFP displayed a punctate distribution in the cytoplasm of developing seed coat epidermal cells but also appeared in enlarged bodies at the stage of mucilage biosynthesis. Unlike TUL1, which is Golgi localized (Reggiori and Pelham, 2002), the majority of FLY1-YFP compartments were distinct from the Golgi marker ST-RFP in seed coat epidermal cells. We also found that, despite occasional overlap, the majority of FLY1-YFP punctae did not colocalize with the TGN/EE marker VHAa1-RFP. In addition, FLY1-YFP does not resemble the reticular distribution of endoplasmic reticulum or the linear distribution of plasma membrane (PM) markers. Thus, FLY1-YFP most likely resides in a compartment between the TGN/EE and the PM, namely, the LE/MVB/PVC and/or the vacuole (Müller et al., 2007). Although FM4-64 and Brefeldin A were not taken up by seed coat epidermal cells, we successfully treated seeds with wortmannin, which induces the fusion of LE/MVB/PVC in plant cells (Wang et al., 2009). Consistent with FLY1-YFP being localized in LE/MVB/PVC, wortmannin-treated seeds displayed fewer punctae and an increased number of large bodies.
A remaining question concerns the manner in which FLY1, a transmembrane E3 ligase that appears to be associated with the post-Golgi endomembrane system, can influence the level of DM in mucilage. Pectin DM is controlled by the activity of PMTs that add methylester groups and PMEs that remove methylester groups (Wolf et al., 2009a). Since fly1 mucilage has a lower DM relative to the wild type, FLY1 could be promoting PMT activity or decreasing PME activity.
As an E3 ligase, FLY1 could mark PMTs with monoubiquitin for retention in the Golgi since monoubiquitin has been identified as a signal for membrane protein localization in yeast, animal, and plant systems (Reggiori and Pelham, 2002; Hicke and Dunn, 2003; Schnell and Hicke, 2003, Barberon et al., 2011; Dowil et al., 2011). Alternatively, the FLY1 E3 ligase could use polyubiquitin chains to target PME proteins for degradation.
Although PMTs are predicted to have a single membrane-spanning domain (Krupková et al., 2007), this may not be sufficient for Golgi localization as the mechanisms for the retention of plant proteins in the Golgi are not well understood (Saint-Jore-Dupas et al., 2004). None of the 67 putative PMTs in the Arabidopsis genome have a confirmed biochemical activity (Harholt et al., 2010), and only a few, such as QUA2 (Mouille et al., 2007), QUA3 (Miao et al., 2011), and CGR3 (Held et al., 2011), are known to impact pectin biosynthesis. Because the putative PMTs studied to date are Golgi localized and FLY1-YFP localization appeared distinct from the Golgi marker ST-RFP, we do not favor the hypothesis that FLY1 targets PMTs.
A second possibility for the role of FLY1 in controlling pectin DM is that FLY1 is needed to recycle PMEs active in seed mucilage. Recycling of PME enzymes by FLY1 in wild-type cells may be essential for establishing the correct level of pectin DM in the mucilage pockets to facilitate proper mucilage extrusion.
PMEs are typically soluble (Pelloux et al., 2007) and lack the cytosolic Lys residues required for ubiquitination, but there are at least two PMEs that are preferentially expressed in the seed coat and have putative ubiquitination sites. Both PME6 and PME16 are classified as Type I/Group 2 PMEs because they also contain a PRO region with PME inhibitor homology and a processing motif for cleavage by subtilisin-like proteases (Pelloux et al., 2007; Wolf et al., 2009b). While PMEs whose PRO region is not cleaved are predicted to be retained in the Golgi (Wolf et al., 2009b), recent data suggest that certain Type I/Group 2 PMEs can be secreted to the cell wall without processing (Mareck et al., 2012). Therefore, FLY1 could be recycling unprocessed PMEs post-Golgi or upon endocytosis from the PM. Interestingly, elevated PME activity during seed coat development was proposed to cause reduced mucilage extrusion and primary wall detachment in the sbt1.7 (Rautengarten et al., 2008) and pmei6 (Saez-Aguayo et al., 2013) mutants. However, unlike fly1 discs, sbt1.7 and pmei6 outer tangential primary walls detach as a large continuous sheet (Rautengarten et al., 2008; Saez-Aguayo et al., 2013), suggesting that FLY1 functions independently of SBT1.7 and PMEI6.
