Abstract
Catalytic metallodrugs were used to oxidatively cleave HIV-1 Rev Response Element RNA (RRE RNA), and the mechanisms of RNA cleavage were studied using a combination of matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), fluorescence spectroscopy, and gel electrophoresis. The metallodrugs, which contained combinations of the transition metals Fe2+, Co2+, Ni2+, and Cu2+ and the Rev-coupled chelators DOTA, DTPA, EDTA, NTA, tripeptide GGH, and tetrapeptide KGHK, bind to and cleave HIV RRE RNA through heretofore unknown oxidative mechanisms. The broad spectrum of metal catalysts and co-reagents provided a means for systematic variation of oxidative reactivity without significant perturbation of binding between catalyst and RNA. Detailed MS analyses were used to monitor formation of RNA fragments containing terminal 2’,3’-cyclic phosphate (2’,3’-cPO4), 3’-phosphate (3’-PO4), 3’-phosphoglycolate (3’-PG), 5’- hydroxyl (5’-OH), 5’- phosphate (5’-PO4) and other nascent overhangs at sites of cleavage. The distinct overhangs corresponded to distinct mechanisms of oxidative hydrogen-abstraction (H abstraction), hydrolysis, and/or endonucleolysis, allowing a dissection of the contributions of various mechanisms of oxidative cleavage. Rapid co-reactant- and catalyst-dependent formation of fragments containing terminal 3’-PG, 3’-PO4 and 5’-PO4 overhangs appeared to be initiated primarily by H abstraction events. The standard thiobarbituric acid (TBA) assay was employed herein in a novel usage to monitor the formation of base 2-hydroxypropenal products produced by 4’-H abstraction in RNA. Formation of an adduct with TBA was monitored by fluorescence, and its quantification correlated with the formation of 3’-PG monitored by MALDI-TOF MS, confirming oxidative 4’-H abstraction as a major mechanism of rapid catalyst-mediated cleavage of RRE RNA. Rapid formation of 3’-PO4 overhangs was most likely a result of 5’-H abstraction. Apparent rates of formation of 3’-PG (a unique product of 4’-H abstraction) at differing nucleotide positions within the RNA were used to triangulate probable 3D positions of metal centers and establish the distance-dependence of 4’-H abstraction for certain catalytic metallodrugs.
Keywords: HIV, RRE, RNA, catalytic metallodrugs, artificial nucleases, cleavage, scission, mechanisms, oxidation, damage, hydrogen abstraction, hydrolysis, endonucleolysis, MALDI-TOF, mass spectrometry, thiobarbituric acid, tartronal, 2-hydroxypropanedial, base 2-hydroxypropenal
INTRODUCTION
Ribonucleic acid (RNA) polymers serve critical roles as messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and even entire genomes in RNA viruses such as hepatitis, influenza, or HIV. Given the critical nature of RNA and its increased exposure to damaging oxidative conditions in the cytosol, there is a strong need for elucidation of the mechanisms of oxidative damage to RNA. Several studies have focused on in vivo damage and/or natural processing of RNA,1–4 with relatively sparse information gained regarding oxidative damage, while others have focused on the development of specific RNA-binding5–12 and RNA-targeting affinity cleavage13–20 reagents, which allow an in vitro evaluation of oxidative damage/cleavage of RNA by artificial catalysts.
While the mechanisms of oxidative damage to duplex DNA have been well characterized for bleomycin, Fenton reagents, copper-containing catalysts, or ionizing radiation,21–27 surprisingly little research has been devoted to elucidating the mechanisms of oxidative damage/cleavage of RNA. Although the only chemical difference between the sugar-phosphate backbones of RNA and DNA is the presence of the 2’-hydroxyl (2’-OH) for RNA, RNA typically adopts a much wider variety of secondary and tertiary structures. This structural variability likely complicates reactivity patterns, relative to those observed for duplex DNA, for which firm relationships have been established between its regular double-helical structure and oxidative reactivity patterns, such as hydrogen abstraction.21 Additionally, it is thought that the 2’-OH may endow RNA with some degree of increased resistance to oxidative damage (although increasing susceptibility to hydrolytic scission),28 as a result of electronic inductive effects within the ribose ring or, more simply, the fact that the ribose ring in RNA is, by definition, already further oxidized than the deoxyribose ring in DNA. However, there has remained a persistent need for detailed characterization of RNA oxidation mechanisms, as well as a characterization of how these mechanisms differ among various oxidants.
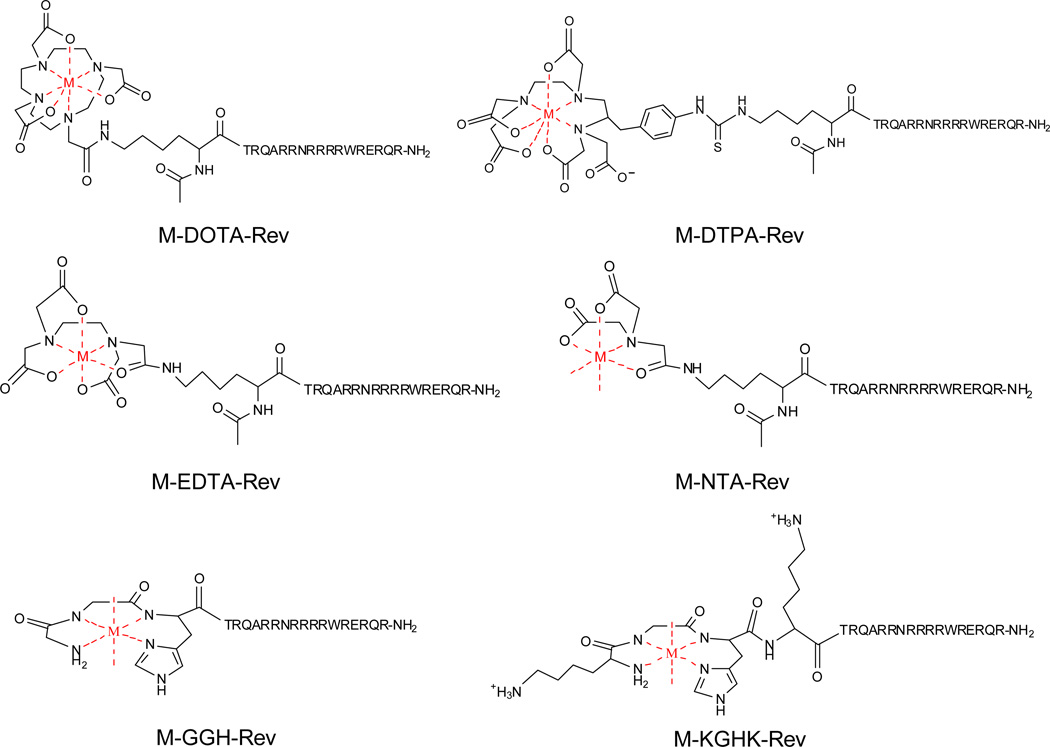
Herein, we report the observed mechanisms of oxidative scission (both qualitatively and semi-quantitatively) of a specific RNA that result from the use of cognate catalytic metallodrugs that target a stem-loop motif of HIV RRE RNA. These metallodrugs contain a variety of oxidative transition metal centers and provide a wide range of oxidative reactivity when used in combination with redox co-reactants. The metallodrugs consist of combinations of Rev-coupled chelators DOTA, DTPA, EDTA, tripeptide GGH, tetrapeptide KGHK, and NTA and the transition metals Fe2+, Co2+, Ni2+, and Cu2+ (Figure 1), for which the synthesis, characterization, and basic reactivity were reported recently, including the influence of sterics, charge and the reduction potential of the metal chelate on the reaction profile.20, 29 The influence of pH on reactivity is not addressed since this influences many of the previously noted parameters. Nevertheless, the catalysts that are the focus of these studies each bind to targeted HIV Rev Response Element (RRE) RNA with high affinity, allowing a controlled screening for catalytic cleavage of the RNA that is unaffected by variability in RNA-binding affinity but is sensitive to systematic variation in the identity of the tethered metal chelate (M-chelate). While our previous studies demonstrated the general reactivity of these catalysts and revealed several interesting relationships, basic limitations of the techniques used prevented any analysis of the mechanisms or specific products of catalyst-mediated RNA cleavage. It is expected that the insights from such studies would be broadly relevant to understanding pathways for oxidative damage of RNA.
Figure 1.
Summary of the metal-chelate-Rev complexes.31, 32 M = Fe2+, Co2+, Ni2+, Cu2+. The sequence of the Rev peptide is shown above. The 7-coordinate geometries shown here for M-DOTA-Rev and M-DTPA-Rev are based on the X-ray crystal structures of [Fe-DOTA]1− and [Fe-DTPA]2−.33, 34
In order to probe the exact mechanisms of oxidative RNA cleavage by these metallodrugs, we used a combination of matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), fluorescence spectroscopy, and gel electrophoresis to monitor cleavage of RRE RNA. The result is a detailed analysis of the mechanisms, kinetics, products, and co-reactant dependence of the RNA cleavage mediated by each of the M-chelate-Rev catalysts. The experiments permitted a dissection of the relative contributions of distinct mechanisms (different modes of oxidative hydrogen abstraction, 2’-hydroxyl-mediated endonucleolysis, and/or hydrolysis) toward observed RNA cleavage, for each of the catalysts. A spatial analysis of the 3D reactivity of each catalyst provided information regarding the status of the reactive oxygen species (ROS) responsible for RNA cleavage. Moreover, a novel application of the standard thiobarbituric acid assay21, 30 is presented that provides a quantitative measure of 4’-hydrogen abstraction from RNA (practically useful for rapid quantification of the extent of cleavage prior to time-consuming gel separations in hydroxyl radical footprinting experiments). The results of this study provide a wealth of information regarding the mechanisms of oxidative scission of RNA.
