In the unicellular green alga Chlamydomonas, small RNAs trigger translation repression of target transcripts at a postinitiation step, without or with only minimal mRNA destabilization. The ribosomes associated with small RNA–repressed transcripts display reduced sensitivity to translation inhibition by certain antibiotics such as cycloheximide.
Abstract
Small RNAs (sRNAs; ∼20 to 30 nucleotides in length) play important roles in gene regulation as well as in defense responses against transposons and viruses in eukaryotes. Their biogenesis and modes of action have attracted great attention in recent years. However, many aspects of sRNA function, such as the mechanism(s) of translation repression at postinitiation steps, remain poorly characterized. In the unicellular green alga Chlamydomonas reinhardtii, sRNAs derived from genome-integrated inverted repeat transgenes, perfectly complementary to the 3′ untranslated region of a target transcript, can inhibit protein synthesis without or with only minimal mRNA destabilization. Here, we report that the sRNA-repressed transcripts are not altered in their polyadenylation status and they remain associated with polyribosomes, indicating inhibition at a postinitiation step of translation. Interestingly, ribosomes associated with sRNA-repressed transcripts show reduced sensitivity to translation inhibition by some antibiotics, such as cycloheximide, both in ribosome run-off assays and in in vivo experiments. Our results suggest that sRNA-mediated repression of protein synthesis in C. reinhardtii may involve alterations to the function/structural conformation of translating ribosomes. Additionally, sRNA-mediated translation inhibition is now known to occur in a number of phylogenetically diverse eukaryotes, suggesting that this mechanism may have been a feature of an ancestral RNA interference machinery.
INTRODUCTION
RNA-mediated silencing is an evolutionarily conserved process in eukaryotes by which small RNAs (sRNAs) induce the inactivation of cognate sequences through a variety of mechanisms, including translation repression, RNA degradation, transcriptional inhibition, and/or, in a few organisms, DNA elimination (Voinnet, 2009; Fabian et al., 2010; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011; Ketting, 2011). Intriguingly, recent studies indicate that these noncoding RNAs may also participate in transcriptional or translational activation (Steitz and Vasudevan, 2009; Younger and Corey, 2009; Fabian et al., 2010). Despite the mechanistic diversity of these processes, in most characterized pathways, sRNAs (∼20 to 30 nucleotides in length) are incorporated into effector complexes containing at their core Argonaute proteins, which include two major subfamilies of polypeptides named after Arabidopsis thaliana ARGONAUTE1 (AGO1) and Drosophila melanogaster P-element induced wimpy testis (PIWI) (Song et al., 2004; Rivas et al., 2005; Hutvagner and Simard, 2008; Fabian et al., 2010; Cenik and Zamore, 2011). Some AGO-PIWI proteins function as sRNA-guided endonucleases (slicers) that cleave complementary transcripts, whereas others lack endonucleolytic activity and repress their targets through other mechanisms (Meister et al., 2004; Hutvagner and Simard, 2008; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011).
Three major classes of sRNAs have been recognized in metazoans: microRNAs (miRNAs), PIWI-interacting RNAs, and small interfering RNAs (siRNAs) (Bartel, 2009; Axtell et al., 2011; Cenik and Zamore, 2011; Ketting, 2011). Land plants and green algae lack PIWI proteins and contain only miRNAs and siRNAs that associate with members of the AGO clade (Cerutti and Casas-Mollano, 2006; Voinnet, 2009; Axtell et al., 2011). miRNAs commonly originate from endogenous, single-stranded, noncoding RNA transcripts or introns that fold into imperfectly paired hairpin structures. They often modulate the expression of genes with roles in development, physiological or metabolic processes, or stress responses (Bartel, 2009; Voinnet, 2009; Axtell et al., 2011; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011; Ketting, 2011). siRNAs are produced from long, nearly perfectly complementary, double-stranded RNAs (dsRNAs) of diverse origins (Voinnet, 2009; Axtell et al., 2011; Cenik and Zamore, 2011; Ketting, 2011). In land plants and algae, these siRNAs play various roles in suppression of viruses and transposable elements, posttranscriptional regulation of gene expression, DNA methylation, and/or heterochromatin formation (Voinnet, 2009; Yu and Wang, 2010; Cerutti et al., 2011). Despite considerable advances in our understanding of the biogenesis and function of sRNAs (Hutvagner and Simard, 2008; Bartel, 2009; Voinnet, 2009; Fabian et al., 2010; Axtell et al., 2011; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011; Ketting, 2011), key mechanistic aspects of their mode of action remain poorly characterized.
The degree of complementarity between a sRNA and its target site has been considered a main determinant of the posttranscriptional repression mechanism (Bartel, 2009; Voinnet, 2009; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011). Highly complementary sRNA-mRNA hybrids, with perfect central pairing, activate Argonaute-mediated endonucleolytic cleavage of target transcripts (Meister et al., 2004; Rivas et al., 2005; Hutvagner and Simard, 2008; Cenik and Zamore, 2011). This is the best-characterized mechanism of posttranscriptional silencing mediated by siRNAs and, in land plants, by many miRNAs (Llave et al., 2002; Schwab et al., 2005; Voinnet, 2009; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011). Conversely, imperfect sRNA-mRNA hybrids, with central bulges or mismatches, enable translational inhibition and/or accelerated exonucleolytic (slicer independent) transcript decay, which are the prevalent modes of repression involving metazoan miRNAs (Hutvagner and Simard, 2008; Bartel, 2009; Fabian et al., 2010; Huntzinger and Izaurralde, 2011). Interestingly, recent evidence indicates that sRNAs perfectly complementary to a target mRNA can also cause translational inhibition without, or with only minimal, transcript destabilization (Rao et al., 2006; Brodersen et al., 2008; Wu et al., 2008; Voinnet, 2009; Yu and Wang, 2010). This outcome may result from the association of sRNAs with Argonautes that lack endonucleolytic activity (Meister et al., 2004; Wu et al., 2008). However, siRNA-programmed AGO proteins, known to possess the predicted catalytic motif, can also fail to cleave (Meister et al., 2004; Hutvagner and Simard, 2008; Cenik and Zamore, 2011), suggesting that our understanding of the determinants of the Argonaute slicer activity is insufficient and/or that associated factors may modulate AGO endonucleolytic activity.
Over the past few years, remarkable progress has been made in our understanding of the mechanism(s) of miRNA-mediated posttranscriptional silencing in metazoans, but no consensus has emerged yet unifying all current observations (Fabian et al., 2010; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011; Djuranovic et al., 2012; Fukaya and Tomari, 2012). Animal miRNAs have been proposed to repress translation in at least four distinct ways: inhibition of translation initiation, inhibition of translation elongation, cotranslational degradation of nascent polypeptides, and premature termination of translation (Maroney et al., 2006; Nottrott et al., 2006; Petersen et al., 2006; Kong et al., 2008; Ding and Grosshans, 2009; Gu et al., 2009; Iwasaki et al., 2009; Fabian et al., 2010; Huntzinger and Izaurralde, 2011; Stadler et al., 2012). miRNAs can also promote sequestration of target mRNAs in discrete cytoplasmic foci, either processing bodies or stress granules (Liu et al., 2005; Sen and Blau, 2005), but this localization may be a consequence of silencing rather than a requirement for translation repression (Eulalio et al., 2007; Detzer et al., 2011; Huntzinger and Izaurralde, 2011). Additionally, genome-wide proteomic and transcriptomic analyses, after the removal or the ectopic expression of miRNAs, have suggested that the slicer-independent degradation of miRNA targets may account for most of the stable repression mediated by miRNAs in mammalian cell cultures (Baek et al., 2008; Selbach et al., 2008; Hendrickson et al., 2009; Guo et al., 2010; Huntzinger and Izaurralde, 2011). One possible explanation for all these disparate and sometimes conflicting observations is that metazoan miRNAs may regulate target transcripts via multiple, interrelated mechanisms that can be modulated by AGO-associated factors and target mRNA effects. Indeed, AGO binding GW-repeat proteins (TNRC6/GW182-like) have been shown to interact with cytoplasmic poly(A) binding protein and with the CCR4-NOT and PAN2-PAN3 deadenylase complexes leading to mRNA deadenylation as well as translation repression (Fabian et al., 2010, 2011; Braun et al., 2011; Chekulaeva et al., 2011; Huntzinger and Izaurralde, 2011; Fukaya and Tomari, 2012), although there is also increasing evidence for miRNA-mediated translation inhibition in a deadenylation-independent manner (Fabian et al., 2010; Fukaya and Tomari, 2011, 2012; Bazzini et al., 2012; Djuranovic et al., 2012; Mishima et al., 2012). Depending on the cell type and/or specific target, mRNAs may be maintained in a translationally repressed state or rapidly degraded (Fabian et al., 2010; Huntzinger and Izaurralde, 2011; Béthune et al., 2012; Bazzini et al., 2012; Djuranovic et al., 2012).
sRNAs can also cause translation repression in land plants. In Arabidopsis, the transcripts of APETALA2, a target of miR172, the SBP-box gene SPL3, a target of miR156/157, and two copper/zinc superoxide dismutases (CSD1 and CSD2), as well as the copper chaperone for superoxide dismutase (CCS1), targets of miR398, were found to be regulated by miRNA-mediated translation inhibition (Aukerman and Sakai, 2003; Chen, 2004; Gandikota et al., 2007; Dugas and Bartel, 2008; Beauclair et al., 2010; Yu and Wang, 2010). Mutations in two genes implicated in sRNA function (encoding the microtubule-severing protein KATANIN and the enhancer of decapping protein VARICOSE) were shown to increase polypeptide levels of several miRNA-regulated genes without causing a corresponding change in the abundance of their mRNAs (Brodersen et al., 2008; Voinnet, 2009). Moreover, Arabidopsis AGO1 and a subset of miRNAs have been demonstrated to associate with polyribosomes, consistent with a role for miRNAs in translation inhibition (Lanet et al., 2009). Indeed, translational regulation may be an important aspect of miRNA function in Arabidopsis based on the phenotypes of loss-of-function mutants of SUO, coding for a large GW-repeat polypeptide involved in miRNA-mediated repression of protein synthesis (Yang et al., 2012). However, SUO does not appear to be an ortholog of animal TNRC6/GW182 and the mechanism(s) by which sRNAs inhibit translation in land plants remains uncharacterized.
Translation inhibition mediated by sRNAs may also operate in unicellular eukaryotes. In the parasitic protozoan Giardia lamblia, sRNAs have been shown to repress the expression of reporter genes containing sRNA target sites in their 3′ untranslated regions (UTRs) without changes in transcript levels (Li et al., 2011; Saraiya et al., 2011). Likewise, in the marine diatom Phaeodactylum tricornutum, transformation with an inverted repeat (IR) transgene, producing dsRNA homologous to a phytochrome gene, did not alter target mRNA amounts but significantly reduced cognate protein abundance (De Riso et al., 2009). These observations are consistent with sRNA-mediated translation inhibition, which also occurs in the unicellular green alga Chlamydomonas reinhardtii. Here, we show that transgenic siRNAs perfectly complementary to a target transcript can repress protein synthesis at a postinitiation step. Moreover, ribosomes associated with an siRNA-repressed transcript display reduced sensitivity to inhibition by the antibiotic cycloheximide, suggesting that the silencing mechanism(s) alters the function/structural conformation of translating ribosomes.
