Abstract
Insulators are defined as a class of regulatory elements that delimit independent transcriptional domains within eukaryotic genomes. The first insulators to be identified were scs and scs', which flank the domain including two heat shock 70 genes. Zw5 and BEAF bind to scs and scs', respectively, and are responsible for the interaction between these insulators. Using the regulatory regions of yellow and white reporter genes, we have found that the interaction between scs and scs' improves the enhancer-blocking activity of the weak scs' insulator. The sequences of scs and scs' insulators include the promoters of genes that are strongly active in S2 cells but not in the eyes, in which the enhancer-blocking activity of these insulators has been extensively examined. Only the promoter of the Cad87A gene located at the end of the scs insulator drives white expression in the eyes, and the white enhancer can slightly stimulate this promoter. The scs insulator contains polyadenylation signals that may be important for preventing transcription through the insulator. As shown previously, scs and scs' can insulate transcription of the white transgene from the enhancing effects of the surrounding genome, a phenomenon known as the chromosomal position effect (CPE). After analyzing many independent transgenic lines, we have concluded that transgenes carrying the scs insulator are rarely inserted into genomic regions that stimulate the white reporter expression in the eyes.
Introduction
Enhancer-mediated activation is a fundamental mechanism of gene regulation in eukaryotes [1], [2]. Enhancers interact with tagged genes by looping out the intervening sequences. The putative ability of enhancers to stimulate unrelated promoters has provided a basis for the model suggesting the existence of a specific class of regulatory elements, named insulators, that form independent transcriptional domains and preclude undesirable interactions between enhancers and promoters [3]–[10]. Insulators have two properties: (1) they prevent enhancers and silencers from communicating with a promoter only when inserted between such regulatory elements and a promoter [11]–[16] and (2) protect gene expression from positive and negative chromatin position effects [17]–[19].
The second property of insulators has been mainly examined using the white reporter in transgenic Drosophila lines [17], [19]–[23]. Flies carrying the white transgene without the upstream regulatory region (mini-white) display a wide variety of eye colors depending on the transgene insertion site, a phenomenon referred to as the chromosomal position effect (CPE) [24], [25]. To explain the high sensitivity of the mini-white gene to chromosomal position effects, it has been suggested that the white promoter can function as an enhancer trap, meaning that enhancers located either 5' or 3' of the transposon are able to stimulate transcription of the mini-white gene. However, we have recently found that, in more than 70% of cases, transcription through the mini-white gene is responsible for positive position effects [26]. Consistently with this finding, transcriptional terminators proved to be efficient in protecting mini-white expression from CPE.
The first Drosophila insulators to be identified were scs and scs', which flank the 14-kb region containing five genes (Figure 1), including two heat shock 70 genes [17], [27], [28]. It has been shown that the scs and scs' insulators protect from CPE [17], [21] and that multiple sequences within scs and scs' are required for their insulator function [29]–[31]. Two proteins, Zw5 and BEAF, bind to and partially confer the insulator function to scs and scs', respectively [30]–[32]. According to the chromosome conformation capture assay, scs and scs' pair with each other in vivo [33]. The Zw5 and BEAF proteins interact in vitro and in vivo, which is evidence for their involvement in the formation of a chromatin loop between the scs and scs' insulators [33]. However, the role of such a chromatin loop in forming an independent chromatin domain has not been demonstrated.
Figure 1. Genomic region containing the hsp70 genes (FlyBase data).
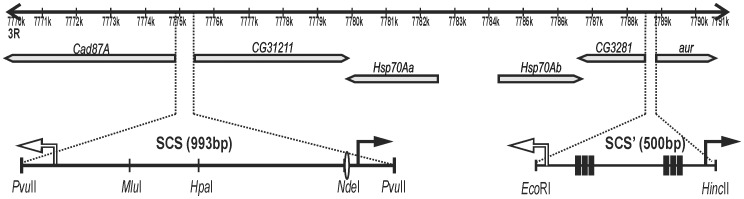
This 15-kb region contains six genes (shown as gray arrows): a pair of divergently transcribed hsp70 genes, Cad87A, CG31211, CG3281, and aurora. Dotted lines show locations of scs (993 bp) and scs' (500 bp). Black arrows indicate positions of the aur and CG31211 promoters. White arrows indicate positions of the Cad87A and CG3281 promoters. The Zw5 binding site within scs is shown as a white oval. Positions of BEAF binding sites within scs' are shown as black rectangles.
