The Tat system uniquely transports fully folded proteins across the thylakoid membrane. Functional domains of the multispanning Tat component cpTatC were dissected with isolated pea chloroplasts onto a three-dimensional model of cpTatC. The results provide insight into folded protein transport and a novel assay for structure-function studies of a membrane protein.
Abstract
Twin arginine translocation (Tat) systems of thylakoid and bacterial membranes transport folded proteins using the proton gradient as the sole energy source. Tat substrates have hydrophobic signal peptides with an essential twin arginine (RR) recognition motif. The multispanning cpTatC plays a central role in Tat operation: It binds the signal peptide, directs translocase assembly, and may facilitate translocation. An in vitro assay with pea (Pisum sativum) chloroplasts was developed to conduct mutagenesis and analysis of cpTatC functions. Ala scanning mutagenesis identified mutants defective in substrate binding and receptor complex assembly. Mutations in the N terminus (S1) and first stromal loop (S2) caused specific defects in signal peptide recognition. Cys matching between substrate and imported cpTatC confirmed that S1 and S2 directly and specifically bind the RR proximal region of the signal peptide. Mutations in four lumen-proximal regions of cpTatC were defective in receptor complex assembly. Copurification and Cys matching analyses suggest that several of the lumen proximal regions may be important for cpTatC–cpTatC interactions. Surprisingly, RR binding domains of adjacent cpTatCs directed strong cpTatC-cpTatC cross-linking. This suggests clustering of binding sites on the multivalent receptor complex and explains the ability of Tat to transport cross-linked multimers. Transport of substrate proteins cross-linked to the signal peptide binding site tentatively identified mutants impaired in the translocation step.
INTRODUCTION
Delivery of cytosolically synthesized proteins to the six different compartments of the chloroplast involves a hierarchical system of at least five protein translocases (enzymes that catalyze protein translocation into or across a membrane) (Cline and Dabney-Smith, 2008; Celedon and Cline, 2013). Most chloroplast precursor proteins are imported across the plastid envelope into the stroma by the Translocon at the outer chloroplast envelope (Toc) and Translocon at the inner chloroplast envelope (Tic) translocases (Kessler and Schnell, 2006). Thylakoid-destined proteins then enter one of the ancestral pathways, chloroplast signal recognition particle (cpSRP), chloroplast Sec system 1 (cpSec1), or chloroplast Tat (cpTat), that derive from the cyanobacterial endosymbiont that gave rise to chloroplasts (Celedon and Cline, 2013). The ∼100 thylakoid lumenal proteins are transported across the thylakoid membrane by cpSec1 or cpTat systems, each system specifically transporting a subset of lumenal proteins (Schubert et al., 2002; Peltier et al., 2004). The cpTat and cpSec systems transport subunits of the photosystem I, photosystem II, and cytochrome bf complexes (Cline, 2003); accordingly, a null mutation in any cpTat component or any cpSec component results in chlorotic seedlings that are photosynthetically inactive and die at the seedling stage (Motohashi et al., 2001; Voelker and Barkan, 1995; Skalitzky et al., 2011).
Twin arginine translocation systems (cpTat in chloroplasts; Tat in prokaryotes) were discovered in plant chloroplasts but are widely represented among prokaryotes (Cline and Theg, 2007; Fröbel et al., 2012; Palmer and Berks, 2012; Celedon and Cline, 2013). Tat substrates are targeted by hydrophobic signal peptides containing an essential Arg-Arg (RR) motif. Tat systems are unique in that they transport folded proteins across sealed membranes using only the proton electrochemical gradient as an energy source. They are unusual because they have a transient translocase that exists only until translocation is complete (Mori and Cline, 2002; Schnell and Hebert, 2003). Tat systems consist of three membrane protein components, called chloroplast TatC (cpTatC), High chlorophyll fluorescence 106 (Hcf106), and Thylakoid assembly 4 (Tha4) in chloroplasts and TatC, TatB, and TatA in prokaryotes. cpTatC-Hcf106 form a large multivalent signal peptide receptor. Binding of the signal peptide triggers assembly of Tha4 (Mori and Cline, 2002). In nontransporting membranes, Tha4 appears to be a homotetramer, whereas in the translocase, it assembles as homooligomers of ∼20 to 25 Tha4 protomers (Leake et al., 2008; Dabney-Smith and Cline, 2009; Celedon and Cline, 2012). For this and other reasons, Tha4 (TatA) is thought to form the protein-conducting structure.
The cpTat receptor complex contains an estimated approximately eight cpTatCs and approximately eight Hcf106s and migrates on blue native polyacrylamide gels at ∼700 kD. cpTatC (TatC) is the primary receptor for the signal peptide (Alami et al., 2003; Gérard and Cline, 2006), although Hcf106 (TatB) also contacts the signal peptide to a lesser extent. Under saturating conditions, each cpTatC within a receptor complex binds a substrate protein (Celedon and Cline, 2012). Kinetic analysis of transport showed that each cpTatC-Hcf106 pair independently transports its bound substrate in an apparently stochastic manner (Celedon and Cline, 2012). In some circumstances, adjacent sites on the same receptor complex can collaborate to efficiently transport oligomeric substrates of at least four signal peptide bearing subunits (Ma and Cline, 2010).
The multispanning cpTatC protein appears to play the central role in all aspects of Tat function. It forms a homooligomer to which Hcf106 (TatB) binds to assemble the receptor complex (Behrendt et al., 2007; Orriss et al., 2007); it binds to the signal peptide (Cline and Mori, 2001; Alami et al., 2003; Gérard and Cline, 2006); it probably serves as the organizing center for Tha4 assembly (Fröbel et al., 2011; Rollauer et al., 2012); and it may provide the motive force for transmembrane protein movement (Brüser and Sanders, 2003; Dabney-Smith et al., 2006; Cline and McCaffery, 2007). Yet the molecular determinants of these different functions are largely unknown. One approach to address this combines mutagenesis with biochemical characterization and protein–protein interaction studies. This approach has been conducted to some extent with the Escherichia coli Tat system (Allen et al., 2002; Buchanan et al., 2002; Barrett et al., 2005; McDevitt et al., 2006; Holzapfel et al., 2007), but not with the plant Tat system owing to the relative difficulty of mutagenesis and gene replacement.
In this study, we conducted mutagenesis, biochemical characterization of the mutants, and Cys matching to fine map the signal peptide RR binding site to the cpTatC N terminus and first stromal loop, receptor complex assembly to lumen-proximal regions, strong cpTatC–cpTatC interactions to the signal peptide binding site and two lumenal loops, and cpTatC–Hcf106 interaction to the third lumenal loop. In addition, we show evidence that transmembrane domain 4 may be involved in the translocation step. This was accomplished with a novel system in which recombinant cpTatC was imported into isolated intact chloroplasts and was analyzed independently of endogenous cpTatC. The results of these studies fit nicely with and extend studies of the bacterial Tat system and map biochemical function onto the recently determined crystal structure for monomeric TatC from Aquifex aeolicus (Rollauer et al., 2012). In addition to providing insight into the roles of cpTatC in folded protein transport, our studies provide a rapid and effective method, unique to chloroplasts, for analyzing the functions of an integral thylakoid membrane protein.
RESULTS
In Vitro–Synthesized cpTatC Is Efficiently Imported into Chloroplasts and Integrated into the Thylakoids Membrane
The cpTatC precursor protein has six transmembrane domains and a 50-residue transit peptide that directs import into the chloroplast. Because the objective was to maximize the amount of imported cpTatC, the high-efficiency transit peptide of the small subunit of ribulose bisphosphate carboxylase oxygenase was fused to the N terminus of the complete precpTatC precursor protein. This construct, with tandem transit peptides, proved to be superior to several other versions both in import yield and in proper processing to mature mcpTatC. All cpTatC precursors in this study contained this tandem transit peptide and are simply referred to as precpTatC. Two modified forms of precpTatC were also used; precpTatC-His contains six His residues and a flexible linker fused to the precpTatC C terminus for use in purification; precpTatCaaa has the three Cys residues of the mature cpTatC replaced with Ala residues and was used in Cys matching studies. Details of the construction of precpTatC, precpTatC-His, and precpTatCaaa are in Methods and Supplemental Figure 1 online.
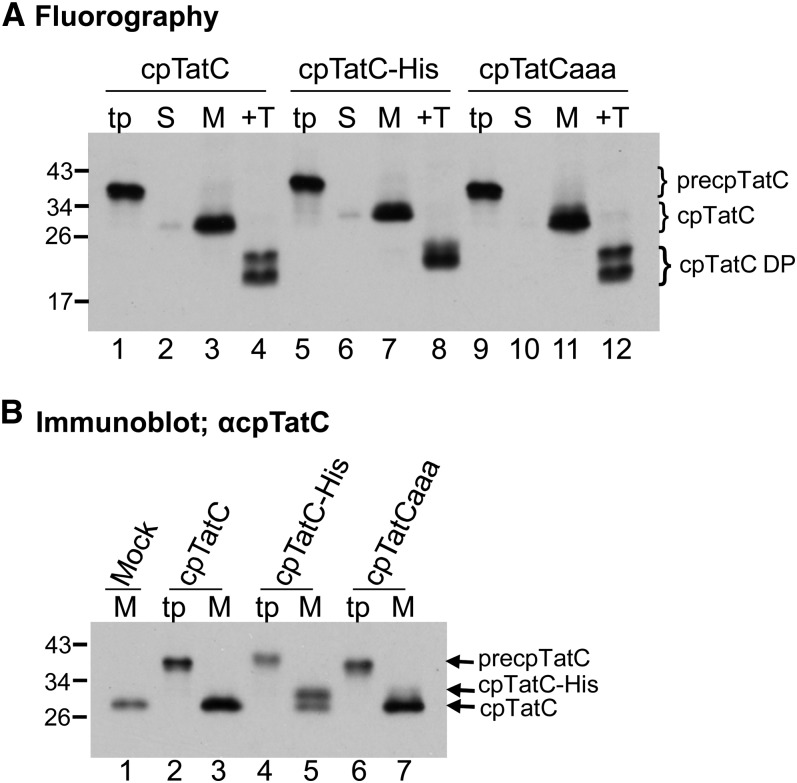
As shown in Figure 1A, the three radiolabeled cpTatC precursors (lanes 1, 5, and 9) were imported into chloroplasts, processed to mature size, and integrated into the membrane (lanes 3, 7, and 11). Proper integration was demonstrated by the characteristic degradation products (lanes 4, 8, and 12) produced by thermolysin treatment of the membranes (Mori et al., 2001). Both degradation products of cpTatC-His (lane 8) are slightly larger than those from cpTatC or cpTatCaaa, supporting our previous assertion that proteolysis occurs strictly on the N terminus of cpTatC (Gérard and Cline, 2006). The thylakoids recovered from import assays were also subjected to immunoblotting analysis with α-cpTatC antibodies (Figure 1B). As is apparent, membranes recovered from import assays (lanes 3 and 7) had darker cpTatC bands than the mock import membranes (lane 1). Examination of the immunoblot of the cpTatC-His membranes allowed a better estimate of the amount of imported cpTatC compared with the endogenous protein (i.e., cpTatC-His migrated at a higher Mr, with the upper band representing imported protein and the lower band the endogenous cpTatC [lane 5]). Densitometry measurements of the bands from two experiments showed that the imported protein was present at ∼120% of the endogenous cpTatC.