RESULTS
Inverted Repeat Transgenes Can Trigger Translation Repression of Homologous Endogenous Transcripts
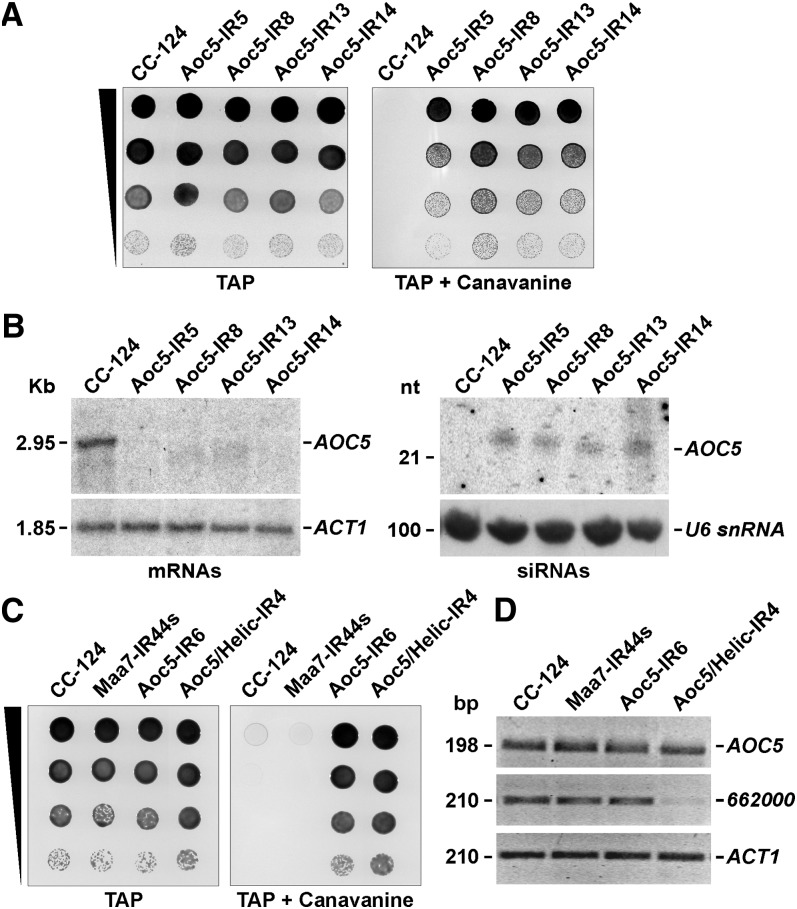
In C. reinhardtii, RNA interference (RNAi) has been achieved, among other approaches, by the production of hairpin dsRNA from genome-integrated IR transgenes (Cerutti et al., 2011). The transcribed dsRNA is processed into siRNAs and, in most cases, reduction in the steady state levels of target mRNAs is observed (Rohr et al., 2004; Schroda, 2006), implying RNAi-induced transcript degradation. For instance, transformation of C. reinhardtii with an IR construct targeting the 3′ UTR of AMINO ACID CARRIER5 (AOC5) (see Supplemental Figure 1A online), encoding a putative basic amino acid permease, resulted in transgenic lines tolerant to the Arg analog l-canavanine (Figure 1A). These strains contained ∼22-nucleotide AOC5 siRNAs and the AOC5 mRNA amount was significantly reduced (Figure 1B).
Figure 1.
RNA-Mediated Silencing of the AOC5 Gene Induced by Expression of AOC5 IR Transgenes in C. reinhardtii Transformants.
(A) Growth and survival of Aoc5-IR transformants on TAP medium without (left) or with (right) 400 μg/mL l-canavanine. CC-124, untransformed wild-type strain.
(B) RNA gel blot analyses of mRNAs and siRNAs in the Aoc5-IR transgenic strains. The left panels correspond to agarose gel–separated total RNA samples sequentially hybridized with 32P-labeled PCR products corresponding to the AOC5 3′ UTR (top panel), to evaluate the degree of mRNA reduction, or the coding sequence of Actin (ACT1) (bottom panel) as a control for equivalent loading of the lanes. The right panels correspond to total RNA samples separated in a 15% denaturing polyacrylamide gel and probed sequentially with the AOC5 3′ UTR sequence (top panel), to detect siRNAs, or the U6 small nuclear RNA sequence (bottom panel), to assess the amount of sample loaded per lane. nt, nucleotides.
(C) Growth and survival of the indicated strains on TAP medium alone or containing 400 μg/mL l-canavanine. Maa7-IR44s, strain containing an IR transgene targeting the 3′ UTR of the MAA7 gene (encoding TSβ). Aoc5/Helic-IR4, strain containing a tandem IR transgene targeting both AOC5 and Cre16.g662000 (encoding a putative RNA helicase).
(D) RT-PCR analyses on total RNA samples from the indicated strains. The panels show reverse images of agarose gel–fractionated RT-PCR products corresponding to AOC5 or Cre16.g662000. Amplification of the mRNA corresponding to ACT1 was used as a control for equal amounts of input RNA and for the efficiency of the RT-PCRs (bottom panel). Reactions using RNA not treated with reverse transcriptase as the template were employed as a negative control (data not shown).
l-canavanine is a nonproteinogenic α-amino acid structurally related to l-Arg. However, its incorporation in place of Arg during protein translation can generate functionally aberrant polypeptides and eventual cell death (Rosenthal, 1986). Suppression of expression of the AOC5 transporter in the C. reinhardtii RNAi strains likely diminishes l-canavanine uptake, allowing cells to survive and grow in the presence of this compound (Figure 1A). Intriguingly, ∼10% of the transgenic lines showed the expected survival on medium containing l-canavanine (e.g., Figure 1C, Aoc5-IR6) but no reduction in the AOC5 mRNA level (e.g., Figure 1D, Aoc5-IR6). These strains were obtained at a frequency much higher than expected for conventional genetic mutation (i.e., natural mutations disrupting the AOC5 gene) and they displayed no obvious alteration of the endogenous AOC5 locus when examined by DNA gel blotting and hybridization with a probe homologous to the AOC5 coding sequence (data not shown). Thus, these observations raised the possibility that IR-mediated suppression of AOC5 gene expression could occur at the translational level in a subset of C. reinhardtii transformants.
To explore whether RNAi was functional in C. reinhardtii strains with no significant alteration in target transcript levels, we used a tandem IR system, previously demonstrated to suppress simultaneously cotargeted genes (Rohr et al., 2004; Kim and Cerutti, 2009). A hairpin-forming construct homologous to part of the coding sequence of Cre16.g662000, encoding a putative RNA helicase, was engineered inside the AOC5 IRs (see Supplemental Figure 1B online). Transformation of C. reinhardtii with this tandem IR transgene and selection on l-canavanine containing medium allowed the recovery of strains showing reduced transcripts levels for both AOC5 and Cre16.g662000 (data not shown). However, as observed before with the single AOC5 IR strains, ∼5 to 10% of the tandem IR transformants were able to grow in the presence of l-canavanine (e.g., Figure 1C, Aoc5/Helic-IR4) without any obvious change in the AOC5 mRNA abundance (e.g., Figure 1D, Aoc5/Helic-IR4). Interestingly, the Cre16.g662000 transcript was considerably downregulated in the same transgenic lines (e.g., Figure 1D, Aoc5/Helic-IR4). Since the tandem IR transgene directs production of siRNAs homologous to both AOC5 and Cre16.g662000 and the reduction in Cre16.g662000 mRNA amount is indicative of functional RNAi, these results are consistent with AOC5 being repressed at the translational level in a subset of transgenic strains. However, we were unable to test this hypothesis directly due to lack of an antibody to assay AOC5 protein abundance.
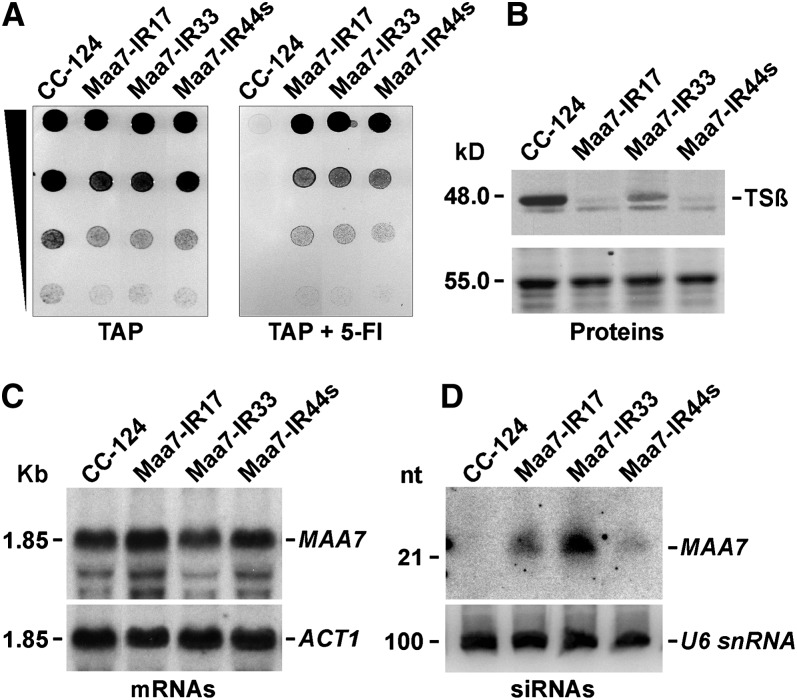
To examine more conclusively whether IR transgenes can suppress gene expression by translation inhibition in C. reinhardtii, we used an alternative system. Trp synthase β-subunit (TSβ; encoded by the METHYL ANTHRANILIC ACID7 —MAA7— gene) is required to convert the indole analog 5-fluoroindole (5-FI) into the toxic Trp analog 5-fluorotryptophan. RNAi-mediated suppression of MAA7 in C. reinhardtii, triggered by dsRNA produced from IR transgenes, results in strains resistant to 5-FI that have reduced MAA7 transcript levels (Rohr et al., 2004). However, ∼10% of the C. reinhardtii transformants containing an IR transgene designed to produce dsRNA homologous to the MAA7 3′ UTR showed tolerance to 5-FI (Figure 2A) and significantly reduced levels of the TSβ protein, as detected by immunoblotting assays (Figure 2B), without any marked change in the MAA7 mRNA amount (Figure 2C; see Supplemental Figure 2A online). Taken together, our observations strongly suggest that IR transgenes can induce translation repression of targeted transcripts in C. reinhardtii, although it remains unexplained why the same construct can trigger primarily either mRNA destabilization or inhibition of protein synthesis in different transgenic lines.
Figure 2.