In contrast to classic insulators, scs and scs' are not neutral chromatin domain boundaries [34] but contain promoter regions that may be involved in the enhancer-blocking activity of these insulators. The scs' insulator sequence (approximately 500 bp) includes the promoters of the CG3281 and aurora genes (FlyBase database). In the scs insulator, the CG31211 and the Cad87A promoters are located at the ends of its 993-bp sequence [35, FlyBase database]. Recent genome-wide studies have identified binding sites for BEAF and Zw5 proteins as preferentially located in promoter regions [36]–[39]. These and other recent data suggest that insulators might have evolved as specialized derivatives of promoters and that the two types of elements employ related mechanisms to mediate their distinct functions [8], [40]. However, functionality of these promoters and their contribution to the activities of scs and scs' insulators have not been examined.
Since characterization of BEAF and ZW5 as insulator proteins is impossible without the results obtained with the scs and scs' insulators themselves, we examined the properties of these insulators in transgenic lines using the model of yellow and white regulatory regions. As a result, we found that scs improves the enhancer-blocking activity of scs', supporting the functional interaction between these insulators. According to an assay in Drosophila S2 cells, both scs and scs' contain functional promoters at their ends, but only the Cad87A promoter of scs can effectively drive white transcription in the eyes. The scs insulator contains terminators that may be important for preventing transcription through the insulator. In addition, it decreases the frequency of integration of the mini-white transgene into genes actively transcribed in the eyes. This may shed some light on the mechanism of scs-mediated blocking of the chromosomal position effects.
Materials and Methods
Generation and Analysis of Transgenic Lines
The study was performed with 993-bp scs, 500-bp scs', zw5×4, zw5×8, and 852-bp Wary fragments, which were obtained as described [41], [42] and cloned between lox or frt sites. The constructs were based on the CaSpeR vector [43]. The Wari insulator located on the 3′ side of the mini-white gene was deleted from CaSpeR to produce plasmid pCaSpeRΔ700. The EcoRI restriction site was inserted at 3' mini-white end for cloning the test elements in some constructs. The constructs with yellow and white reporter genes for testing enhancer-blocking activity was made as described previously. The test insulator fragments were cloned at –893 relative to the yellow transcription start site. Details of plasmid construction and their schemes are available upon request.
The construct and P25.7 wc plasmid were injected into yacw1118 preblastoderm embryos [44]. The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color. The lines with DNA fragment excisions were obtained by crossing transposon-bearing flies with the Flp (w1118; S2CyO, hsFLP, ISA/Sco;+) or Cre (yw; Cyo, P[w+,cre]/Sco;+) recombinase-expressing lines. The Cre recombinase induces 100% excisions in the next generation [45]. A high level of Flp recombinase was produced by heat shock treatment for 2 h during the first 3 days after hatching [46]. All excisions were confirmed by PCR analysis. Details of the crosses and primers used for genetic analysis and excision of functional elements are available upon request.
To induce GAL4 expression, we used the modified yw1118; P[w–, tubGAL4]117/TM3,Sb line (Bloomington Center #5138) in which the marker mini-white gene was deleted as described [47].
To estimate the levels of yellow and white expression, we visually determined the degree of pigmentation in the abdominal cuticle and wing blades (yellow) and in the eyes (white) of 3- to 5-day-old males developing at 25°C, with reference to standard color scales. In the five-grade scale for yellow, grade 5 corresponds to wild type, and grade 1, to the total loss of yellow expression. Identical data were obtained for the wing and body pigmentation in all experiments. In the nine-grade scale for white, brick red (R) eyes correspond to wild type, and white eyes (W), to the total loss of white expression. Intermediate levels of eye pigmentation, in order of decreasing gene expression, are brownish red (BrR), brown (Br), dark orange (dOr), orange (Or), dark yellow (dY), yellow (Y) and pale yellow (pY).