RESULTS
Analysis of RNA Cleavage by MALDI-TOF MS
The HIV RRE RNA stem loop IIB construct (Fl-RRE RNA) was incubated with each M-chelate-Rev catalyst (Figure 1) under aerobic conditions, with various combinations of added redox co-reactants: 1 mM ascorbate + 1 mM H2O2; 1 mM ascorbate; or no co-reactants. Control experiments were performed that contained no catalyst, no co-reactants, or both; additional control experiments were performed using M-chelates and free metal ions lacking Rev, so that the effect of targeting by Rev could be assessed, as discussed in later sections. Time-dependent experiments were performed for all M-chelate-Rev catalysts and free metal ions.
Following incubation, reaction mixtures were quenched and desalted using C18 zip-tips and analyzed by MALDI-TOF MS. The identities of primary cleavage products (resulting from single-cleavage events) were readily determined by automated mass-matching of observed MS peaks with the masses expected for each primary cleavage product at each position within the RNA sequence, using a mass-matching tolerance of 200 ppm (0.02 % of m/z) and a detection threshold (Figures SM31-SM32, Supporting Information).35 The apparent abundances of each species were semi-quantitatively determined using the MS peak areas (abundance expressed as MS peak area fraction); the resulting data were highly reproducible for individual peak area fractions above 0.02 (Figure SM37, Supporting Information).
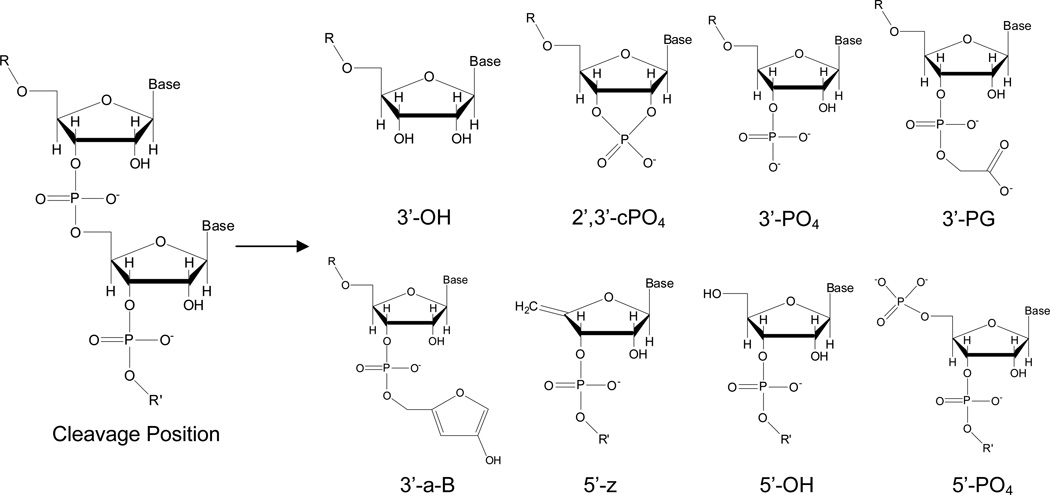
Following RNA strand scission, cleavage fragments of differing masses were observed that corresponded accurately to differing nucleotide lengths and/or differing nascent terminal overhangs (Figure 2, Figure 3). Differing nucleotide lengths corresponded to differing sites of cleavage, and differing nascent terminal overhangs corresponded to differing mechanisms of catalyst-mediated cleavage and/or MALDI-induced background fragmentation at each site (Table 1). At the nascent 3’ termini of 5’ fragments, the monitored overhangs were 3’-hydroxyl (3’OH), 2’,3’-cyclic phosphate (2’,3’-cPO4), 3’-phosphate (3’-PO4), 3’-phosphoglycolate (3’-PG), and 3’-phospho-5-methylene-3-hydroxy-furan (3’-a-B) functional groups; at the nascent 5’ termini of 3’fragments, the monitored overhangs were 4’,5’-alkene (5’-z), 5’-OH, and 5’-PO4 functional groups (summarized in Figure 2).
Figure 2.
RNA fragments monitored by MALDI-TOF MS that resulted from catalyst-mediated scission and/or background fragmentation. R = 5’ fragment. R’ = 3’ fragment.
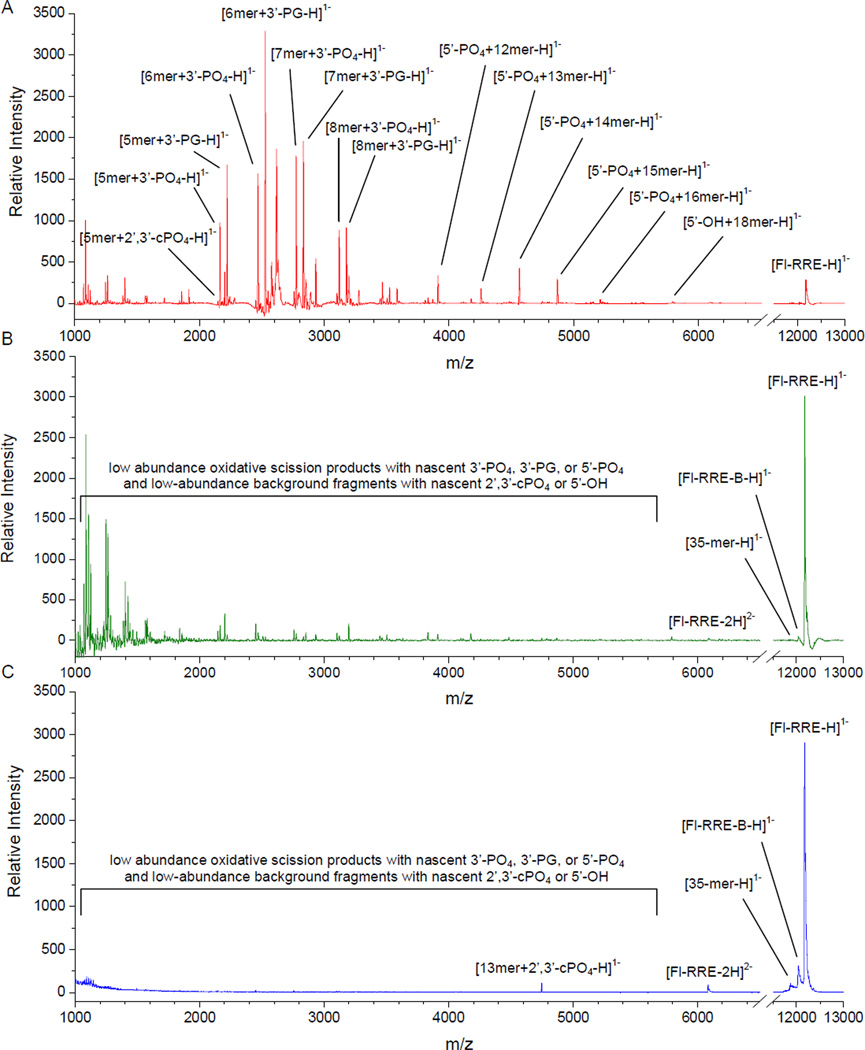
Figure 3.
MALDI-TOF mass spectra. (A) Targeted cleavage of Fl-RRE RNA by Fe-EDTA-Rev, (B) non-targeted cleavage of Fl-RRE RNA by Fe-EDTA lacking Rev, or (C) lack of cleavage in the absence of catalyst, each after a 1 h incubation in the presence of 1 mM ascorbate/H2O2. Mass-matching was performed with a matching tolerance of 200 ppm (0.02 % of the m/z). For predicted masses, see Table SM3 of the Supporting info. Only a small fraction of the product assignments are shown here.
Table 1.
RNA fragments monitored by MALDI-TOF MS, masses of product overhangs, and proposed mechanisms of formation
| terminal overhanga | Δ mass (amu) b | proposed mechanisms of formation |
|---|---|---|
| 3’-OH | 0.00 | hydrolysis,c MALDI d |
| 2’,3’-cPO4 | 61.96 | endonucleolysis,e MALDI d |
| 3’-PO4 | 79.98 | 1’-H, 2’-H, 3’-H, 4’-H, or 5’-H abstraction,f hydrolysis,c MALDI d |
| 3’-PG | 138.02 | 4’-H abstraction f |
| 3’-a-B | 176.06 | MALDI d |
| 5’-z | −18.02 | MALDI d |
| 5’-OH | 0.00 | endonucleolysis,e hydrolysis,c MALDI d |
| 5’-PO4 | 79.98 | 1’-H, 2’-H, 3’-H, 4’-H, or 5’-H abstraction,f hydrolysis,cMALDI d |
See Figure 2.
Relative to the nascent terminal hydroxyl group.
The 3’-OH and 5’-PO4 overhangs are expected as minor hydrolysis products, while hydrolysis is expected to give primarily 3’-PO4 (or 2’-PO4) and 5’-OH overhangs.