RNA-Mediated Translation Repression of the MAA7 Transcript (Coding for TSβ), Induced by Expression of MAA7 IR Transgenes in C. reinhardtii Transformants.
(A) Growth and survival of Maa7-IR transformants on TAP medium without (left) or with (right) 7 μM 5-FI. CC-124, untransformed wild-type strain.
(B) Immunoblot analysis of TSβ levels. The smaller cross-reacting antigen is likely a TSβ degradation product and was not consistently detected in replicate blots. Coomassie blue staining of an equivalent gel is shown as a control for similar loading of the lanes (bottom panel).
(C) RNA gel blot analysis of agarose gel–separated total RNA samples sequentially hybridized with 32P-labeled PCR products corresponding to the coding sequence of MAA7 (top panel), to examine the degree of mRNA reduction, or the coding sequence of ACT1 (bottom panel), to assess the amount of sample loaded per lane.
(D) Detection of siRNAs in transgenic strains undergoing MAA7 silencing. Total cell RNA was separated in a 15% denaturing polyacrylamide gel, electroblotted onto a nylon membrane, and hybridized with the MAA7 3′ UTR sequence (top panel). The same filter was reprobed with the U6 small nuclear RNA sequence (bottom panel) as a control for equivalent loading of the lanes. nt, nucleotides.
siRNAs Are Required for the Translation Repression Mediated by IR Transgenes
The Maa7-IR transgenic lines with marked reduction of the TSβ protein content without changes in MAA7 transcript levels contained detectable amounts of MAA7 siRNAs (Figure 2D). Moreover, in the case of the Maa7-IR44s strain, deep sequencing of sRNAs revealed that siRNAs homologous to the MAA7 mRNA matched exclusively to its 3′ UTR, as expected for Dicer processing of the hairpin dsRNA transcribed from the introduced IR transgene (see Supplemental Figure 2B and Supplemental Data Set 1 online). Interestingly, sense siRNAs that would have no functional consequence on MAA7 expression seemed more abundant than antisense siRNAs, but, in both cases, sequenced siRNAs appeared to correspond to nearly every position in the MAA7 3′ UTR (see Supplemental Figure 2B and Supplemental Data Set 1 online). The lower abundance of antisense MAA7 siRNAs is consistent with recent reports suggesting that extensive pairing with a target transcript may destabilize sRNAs in flies and mammals (Ameres et al., 2010), but we did not explore this phenomenon in C. reinhardtii.
To test whether siRNAs are required for the observed suppression of TSβ protein production in C. reinhardtii, we identified a deletion mutant of Exportin 5 (Cre10.g420400) (see Supplemental Figure 3 online) by screening a library of insertional mutants generated in the Maa7-IR44s background. In metazoans, Exportin 5 (EXP5), a member of the importin-β/karyopherin family of proteins, mediates the nuclear export of miRNA precursors (pre-miRNAs), and its depletion results in diminished miRNA amounts (Yi et al., 2003; Lund et al., 2004). The Arabidopsis ortholog of EXP5, HASTY, also appears to be required for the biogenesis (presumably through the nuclear export of Dicer-processed duplex sRNAs) and/or the stability of some miRNAs since mutant plants show a general reduction in miRNA levels (Park et al., 2005). Likewise, in C. reinhardtii, depletion of the EXP5 ortholog caused a decrease in the abundance of many miRNAs, albeit to different extents [see Supplemental Figure 3C online; Maa7-IR44s(exp5)]. This is likely a null phenotype since almost the entire EXP5 gene was deleted in the C. reinhardtii mutant (see Supplemental Figure 3A online) and no EXP5 transcript was detected in RT-PCR assays [see Supplemental Figure 3B online; Maa7-IR44s(exp5)].
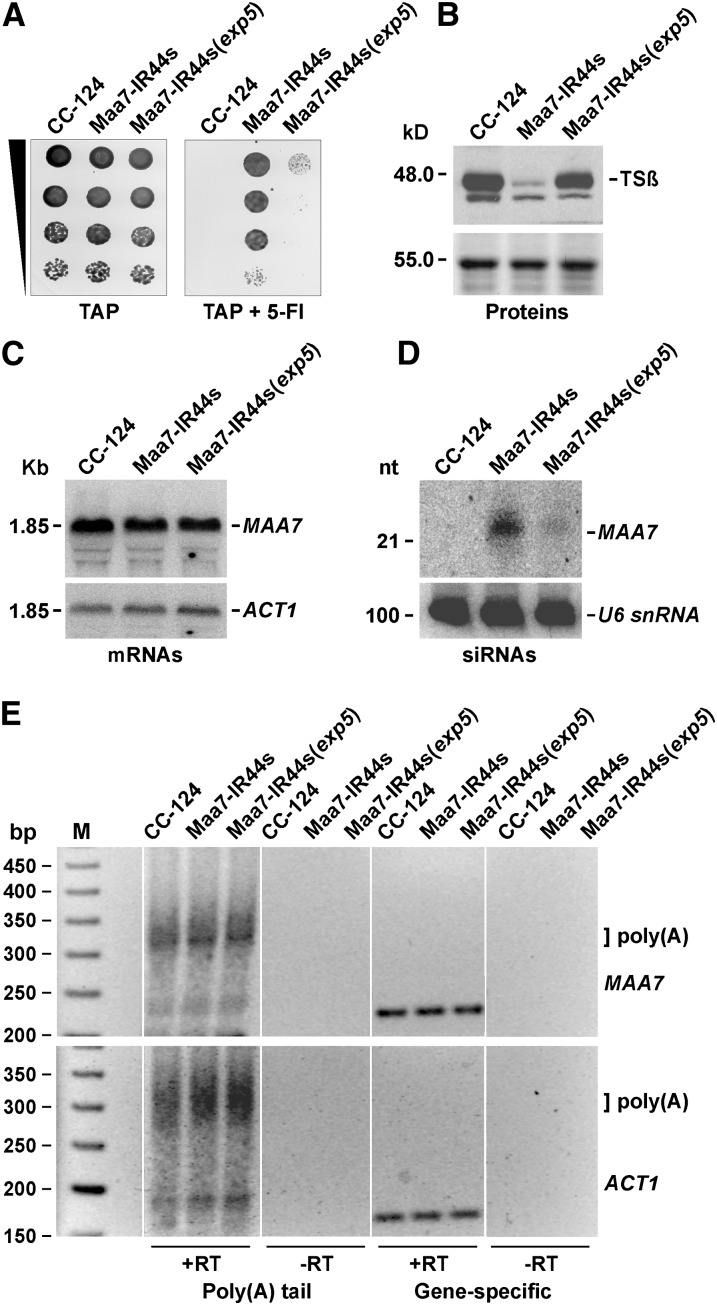
The EXP5-deleted C. reinhardtii strain Maa7-IR44s(exp5) was sensitive to 5-FI, as would be expected for a strain defective in RNAi-mediated downregulation of MAA7 expression (Figure 3A). Moreover, the mutant contained TSβ protein amounts quite similar to those in the wild-type strain (Figure 3B) without appreciable changes in the MAA7 transcript abundance (Figure 3C). Notably, MAA7 siRNA levels were greatly reduced in the mutant background and barely detectable even after prolonged exposure to a phosphor imager screen (Figure 3D). EXP5 does not appear to play a direct role in protein translation, and, if anything, its depletion might have an overall detrimental effect on protein synthesis due to EXP5’s role in the nuclear export of tRNAs (Yi et al., 2003; Lund et al., 2004). Thus, the observed accumulation of TSβ protein in the C. reinhardtii exp5 mutant is most likely a reflection of the requirement for siRNAs (much diminished in the mutant background) as effectors of the translation repression triggered by IR transgenes.
Figure 3.
Translation Repression of the MAA7 Gene Is Greatly Diminished in a C. reinhardtii Mutant Deleted for the EXP5 Ortholog (Encoded by Cre10.g420400) and Does Not Involve Transcript Deadenylation.
(A) Growth and survival of the indicated strains on TAP medium without (left) or with (right) 7 μM 5-FI. Maa7-IR44s(exp5), Maa7-IR44s strain containing a deletion of the Cre10.g420400 gene.
(B) Immunoblot analysis of TSβ abundance. Coomassie blue staining of an equivalent gel is shown as a control for similar loading of the lanes (bottom panel).
(C) RNA gel blot analysis of agarose gel–separated total RNA samples sequentially hybridized with 32P-labeled PCR products corresponding to the coding sequence of MAA7 (top panel), to evaluate the degree of mRNA reduction, or the coding sequence of ACT1 (bottom panel), to assess the amount of sample loaded per lane.
(D) Detection of siRNAs in the Maa7-IR transgenic strains. Total cell RNA was separated in a 15% denaturing polyacrylamide gel, electroblotted onto a nylon membrane, and hybridized with the MAA7 3′ UTR sequence (top panel). The same filter was reprobed with the U6 small nuclear RNA sequence (bottom panel) as a control for equivalent loading of the lanes. nt, nucleotides.
(E) Analysis of polyadenylated tail lengths of the MAA7 and ACT1 transcripts in the indicated strains. Poly(A) tail lengths were examined using a G/I tailing protocol and RT-PCR assays (see Supplemental Figure 4 online). Reactions were performed as described in the Methods in the presence (+RT) or absence (−RT) of reverse transcriptase.
siRNA-Mediated Translation Repression of the MAA7 Transcript Occurs at a Postinitiation Step
In metazoans, mRNA deadenylation is a widespread (although not universal) consequence of miRNA regulation (Huntzinger and Izaurralde, 2011; Bazzini et al., 2012; Béthune et al., 2012; Fukaya and Tomari, 2012; Mishima et al., 2012). Thus, to begin addressing the mechanism of translation repression mediated by IRs in C. reinhardtii, we examined first whether the poly(A) tail length is reduced in the siRNA-repressed MAA7 transcripts. However, we could find no change in the polyadenylation status of this mRNA and of a control transcript encoding actin (ACT1) in Maa7-IR44s in comparison with the wild type and the Maa7-IR44s(exp5) strains (Figure 3E; see Supplemental Figure 4 online). Thus, siRNA mediated translation inhibition of MAA7 in C. reinhardtii seems to occur in a deadenylation-independent manner.