Figure 1.
Imported cpTatC Faithfully Integrates into Thylakoids in Amounts Greater Than Endogenous cpTatC.
(A) Radiolabeled precpTatC (lanes 1 to 4), precpTatC-His (lanes 5 to 8), and precpTatCaaa (lanes 9 to 12) were incubated with isolated chloroplasts in protein import assays. The chloroplasts were then fractionated to stroma (S) and membranes (M). Membranes were treated with thermolysin (+T). Lanes labeled “tp” contain in vitro–translated cpTatC precursors used. Samples were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography. DP, thermolysin degradation product.
(B) The abundance of imported cpTatC is comparable to endogenous cpTatC. cpTatC precursors (tp) and the membrane samples recovered from mock import and precpTatC import assays (M) were analyzed by immunoblotting with α-cpTatC antibody. Mock import underwent all assay procedures but received no precursor protein.
Imported cpTatC Protein Assembles into New cpTat Receptor Complexes That Are Functional for Substrate Binding
For this approach to be useful, the imported cpTatC should exhibit all of the characteristics of endogenous cpTatC and faithfully report on disabling mutations. The ability of imported cpTatCs to assemble into a cpTat receptor complex with Hcf106 and to bind substrate proteins was tested in the experiment in Figure 2. As proof of concept for mutagenesis studies, this experiment also included three pea (P. sativum) cpTatC mutants based on E. coli TatC mutants impaired in substrate binding (F94A and E103A) or TatC–TatB interaction (D211A) (Buchanan et al., 2002; Holzapfel et al., 2007). The comparable cpTatC Ala substitutions are F158A, E167A, and D276A. Because of the apparent redundancy of Glu at residues 166 and 167, both Glu residues were mutated to Ala (see below for location on cpTatC).
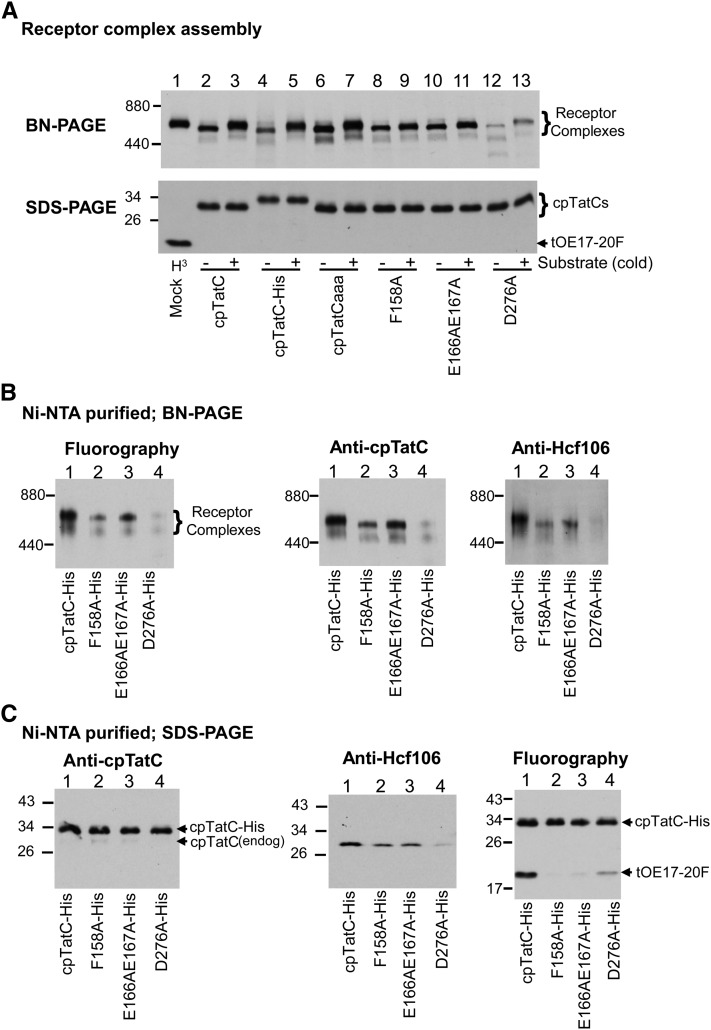
Figure 2.
Imported cpTatC Assembles into New Receptor Complexes That Bind Substrate.
(A) Imported cpTatCs are assembled into cpTat receptor complexes. Mock, radiolabeled precpTatC, precpTatC-His, precpTatCaaa, and precursors to cpTatC mutants F158A, E166AE167A, and D276A were incubated with chloroplasts in import assays. Thylakoids obtained from mock import chloroplasts were incubated with radiolabeled tOE17-20F (lane 1), and thylakoids from cpTatC import chloroplasts were incubated without (−) or with (+) unlabeled tOE17-20F (lanes 2 to 13). Recovered thylakoids were then solubilized with digitonin and analyzed by BN-PAGE and fluorography (top panel). The same samples were subjected to SDS-PAGE (bottom panel).
(B) Ni-NTA–purified cpTat complexes formed with imported His-tagged cpTatCs contain Hcf106. Chloroplast import was conducted with radiolabeled precursors to the His-tagged versions of cpTatC and the above mutants. Thylakoids recovered from import chloroplasts were incubated with radiolabeled substrate tOE17-20F and then solubilized with digitonin and purified on magnetic Ni-NTA beads. The purified cpTatC complexes were analyzed by BN-PAGE/fluorography (left panel) or BN-PAGE/immunoblotting with antibodies to cpTatC (middle panel) or Hcf106 (right panel).
(C) Hcf106 and bound substrate but only trace endogenous cpTatC are associated with imported wild-type cpTatC-His. Purified cpTatC complexes from (B) were separated by SDS-PAGE and analyzed by immunoblotting with α-cpTatC (left panel) or α-Hcf106 (middle panel) or by fluorography (right panel).
Membranes recovered from import assays were incubated with or without unlabeled substrate tOE17-20F, which has a signal peptide that provides very high affinity for the cpTat receptor complex (Gérard and Cline, 2007; Celedon and Cline, 2012). Recovered membranes were dissolved with digitonin and analyzed by blue native (BN) PAGE and fluorography. As can be seen in Figure 2A (top panel), imported cpTatC and its variants cpTatC-His and cpTatCaaa migrated at the expected ∼700 kD location of fully assembled receptor complex. When prebound with unlabeled tOE17-20F, the cpTatC exhibited the molecular mass band shift to ∼750 kD, characteristic of receptor complex with bound substrate (lanes 2 to 7, compare with lane 1, mock import membranes incubated with radiolabeled substrate). Imported cpTatC mutants F158A and E166AE167A also produced an ∼700-kD band (lanes 8 and 10). We could not detect a substrate-induced band shift for the F158A mutant (lane 9). There might be a very slight substrate-induced shift of the E166AE167A cpTatC mutant band (lane 11), although accurate measurements in this region of the gel are difficult. Imported mutant D276A produced a weak ladder of bands at and below ∼700 kD (lane 12), indicative of impaired receptor complex assembly. The SDS-PAGE fluorogram of samples used for the BN gel (Figure 2A, bottom panel) show that cpTatC D276A was imported as efficiently as the other cpTatC variants. Interestingly, binding of the substrate protein shifted the D276A complexes into a mostly single band at ∼750 kD (lane 13), suggesting that substrate binding per se was not directly affected in this mutant and supporting previous observations that substrate binding stabilizes the receptor complex during BN-PAGE (Fincher et al., 2003).
In a separate experiment, radiolabeled cpTatC-His and the above mutant cpTatCs, equipped with His tags, were imported into chloroplasts and recovered thylakoids were then incubated with radiolabeled tOE17-20F and solubilized with digitonin. The solubilized complexes were affinity purified on magnetic Ni 2+-nitrilotriacetic acid (Ni-NTA) beads and analyzed by BN-PAGE/fluorography (Figure 2B, left panel). The purified cpTatC-His (lane 1) migrated at ∼750 kD and mutants F158A-His and E166AE167A (lanes 2 and 3) at ∼700 kD, characteristic of receptor complex without bound precursor. Also note that radiolabeled substrate and radiolabeled cpTatC-His contribute to the intensity of the 750-kD band (i.e., accounting for the lower intensity of the nonbinding mutants). The purified D276A-His showed very weak banding at ∼750 and ∼500 kD. Immunoblots of the BN-PAGE gels of purified samples showed that both cpTatC (middle panel) and Hcf106 (right panel) were present in the ∼700- to 750-kD band, with the exception of D276A-His, which gave very weak bands for each.
The total amounts of cpTatC, Hcf106, and the bound substrate present in purified samples were determined by SDS-PAGE and immunoblotting or fluorography (Figure 2C). As expected, bound substrate was recovered from the eluates with wild-type cpTatC-His and a small amount with D276-His; only trace amounts of substrate protein were recovered with F158A-His and E166AE167A-His (right panel). Comparable amounts of imported cpTatC were present in the wild type and mutants (left panel) and endogenous Hcf106 copurified with the imported cpTatC-His, F158A-His, and E166AE167A-His but was markedly reduced with D276A-His, as expected for an assembly mutant (middle panel). Importantly, very low amounts of endogenous cpTatC (the lower bands of left panel) copurified with imported cpTatC. This is significant because it indicates that imported cpTatC assembles new receptor complexes with a free pool of Hcf106 (Cline and Mori, 2001) and does not exchange with endogenous cpTatC into preexisting receptor complexes. Thus, the behavior of imported cpTatC can be examined independently of the influence of endogenous cpTatC.
The amount of substrate bound per imported cpTatC was quantified by conducting saturation binding assays with thylakoids recovered from chloroplast import assays or a mock import assay (Figure 3A). The numbers of imported cpTatC and bound substrate were determined by radiolabel counting of extracted gel slices. The binding stoichiometry for each imported cpTatC was calculated as the increase in bound substrate above that bound by membranes from mock import chloroplasts divided by the number of imported cpTatC proteins (Figure 3B). The binding stoichiometry of wild-type cpTatC and cpTatCaaa was approximately one substrate/per cpTatC, indicating that the imported cpTatC was as functional for substrate binding as the endogenous cpTatC (Celedon and Cline, 2012). cpTatC-His was slightly less effective at binding. The three mutant cpTatCs exhibited little binding above the amount bound by endogenous cpTatC. The ∼10% binding by cpTatC E166AE167A might explain the slight band shift seen for this mutant in Figure 2A. In experiments conducted here and in experiments associated with Figure 4, the estimated amount of imported cpTatC was ∼70% of the endogenous cpTatC.
