Abstract
Specific combinations of transcription-factor binding sites in the promoter regions of genes regulate gene expression, and thus key functional processes in cells. Analysis of such promoter regions in specific functional contexts can be used to delineate novel disease-associated genes based on shared phenotypic properties. The aim of this study was to utilize promoter analysis to predict cell proliferation-associated genes and to test this method in colon cancer cell lines. We used freely-available bioinformatic techniques to identify cell-proliferation-associated genes expressed in colon cancer, extract a shared promoter module, and identify novel genes that also contain this module in the human genome. An EGRF/ETSF promoter module was identified as prevalent in proliferation-associated genes from a colon cancer cDNA library. We detected 30 other genes, from the known promoters of the human genome, which contained this proliferation-associated module. This group included known proliferation-associated genes, such as HERG1 and MCM7, and a number of genes not previously implicated in cell proliferation in cancer, such as TSPAN3, Necdin and APLP2. Suppression of TSPAN3 and APLP2 by siRNA was performed and confirmed by RT-PCR. Inhibition of these genes significantly inhibited cell proliferation in colon cancer cell lines. This study demonstrates that promoter analysis can be used to identify novel cancer-associated genes based on shared functional processes.
Keywords: colorectal cancer, cell proliferation, promoter modules
Background
The methods of analysis of the colon cancer transcriptome described thus far produce large quantities of data in their output (Alon et al. 1999; Saha et al. 2001). Given the often arbitrary nature of the statistical thresholds for determining disease association, the functional relevance of many “over-expressed” genes is often unclear (Kothapalli et al. 2002; Troyanskaya, 2005). The absence of hypothesis in many microarray papers has yielded as many questions as answers (Shih et al. 2005).
One approach to this “data overload” is to focus on specific biological processes rather than individual genes that are altered in malignant cells. Such processes are driven by transcription factors that are common to genes which share similar functional contexts e.g. proliferation, invasion (Qiu, 2003; Werner, 2001). The promoter regions of these genes contain patterns of transcription factor binding sites (promoter modules) that form the basis for such co-regulation. These modules contain at least two transcription factor binding sites separated by a defined distance (Fessele et al. 2002). By identifying the promoter modules prevalent in genes that are known to share a common biological function, one can use these as a starting point to detect previously unknown genes that are involved in this process (Werner, 2001). The presence of these modules in a genes’ promoter region can positively or negatively influence functional processes. In this manner, a network of co-regulated genes can be determined that are implicated in specific processes (Liu et al. 2003). This approach has been used successfully in detecting interferon-responsive genes in inflammation, and novel cell-junction associated proteins (Cohen et al. 2006; Klingenhoff et al. 1999).
The purpose of this study was to use bioinformatic techniques to determine promoter modules common to those genes in the colon cancer transcriptome that are involved in cell proliferation. In this paper we utilize an integrated bioinformatics pathway to identify novel genes associated with cell proliferation in colon cancer, and validate this approach in an in vitro model.
Methods
Bioinformatic techniques
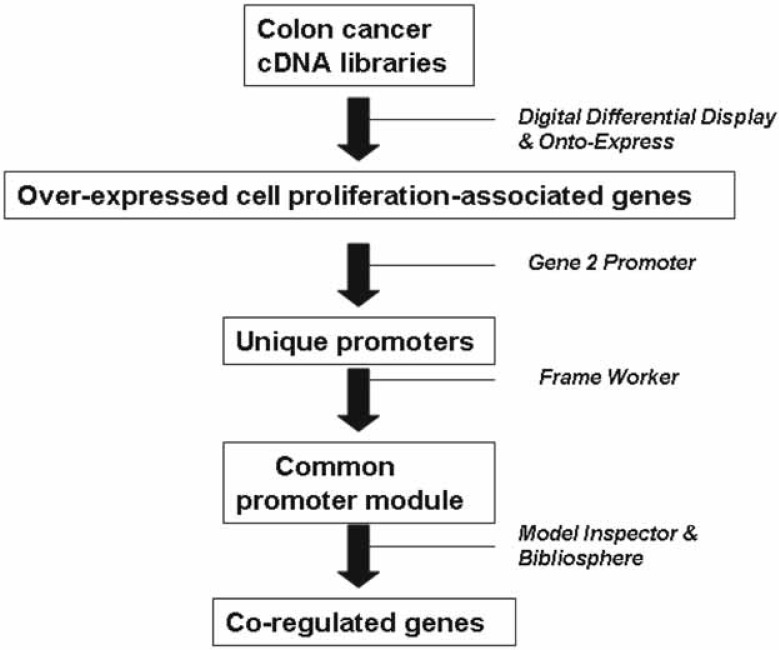
An outline of the bioinformatics pipeline is illustrated in Figure 1. A transcriptional profile of colorectal cancer was produced by comparing cDNA libraries obtained from normal colon and colon carcinoma with Digital Differential Display (DDD), as previously described (Moss et al. 2006). Briefly, the relative abundance of ESTs in colon cancer libraries was compared to normal tissue libraries, and those genes significantly over-expressed in colon cancer were extracted. The output was ontologically classified using Onto-Express to select those transcripts associated with cell proliferation (Khatri et al. 2002). The accession numbers of these transcripts were uploaded to Gene2Promoter (Genomatix Software GmbH), a software program that allowed identification of promoter regions based on the individual transcripts in a gene expression profile (Werner, 2001). The promoter sequences from Gene2Promoter were submitted to Frame-Worker, (FrameWorker, 2006) and once a model common to the input promoters was identified, its presence was screened for in known promoters of the human genome using Model Inspector (Model Inspector 2006). Briefly, all matches for individual elements of the module which score above a pre-set threshold are located in the promoter database. These individual elements are combined to match the organization (element order and distances) of the input module, to evaluate the fit of the model. Finally, Bibliosphere was utilized to examine the characteristics of selected genes based on the published literature (Scherf et al. 2005).
Figure 1.
Summary of bioinformatics methods used. References for each method contained in text.
Gene expression
Public gene expression repositories derived from microarray data from normal colon, colonic cancers and colon cancer cell lines, were interrogated for genes of interest. The normal colon microarray profile originated from pooled samples from normal colonic tissue (Gene Expression Omnibus tissue GSM44680) hybridized to the Affymetrix GeneChip Human Genome U133 Array (Ge et al. 2005). The results are expressed in log2 of user-provided counts for comparison to other normal tissues. Colon cancer tissue expression profile was obtained from the transcriptome of 10 colorectal adenocarcinomas hybridized to the U95a Affymetrix GeneChip and compared to other human cancers (Su et al. 2001). Finally, the microarray data from a primary colon cancer (SW480) and a metastatic colon cancer cell line (SW620) hybridized to the Affymetrix GeneChip Human Genome U133 Array was surveyed (Provenzani et al. 2006). The results are expressed in log2 of user-provided counts for comparison between the cell lines.
Cell lines
The Caco2 human colonocyte cell line was purchased form ATCC (LGC Promochem, U.K.) and the T84 cells were a kind gift from Dr. Cormac Taylor, UCD. Cell lines were cultured in minimum essential medium (Caco2) or mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium under standard conditions (T84).
siRNA transfection
Prior to transfection 1×105 cells were seeded in 500 μl of medium in each well of a 24 well plate and cultured until 50–80% confluent (∼24 hours). For transfection, 0.5 μg of custom-designed siRNA (Dharmacon, IL, U.S.A.) was diluted in 100 μl medium and 1.5 μl RNAifect transfection reagent added (Qiagen, U.K.) at a 1:3 ratio and added to each well as per protocol. Three controls were used for each experiment; a positive control of laminin siRNA for mRNA quantification, a positive control of fluorescent-labeled siRNA for microscopy, and negative controls of medium only, transfection reagent only and scrambled siRNA only. The transfected cells were incubated for 24 hours under normal conditions.
RT-PCR
RNA extraction was subsequently performed from cells using the RNeasy kit (Qiagen, U.K.), and reverse transcribed using SuperScript II (Promega, U.K.). Quantitative PCR was performed using an ABIPrism Taqman PCR machine. Expression levels of individual genes were normalized to 18s RNA.
Cell proliferation assay
In order to determine the effect of siRNA on cell proliferation rates, transfected CaCO2 cells were seeded into 96-well plates at a concentration of 1×104 cells in 100 μl per well and allowed to adhere overnight. The MTS cell proliferation assay (Promega, U.K.) was used to assess proliferation rates at 48 hours, based on absorbance at 490 nm in an ELISA plate reader. Proliferation ratios were based on comparison of mean absorbance values for transfected and untransfected wells using one-way ANOVA.
Statistical analysis
Statistical analysis of laboratory results was performed using StatView software (SAS Institute, Cary, NC). Normalised gene expression was analysed using ANOVA, after testing for equality of variance. A p < 0.05 was considered significant. The differential expression profiles, promoter analysis and module detection all contain integral statistical thresholds for results as described in the results section.
Results
An EGRF/ETSF transcription factor module is prevalent in cell proliferation-associated genes over-expressed in colorectal cancer
Digital Differential Display comparison of normal colon to colorectal cancer cDNA libraries identified 163 transcripts differentially expressed in colon cancer, of which 16 were classified as involved in cellular proliferation (supplementary 1)(Moss et al. 2006). These 16 genes were the source material for promoter screening. The loci of these 16 genes were entered into Gene2Promoter, which detected 30 unique promoters assigned to 30 transcripts in the mapped regions; all transcripts with at least one exon identical to one of the mapped exons and their promoters were listed (Table 1). Fifteen of these promoters had been experimentally verified, and the other 15 were computational predictions based in sequence location and content.