The identification of FLY1, a transmembrane E3 ubiquitin ligase that controls the pectin DM in seed mucilage and is required for proper seed mucilage extrusion, is an important step forward in unraveling how plant cells synthesize, secrete, and modify pectin polymers. Detailed characterization of the fly1 seed coat phenotype revealed an unexpected scenario where primary cell wall detachment appears to result from the extrusion of mucilage with stronger gelling properties and an increased number of HG cross-links via calcium bridges. The discovery of the targets of the FLY1 protein and the elucidation of role of ubiquitination in the regulation of pectin DM may provide new perspectives on pectin modification and on the regulation of cell wall enzymes in the endomembrane system. This protein also offers a great opportunity to identify components of the pectin methylesterification pathway that are active in Arabidopsis seed mucilage. Finally, since FLY1 and FLY2 are both expressed in the xylem, it will be of interest to examine the development of this tissue in fly1 fly2 double mutants.
METHODS
Plant Materials and Growth Conditions
The Col-2 ecotype was propagated from a Columbia-0 (Col-0) seed through five generations of single seed descent by Shauna Somerville. The fly1-1 line was isolated from an EMS-mutagenized Col-2 population and was backcrossed to Col-2 four times to remove background EMS mutations. The fly1-1 mutant line (backcrossed twice to Col-2) was crossed to the Landsberg erecta (Ler) ecotype to generate an F2 mapping population. Map-based cloning was conducted using insertion/deletion polymorphisms between Col-0 and Landsberg erecta (Jander et al., 2002). Col-0 seeds bearing SALK T-DNA insertions in exons (Alonso et al., 2003), 5′ untranslated regions (UTRs), or introns (in decreasing order of preference) were selected using the SIGnAL T-DNA Express site (http://signal.salk.edu/cgi-bin/tdnaexpress) and obtained from the ABRC in Columbus, Ohio.
Seeds were germinated on plates with AT medium (Haughn and Somerville, 1986) and 7% (w/v) agar, and seedlings were transferred to soil (Sunshine Mix 4; SunGro) after 7 to 10 d. Plants were grown with continuous fluorescent illumination of 80 to 140 μE m−2 s−1 at 20 to 22°C. Developing seeds were staged as previously described (Western et al., 2001).
Preparation of Seed Coat Sections
Developing seeds were fixed using high-pressure freezing and freeze substitution, embedded in Spurr’s resin, and sectioned according to previous methods (Rensing et al., 2002; Young et al., 2008; Mendu et al., 2011). Well-preserved seed coats were examined by light microscopy.
Light Microscopy
Mature dry seeds were hydrated in distilled water, 50 mM CaCl2, or 50mM EDTA, pH 8.0, for 1 to 2 h, rinsed with once with water, and stained with 0.01% (w/v) RR (Sigma-Aldrich) for 30 to 60 min while shaking on a rotator. Bright-field micrographs of stained samples were taken with QCapture software and a digital camera (QImaging) equipped on a Zeiss AxioSkop 2 upright light microscope (Carl Zeiss). Contrast of unstained seeds was enhanced with phase contrast or differential interference contrast.
The effect of buffer pH on fly1-1 mucilage extrusion was tested using 50 mM EDTA and 50 mM HEPES buffers ranging from pH 6.0 to 8.0. Seeds were shaken in buffer for 1 h before rinsing with water and RR staining.
Live, developing seeds at 12 DPA were imaged using transmitted light on a Perkin-Elmer Ultraview VoX Spinning Disk Confocal system.
For cellulose staining, seeds were shaken in water for 2 h and then mixed with 0.01% (w/v) S4B (Sigma-Aldrich Rare Chemical Library) and 50 to 150 mM NaCl for 1 h (Anderson et al., 2010; Mendu et al., 2011). Seeds were rinsed twice with distilled water and imaged using a 561-nm laser on either a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss) or a Perkin-Elmer Ultraview VoX spinning disk confocal system. Xylem cells were analyzed in stem base cross sections stained with one drop of saturated phloroglucinol (Sigma-Aldrich) in 20% HCl solution.
Immunohistochemistry
Whole-seed immunolabeling was conducted according to a published method (Harpaz-Saad et al., 2011). The specificities of JIM5, JIM7 (CarboSource), and 2F4 (PlantProbes) primary antibodies have been extensively described (Liners et al., 1989; Knox et al., 1990; Knox, 1997; Willats et al., 2001b; Macquet et al., 2007a; Pattathil et al., 2010; Xu et al., 2011). The 2F4 antibody (PlantProbes) does not work with the conventional phosphate buffer (Liners et al., 1989) and was used with the following buffer: 20 mM Tris-HCl, pH 8.2, 0.5 mM CaCl2, and 150 mM NaCl. Goat-anti-rat secondary antibody conjugated to AlexaFluor488 was used against JIM5 and JIM7, while goat-anti-mouse conjugated to AlexaFluor488 (Molecular Probes, Invitrogen) was used against 2F4. The protocol was performed without primary antibody as a negative control. Seeds were imaged using a 488-nm laser (antibody fluorescence) and 561-nm laser (seed intrinsic fluorescence, background) on a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss) or a Perkin-Elmer Ultraview VoX spinning disk confocal system. All confocal micrographs were processed with ImageJ (Abramoff et al., 2004). Images containing signals from multiple optical stacks were rendered using the Z-project maximum intensity method.