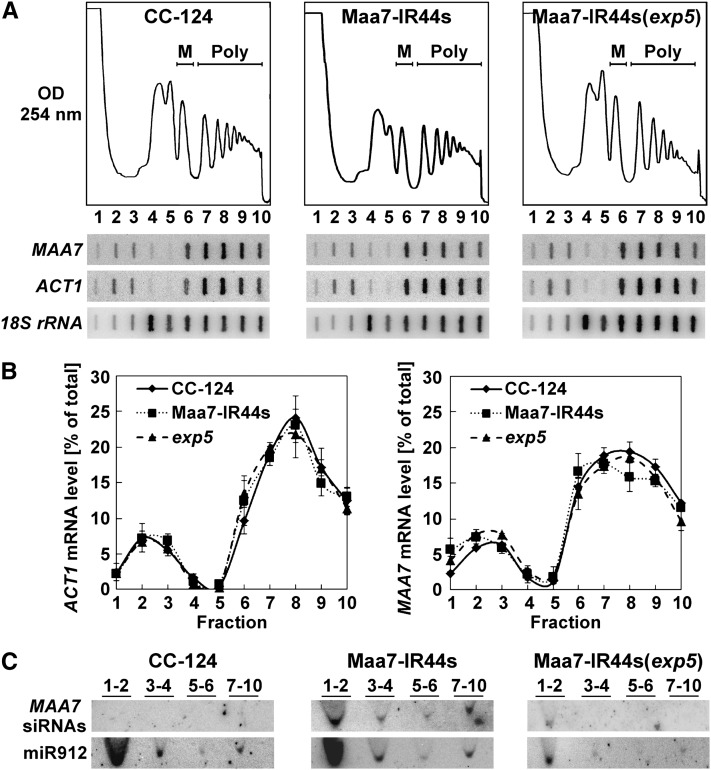
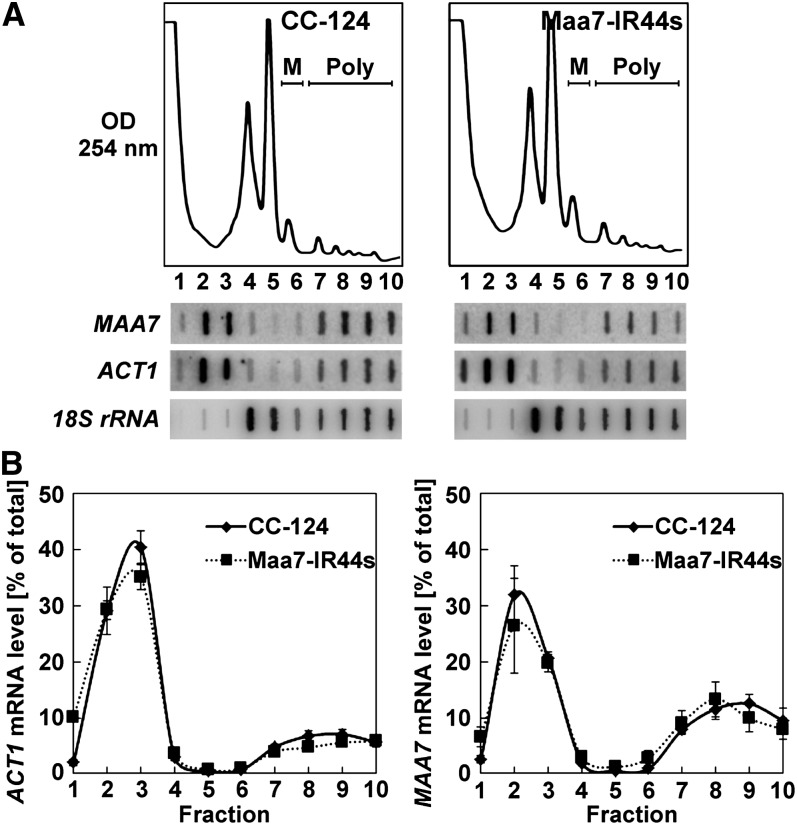
We next performed polyribosome profiling to examine whether translation was being repressed at initiation or postinitiation steps. If translation were inhibited at initiation, the MAA7 transcript would be expected to shift to lighter fractions (with fewer or no ribosomes) when separating Maa7-IR44s cell extracts on Suc sedimentation gradients. By contrast, if protein synthesis were inhibited after initiation, the MAA7 mRNA would be expected to associate with heavier polyribosomal fractions in the translationally repressed transgenic line, although the exact distribution would vary depending on the specific translation step being affected. Cells from the wild type, Maa7-IR44s, and Maa7-IR44s(exp5) strains were treated with a high concentration of cycloheximide (150 μg/mL) to arrest translating ribosomes, resuspended in lysis buffer containing the antibiotic, and broken by one passage through a French press. Lysates were then fractionated by Suc density gradient ultracentrifugation to separate free mRNAs from those associated with varying numbers of ribosomes. The presence of the MAA7 transcript, the control ACT1 mRNA, and the 18S rRNA in each fraction of the gradient was assayed by slot blot hybridization (Figure 4A).
Figure 4.
Translationally Repressed MAA7 Transcripts Comigrate with Polyribosomes in Suc Density Gradients.
(A) Typical polyribosome profiles of the indicated strains treated with 150 μg/mL cycloheximide throughout lysis and ultracentrifugation procedures (top panels). M, monoribosomes; Poly, polyribosomes. The distribution of the MAA7, ACT1, and 18S rRNA transcripts in the gradient fractions was examined by slot blot hybridization (bottom panels).
(B) Distribution of ACT1 and MAA7 mRNAs across polyribosome profiles of the CC-124, Maa7-IR44s, and Maa7-IR44s(exp5) strains. The values represent the average of three independent experiments ± se.
(C) Distribution of MAA7 siRNAs and of an endogenous miRNA (miR912) in Suc density gradients of the indicated strains, assessed by RNA gel blot hybridization. Numbers above the lanes indicate pooled gradient fractions.
Interestingly, MAA7 transcripts were found similarly associated with polyribosomal fractions in the three strains examined (Figures 4A and 4B), regardless of the TSβ protein accumulation (Figure 3B) or the MAA7 siRNAs content (Figure 3D). However, consistent with siRNA-mediated translation repression, some of the MAA7 siRNAs comigrated with polyribosomes in the Maa7-IR44s samples, whereas these siRNAs were practically undetectable in the heavier gradient fractions of the Maa7-IR(exp5) strain (Figure 4C, lanes 7 to 10). In Maa7-IR44s, the proportion of MAA7 siRNAs associated with polyribosomal fractions (relative to the total MAA7 siRNA amount) was much larger than that of an endogenous C. reinhardtii miRNA, such as miR912 (Figure 4C). By contrast, in a previously characterized transgenic strain, Maa7-IR5, containing the same MAA7 IR construct as Maa7-IR44s but inducing target transcript destabilization rather than translation repression (Rohr et al., 2004), the MAA7 siRNAs were more abundant in the ribosome-free portion of the gradient and virtually absent from the polyribosomal fractions (see Supplemental Figure 5 online).
To determine further whether the fast-sedimenting mRNAs and siRNAs were indeed associated with polyribosomes, we treated lysates with EDTA, known to chelate Mg2+ and dissociate translating cytosolic ribosomes into their 40S and 60S subunits (Nolan and Arnstein, 1969). This caused, as expected, redistribution of all tested RNAs to the subpolysomal region of the gradient (see Supplemental Figure 6 online). As EDTA may also disrupt some nonribosomal ribonucleoprotein complexes, we also treated cells with puromycin, prior to cell breakage, in an attempt to induce premature termination of elongating peptide chains and specific disassembly of translating ribosomes (Blobel and Sabatini, 1971; Kong et al., 2008; Ding and Grosshans, 2009). However, this treatment caused only a minor reduction in the polyribosomal fractions in C. reinhardtii (similarly in the three strains examined), presumably because of poor drug uptake (data not shown). The sedimentation patterns of the MAA7 and ACT1 transcripts in puromycin-treated cells were again indistinguishable among the wild type, Maa7-IR44s, and Maa7-IR44s(exp5) (data not shown). Thus, the MAA7 mRNA appears to associate predominantly with bona fide translating ribosomes in the three examined strains (see Supplemental Figure 7A online). These observations led us to conclude that MAA7 siRNAs repress translation of the target transcript primarily at a postinitiation stage in the C. reinhardtii Maa7-IR44s transgenic line.
Ribosomes Subjected to siRNA-Mediated Translation Repression Show Lower Sensitivity to Inhibition by Cycloheximide
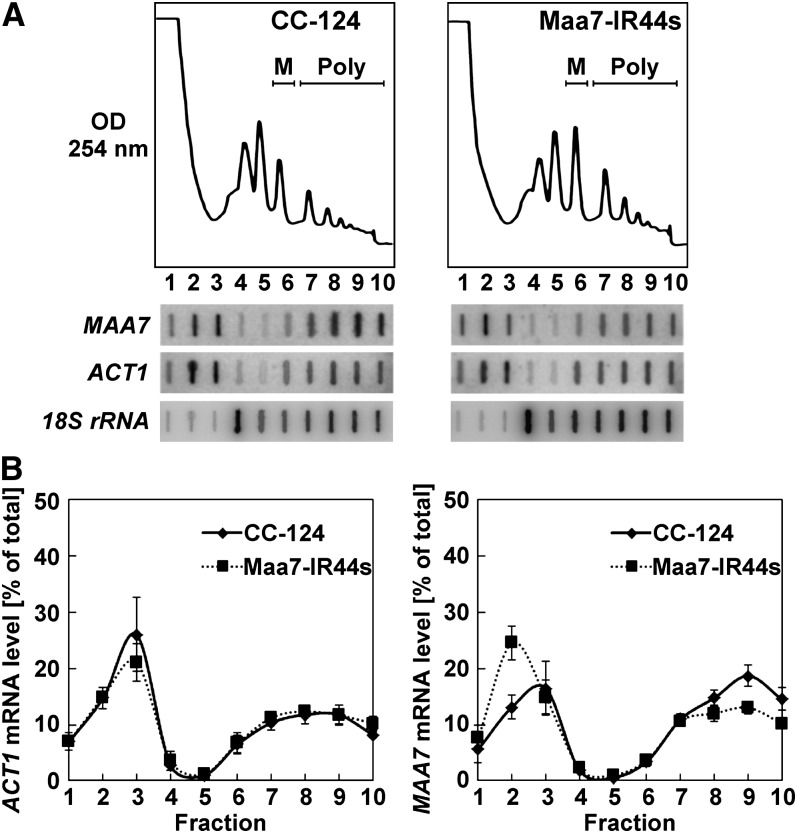
The association of MAA7 transcripts with polyribosomes in the Maa7-IR44s strain suggested several possible mechanisms of siRNA-mediated protein synthesis inhibition, including effects on translation elongation, termination, and/or degradation of nascent polypeptides. We therefore attempted to assess functional differences, between the wild-type and Maa7-IR44s strains, in the ribosomes associated with the MAA7 mRNA. In the absence of cycloheximide, protein translation proceeds for a short time in cell extracts, partly depleting ribosomes from mRNA templates (Saini et al., 2009). This ribosome run-off assay can be used to evaluate the stability of ribosome association with transcripts, which will depend on the elongation rate and susceptibility to premature termination. Cell extracts from the wild type and Maa7-IR44s strains were prepared in lysis buffer lacking cycloheximide and containing 150 mM KCl and 5 mM Mg2+, ionic conditions near the optimum for in vitro protein synthesis (Shenvi et al., 2005). As expected, upon Suc density gradient ultracentrifugation, the polyribosomal component in these extracts was much reduced (Figure 5A) in comparison with that observed in high cycloheximide-treated cells (Figure 4A). However, the new experimental conditions were uninformative as to the function of siRNA-repressed ribosomes since we found no significant difference in the MAA7 transcript (and the control ACT1 mRNA) gradient distribution between the two examined strains (Figures 5A and 5B; see Supplemental Figure 7B online).
