Abstract
The oncogene MCTS1, discovered as an amplified product in a subset of T-cell lymphoma lines, has been implicated in cell cycle progression and conferring a growth advantage in lymphomas and breast cancer. Recent research shows that it modulates the MAPK pathway and acts as a translational activator both in vivo and in vitro. In breast cancer cells, expression of MCTS1 confers aggressive properties and inhibits apoptosis. This article will review these data and its implications on our understanding of cancer.
Keywords: oncogene, cell cycle, DNA damage, angiogenesis, pseudogene
Introduction
The neoplastic transformation of a normal cell, or oncogenesis, can be induced through the dysregulation of specific genes. This usually involves a gain of function in an oncogene and/or loss of function in a tumor suppressor gene. The stringent control of essential cellular mechanisms involving proliferation, cycle progression and apoptosis defines normal cellular behavior. Oncogenes are at the center of controlling these mechanisms through signal transduction wherein their proteins relay extra-cellular messages to the nucleus, causing changes in the cellular transcriptional patterns. Genetic modifications that dysregulate oncogenes or tumor suppressors, such as gene mutation, amplification and translocation, can perturb cellular signaling pathways resulting in the exhibition of neoplastic properties.
While gene amplification is a common mechanism of oncogenesis in solid tumors, chromosomal translocations are often found in hematologic malignancies and play a major role in lymphomagenesis. The role of gene amplification in lymphomagenesis remained poorly established, until newer genomic techniques became available. MCT-1 or multiple copies in a T-cell malignancy is a newly identified oncogene that is amplified in a variety of T-cell and B-cell lymphomas. The discovery of the MCT-1 gene, its oncogenic properties and its proposed mechanism of action at the cellular level will be reviewed in this article. Since the MCT-1 acronym is also shared by the monocarboxylate transporters, the oncogene was renamed MCTS1 in both the NCBI and Ensembl databases.
The Oncogene Mcts1
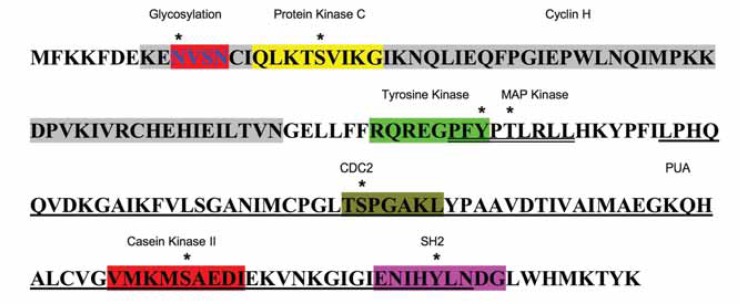
MCTS1 was discovered in 1998 using arbitrary-primed-PCR (AP-PCR), a technique that employs random primers in order to amplify unknown stretches of DNA (Prosniak et al 1998). By screening a panel of T-cell leukemia/lymphoma lines through this approach an amplified DNA sequence encoding a novel gene was identified in the HUT 78 T-cell line, thus the name multiple copies in a T-cell malignancy (MCT-1). Comparative studies showed no significant homology of MCTS1 to known sequences in the GenBank database, and expression analysis of a variety of normal human tissues revealed low level ubiquitous expression of the newly sequenced gene. MCTS1 was mapped to the long arm of chromosome Xq22–24. The gene spans 8.5 kbp of the human genome with a transcript length of 1018 that contains 6 exons. The 5′ and 3′ ends of the transcript have untranslated regions with respective sizes of 257 and 215 bp. The translated protein has 181 amino acids that form a number of post-translational modification sites. The most studied of these are the MAPK, CDC2 and the PUA domains. The PUA domain, named after PseudoUridine synthase and Archaeosine-specific transglycosylase, is a domain widely conserved in eukaryotic as well as archaeal RNA modification enzymes (Aravind and Koonin, 1999). This domain is found in a conserved family of translation factors. These are detailed in Figure 1.
Figure 1.
The different putative and established domains in the MCTS1 protein are shown. The gray shaded area is the Cyclin H homology domain. The tyrosine kinase (green) and the MAPK domain (double underlined) overlap by 3 amino acids. The PUA domain is single underlined and encompasses CDC2 (dark yellow) Casein Kinase II (red) and SH2 (purple).