Scanning Electron Microscopy
Dry seeds were mounted on stubs, coated with gold-palladium in a SEMPrep2 sputter coater (Nanotech), and examined with a Hitachi S4700 scanning electron microscope (Hitachi High-Technologies). For cryo-scanning electron microscopy, seeds were hydrated with a drop of distilled water and were quickly transferred to stubs topped with Tissue-Tek mounting medium (Sakura Finetek) and small squares of filter paper to absorb excess water. Once mounted on the stubs, seeds were immediately frozen in liquid nitrogen. Samples were analyzed under high vacuum at liquid nitrogen temperatures with a Hitachi S-4700 field emission scanning electron microscope (Hitachi High-Technologies) equipped with a Leica VCT 100 cryo transfer system and cryo stage control (Leica).
Light and electron micrographs were processed and measured with ImageJ (Abramoff et al., 2004). Image panels were made using Photoshop (Adobe Systems).
Determination of Monosaccharide Composition by HPAEC
The average weight of Col-2 and fly1-1 seeds was determined by weighing three replicates of 100 seeds for each genotype. For sequential mucilage extractions, four technical replicates of 25 mg of Col-2 or fly1-1 seeds (exact weight recorded) were mixed with 1.4 mL of distilled water and 10 μL of 5 mg/mL d-erythritol (internal standard). These samples were gently shaken using a tube rotator for 1 h. The mucilage in the supernatant (1 mL) of the first extraction was transferred to a glass tube and dried at 60°C under nitrogen gas. The same seeds were then rinsed twice with 700 μL of water and shaken vigorously with a vortex mixer on the highest setting for 2 h, in 1.4 mL of water and 10 μL of 5 mg/mL d-erythritol. The supernatant was transferred to a new glass tube and dried as described for the first extraction. Whole seeds, sugar standards dissolved in water, and the dried mucilage samples were processed according to published methods (Dean et al., 2007; Mendu et al., 2011).
Biochemical Determination of Pectin DM
Mucilage was extracted by vigorously shaking 5 mg of seeds (exact weight measured) with a vortex mixer at high speed in 300 μL of 50 mM EDTA for 60 min. After the seeds settled for 2 min, 100 μL of supernatant was transferred to a new tube and saponified with 0.25 M NaOH for 60 min on an orbital shaker. The reaction was neutralized with 0.25 M HCl (to give a total volume of 300 µL) and centrifuged for 5 min at 10,000g. Fifty microliters of the supernatant was transferred to a 96-well plate for the methanol release after saponification assay (Lionetti et al., 2007), using 0.5 units of alcohol oxidase (Sigma-Aldrich A2404) enzyme per sample instead of 0.03 units. Absorbance was measured at 412 nm and quantified using a methanol standard curve (Klavons and Bennett, 1986).
Forty microliters of the remaining 250 μL mucilage saponification solution was transferred to a 96-well plate, and the uronic acid assay was performed as described by van den Hoogen et al. (1998). Absorbance was measured at 525 nm, before and after addition of the dye, and quantified using a d-(+)-galacturonic acid monohydrate (Sigma-Aldrich) standard curve.
For whole seed analysis, 5 mg of seeds were frozen in liquid nitrogen and ground to a find powder using pestles in 1.7-mL tubes. Ground seeds were washed twice with 500 μL of 70% ethanol on an orbital shaker and centrifuged for 2 min at 10,000g. Ethanol was removed again and the pellet was dried under nitrogen gas at 60°C. One additional methanol-chloroform (1:1) wash and three acetone washes were performed under similar conditions. After the final wash, samples were dried under nitrogen gas at 60°C and the cell wall pellet was saponified using 200 µL of 0.25 M NaOH for 60 min on an orbital shaker. Samples were neutralized with 200 µL of 0.25 M HCl and centrifuged for 5 min at 10,000g. Fifty microliters of the supernatant was used for the 96-well plate methanol assay (Lionetti et al., 2007). Background corrections for methanol assay were made using mucilage and whole seed samples that were not saponified.