Two experts separately inspected 30–50 flies from each of two independent crosses for every transgenic line. Each line thus assessed contributed a unit to the corresponding cell of the scoring table. Hence, each numerical entry in the distributions shown in the figures under the scales is the number of fly lines with the specified pigmentation grade (corresponding to the gene expression level decreasing from left to right).
Construct insertion sites in transgenic lines were determined with inverse PCR technique. Genomic DNA extracted from transgenic flies was treated with RsaI or MboI endonuclease. The cleaved DNA was ligated and PCR-amplified with primers 5′-aagattcgcagtggaaggctgcac-3′and 5′-tccgcacacaacctttcctctcaac-3′ (after RsaI cleavage) or 5′-cccttagcatgtccgtggggtttg-3′ and 5′-cgctgtctcactcagactcaatacgacac-3′ (after MboI cleavage). The PCR products were sequenced, and the coordinates and directions of insertions were determined with the Flybase R5.13 database.
Construction of Plasmid Reporter System and Dual Luciferase Assay
Constructs for promoter and terminator assays were generated in pAc5.1/V5-His B (Invitrogen). The firefly and Renilla luciferase sequences were taken from pGL3Basic and pRL-CMV vectors (Promega), respectively. In the control plasmid, the firefly luciferase ORF without the promoter sequence was used. Potential promoter elements were inserted upstream of the firefly ORF. To normalize the firefly data, the promoter assay was performed with the plasmid containing the Renilla luciferase ORF under actin promoter. For terminator assay, we generated a bicistronic system with Renilla and firefly luciferases sequentially located downstream of the general actin promoter. For the basic construct, the reaper gene IRES was amplified from genomic DNA and cloned between the luciferase sequences. The SV40 terminator sequence was taken from pAc5.1/V5-His B vector. SV40 terminator and scs insulator were inserted upstream of IRES.
Drosophila Schneider 2 cells were grown in SFX medium (HyClone) at 25°C. Their transfection with plasmids was performed using the Cellfectin II reagent (Invitrogen) according to the manufacturer's instructions, in six-well plates at a density of 106 cells/ml, with the cells being grown for 24–48 hours before harvesting. The firefly luciferase data were normalized relative to the Renilla luciferase data. The dual luciferase assay was performed with the Firefly & Renilla Luciferase Assay Kit (Biotium). At least three independent experiments were performed for three independent transfection procedures.
Results
Testing the scs and scs' Insulators for Enhancer Blocking Activity
The scs and scs' insulators were previously tested in the transgenes carrying the mini-white gene as a reporter or selection marker [13], [28], [29], [48]–[51]. We found that the endogenous insulator, named Wari, was located at the 3' end of the endogenous white gene and the mini-white gene used in the constructs [41] and showed that Wari improved the enhancer-blocking activity of Su(Hw)-dependent insulators. To find out whether the Wari insulator is required for the enhancer-blocking activity of scs and scs' insulators, we used the previously described model system with two reporter genes, yellow and white (Figure 2). The yellow gene accounts for dark pigmentation of larval and adult cuticle and its derivatives, with two upstream enhancers being responsible for its activation in the body cuticle and wing blades [52]. The white gene is responsible for eye pigmentation, and its expression in the eyes is activated by a specific enhancer [53]. In our experiments, the eye enhancer was inserted between the wing and body enhancers (collectively designated as Eye, Figure 2). All enhancers flanked by frt sites were inserted in front of the yellow gene. The white gene was inserted on the 3' side of the yellow gene. In this configuration, the eye enhancer–white promoter communication was partially attenuated by the yellow promoter (data not shown). The endogenous Wari insulator was deleted from the constructs, flanked by lox sites, and reinserted at the same place.