Figure 5.
The Fraction of MAA7 Transcripts Comigrating with Polyribosomes Is Reduced after Suc Density Gradient Ultracentrifugation of Cell Extracts Subjected to Ribosome Run-Off in the Absence of Cycloheximide.
(A) Typical polyribosome profiles of the indicated strains (top panels). M, monoribosomes; Poly, polyribosomes. The distribution of the MAA7, ACT1, and 18S rRNA transcripts in the gradient fractions was examined by slot blot hybridization (bottom panels).
(B) Distribution of ACT1 and MAA7 mRNAs across polyribosome profiles of the CC-124 and Maa7-IR44s strains. The values represent the average of three independent experiments ± se.
Cycloheximide is a potent inhibitor of protein synthesis with high specificity for eukaryotic ribosomes. This antibiotic has been reported to inhibit translation elongation by binding to the large ribosomal subunit exit (E) site, stalling translocation as a consequence of the occupation of the E site by both cycloheximide and a deacylated tRNA (Pestova and Hellen, 2003; Schneider-Poetsch et al., 2010; Klinge et al., 2011). We next tested low concentrations of cycloheximide (30 μg/mL) to reduce the rate of elongation in the ribosome run-off assay rather than totally inhibit this process. We reasoned that slowing down elongation might increase polyribosomal association if a transcript was being translated by ribosomes already partly repressed at the elongation step, whereas a normally translated mRNA might be affected to a lesser degree. Under these low cycloheximide conditions, the overall abundance of polyribosomes (Figure 6A) was intermediate between the high cycloheximide (Figure 4A) and the no cycloheximide (Figure 5A) treatments for both tested strains. Intriguingly, in the low cycloheximide ribosome run-off experiments, the MAA7 transcript was moderately but consistently depleted from the polyribosomal fractions in the translationally repressed Maa7-IR44s strain relative to the wild type (Figures 6A and 6B; see Supplemental Figure 7C online). As a control, the distribution of the ACT1 mRNA in the Suc density gradients was virtually identical in both examined strains (Figures 6A and 6B; see Supplemental Figure 7C online). These results, although unexpected based on our initial reasoning, provided the first evidence for a functional difference(s) between the ribosomes associated with the MAA7 transcript in the wild-type strain (translationally competent) and the Maa7-IR44s strain (translationally inhibited by an siRNA-dependent mechanism). Similarly, when cells were treated with low concentrations of cycloheximide, the AOC5 mRNA was also moderately depleted from polyribosomal fractions in the repressed Aoc5/Helic-IR4 strain relative to the CC-124 control (see Supplemental Figure 8 online), suggesting that these observations are indicative of a general feature of siRNA-inhibited ribosomes.
Figure 6.
siRNA-Repressed MAA7 Transcripts Are Moderately Depleted from Polyribosomal Fractions after Ribosome Run-Off Assays in the Presence of Low Concentrations of Cycloheximide.
(A) Typical polyribosome profiles of the indicated strains treated with 30 μg/mL cycloheximide throughout lysis and ultracentrifugation procedures (top panels). M, monoribosomes; Poly, polyribosomes. The distribution of the MAA7, ACT1, and 18S rRNA transcripts in the gradient fractions was examined by slot blot hybridization (bottom panels).
(B) Distribution of ACT1 and MAA7 mRNAs across polyribosome profiles of the CC-124 and Maa7-IR44s strains. The values represent the average of three independent experiments ± se.
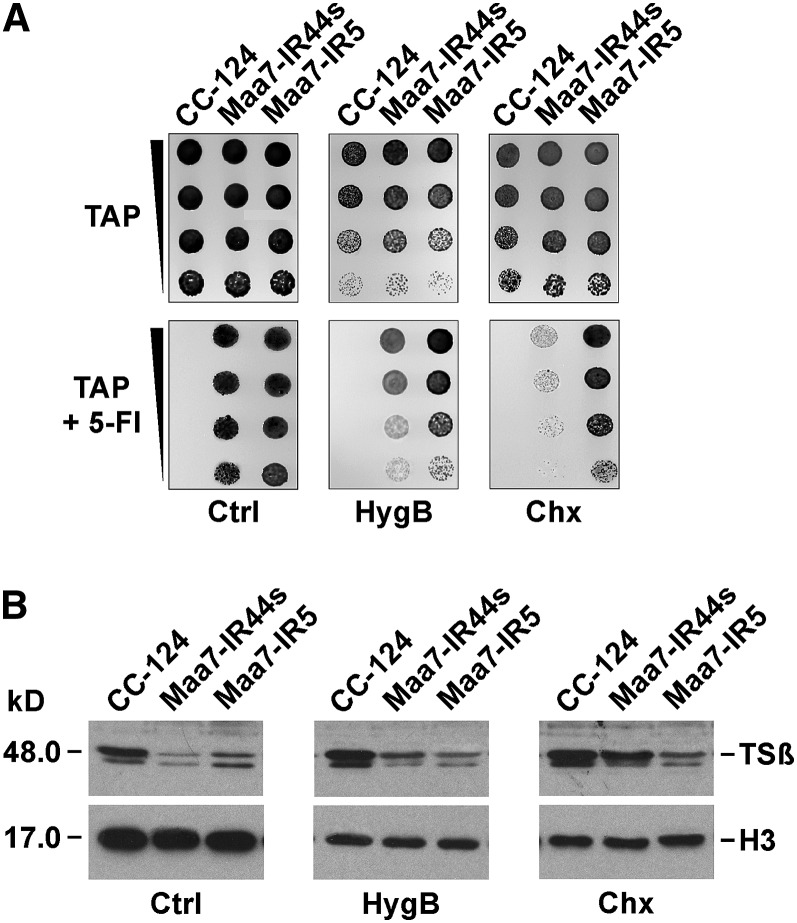
To gain further insight on the function of siRNA-repressed ribosomes, we examined the effect of different antibiotics and growing conditions on the accumulation of the TSβ protein in vivo. We were particularly interested in testing whether siRNA-mediated translation repression was altered by antibiotics with dissimilar modes of action, inhibiting distinct ribosome functions. A sublethal concentration of cycloheximide severely compromised survival of the Maa7-IR44s strain in medium containing 5-FI (Figure 7A), consistent with greater MAA7 expression. By contrast, this treatment had little effect on the phenotype of the previously characterized Maa7-IR5 strain containing the same MAA7 IR construct integrated into its genome but inducing target transcript destabilization rather than translation repression (Rohr et al., 2004). In cells growing in liquid medium, exposure to low cycloheximide for 18 h (see Methods) had a similar inhibitory effect on overall translation in the three examined strains, the wild type, Maa7-IR44s, and Maa7-IR5, as reflected by a comparable reduction in histone H3 levels (Figure 7B, cf. Ctrl and Chx panels). However, the TSβ protein amount uniquely increased in Maa7-IR44s subjected to sublethal cycloheximide concentrations (Figure 7B, cf. Ctrl and Chx panels), whereas the antibiotic did not affect TSβ protein levels (a fairly stable polypeptide) in the wild type or the Maa7-IR5 strains. This suggested that cycloheximide treatment is of no consequence for RNAi-triggered mRNA destabilization in Maa7-IR5. No change in the original MAA7 mRNA abundance was observed in any of these strains during the 18-h experimental period (data not shown).
Figure 7.
TSβ Protein Synthesis from the MAA7 Transcript, Subjected to siRNA-Mediated Translation Repression, Shows Lower Sensitivity to Inhibition by Cycloheximide.
(A) Growth and survival of the indicated strains on TAP medium without (top panels) or with (bottom panels) 7 μM 5-FI supplemented with solvent (Ctrl) or sublethal concentrations of hygromycin B (HygB; 4.0 μg/mL) or cycloheximide (Chx; 2.0 μg/mL). Maa7-IR5, strain expressing a MAA7 IR transgene that induces target mRNA degradation (Rohr et al., 2004).
(B) Immunoblot analyses of TSβ and histone H3 protein levels. Cells from the indicated strains were cultured for 18 h in liquid TAP medium alone (Ctrl) or containing sublethal concentrations of antibiotics (2.0 μg/mL of hygromycin B or 1.0 μg/mL of cycloheximide). Proteins corresponding to equal numbers of cells were loaded per lane.
Paromomycin, an aminoglycoside antibiotic that influences the decoding center of the ribosome, translation fidelity, and perhaps an early stage of translation after initiation (Eustice and Wilhelm, 1984; Borovinskaya et al., 2008), did not alter the survival on 5-FI–containing medium or the TSβ protein amount in Maa7-IR44s (see Supplemental Figures 9A and 9B online), despite being as effective at inhibiting histone H3 accumulation as cycloheximide (see Supplemental Figure 9B online). Likewise, anisomycin, an antibiotic that binds to the aminoacyl site of the large ribosomal subunit and inhibits translation elongation by competing with the binding of aminoacyl-tRNA to the peptidyltransferase center (Blaha et al., 2008; Rakauskaite and Dinman, 2008), did not affect survival on 5-FI or TSβ protein levels of Maa7-IR44s (see Supplemental Figures 9C and 9D online). Merely reducing growth rate (and overall protein synthesis) by culturing cells in minimal medium also had no apparent consequence on TSβ protein expression in Maa7-IR44s, based on the strain’s ability to survive and grow in the presence of 5-FI (see Supplemental Figure 9E online). Treatment with sublethal concentrations of a fourth antibiotic, hygromycin B, slightly increased TSβ protein accumulation in Maa7-IR44s (Figure 7B) and reduced to some extent survival of the strain on 5-FI–containing medium (Figure 7A). Interestingly, hygromycin B seems to have a mode of translation inhibition that differs from other aminoglycoside antibiotics. In addition to affecting decoding fidelity, it appears to have, like cycloheximide, an inhibitory effect on the translocation of mRNA and tRNAs on the ribosome (Eustice and Wilhelm, 1984; Borovinskaya et al., 2008).
The observed effects of cycloheximide, and to a much lower degree of hygromycin B, on TSβ protein accumulation in the Maa7-IR44s strain are unlikely to be indirect, such as destabilization of a short-lived protease required for TSβ degradation or of a polypeptide specifically involved in siRNA-mediated translation repression, since, if this were the case, paromomycin and anisomycin would be expected to have a similar consequence on TSβ protein content. Additionally, since MAA7 suppression by target mRNA destabilization in the Maa7-IR5 strain was not affected by the cycloheximide or hygromycin B treatments, general depletion of core components of the RNAi machinery also appears unlikely. Moreover, exposure to low concentrations of cycloheximide distinctly altered ribosome association with MAA7 or AOC5 transcripts, assessed by Suc density gradient ultracentrifugation, when comparing Maa7-IR44s or Aoc5/Helic-IR4 with the wild-type strain (Figures 6A and 6B; see Supplemental Figures 7C and 8 online). These observations, taken together, suggest that ribosomes translationally repressed by siRNAs are differentially (less) sensitive to inhibition by cycloheximide and to some extent hygromycin B, strongly implying that the siRNA machinery imposes some alteration on normal ribosome function/structure.