Table 1.
Genes associated with cell proliferation in colon cancer that had promoter regions identified (verified = published experimental verification, predicted = transcript with 5’ end confirmed by Gene2Promoter 2004).
| mRNA | Locus | Transcript/TSS | Quality Level |
|---|---|---|---|
| NM_002394 | SLC3A2 (Loc 6520) | AK090758_1 | Verified |
| AK094620_1 | Verified | ||
| NM_002394 | Predicted | ||
| NM_005916 | MCM7 | AK055379_1 | Verified |
| AK096959_1 | Verified | ||
| NM_005916 | Predicted | ||
| NM_014865 | CNAP1 | AK022511_1 | Verified |
| AK125155_1 | Verified | ||
| AK128354_1 | Verified | ||
| NM_001034 | RRM2 | AK092671_1 | Verified |
| AK123010_1 | Verified | ||
| NM_001034 | Predicted | ||
| NM_002707 | PPM1G | AK127593_1 | Verified |
| NM_002707 | Predicted | ||
| NM_177983 | Predicted | ||
| NM_000077 | CDKN2A | NM_058195 | Predicted |
| NM_016343 | CENPF | NM_016343 | Predicted |
| NM_002447 | MST1R | NM_002447 | Predicted |
| NM_001255 | CDC20 | NM_001255 | Predicted |
| NM_004526 | MCM2 | AK128291_1 | Verified |
| NM_005186 | CAPN1 | AK025380_1 | Verified |
| AK097277_1 | Verified | ||
| NM_005186 | Predicted | ||
| NM_004494 | HDGF | AK096411_1 | Verified |
| NM_004494 | Predicted | ||
| NM_003334 | UBE1 | AK097343_1 | Verified |
| NM_003334 | Predicted | ||
| NM_002335 | LRP5 | NM_002335 | Predicted |
| NM_002032 | FTH1 | NM_002032 | Predicted |
| NM_005030 | PLK | NM_005030 | Predicted |
The identified promoter sequences were investigated using FrameWorker software, which detects patterns in transcription factor binding sites (TFBS). We searched for modules containing at least 2 elements (TFBS), at a distance of 5–50 nucleotides apart, and adjusted the quorum constraint (prevalence threshold) until the program identified a common module. No individual module was common to all the input promoter sequences. However, one complex module, containing members of the EGRF and ETSF transcription factor binding site families, was present in 18/30 (60%) of the input promoters (Fig. 2). The specificity score of this model had a p-value of 0.0059 e.g. the probability that an equal or greater number of sequences with a model match would be obtained in a randomly drawn sample of the same size as the input sequence set. The relative occurrence of individual model matches in a background promoter sequence set of 5000 human promoters scanned with this module was 0.27 and 0.50 for EGRF and ETSF respectively.
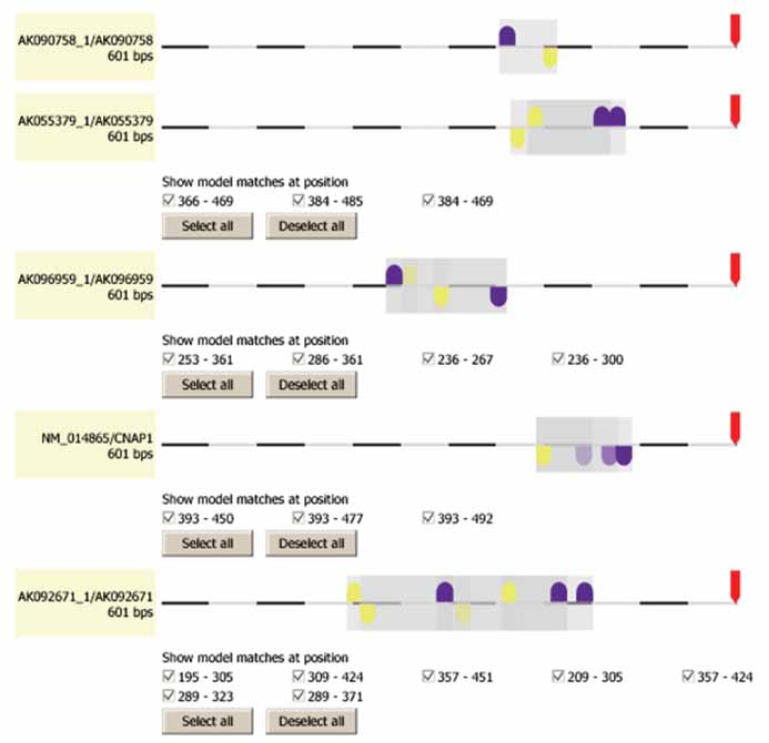
Figure 2.
EGRF/ETSF module is common to proliferation genes expressed in colon cancer. EGRF element (purple), ETSF element (yellow) and combined module (grey) location in promoter region of representative sample of input loci relative to transcription start site (TSS, red arrow). Graphical output generated by FrameWorker software.
This EGRF/ETSF module contains members of the Early Growth Response Factor family and the ETS factor family at a distance of 6–44 base pairs between elements. The matrices (transcription factors) of the EGRF family were EGR1, EGR2, EGR3, EGR4 and Wilms tumour suppressor. The re-value, an expectation value of the number of matches per 1000 base pairs of random DNA sequence for each individual matrix, ranged from 0.03–0.35 for the EGRF elements. ETS1, ETS2, ELK1 and NRF2 were the components of the ETSF element, with re-values of <0.01–2.05. The free version of the software does not detail the exact sequences of the modules, as it is their relative location, rather than sequence, that determines a module’s functional activity.
The EGRF/ETSF module identifies novel proliferation-associated genes
The known promoters of the human genome were screened for the EGRF/ETSF module using Model Inspector, based not on sequence alignment, but detection of individual elements and their position relative to each other. At the time of the experiment, the database contained 46,119 promoters with known transcripts. A total of 102 matches for the selected proliferation-associated module were detected in 30 genes (Table 2). All matches contained a model score of ≥85% specificity. The chromosomal locations of these genes were widely dispersed throughout the genome, excluding the possibility of co-regulation due to overlapping sequences (data not shown).
Table 2.
Identified genes that contain the EGRF/ETSF promoter module in their promoter regions.
| Accession | Genea | Model score | Effect on proliferationb | Expression in colon cancerc | Public microarray datad | References |
|---|---|---|---|---|---|---|
| AB000381 | GML | 89% | negative | Yes - in cell lines | No | Oncogene 1996; 13 (9) 1965–7. Int J Clin Oncol. 2001 Apr; 6(2):90–6 |
| AB001517 | TMEM1 | 90% | ? | ? | No | |
| AB001523 | PWP2 | 90% | ? | ? | No | |
| AB003173 | WRN | 90% | positive | Yes – in unmethylated tumours | Yes | DNA Repair 2004; 3(5): 475–482 Proc Natl Acad Sci USA. 2006 Jun 6; 103(23):8822–7 |
| AB003469 | MCM5 | 90% | ? | Yes | Yes | Clin Cancer Res. 1999 Aug; 5(8):2121–32 |
| AB004270 | MCM7 | 90% | positive | ? | Yes | Oncogene 2006; 25(7): 1090–9 |
| AB005647 | NPR2 | 90% | ? | ? | No | |
| AB006075 | HMG-CoA synthase | 89% | ? | Yes | No | Mol Carcinog. 2001 Nov; 32(3):154–66. |
| AB006684 | AIRE | 91% | ? | ? | No | |
| AB007828 | NDN | 88% | negative | ? | No | Gene 1998; 213(1–2): 65–72 |
| AB008496 | COL4A3 | 90% | negative | ? | No | J Biol Chem 2000; 275 (28):21340–8 |
| AB008502 | TLX2 | 90% | ? | ? | No | |
| AB008681 | ACVR2B | 92% | positive | Yes – in cell lines | No | Dev Biol 2004; 266(2): 334–45 Gut. 2001 Sep; 49(3):409–17 |
| AB008822 | TNFRSF1 1B | 92% | negative | ? | No | J Clin Invest 2001; 107(10):1235–4 |
| AB009071 | KCNH2 | 88% | positive | Yes | Yes | J Biol Chem 2003; 278(5):2947–55 Cancer Res. 2004 Jan 15; 64(2):606–11 |
| AB009667 | Klotho | 95% | ? | ? | No | |
| AB009777 | NID2 | 85% | ? | ? | No | |
| AB012286 | ITGB4 | 90& | positive | ? | Yes | Cancer Res 2005; 65(23):10674–9 |
| AB012668 | hFUCT-7 | 91% | ? | ? | No | |
| AB015751 | APLP2 | 96% | ? | ? | No | |
| AB016243 | SLC9A3R2 | 91% | ? | ? | No | |
| AB016656 | LIMK2b | 91% | ? | ? | No | |
| AB016767 | TERT | 95% | ? | ? | No | |
| AB017018 | HNRPDL | 94% | ? | ? | Yes | |
| AB017547 | SPR | 93% | ? | ? | Yes | |
| AB017567 | LIPT1 | 95% | ? | ? | No | |
| AB017602 | PDE9A | 90% | ? | ? | Yes | |
| AB018192 | PHC1 | 95% | ? | ? | No | |
| AB018401 | DHH | 90% | positive | ? | No | Development 2004 Oct; 131(20):5009–19 |
| AK001326 | TSPAN3 | 90% | positive | ? | Yes | J Cell Biol 153:295–305 |
HUGO accepted gene name
Published experimental evidence of effect on cell proliferation
Experimental evidence of increased protein or mRNA expression in colon cancer
Upregulation of gene in public microarray database of expression relative to normal (Diehn et al. 2003)
The products of the 30 genes were entered into Bibliosphere (Bibliosphere, 2006) to determine their functional context based on the scientific literature e.g. published experimental evidence of a role in affecting cellular proliferation (Table 2). Eleven of these genes (37%) have been implicated in cell proliferation in the literature, including KCNH2 and MCM7 (Lastraioli et al. 2004; Yoshida et al. 2003) (Table 2). Six of the genes have been described in the literature as expressed in colonic neoplasia based on experimental data, and nine are up-regulated in colon cancer gene expression profiles in public databases (Table 2) (Diehn et al. 2003). As a control functional context, a common disease process, inflammation, was explored in the 30 identified genes using Bibliosphere; only one (TLX2) has been associated with inflammation (data not shown).