From 2’-OH-mediated transesterification.
Reported for the analogous oxidative scission of DNA.21
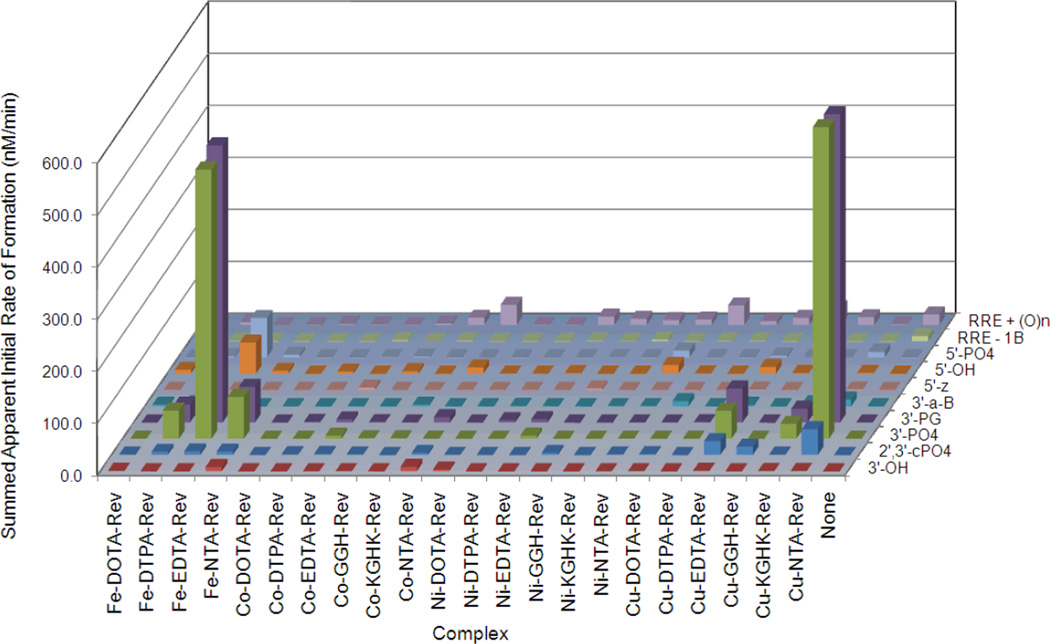
Catalysts that promoted rapid oxidative cleavage of RRE RNA were observed to give cleavage fragments with primarily 3’-PO4, 3’-PG, or 5’-PO4 as nascent terminal overhangs (Figures 2 and 3), consistent with various modes of oxidative hydrogen abstraction (Table 1), as discussed in a later section. Oxidative cleavage, monitored by formation of these overhangs, was most rapid for Cu- or Fe-containing catalysts, especially Cu-NTA-Rev (Figure 6, later), Fe-EDTA-Rev, Cu-KGHK-Rev, Cu-EDTA-Rev, Fe-NTA-Rev, and Fe-DTPA-Rev, although mild levels of oxidative cleavage were also observed for Co- and Ni-EDTA-Rev, as well as Co-NTA-Rev (Figure 7, later).
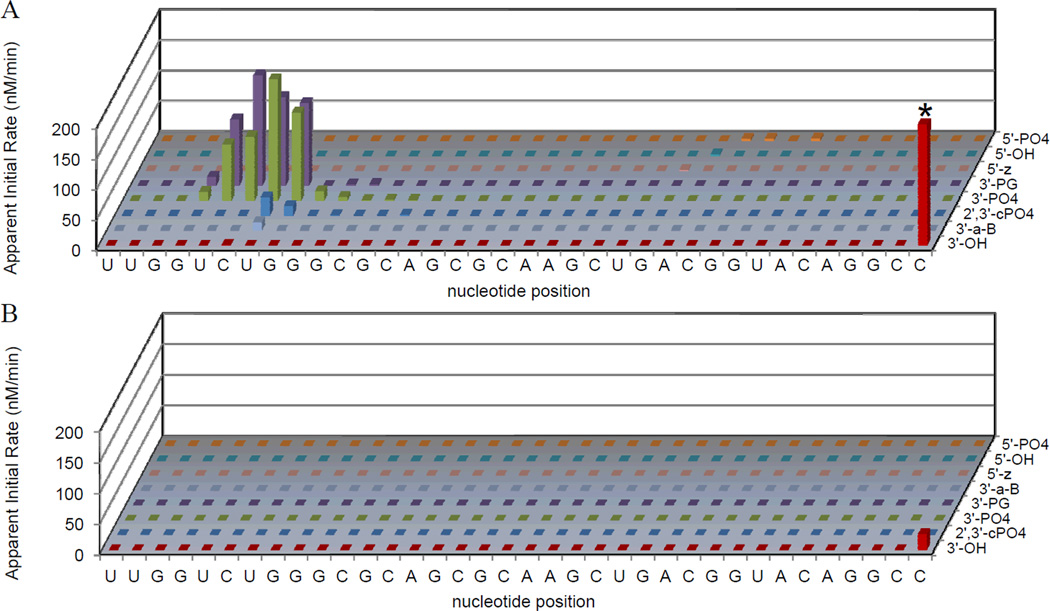
Figure 6.
Apparent initial rates of formation of cleavage products were determined for cleavage at all nucleotide positions within the RRE RNA, and for fragments with each type of nascent terminal overhang, following time-dependent cleavage of Fl-RRE RNA by (A) Cu-NTA-Rev, or (B) no catalyst, each in the presence of co-reactants (1 mM ascorbate/H2O2). Time-dependent experiments were monitored by MALDI-TOF MS (~ 8 mass spectra per catalyst). Rates for all other M-chelate-Rev complexes are shown in the same format in Figures SM1 to SM12 of the Supporting Information. *The apparent initial rate of disappearance of the full-length Fl-RRE RNA is truncated in the figure for Cu-NTA-Rev.
Figure 7.
Breakdown of the apparent initial rates of formation of RNA cleavage fragments containing each type of nascent terminal overhang, resulting from cleavage of Fl-RRE RNA by each M-chelate-Rev complex, in the presence of co-reactants (1 mM ascorbate/H2O2). Time-dependent experiments were monitored by MALDI-TOF MS (typically 8 mass spectra per catalyst). Also included were the apparent initial rates of formation of full-length RNA with either missing bases or one or more additions of oxygen. The quantified nascent terminal overhang types were 3’-OH, 2’,3’-cPO4, 3’-PO4, 3’-PG, 3’-a-B, 5’-z, 5’OH, and 5’-PO4. In order to obtain the apparent initial rate of formation for a given overhang type, the apparent rates of formation for that product type at all positions within the RNA sequence were summed. All rates are listed in Table SM1 of the Supporting Information.
The dependence of oxidative reactivity (H-abstraction events) on the identity of the base immediately attached to the cleaved ribose was analyzed; the only significant relationship was that relatively few oxidative cleavage events occurred at positions containing adenosine (Table SM5, Supporting Information). A similar analysis was conducted for 2’-OH-mediated transesterification reactions, and the only significant dependence on base composition was that relatively few 2’-OH-mediated transesterification events occurred at 3’-phosphates of adenosines. However, it remains unclear whether the apparent reduction in reactivity at adenosine positions was due to either a decreased intrinsic reactivity for adenosine nucleotides or possible skewing by the chance positioning of the bases relative to the catalytic metal centers of bound M-chelate-Rev complexes.
The MALDI-induced background fragments containing 2’,3’-cPO4, 5’-OH, 3’-a-B, and 5’-z overhangs were observed irrespective of whether a catalyst or co-reactants were present. However, several catalysts promoted the formation of identical 2’-OH-mediated transesterification products, containing nascent terminal 2’,3’-cPO4 and 5’-OH, and the extents to which formation of these products was catalyst-mediated were determined by use of time-dependent experiments. Such transesterification reactions appeared most rapid for several of the Cu- and Fe-containing catalysts, as well as free metal ions (Supporting Information). The high co-reactant-dependence of the formation of 3-PO4-containing products suggests that, in the presence of co-reactants, oxidative cleavage occurred at much faster rates than hydrolytic cleavage (both hydrolysis and several oxidative hydrogen-abstraction mechanisms are proposed to give 3’-PO4 overhangs).
The masses of fragments containing nascent 3’ overhangs were observed to increase as the cleavage site moved in the direction 5’ to 3’, in increments corresponding to the mass of each nucleotide, while the masses of fragments containing nascent 5’ overhangs conversely increased in mass as the cleavage site moved in the direction 3’ to 5’. The apparent abundance of each fragment was considered semi-quantitative as a result of the possible skewing of relative abundances during purification, as well as the commonly observed mass bias that gave artificially low abundances for larger fragments, due to an increased susceptibility to MALDI-induced background fragmentation and/or decreased efficiency of desorption/ionization for larger fragments. Mass bias was partially accounted for by the inclusion of MALDI-induced background fragments in calculations of MS peak area fractions. However, the appearance of higher abundances, and higher rates of formation, of 5’-fragments (containing 3’-terminal nascent overhangs), relative to 3’-fragments, most likely resulted from the presence of the 5’-fluorescein label (used for quantification in gel electrophoresis), which may have improved zip-tip retention during desalting and/or improved the efficiency of desorption/ionization for the 5’-fragments (these processes may also be influenced to some degree by the identity of the nascent overhang). This should be considered when comparing rates or abundances between the 5’ and 3’ fragments.