DISCUSSION
RNAi has been developed as a practical tool to study gene function in a few algal species (Cerutti et al., 2011). In C. reinhardtii, hairpin-forming transcripts produced from genome-integrated IR transgenes have been successfully used to downregulate the expression of a number of endogenous genes (Schroda, 2006; Cerutti et al., 2011). In most cases, reduction of the steady state level of targeted mRNAs was observed, implying RNAi-mediated transcript degradation (Rohr et al., 2004; Schroda, 2006; Cerutti et al., 2011). However, in a few instances, discrepancies between protein and mRNA amounts, suggestive of inhibitory effects on translation, have also been reported. For example, a C. reinhardtii transgenic line containing an IR transgene designed to suppress expression of Chlamyopsin, coding for an opsin-related protein, displayed a 50-fold reduction in protein abundance but only a threefold decrease in transcript amount, in comparison with the wild-type strain (Fuhrmann et al., 2001). In land plants and animals, there is convincing evidence that siRNAs perfectly complementary to a target mRNA can mediate translation repression in addition to mRNA degradation (Rao et al., 2006; Brodersen et al., 2008; Wu et al., 2008; Yu and Wang, 2010). Our results indicate that this phenomenon also occurs in the unicellular green alga C. reinhardtii, triggered by MAA7 IR transgenes (Figures 2 and 3) and by AOC5 IR transgenes (Figure 1D; see Supplemental Figure 8 online) designed to produce hairpin dsRNA homologous to the 3′ UTR of target transcripts.
Interestingly, in C. reinhardtii, the same IR construct (homologous to a 3′ UTR) can induce predominantly either target mRNA degradation or translation repression in different transgenic lines. One obvious difference among the RNAi strains is the site of integration of the IR transgene in the algal nuclear genome. In mammalian cells, it has been recently demonstrated that the promoter driving transcription of an mRNA influences the type of miRNA-mediated translation repression. Transcripts derived from the Simian Virus 40 promoter, containing let-7 target sites in their 3′ UTRs, are repressed at the initiation state of translation, whereas identical mRNAs derived from the Thymidine Kinase promoter are repressed at a postinitiation step (Kong et al., 2008). Bushell and colleagues proposed that a nuclear event linked to the promoter, such as cotranscriptional loading of factors onto the nascent mRNA, might determine the type of miRNA-mediated translation repression (Kong et al., 2008). Similarly, we speculate that the site of integration of an IR transgene in the C. reinhardtii genome may influence its transcriptional activity, site of hairpin dsRNA processing to siRNAs (nuclear versus cytoplasmic), and the eventual association of a factor(s) with siRNA-loaded AGOs that may modulate the type of repression. However, addressing the actual molecular mechanism(s) determining this choice will require further investigation.
In metazoans, the mechanism(s) of miRNA-mediated silencing has been the subject of extensive research (see Introduction). However, since translation repression, deadenylation, and transcript decay are closely linked processes, delineating a unifying model of silencing has been difficult (Fabian et al., 2010; Cenik and Zamore, 2011; Huntzinger and Izaurralde, 2011; Béthune et al., 2012; Djuranovic et al., 2012; Fukaya and Tomari, 2012; Ricci et al., 2013; Stadler et al., 2012). Recent studies examining the relative timing of different events suggest that miRNA targets in Danio rerio, Drosophila, and HeLa cell lines are first subject to translational inhibition, followed by effects on deadenylation and mRNA degradation (Bazzini et al., 2012; Béthune et al., 2012; Djuranovic et al., 2012; Mishima et al., 2012). In this context, deadenylation may consolidate the initial translational inhibition (Huntzinger and Izaurralde, 2011), which apparently occurs at the initiation level (Bazzini et al., 2012; Djuranovic et al., 2012; Ricci et al., 2013). However, in animal systems, there is also evidence for some repressed mRNAs remaining associated with polyribosomes, a strong argument in support of postinitiation translation inhibition (Gu et al., 2009; Fabian et al., 2010; Huntzinger and Izaurralde, 2011; Stadler et al., 2012). miRNAs have been proposed to slow down translation elongation (Maroney et al., 2006), promote premature termination (ribosome drop-off) (Petersen et al., 2006), or induce rapid proteolysis of nascent polypeptides (Nottrott et al., 2006). In addition, in Drosophila, which contains two AGO subfamily proteins, both Argonautes can inhibit translation but by different mechanisms. AGO2 specifically represses the cap recognition step, whereas AGO1 induces deadenylation of target mRNAs and, secondarily, blocks translation downstream from cap recognition (Iwasaki et al., 2009; Fukaya and Tomari, 2012). Indeed, alternative sRNA effector complexes, including AGO-PIWI polypeptides and associated factors such as GW-repeat proteins, as well as specific features of the sRNA binding site and the proteins associated with a given target transcript may determine the actual mode of sRNA-mediated repression (Bartel, 2009; Gu et al., 2009; Fabian et al., 2010; Huntzinger and Izaurralde, 2011).
Despite these advances, our mechanistic understanding of the sRNA-mediated inhibition of translation at postinitiation steps is still very limited. Several ribosomal proteins have been implicated in sRNA-triggered silencing (Pinon et al., 2008; Zhou et al., 2008; Chan and Slack, 2009), although, in mammalian cells, relief of miRNA repression of translation by depletion of ribosomal proteins may be caused indirectly by activation of the p53 pathway (Janas et al., 2012). Nonetheless, certain ribosomal proteins have been demonstrated to coimmunoprecipitate with Argonautes and other components of sRNA effector complexes (Ishizuka et al., 2002; Landthaler et al., 2008; Zhou et al., 2008; Chan and Slack, 2009). In nematodes and mammals, it was recently demonstrated that the RECEPTOR FOR ACTIVATED C-KINASE1, an integral component of the 40S ribosomal subunit, is required for the association of miRNA effector complexes with translating ribosomes and may contribute to silencing at a postinitiation step (Jannot et al., 2011). Similarly, Argonaute proteins can form a complex with PUF (Pumilio/FBF) RNA binding proteins and with eukaryotic translation elongation factor 1A, reducing its ability to hydrolyze GTP (Friend et al., 2012). This complex attenuates translation elongation perhaps by interfering with the proper delivery of aminoacylated tRNAs to the ribosome (Friend et al., 2012). However, in this experimental system, target specificity was conferred by the PUF proteins and it remains to be examined whether sRNA-guided AGO proteins can recruit a similar complex to mRNA targets and elicit the same regulatory mechanism. Nevertheless, these studies, taken together, indicate that AGOs or sRNA-guided effector complexes may interact with ribosomes and/or other components of the translation machinery to bring about translation inhibition at postinitiation steps in metazoans.
In land plants and algae, repression of protein synthesis by sRNAs remains poorly characterized (Lanet et al., 2009; Voinnet, 2009; Yu and Wang, 2010; Cerutti et al., 2011; Yang et al., 2012). Our observations suggest that, in C. reinhardtii, siRNA-mediated translation inhibition of the MAA7 transcript occurs in a deadenylation-independent manner (Figure 3E). Suc density gradient ultracentrifugation analysis, separating mRNAs according to the number of associated ribosomes, has been the main experimental technique used to support either the initiation or postinitiation modes of sRNA repression (Huntzinger and Izaurralde, 2011). In this approach, cells/organisms are commonly treated with cycloheximide to freeze translating ribosomes on mRNAs. After the addition of a high molar excess of cycloheximide, the first ribosome initiating on a mRNA becomes locked over the start codon, preventing the loading of additional ribosomes and elongating ribosomes become blocked on their progression (Pestova and Hellen, 2003; Schneider-Poetsch et al., 2010). Hence, this experimental condition should ideally reflect the in vivo ribosome occupancy and ribosome density on a given transcript (Hendrickson et al., 2009; Huntzinger and Izaurralde, 2011). Since MAA7 repressed transcripts were found associated with bona fide translating ribosomes in the high cycloheximide Suc density gradient assays (Figure 4), siRNA-mediated inhibition of protein synthesis in C. reinhardtii appears to occur at a postinitiation step. Interestingly, in ribosome run-off experiments in the presence of low concentrations of cycloheximide (unable to cause total elongation block), the MAA7 and the AOC5 transcripts were moderately but consistently depleted from the polyribosomal fractions in translationally repressed strains in comparison with the wild type (Figure 6; see Supplemental Figure 8 online). The simplest interpretation of these results suggests that ribosome run-off (i.e., elongation and normal termination) and/or drop-off (i.e., premature, abnormal termination) can still occur on the siRNA inhibited MAA7 and AOC5 mRNAs in the presence of a low dose of the antibiotic.
The ribosome run-off assays imply that siRNA-repressed ribosomes are more active (less inhibited) than normal ones under low concentrations of cycloheximide. Moreover, in in vivo 18-h experiments with sublethal concentrations of cycloheximide, accumulation of TSβ is blocked in the translationally competent CC-124 strain, without a significant decrease in TSβ abundance since this is a long-lived protein (Figure 7B). By contrast, in the translationally repressed Maa7-IR44s strain, TSβ protein levels increase in the presence of low concentrations of cycloheximide, indicating that the ribosomes can still translate the MAA7 transcript (Figure 7B). The loading of the lanes in Figure 7B is normalized for equal number of cells and is therefore indicative of average protein content per cell at the end of the 18-h experimental period. Since antibiotic-treated cells, as previously reported (Bulté and Bennoun, 1990), are arrested in growth and division, the observed changes in protein content largely reflect what occurs in the initially inoculated cells. However, C. reinhardtii in control medium undergoes one or two rounds of cell division during the 18-h experimental period and the corresponding increase in culture protein accumulation (as a consequence of an increase in cell numbers) is not displayed in Figure 7B. Taking these technical aspects into consideration, the results suggest that siRNA-repressed ribosomes have reduced sensitivity to inhibition by cycloheximide, allowing translation of the TSβ protein from the MAA7 transcript in the presence of sublethal concentrations of the antibiotic. Interestingly, pretreatment of the mammalian ECV-304 cell line with cycloheximide also partly relieves the miRNA-mediated repression of a Renilla luciferase reporter (Detzer et al., 2011).
In the recently solved crystal structure of the Tetrahymena thermophila 60S ribosomal subunit, cycloheximide was shown to bind in a tight pocket of the E site, previously identified as the binding site for 3′-terminal nucleotides of deacylated tRNAs in the archaeal ribosome (Klinge et al., 2011). This is in agreement with observations in C. reinhardtii where substitutions of a Pro residue in ribosomal protein L41 (named L36a in higher eukaryotes), which is a conserved component of the T. thermophila cycloheximide binding pocket (Klinge et al., 2011), confer resistance to cycloheximide (Stevens et al., 2001). Occupation of the E site by both cycloheximide and a deacylated tRNA, effectively trapping deacylated tRNA at the E site, is thought to block eukaryotic ribosome translocation (Pestova and Hellen, 2003; Schneider-Poetsch et al., 2010; Budkevich et al., 2011). Antibiotics binding to the aminoacyl site, such as anisomycin, or to the decoding center, such as paromomycin, of the ribosome have no effect on siRNA-mediated translation repression in C. reinhardtii (see Supplemental Figure 9 online). Thus, our findings are consistent with a fairly specific alteration(s) of the function/structural conformation of translating ribosomes, mediated by siRNA-effector complexes, which may also affect the binding and/or the action of certain antibiotics such as cycloheximide. However, the exact mechanism of translation repression induced by sRNAs in C. reinhardtii remains to be elucidated.