The whole seed sugars in the leftover saponification reaction were precipitated by adding 1 mL of 100% ethanol, vortexing briefly, and centrifuging for 2 min at 10,000g. The supernatant was removed and the cell wall pellet was dried under nitrogen gas at 60°C. The cell wall was fully resuspended in 1 mL distilled water. Each sample was sonicated for 20 s with a Branson Sonifier 150 (Branson Ultrasonics), setting 3, to create a homogenous cell wall suspension. Twenty-microliter aliquots of each sample, diluted with 20 µL water, were used for the 96-well plate uronic acid assay (van den Hoogen et al., 1998).
DNA Isolation and T-DNA Genotyping
DNA was isolated from rosette leaves using a published one-step protocol (Kasajima et al., 2004), except that samples were quickly ground using 1.0-mm zirconia/silica beads (Biospec Products) and a Precellys 24 tissue homogenizer (Bertin Technologies) (Verollet, 2008) instead of homogenization using plastic pestles. The supernatant was used immediately (1 μL of DNA solution in 20 μL PCR reaction) or was stored at −20°C until needed.
SALK T-DNA insertions were genotyped by PCR using the LBb1.3 insert-specific primer (5′-ATTTTGCCGATTTCGGAAC-3′) and gene-specific primers selected using the SALK T-DNA Primer Design tool (http://signal.salk.edu/tdnaprimers.2.html; see Supplemental Table 1 online).
Protein Bioinformatics
All genes in this study were first analyzed using The Arabidopsis Information Resource (http://Arabidopsis.org; Lamesch et al., 2012). The SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP) was used for signal peptide analysis (Petersen et al., 2011). Transmembrane α-helices were predicted using the ARAMEMNON database (http://aramemnon.botanik.uni-koeln.de), which integrates the predictions of 18 individual programs (Schwacke et al., 2003). FLY1 homologs identified using BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were verified using the Phytozome (http://www.phytozome.net; Goodstein et al., 2012), PLAZA (http://bioinformatics.psb.ugent.be/plaza; Van Bel et al., 2012), and the InParanoid 7 databases (http://InParanoid.sbc.su.se; Ostlund et al., 2010).
Functional domains were identified using the InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan; Quevillon et al., 2005) and PROSITE databases (http://prosite.expasy.org; Sigrist et al., 2010). FLY1, FLY2, and TUL1 protein sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2; Larkin et al., 2007).
Recombinant Protein Expression and Purification
The soluble C-terminal FLY1 region (after the seventh transmembrane domain and before the stop codon) containing the RING domain was amplified using a left primer (LP) plus EcoRI site (5′-tgaGAATTCCTCACGCTGCTTTGTTCCCCGTCAGA-3′) and a right primer (RP) plus XhoI site (5′-tatCTCGAGCTATGCTGGAGGAAGAGACCGCCGA-3′) using Col-2 seed coat cDNA. The resulting 243-bp Fly1RING amplicon was fused with GST using an EcoRI-XhoI cassette on the pGEX-4T-3 vector (GE Healthcare). The plasmid was propagated in Escherichia coli DH5α cells, verified by sequencing, and transformed in E. coli BL21(DE3)-pLysS for protein expression. A 50 mL culture was grown until OD600 = 0.5, when it was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown for additional 4 h. Cells were harvested by centrifugation and resuspended in 2 mL lysis buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.01% [v/v] Triton-X, and one protease inhibitor tablet [Roche]) and sonicated three times at 20% amplitude and 20 mA for 30 s. Lysate was cleared by centrifugation at 12,000g for 15 min at 4°C followed by filtration through a 0.45 mM filter. For purification, 200 μL of 50% glutathione agarose slurry (Sigma-Aldrich) was added to the cleared lysate and incubated under rotation at 4°C for 2 h. Sample was washed four times with 1 mL wash buffer (25 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 0.01% [v/v] Triton-X) and purified protein eluted in 100 mL elution buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.01% [v/v] Triton-X, and 15 mM reduced glutathione); glycerol was added to a final concentration of 40% (v/v).
The 6xHis-UBC8 and 6xHis-UBA1 Arabidopsis thaliana recombinant proteins were expressed in E. coli BL21-AI (Invitrogen) and induced with 0.2% (w/v) arabinose at an OD600 = 0.5. Proteins were purified on a nickel-nitrilotriacetic acid (Ni-NTA) column according to the manufacturer's instructions (Qiagen) with lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, and 10 mM imidazole). In wash buffer, the concentration of imidazole was increased to 20 mM and the elution buffer contained 250 mM imidazole.