Targeted vs Non-Targeted Cleavage
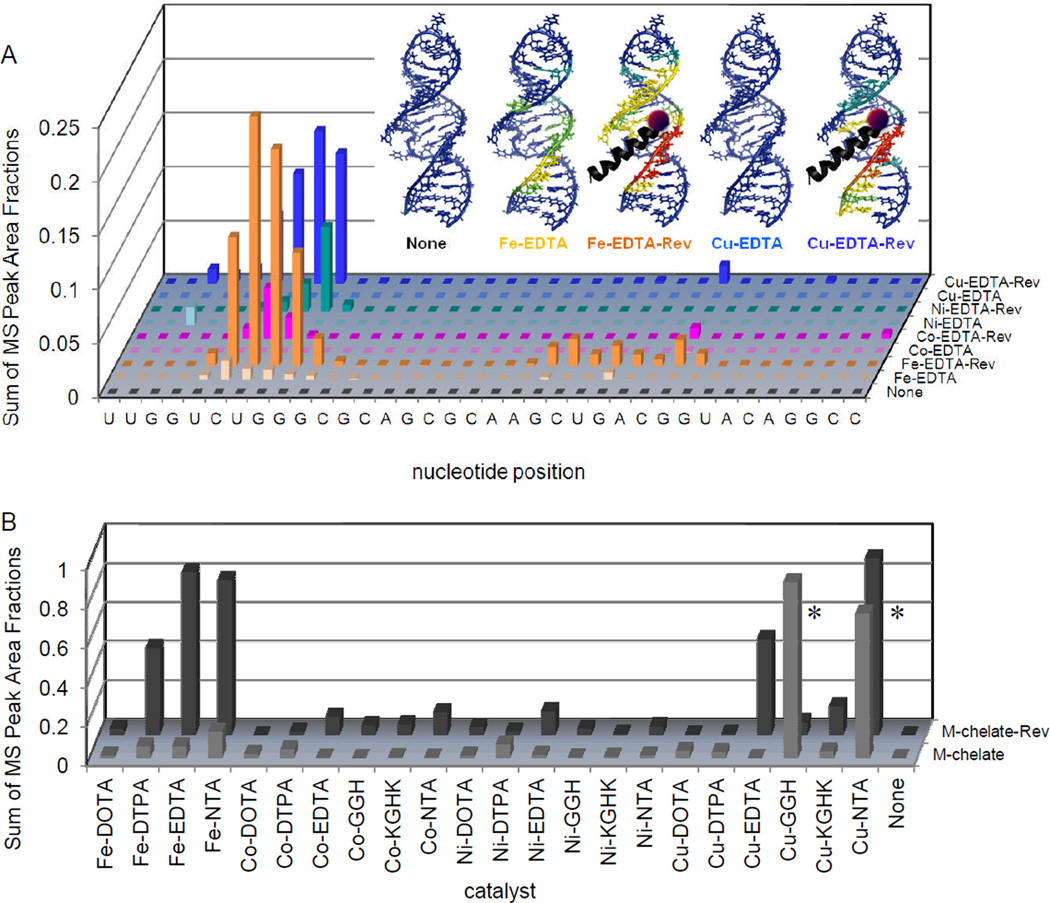
Comparison of the RNA-cleavage reactivity of M-chelate-Rev catalysts vs M-chelates lacking Rev demonstrated less random and much higher rates of RNA cleavage for catalysts containing Rev, due to targeting of the RRE RNA through high-affinity binding (KD ~ 1 nM).20 The MALDI-TOF mass spectra in Figure 3 show the distributions and relative abundances of RNA cleavage products resulting from incubation of RRE RNA with co-reacants and either Fe-EDTA-Rev, Fe-EDTA lacking Rev, or no catalyst—cleavage was clearly more efficient for Fe-EDTA-Rev (Figure 3A) than for Fe-EDTA (Figure 3B) and obviously depended on the presence of catalyst. Figure 4A shows the results of MALDI-TOF MS analysis for each M-EDTA-Rev catalyst vs each M-EDTA catalyst lacking Rev, with sums of peak area fractions shown for fragments containing 3’-PO4, 3’-PG, or 5’-PO4 overhangs that derive from each illustrated site of initial hydrogen abstraction (mechanisms of hydrogen-abstraction are provided later in Scheme 1 and Supporting Information). Furthermore, mapping of the oxidative reactivity of M-chelate-Rev catalysts onto the NMR solution structure of the RRE RNA (based on the complex with Rev peptide)10 clearly indicates that the observed sites of cleavage (two regions) align with predicted sites for each Rev-coupled metal center (see Figure 4A), consistent with our energy-minimized 3D models of the M-chelate-Rev/RRE complexes.20 Comparisons of each M-chelate-Rev catalyst vs each respective M-chelate lacking Rev are summarized in Figure 4B and Figures SM15-SM17 of the Supporting Information. Exceptions were observed for the complexes Cu-GGH, Cu-NTA, and free Cu, and the relatively high activity of these complexes lacking Rev was due to the combination of a low-affinity Cu-binding site within the RRE RNA (Figures SM26 and SM27, Supporting Information), and the high concentrations of catalyst and RNA required for MALDI-TOF MS detection, as discussed later.
Figure 4.
Targeted cleavage of RRE RNA required attachment of catalysts to the Rev peptide in order to achieve efficient cleavage, as seen by differences in reactivity between the M-chelate-Rev catalysts and M-chelates lacking Rev. After 1 h incubation of each catalyst (10 µM) and co-reactants (1 mM ascorbate/H2O2) with the Fl-RRE RNA (10 µM), the apparent abundances of individual RNA cleavage fragments were quantified. (A) The data correspond to the abundances (MS peak area fractions) of oxidation products (cleavage fragments containing nascent terminal 3’-PO4, 3’-PG, or 5’-PO4 overhangs), at positions adjacent to each illustrated site of H-abstraction, after 1h reaction. Inset: The same data were mapped onto the 3D solution structure of the RRE RNA,10 for several catalysts and the control catalysts lacking the Rev targeting domain (red corresponds to highest reactivity; blue corresponds to reactivity below background). (B) Comparison of the reactivity of each M-chelate-Rev catalyst vs each M-chelate lacking Rev, in the presence of co-reactants, after 1h reaction. Plots similar to that shown in (A) are shown for all catalysts in Figures SM15 to SM17 of the Supporting Information. * The relatively high activity of the Cu-GGH and Cu-NTA chelates lacking Rev was the result of the combination of a low-affinity Cu-binding site within the RRE RNA and the high concentrations of catalyst and RNA required for MALDI-TOF MS detection.
Scheme 1.
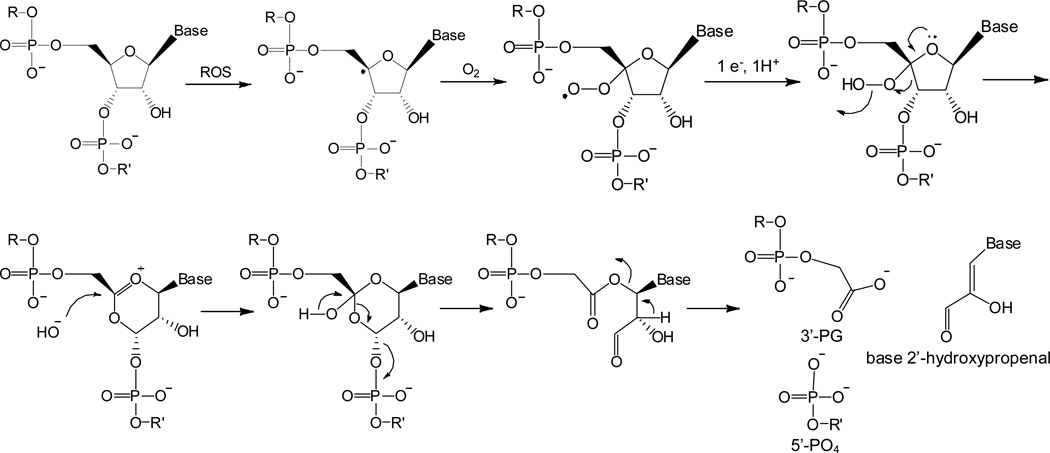
Proposed mechanism of 4’-hydrogen abstraction from RNA, forming base 2-hydroxylpropenals and cleavage fragments containing nascent 3’-PG and 5’-PO4 terminal overhangs (analogous to proposed mechanisms of DNA scission initiated by 4’-H abstraction).39 Other mechanistic variations are possible that give the same products, such as one involving the formation of a tetraoxide intermediate.21 The ROS responsible for 4’-H abstraction were most likely hydroxyl and/or superoxide radicals.