We previously argued, based on phylogenetic and taxonomic distribution analyses, that fairly complex RNAi machinery was already present in the last common ancestor of eukaryotes (Cerutti and Casas-Mollano, 2006). This ancestral RNAi machinery may have been capable of both sRNA-guided transcript degradation as well as transcriptional repression, both widespread sRNA-mediated processes among living eukaryotes (Cerutti and Casas-Mollano, 2006; Voinnet, 2009; Cenik and Zamore, 2011). By contrast, reports of sRNA-induced translation repression were initially limited to animals and land plants, suggestive of a more recently evolved mechanism confined to certain lineages (Voinnet, 2009; Fabian et al., 2010; Yu and Wang, 2010; Huntzinger and Izaurralde, 2011). However, current evidence indicates that sRNAs can inhibit translation in a much wider range of eukaryotes, including the protozoan parasite G. lamblia, the diatom P. tricornutum, and the green alga C. reinhardtii (this work; De Riso et al., 2009; Li et al., 2011; Saraiya et al., 2011). Additionally, there is experimental support for the association of sRNAs and of Argonautes with polyribosomes in the parasites Trypanosoma brucei and Toxoplasma gondii (Djikeng et al., 2003; Shi et al., 2009; Braun et al., 2010). Given the much wider taxonomic distribution of sRNA-mediated translation repression, it is tempting to speculate that a basic process of protein synthesis inhibition may have been another feature of an ancestral RNAi machinery.
METHODS
Transgenic Strains, Mutants, and Culture Conditions
Chlamydomonas reinhardtii transgenic strains containing IR constructs homologous to AOC5, AOC5/Cre16.g662000, or MAA7 were generated as previously described (Rohr et al., 2004; Kim and Cerutti, 2009). DNA fragments for building the IR constructs were generated by RT-PCR amplification with the following primers: for AOC5, AA-Per-1 (5′-GCTGACGAGTCTGTGGAGACG-3′) and AA-Per-2 (5′-CTTACTCACGCCCAGCAGAGA-3′); and for Cre16.g662000, Helic-F1 (5′-GGATGACGTGATCGCCAAG-3′) and Helic-R2 (5′-GGCCTGAATCCCATGTCTAGC-3′). The AOC5 primers amplify a 930-bp fragment that was digested with NheI to generate a 3′ segment of 380 bp used to build the IR transgene (see Supplemental Figure 1 online). The IR construct targeting the MAA7 3′ UTR has already been described (Rohr et al., 2004). The Cre10.g420400-deleted strain, lacking EXP5, was obtained in an insertional mutagenesis screen designed to isolate mutants defective in RNAi in C. reinhardtii (Ibrahim et al., 2006, 2010). Unless noted otherwise, C. reinhardtii cells were grown photoheterotrophically in Tris-Acetate-Phosphate (TAP) medium or photoautotrophically in minimal high salt medium (Harris, 1989). For phenotypic analyses, cells grown to logarithmic phase in TAP or high salt media were serially diluted, spotted on plates of the appropriate media (see figure legends), and incubated for 7 to 15 d under dim lights (Rohr et al., 2004). The antibiotic concentrations used in in vivo experiments were previously demonstrated to be inhibitory of protein synthesis in C. reinhardtii (Bulté and Bennoun, 1990).
RNA Analyses
Total cell RNA was purified with TRI reagent (Molecular Research Center), following the manufacturer’s instructions. For RNA gel blot analyses of mRNAs, the isolated RNA was separated by agarose/formaldehyde gel electrophoresis, blotted onto nylon membranes, and hybridized with 32P-labeled probes (Sambrook and Russell, 2001; Rohr et al., 2004). For sRNA analyses, total RNA samples were resolved on 15% polyacrylamide/7 M urea gels and electroblotted to Hybond-XL membranes (GE Healthcare) (Sambrook and Russell, 2001; Rohr et al., 2004). Blots were hybridized with 32P-labeled DNA probes at 40°C for 48 h using the High Efficiency Hybridization System (Molecular Research Center). Specific miRNAs were detected by hybridization with DNA oligonucleotides labeled at their 5′ termini with [γ-32P]ATP and T4 Polynucleotide Kinase (New England Biolabs). The poly(A) tail length of specific transcripts was estimated using a G/I tailing protocol followed by RT-PCR analysis (Kusov et al., 2001), according to a commercially available kit (USB; Affymetrix). The primer sequences for the poly(A) tail analyses were as follows: for ACT1, ACT-3′UTR-PF4 (5′-AAGATATGAGGAGCGGGTCA-3′) and ACT-3′UTR-PR2 (5′-AAATGGTCCGAGCAGGTTTT-3′); and for MAA7, MAA7-3′UTR-PF1 (5′-GTGATTGAAAGGGGAGCGTA-3′) and MAA7-3′UTR-PR1 (5′-ACATGCGATTGGTAGCAACA-3′).
Immunoblot Analyses
The C. reinhardtii TSβ protein was immunodetected, following a standard procedure (Rohr et al., 2004), by overnight incubation at 4°C with a 1:5000 dilution of a rabbit antibody raised against the full-length recombinant protein (GenScript). A modification-insensitive polyclonal antibody (Abcam ab1791) was used to detect histone H3.
RT-PCR Analyses
Total RNA was isolated with TRI reagent, and contaminant DNA was removed by DNase-I treatment (Ambion). First-strand cDNA synthesis and PCR reactions were performed as previously described (Sambrook and Russell, 2001; Rohr et al., 2004). PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining (Sambrook and Russell, 2001). The number of cycles showing a linear relationship between input RNA and the final product was determined in preliminary experiments. Controls included the use as template of reactions without RT and verification of PCR products by hybridization with specific probes (data not shown). The primer sequences were as follows: for AOC5, AA-Per-3(F) (5′-CTTCAAAGTGCCGCTGTACCC-3′) and AA-Per-4(R) (5′-GTCTCCACAGACTCGTCAGCA-3′); for EXP5, Mut3-cod-F1 (5′-ACAGGGACGCAGTCAAGG-3′) and Mut3-cod-R2 (5′-CCAGGCTCAGGACCATGTAG-3′); and for ACT1, ACT-cod-F (5′-GACATCCGCAAGGACCTCTAC-3′) and ACT-cod-R (5′-GATCCACATTTGCTGGAAGGT-3′). The Cre16.g662000 gene, encoding a putative RNA helicase, has a very close paralog in the C. reinhardtii genome (Cre16.g661900). Thus, to avoid amplification of the related transcript, reverse transcription was performed with a Cre16.g662000-specific primer (Helic-R7, 5′-CACATCCGAGCTGAACATGAC-3′) and then PCR was performed with Helic-F2 (5′-CCAAATTTCCAAGATCCTCAGC-3′) and Helic-R4 (5′-AGCATGACGTCGCGCTTG-3′).
Sequencing of sRNAs Matching the MAA7 Transcript
The sRNA fraction was precipitated from 300 µg of total RNA isolated from strain Maa7-IR44s, and a cDNA library was constructed and sequenced on an Illumina Genome Analyzer II as previously described (Lu et al., 2007; Ibrahim et al., 2010). After sequencing, the adapter sequences were removed and sequences shorter than 18 nucleotides or longer than 28 nucleotides were discarded. Sequence reads were compared with the C. reinhardtii genome version 4.0, ESTs, predicted transcripts, transposons, and bonus reads (http://genome.jgi-psf.org/Chlre4/Chlre4.download.ftp.html). Sequences matching tRNAs, rRNAs, small nuclear RNAs, small nucleolar RNAs, the chloroplast, or the mitochondrial genomes were removed. The remaining sRNAs were normalized to transcripts per million (Ibrahim et al., 2010). All sequences matching the MAA7 mRNA (see Supplemental Data Set 1 online), with one mismatch allowed, were then extracted from this population.
Polyribosome Profile Analyses
C. reinhardtii strains were grown to mid logarithmic phase in liquid TAP medium, ∼8 × 108 cells pelleted by centrifugation, and resuspended in 10 mL of the same medium. Resuspended cells were incubated under dim lights and constant shaking for 15 min in the presence of 150 μg/mL cycloheximide, 30 μg/mL cycloheximide, or 300 μg/mL puromycin. In the no-antibiotic experiments, the TAP medium contained an amount of ethanol (solvent) equivalent to that added with the antibiotics. Cells were then pelleted again and resuspended in 10 mL of lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM KCl, 5 mM MgCl2, and 1 mM DTT) containing the same antibiotics or solvent control and supplemented with an EDTA-free protease inhibitor cocktail (Sigma-Aldrich). From this step on, cells and lysates were always kept on ice. Cells were broken by one passage through a French press at a pressure of ∼2000 p.s.i. To complete cell lysis, 0.1 volume of 5% sodium deoxycholate, pH 8.0, was added to the lysates and mixed gently for ∼5 min. Cell extracts were then centrifuged at 10,000g for 10 min at 4°C. Supernatant concentrations were normalized by measuring absorbance at 254 nm, and a 0.125 volume of 10% Triton X-100 was added gently. Finally, ∼600 μL of the clarified cell extracts were layered on 4.5 to 45% (w/v) Suc gradients prepared in 25 mM Tris-HCl, pH 7.5, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, and 0.5 mg/mL heparin and then centrifuged for 2.5 h at 260,000g in a Beckman SW41 rotor. Gradients were fractionated with an ISCO system while monitoring absorbance at 254 nm. Total RNA was purified from each fraction by two phenol/chloroform extractions and ethanol precipitation. Specific transcripts were detected by slot blot hybridization of RNA treated with DNase-I (Ambion) to remove any contaminating DNA (particularly in subpolyribosomal fractions). sRNAs were isolated as previously described (Djikeng et al., 2003). For EDTA treatment, antibiotics were omitted and EDTA was added to 50 mM in the lysis buffer and to 10 mM in the Suc gradients.
Accession Numbers
Sequence data for the genes discussed in this article can be found in Phytozome v9.0 (www.phytozome.net; C. reinhardtii v5.3.1) under the following accession numbers: Cre03.g161400.t1.2 (MAA7), Cre07.g329050.t1.2 (AOC5), Cre10.g420400.t1.2 (EXP5), Cre13.g603700.t1.2 (ACT1), and Cre16.g662000.t1.2 (putative ATP-dependent RNA helicase).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Diagrams of Inverted Repeat Transgenes Used to Induce RNA Interference in Chlamydomonas.