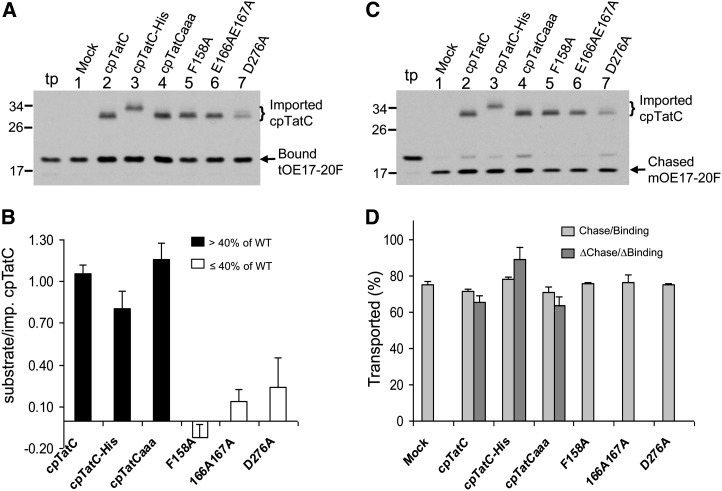
Figure 3.
Each Imported cpTatC Binds One Substrate Protein.
(A) cpTatC import and substrate binding assays. Chloroplast import was conducted for precpTatC and its variants. The recovered thylakoids were incubated with radiolabeled substrate tOE17-20F for binding assays. Samples were analyzed by SDS-PAGE and fluorography. Lane tp, tOE17-20F translation product.
(B) Quantification of bound substrate and imported cpTatC. Radioactivity in bands of (A) was determined (see Methods) and the increase in bound substrate due to the imported cpTatC calculated and expressed as substrate/imported cpTatC average + se (n = 3). WT, the wild type.
(C) Portions of washed thylakoids from binding reactions of (A) were further incubated in chase assays. Samples were analyzed by SDS-PAGE and fluorography.
(D) The bands of (C) were quantified. The percentages of total bound substrates that were transported in the chase assay are displayed (gray bars) as average + se (n = 3). The percentages of net chase per net binding (ΔChase/ΔBinding) due to the imported cpTatCs were plotted (dark gray bars) as average + se (n = 3).
Figure 4.
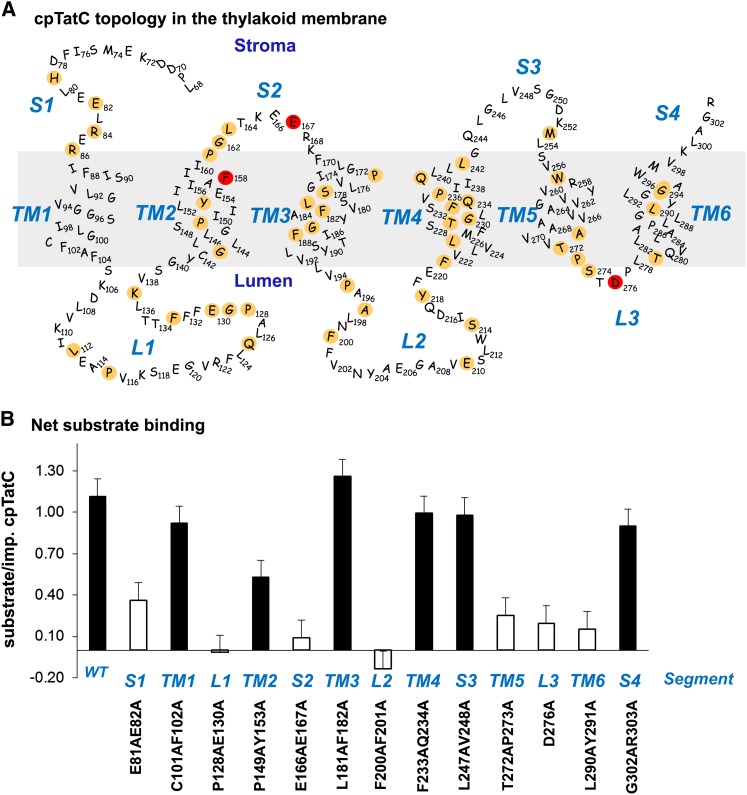
Site-Directed Mutagenesis of cpTatC Identifies Regions Involved in Binding.
(A) Predicted structure and topology of the pea cpTatC protein. The nonconserved 67 residues of the N terminus are not shown. cpTatC is divided into stromal (S), transmembrane (TM), and lumenal (L) segments by topological location. Conserved residues identified by SIFT are colored tan. Residues Phe-158, Glu-167, and Asp-276, which correspond to the E. coli TatC essential residues Phe-94, Glu-103, and Asp-211, are colored red.
(B) Binding assays of the mutants in each segment of cpTatC. Wild-type (WT) and mutant precpTatC with Ala substitutions of pairs of conserved residues, as designated below the chart, were imported into chloroplasts. Recovered thylakoids were incubated with tOE17-20F for binding assays. The numbers of bound substrate per imported cpTatC are displayed as average + se (n = 3). Closed bars represent binding stoichiometry >40% of wild-type cpTatC; open bars represent ≤40% of the wild type.
In order to determine if the substrates bound by imported cpTatC were as efficiently transported as those bound by endogenous cpTatC, the substrate-bound membranes were subjected to chase assays (transport from the bound state; Figure 3C). The ratio of the total transported substrate to total bound substrate was essentially the same for thylakoids with imported cpTatC as for mock import thylakoids (Figure 3D, light-gray bars). This indicates that imported cpTatC is as functional for transport as endogenous cpTatC. In addition, it indicates that the mutant cpTatCs do not have a dominant-negative effect on endogenous cpTatC transport. A separate calculation was made to specifically determine the efficiency of transport of substrate bound to imported cpTatC (Figure 3D, dark-gray bars). This calculation subtracted the bound substrate and the transported substrate of mock thylakoids from the bound substrate and transported substrate of the imported cpTatC samples. As expected, comparable efficiencies of transport were obtained compared with mock import membranes.
In summary, the results of Figures 1 to 3 demonstrate that imported cpTatC protein exhibits all of the characteristics of endogenous cpTatC, including integration into the thylakoid membrane, assembly with Hcf106 to form the large cpTat receptor complex, binding of substrate protein in the expected 1:1 stoichiometry, and transport of bound substrate protein. Our results indicate that imported cpTatC forms new receptor complexes rather than exchanging into preexisting complexes. Finally, our assay faithfully reports the phenotypes of cpTatC mutants defective in substrate binding and receptor complex assembly.
Mutagenesis of cpTatC Identifies Regions Required for Substrate Binding and Receptor Complex Assembly
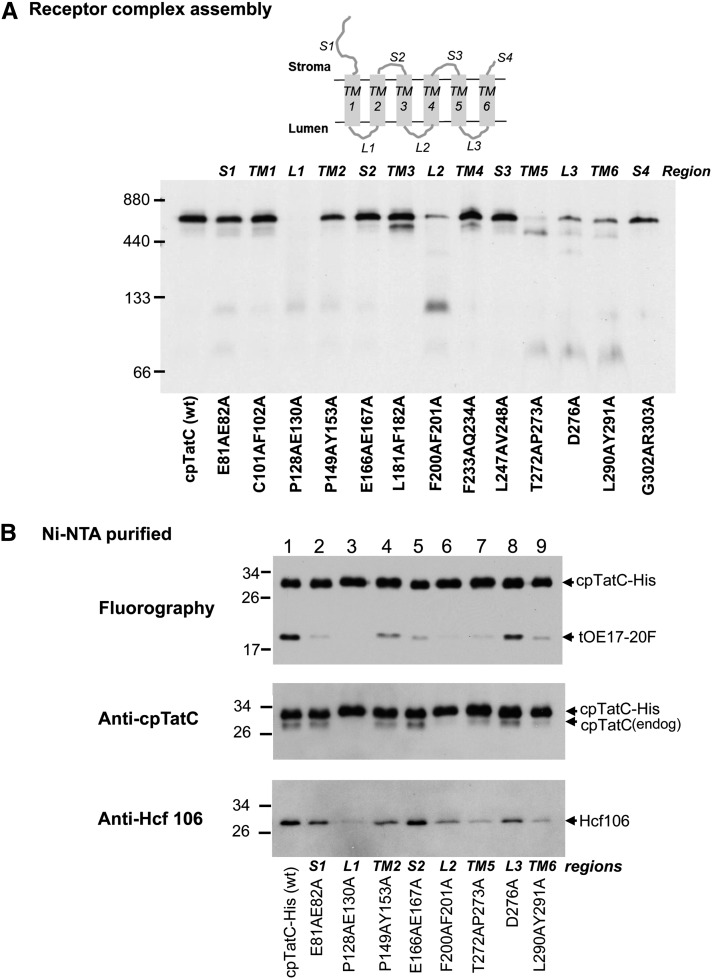
A limited Ala scanning mutagenesis was conducted to identify regions and residues required for substrate protein binding. In order to guide the mutagenesis studies, the pea cpTatC sequence was submitted to Sorting Intolerant from Tolerant analysis, an algorithm that predicts the likelihood that an amino acid substitution affects protein function based on sequence similarity and the physical properties of amino acids (Ng and Henikoff, 2001; Ng and Henikoff, 2003; Kumar et al., 2009). Because the transit peptide and first ∼40 resides of plant cpTatCs are not conserved among TatC proteins, 100 sequences 80% or less identical to pea precpTatC were analyzed to minimize redundancy of cpTatC proteins from highly related plant species. These analyzed sequences consisted primarily of plant and algal chloroplast TatC proteins and cyanobacterial TatC proteins. The cpTatC protein was arbitrarily divided into 13 regions that represent stroma-exposed (S1 to S4), transmembrane helix (TM1 to TM6), and lumen-exposed (L1 to L3) segments. All regions of cpTatC, except for TM1, S3, and S4, contained multiple residues that are either absolutely or predominantly conserved (Figure 4A).
To speed screening, pairs of conserved residues of each segment except segment L3 were mutated to Ala residues. The mutated precpTatC proteins were individually imported into chloroplasts, and the recovered membranes were assayed for substrate binding. All mutant cpTatC precursors were efficiently imported into chloroplasts, processed to mature size, and integrated into the membranes (see Supplemental Figure 2 online). However, mutant cpTatCs exhibited a large range of substrate binding capability (Figure 4B). Mutations in seven different segments (plus the F158A substitution in Figure 3) resulted in binding stoichiometries of ≤0.4 substrates/per cpTatC.