In humans, MCTS1 is normally expressed in almost all tissues except the lungs where it is significantly absent (Prosniak et al. 1998). Elevated levels of MCTS1 protein were particularly found in IL-2-independent T-cell lines, in contrast to the IL-2-dependent T-cell lines. Also, increased levels of MCTS1 protein were found in a panel of transformed B-cell lines derived from patients with Non-Hodgkin Lymphoma (NHL) and a subset of primary Diffuse Large B-Cell Lymphoma (DLBCL) (Shi et al. 2003).
MCTS1 has several hallmarks of an oncogene. In NIH 3T3 fibroblasts, MCTS1 decreased cell-doubling time, dramatically shortened the duration of G1 transit time and/or G1-S transition as compared to controls. MCTS1 transformed NIH 3T3 cells in a colony forming assay, and promoted anchorage-independent growth with formation of colonies in a soft-agar assay in transfected NIH 3T3 cells and MCF10A cells. It also protected IL-2 dependent T-cell line (EC155) from apoptosis in annexin V binding assay, suggesting that MCTS1 may activate survival-related pathways (Prosniak et al. 1998; Dierov et al. 1999; Hsu et al. 2005).
The Cell Cycle and Mcts1
Deregulation of the cell cycle is one of the most important contributing factors to oncogenesis (Deshpande et al. 2005; Lee and Yang, 2003). The eukaryotic cell cycle is controlled by cyclins and their associated kinases (cdk) whose activity is regulated by phosphorylation and dephosphorylation (Collins and Garrett, 2005; Sherr and Roberts 1999). While the level of cdks remains constant during the cell cycle, that of their corresponding cyclins varies at critical checkpoints. This process assures the appropriate entry of a cell into the various cycle phases. Dysregulation of a cell cycle checkpoint can thus result in uncontrolled growth and compromise the integrity of DNA replication (Owa et al. 2001). Cyclin D1 along with cdk4 and cdk6 controls the G1/S checkpoint. In early G1 phase, growth factors and mitogens through several signaling pathways activate the synthesis of D cyclins (D1, D2 and D3) which then bind cdk4 and cdk6 (Michalides et al. 2002). The cyclin D-cdk complexes then translocate to the nucleus where they are phosphorylated and thus form activating complexes. These phosphorylate Retinoblastoma protein (Rb) which in turn releases transcription factors of the E2F family. At late G1, cyclin E and cyclin A forms complexes with cdk2 that promotes progression into the synthesis (S) phase of the cell cycle, a review of which is beyond the scope of this article. However, this progression is also negatively regulated by two groups of proteins: the Kinase Inhibiting Proteins (KIP)/Cyclin Inhibiting Proteins (CIP) and the Inhibitors of cdk4 (Ink4). The protein p27Kip1 can inactivate the D cyclins by interacting with their binding to cdk4 and cdk6. Ink4 proteins like p16INK4a, p15INK4b, p18INK4c and p19INK4d that bind only to cdk4 and cdk6 prevent phosphorylation of Rb, thus inhibiting the release of the E2F factors (Michalides et al. 2002). Both result in blocking the progression through the G1 phase. It is therefore not surprising that aberrations in the G1/S checkpoint control result in tumorigenesis. This can occur through either inactivating mutations of the negative regulators or activating mutations of the positive signals. Indeed a number of studies have reported amplification of the cyclin D1 gene in breast cancers, non-small cell lung carcinoma (Owa et al. 2001) and head and neck cancers (Michalides et al. 2002). Additionally, cyclin D3 has been found to be overexpressed in several B-cell malignancies such as diffuse large B-cell lymphomas and multiple myelomas (Deshpande et al. 2005).
Initial studies with MCTS1 revealed that it shortened the G1/S transition time (Prosniak et al. 1998). It also increased the percentage of cells in the S phase while reducing that in the G1 phase, suggesting a compromise of the G1/S checkpoint. Overexpression of MCTS1 resulted in increased level of cyclin D1 as well as increased complex formation of both cyclin D1-cdk4 and cyclin D1-cdk6 in cell lines. This data was also confirmed by showing enhanced cdk4 and cdk6 kinase activity in vitro with secondary increased phosphorylation of Rb protein (Dierov et al. 1999).