Suppression of TSPAN3 and APLP2 inhibits cell proliferation in colorectal cell lines
The experiments above identified genes containing a promoter module that is frequently present in genes associated with cell proliferation in colon cancer. In order to determine the functional significance of the presence of this module in these genes, the role of their knock-down by siRNA on cell proliferation was determined. We screened the identified genes using Bibliosphere for 1) reports of expression in colon cancer, 2) reports of a role in cell proliferation (Table 2). As our interest was in novel proliferation-associated genes which may be relevant to colon cancer, we selected three genes not previously reported as altered in colon cancer; TSPAN3, NDN and APLP2. They have been described as having positive, negative and unknown roles in cell proliferation (Taniura et al. 1999; Tiwari-Woodruff et al. 2001). The TSPAN3 gene contains the EGRF/ETSF module at position 491-418 on the negative strand at 15q24.3. The APLP2 gene contained the EGRF/ETSF module at position 2492–2532 on the positive strand at 11q24. The NDN gene contained the EGRF/ETSF module at position 1268-1179 on the negative strand at 15q11.2-q12.
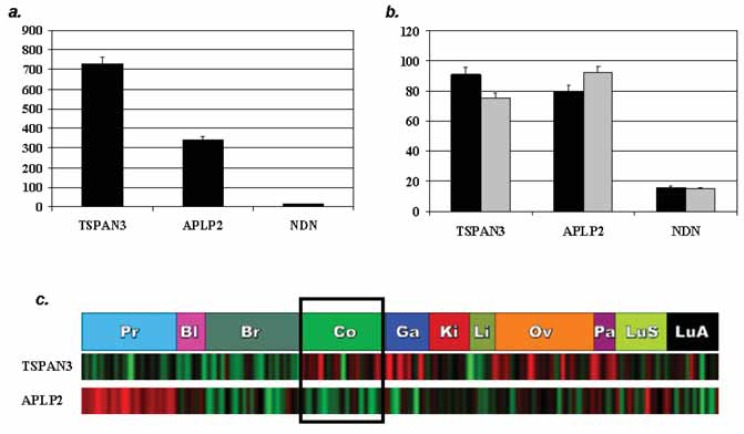
Gene expression of each gene in colon cancer was first measured from 3 diverse microarray databases; one from 36 types of normal tissue, one from 174 epithelial tumors that included 10 colorectal adenocarcinomas, and a third from primary and metastatic colorectal cell lines (Ge, Yamamoto, Tsutsumi, Midorikawa, Ihara, Wang and Aburatani, 2005; Provenzani, Fronza, Loreni, Pascale, Amadio and Quattrone, 2006; Su, Welsh, Sapinoso, Kern, Dimitrov, Lapp, Schultz, Powell, Moskaluk, Frierson, Jr. and Hampton, 2001). Both TSPAN3 and APLP2 were expressed in normal colon, and colon cancer cell lines, although only TSPAN3 was relatively over-expressed in colonic adenocarcinoma tissue relative to other tumours (Figure 3). NDN was not expressed in normal colon, adenocarcinoma or colon cancer cell lines.
Figure 3.
TSPAN3 and APLP2 are expressed in normal and neoplastic colon. Expression of TSPAN3, APLP2 and NDN in microarray profiles of: (a) normal colon (Log2 of user-provided count of gene expression on oligonucleotide microarray (Affymetrix U133) using pooled RNA) (b) primary colon cancer cell line (black bars) and metastatic colon cancer cell line (grey bars) (Log2 of user-provided count of gene expression on oligonucleotide microarray (Affymetrix U133) using pooled RNA) (c) 174 human epithelial tumors (Co; colon samples, red, increased gene expression; green, decreased expression; black, median level of gene expression. The color intensity is proportional to the hybridization intensity of a gene from its median level across all samples.
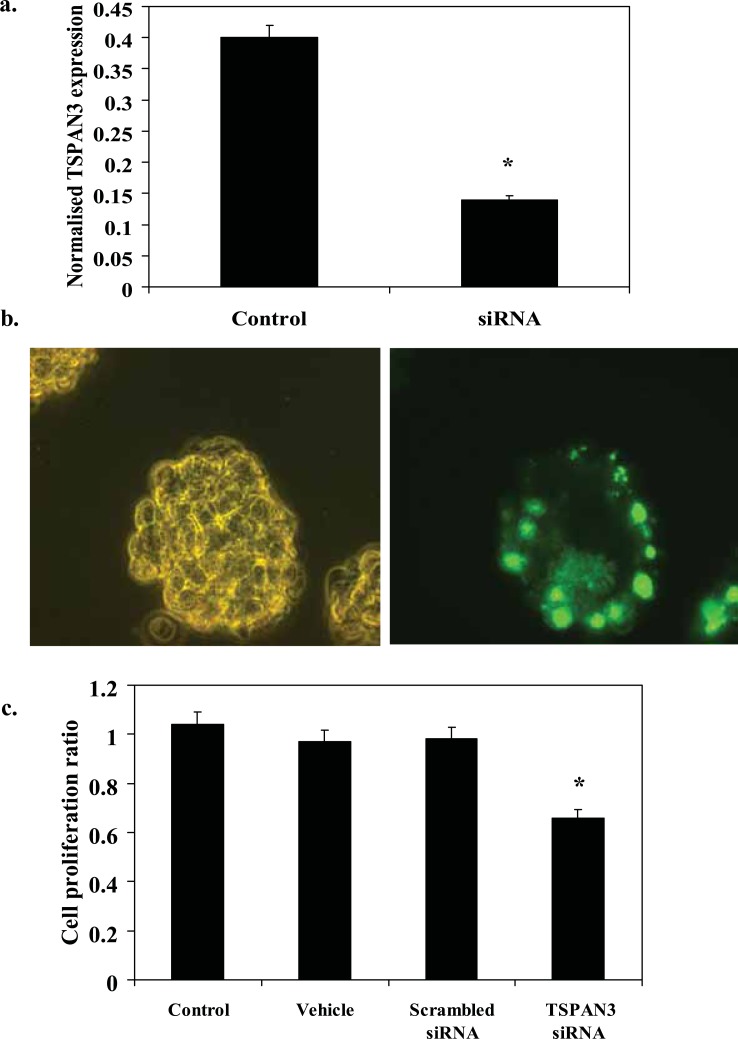
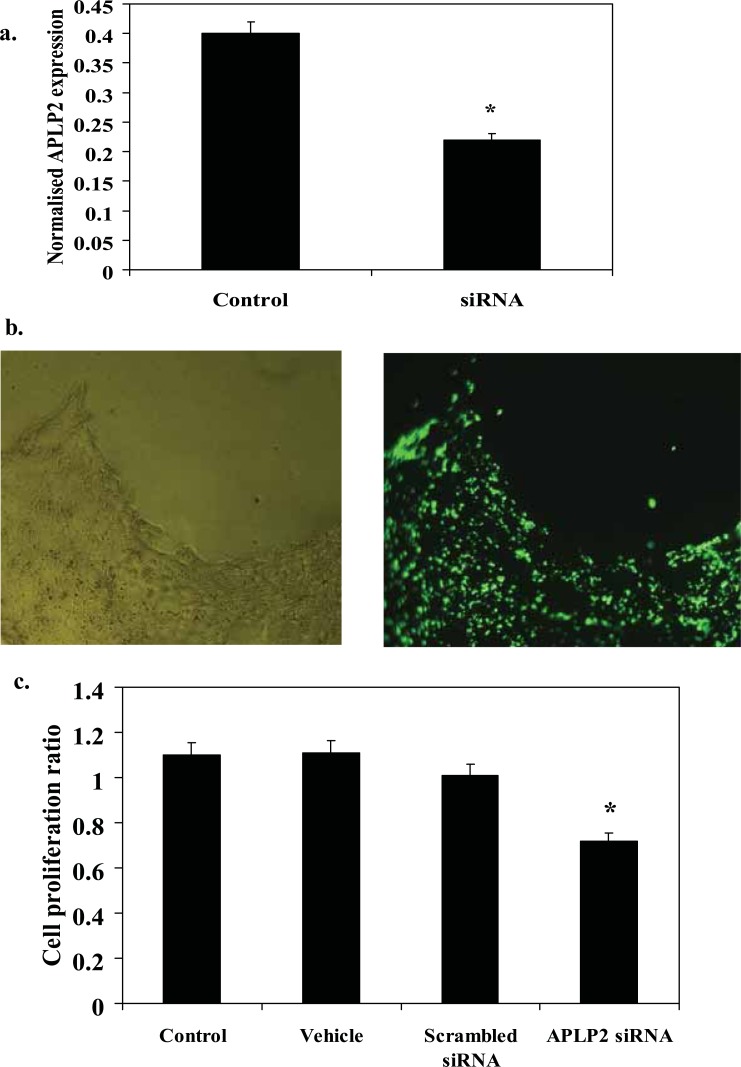
Cells were transfected with siRNA designed to provide at least 70% silencing of expression, and mRNA levels and cell proliferation quantified (Reynolds et al. 2004). TSPAN3 expression in T84 colon cancer cell line was confirmed by RT-PCR (Fig. 4a). siRNA caused a 62% inhibition of TSPAN3 expression at 24 hours (Fig. 4a, p < 0.05). Confirmation of cellular uptake was observed using the labeled fluorescent siRNA (Fig. 4b). This led to a 40% reduction in cellular proliferation at 48 hours in T84 cells (Fig. 4c, p < 0.05). Neither scrambled siRNA or transfection agent alone affected cell proliferation. APLP2 expression in colon cancer cells was confirmed by RT-PCR (Fig. 5a). siRNA caused a 45% inhibition of APLP2 expression at 24 hours (Fig. 5a, p < 0.05). Confirmation of cellular uptake was observed using the labeled fluorescent siRNA (Fig. 5b). This inhibition led to a 40% reduction in cellular proliferation at 48 hours in CaCo2 cells (Fig. 5c, p < 0.05). Neither scrambled siRNA or transfection agent alone affected cell proliferation.