Dependence of Reactivity on Co-reactants
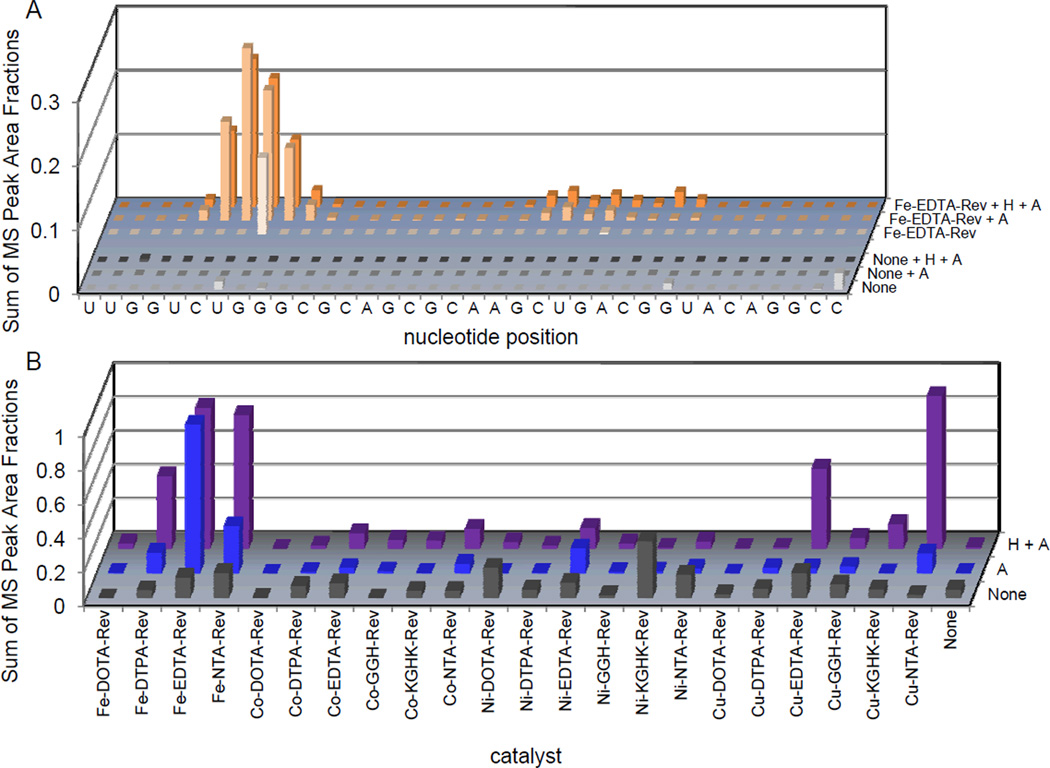
Oxidative cleavage of the RRE RNA by M-chelate-Rev catalysts was highly dependent on the presence of co-reactants, as shown in Figure 5A. However, the co-reactant dependence varied from catalyst to catalyst, as shown in Figure 5B. Oxidative cleavage was typically fastest when both H2O2 and ascorbate were present. However, certain catalysts, such as Fe-EDTA-Rev produced approximately the same abundance of oxidative cleavage fragments after 1h reaction, irrespective of whether H2O2 was present.
Figure 5.
Catalyst-mediated oxidative cleavage of the Fl-RRE RNA was dependent on the presence of redox co-reactants. (A) The dependence of the abundance (MS peak area fractions) of oxidation products (cleavage fragments containing nascent terminal 3’-PO4, 3’-PG, or 5’-PO4 overhangs at positions adjacent to each illustrated site of H-abstraction) on the co-reactant combinations used (1 mM each co-reactant), for Fe-EDTA-Rev and for the control reactions lacking catalysts, after 1h reaction. (B) The dependence of the total relative abundance of oxidation products (sequence-summed) on the co-reactant combinations used, for all complexes, after 1h reaction. Three different co-reactant combinations were used: 1 mM H2O2 + 1 mM ascorbate (H + A), 1 mM ascorbate (A), or no co-reactants (none). Co-reactant-dependence plots are shown for other catalysts in Figures SM18 to SM25 of the Supporting Information.
Time-Dependence of Cleavage and Analysis of Product Distributions
Time-dependent experiments were used to obtain apparent initial rates of formation of RNA cleavage products for reactions containing redox co-reactants and each of the M-chelate-Rev catalysts, as well as the free metal ions. For each time-point in an incubation, a MALDI-TOF mass spectrum was used to identify and semi-quantitatively determine the abundance of each RNA cleavage product; the use of ~ 8 time-points (8 mass spectra) allowed apparent initial rates of formation to be established for each product. An example of the time-dependent cleavage of RRE RNA by Cu-NTA-Rev (as well as the control lacking catalyst) and co-reactants is shown in Figure 6. The use of apparent initial rates of product formation gave a robust comparison between fragments of differing nascent overhang types. Time-dependent cleavage of Fl-RRE was monitored by both MALDI-TOF MS and PAGE analysis, and time-dependent modification of AP-RRE, which contains an internal 2-aminopurine label,9, 20 was monitored by real-time fluorimetry—all rates are compared in Table SM2 of the Supporting Information.
For each catalyst, the rates of formation of each type of overhang were summed across the whole RRE RNA sequence, allowing a detailed analysis of the distributions of the apparent initial rates of formation for differing overhang types (Figure 7). These data provided useful mechanistic insights, including an approximation of the relative rates of hydrogen abstraction, hydrolysis, 2’-OH-mediated transesterification, and/or other mechanisms. Additionally, the data provide a comparison between catalysts of the overall rates of RNA cleavage. Importantly, the rates shown in Figure 7 are relatively unaffected by mass bias, since they were summed across the entire RNA sequence, although care should be exercised when comparing rates between 5’- and 3’- fragments.
Detection of Base 2’-hydroxypropenals
Upon reaction with the RRE RNA, many of the M-chelate-Rev catalysts were observed to give rise to 3’-PG overhangs at cleavage positions (Figure 7), a unique product of 4’-H abstraction (Scheme 1). Also predicted to arise from 4’-H abstraction from RNA are base 2-hydroxypropenals (analogous to base propenals formed upon 4’-H abstraction from DNA) and 5’-PO4 overhangs, as shown in Scheme 1.
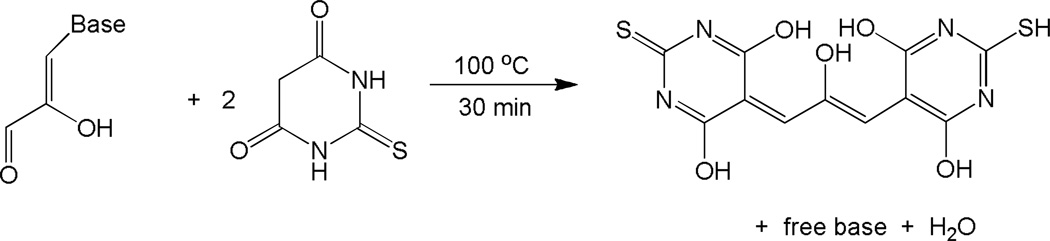
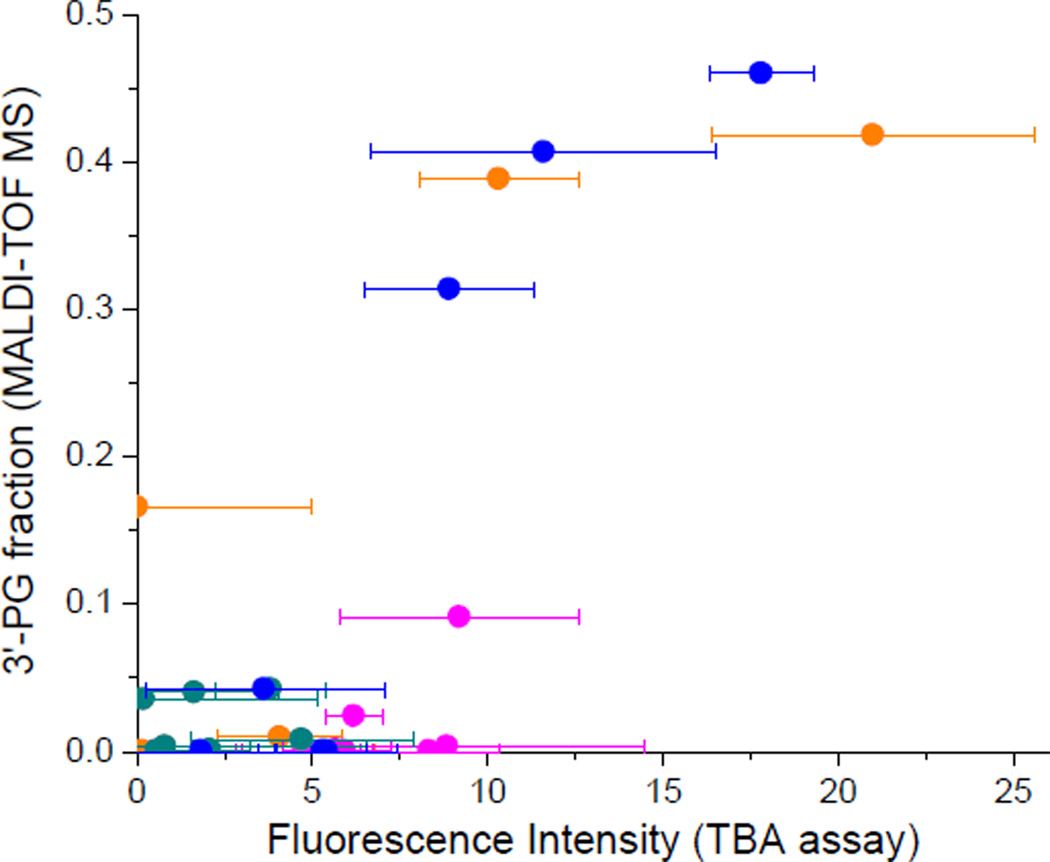
The thiobarbituric acid (TBA) assay was used to determine the relative abundance of base 2-hydroxypropenals formed after 1 h incubation of RRE RNA with each M-chelate-Rev catalyst and co-reactants, under conditions similar to those used for MALDI-TOF-monitored cleavage. After each 1 h incubation, the reaction mixture was boiled for 30 min in the presence of TBA under acidic conditions, resulting in the decomposition of base 2-hydroxypropenals and ultimately forming a colored/fluorescent 2-hydroxypropene-bis(thiobarbituric acid) adduct (Scheme 2). This adduct was quantified by fluorescence detection to determine the relative abundance of base 2-hydroxypropenals that formed after 1 h incubation of RRE RNA with each M-chelate-Rev catalyst and co-reactants. A correlation was observed between the abundance of base 2-hydroxypropenals (monitored by TBA assay) and the abundance of 3’-PG overhangs (monitored by MS peak area fractions) that formed after 1 h incubation, confirming 4’-H abstraction as a major mechanism of oxidative scission of the RRE RNA by M-chelate-Rev catalysts (Figure 8). Formation of the fluorescent TBA adduct was further verified by reaction of 2-hydroxypropanedial with TBA under the same reaction conditions to give a colored/fluorescent product with the expected mass of 339 amu (verified by ESI-MS, Supporting Information).