The levels of MCTS1 protein during the various phases of the cell cycle were examined in T-cell lines and were found to be invariant and predominantly localized to the cytoplasm, suggesting an indirect role of MCTS1 in the G1/S activation (Herbert et al. 2001). T-cells normally do not divide, but when stimulated by antigens, T-cells upregulate their expression of IL-2Rα to which IL-2 binds and stimulates T-cell division (Smith, 1988). While some T-cell lines require IL-2 in order to divide and proliferate, others are growth factors independent. Analyses of several T-cell lines reveal that those that express MCTS1 are independent of IL-2 (Dierov et al. 1999). Thus MCTS1 might confer a mitotic advantage to T-cells rendering them IL-2 independent.
The reasons behind the inappropriate activation of the G1/S phase of the cell cycle by MCTS1 are still unfolding, but evidence suggests an indirect role which is independent of mitogens, but affecting the delicate balance between the activators and inhibitors of the G1 cyclins.
Mcts1, Mapk and Dna Damage
It has been reported that there is an increase in expression of cyclin D1 following DNA damage. MCTS1 protein levels in PBLs also increase after treatment with Adriamycin or 10 Gy radiation in as little as 2 h post treatment (Herbert et al. 2001). MCTS1 allows bypassing the G1/S checkpoint and pushes cells into the S phase following DNA damage. This was observed in MCF-7 breast cancer cells overexpressing MCTS1 (Hsu et al. 2005). Upon DNA damage, a number of checkpoints are activated in these cells, notably the phosphorylation of H2AX (Fillingham et al. 2006). Histone H2AX is the immediate downstream target of DNA damage activated ATM. Phosphorylation of H2AX (γ-H2AX) is a measure of double strand breaks (DSB) in the DNA (Fillingham et al. 2006; Hsu et al. 2005). Following DNA damage, there is a rapid induction of γ-H2AX at DNA breakpoints. This decreases over time as these DSBs are repaired (Fillingham et al. 2006). The number of γ-H2AX foci in the MCTS1 overexpressing cells was higher than in the control cells and was present at later time points compared to the control. This increased response to DSB in MCTS1 cells could potentially promote genomic instability (Aten et al. 2004). The γ-H2AX induction was also observed in IL-2 dependent T-cell line EC155 overexpressing MCTS1 showing cell-type independency.
The increase of MCTS1 protein level following DNA damage is due to post-translational modification as no corresponding increase in RNA or protein synthesis is observed (Herbert et al. 2001). Phosphorylation is the most common mechanism for post-translational control of protein activity. MCTS1 protein contains a number of putative phosphorylation sites. Of these, the MAPK phosphorylation site seems to be the most relevant and important. A point mutation at this site abrogated the phosphorylation at this site as evidenced by an in vitro kinase assay. A growth assay in NIH3T3 cells and in Human Dermal Fibroblasts that had stably integrated wtMCTS1 and this MAPK mutant (hereafter termed T81A; Nandi et al. 2006a), showed that the MAPK mutant grows at a significantly slower rate almost at the base vector only growth level (Nandi et al. 2006a). The basal level of this protein is also low compared to the wt MCTS1. This mutation is therefore critical as it takes away the proliferative advantage of MCTS1 and probably inhibits it action as a G1/S mitogen. Response to DNA damaging radiation was also different in wt MCTS1 and its MAPK mutant. In the Burkitt lymphoma cell line Daudi, stably expressing MCTS1, the level of MCTS1 increased in as little as 2 h with a significant increase in 24 h after irradiation. Using MEK inhibitors U0126 and PD98059, this increase was dramatically curtailed in this cell line. In NIH3T3 cell lines, stably over-expressing MCTS1 and the mutant, the MAPK mutant showed low levels initially, as described earlier, that decreased further in response to γ-irradiation. Also, the levels of phospho-MCTS1 increased after irradiation in the wt protein but not in the MAPK mutant. The same effects were observed in the T-cell line Jurkat, which has a high endogenous level of MCTS1 (Nandi et al. 2006a). These experiments show that the MAPK kinase pathway is critical for the activation of MCTS1 and its stabilization after DNA damage. It is possible that following DNA damaging irradiation, phospho-MCTS1 levels increase to protect cells against G1/S arrest by acting on cyclin D1 and thus pushing cells through the cell cycle. Inhibition of MCTS1 phosphorylation either through mutation or by inhibitors prevents this cell-cycle mitogenic activity. The exact mechanism remains to be investigated further.