Figure 4.
Inhibition of TSPAN3 expression by siRNA inhibits colon cell line proliferation. (a) RNA extracted from transfected and untransfected T84 cells after 24 hours was reverse transcribed to cDNA and probed for TSPAN3 using Taqman PCR (expressed in arbitrary units normalised to 18s RNA). (b) T84 cells in a 96-well plate were transfected with a control fluorescent-labeled siRNA to confirm transfection efficiency. (c) Proliferation of transfected T84 cells in a 96-well plate was assessed after 24 hours using the MTS Cell Proliferation Assay. Control = media only, vehicle = media and transfection agent (Lipofectamine), scrambled siRNA = transfection agent and scrambled siRNA, and TSPAN3 siRNA = custom-designed TSPAN3 siRNA (10 nm)
Figure 5.
Inhibition of APLP2 expression by siRNA inhibits colon cell line proliferation. (a) RNA extracted from transfected and untransfected T84 cells after 24 hours was reverse transcribed to cDNA and probed for APLP2 using Taqman PCR (expressed in arbitrary units normalised to 18s RNA) (b) T84 cells in a 96-well plate were transfected with a control fluorescent-labeled siRNA to confirm transfection efficiency (c) Proliferation of transfected T84 cells in a 96-well plate was assessed after 24 hours using the MTS Cell Proliferation Assay. Control = media only, vehicle = media and transfection agent (Lipofectamine), scrambled siRNA = transfection agent and scrambled siRNA, and APLP2 siRNA = custom-designed APLP2 siRNA (10 nm)
Discussion
This study has demonstrated the use of promoter modules as bioinformatic “bait” to delineate key regulatory networks in colon cancer, and to identify novel biological players in cell proliferation. It is based on the premise that genes expressed in similar disease states share a common “footprint” of transcriptional regulatory processes for specific functional activities. The relative order and spacing of these transcription factor (TF) binding sites (TFBSs) within a module are often highly conserved through evolution, highlighting their importance in regulation. This conservation allows us to use computational searching to pinpoint these clusters of known TF binding sites rather than specific nucleotide sequences (Berman et al. 2002). Although the shared process selected for this study, proliferation, is not unique to cancer cells, it is a dominant process that partially defines this disease state. The presence of the EGRF/ETSF module in the promoter region of genes associated with cell proliferation suggests a role for this module in the regulation of cellular proliferative activity (Lantingavan, I et al. 2005). Although this influence could be positive or negative, its frequency in the promoter regions of over-expressed genes in colon cancer cDNA libraries, compared to random promoter sets, suggests a pro-proliferative effect in this setting. The experimental data clearly suggests a role for the studied genes in cell proliferation, although whether this is actually dependent on the identified promoter module, would require further studies.
This strategy predicted a role for TSPAN3 in cell proliferation in colorectal cancer that has not previously been described. TSPAN3 is a member of the tetraspanin family of cell surface receptors that have been implicated in the cell proliferation process in oligodendrocytes (Tiwari-Woodruff, Buznikov, Vu, Micevych, Chen, Kornblum and Bronstein, 2001). Our data demonstrating its expression in normal and neoplastic colon, and the negative effects of its inhibition on cell proliferation in colon cancer cell lines, confirming the predicted role based on promoter analysis. Further work will be required to determine whether this effect is unique to colon cancer cells, and which component of proliferation is involved. APLP2 is an amyloid-like protein precursor that plays a role in G-coupled signaling. It may be required for epithelial cell growth in wounds (Siemes et al. 2006). This study suggests a role for APLP2 in colon cancer cell proliferation, which may be due to its key function in genomic segregation (von der et al. 1994).
Although this study validates this approach in identifying co-regulated genes, the activation of the promoters involved has not been experimentally tested. As this work focuses on functionally relevant associations between genes and disease, we sought to examine the functional end-point primarily. The confirmation of alterations in expression and proliferation validates the computational predictions. We have not focused on the descriptive aspects of the module discussed e.g. sequence, as it is the strategic organization of the EGRF/ETSF matrix in the promoters of interest, rather than sequence composition, that confers its functional properties (Dohr et al. 2005). Our intention was proof-of-concept evidence that could validate this bioinformatic approach.
In conclusion, this study demonstrates that an integrated in silico promoter analysis approach can be used to delineate novel cancer-associated genes. We have described a previously unreported role for TSPAN3 and APLP2, in cell proliferation in colon cancer based on a common promoter module. Further study of this module may provide increased understanding of this regulatory network.
Acknowledgments
Anne-Marie Griffin, Conway Institute, UCD for assistance with cell line work. This research was funded by Cancer Research Ireland.
List of abbreviations:
- siRNA
small interfering RNA,
- EGRF
early growth response family,
- ETSF
E26 transformation-specific family.
Footnotes
Competing interests
The authors have no competing interests to disclose.
Supplementary 1
| Accession | Cluster | Name | Expression | Fold Diff |
|---|---|---|---|---|
| NM_001644.2 | 560 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) | Exclusive | 54 |
| NM_001804.1 | 1545 | caudal type homeo box transcription factor 1 (CDX1) | Exclusive | 14 |
| NM_005814 | 143131 | glycoprotein A33 (transmembrane) (GPA33) | Exclusive | 11 |
| NM_001986.1 | 77711 | ets variant gene 4 (E1A enhancer binding protein, E1AF) (ETV4) | Significant | 20 |
| NM_001265.2 | 77399 | caudal type homeo box transcription factor 2 (CDX2) | Significant | 20 |
| NM_138768.1 | 116051 | myeloma overexpressed gene positive multiple myelomas) (MYEOV) | Significant | 14 |
| NM_004963.1 | 1085 | guanylate cyclase 2C (heat stable enterotoxin receptor) (GUCY2C) | Significant | 13 |
| NM_024017.3 | 86327 | homeo box B9 (HOXB9) | Significant | 6 |
| XM_032721.3 | 109358 | ATPase, Class V, type 10B (ATP10B) | Significant | 5 |
| NM_033266.1 | 114905 | ER to nucleus signalling 2 (ERN2) | Significant | 4 |
| NM_019010.1 | 84905 | cytokeratin 20 (KRT20) | Significant | 4 |
| NM_005310.1 | 86859 | growth factor receptor-bound protein 7 (GRB7) | Significant | 4 |
| NM_001738.1 | 23118 | carbonic anhydrase I (CA1) | Significant | 4 |
| NM_004306.1 | 181107 | annexin A13 (ANXA13) | Significant | 3 |
| NM_007028.2 | 91096 | tripartite motif-containing 31 (TRIM31) | Significant | |
| NM_001500.1 | 1054435 | GDP-mannose 4,6-dehydratase (GMDS) | Preferential | 39 |
| NM_005628.1 | 183556 | solute carrier family 1 (neutral amino acid transporter), member 5 (SLC1A5) | Preferential | 36 |
| NM_002276.2 | 182265 | keratin 19 (KRT19) | Preferential | 33 |
| NM_001569.2 | 182018 | interleukin-1 receptor-associated kinase 1 (IRAK1) | Preferential | 23 |
| NM_002295 | 356261 | laminin receptor 1 (67kD, ribosomal protein SA) (LAMR1) | Preferential | 19 |
| NM_001402 | 493552 | eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) | Preferential | 19 |
| NM_002087 | 180577 | granulin (GRN) | Preferential | 19 |
| NM_006597 | 180414 | heat shock 70kD protein 8 (HSPA8) | Preferential | 18 |
| NM_005507 | 170622 | cofilin 1 (non-muscle) (CFL1) | Preferential | 17 |
| NM_001903 | 254321 | catenin (cadherin-associated protein), alpha 1 (102kD) (CTNNA1) | Preferential | 17 |
| NM_002819 | 172550 | polypyrimidine tract binding protein 1 (PTBP1) | Preferential | 17 |