Protein purity was analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining, and concentration was measured by Bradford assay (Bio-Rad). The integrity of fusion proteins was checked by immunoblot analysis using anti-GST and anti-His antibodies (both Sigma-Aldrich).
In Vitro Ubiquitination Assay
In vitro ubiquitination assays were performed as previously described in Hardtke et al. (2002). In 30-μL reactions (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM ATP, 0.05 mM ZnCl2, 0.2 mM DTT, 10 mM phosphocreatine, 0.1 units of phosphocreatine kinase), 100 ng E1 (6xHis-UBA1), 250 ng E2 (6xHis-UBC8), 2 µg ubiquitin (BostonBiochem), and 250 ng GST-Fly1RING were added for the complete reaction. For control reactions, the E2, E3, or ubiquitin components were omitted. The reactions were incubated at 30°C for 2 h and stopped by addition of 3.3 μL 10× reducing loading buffer. Samples were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and probed with anti-ubiquitin (Enzo Life Sciences).
Transcript Analysis
Expression patterns in specific Arabidopsis organs and cell types were visualized with the eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007) and corroborated with GENEVESTIGATOR (https://www.genevestigator.com/gv/plant.jsp; Hruz et al., 2008).
For RT-PCR, RNA was extracted from Col-2 tissues using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. RNA quantification was performed using a NanoDrop 8000 (Thermo Scientific). Five hundred nanograms of total RNA treated with DNaseI (Fermentas) was used for first-strand cDNA synthesis along with iScript RT supermix (Bio-Rad). RT-PCR was conducted using a typical PCR reaction containing Taq polymerase (Invitrogen) and gene-specific pairs of intron-spanning primers (see Supplemental Table 1 online). Amplicons of ∼200 bp were expected after intron splicing. Transcript levels were analyzed after 23 amplification cycles, and GAPC was used as a loading control.
FLY1pro:FLY1-YFP Cloning
FLY1 was amplified from Col-2 DNA using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific), an LP plus a KpnI site (5′-gcgGGTACCtctcacaaacaaacatttctcactc-3′) and an RP including an XhoI site (5′-tactcgagTGCTGGAGGAAGAGACCGCCGACAA-3′). The 5156-bp FLY1pro:FLY1 amplicon starts 690 bp upstream of the 5′ UTR of FLY1 (123 bp away from the At4g28365 5′ UTR, its upstream neighbor) and includes the complete FLY1 coding region, except for the translation stop codon.
Citrine YFP was selected due to its superior photostability in the acidic environment of the apoplast. The FLY1pro:FLY amplicon was introduced as a KpnI-XhoI cassette into a pAD vector with Citrine YFP (DeBono, 2012). The plasmid was propagated in E. coli DH5α cells and verified using BglII restriction enzyme digestion and sequencing. The FLY1pro:FLY-YFP construct was introduced in Arabidopsis fly1-1 plants by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101 pMP90 plus pSOUP (Hellens et al., 2000).
FLY1-YFP, ST-RFP, and VHAa1-RFP Subcellular Localization
Transgenic plants were selected by spraying the leaves of plants germinated on soil with 200 mM Basta herbicide solution every 2 to 3 d for 2 weeks. Seeds were removed from developing siliques of Basta-resistant transgenic plants. FLY1-YFP and ST-RFP/VHAa1-RFP were excited using 514- and 561-nm lasers, respectively, on a Perkin-Elmer Ultraview VoX spinning disk confocal system. FLY1-YFP expression in dissected seed coats, embryos, and other tissues was examined with a ×20 oil immersion objective, while subcellular localization of the FLY1-YFP signal was investigated with a ×63 glycerol immersion objective. The controls used were untransformed Col-2 and fly1-1 seeds and transgenic lines expressing a single fluorescent construct. In addition, 7 DPA seeds expressing FLY1pro:FLY-YFP were dissected from siliques and treated for 2 h with 20 μM wortmannin (diluted in water from a 1 mM DMSO stock) or with water containing an equivalent amount of DMSO, before imaging.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At4g28370 (FLY1) and At2g20650 (FLY2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of fly1-1 Mucilage Extrusion and Primary Wall Attachment.
Supplemental Figure 2. Mucilage Extractions and Total Monosaccharide Composition.
Supplemental Figure 3. Time-Course Analysis of Mucilage Release from Seeds upon Hydration.
Supplemental Figure 4. Analysis of the Effect of Buffer pH on fly1-1 Mucilage Defects.
Supplemental Figure 5. Immunolabeling of Partially Methylesterified HG in Seeds.