It is interesting to note that MCTS1 also has a Cyclin H domain (Prosniak et al. 1998) with cdk7 as its kinase. Cdk7 forms heterodimeric complex with Cyclin H and activates other cell cycle cyclins, notably cdk1 and cdk2 by phosphorylation. The cyclinH/cdk7 complex has also been implicated in the transcriptional machinery. It is a member of the Transcription Factor IIH (TFIIH) complex and has been shown to be essential for phosphorylation of the C-terminal Domain of RNA polymerase II (Feaver et al. 1994; Shiekhattar et al. 1995). TFIIH is also part of the DNA repair machinery and phosphorylates both cdc2 and cdk2 (Shiekhattar et al. 1995). The role of MCTS1 in these processes has not yet been investigated.
The Ras/Raf/MAPK pathway has been shown to be involved in activating cyclin D1 and the G1/S transition machinery (Lukas et al. 1996). It is possible that phospho-MCTS1 activates cyclin D1 through a MAPK dependent pathway, the exact mechanism of which needs to be determined. It has been shown that cyclin D1 transgenic mice have lymphopenia, and that double transgenic mice (with myc) frequently have full-blown lymphomas (Bodrug et al. 1994; Lovec et al. 1994). It will be interesting to see if MCTS1 and cyclin D1 co-operate in the same way towards lymphomagenesis.
Mcts1 and Translation
The mechanisms that allow MCTS1 to display such a wide array of activity are still unfolding. The search through the conserved domain database revealed that MCTS1 protein contained a recently described RNA-binding domain designated PUA domain. The PUA domain is widely conserved in several tRNA and rRNA modification enzymes suggesting that MCTS1 might have an RNA-binding function (Aravind and Koonin, 1999; Reinert et al. 2006). The PUA domain can also be found in several eukaryotic proteins that contain SUI1, a domain homologous to the eukaryotic initiation factor eIF1, inferring that PUA containing proteins may have a role in translational regulation. In fact, translational control is an important step in the regulation of gene expression and subsequent control of cell growth, differentiation, or cell death.
Using a yeast two-hybrid system, a human HeLa cell cDNA library was screened with MCTS1 as a bait to determine which proteins MCTS1 physically interacts with. A total of 16 positive clones were fished out, all of which encoded for the same protein known as Density-regulated protein (DENR/DRP). DENR has a translation initiation domain known EIF1/SUI1 that is required for fidelity in recognition of the start codon (He et al. 2003). Since initiation is the rate limiting step in mRNA translation and thus is commonly the primary target for translational control, the MCTS1-DENR protein complex may be involved in the regulation of translation initiation, affecting the efficiency of expression of key proteins involved in the induction of cell proliferation and activation of survival-related pathways.
Initially, in vitro experiment showed that MCTS1 was able to bind the cap complex present at the 5′ ends of mRNA, but only in the presence of eIF4E, which is an essential component of the translation machinery (Gingras et al. 1999). Reinert et al. (2006) showed that the PUA domain of MCTS1 was necessary for interaction with the cap complex but the full length MCTS1 was required for interaction of DENR with the cap. Further support to the putative role of the MCTS1/DENR complex in controlling translation initiation came from polysome isolation assays demonstrating that that MCT-1 and DENR co-sedimented in the same fractions as the eIF2 component of the translation initiation complex (Reinert et al. 2006).
In vivo, the effect of MCTS1 overexpression was analyzed using a microarray based assay that used polysomal RNA from MCTS1 transfected cells compared with vector control cells. Western blot analyses confirmed that protein levels of selectively recruited mRNAs to polysomes were also increased accordingly. Real-time PCR on total mRNA form MCTS1 transfected cells as compared to vector control cells verified that the increased protein levels of these genes was a reflection of enhanced translational efficiency rather than increased transcription. Among these genes, cyclin D1, MRE11A, BCL2L and E2F1 were the most notable ones. These findings were also confirmed reversely in a cell-based assay by utilizing an MCTS1- siRNA knock-down approach (Reinert et al. 2006). A global array needs to be looked into for further characterizing the translational effects of MCTS1.