Defective substrate binding could result from alteration of the binding site, from improper integration into the membrane, or from failure to assemble into the receptor complex. All of the mutants produced the characteristic thermolysin degradation products of properly integrated cpTatC (see Supplemental Figure 2 online). By contrast, there was a wide range of receptor complex abundance among the mutants (Figure 5A). The cpTatC with mutations in L1, L2, L3, TM5, and TM6 showed much reduced receptor complex. The L1 mutant was consistently devoid of receptor complex; mutant L2 varied from moderate loss to near complete loss of receptor complex; and TM5, L3, and TM6 mutants consistently showed moderate loss of receptor. It is likely that the defective binding phenotypes of these five mutants are secondary effects of defective receptor assembly. The cpTatC variants with mutations in segments S1 and S2 (Figure 5A) formed receptor complex, and their failure to bind substrate is likely due to a direct effect on the binding site. This was explored in more detail by single residue substitutions in these segments. Segment S1 residues H79A and E82A and segment S2 residues P161A, L163A, and E166A/E167A were defective in substrate binding (see Supplemental Figure 3 online; Figure 2C). A separate experiment showed that the Ala substitution of the conserved Glu-167 residue alone eliminated binding, whereas Ala substitution of Glu-166 was without effect.
Figure 5.
cpTatC Segments Important for Receptor Complex Assembly.
(A) Mutants defective in assembling the receptor complex. Radiolabeled wild-type (wt) precpTatC and its mutants were imported into chloroplasts. Recovered thylakoid membranes were solubilized with digitonin and analyzed by BN-PAGE and fluorography. A diagram of cpTatC with segment designations is displayed above the panel.
(B) cpTatC mutants were defective in binding to endogenous cpTatC and Hcf106. Radiolabeled precpTatC-His (wild type [wt]) and its mutants with Ala substitutions in different segments and positions as indicated below the panel were imported into chloroplasts. Recovered thylakoids were incubated with radiolabeled tOE17-20F for binding assays. The thylakoid membranes were then solubilized with digitonin and purified on magnetic Ni-NTA beads. The purified cpTatC variants were separated by SDS-PAGE and analyzed by fluorography (top panel), by immunoblotting with α-cpTatC (middle panel), or with α-Hcf106 (bottom panel).
Lumen-Proximal Regions Are Important for cpTatC–cpTatC and cpTatC–Hcf106 Interactions
To gain more insight into the cause of assembly defects, His-tagged versions of the mutant cpTatCs were prepared and imported into chloroplasts, and the thylakoids were used for binding assays. After solubilization of recovered thylakoids with digitonin, the cpTatC-His was purified on Ni-NTA magnetic beads (Figure 5B). The substrate protein tOE17-20F associated with the mutants (top panel) reflected the quantitative measure of substrate binding in Figure 4B. Comparable amounts of cpTatC were recovered in all of the samples. There was a noticeable absence of copurifying endogenous cpTatC for cpTatC with mutations in L1 and L2, suggesting that these mutants might be defective in cpTatC–cpTatC interactions. Variable amounts of Hcf106 were associated with purified cpTatC, with the least amount with cpTatC with mutations in segments L1, L2, TM5, and TM6 (bottom panel).
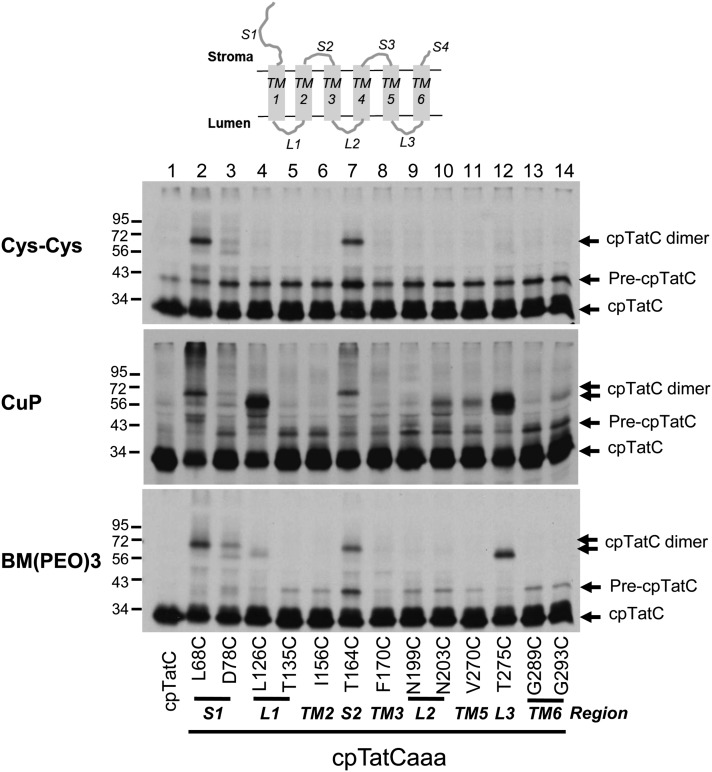
As an additional approach to analyzing cpTatC–cpTatC interactions, active single Cys variants of cpTatCaaa were prepared and imported into chloroplasts, and the recovered thylakoids were subjected to Cys-Cys cross-linking (Figure 6), either by unassisted disulfide formation (top panel), by copper phenanthroline (CuP)-catalyzed disulfide formation (middle panel), or with the bismaleimide cross-linker 1, 8-bis-maleimidotriethyleneglycol (BM[PEO]3) (bottom panel). Interestingly, strong cpTatC dimers were formed when Cys residues were in segments S1 and S2 by all three methods. Because substitutions in S1 and S2 segments affect binding, but not receptor complex assembly, the cross-linking likely reflects proximity of neighboring cpTatC subunits rather than critical interactions. The oxidant CuP promoted substantial amounts of cpTatC-cpTatC cross-linking directed from L1 and L3 segment Cys residues and small amounts from L2, TM5, and TM6. Dimers directed from different regions of the cpTatC protein displayed slightly different electrophoretic mobilities. Such differences likely result from different geometries of the branched adducts produced by cross-linking from different positions in the cpTatC molecule. Similar behavior has been observed for preSufI-TatB cross-linking products (Maurer et al., 2010).
Figure 6.
cpTatC Proteins with Cys Residues in S1, S2, L1, or L3 Form Disulfide Linked Dimers.
Radiolabeled precpTatC with three endogenous Cys residues (lane 1) and precpTatCaaa variants with a single Cys substitution at varying positions as indicated were imported into chloroplasts. Recovered thylakoids were washed in DTT-free buffer (top panel) or treated with the oxidant CuP to promote disulfide bond formation (middle panel) or subjected to cross-linking with BM(PEO)3 (bottom panel) and then analyzed by nonreducing SDS-PAGE and fluorography. The different protein species are indicated to the right of the panels.
cpTatC Segments S1 and S2 Interact with the RR Domain of the Substrate
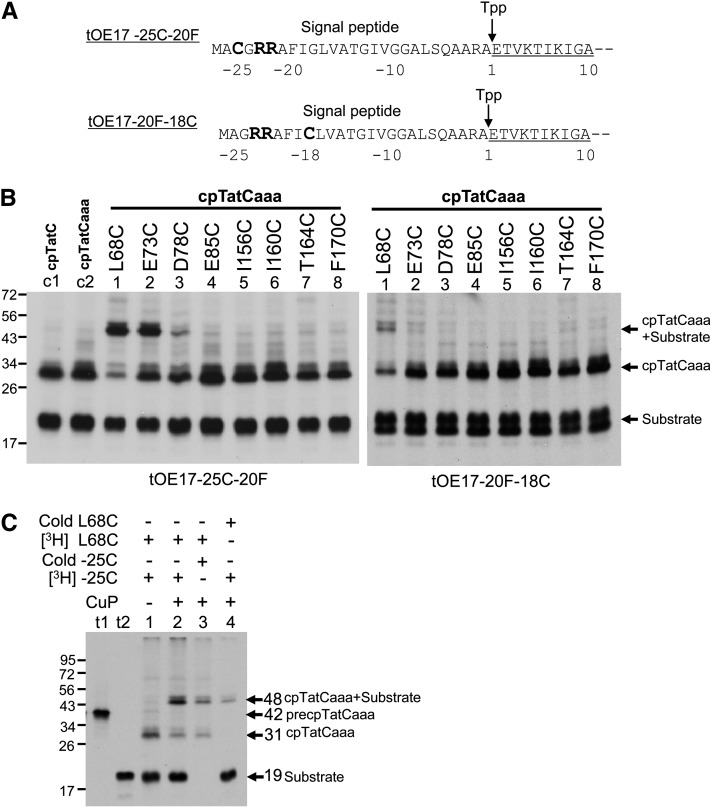
If cpTatC segments S1 and S2 compose the signal peptide binding site, then these regions should come in close contact with the signal peptide. Direct contacts to substrate were explored by engineering single Cys substitutions for use in Cys-Cys cross-linking (called Cys matching). The cpTatCaaa variant carried a site-specific Cys on S1 or S2 and the substrate variants carried a Cys in RR proximal residues −25 or −18 (i.e., tOE17-25C-20F or tOE17-20F-18C as shown in Figure 7A). Independent assays verified that Cys-substituted substrate and Cys-substituted cpTatC variants are functional for binding and transport (see Supplemental Figure 4 online).
Figure 7.
The RR-Proximal Domain of Substrates Contacts cpTatC Segment S1 as Shown by Disulfide Matching.
(A) The tOE17-25C-20F and tOE17-20F-18C substrates with RR-proximal Cys residues. Amino acids are numbered relative to the thylakoid signal peptidase (Tpp) cleavage site. The signal peptide and the first 10 residues of the N terminus of the mature domain (underlined) are depicted.
(B) Disulfide cross-linking between substrate and cpTatC. Radiolabeled precursors to cpTatC (lane c1), cpTatCaaa (lane c2), and cpTatCaaa variants with single Cys substitutions at positions indicated (lanes 1 to 8) were imported into chloroplasts. Recovered thylakoids were incubated with radiolabeled tOE17-25C-20F or tOE17-20F-18C in binding assays. Washed thylakoids were then treated with the oxidant CuP to promote disulfide bond formation. Samples were analyzed by nonreducing SDS-PAGE and fluorography.
(C) The ∼48-kD cross-linked product is an adduct of cpTatC and its substrate. Radiolabeled cpTatCaaa L68C (lanes 1 to 3) or unlabeled cpTatCaaa L68C (lane 4) were imported into chloroplasts. Recovered thylakoids were incubated with radiolabeled tOE17-25C-20F (lanes 1, 2, and 4) or unlabeled tOE17-25C-20F (lane 3) for binding assays. Washed thylakoids were treated with CuP (lanes 2 to 4) or mock treated (lane 1). Membrane samples were analyzed by nonreducing SDS-PAGE and fluorography. In vitro–translated precpTatCaaa L68C and tOE17-25C-20F are in lanes t1 and t2, respectively.