Supplemental Figure 6. Analysis of fly1 and fly2 Mucilage Extrusion.
Supplemental Figure 7. FLY2 Gene Structure and Mutations.
Supplemental Figure 8. FLY1 and FLY2 May Be Involved in Xylem Development.
Supplemental Figure 9. Alignment of the RING-H2 Domains of FLY1, FLY2, and TUL1.
Supplemental Figure 10. Analysis of FLY1pro:FLY1-YFP Expression in Developing Seeds.
Supplemental Figure 11. Wortmannin Induces Fusions of FLY1-YFP Punctae.
Supplemental Table 1. Gene-Specific Primers Used for T-DNA Genotyping and RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank the staff of the University of British Columbia BioImaging Facility for invaluable help with microscopy. We are grateful for the assistance of Shawn Mansfield University of British Columbia (UBC) with monosaccharide analysis. Gabriel Lévesque-Tremblay (UBC) provided cDNA samples. The pAD vector was a gift from Allan DeBono (UBC). The 35S:ST-RFP construct was a gift from Chris Hawes (Oxford Brookes University), and 35S:VHAa1-RFP transgenic seeds were provided by Miki Fujita (University of British Columbia). We thank Diana Starr Young (UBC), Andrew Karpov (UBC), Golya Mirderikvand (UBC), Kuljit Dhaliwal (UBC), and Tiffany Ngai (UBC) for technical assistance with mapping. This research was funded by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to G.W.H. and by a grant from the National Science Foundation (MCB 0929100), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation to M.E.
AUTHOR CONTRIBUTIONS
C.V. designed and conducted research, analyzed data, and wrote the article. G.H.D., J.S.G., K.K., and T.L.W. designed and conducted research and analyzed data. A.G., G.D., and Y.T.H. conducted research. M.E. designed research and analyzed data. G.W.H. designed research, analyzed data, and wrote the article.
Glossary
- HG
homogalacturonan
- DM
degree of methylesterification
- GalA
(1→4)-α-d-galacturonic acid
- RG-I
rhamnogalacturonan-I
- Rha
(1→2)-α-l-rhamnose
- PMT
pectin methyltransferase
- PME
pectin methylesterase
- DPA
days postanthesis
- Col-2
Columbia-2
- EMS
ethyl methanesulfonate
- RR
Ruthenium Red
- S4B
Pontamine Fast Scarlet 4B
- HPAEC
high-performance anion-exchange chromatography
- YFP
yellow fluorescent protein
- GST
glutathione S-transferase
- RFP
red fluorescent protein
- TGN/EE
trans-Golgi network/early endosome
- LE/MVB/PVC
late endosome/multivesicular body/prevacuolar compartment
- PM
plasma membrane
- Col-0
Columbia-0
- UTR
untranslated region
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. (2004). Image Processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson C.T., Carroll A., Akhmetova L., Somerville C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski A.A., Popma T.M., Haughn G.W., Carpita N.C., McCann M.C., Western T.L. (2009). AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo M.A., Sakuragi Y., Zhu X., Burrell A.J., Mohanty S.S., Atwood J.A., III, Orlando R., Scheller H.V., Mohnen D. (2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A., Ahad A., Wang S., Mansfield S.D., Haughn G.W., Douglas C.J., Ellis B.E. (January 18, 2013). The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta, in press. [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T., De Rycke R., Viane R., Inzé D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. V113: 139–148 [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.-Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Fricker M., Hawes C. (2002). A greener world: The revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 3: 520–530 [DOI] [PubMed] [Google Scholar]
- Caffall K.H., Pattathil S., Phillips S.E., Hahn M.G., Mohnen D. (2009). Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol. Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dean G.H., Zheng H., Tewari J., Huang J., Young D.S., Hwang Y.T., Western T.L., Carpita N.C., McCann M.C., Mansfield S.D., Haughn G.W. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono, A.G. (2012). The Role and Behavior of Arabidopsis thaliana Lipid Transfer Proteins during Cuticular Wax Deposition. PhD dissertation (Vancouver, Canada: University of British Columbia). [Google Scholar]
- Derbyshire P., McCann M.C., Roberts K. (2007). Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol. 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowil R.T., Lu X., Saracco S.A., Vierstra R.D., Downes B.P. (2011). Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J. Biol. Chem. 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Ciechanover A. (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Goldberg R., Morvan C., Jauneau A., Jarvis M.C. (1996). Methyl-esterification, de-esterification and gelation of pectins in the primary cell wall. In Pectins and Pectinases, Visser J., Voragen A.G.J., eds (Amsterdam: Elsevier Science BV), pp. 151–172 [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40 (Database issue): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.M.U., Simpson D.J., Hyman S.C., Ndimba B.K., Slabas A.R. (2003). Ara12 subtilisin-like protease from Arabidopsis thaliana: Purification, substrate specificity and tissue localization. Biochem. J. 370: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Okamoto H., Stoop-Myer C., Deng X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30: 385–394 [DOI] [PubMed] [Google Scholar]