Scheme 2.
Reaction of base 2-hydroxypropenals with thiobarbituric acid to form the fluorescent 2-hydroxypropene-bis(thiobarbituric acid) adduct.
Figure 8.
Correlation between the abundance of 3’-PG overhangs and the abundance of base 2-hydroxypropenals, following 4’-hydrogen abstraction from RNA. Following 1 h reaction of RRE RNA (10 µM) with each M-chelate-Rev catalyst (10 µM) and the co-reactants ascorbate and H2O2 (1 mM each), the apparent abundance of fragments containing nascent 3’-PG overhangs was determined by MALDI-TOF MS (y-axis), and the relative abundance of 2-hydroxypropenals was determined by the TBA assay (x-axis). Each point above represents a unique catalyst, and the colors shown correspond to the metal in each catalyst: orange, Fe; pink, Co; green, Ni; blue, Cu. The limit of detection for the TBA assay was 3.6 fluorescence intensity units.
Reactivity of Cu2+(aq) and Coordinatively-Unsaturated Copper Complexes
Binding of free metal ions to RRE RNA was monitored by titration of AP-RRE, which contains an internal 2-aminopurine (2-AP) substitution, by free metal ions (and Cu-NTA and Cu-GGH); the fluorescence of AP-RRE was monitored throughout each titration. Among the free metal ions, free Cu2+(aq) appeared to possess the highest affinity for AP-RRE, with an apparent KD ~ 5 µM. Binding of AP-RRE by free Cu2+(aq) appeared to occur with high cooperativity, with apparent binding of multiple Cu2+ ions to each AP-RRE molecule. Cooperativity of Cu-binding is supported by both titration of AP-RRE with free Cu2+ (Figure SM26, Supporting Information) and stopped-flow analysis of the kinetics of free Cu2+ binding to AP-RRE (fit to a pentaphasic first order kinetic model, Figure SM26). Stopped-flow analysis revealed five discreet events during the binding interaction, and the titration revealed a sigmoidal titration response curve. These results suggest initial binding of free Cu2+(aq) to at least one site that did not alter the fluorescence of the 2-aminopurine probe (2-AP), with one or more subsequent cooperative bindings of Cu2+ that resulted in quenching of the fluorescence of the 2-AP probe. Relative to free Cu2+, lower-affinity binding was observed for Cu-NTA, free Fe2+, free Co2+, and free Ni2+. Although no binding was directly observed for Cu-GGH upon titration into AP-RRE, it is likely (based on cleavage studies) that Cu-GGH bound AP-RRE in a manner that did not alter the fluorescence of the AP probe. The distributions of RNA cleavage products that arose following cleavage of RRE RNA by free Cu2+(aq), Cu-GGH, and Cu-NTA, which each possess non-chelated Cu-coordination sites, are consistent with Cu-binding within the bulge of stem loop IIB of the RRE RNA (Figures SM24, SM25, and SM28).
DISCUSSION
Mechanisms of RNA Cleavage
The selection of co-reagents and concentrations for these studies was based on several factors. First, the choice allows a careful comparison with the results of previous studies that employed the same co-reactant concentrations. Moreover, classical hydroxyl radical footprinting studies typically employ a concentration of H2O2 and/or ascorbic acid of ~ 1 mM, the same as used in this study, thereby increasing the broad relevance of our findings. Second, both ascorbic acid (vitamin C) and H2O2 are present physiologically, on the order of 1 mM and 1 µM, 40, 41 respectively, and are expected to be available for any chemistry that occurs in vivo, although other oxidants/reductants could also be involved. While the metallodrug catalyst can mediate the production of peroxide from ascorbate and dioxygen, 20, 29 we typically add peroxide to promote a more rapid reaction (along the lines of a peroxide “shunt” commonly used in studies of heme peroxidase mimics 42, 43).
The cleavage products identified by MALDI-TOF MS analysis were indicative of a variety of mechanisms of RNA cleavage, including oxidative pathways initiated by hydrogen abstraction, as well as hydrolysis, 2’-OH-mediated endonucleolysis, and various types of MALDI-induced background fragmentation. Table 1 summarizes the most likely mechanisms of formation of each of the monitored overhang products, based on a pool of previous studies of DNA/RNA cleavage,21, 26, 27, 39 and mass spectrometric analysis of RNA.35–38 In our studies, the most rapid mechanisms of catalyst-mediated RNA cleavage were those initiated by oxidative hydrogen abstraction. Each hydrogen abstraction event was most likely mediated by an intermediate reactive oxygen species (ROS), produced at the metal center of reactive catalysts, by single-electron reduction of either dioxygen or peroxide. Ascorbic acid, a single-electron reducing agent, functioned to reduce the oxidized metal centers, following each formation of ROS and allowing multiple redox cycles to occur.29, 44, 45
Oxidative Hydrogen Abstraction from RNA
It is clear from our studies that a major pathway for oxidative RNA cleavage by the M-chelate-Rev catalysts is via 4’-H abstraction, as evidenced by the formation of signature 4’-H abstraction products: cleavage fragments terminated with nascent 3’-PG overhangs and base 2-hydroxypropenals. The observed correlation between their relative abundances following cleavage reactions further punctuates this point. Additionally, the non-unique products of 4’-H abstraction (5’-PO4 overhangs) were also observed. A proposed mechanism of 4’-hydrogen abstraction from RNA by ROS produced at catalytic metal centers is shown in Scheme 1; all of the proposed products (3’-PG, 5’-PO4, and 2’-hydroxypropenal) were observed.
In addition to clear evidence for 4’-H abstraction from RNA, the rapid formation of fragments with nascent terminal 3’-PO4 and 5’-PO4 overhangs is consistent with the possible abstraction of other hydrogen atoms (1’, 2’, 3’, and 5’), based on analogous studies of hydrogen abstraction from DNA (Table 1). However, neither of these overhangs are known to be unique to any single H-abstraction pathway, unlike the products of 4’-H abstraction. Among the possible remaining catalyst-mediated H-abstraction pathways for RNA, 5’-H abstraction is most likely (proposed mechanism shown in Figure SM34, Supporting Information). Consistent with this claim, previous studies have indicated that hydroxyl radicals generated in bulk solution by either gamma radiolysis or Fenton reagents react with DNA to generate primarily 3’-PO4, 3’-PG, and 5’-PO4 overhangs26, 27 and show a preference for hydrogen abstraction in the order 5’-H > 4’-H > 3’-H ~ 2’-H > 1’-H.21 The presence of two hydrogen atoms at each 5’ position vs only one at each of the other ribose positions, as well as the relatively high solvent-accessibility of 5’-H atoms, increases the likelihood of 5’-H abstraction as a major mechanism of RNA cleavage.
Although it is theoretically possible that 3’-PO4 overhangs could arise as a side reaction of 4’-H abstraction, this is unlikely to be the predominant cause of 3’-PO4 overhangs in our studies, since the ratio of 3’-PG and 3’-PO4 overhangs at each position typically appeared to vary with the helicity of the RNA—in Figure 6A, note that the rate of formation of 3’-PO4 overhangs upon reaction with Cu-NTA-Rev and co-reactants was highest at nucleotide position U7, whereas the rate of formation of 3’-PG overhangs was fastest at position C6. Variation in the ratio of rates of formation of 3’-PG and 3’-PO4 overhang products as a function of RNA sequence position (and helicity) most likely arises from the fact that at any given position, either the 4’-H atom or one of the two 5’-H atoms is closer to the metal-associated ROS, and depending on which is closer, the corresponding overhang is generated more rapidly at that position. In summary, oxidative scission of HIV RRE RNA by M-chelate-Rev complexes appears to result from primarily 4’-H and 5’-H abstraction events, and these hydrogen abstraction events were mediated by ROS, such as hydroxyl and/or superoxide radicals.