For Cys matching, the substrates were bound to the thylakoids recovered from import assays with cpTatCaaa-Cys variants and subjected to oxidation with CuP to induce disulfide bond formation. As shown in Figure 7B, left panel, an additional band with a molecular mass of ∼48 kD was produced from the incubation of tOE17-25C-20F with the membranes containing cpTatCaaa L68C, E73C, or D78C residues of S1. The same band was also formed from the incubation of tOE17-20F-18C and the membranes containing cpTatCaaa L68C (right panel). Only a trace amount of radioactivity was present at the same location when the imported cpTatC lacked all Cys residues (left panel, lane c2). The sum of the molecular masses of substrate protein and cpTatC is ∼48 kD. The identity of the band as a cpTatC substrate adduct was verified by an experiment in which only one protein, either substrate or cpTatC, was radiolabeled (Figure 7C). The 48-kD band was produced in both cases, but with the expected reduced radiolabel intensity.
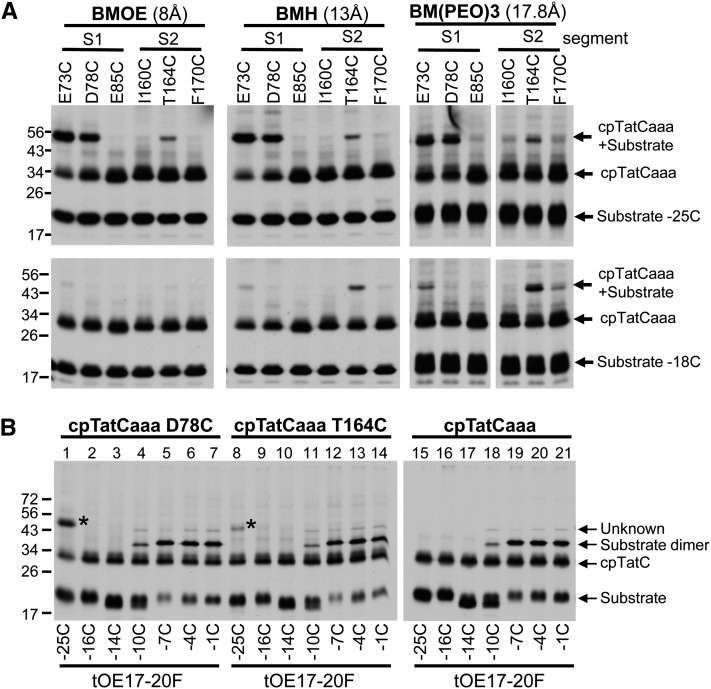
To gauge the distance between Cys residues in the substrate and cpTatC, a similar experiment was conducted with cross-linking mediated by bismaleimide compounds with increasing spacer lengths (Figure 8A). Cross-linking to tOE17-20F-25C substrate was obtained with cpTatC S1 variants E73C and D78C but also with S2 residue T164C with either bis-maleimidoethane (BMOE) (8-Å spacer) or bis-maleimidohexane (BHA) (13-Å spacer) (Figure 8A, top panels). Additional cross-links with S1 and S2 resides were obtained with BM(PEO)3 (17.8-Å spacer). When tOE17-20F-18C was used as substrate, cross-links to the S1 segment were diminished and cross-links to the S2 segment were enhanced (Figure 8A, bottom panels). cpTatC with Cys residues located in a number of other segments did not form cross-links to substrate with Cys residues in the RR proximal region of the signal peptide (see Supplemental Figure 5 online).
Figure 8.
Mapping the Interactions of the Substrate Signal Peptide and cpTatC.
(A) S1 is closer to the amino-proximal side of RR and S2 closer to the carboxyl-proximal side. Radiolabeled precpTatC Cys variants were imported into chloroplasts and recovered thylakoids incubated with radiolabeled tOE17-25C-20F (top panels) or tOE17-20F-18C (bottom panels). Washed thylakoids were subjected to cross-linking with bis-maleimide agents that vary in spacer length: BMOE (left panels), BMH (middle panels), or BM(PEO)3 (right panels).
(B) S1 and S2 do not contact downstream residues of the signal peptide. Radiolabeled precpTatCaaa D78C (S1; lanes 1 to 7), precpTatCaaaT164C (S2; lanes 8 to 14), and precpTatCaaa (lanes 15 to 21) were imported into chloroplasts and recovered thylakoids incubated with radiolabeled substrate tOE17-20F with single Cys substitutions at varying positions as indicated below the panels. Washed thylakoids were then subjected to cross-linking with BM(PEO)3. The cpTatC-substrate adducts are marked with an asterisk.
cpTatCaaa Cys-substituted residues D78C and T164C were also examined for cross-linking to tOE17-20F with Cys substitutions in downstream residues of the signal peptide (Figure 8B). No significant cross-links were detected with any other residues, suggesting that the N terminus and first stromal loop of cpTatC recognize only the RR proximal region of the signal peptide. This is consistent with and extends results of site-directed photo-cross-linking directed from different regions of the signal peptide (Alami et al., 2003; Gérard and Cline, 2006). Substrates with Cys substitutions from −10 to −1 contained small amounts of a cross-linking product to an unknown protein; this protein was present even when the imported cpTatC variant lacked a Cys residue (right-hand panel). Surprisingly, a cross-linked substrate dimer product was a major band for substrate proteins with Cys residues from −7 to −1. In a separate experiment, the substrate-substrate cross-links directed by Cys residues from −7 to −1 were also obtained by CuP-catalyzed disulfide cross-linking and even without chemical oxidants, indicating that signal peptides of adjacently bound substrates are in close proximity (see Supplemental Figure 6 online). This extends previous observations (Ma and Cline, 2010) that bound substrate proteins are in very close contact throughout the amino proximal region of their mature domains.
Transport of Substrate Proteins Cross-Linked to Imported cpTatC
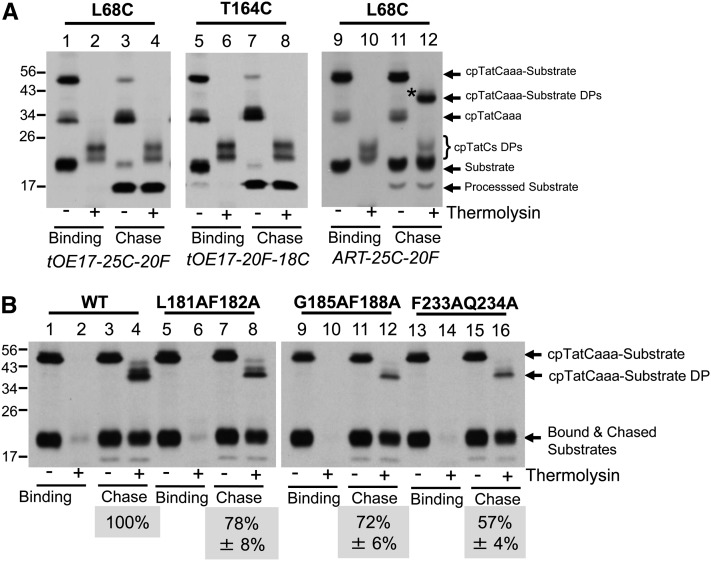
Chase reactions of cross-linked substrate were used to assess the functional relevance of contacts between the RR motif of the signal peptide and segments S1 and S2. As shown in Figure 9A, thylakoids containing cpTatC with Cys substitutions in segment S1 (L68C) or segment S2 (T164C) were used for binding assays with substrate containing a Cys residue either at the −25 position (with L68C; lanes 1 to 4) or the −18 position (with T164C; lanes 5 to 8). Cross-linking was mediated by BM(PEO)3, and the membranes were then energized for transport of bound substrates. In both cases, the cross-linked substrate-cpTatC product largely disappeared during the chase reaction (Figure 9A, lanes 3 and 7), indicating that the substrate protein was transported and the signal peptide cleaved. In order to verify this conclusion, the cleavage site of the substrate signal peptide was modified from A-R-A to A-R-T to prevent signal peptidase-mediated cleavage. In this case, the cross-linked substrate-cpTatC band did not disappear upon a chase reaction (lane 11). Nevertheless, the substrate protein was transported as evidenced by the fact that it was largely protected by thermolysin treatment of the membranes (lane 12), producing a degradation product with only the N terminus of the cpTatC moiety cleaved by the protease as described (Gérard and Cline, 2006).
Figure 9.
cpTatC Mutants with Reduced Translocation.
(A) Substrates cross-linked to cpTatC S1 or S2 are transported. Radiolabeled precpTatCaaa Cys variants L68C or T164C were imported into chloroplasts. Chloroplasts were pelleted and washed. Recovered thylakoids were incubated with radiolabeled tOE17-25C-20F (left panel) or tOE17-20F-18C (middle panel) or the noncleavable tOE17ART-25C-20F (right panel). Washed thylakoids were cross-linked with BM(PEO)3 (left and middle panels) or BMOE (right panel). Thylakoids were then analyzed directly (lanes 1, 2, 5, 6, 9, and10) or first subjected to transport conditions (chase; lanes 3, 4, 7, 8, 11, and 12). Thylakoid aliquots were treated with thermolysin (lanes 2, 4, 6, 8, 10, and 12).
(B) TM mutants of cpTatC with reduced translocation. Unlabeled precpTatCaaa L68C wild type (WT; lanes 1 to 4) or containing Ala substitutions in TM3 L181AF182A (lanes 5 to 8), G185AF188A (lanes 9 to 12), or TM4 F233AQ234A (lanes 13 to 16) were imported into chloroplasts. Recovered thylakoids were incubated with radiolabeled ART-25C-20F and subjected to BMOE cross-linking. The thylakoids were analyzed directly (binding; lanes 1, 2, 5, 6, 9, 10, 13, and14) or subjected to transport conditions (chase; lanes 3, 4, 7, 8, 11, 12, 15, and 16). Aliquots of the recovered thylakoids were treated with the thermolysin (lanes 2, 4, 6, 8, 10, 12, 14, and 16). The ratio of protease-protected cross-linked substrate from the chase reaction to the amount of cross-linked bound substrate is the fraction transported. Transport efficiencies were normalized to wild-type cpTatCaaa L68C, which was arbitrarily set at 100%. The actual transport efficiency of substrate cross-linked wild-type cpTatCaaa L68C was 59.3% ± 4.3% for the three experiments. The efficiencies shown are the average ± se (n = 3) or average ± range (n = 2) for L181AF182A). DP, thermolysin degradation product.