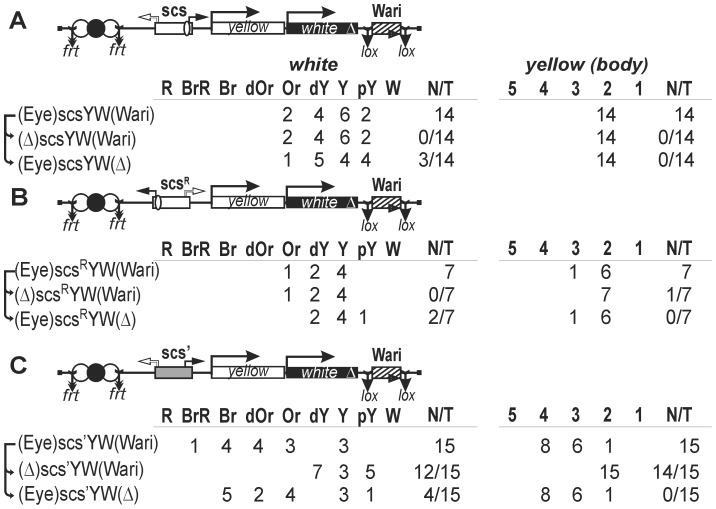
Figure 2. The role of the Wari insulator in the enhancer-blocking activity of scs and scs' insulators in transgenic lines.
Tests were conducted for the functional interaction of Wari with the scs insulator inserted in (A) direct or (B) reverse orientation and (C) with the scs' insulator. In the reductive scheme of the transgenic construct used in the assay, the white and yellow genes are shown as white and black boxes, respectively, with an arrow indicating the direction of transcription; the delta sign (Δ) indicates deletion of Wari located at the 3' end of the white gene; downward arrows indicate target sites for Flp recombinase (frt) or Cre recombinase (lox); the same sites in construct names are denoted by parentheses; the eye enhancer is shown as black oval; the yellow wing and body enhancers are shown as white ovals. The “white” column shows the number of transgenic lines with different levels of eye pigmentation. Arrows indicate the excision of an element to produce the derivative transgenic lines. Wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging from pale yellow (pY), through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr), and brown (Br) to brownish red (BrR), reflect the increasing levels of white expression. The “yellow” column shows the numbers of transgenic lines with the yellow pigmentation level in the abdominal cuticle (reflecting the activity of the body enhancer); in most of the lines, the pigmentation level in wing blades (reflecting the activity of the wing enhancer) closely correlated with these scores. The level of pigmentation (i.e., of y expression) was estimated on an arbitrary five-grade scale, with wild-type expression and the absence of expression assigned scores 5 and 1, respectively. N is the number of lines in which flies acquired a new white or yellow phenotype after deletion (Δ) of the specified DNA fragment; T is the total number of lines examined for each particular construct. Other designations are as in Figure 1.
The 993-bp scs insulator was inserted in either direct (Figure 2A) or reverse orientation (Figure 2B) between the enhancers and the yellow promoter. In all transgenic lines, flies had yellow pigmentation of wing blades and body cuticle, and eye pigmentation ranged from pale yellow to orange, indicating that the enhancer were unable to activate the reporter genes. This conclusion was supported by the fact that deletion of the enhancers resulted in only a slight decrease in yellow and white expression. Deletion of the Wari insulator led to reduction of eye pigmentation in five transgenic lines but did not affect yellow expression (Figures 2A, 2B). In the light of our previous observations [54], we consider that the slight positive effect of the Wari insulator results from a positive influence on the white promoter rather than from an interaction with scs. Taken together, these results show that the Wari insulator is not required for the enhancer-blocking activity of the strong scs insulator.
Next, we inserted the scs' insulator between the enhancers and the yellow promoter (Figure 2C). In transgenic lines, flies had a moderate level of wing and body pigmentation, suggesting partial activation of the yellow promoter by the enhancers. Likewise, transgenic flies had the eye color ranging from yellow to brown-red, which was indicative of white stimulation by the eye enhancer in some transgenic lines. Indeed, deletion of the enhancers proved to considerably reduce the yellow and white expression. Thus, the results of these experiments confirmed previous observations that scs' is a relatively weak insulator [48], [49]. Once again, deletion of the Wari insulator did not affect the enhancer-blocking activity of the scs’ insulator. Taken together, these results provide evidence that the scs and scs' insulators do not functionally interact with the Wari insulator.