Mcts1 and Angiogenesis
As mentioned earlier, MCTS1 was studied in the context of DNA damage in MCF7 breast cancer cells (Hsu et al. 2005). Levenson et al. (2005) also studied the role of MCTS1 in breast cancer in detail. Breast cancer cells that express the Estrogen Receptor (ER) are in general less invasive and have better prognosis. MCF7 cells fall in this category. Screening several different breast cancer cell lines with known ER status, it was found that MCTS1 is endogenously expressed at robust levels in ER–negative lines and that this expression correlates with aggressiveness. MCF7 cells have a very low expression of MCTS1, consistent with their ER-positive status. However, the expression of MCTS1 in MCF-7 cells conferred aggressive behavior as demonstrated in an in vitro migration assay. Compared to vector control, the invasiveness of the MCTS1 expressing clone was very high and was almost equivalent to the ER-negative cell line MDA-MB-231. The functionality of the ER was however not lost as it could be stimulated and inhibited by Estradiol and Fulvestrant respectively. In vivo tumor formation assay on ovarectomized nude (athymic) mice resulted in larger tumors than the control group. MCTS1 expressing cells resulted in larger tumors that had increased microvasculature with tightly packed cells and very few apoptotic cells as compared to the control group. The levels of angiogenesis inhibitor, thrombospondin1 (TSP1) was decreased in these MCTS1 expressing cells both at the protein and the mRNA level, whereas the pro-angiogenic factor VEGF had higher mRNA but same levels of the protein. It has been reported that overexpression of ErbB2/Her2/Neu is most commonly associated with aggressive tumors ( Badache and Gonçalves, 2006). The ErbB2 signalling pathway activates the Raf/MEK/ERK signaling pathway among others ( Badache and Gonçalves, 2006). It would be interesting to delineate the status of the Her2/Neu pathway under MCTS1 overexpressing conditions and determine whether MCTS1 is capable of activating a latent Her2/Neu signaling mechanism.
Mcts1: Pseudogene and Evolution
One interesting facet of cancer is that there is hardly any report of oncogenesis outside the mammalian class (Bubanovic and Najman, 2005). Oncogenes and tumour suppressors have not been studied in much detail from an evolutionary point of view. Using a comparative genomics approach, an ortholog of MCTS1 has been identified in archaebacteria (Matte-Taillez et al. 2000; Nandi et al. 2006b). MCTS1 is conserved throughout evolution and is present in several organisms including worms, flies, fish, rodents and non-human primates. At the genomic level, MCTS1 has a fully processed (Type II) pseudogene. This pseudogene is present in a highly amplified region of the genome in Chromosome 20. This region of the genome is frequently amplified in breast and ovarian cancers (Nandi et al. 2006b). Unlike the MCTS1 protein sequence, the pseudogene is present in its entirety only in the non-human primates genome (chimp and rhesus monkey), and partially (∼65%) in the rodent and dog genome (Nandi et al. 2006b). This implies that the insertion of the pseudogene is a relatively recent event (probably not earlier than 35mya). Insertion of a Type II pseudogene most likely indicates a high transcriptional activity, suggesting that this gene has been highly active transcriptionally since that period in time.
Conclusion
A single mutation does not cause cancer, but may give a subpopulation of cells an unrestrained proliferative advantage by abrogating the cell cycle checkpoints. In such uncontrolled environment more genetic mutations will accumulate, conferring these cells additional properties like inhibition of apoptosis and induction of angiogenesis resulting in malignant transformation. MCTS1 is an unique oncogene that acts on a subpopulation of cells in different ways: by deregulating the cell cycle, inappropriately activating kinases, causing DNA damage and activating other oncogenes at the translation level, possibly in that order. Further studies are required to determine the critical mechanistic pathway for its oncogenic activity.