Thus, this approach not only verifies that contacts of the signal peptide with cpTatC segments S1 and S2 are productive for transport but also provides a definitive assay for determining the transport competence of other cpTatC mutants that are unaffected in receptor complex assembly and substrate binding. The Ala substitutions of conserved residues in TM3 and TM4 (cpTatC L181AF182A and cpTatC F233AQ234A, respectively) resulted in cpTatC proteins that formed receptor complex and bound substrate at wild-type levels (Figures 4 and 5). The L68C mutation was introduced into these Ala substitution variants and also into TM3 variant cpTatC G185AF188A that was not previously tested for substrate binding. The resulting proteins were imported into chloroplasts, and recovered thylakoids were incubated with tOE17ART-25C-20F followed by cross-linking with BMOE. In the experiment shown in Figure 9B, the substrate was radiolabeled and the cpTatC was unlabeled, allowing for a direct quantification of bound substrate and transported substrate. As can be seen, all three mutant cpTatC mutants efficiently cross-linked to bound substrate but exhibited moderate reductions in translocation (lanes 7 and 8, 11 and 12, and 15 and 16) with the greatest reduction in cpTatC F233AQ234A.
DISCUSSION
Unraveling the mechanism of Tat protein transport is being pursued in two systems, plant thylakoids and E. coli. Thylakoids provide a robust in vitro transport assay with native levels of cpTat components. Here, the thylakoid system was adapted to allow both mutagenesis and characterization of mutants in vitro. Because cpTatC does not integrate into isolated thylakoids (Fincher et al., 2003; Martin et al., 2009), the cpTatC variants were imported into intact chloroplasts to achieve efficient thylakoid integration. The analysis of exogenous cpTatC variants was enabled by a large free pool of assembly-competent Hcf106 (Cline and Mori, 2001) (Figure 2) and the preferential formation of new receptor complexes by imported cpTatC (Figure 2). This allowed analysis of the recombinant cpTatC without interference from endogenous cpTatC. This is important because in E. coli, wild-type Tat function could be restored by coexpression of two different nonfunctional mutant TatCs (Buchanan et al., 2002). This presumably means that the phenotype of a mutant cpTatC might be masked by the presence of wild-type cpTatC. However, this was not the case in our assay. Several cpTatC mutants analogous to previously characterized E. coli TatC mutants (Buchanan et al., 2002; Holzapfel et al., 2007) gave essentially identical phenotypes with regards to substrate binding and receptor complex assembly (Figures 2 and 3).
The finding that imported cpTatC does not significantly associate with endogenous cpTatC provides additional support for the existence of cpTatC and Hcf106 as a large multimeric complex in the membrane. Most studies analyzed detergent-solubilized receptor complexes. However, there is always the possibility that the detergent could stabilize an oligomeric state of proteins that was not present in situ. This appears to be the case for E. coli TatA, whereby detergent solubilization results in large homooligomers (Gohlke et al., 2005) that may not exist in vivo (Leake et al., 2008). If cpTatC-Hcf106 existed in the membranes as heterodimers and formed the large complex only upon solubilization, a substantial amount of endogenous cpTatC would be expected in the Ni-NTA–purified receptor complex (Figure 2).
Identification of the cpTatC signal peptide binding site employed targeted Ala substitutions of highly conserved residues. Eight of 13 segments of cpTatC were defective in substrate binding. Of these, only three segments appear to be specifically important for signal peptide recognition: the N terminus (S1), the first stromal loop (S2), and the Phe-158 residue that borders the first stromal loop. Single and double mutations showed that His-79 and Glu-82 in the N terminus (S1) and Pro-161, Leu-163, and Glu-167 in the first stromal loop (S2) are important for binding (Figures 4 and 5). Interestingly, these experimental results are consistent with an early prediction of the twin Arg recognition motifs based on sequence conservation alone. Mori and Cline (2001) speculated that conserved Glu residues in motifs present in the N terminus H-[IL]-x-E-[FIL]-[KR]-x-R (S1) and the first stromal loop P-[AG]-[LM]-x-x-x-E-[KRT]-[KRNG] (S2) were the likely binding sites for the twin arginine motif of the signal peptide. Zoufaly et al. (2012), using site-directed photo-cross-linking, recently showed that the E. coli TatC N terminus and first cytoplasmic loop are in close contact with bound substrate. They suggested but did not demonstrate that the observed interactions were with the twin Arg motif of signal peptide. Here, we conclusively demonstrate that S1 and S2 bind only to the RR-proximal motif with a method called Cys matching, which permitted simultaneous identification of interacting sites on substrate and cpTatC (Figures 7 and 8). S1 and S2 Cys-substituted residues cross-linked strongly to RR-proximal Cys residues but did not cross-link with other Cys-substituted regions of the signal peptide (Figure 8B), nor did other cpTatC segments cross-link to RR-proximal Cys residues (see Supplemental Figure 5 online). Thus, our results provide a map of the domains and residues involved in signal peptide binding. As such, they are consistent with and extend previous photo-cross-linking studies (Alami et al., 2003; Gérard and Cline, 2006; Zoufaly et al., 2012) as well as recent isothermal calorimetric analysis (Rollauer et al., 2012).
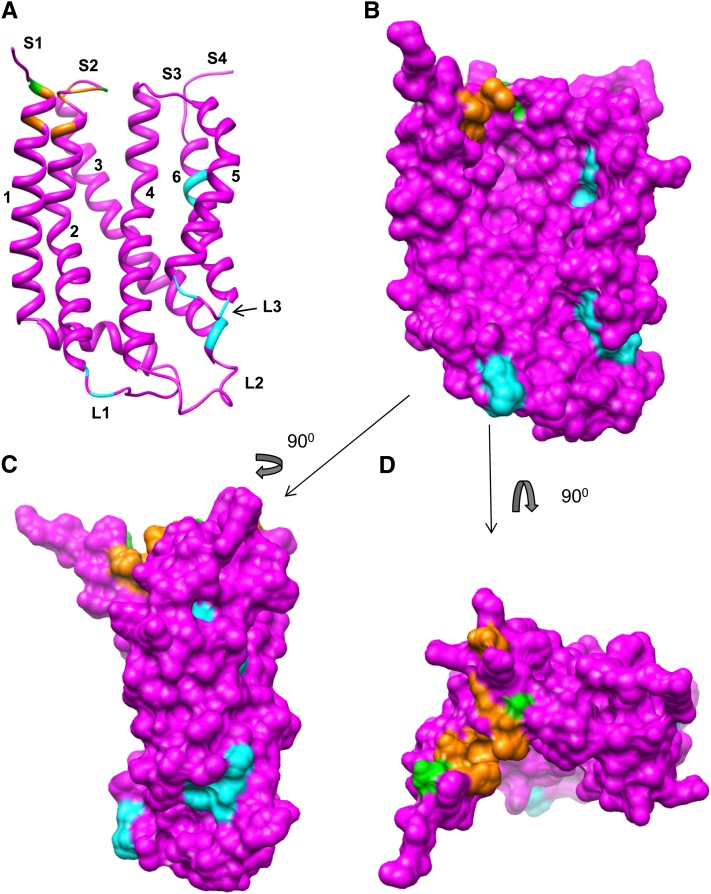
A logical inference could be made that Glu residues in S2 and S1 coordinate the guanidinium groups of the twin arginine and cpTatC F158 π-bonds the RR+2 Phe that is important for tight binding (Gérard and Cline, 2007). On a map of the recently determined crystal structure of TatC from A. aeolicus (Rollauer et al., 2012), all of the residues shown here to be important for binding are located on the surface of the membrane. A model of cpTatC based on homology threading (Figure 10) depicts the residues important for binding (in orange) and those that form cross-links to the RR proximal region (in green). As is apparent, the relevant residues form a tightly grouped stroma-exposed surface patch. This patch is similar in position and shape to a patch of amino acid conservation on the A. aeolicus TatC structure (Rollauer et al., 2012).
Figure 10.
Residues Involved in Substrate Binding and Those Important for Receptor Complex Assembly Mapped to a Model of the cpTatC Structure.
A structural model of cpTatC was generated with the Modeler tool of Chimera. Residues colored orange are those that resulted in impaired substrate binding when mutated to Ala. Residues in green are those that cross-linked to the RR region of the signal peptide. Residues in cyan, when mutated to Ala, resulted in defective assembly of the receptor complex.
(A) Ribbon depiction showing the concave face.
(B) Surface depiction with the same perspective as in (A).
(C) Side view. Note the grouping of mutants defective in assembly of receptor complexes at the lumenal side centered near L3 ([A] to [C]).
(D) Top view showing the residues of the RR signal peptide binding site.
The integrity of the receptor complex is essential for function; the results of mutagenesis in this work make that clear as mutations that impaired receptor complex assembly also impaired substrate binding (Figures 4 and 5). In addition, signal peptide binding to the isolated A. aeolicus TatC, although being RR specific, displayed a very low affinity [compare Rollauer et al. [2012] with Celedon and Cline [2012]). Observations with the E. coli Tat system suggest that a TatC (cpTatC) oligomer forms a 250-kD core complex to which TatB (Hcf106) is added to form the receptor complex (Behrendt et al., 2007; Orriss et al., 2007). Comparable experiments with the thylakoid system have not been done, but solubilization of thylakoid membranes with the stronger detergent dodecyl maltoside separated the receptor complex into an ∼250-kD cpTatC complex and an ∼230-kD Hcf106 complex (K. Cline, unpublished data). The combined observations point to cpTatC (TatC) oligomerization as a critical first step in the assembly process. The crystal structure of the A. aeolicus TatC did not yield insight into the arrangement of TatC subunits in the complex (Rollauer et al., 2012). Thus, an important first step is to define the regions and residues important for receptor complex assembly as well as regions that make contact with other subunits.