| NM_007363 | 355861 | non-POU domain containing, octamer-binding (NONO) | Preferential | 17 |
| NM_002568 | poly(A) binding protein, cytoplasmic 1 (PABPC1) | Preferential | 16 | |
| NM_006516 | 169902 | solute carrier family 2 (facilitated glucose transporter), member 1 (SLC2A1) | Preferential | 16 |
| NM_002046 | glyceraldehyde-3-phosphate dehydrogenase (GAPD) | Preferential | 16 | |
| NM_003906 | 389037 | MCM3 minichromosome maintenance deficient 3 protein (MCM3AP) | Preferential | 15 |
| NM_004433 | 67928 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) (ELF3) | Preferential | 14 |
| NM_007127 | 166068 | villin 1 (VIL1) | Preferential | 14 |
| NM_000218 | 367809 | potassium voltage-gated channel, KQT-like subfamily, member 1 (KCNQ1) | Preferential | 13 |
| NM_003379 | 403997 | villin 2 (ezrin) (VIL2) | Preferential | 13 |
| NM_001084 | 153357 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (PLOD3) | Preferential | 12 |
| NM_005789 | 152978 | proteasome (prosome, macropain) activator subunit 3 (PSME3) | Preferential | 12 |
| NM_005561 | 150101 | lysosomal-associated membrane protein 1 (LAMP1) | Preferential | 11 |
| NM_005080 | 437638 | X-box binding protein 1 (XBP1) | Preferential | 11 |
| NM_002105 | H2A histone family, member X (H2AFX) | Preferential | 11 | |
| NM_004429 | 144700 | ephrin-B1 (EFNB1) | Preferential | 10 |
| NM_014498 | golgi phosphoprotein 4 (GOLPH4) | Preferential | 9 | |
| 139800 | high-mobility group (nonhistone chromosomal) protein isoforms (HMGIY) | Preferential | 9 | |
| NM_007052 | 132370 | NADPH oxidase 1 (NOX1) | Preferential | 9 |
| NM_001416 | 129673 | eukaryotic translation initiation factor 4A, isoform 1(EIF4A1) | Preferential | 9 |
| NM_004655 | 127337 | axin 2 (conductin, axil) (AXIN2) | Preferential | 9 |
| NM_004442 | 125124 | EphB2 (EPHB2) | Preferential | 9 |
| NM_000967 | 119598 | ribosomal protein L3 (RPL3) | Preferential | 8 |
| NM_005063 | 119597 | stearoyl-CoA desaturase (delta-9-desaturase) (SCD) | Preferential | 8 |
| NM_000090 | 443625 | collagen, type III, alpha 1 (COL3A1) | Preferential | 8 |
| NM_012423 | 419535 | ribosomal protein L13a (RPL13A) | Preferential | 8 |
| Preferential | ||||
| NM_006026 | 75307 | H1 histone family, member X (H1FX) | Preferential | 8 |
| NM_001923 | 290758 | damage-specific DNA binding protein 1 (127kD) (DDB1) | Preferential | 8 |
| NM_032044 | 105484 | regenerating gene type IV (REG-IV) | Preferential | 8 |
| NM_003258 | 105097 | thymidine kinase 1, soluble (TK1) | Preferential | 7 |
| XM_039877 | 102482 | mucin 5, subtype B, tracheobronchial (MUC5B) | Preferential | 7 |
| NM_005724 | 100090 | tetraspan 3 (TSPAN-3) | Preferential | 7 |
| NM_000972 | 416801 | ribosomal protein L7a (RPL7A) | Preferential | 7 |
| NM_018952 | 98428 | homeo box B6 (HOXB6) | Preferential | 7 |
| NM_015925 | 312129 | Similar to liver-specific bHLH-Zip transcription factor | Preferential | 7 |
| NM_000075 | 95577 | cyclin-dependent kinase 4 (CDK4) | Preferential | 6 |
| NM_006408 | 226391 | anterior gradient 2 homolog (Xenepus laevis) (AGR2) | Preferential | 6 |
| NM_004044 | 90280 | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (ATIC) | Preferential | 6 |
| NM_004494 | 89525 | hepatoma-derived growth factor (high-mobility group protein 1-like) (HDGF) | Preferential | 6 |
| NM_004063 | 89436 | cadherin 17, LI cadherin (liver-intestine) (CDH17) | Preferential | 6 |
| NM_000213 | 85266 | integrin, beta 4 (ITGB4) | Preferential | 6 |
| NM_001730 | 84728 | Kruppel-like factor 5 (intestinal) (KLF5) | Preferential | 6 |
| NM_001255 | 82906 | CDC20 cell division cycle 20 homolog (S. cerevisiae) (CDC20) | Preferential | 5 |
| NM_001747 | 82422 | capping protein (actin filament), gelsolin-like (CAPG) | Preferential | 5 |
| NM_002534 | 442936 | 2′,5′-oligoadenylate synthetase 1 (40–46 kD) (OAS1) | Preferential | 5 |
| NM_000178 | 82327 | glutathione synthetase (GSS) | Preferential | 5 |
| NM_000903 | 406515 | NAD(P)H dehydrogenase, quinone 1 (NQO1) | Preferential | 5 |
| NM_002394 | 79748 | solute carrier family 3 member 2 (SLC3A2) | Preferential | 5 |
| NM_005567 | 79339 | lectin, galactoside-binding, soluble, 3 binding protein (LGALS3BP) | Preferential | 5 |
| NM_000404 | galactosidase, beta 1 (GLB1) | Preferential | 5 | |
| NM_006907 | 458332 | pyrroline-5-carboxylate reductase 1 (PYCR1) | Preferential | 5 |
| NM_000291 | 78771 | phosphoglycerate kinase 1 (PGK1) | Preferential | 5 |
| NM_002635 | 290404 | solute carrier family 25 member 3 (SLC25A3) | Preferential | 5 |
| NM_001640 | 221589 | N-acylaminoacyl-peptide hydrolase (APEH) | Preferential | 5 |
| NM_005030 | 329989 | polo-like kinase (Drosophila) (PLK) | Preferential | 5 |
| NM_002224 | 77515 | inositol 1,4,5-triphosphate receptor, type 3 (ITPR3) | Preferential | 4 |
| NM_002668 | 77422 | proteolipid protein 2 (colonic epithelium-enriched) (PLP2) | Preferential | 4 |
| NM_016343 | 77204 | centromere protein F (350/400kD, mitosin) (CENPF) | Preferential | 4 |
| NM_005916 | 438720 | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) (MCM7) | Preferential | 4 |
| NM_001006 | 356572 | ribosomal protein S3A (RPS3A) | Preferential | 4 |
| NM_000701 | 371889 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (ATP1A1) | Preferential | 4 |
| NM_000990 | 356542 | ribosomal protein L27a (RPL27A) | Preferential | 4 |
| NM_015379 | 410497 | brain protein I3 (BRI3) | Preferential | 4 |
| NM_012408 | 191990 | protein kinase C binding protein 1 (PRKCBP1) | Preferential | 4 |
| NM_002773 | 75799 | protease, serine, 8 (prostasin) (PRSS8) | Preferential | 4 |
| NM_002951 | 406532 | ribophorin II (RPN2) | Preferential | 4 |
| NM_001673 | 446546 | asparagine synthetase (ASNS) | Preferential | 4 |
| NM_002862 | 145820 | phosphorylase, glycogen; brain (PYGB) | Preferential | 4 |
| NM_000918 | 410578 | procollagen-proline, 2-oxoglutarate 4-dioxygenase (P4HB) | Preferential | 3 |
| NM_000228 | 436983 | laminin, beta 3 (nicein (125kD), kalinin (140kD), BM600 (125kD) (LAMB3) | Preferential | 3 |
| NM_001034 | 226390 | ribonucleotide reductase M2 polypeptide (RRM2) | Preferential | 3 |
| NM_003217 | 35052 | testis enhanced gene transcript (BAX inhibitor 1) (TEGT) | Preferential | 3 |
| NM_001658 | 286221 | ADP-ribosylation factor 1 (ARF1) | Preferential | 3 |
| NM_000014 | alpha-2-macroglobulin (A2M) | Preferential | 3 | |
| NM_007355 | 74335 | heat shock 90kD protein 1, beta (HSPCB) | Preferential | 3 |
| NM_001288 | 414565 | chloride intracellular channel 1 (CLIC1) | Preferential | 3 |
| Preferential | ||||
| NM_007367 | 74111 | RNA binding protein (RALY) | Preferential | 3 |
| NM_002483 | 436718 | carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) | Preferential | 3 |
| NM_021220 | 386387 | zinc finger protein 339 (ZNF339) | Preferential | 3 |
| NM_001202 | 68879 | bone morphogenetic protein 4 (BMP4) | Preferential | 3 |
| NM_000224 | 406013 | keratin 18 (KRT18) | Preferential | 3 |
| NM_019894 | 414005 | transmembrane protease, serine 4 (TMPRSS4) | Preferential | 3 |
| NM_002032 | 448738 | ferritin, heavy polypeptide 1 (FTH1) | Preferential | 3 |
| NM_016276 | 62863 | serum/glucocorticoid regulated kinase 2 (SGK2) | Preferential | 3 |
| NM_003756 | 127149 | eukaryotic translation initiation factor 3, subunit 3 (gamma, 40kD) (EIF3S3) | Preferential | 3 |
| NM_003751 | 371001 | eukaryotic translation initiation factor 3, subunit 9 (eta, 116kD) (EIF3S9) | Preferential | 3 |
| NM_004526 | 57101 | MCM2 minichromosome maintenance deficient 2, (S. cerevisiae) (MCM2) | Preferential | 3 |
| NM_021978 | 56937 | suppression of tumorigenicity 14 (colon carcinoma, epithin) (ST14) | Preferential | 3 |
| NM_006187 | 129895 | 2′-5′-oligoadenylate synthetase 3 (100 kD) (OAS3) | Preferential | 3 |
| NM_003753 | 55682 | eukaryotic translation initiation factor 3, subunit 7 (zeta, 66/67kD) (EIF3S7) | Preferential | 3 |
| NM_001712 | 512682 | carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) | Preferential | 3 |