- Harholt J., Suttangkakul A., Vibe Scheller H. (2010). Biosynthesis of pectin. Plant Physiol. 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S., McFarlane H.E., Xu S., Divi U.K., Forward B., Western T.L., Kieber J.J. (2011). Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J. 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Haughn G.W., Somerville C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Haughn G.W., Western T.L. (2012). Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front. Plant Sci. 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M.A., Be E., Zemelis S., Withers S., Wilkerson C., Brandizzi F. (2011). CGR3: A Golgi-localized protein influencing homogalacturonan methylesterification. Mol. Plant 4: 832–844 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R. (2003). Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Miller W. (1991). A time-efficient linear-space local similarity algorithm. Adv. Appl. Math. 12: 337–357 [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M.C. (1984). Structure and properties of pectin gels in plant cell walls. Plant Cell Environ. 7: 153–164 [Google Scholar]
- Jolie R.P., Duvetter T., Van Loey A.M., Hendrickx M.E. (2010). Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 345: 2583–2595 [DOI] [PubMed] [Google Scholar]
- Kasajima I., Ide Y., Ohkama-Ohtsu N., Hayashi H., Yoneyama T., Fujiwara T. (2004). A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 22: 49–52 [Google Scholar]
- Klavons J.A., Bennett R.D. (1986). Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J. Agric. Food Chem. 34: 597–599 [Google Scholar]
- Knox J.P. (1997). The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 171: 79–120 [DOI] [PubMed] [Google Scholar]
- Knox J.P., Linstead P.J., King J., Cooper C., Roberts K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Krupková E., Immerzeel P., Pauly M., Schmülling T. (2007). The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 50: 735–750 [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40 (Database issue): D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Min M.K., Lee Y.J., Jin J.B., Shin D.H., Kim D.H., Lee K.-H., Hwang I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F., Letesson J.J., Didembourg C., Van Cutsem P. (1989). Monoclonal antibodies against pectin: Recognition of a conformation induced by calcium. Plant Physiol. 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Raiola A., Camardella L., Giovane A., Obel N., Pauly M., Favaron F., Cervone F., Bellincampi D. (2007). Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord E.M., Mollet J.-C. (2002). Plant cell adhesion: A bioassay facilitates discovery of the first pectin biosynthetic gene. Proc. Natl. Acad. Sci. USA 99: 15843–15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A., Ralet M.-C., Kronenberger J., Marion-Poll A., North H.M. (2007a). In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48: 984–999 [DOI] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Emr S.D. (2012). Ubiquitin and membrane protein turnover: From cradle to grave. Annu. Rev. Biochem. 81: 231–259 [DOI] [PubMed] [Google Scholar]
- Macquet A., Ralet M.-C., Loudet O., Kronenberger J., Mouille G., Marion-Poll A., North H.M. (2007b). A naturally occurring mutation in an Arabidopsis accession affects a beta-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareck A., Lamour R., Schaumann A., Chan P., Driouich A., Pelloux J., Lerouge P. (2012). Analysis of LuPME3, a pectin methylesterase from Linum usitatissimum, revealed a variability in PME proteolytic maturation. Plant Signal. Behav. 7: 59–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V., Griffiths J.S., Persson S., Stork J., Downie A.B., Voiniciuc C., Haughn G.W., DeBolt S. (2011). Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Li H.-Y., Shen J., Wang J., Jiang L. (2011). QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 62: 5063–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Mouille G., Ralet M.-C., Cavelier C., Eland C., Effroy D., Hématy K., McCartney L., Truong H.N., Gaudon V., Thibault J.-F., Marchant A., Höfte H. (2007). Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Moustacas A.M., Nari J., Borel M., Noat G., Ricard J. (1991). Pectin methylesterase, metal ions and plant cell-wall extension. The role of metal ions in plant cell-wall extension. Biochem. J. 279: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Mettbach U., Menzel D., Samaj J. (2007). Molecular dissection of endosomal compartments in plants. Plant Physiol. 145: 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Debolt S., Persson S. (2008). Cellulose synthesis: A complex complex. Curr. Opin. Plant Biol. 11: 252–257 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Nishida K., Kasahara K., Kinoshita K. (2011). ATTED-II updates: Condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Nemoto T., Jigami Y. (2007). Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-d-glucose to UDP-l-rhamnose conversion. J. Biol. Chem. 282: 5389–5403 [DOI] [PubMed] [Google Scholar]