Five of 13 Ala substitutions resulted in impaired receptor complex assembly. Mutations in L3, TM5, and TM6 shared similar characteristics; their disruption of receptor complex assembly was moderate, they possessed some ability to bind substrate, and they showed reduced association with Hcf106 (Figures 4 and 5). The E. coli mutation comparable to the L3 D276A was previously shown to disrupt TatC–TatB interactions (Buchanan et al., 2002). In addition, the homologous E. coli TM5 residues adjacent to TM5 mutations T272AP273A described here direct Cys-Cys cross-links with the TatB transmembrane domain (Kneuper et al., 2012; Rollauer et al., 2012). These thylakoid and E. coli TM5 residues are on the lumenal (periplasmic) end of TM5 close to L3 (P3). This is also a region of high sequence conservation (Rollauer et al., 2012). Thus, it is likely that the TM5 and L3 mutations disrupt cpTatC–Hcf106 interaction. The question of whether the TM6 mutation disrupts interaction with Hcf106 remains to be determined. The L2 mutation F200AF201A also disrupts receptor complex assembly moderately (Figure 5) to severely depending on the particular experiment. On the two-dimensional map of cpTatC (Figure 4), the mutated residues appear quite distant from the described lumenal TM5-L3 region. However, when modeled onto the three-dimensional TatC structure, they are on the lumenal end of TM3, very near to the TM5-L3 region, describing a continuous patch in the surface depiction (Figure 10). Thus, it is possible that these three regions, containing mutants F200AF201A, D276, and T272AP273A, define a hotspot for Tat component interactions. Consistent with this general conclusion is the fact that the L3 residue Thr-275 directed strong cpTatC-cpTatC cross-linking (Figure 6), the E. coli TatC P3 residue Asp-211 directed photo-cross-linking to TatA (Tha4 ortholog) (Zoufaly et al., 2012), and the TM3-proximal P2 region residue 150 of E. coli TatC directed cross-linking to TatB and TatA (Zoufaly et al., 2012)
Mutations in L1 resulted in complete loss of receptor complex and absence of any endogenous cpTatC in the purified mutant cpTatC (Figure 5). The E. coli TatC periplasmic loop P1 (comparable to L1) is also hypersensitive to mutation (Kneuper et al., 2012) and at least one mutant in P1, P48A, resulted in loss of receptor complex (Barrett et al., 2005). Also of interest is that L1 Cys substitution directed strong cpTatC-cpTatC cross-linking (Figure 6), and a P1 residue of E. coli TatC directed photo-cross-linking to adjacent TatC (Zoufaly et al., 2012). It is tempting to conclude that L1 (P1) is involved cpTatC–cpTatC interactions. However, L1 is certainly is not the sole site of contact between neighboring cpTatC (TatC) because all three lumenal (periplasmic) domains direct such cross-linking. Punginelli et al. (2007) observed strong E. coli TatC-TatC cross-linking from the P2 loop residue G144C, the L2 residue N203C directed a small amount of cpTatC-cpTatC cross-linking (Figure 6), and, as mentioned, the L3 T275C directed strong cpTatC-cpTatC cross-linking (Figure 6). An alternative explanation for the effects of mutations in L1 and L2 is that such mutations disrupt the cap structure formed by the two loops that appears to stabilize the overall arrangement of transmembrane domains of TatC (Rollauer et al., 2012). Additional experiments are required to resolve this issue.
One very interesting result of the cpTatC-cpTatC cross-linking studies is that residues in segments S1 and S2 cross-linked with their counterparts on adjacent cpTatC subunits, even without chemical oxidants or bismaleimide cross-linkers (Figure 6). This indicates that the RR binding sites of adjacent cpTatCs are in close contact in the receptor complex. In previous work, Ma and Cline (2010) found that receptor-bound precursors cross-linked to each other to form dimers and tetramers without chemical oxidants through the N termini of their mature domains. Here, we extend that result to include the middle and carboxyl proximal signal peptide domains of bound substrates (Figure 8B). Taken together, these studies portray a receptor complex in which signal peptide binding sites are clustered. The spatial significance of grouping signal peptide binding domains of cpTatC in the receptor complex is not entirely clear, but it coincides with the interesting observation that cross-linked dimeric and tetrameric substrates are transported as readily as monomeric substrates (Ma and Cline, 2010).
Rollauer et al. (2012) recently proposed a TatA (Tha4) assembly site on TatC. They suggested that initial recruitment of TatA occurs on the concave face of the TatC structure, mediated by a conserved residue, Glu-165 in A. aeolicus TatC near the middle of that face. Ala substitution of the homologous E. coli TatC residue Glu-170 does not affect substrate binding or receptor complex assembly but does reduce transport to ∼10% of the wild type (Buchanan et al., 2002; Holzapfel et al., 2007). The proposed recruitment would be mediated by hydrogen bonding of the polar group of Glu-170 to the essential transmembrane Gln of TatA. The comparable scenario in thylakoids would involve cpTatC residue Gln-234 in TM4 and the Tha4 transmembrane Glu-10, which along with the proton gradient is essential for Tha4 assembly (Mori and Cline, 2002; Dabney-Smith et al., 2003; Dabney-Smith and Cline, 2009). In our experiments, we found that Ala substitution of Gln-234 did result in reduced translocation efficiency without affecting receptor complex assembly or substrate binding. However, the reduction in translocation was less severe than the E. coli TatC E170A substitution (Figure 9B). Nevertheless, the system described here with Cys matching and a controlled translocase assembly step is ideal for experimentally determining the site of Tha4 assembly.
In summary, we developed an in vitro method for structure-function studies of the chloroplast Tat system. The combined Ala scanning mutagenesis, biochemical characterization, and Cys matching interaction experiments have mapped the signal peptide binding site in detail. Furthermore, they conclusively showed that this site recognizes only the RR-proximal signal peptide domain and that other regions of cpTatC do not recognize the RR motif. Several segments were also identified as important for receptor complex assembly and cpTatC-cpTatC (TatC-TatC) contact. Of these, the most interesting was the signal peptide binding site. Combined with a previous observation (Ma and Cline, 2010), the results here suggest that the binding sites of adjacent cpTatCs are clustered in the receptor complex.
METHODS
Plasmid Construction and Mutagenesis
The clone pSSU13precpTatC for the precpTatC protein in this study was constructed by splice overlap extension (Horton et al., 1989) and contains the transit peptide and first 13 residues of the mature domain of the pea (Pisum sativum) small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase fused to the N terminus of the complete precursor sequence for pea cpTatC. The His-tagged precpTatC precursor was constructed by splice overlap extension with the coding sequence for GGGGSGGGGSGGGGSHHHHHH fused to the precpTatC carboxyl terminus. The transcription plasmid for substrate tOE17-20F (Gérard and Cline, 2007) is as described. The cpTatC and tOE17-20F variants with amino acid substitutions or insertions were constructed by PCR mutagenesis with the QuikChange mutagenesis kit (Stratagene). The Cys substitutions were obtained by replacing the relevant codons with TGC, and Ala substitutions with GCG, GCA, GCT, or GCC. The noncleavable tOE17ART-20F changed the Ala at the −1 position to Thr.
In Vitro Synthesis of Proteins
Capped RNAs were transcribed in vitro with SP6 polymerase (Promega) and were translated in vitro in the presence of [3H]-Leu or unlabeled Leu with a homemade wheat germ translation system (Cline, 1986). Before use, the translation products were diluted 1:1 and adjusted to import buffer (IB; 50 mM HEPES/KOH, pH 8.0, and 0.33 M sorbitol) containing 30 mM Leu.
Preparation of Chloroplasts, Thylakoids, and Stromal Extract
Intact chloroplasts were isolated from 9- to 10-d-old pea seedlings (Cline et al., 1993) and resuspended in IB at 1 mg of chlorophyll per mL before use. For preparation of thylakoids and stroma, chloroplasts were lysed hypotonically in 10 mM HEPES/KOH, pH 8.0, containing 10 mM MgCl2 at 0°C for 6 min and adjusted to IB, 5 mM MgCl2. Thylakoids were obtained from the lysates by centrifugation at 3200g for 8 min, were washed with IB, and then resuspended in IB at 1 mg of chlorophyll per mL before use. Stromal extract was obtained from the lysate supernatant by additional centrifugation at 12,000g for 10 min to pellet the envelope membranes. Chlorophyll concentrations were determined according to Arnon (1949).
Chloroplast Protein Import, Substrate Binding, and Chase Assays
Radiolabeled precpTatC was incubated with isolated chloroplasts (0.33 mg chlorophyll/mL) and 5 mM Mg-ATP in IB with 120 μE/m2/s of light in a 25°C water bath for 40 min. Following import, intact chloroplasts were repurified by centrifugation through 35% Percoll in IB and washed with IB (Cline et al., 1993). For experiments that involved cross-linking or as noted in the figure legends, chloroplasts were recovered from assays by pelleting, were washed, and then used to prepare thylakoids. The thylakoid membranes were prepared from the chloroplasts by lysis and centrifugation and were analyzed directly or first treated with thermolysin (Cline et al., 1993). Substrate binding assays were conducted with apyrase-treated in vitro–translated substrate and thylakoids, in IB and 3 mM MgCl2 as described (Ma and Cline, 2010), for 30 min at 0°C in darkness. Thylakoid membranes were then recovered by centrifugation, washed twice with IB, and analyzed directly as binding samples. They were used for cross-linking (below) or subjected to transport conditions for chase assays. Chase assays generally contained 20 μg chlorophyll of substrate-bound thylakoids, a stoichiometric amount of stromal extract, and 5 mM Mg-ATP in IB containing 3 mM MgCl2. Assays were incubated for 15 min at 25°C in ∼120 μmol/m2/s white light. Thylakoid membranes were recovered by centrifugation and analyzed directly or first treated with thermolysin (Ma and Cline, 2010).
Cys Cross-Linking
Cys-Cys disulfide formation was accelerated by treating thylakoid samples with 1 mM CuP oxidant (Dabney-Smith et al., 2006) for 2 min at room temperature. Thylakoids were then recovered by centrifugation and resuspended in IB, 1 mM EDTA, and 4 mM N-ethyl maleimide to terminate disulfide formation. For cross-linking with bismaleimide cross-linkers, the thylakoid samples were washed with IB plus 0.5 mM tris-(2-carboxyethyl) phosphine (Invitrogen) and resuspended in IB, pH 6.8, 2 mM EDTA, and 0.5 mM tris-(2-carboxyethyl) phosphine. Cross-linking was initiated with 1 mM cross-linker [i.e., BMOE, BMH, or BM(PEO)3; Pierce Chemical] dispensed from 40 mM stocks in DMSO. Cross-linking was terminated after 8 min at 25°C by pelleting of thylakoids and washing with IB and 2 mM DTT. For chase assays following bismaleimide cross-linking, the thylakoids were incubated with unlabeled, in vitro–translated Tha4 at 0.33 mg chlorophyll per mL in IB and 3.3 mM MgCl2 for 15 min at room temperature. Thylakoids were recovered by centrifugation and subjected to chase assays as described above.
Metal Ion Affinity Purification of Complexes Formed by Imported cpTatC-His
Washed thylakoids with imported cpTatC-His, with or without bound substrate, were resuspended at 1 mg of chlorophyll per mL in solubilization buffer (0.25× IB, 20% glycerol, and 0.5 M amino-caproic acid) containing 1% digitonin and 1 mM phenylmethylsulfonyl fluoride. The samples were incubated on ice for 1 h and then centrifuged at 150,000g at 2°C for 30 min. The supernatants were adjusted to binding buffer (20 mM HEPES, pH 7.8, 150 mM NaCl, and 20 mM imidazole) and mixed with Ni-NTA magnetic agarose beads (Qiagen) end-over-end for ≥4 h at 4°C. The beads were washed with binding buffer containing 0.2% digitonin, and the bound proteins were eluted with 0.5× binding buffer containing 100 mM EDTA and 0.2% digitonin.