Testing for the Functional Interaction between the scs and scs' Insulators
There is evidence that scs and scs' interact in vivo [33], but the functional role of their interaction has not been demonstrated. Therefore, we then used the same transgenic assay with the yellow and white genes to find out if a chromatin loop formed by the scs and scs' insulators could improve enhancer blocking. Since a single copy of scs completely blocked the yellow and white enhancers, we inserted the weak scs' insulator between the enhancers and the yellow gene (Figure 3A). The scs insulator flanked by lox sites was inserted instead of the Wari insulator downstream of the white gene (Figure 3A). As a result, the scs and scs' insulators formed a 9226-bp chromatin domain including two reporter genes, which corresponded to the distance between these insulators in their endogenous positions at the ends of the domain containing the heat shock 70 genes [27].
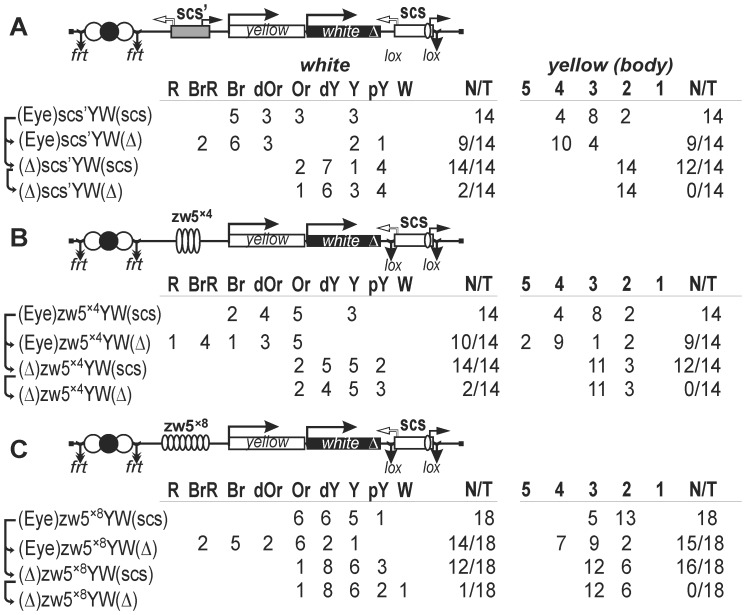
Figure 3. Testing for the functional interaction between (A) scs' or (B–C) Zw5 binding sites and the scs insulator located on the 3' side of the white gene.
In 14 transgenic lines, y and w phenotypes of flies indicated that the enhancers were partially active (Figure 3A). Deletion of the scs insulator resulted in an enhancement of eye pigmentation in seven transgenic lines and of wing and body pigmentation in nine transgenic lines. Thus, scs partially improved the enhancer-blocking activity of the scs' insulator.
The scs insulator contains a binding site for the Zw5 protein, which is required for the enhancer-blocking activity [31]. It was shown that four Zw5 binding sites partially blocked the eye enhancer, and we previously found that Zw5 binding sites supported distant interactions between regulatory elements in transgenic lines [42]. To test if scs can improve the enhancer-blocking activity of Zw5 binding sites, we inserted either four (Figure 3B) or eight such sites (Figure 3C) between the enhancers and the yellow promoter. The enhancer-blocking activity proved to be stronger in transgenic lines with eight, rather than four, Zw5 binding sites. In both cases, deletion of the scs insulator considerably improved yellow and white expression, suggesting that the scs insulator functionally interact with the Zw5-binding regions in blocking the enhancer activity.
Testing for the Promoter and Transcription Terminator Activity of scs and scs' Insulators in S2 Cells
Previously, promoters were mapped at the ends of the scs' and scs sequences [34], [35], [49] (Figure 1). To check if the scs and scs' insulators used in the studies included all sequences necessary for promoter activity, we tested them for this activity in S2 cells using a luciferase reporter assay. As a result, we found that both ends of the scs and scs' insulators contained functional promoters that could drive luciferase transcription at a level comparable to that of the hsp70 promoter (Figure 4A).