- Ostlund G., Schmitt T., Forslund K., Köstler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.L. (2010). InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38 (Database issue): D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J., Rustérucci C., Mellerowicz E.J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Penfield S., Meissner R.C., Shoue D.A., Carpita N.C., Bevan M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33 (Web Server issue): W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Usadel B., Neumetzler L., Hartmann J., Büssis D., Altmann T. (2008). A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R.B. (2002). A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4: 117–123 [DOI] [PubMed] [Google Scholar]
- Rensing K.H., Samuels A.L., Savidge R.A. (2002). Ultrastructure of vascular cambial cell cytokinesis in pine seedlings preserved by cryofixation and substitution. Protoplasma 220: 39–49 [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13: 405–408 [DOI] [PubMed] [Google Scholar]
- Romano, J.M., et al. (2012). AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 195: 774–786. [DOI] [PubMed]
- Saint-Jore-Dupas C., Gomord V., Paris N. (2004). Protein localization in the plant Golgi apparatus and the trans-Golgi network. Cell. Mol. Life Sci. 61: 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schnell J.D., Hicke L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.-I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J.A., Cerutti L., de Castro E., Langendijk-Genevaux P.S., Bulliard V., Bairoch A., Hulo N. (2010). PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38 (Database issue): D161–D166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriamornsak S. (2003). Chemistry of pectin and its pharmaceutical uses: A review. Silpakorn University International Journal. 3: 206–228 [Google Scholar]
- Sterling C. (1970). Crystal-structure of ruthenium red and stereochemistry of its pectic stain. Am. J. Bot. 57: 172–175 [Google Scholar]
- Sterling J.D., Atmodjo M.A., Inwood S.E., Kumar Kolli V.S., Quigley H.F., Hahn M.G., Mohnen D. (2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S., Ralet M.C., Berger A., Botran L., Ropartz D., Marion-Poll A., North H.M. (2013). PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork J., Harris D., Griffiths J., Williams B., Beisson F., Li-Beisson Y., Mendu V., Haughn G., Debolt S. (2010). CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol. 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S., Ralet M.-C., Berger A., Diatloff E., Bischoff V., Gonneau M., Marion-Poll A., North H.M. (2011). CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol. 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Kuschinsky A.M., Rosso M.G., Eckermann N., Pauly M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbosch K.A., Bradley D.J., Knox J.P., Perotto S., Butcher G.W., Brewin N.J. (1989). Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J. 8: 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.M., van Weeren P.R., Lopes-Cardozo M., van Golde L.M.G., Barneveld A., van de Lest C.H.A. (1998). A microtiter plate assay for the determination of uronic acids. Anal. Biochem. 257: 107–111 [DOI] [PubMed] [Google Scholar]
- Verollet R. (2008). A major step towards efficient sample preparation with bead-beating. Biotechniques 44: 832–833 [DOI] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Burn J., Tan W.L., Skinner D.J., Martin-McCaffrey L., Moffatt B.A., Haughn G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Young D.S., Dean G.H., Tan W.L., Samuels A.L., Haughn G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor J.B., Symonds V.V., Mendenhall J., Lloyd A.M. (2000). Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Willats W.G., McCartney L., Mackie W., Knox J.P. (2001a). Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 47: 9–27 [PubMed] [Google Scholar]
- Willats W.G.T., McCartney L., Knox J.P. (2001b). In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., Orfila C., Limberg G., Buchholt H.C., van Alebeek G.-J.W.M., Voragen A.G.J., Marcus S.E., Christensen T.M.I.E., Mikkelsen J.D., Murray B.S., Knox J.P. (2001c). Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Mouille G., Pelloux J. (2009a). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2: 851–860 [DOI] [PubMed] [Google Scholar]
- Wolf S., Rausch T., Greiner S. (2009b). The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J. 58: 361–375 [DOI] [PubMed] [Google Scholar]
- Xu C., Zhao L., Pan X., Šamaj J. (2011). Developmental localization and methylesterification of pectin epitopes during somatic embryogenesis of banana (Musa spp. AAA). PLoS ONE 6: e22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.E., McFarlane H.E., Hahn M.G., Western T.L., Haughn G.W., Samuels A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.F., Staehelin L.A. (1992). Functional compartmentation of the Golgi apparatus of plant cells: Immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 99: 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.