| NM_005727 | tetraspan 1 (TSPAN-1) | Preferential | 3 | |
| NM_021102 | 31439 | serine protease inhibitor, Kunitz type, 2 (SPINT2) | Preferential | 3 |
| NM_007183 | 26557 | plakophilin 3 (PKP3) | Preferential | 3 |
| NM_001306 | 25640 | claudin 3 (CLDN3) | Preferential | 3 |
| NM_004572 | 25051 | plakophilin 2 (PKP2) | Preferential | 3 |
| NM_004289 | 404741 | nuclear factor (erythroid-derived 2)-like 3 (NFE2L3) | Preferential | 3 |
| NM_003627 | 444159 | SLC43A1 | Preferential | 2 |
| NM_005498 | 18894 | adaptor-related protein complex 1, mu 2 subunit (AP1M2) | Preferential | 2 |
| NM_005558 | 18141 | ladinin 1 (LAD1) | Preferential | 2 |
| NM_002707 | 17883 | protein phosphatase 1G magnesium-dependent, gamma isoform (PPM1G) | Preferential | 2 |
| NM_020384 | 16098 | claudin 2 (CLDN2) | Preferential | 2 |
| NM_001614 | 14376 | actin, gamma 1 (ACTG1) | Preferential | 2 |
| NM_052854 | 405961 | old astrocyte specifically induced substance (OASIS) | Preferential | 2 |
| NM_016234 | 11638 | fatty-acid-Coenzyme A ligase, long-chain 5 (FACL5) | Preferential | 2 |
| NM_021107 | 411125 | mitochondrial ribosomal protein S12 (MRPS12) | Preferential | 2 |
| NM_002335 | 6347 | low density lipoprotein receptor-related protein 5 (LRP5) | Preferential | 2 |
| NM_022085 | 430169 | thioredoxin related protein (MGC3178) | Preferential | 2 |
| NM_033049 | 5940 | mucin 13, epithelial transmembrane (MUC13) | Preferential | 2 |
| NM_014865 | chromosome condensation-related SMC-associated protein 1 (CNAP1) | Preferential | 2 | |
| NM_006098 | 5662 | guanine nucleotide binding protein beta polypeptide 2-like 1 (GNB2L1) | Preferential | 2 |
| NM_024526 | 5366 | epidermal growth factor receptor pathway related protein 3 (EPS8R3) | Preferential | 1 |
| NM_006149 | 5302 | lectin, galactoside-binding, soluble, 4 (galectin 4) (LGALS4) | Preferential | 1 |
| NM_014275 | 437277 | mannosyl (alpha-1,3-) (MGAT4B) | Preferential | 1 |
| NM_003752 | 388163 | eukaryotic translation initiation factor 3, subunit 8 (110kD) (EIF3S8) | Preferential | |
| 3989 | plexin B2 (PLXNB2) | Preferential | ||
| NM_002447 | 2942 | macrophage stimulating 1 receptor (c-met-related tyrosine kinase) (MST1R) | Preferential | |
| NM_001038 | sodium channel, nonvoltage-gated 1 alpha (SCNN1A) | Preferential | ||
| NM_002083 | 2704 | glutathione peroxidase 2 (gastrointestinal) (GPX2) | Preferential | |
| NM_005186 | 356181 | calpain 1, (mu/I) large subunit (CAPN1) | Preferential | |
| NM_001404 | 256184 | eukaryotic translation elongation factor 1 gamma (EEF1G) | Preferential | |
| NM_003334 | 406683 | ubiquitin-activating enzyme E1 (UBE1) | Preferential | |
| NM_005998 | 1708 | chaperonin containing TCP1, subunit 3 (gamma) (CCT3) | Preferential | |
| NM_012073 | 1600 | chaperonin containing TCP1, subunit 5 (epsilon) (CCT5) | Preferential | |
| NM_000077 | 421349 | cyclin-dependent kinase inhibitor 2A (melanoma (CDKN2A) | Preferential | |
| NM_002014 | 848 | FK506 binding protein 4 (59kD) (FKBP4) | Preferential | |
| NM_004502 | 436181 | homeo box B7 (HOXB7) | Preferential | |
| NM_004966 | 808 | heterogeneous nuclear ribonucleoprotein F (HNRPF) | Preferential | |
| NM_002354 | 692 | tumor-associated calcium signal transducer 1 (TACSTD1) | Preferential | |
| NM_005435 | 334 | Rho guanine nucleotide exchange factor (GEF) 5 (ARHGEF5) | Preferential | |
| NM_002457 | 458274 | mucin 2, intestinal/tracheal (MUC2) | Preferential | |
| NM_000968 | 186350 | ribosomal protein L4 (RPL4) | Preferential |
References
- Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. “Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays.”. Proc. Natl. Acad. Sci. U.S.A. 1999;96(12):6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. “Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome.”. Proc. Natl. Acad. Sci. U.S.A. 2002;99(2):757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibliosphere. http://www.genomatix.de/products/Bibliosphere/index.html. 2006. Ref Type: Computer Program.
- Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, Schmid H, Merkle M, Saleem MA, Koller KP, Werner T, Grone HJ, Nelson PJ, Kretzler M. “Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins.”. Proc. Natl. Acad. Sci. U.S.A. 2006;103(15):5682–5687. doi: 10.1073/pnas.0511257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. “SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data.”. Nucleic Acids Res. 2003;31(1):219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digital Differential Display. http://www.ncbi.nlm.nih.gov/UniGene/info_ddd.shtml. NCBI. 2006. Ref Type: Computer Program.
- Dohr S, Klingenhoff A, Maier H, Hrabe dA, Werner T, Schneider R. “Linking disease-associated genes to regulatory networks via promoter organization.”. Nucleic Acids Res. 2005;33(3):864–872. doi: 10.1093/nar/gki230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessele S, Maier H, Zischek C, Nelson PJ, Werner T. “Regulatory context is a crucial part of gene function.”. Trends Genet. 2002;18(2):60–63. doi: 10.1016/s0168-9525(02)02591-x. [DOI] [PubMed] [Google Scholar]
- FrameWorker. http://www.genomatix.de/products/FrameWorker/index.html. 2006. Ref Type: Computer Program.
- Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, Aburatani H. “Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues”. Genomics. 2005;86(2):127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Khatri P, Draghici S, Ostermeier GC, Krawetz SA. “Profiling gene expression using onto-express”. Genomics. 2002;79(2):266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- Klingenhoff A, Frech K, Quandt K, Werner T. “Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity,”. Bioinformatics. 1999;15(3):180–186. doi: 10.1093/bioinformatics/15.3.180. [DOI] [PubMed] [Google Scholar]
- Kothapalli R, Yoder SJ, Mane S, Loughran TP., Jr “Micro array results: how accurate are they?”. BMC Bioinformatics. 2002;3(22) doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantingavan LI, Leonhard WN, Dauwerse H, Baelde HJ, van Oost BA, Breuning MH, Peters DJ. “Common regulatory elements in the polycystic kidney disease 1 and 2 promoter regions.”. EurJHumGenet. 2005 doi: 10.1038/sj.ejhg.5201392. [DOI] [PubMed] [Google Scholar]
- Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H, Bencini L, Calistri M, Messerini L, Scatizzi M, Moretti R, Wanke E, Olivotto M, Mugnai G, Arcangeli A. “herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells.”. Cancer Res. 2004;64(2):606–611. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- Liu R, McEachin RC, States DJ. “Computationally identifying novel NF-kappa B-regulated immune genes in the human genome.”. Genome Res. 2003;13(4):654–661. doi: 10.1101/gr.911803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model Inspector. http://www.genomatix.de/products/ModelInspector/index.html. 2006. Ref Type: Computer Program. [Google Scholar]
- Moss AC, Lawlor G, Murray D, Tighe D, Madden SF, Mulligan AM, Keane CO, Brady HR, Doran PP, Macmathuna P. “ETV4 and Myeov knockdown impairs colon cancer cell line proliferation and invasion.”. Biochem. Biophys. Res. Commun. 2006;345(1):216–221. doi: 10.1016/j.bbrc.2006.04.094. [DOI] [PubMed] [Google Scholar]
- Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. “Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis”. Carcinogenesis. 2006;27(7):1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- Qiu P. “Recent advances in computational promoter analysis in understanding the transcriptional regulatory network.”. Biochem. Biophys. Res. Commun. 2003;309(3):495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. “Rational siRNA design for RNA interference.”. Nat. Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St CB, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. “A phosphatase associated with metastasis of colorectal cancer”. Science. 2001;294(5545):1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- Scherf M, Epple A, Werner T. “The next generation of literature analysis: integration of genomic analysis into text mining.”. Brief Bioinform. 2005;6(3):287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- Shih W, Chetty R, Tsao MS. “Expression profiling by microarrays in colorectal cancer (Review).”. Oncol. Rep. 2005;13(3):517–524. [PubMed] [Google Scholar]