Distance-Dependence of 4’-Hydrogen Abstraction
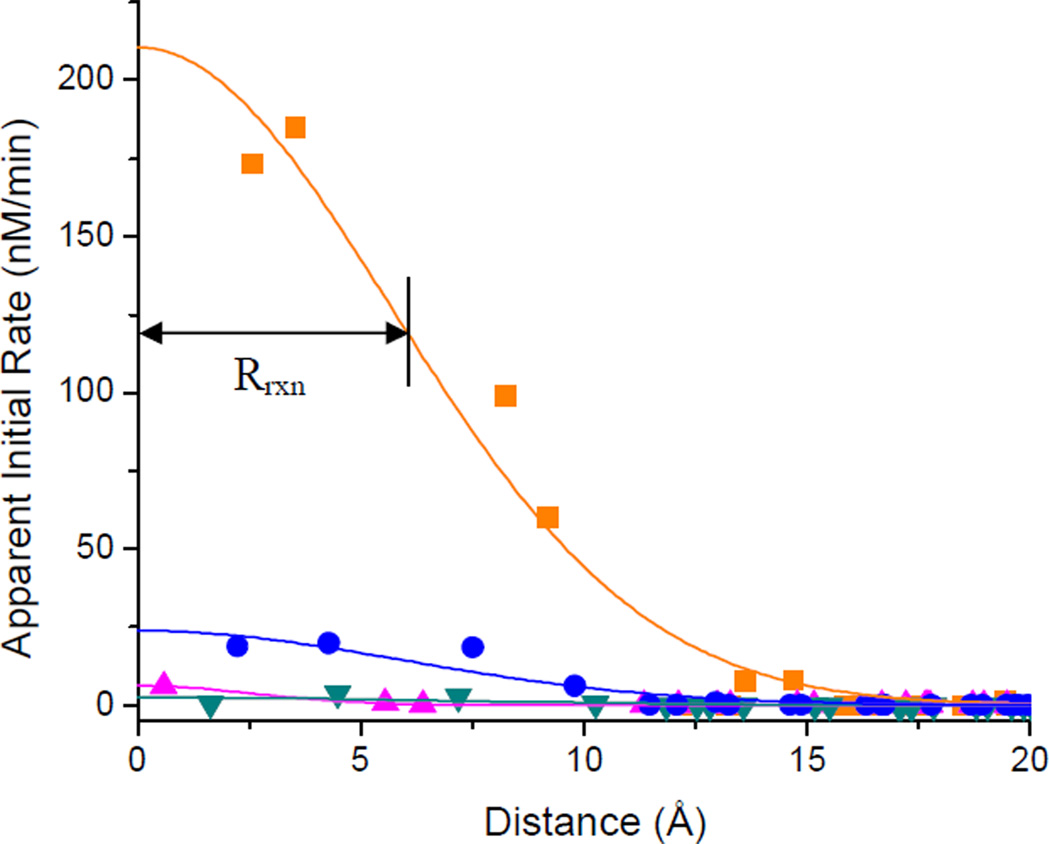
The ROS responsible for hydrogen abstraction originated at the metal center of each redox-active M-chelate-Rev catalyst. In order to characterize the diffusion radius of these ROS, the degree to which these ROS were metal-associated vs metal-dissociated, and/or the flexibility of the M-chelate-Rev scaffold, we established a relationship between the distance of each 4’-position from the metal center and the apparent initial rate of 4’-H abstraction at each respective 4’-position in the RRE RNA, separately for each M-chelate-Rev catalyst, as described in the Supporting Information. The position of the metal center was calculated by use of a weighted average of the Cartesian coordinates of each 4’-position in the RRE/Rev NMR solution structure, using the initial rates of formation of corresponding 3’-PG overhangs as weights (it was assumed that the attachment of M-chelates to Rev did not significantly perturb the RRE/Rev structure). The apparent initial rates of formation of 3’-PG overhangs were then plotted as a function of the distance between the metal center and corresponding 4’-positions, and the resulting curve was fit to a Gaussian equation; from this curve a Gaussian radius (Rrxn) was calculated (Figure 9). The apparent initial rates of 4’-H abstraction within this radius accounted for ~70% of the total apparent rate of 4’-H abstraction (total shown in Figure 7 for each M-chelate-Rev catalyst). The resulting radii of reactivity are summarized in Table 2. On a related note, no dependence was observed on the metals’ ionic radii (Table SM6, Supporting information), as expected.
Figure 9.
Dependence of the apparent rate of 4’-H abstraction on the distance from the metal center for each M-chelate-Rev catalyst (with 1 mM ascorbate/H2O2). A Gaussian radius (Rrxn) was calculated for each curve. The M-EDTA-Rev species shown here are: Fe-EDTA-Rev, orange ■; Co-EDTA-Rev, pink ▲; Ni-EDTA-Rev, green ▼; Cu-EDTA-Rev, blue ●.
Table 2.
Summary of the distance-dependence of catalyst-promoted 4’-hydrogen abstraction.
| complex | Rrxn (Å)a |
|---|---|
| Fe-DOTA-Rev | 2 ± 1 |
| Fe-DTPA-Rev | 5 ± 1 |
| Co-DTPA-Rev | < 5 |
| Ni-DTPA-Rev | < 6 |
| Cu-DTPA-Rev | < 6 |
| Fe-EDTA-Rev | 6 ± 1 |
| Co-EDTA-Rev | 3 ± 1 |
| Ni-EDTA-Rev | 7 ± 1 |
| Cu-EDTA-Rev | 6 ± 1 |
| Cu-KGHK-Rev | 5 ± 1 |
| Fe-NTA-Rev | 5 ± 1 |
| Co-NTA-Rev | 4 ± 1 |
| Cu-NTA-Rev | 6 ± 1 |
Radius obtained by fitting to a Gaussian distribution; the apparent rates of 4’-H abstraction within this radius accounted for ~70% of the total apparent rate of 4’-H abstraction for each M-chelate-Rev catalyst. A larger radius is attributed to either a metal-dissociated ROS as the reactive intermediate or greater flexibility of the linker between the metal center and Rev. The reactions contained both H2O2 and ascorbate (1 mM each).
Reported diffusion radii for hydroxyl radicals have ranged from ~ 10 Å, prior to reaction with alcohol-containing radical scavengers and/or DNA,46 to ~ 100 Å or ~10000 Å if scavenging species (organics, other radicals, or H2O2) were present at 10−3 or 10−6 M, respectively.47 In our experiments, the observed radii for 4’-H abstraction by M-chelate-Rev catalysts varied from ~2 Å to ~7 Å, demonstrating the high probability that the ROS responsible for 4’-H abstraction were either metal-associated or had very short diffusion pathlengths (< 7 Å) and were distinct from bulk solution ROS, especially considering the flexibility of each Rev-tethered M-chelate (several Å). This conclusion is consistent with the requirement for attachment of M-chelates to Rev to achieve rapid cleavage of RRE RNA.
Reactivity with Co-Reactants
Comparison of the apparent initial rates of catalyst-mediated RNA cleavage obtained for 10 µM catalyst:RNA complex (monitored by MALDI-TOF MS) with those obtained previously for 0.1 µM catalyst:RNA complex (monitored by PAGE)20 provided a glimpse of the variation in second-order reactivity between catalysts and fixed concentrations of the co-reactants ascorbate and H2O2. Upon a 100-fold increase in catalyst/RNA concentration, the initial rate of RNA cleavage was expected to increase by 100-fold to 10000-fold, assuming an absence of scavenging of ROS and/or other complications. These increases were expected due to the 100-fold higher concentration of catalyst:RNA complex (baseline increase) in combination with an extra 1- to 100-fold increase resulting from variable second-order reactivity of catalysts with the co-reactants H2O2 and/or ascorbate, which depended on the rate limiting step of the redox cycle and/or subsequent RNA cleavage for each catalyst. In fact, a 100-fold to 1000-fold increase in the apparent initial rates of RNA cleavage was most common (Table SM4, Supporting Information). However, an increase of ≥10,000-fold was observed for Fe-EDTA-Rev, which implies scavenging of ROS at low catalyst concentration (low flux of ROS), most likely by ascorbate or HEPES, that became less efficient at higher catalyst concentration due to a higher flux of ROS. The distinct behavior for Fe-EDTA-Rev suggests that the ROS produced by the Fe-EDTA center were more metal-dissociated (distinct from bulk solution ROS), rather than metal-associated, leaving a greater opportunity for scavenging by ascorbate (or other non-RNA species) than for the other catalysts, consistent with previous studies.20, 29, 31 This scavenging effect is complementary to the previously reported concentration-dependent shift of ascorbate from pro-oxidant (low concentration of ascorbate) to anti-oxidant (high concentration of ascorbate), in the presence of redox-active transition metals.44 The dependence of the initial rate of RNA cleavage on the concentration of catalyst was studied in further detail for both Fe-EDTA-Rev and Cu-NTA-Rev, with monitoring by PAGE analysis (Figure SM33).
Binding and Cleavage of RRE RNA by Coordinatively-Unsaturated Copper Complexes
Another effect of the elevated concentrations of catalyst/RNA used for MS-monitored experiments was that, in the presence of both H2O2 and ascorbate, the catalysts Cu-GGH, Cu-NTA, and free Cu2+(aq) were each observed to rapidly cleave HIV RRE RNA, even without attachment to the Rev peptide. This non-targeted cleavage occurred because the relatively high concentrations of catalyst and RNA used for reactions (10 µM) were higher than the KD of a low-affinity Cu-binding site (KD ~ 5 µM) within the stem loop bulge of the RRE RNA. This binding was detected by fluorescence-monitored titrations and stopped-flow experiments (Figure SM26, Supporting Information), as well as by analysis of cleavage data (Figure SM27).