Supplemental Figure 2. Abundance of the MAA7 (Encoding Trp Synthase β-Subunit) mRNA and siRNAs in Transgenic Strains Undergoing RNA-Mediated Silencing.
Supplemental Figure 3. A Chlamydomonas Exportin5 Deletion Mutant Shows Reduced Steady State Levels of Several Endogenous MicroRNAs.
Supplemental Figure 4. Schematic Diagram of the G/I Tailing Protocol Used to Examine mRNA Poly(A) Tail Length.
Supplemental Figure 5. Distribution of MAA7 siRNAs in Suc Density Gradients of the Maa7-IR44s and Maa7-IR5 Strains.
Supplemental Figure 6. Migration of MAA7 and ACT1 Transcripts in Suc Density Gradients When Examining Cell Extracts in the Presence of 50 mM EDTA.
Supplemental Figure 7. Ribosome Occupancy, the Fraction of a Specific mRNA Associated with Ribosomes, for the MAA7 and ACT1 Transcripts after Separation on Suc Density Gradients.
Supplemental Figure 8. IR-Repressed AOC5 Transcripts Are Moderately Depleted from Polyribosomal Fractions after Ribosome Run-Off Assays in the Presence of Low Concentrations of Cycloheximide.
Supplemental Figure 9. TSβ Protein Synthesis from the MAA7 Transcript, Subjected to siRNA-Mediated Translation Repression, Is Not Affected by Treatment with Paromomycin, Anisomycin, or by Slow Growth on Minimal Medium.
Supplemental Data Set 1. Small Interfering RNAs Matching the MAA7 Transcript in Strain Maa7-IR44s.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Science Foundation (to H.C.). We acknowledge the support of the Nebraska Experimental Program to Stimulate Competitive Research. We thank the Atkin lab (University of Nebraska–Lincoln) for the use of gradient fractionation equipment.
AUTHOR CONTRIBUTIONS
H.C. and X.M. conceived the project and designed the experiments. X.M., E.-J.K., and I.K. performed the research. F.M., A.V., and E.M. contributed to the analyses of sRNA sequences and to statistical evaluations. H.C. and X.M. wrote the article. All authors contributed to the analysis of results and edited the article.
Glossary
- sRNA
small RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- dsRNA
double-stranded RNA
- UTR
untranslated region
- RNAi
RNA interference
- IR
inverted repeat
- 5-FI
5-fluoroindole
- TAP
Tris-Acetate-Phosphate
References
- Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Westholm J.O., Lai E.C. (2011). Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. (2008). The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A.A., Lee M.T., Giraldez A.J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L., Yu A., Bouché N. (2010). microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62: 454–462 [DOI] [PubMed] [Google Scholar]
- Béthune J., Artus-Revel C.G., Filipowicz W. (2012). Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 13: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G., Gürel G., Schroeder S.J., Moore P.B., Steitz T.A. (2008). Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 379: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Sabatini D. (1971). Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. USA 68: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya M.A., Shoji S., Fredrick K., Cate J.H. (2008). Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 14: 1590–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J.E., Huntzinger E., Fauser M., Izaurralde E. (2011). GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Braun L., Cannella D., Ortet P., Barakat M., Sautel C.F., Kieffer S., Garin J., Bastien O., Voinnet O., Hakimi M.A. (2010). A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 6: e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Budkevich T., Giesebrecht J., Altman R.B., Munro J.B., Mielke T., Nierhaus K.H., Blanchard S.C., Spahn C.M.T. (2011). Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 44: 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulté L., Bennoun P. (1990). Translational accuracy and sexual differentiation in Chlamydomonas reinhardtii. Curr. Genet. 18: 155–160 [DOI] [PubMed] [Google Scholar]
- Cenik E.S., Zamore P.D. (2011). Argonaute proteins. Curr. Biol. 21: R446–R449 [DOI] [PubMed] [Google Scholar]
- Cerutti H., Casas-Mollano J.A. (2006). On the origin and functions of RNA-mediated silencing: From protists to man. Curr. Genet. 50: 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Ma X., Msanne J., Repas T. (2011). RNA-mediated silencing in Algae: Biological roles and tools for analysis of gene function. Eukaryot. Cell 10: 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.P., Slack F.J. (2009). Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans. Dev. Biol. 334: 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. (2011). MiRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detzer A., Engel C., Wünsche W., Sczakiel G. (2011). Cell stress is related to re-localization of Argonaute 2 and to decreased RNA interference in human cells. Nucleic Acids Res. 39: 2727–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.C., Grosshans H. (2009). Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 28: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Shi H., Tschudi C., Shen S., Ullu E. (2003). An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9: 802–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A., Green R. (2012). MiRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas D.V., Bartel B. (2008). Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 67: 403–417 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice D.C., Wilhelm J.M. (1984). Mechanisms of action of aminoglycoside antibiotics in eucaryotic protein synthesis. Antimicrob. Agents Chemother. 26: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., Sonenberg N. (2011). miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Friend K., Campbell Z.T., Cooke A., Kroll-Conner P., Wickens M.P., Kimble J. (2012). A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M., Stahlberg A., Govorunova E., Rank S., Hegemann P. (2001). The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for phototaxis and photophobic responses. J. Cell Sci. 114: 3857–3863 [DOI] [PubMed] [Google Scholar]
- Fukaya T., Tomari Y. (2011). PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J. 30: 4998–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., Tomari Y. (2012). MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol. Cell 48: 825–836 [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gu S., Jin L., Zhang F., Sarnow P., Kay M.A. (2009). Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 16: 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed] [Google Scholar]
- Hendrickson D.G., Hogan D.J., McCullough H.L., Myers J.W., Herschlag D., Ferrell J.E., Brown P.O. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. (2011). Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. (2008). Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Ibrahim F., Rohr J., Jeong W.J., Hesson J., Cerutti H. (2006). Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314: 1893. [DOI] [PubMed] [Google Scholar]
- Ibrahim F., Rymarquis L.A., Kim E.-J., Becker J., Balassa E., Green P.J., Cerutti H. (2010). Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 107: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M.C., Siomi H. (2002). A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Kawamata T., Tomari Y. (2009). Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell 34: 58–67 [DOI] [PubMed] [Google Scholar]
- Janas M.M., et al. (2012). Reduced expression of ribosomal proteins relieves microRNA-mediated repression. Mol. Cell 46: 171–186 [DOI] [PubMed] [Google Scholar]
- Jannot G., Bajan S., Giguère N.J., Bouasker S., Banville I.H., Piquet S., Hutvagner G., Simard M.J. (2011). The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 12: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F. (2011). The many faces of RNAi. Dev. Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Kim E.-J., Cerutti H. (2009). Targeted gene silencing by RNA interference in Chlamydomonas. Methods Cell Biol. 93: 99–110 [DOI] [PubMed] [Google Scholar]
- Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. (2011). Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Kong Y.W., Cannell I.G., de Moor C.H., Hill K., Garside P.G., Hamilton T.L., Meijer H.A., Dobbyn H.C., Stoneley M., Spriggs K.A., Willis A.E., Bushell M. (2008). The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl. Acad. Sci. USA 105: 8866–8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusov Y.Y., Shatirishvili G., Dzagurov G., Gauss-Müller V. (2001). A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res. 29: E57–E7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T. (2008). Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Saraiya A.A., Wang C.C. (2011). Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl. Trop. Dis. 5: e1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J. (2005). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lu C., Meyers B.C., Green P.J. (2007). Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117 [DOI] [PubMed] [Google Scholar]
- Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. (2004). Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Maroney P.A., Yu Y., Fisher J., Nilsen T.W. (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 13: 1102–1107 [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Mishima Y., Fukao A., Kishimoto T., Sakamoto H., Fujiwara T., Inoue K. (2012). Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc. Natl. Acad. Sci. USA 109: 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan R.D., Arnstein H.R.V. (1969). The dissociation of rabbit reticulocyte ribosomes with EDTA and the location of messenger ribonucleic acid. Eur. J. Biochem. 9: 445–450 [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D. (2006). Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13: 1108–1114 [DOI] [PubMed] [Google Scholar]
- Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen C.U. (2003). Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. (2006). Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21: 533–542 [DOI] [PubMed] [Google Scholar]
- Pinon V., Etchells J.P., Rossignol P., Collier S.A., Arroyo J.M., Martienssen R.A., Byrne M.E. (2008). Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324 [DOI] [PubMed] [Google Scholar]
- Rao M.K., Pham J., Imam J.S., MacLean J.A., Murali D., Furuta Y., Sinha-Hikim A.P., Wilkinson M.F. (2006). Tissue-specific RNAi reveals that WT1 expression in nurse cells controls germ cell survival and spermatogenesis. Genes Dev. 20: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakauskaite R., Dinman J.D. (2008). rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res. 36: 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci E.P., Limousin T., Soto-Rifo R., Rubilar P.S., Decimo D., Ohlmann T. (2013). MiRNA repression of translation in vitro takes place during 43S ribosomal scanning. Nucleic Acids Res. 41: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Rohr J., Sarkar N., Balenger S., Jeong B.R., Cerutti H. (2004). Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40: 611–621 [DOI] [PubMed] [Google Scholar]
- Rosenthal G.A. (1986). Biochemical insight into insecticidal properties of L-canavanine, a higher plant protective allelochemical. J. Chem. Ecol. 12: 1145–1156 [DOI] [PubMed] [Google Scholar]
- Saini P., Eyler D.E., Green R., Dever T.E. (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Saraiya A.A., Li W., Wang C.C. (2011). A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA 17: 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. (2010). Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. (2006). RNA silencing in Chlamydomonas: Mechanisms and tools. Curr. Genet. 49: 69–84 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. (2005). Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Shenvi C.L., Dong K.C., Friedman E.M., Hanson J.A., Cate J.H. (2005). Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations—Implications for the study of ribosome dynamics. RNA 11: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Chamond N., Djikeng A., Tschudi C., Ullu E. (2009). RNA interference in Trypanosoma brucei: Role of the N-terminal RGG domain and the polyribosome association of argonaute. J. Biol. Chem. 284: 36511–36520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Stadler M., Artiles K., Pak J., Fire A. (2012). Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 22: 2418–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J.A., Vasudevan S. (2009). miRNPs: Versatile regulators of gene expression in vertebrate cells. Biochem. Soc. Trans. 37: 931–935 [DOI] [PubMed] [Google Scholar]
- Stevens D.R., Atteia A., Franzén L.G., Purton S. (2001). Cycloheximide resistance conferred by novel mutations in ribosomal protein L41 of Chlamydomonas reinhardtii. Mol. Gen. Genet. 264: 790–795 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. (2008). Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr. Biol. 18: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wu G., Poethig R.S. (2012). Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., Cullen B.R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger S.T., Corey D.R. (2009). The puzzle of RNAs that target gene promoters. ChemBioChem 10: 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Wang H. (2010). Translational inhibition by microRNAs in plants. Prog. Mol. Subcell. Biol. 50: 41–57 [DOI] [PubMed] [Google Scholar]
- Zhou R., Hotta I., Denli A.M., Hong P., Perrimon N., Hannon G.J. (2008). Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell 32: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.