References
- Aravind L, Koonin EV. Novel predicted RNA-binding domains associated with the translational machinery. J. Mol. Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Badache A, Gonçalves A. The ErbB2 signalling network as a target for breast cancer therapy. J. Mammary Gland Biol. Neoplasia. 2006;11:13–25. doi: 10.1007/s10911-006-9009-1. [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Warner BJ, Bath ML, et al. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubanovic I, Najman S. Comparative oncology and comparative tumor immunology. J. Biol. Sci. 2005;5:114–118. [Google Scholar]
- Collins I, Garrett MD. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr. Op. Pharm. 2005;5:366–373. doi: 10.1016/j.coph.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24:2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- Dierov J, Prosniak M, Gallia G, et al. Increased G1 cyclin/cdk activity in cells overexpressing the candidate oncogene MCT-1. J. Cell Biochem. 1999;74:544–550. [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, et al. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Fillingham J, Keogh M-C, Krogan NJ. γH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh R, et al. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell Biol. 2003;23:5431–5445. doi: 10.1128/MCB.23.15.5431-5445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert GB, Shi B, Gartenhaus RB. Expression and stabilization of the MCT-1 protein by DNA damaging agents. Oncogene. 2001;20:6777–6783. doi: 10.1038/sj.onc.1204881. [DOI] [PubMed] [Google Scholar]
- Hsu H-L, Shi B, Gartenhaus RB. The MCT-1 oncogene product impairs cell cycle checkpoint control and transforms human mammary epithelial cells. Oncogene. 2005;24:4956–4964. doi: 10.1038/sj.onc.1208680. [DOI] [PubMed] [Google Scholar]
- Lee M-H, Yang H-Y. Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev. 2003;22:435–449. doi: 10.1023/a:1023785332315. [DOI] [PubMed] [Google Scholar]
- Levenson AS, Thurn KE, Simons LA, et al. MCT-1 oncogene contributes to increased in vivo tumorigenicity of MCF7 cells by promotion of angiogenesis and inhibition of apoptosis. Cancer Res. 2005;65:10651–10656. doi: 10.1158/0008-5472.CAN-05-0845. [DOI] [PubMed] [Google Scholar]
- Lovec H, Grzeschiczek A, Kowalski M-B, et al. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Bartkova J, Bartek J. Convergence of Mitogenic signaling cascades from Diverse classes of receptors at the cyclin D-cyclin dependent kinase-pRb-controlled checkpoint. Mol. Cell Biol. 1996;16:6917–6925. doi: 10.1128/mcb.16.12.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte-Taillez O, Zivanovic Y, Forterre P. Mining archaeal proteomes for eukaryotic proteins with novel functions: the PACE case. Trends Genet. 2000;16:533–536. doi: 10.1016/s0168-9525(00)02137-5. [DOI] [PubMed] [Google Scholar]
- Michalides RJAM, van de Brekel M, Balm F. Defects in G1-S cell cycle control in head and neck cancer: A review. Head Neck. 2002;24:694–704. doi: 10.1002/hed.10109. [DOI] [PubMed] [Google Scholar]
- Nandi S, Reinert LS, Hachem A, et al. Phosphorylation of MCT-1 by p44/42 MAPK is required for its stabilization in response to DNA damage. Oncogene. 2006a Oct 2; doi: 10.1038/sj.onc.1210030. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Nandi S, Shi B, Perreault J, et al. Characterization of the MCT-1 pseudogene: Identification and implication of its location in a highly amplified region of chromosome 20. Biochim. Biophys. Acta. 2006b;1759:234–239. doi: 10.1016/j.bbaexp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Owa T, Yoshino H, Yoshimatsu K, et al. Cell cycle regulation in the G1 phase: A promising target for the development of new chemotherapeutic anticancer agents. Curr. Med. Chem. 2001;8:1487–1503. doi: 10.2174/0929867013371996. [DOI] [PubMed] [Google Scholar]
- Prosniak M, Dierov J, Okami K, et al. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. 1998;58:4233–4237. [PubMed] [Google Scholar]
- Reinert LS, Shi B, Nandi S, et al. MCT-1 protein interacts with the cap complex and modulates messenger RNA translation profiles. Cancer Res. 2006;66:8994–9001. doi: 10.1158/0008-5472.CAN-06-1999. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shi B, Hsu H-L, Evens AM, et al. Expression of the candidate MCT-1 oncogene in B- and T-cell lymphoid malignancies. Blood. 2003;102:297–302. doi: 10.1182/blood-2002-11-3486. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R, Mermeistein F, Fisher RP, et al. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- Smith KA. Interleukin-2: Inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]