- Siemes C, Quast T, Kummer C, Wehner S, Kirfel G, Muller U, Herzog V. “Keratinocytes from APP/APLP2-deficient mice are impaired in proliferation, adhesion and migration in vitro.”. ExpCell Res. 2006 doi: 10.1016/j.yexcr.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. “Molecular classification of human carcinomas by use of gene expression signatures.”. Cancer Res. 2001;61(20):7388–7393. [PubMed] [Google Scholar]
- Taniura H, Matsumoto K, Yoshikawa K. “Physical and functional interactions of neuronal growth suppressor necdin with p53.”. J. Biol. Chem. 1999;274(23):16242–16248. doi: 10.1074/jbc.274.23.16242. [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, Bronstein JM. “OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes.”. J.Cell Biol. 2001;153(2):295–305. doi: 10.1083/jcb.153.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya OG. “Putting microarrays in a context: integrated analysis of diverse biological data.”. Brief Bioinform. 2005;6(1):34–43. doi: 10.1093/bib/6.1.34. [DOI] [PubMed] [Google Scholar]
- von der KH, Hanes J, Klaudiny J, Scheit KH. “A human amyloid precursor-like protein is highly homologous to a mouse sequence-specific DNA-binding protein.”. DNA Cell. Biol. 1994;13(11):1137–1143. doi: 10.1089/dna.1994.13.1137. [DOI] [PubMed] [Google Scholar]
- Werner T. “Target gene identification from expression array data by promoter analysis.”. Biomol. Eng. 2001;17(3):87–94. doi: 10.1016/s1389-0344(00)00071-x. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Inoue I. “Conditional expression of MCM7 increases tumor growth without altering DNA replication activity.”. FEBS Lett. 2003;553(1–2):213–217. doi: 10.1016/s0014-5793(03)01018-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| Accession | Cluster | Name | Expression | Fold Diff |
|---|---|---|---|---|
| NM_001644.2 | 560 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) | Exclusive | 54 |
| NM_001804.1 | 1545 | caudal type homeo box transcription factor 1 (CDX1) | Exclusive | 14 |
| NM_005814 | 143131 | glycoprotein A33 (transmembrane) (GPA33) | Exclusive | 11 |
| NM_001986.1 | 77711 | ets variant gene 4 (E1A enhancer binding protein, E1AF) (ETV4) | Significant | 20 |
| NM_001265.2 | 77399 | caudal type homeo box transcription factor 2 (CDX2) | Significant | 20 |
| NM_138768.1 | 116051 | myeloma overexpressed gene positive multiple myelomas) (MYEOV) | Significant | 14 |
| NM_004963.1 | 1085 | guanylate cyclase 2C (heat stable enterotoxin receptor) (GUCY2C) | Significant | 13 |
| NM_024017.3 | 86327 | homeo box B9 (HOXB9) | Significant | 6 |
| XM_032721.3 | 109358 | ATPase, Class V, type 10B (ATP10B) | Significant | 5 |
| NM_033266.1 | 114905 | ER to nucleus signalling 2 (ERN2) | Significant | 4 |
| NM_019010.1 | 84905 | cytokeratin 20 (KRT20) | Significant | 4 |
| NM_005310.1 | 86859 | growth factor receptor-bound protein 7 (GRB7) | Significant | 4 |
| NM_001738.1 | 23118 | carbonic anhydrase I (CA1) | Significant | 4 |
| NM_004306.1 | 181107 | annexin A13 (ANXA13) | Significant | 3 |
| NM_007028.2 | 91096 | tripartite motif-containing 31 (TRIM31) | Significant | |
| NM_001500.1 | 1054435 | GDP-mannose 4,6-dehydratase (GMDS) | Preferential | 39 |
| NM_005628.1 | 183556 | solute carrier family 1 (neutral amino acid transporter), member 5 (SLC1A5) | Preferential | 36 |
| NM_002276.2 | 182265 | keratin 19 (KRT19) | Preferential | 33 |
| NM_001569.2 | 182018 | interleukin-1 receptor-associated kinase 1 (IRAK1) | Preferential | 23 |
| NM_002295 | 356261 | laminin receptor 1 (67kD, ribosomal protein SA) (LAMR1) | Preferential | 19 |
| NM_001402 | 493552 | eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) | Preferential | 19 |
| NM_002087 | 180577 | granulin (GRN) | Preferential | 19 |
| NM_006597 | 180414 | heat shock 70kD protein 8 (HSPA8) | Preferential | 18 |
| NM_005507 | 170622 | cofilin 1 (non-muscle) (CFL1) | Preferential | 17 |
| NM_001903 | 254321 | catenin (cadherin-associated protein), alpha 1 (102kD) (CTNNA1) | Preferential | 17 |
| NM_002819 | 172550 | polypyrimidine tract binding protein 1 (PTBP1) | Preferential | 17 |
| NM_007363 | 355861 | non-POU domain containing, octamer-binding (NONO) | Preferential | 17 |
| NM_002568 | poly(A) binding protein, cytoplasmic 1 (PABPC1) | Preferential | 16 | |
| NM_006516 | 169902 | solute carrier family 2 (facilitated glucose transporter), member 1 (SLC2A1) | Preferential | 16 |
| NM_002046 | glyceraldehyde-3-phosphate dehydrogenase (GAPD) | Preferential | 16 | |
| NM_003906 | 389037 | MCM3 minichromosome maintenance deficient 3 protein (MCM3AP) | Preferential | 15 |
| NM_004433 | 67928 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) (ELF3) | Preferential | 14 |
| NM_007127 | 166068 | villin 1 (VIL1) | Preferential | 14 |
| NM_000218 | 367809 | potassium voltage-gated channel, KQT-like subfamily, member 1 (KCNQ1) | Preferential | 13 |
| NM_003379 | 403997 | villin 2 (ezrin) (VIL2) | Preferential | 13 |
| NM_001084 | 153357 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (PLOD3) | Preferential | 12 |
| NM_005789 | 152978 | proteasome (prosome, macropain) activator subunit 3 (PSME3) | Preferential | 12 |
| NM_005561 | 150101 | lysosomal-associated membrane protein 1 (LAMP1) | Preferential | 11 |
| NM_005080 | 437638 | X-box binding protein 1 (XBP1) | Preferential | 11 |
| NM_002105 | H2A histone family, member X (H2AFX) | Preferential | 11 | |
| NM_004429 | 144700 | ephrin-B1 (EFNB1) | Preferential | 10 |
| NM_014498 | golgi phosphoprotein 4 (GOLPH4) | Preferential | 9 | |
| 139800 | high-mobility group (nonhistone chromosomal) protein isoforms (HMGIY) | Preferential | 9 | |
| NM_007052 | 132370 | NADPH oxidase 1 (NOX1) | Preferential | 9 |
| NM_001416 | 129673 | eukaryotic translation initiation factor 4A, isoform 1(EIF4A1) | Preferential | 9 |
| NM_004655 | 127337 | axin 2 (conductin, axil) (AXIN2) | Preferential | 9 |
| NM_004442 | 125124 | EphB2 (EPHB2) | Preferential | 9 |
| NM_000967 | 119598 | ribosomal protein L3 (RPL3) | Preferential | 8 |
| NM_005063 | 119597 | stearoyl-CoA desaturase (delta-9-desaturase) (SCD) | Preferential | 8 |
| NM_000090 | 443625 | collagen, type III, alpha 1 (COL3A1) | Preferential | 8 |
| NM_012423 | 419535 | ribosomal protein L13a (RPL13A) | Preferential | 8 |
| Preferential | ||||
| NM_006026 | 75307 | H1 histone family, member X (H1FX) | Preferential | 8 |
| NM_001923 | 290758 | damage-specific DNA binding protein 1 (127kD) (DDB1) | Preferential | 8 |
| NM_032044 | 105484 | regenerating gene type IV (REG-IV) | Preferential | 8 |
| NM_003258 | 105097 | thymidine kinase 1, soluble (TK1) | Preferential | 7 |
| XM_039877 | 102482 | mucin 5, subtype B, tracheobronchial (MUC5B) | Preferential | 7 |
| NM_005724 | 100090 | tetraspan 3 (TSPAN-3) | Preferential | 7 |
| NM_000972 | 416801 | ribosomal protein L7a (RPL7A) | Preferential | 7 |
| NM_018952 | 98428 | homeo box B6 (HOXB6) | Preferential | 7 |
| NM_015925 | 312129 | Similar to liver-specific bHLH-Zip transcription factor | Preferential | 7 |
| NM_000075 | 95577 | cyclin-dependent kinase 4 (CDK4) | Preferential | 6 |
| NM_006408 | 226391 | anterior gradient 2 homolog (Xenepus laevis) (AGR2) | Preferential | 6 |
| NM_004044 | 90280 | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (ATIC) | Preferential | 6 |
| NM_004494 | 89525 | hepatoma-derived growth factor (high-mobility group protein 1-like) (HDGF) | Preferential | 6 |
| NM_004063 | 89436 | cadherin 17, LI cadherin (liver-intestine) (CDH17) | Preferential | 6 |
| NM_000213 | 85266 | integrin, beta 4 (ITGB4) | Preferential | 6 |