Cu-GGH and Cu-NTA are each similar to free Cu2+ in that these complexes each possess non-chelated metal-coordination sites, which likely allow coordination to the RRE RNA, unlike the other Cu-chelates. The non-targeted reactivity of these Cu2+ complexes was highly dependent on the presence of H2O2; in the absence of H2O2, cleavage by Cu-NTA and Cu-GGH was minor (Figure SM25, Supporting Information), consistent with previous observations by our laboratory.18
CONCLUSIONS
Herein we have provided a detailed analysis of the kinetics and mechanisms of oxidative cleavage of HIV RRE RNA by a series of M-chelate-Rev catalysts (members of a class of reagents that we have defined as catalytic metallodrugs17, 48). By use of MALDI-TOF MS and other techniques, we directly observed RNA cleavage products corresponding to catalyst-mediated oxidative hydrogen abstraction, hydrolysis, and/or 2’-OH-mediated endonucleolysis. Among the oxidation pathways, 4’-hydrogen abstraction was identified as a major mechanism of hydrogen abstraction from RNA, as evidenced by the rapid co-reactant-dependent formation of RNA cleavage fragments containing nascent terminal 3’-phosphoglycolate overhangs and production of base 2-hydroxypropenals (both unique to 4’-H abstraction), which were detected by a novel use of the standard thiobarbituric acid assay for detection of RNA cleavage products. Oxidative abstraction of 5’-H atoms was also highly likely, given the rapid co-reactant-dependent formation of RNA cleavage fragments containing nascent terminal 3’-phosphate or 5’-phosphate overhangs. Quantitative evidence is provided that the ROS responsible for RNA cleavage were primarily metal-associated, based on the distance-dependence of 4’-H abstraction. Comparison of all M-chelate-Rev catalysts demonstrated that the highest RNA cleavage activity was observed for Fe- and Cu-containing catalysts, and the oxidative mechanism(s) employed by these catalysts were highly dependent on the presence of redox co-reactants, such as the mild reducing agent ascorbic acid and/or the oxidants O2 or H2O2. Evidence was also provided for mild levels of catalyst-mediated hydrolytic and/or 2’-OH-mediated endonucleolytic scission. The results of this study provide broadly relevant insights into the mechanisms of transition metal-mediated oxidative cleavage of RNA.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [HL093446 and AA016712]. Jeff Joyner was supported by an NIH Chemistry/Biology Interface training grant (T32 GM08512). The Bruker Microflex instrument used for all MALDI-TOF analysis was provided by a grant from the Ohio BioProducts Innovation Center.
Footnotes
Supporting Information Available. Materials and methods, apparent initial rates of RNA cleavage for all M-chelate-Rev catalysts and free metal ions (organized by overhang type and nucleotide position), comparison of RNA cleavage for all M-chelate-Rev catalysts vs M-chelates lacking Rev, comparison of RNA cleavage for all co-reactant conditions for all M-chelate-Rev catalysts, characterization of the Cu-binding site in RRE RNA by fluorescence-monitored titrations, stopped-flow analyses, and RNA cleavage rates, characterization of the 2-hydroxypropanedial(TBA)2 adduct, concentration dependence experiments, and rate law for catalyst-mediated RNA cleavage.
REFERENCES
- 1.Shan X. PhD Dissertation. The Ohio State University; 2005. [Google Scholar]
- 2.Malim MH, Hauber J, Le S, Maizel J, Cullen B. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 3.Wurtmann E, Wolin S. Crit. Rev. Biochem. Mol. Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Chock B, Stadtman E. Proc. Natl. Acad. Sci. USA. 2006;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei B, Wei Y, Zhang K, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L, Zhang R, Li J. Biomed. Pharmacother. 2009;63:313–318. doi: 10.1016/j.biopha.2008.07.086. [DOI] [PubMed] [Google Scholar]
- 6.Potenza N, Moggio L, Milano G, Salvatore V, Blasio BD, Russo A, Messere A. Int. J. Mol. Sci. 2008;9:299–315. doi: 10.3390/ijms9030299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luedtke NW, Tor Y. Biopolymers. 2003;70:103–119. doi: 10.1002/bip.10428. [DOI] [PubMed] [Google Scholar]
- 8.Long EC. Acc. Chem. Res. 1999;32:827–836. [Google Scholar]
- 9.Lacourciere KA, Stivers JT, Marino JP. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 10.Battiste J, Mao H, Rao N, Tan R, Muhandiram D, Kay L, Frankel A, Williamson J. Science. 1996;273:1547. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 11.Cowan JA, Ohyama T, Wang D, Natarajan K. Nucl. Acids Res. 2000;28:2935–2942. doi: 10.1093/nar/28.15.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondi CS, Rao JS. J. Cell Physiol. 2009;220:285–291. doi: 10.1002/jcp.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura K, Komiyama M. J. Biochem. 1997;122:387–394. doi: 10.1093/oxfordjournals.jbchem.a021765. [DOI] [PubMed] [Google Scholar]
- 14.Iranzo O, Elmer T, Richard J, Morrow J. Inorg. Chem. 2003;42:7737–7746. doi: 10.1021/ic030131b. [DOI] [PubMed] [Google Scholar]
- 15.Bradford S, Kawarasaki Y, Cowan JA. J. Inorg. Biochem. 2009;103:871–875. doi: 10.1016/j.jinorgbio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Cowan JA. Pure Appl. Chem. 2008;80:1799–1810. [Google Scholar]
- 17.Hocharoen L, Cowan JA. Chem. Eur. J. 2009;15:8670–8676. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y, Cowan JA. J. Am. Chem. Soc. 2006;128:410–411. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, Cowan JA. J. Biol. Inorg. Chem. 2007;12:637–644. doi: 10.1007/s00775-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 20.Joyner JC, Cowan JA. J. Am. Chem. Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogozelski W, Tullius T. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 22.Cooke M, Evans M, Dizdaroglu M, Lunec J. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y, Cowan JA. J. Am. Chem. Soc. 2005;127:8408–8415. doi: 10.1021/ja0503985. [DOI] [PubMed] [Google Scholar]
- 24.Angeloff A, Dubey I, Pratviel G, Bernadou J, Meunier B. Chem. Res. Toxicol. 2001;14:1413–1420. doi: 10.1021/tx0100800. [DOI] [PubMed] [Google Scholar]
- 25.Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. Mutat. Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Henner WD, Grunberg SM, Haseltine WA. J. Biol. Chem. 1982;257:11750–11754. [PubMed] [Google Scholar]
- 27.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 28.Thorp HH. Chem. Biol. 2000;7:R33–R36. doi: 10.1016/s1074-5521(00)00080-6. [DOI] [PubMed] [Google Scholar]
- 29.Joyner JC, Reichfield J, Cowan JA. J. Am. Chem. Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janero DR. Free Radical Biol. Med. 1990;9:515–540. doi: 10.1016/0891-5849(90)90131-2. [DOI] [PubMed] [Google Scholar]
- 31.Joyner JC, Keuper KD, Cowan JA. Dalton Trans. 2012;41:6567–6578. doi: 10.1039/c2dt00026a. [DOI] [PubMed] [Google Scholar]
- 32.Joyner JC, Cowan JA. J. Am. Chem. Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finnen D, Pinkerton A, Dunham W, Sands R, Funk M. Inorg. Chem. 1990;30:3960–3964. [Google Scholar]
- 34.Viola-Villegas N, Doyle R. Coordin. Chem. Rev. 2009;253:1906–1925. [Google Scholar]
- 35.Joyner JC, Keuper K, Cowan JA. Nucl. Acids Res. 2012 doi: 10.1093/nar/gks811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, McLuckey SA. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 37.Andersen TE, Kirpekar F, Haselmann KF. J. Am. Soc. Mass Spectr. 2006;17:1353–1368. doi: 10.1016/j.jasms.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Huang T-Y, Kharlamova A, Liu J, McLuckey SA. Am. Soc. Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Breen AP, Murphy JA. Free Radical Bio. Med. 1994;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 40.Koliakos GG, Konstas AGP, Schlotzer-Schrehardt U, Hollo G, Katsimbris IE, Georgiadis N, Ritch R. Br. J. Ophtham. 2003;87:353–356. doi: 10.1136/bjo.87.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller S, Riedel H, Stremmel W. Anal. Biochem. 1997;245:55–60. doi: 10.1006/abio.1996.9939. [DOI] [PubMed] [Google Scholar]
- 42.Ullrich R, Hofrichter M. Cell Mol Life Sci. 2007;64:271–293. doi: 10.1007/s00018-007-6362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rittle J, Green MT. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 44.Buettner GR, Jurkiewicz BA. Radiat. Res. 1996;145:532–541. [PubMed] [Google Scholar]
- 45.Chiou S-H. J. Biochem. 1984;96:1307–1310. doi: 10.1093/oxfordjournals.jbchem.a134951. [DOI] [PubMed] [Google Scholar]
- 46.Roots R, Okada S. Radiat. Res. 1975;64:306–320. [PubMed] [Google Scholar]
- 47.Turchi CS, Ollis DF. J. Catal. 1990;122:178–192. [Google Scholar]
- 48.Bradford S, Cowan JA. Chem. Comm. 2012;48:3118–3120. doi: 10.1039/c2cc17377h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.