Sample Analysis
Most samples were analyzed either by BN-PAGE or by SDS-PAGE and then fluorography as described (Ma and Cline, 2010). Samples separated by SDS-PAGE were prepared in SDS sample buffer containing 4 M urea at room temperature for 30 min. Radiolabeled proteins were extracted from excised gel bands and quantified by scintillation counting with corrections for counting efficiency and gel extraction efficiencies for the individual protein species (Cline, 1986; Celedon and Cline, 2012). For immunoblotting, SDS-PAGE or BN-PAGE gels were electroblotted to 0.2-μm pore-sized nitrocellulose membranes. After incubation with antibodies, the blots were developed with the ECL method (GE Healthcare) (Mori et al., 1999, 2001).
Model for the Structure of cpTatC from Pea
The sequence of cpTatC from pea was homology modeled onto the three-dimensional structure of the A. aqueoris TatC protein PDB 4b4a using the Modeler tool of Chimera (Pettersen et al., 2004; Eswar et al., 2006), guided by an alignment between the last 230 amino acids of pea cpTatC and the sequence of A. aqueoris TatC (GI: 427930763) conducted by Multalin (http://multalin.toulouse.inra.fr/multalin/multalin.html).
Accession Numbers
Sequence data for proteins analyzed in this study can be found in the GenBank/EMBL data libraries under the following accession numbers: cpTatC from pea, AAK93948; Hcf106 from pea, AAK93949; Tha4 from pea, AAD33943; and OE17 from maize (Zea mays) NP_001105348.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The cpTatC Precursors in This Study Contain Two Transit Peptides That Are Cleaved by the Stromal Processing Peptidase upon Import into the Chloroplast.
Supplemental Figure 2. cpTatC Double Ala Mutants Are Correctly Integrated into Thylakoids.
Supplemental Figure 3. Single Ala Mutations in cpTatC Segments S1 and S2 Exhibit a Range of Substrate Binding Capability.
Supplemental Figure 4. Cys-Substituted Substrate and cpTatC Variants Are Functional for Transport or Binding.
Supplemental Figure 5. No Interactions Were Detected between the Signal Peptide RR Domain and cpTatC Segments, Other Than S1 and S2, That Exhibited Substrate Binding Defects.
Supplemental Figure 6. Bound Substrate Proteins Are in Close Contact throughout Downstream Residues of the Signal Peptide.
Supplementary Material
Acknowledgments
We thank Cassie Patricia Aldridge, Yubing Li, and Michael McCaffery for critical review of the article. We also thank Michael McCaffery for excellent technical assistance. This work was supported in part by National Institutes of Health Grant R01 GM46951 to K.C.
AUTHOR CONTRIBUTIONS
K.C. and X.M. designed the experiments, and X.M. conducted the experiments. X.M. and K.C. analyzed the data and wrote the article.
Glossary
- RR
Arg-Arg
- Tat
twin Arg translocation
- BN
blue native
- Ni-NTA
Ni 2+-nitrilotriacetic acid
- BMOE
bis-maleimidoethane
- BMH
bis-maleimidohexane
- IB
import buffer
- CuP
copper phenanthroline
- BM(PEO)3
(1, 8-bis-maleimidotriethyleneglycol)
References
- Alami M., Lüke I., Deitermann S., Eisner G., Koch H.G., Brunner J., Müller M. (2003). Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12: 937–946 [DOI] [PubMed] [Google Scholar]
- Allen S.C., Barrett C.M., Ray N., Robinson C. (2002). Essential cytoplasmic domains in the Escherichia coli TatC protein. J. Biol. Chem. 277: 10362–10366 [DOI] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C.M., Mangels D., Robinson C. (2005). Mutations in subunits of the Escherichia coli twin-arginine translocase block function via differing effects on translocation activity or tat complex structure. J. Mol. Biol. 347: 453–463 [DOI] [PubMed] [Google Scholar]
- Behrendt J., Lindenstrauss U., Brüser T. (2007). The TatBC complex formation suppresses a modular TatB-multimerization in Escherichia coli. FEBS Lett. 581: 4085–4090 [DOI] [PubMed] [Google Scholar]
- Brüser T., Sanders C. (2003). An alternative model of the twin arginine translocation system. Microbiol. Res. 158: 7–17 [DOI] [PubMed] [Google Scholar]
- Buchanan G., de Leeuw E., Stanley N.R., Wexler M., Berks B.C., Sargent F., Palmer T. (2002). Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43: 1457–1470 [DOI] [PubMed] [Google Scholar]
- Celedon J., Cline K. (2012). Stoichiometry for binding and transport by the twin arginine translocation system. J. Cell Biol. 197: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon J.M., Cline K. (2013). Intra-plastid protein trafficking: How plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta. 1833: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. (1986). Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J. Biol. Chem. 261: 14804–14810 [PubMed] [Google Scholar]
- Cline, K. (2003). Biogenesis of green plant thylakoid membranes. In Light- Harvesting Antennas in Photosynthesis. Advances in Photosynthesis and Respiration, Vol. 13, B. Green and W. Parson, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 353–372. [Google Scholar]
- Cline K., Dabney-Smith C. (2008). Plastid protein import and sorting: Different paths to the same compartments. Curr. Opin. Plant Biol. 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Henry R., Li C., Yuan J. (1993). Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 12: 4105–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., McCaffery M. (2007). Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 26: 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Mori H. (2001). Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., and Theg, S. (2007). The Sec and Tat protein translocation pathways in chloroplasts. In The Enzymes: Molecular Machines Involved in Protein Transport across Cellular Membranes, Vol. XXV, R.E. Dalbey, C.M. Koehler, and F. Tamanoi, eds (New York: Academic Press), pp. 463–492. [Google Scholar]
- Dabney-Smith C., Cline K. (2009). Clustering of C-terminal stromal domains of Tha4 homo-oligomers during translocation by the Tat protein transport system. Mol. Biol. Cell 20: 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C., Mori H., Cline K. (2003). Requirement of a Tha4-conserved transmembrane glutamate in thylakoid Tat translocase assembly revealed by biochemical complementation. J. Biol. Chem. 278: 43027–43033 [DOI] [PubMed] [Google Scholar]
- Dabney-Smith C., Mori H., Cline K. (2006). Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281: 5476–5483 [DOI] [PubMed] [Google Scholar]
- Eswar, N., Webb, B., Marti-Renom, M.A., Madhusudhan, M.S., Eramian, D., Shen, M.Y., Pieper, U., and Sali, A. (2006). Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 5: Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher V., Dabney-Smith C., Cline K. (2003). Functional assembly of thylakoid deltapH-dependent/Tat protein transport pathway components in vitro. Eur. J. Biochem. 270: 4930–4941 [DOI] [PubMed] [Google Scholar]
- Fröbel J., Rose P., Müller M. (2011). Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286: 43679–43689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbel J., Rose P., Müller M. (2012). Twin-arginine-dependent translocation of folded proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard F., Cline K. (2006). Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281: 6130–6135 [DOI] [PubMed] [Google Scholar]
- Gérard F., Cline K. (2007). The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J. Biol. Chem. 282: 5263–5272 [DOI] [PubMed] [Google Scholar]
- Gohlke U., Pullan L., McDevitt C.A., Porcelli I., de Leeuw E., Palmer T., Saibil H.R., Berks B.C. (2005). The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. USA 102: 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel E., Eisner G., Alami M., Barrett C.M., Buchanan G., Lüke I., Betton J.M., Robinson C., Palmer T., Moser M., Müller M. (2007). The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46: 2892–2898 [DOI] [PubMed] [Google Scholar]
- Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. (1989). Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D.J. (2006). The function and diversity of plastid protein import pathways: A multilane GTPase highway into plastids. Traffic 7: 248–257 [DOI] [PubMed] [Google Scholar]
- Kneuper H., Maldonado B., Jäger F., Krehenbrink M., Buchanan G., Keller R., Müller M., Berks B.C., Palmer T. (2012). Molecular dissection of TatC defines critical regions essential for protein transport and a TatB-TatC contact site. Mol. Microbiol. 85: 945–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Leake M.C., Greene N.P., Godun R.M., Granjon T., Buchanan G., Chen S., Berry R.M., Palmer T., Berks B.C. (2008). Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. USA 105: 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Cline K. (2010). Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J. 29: 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.R., Harwood J.H., McCaffery M., Fernandez D.E., Cline K. (2009). Localization and integration of thylakoid protein translocase subunit cpTatC. Plant J. 58: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer C., Panahandeh S., Jungkamp A.C., Moser M., Müller M. (2010). TatB functions as an oligomeric binding site for folded Tat precursor proteins. Mol. Biol. Cell 21: 4151–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt C.A., Buchanan G., Sargent F., Palmer T., Berks B.C. (2006). Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels. FEBS J. 273: 5656–5668 [DOI] [PubMed] [Google Scholar]
- Mori H., Cline K. (2001). Post-translational protein translocation into thylakoids by the Sec and DeltapH-dependent pathways. Biochim. Biophys. Acta 1541: 80–90 [DOI] [PubMed] [Google Scholar]
- Mori H., Cline K. (2002). A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid [Delta]pH/Tat translocase. J. Cell Biol. 157: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Summer E.J., Cline K. (2001). Chloroplast TatC plays a direct role in thylakoid (Delta)pH-dependent protein transport. FEBS Lett. 501: 65–68 [DOI] [PubMed] [Google Scholar]
- Mori H., Summer E.J., Ma X., Cline K. (1999). Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J. Cell Biol. 146: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi R., Nagata N., Ito T., Takahashi S., Hobo T., Yoshida S., Shinozaki K. (2001). An essential role of a TatC homologue of a Delta pH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 10499–10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orriss G.L., Tarry M.J., Ize B., Sargent F., Lea S.M., Palmer T., Berks B.C. (2007). TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 581: 4091–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer T., Berks B.C. (2012). The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10: 483–496 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg A.J., Sun Q., van Wijk K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Punginelli C., Maldonado B., Grahl S., Jack R., Alami M., Schröder J., Berks B.C., Palmer T. (2007). Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC Component. J. Bacteriol. 189: 5482–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollauer S.E., et al. (2012). Structure of the TatC core of the twin-arginine protein transport system. Nature 492: 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Hebert D.N. (2003). Protein translocons: Multifunctional mediators of protein translocation across membranes. Cell 112: 491–505 [DOI] [PubMed] [Google Scholar]
- Schubert M., Petersson U.A., Haas B.J., Funk C., Schröder W.P., Kieselbach T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Skalitzky C.A., Martin J.R., Harwood J.H., Beirne J.J., Adamczyk B.J., Heck G.R., Cline K., Fernandez D.E. (2011). Plastids contain a second sec translocase system with essential functions. Plant Physiol. 155: 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R., Barkan A. (1995). Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly S., Fröbel J., Rose P., Flecken T., Maurer C., Moser M., Müller M. (2012). Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. 287: 13430–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.