| NM_001730 | 84728 | Kruppel-like factor 5 (intestinal) (KLF5) | Preferential | 6 |
| NM_001255 | 82906 | CDC20 cell division cycle 20 homolog (S. cerevisiae) (CDC20) | Preferential | 5 |
| NM_001747 | 82422 | capping protein (actin filament), gelsolin-like (CAPG) | Preferential | 5 |
| NM_002534 | 442936 | 2′,5′-oligoadenylate synthetase 1 (40–46 kD) (OAS1) | Preferential | 5 |
| NM_000178 | 82327 | glutathione synthetase (GSS) | Preferential | 5 |
| NM_000903 | 406515 | NAD(P)H dehydrogenase, quinone 1 (NQO1) | Preferential | 5 |
| NM_002394 | 79748 | solute carrier family 3 member 2 (SLC3A2) | Preferential | 5 |
| NM_005567 | 79339 | lectin, galactoside-binding, soluble, 3 binding protein (LGALS3BP) | Preferential | 5 |
| NM_000404 | galactosidase, beta 1 (GLB1) | Preferential | 5 | |
| NM_006907 | 458332 | pyrroline-5-carboxylate reductase 1 (PYCR1) | Preferential | 5 |
| NM_000291 | 78771 | phosphoglycerate kinase 1 (PGK1) | Preferential | 5 |
| NM_002635 | 290404 | solute carrier family 25 member 3 (SLC25A3) | Preferential | 5 |
| NM_001640 | 221589 | N-acylaminoacyl-peptide hydrolase (APEH) | Preferential | 5 |
| NM_005030 | 329989 | polo-like kinase (Drosophila) (PLK) | Preferential | 5 |
| NM_002224 | 77515 | inositol 1,4,5-triphosphate receptor, type 3 (ITPR3) | Preferential | 4 |
| NM_002668 | 77422 | proteolipid protein 2 (colonic epithelium-enriched) (PLP2) | Preferential | 4 |
| NM_016343 | 77204 | centromere protein F (350/400kD, mitosin) (CENPF) | Preferential | 4 |
| NM_005916 | 438720 | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) (MCM7) | Preferential | 4 |
| NM_001006 | 356572 | ribosomal protein S3A (RPS3A) | Preferential | 4 |
| NM_000701 | 371889 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (ATP1A1) | Preferential | 4 |
| NM_000990 | 356542 | ribosomal protein L27a (RPL27A) | Preferential | 4 |
| NM_015379 | 410497 | brain protein I3 (BRI3) | Preferential | 4 |
| NM_012408 | 191990 | protein kinase C binding protein 1 (PRKCBP1) | Preferential | 4 |
| NM_002773 | 75799 | protease, serine, 8 (prostasin) (PRSS8) | Preferential | 4 |
| NM_002951 | 406532 | ribophorin II (RPN2) | Preferential | 4 |
| NM_001673 | 446546 | asparagine synthetase (ASNS) | Preferential | 4 |
| NM_002862 | 145820 | phosphorylase, glycogen; brain (PYGB) | Preferential | 4 |
| NM_000918 | 410578 | procollagen-proline, 2-oxoglutarate 4-dioxygenase (P4HB) | Preferential | 3 |
| NM_000228 | 436983 | laminin, beta 3 (nicein (125kD), kalinin (140kD), BM600 (125kD) (LAMB3) | Preferential | 3 |
| NM_001034 | 226390 | ribonucleotide reductase M2 polypeptide (RRM2) | Preferential | 3 |
| NM_003217 | 35052 | testis enhanced gene transcript (BAX inhibitor 1) (TEGT) | Preferential | 3 |
| NM_001658 | 286221 | ADP-ribosylation factor 1 (ARF1) | Preferential | 3 |
| NM_000014 | alpha-2-macroglobulin (A2M) | Preferential | 3 | |
| NM_007355 | 74335 | heat shock 90kD protein 1, beta (HSPCB) | Preferential | 3 |
| NM_001288 | 414565 | chloride intracellular channel 1 (CLIC1) | Preferential | 3 |
| Preferential | ||||
| NM_007367 | 74111 | RNA binding protein (RALY) | Preferential | 3 |
| NM_002483 | 436718 | carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) | Preferential | 3 |
| NM_021220 | 386387 | zinc finger protein 339 (ZNF339) | Preferential | 3 |
| NM_001202 | 68879 | bone morphogenetic protein 4 (BMP4) | Preferential | 3 |
| NM_000224 | 406013 | keratin 18 (KRT18) | Preferential | 3 |
| NM_019894 | 414005 | transmembrane protease, serine 4 (TMPRSS4) | Preferential | 3 |
| NM_002032 | 448738 | ferritin, heavy polypeptide 1 (FTH1) | Preferential | 3 |
| NM_016276 | 62863 | serum/glucocorticoid regulated kinase 2 (SGK2) | Preferential | 3 |
| NM_003756 | 127149 | eukaryotic translation initiation factor 3, subunit 3 (gamma, 40kD) (EIF3S3) | Preferential | 3 |
| NM_003751 | 371001 | eukaryotic translation initiation factor 3, subunit 9 (eta, 116kD) (EIF3S9) | Preferential | 3 |
| NM_004526 | 57101 | MCM2 minichromosome maintenance deficient 2, (S. cerevisiae) (MCM2) | Preferential | 3 |
| NM_021978 | 56937 | suppression of tumorigenicity 14 (colon carcinoma, epithin) (ST14) | Preferential | 3 |
| NM_006187 | 129895 | 2′-5′-oligoadenylate synthetase 3 (100 kD) (OAS3) | Preferential | 3 |
| NM_003753 | 55682 | eukaryotic translation initiation factor 3, subunit 7 (zeta, 66/67kD) (EIF3S7) | Preferential | 3 |
| NM_001712 | 512682 | carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) | Preferential | 3 |
| NM_005727 | tetraspan 1 (TSPAN-1) | Preferential | 3 | |
| NM_021102 | 31439 | serine protease inhibitor, Kunitz type, 2 (SPINT2) | Preferential | 3 |
| NM_007183 | 26557 | plakophilin 3 (PKP3) | Preferential | 3 |
| NM_001306 | 25640 | claudin 3 (CLDN3) | Preferential | 3 |
| NM_004572 | 25051 | plakophilin 2 (PKP2) | Preferential | 3 |
| NM_004289 | 404741 | nuclear factor (erythroid-derived 2)-like 3 (NFE2L3) | Preferential | 3 |
| NM_003627 | 444159 | SLC43A1 | Preferential | 2 |
| NM_005498 | 18894 | adaptor-related protein complex 1, mu 2 subunit (AP1M2) | Preferential | 2 |
| NM_005558 | 18141 | ladinin 1 (LAD1) | Preferential | 2 |
| NM_002707 | 17883 | protein phosphatase 1G magnesium-dependent, gamma isoform (PPM1G) | Preferential | 2 |
| NM_020384 | 16098 | claudin 2 (CLDN2) | Preferential | 2 |
| NM_001614 | 14376 | actin, gamma 1 (ACTG1) | Preferential | 2 |
| NM_052854 | 405961 | old astrocyte specifically induced substance (OASIS) | Preferential | 2 |
| NM_016234 | 11638 | fatty-acid-Coenzyme A ligase, long-chain 5 (FACL5) | Preferential | 2 |
| NM_021107 | 411125 | mitochondrial ribosomal protein S12 (MRPS12) | Preferential | 2 |
| NM_002335 | 6347 | low density lipoprotein receptor-related protein 5 (LRP5) | Preferential | 2 |
| NM_022085 | 430169 | thioredoxin related protein (MGC3178) | Preferential | 2 |
| NM_033049 | 5940 | mucin 13, epithelial transmembrane (MUC13) | Preferential | 2 |
| NM_014865 | chromosome condensation-related SMC-associated protein 1 (CNAP1) | Preferential | 2 | |
| NM_006098 | 5662 | guanine nucleotide binding protein beta polypeptide 2-like 1 (GNB2L1) | Preferential | 2 |
| NM_024526 | 5366 | epidermal growth factor receptor pathway related protein 3 (EPS8R3) | Preferential | 1 |
| NM_006149 | 5302 | lectin, galactoside-binding, soluble, 4 (galectin 4) (LGALS4) | Preferential | 1 |
| NM_014275 | 437277 | mannosyl (alpha-1,3-) (MGAT4B) | Preferential | 1 |
| NM_003752 | 388163 | eukaryotic translation initiation factor 3, subunit 8 (110kD) (EIF3S8) | Preferential | |
| 3989 | plexin B2 (PLXNB2) | Preferential | ||
| NM_002447 | 2942 | macrophage stimulating 1 receptor (c-met-related tyrosine kinase) (MST1R) | Preferential | |
| NM_001038 | sodium channel, nonvoltage-gated 1 alpha (SCNN1A) | Preferential | ||
| NM_002083 | 2704 | glutathione peroxidase 2 (gastrointestinal) (GPX2) | Preferential | |
| NM_005186 | 356181 | calpain 1, (mu/I) large subunit (CAPN1) | Preferential | |
| NM_001404 | 256184 | eukaryotic translation elongation factor 1 gamma (EEF1G) | Preferential | |
| NM_003334 | 406683 | ubiquitin-activating enzyme E1 (UBE1) | Preferential | |
| NM_005998 | 1708 | chaperonin containing TCP1, subunit 3 (gamma) (CCT3) | Preferential | |
| NM_012073 | 1600 | chaperonin containing TCP1, subunit 5 (epsilon) (CCT5) | Preferential | |
| NM_000077 | 421349 | cyclin-dependent kinase inhibitor 2A (melanoma (CDKN2A) | Preferential | |
| NM_002014 | 848 | FK506 binding protein 4 (59kD) (FKBP4) | Preferential | |
| NM_004502 | 436181 | homeo box B7 (HOXB7) | Preferential | |
| NM_004966 | 808 | heterogeneous nuclear ribonucleoprotein F (HNRPF) | Preferential | |
| NM_002354 | 692 | tumor-associated calcium signal transducer 1 (TACSTD1) | Preferential | |
| NM_005435 | 334 | Rho guanine nucleotide exchange factor (GEF) 5 (ARHGEF5) | Preferential | |
| NM_002457 | 458274 | mucin 2, intestinal/tracheal (MUC2) | Preferential | |
| NM_000968 | 186350 | ribosomal protein L4 (RPL4) | Preferential |