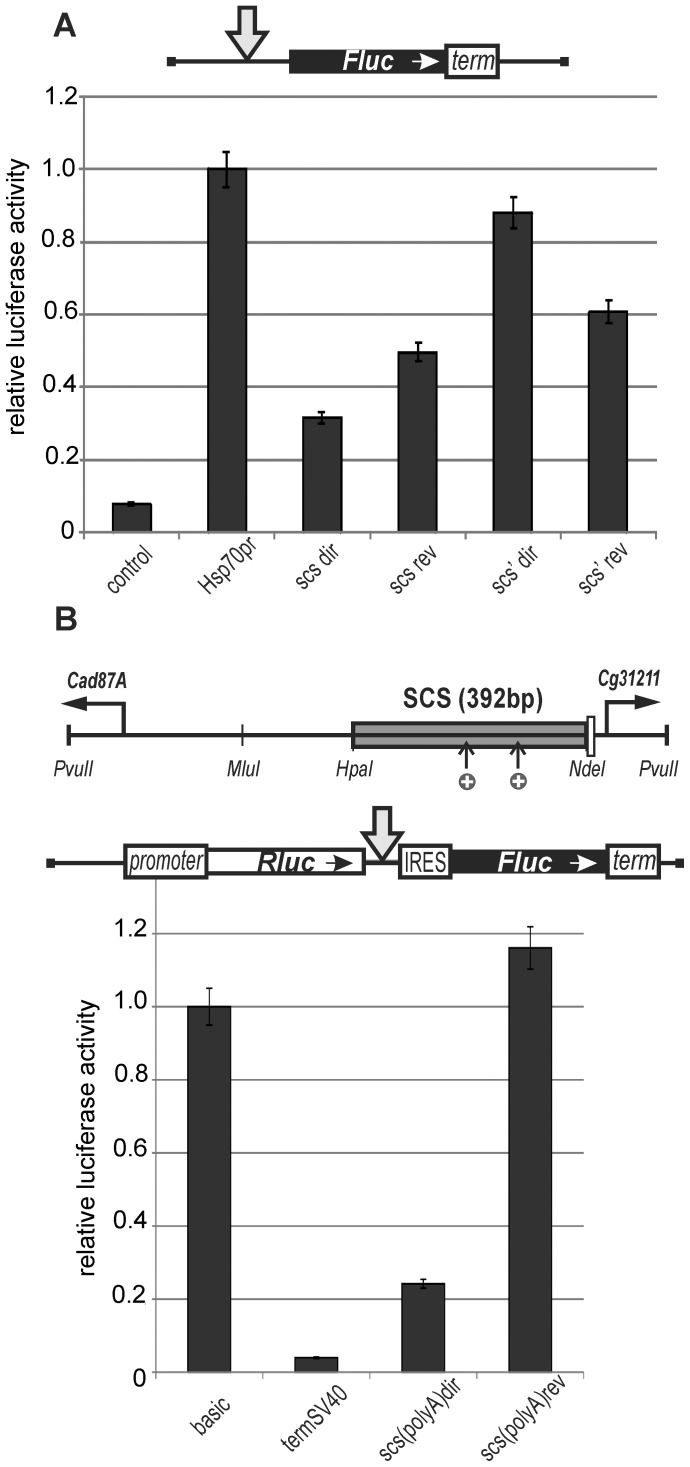
Figure 4. Testing elements for (A) promoter and (B) terminator activities in S2 cells.
In the structural scheme of the scs insulator (PvuII–PvuII fragment), the Zw5 site is shown as a white rectangle; identified polyadenylation sites (PAS), as black circles with “+” sign indicating their direct orientation. Restriction sites HpaI–NdeI indicate the boundaries of the element used in the terminator assay. In the reductive schemes of transgenic constructs, the ORFs encoding Renilla (Rluc) and firefly (Fluc) luciferases are shown as white and black boxes with arrows indicating the direction of transcription; the white rectangle marked “term” is the SV40 terminator. The bicistronic plasmid also contained the actin promoter and rpr IRES (rectangles with corresponding marks). Thick downward arrows indicate insertion sites for the hsp70 promoter (Hsp70 pr), scs (scs dir, scs rev), and scs' (scs' dir, scs' rev) in the promoter assay and for late SV40 (termSV40) and scs PASs (scs(polyA)dir, scs(polyA)rev) in the terminator assay. The reporter system used in the assays is based on measurement of Fluc versus Rluc activity. The Fluc/Rluc ratios for the test constructs are shown in histograms. Error bars show standard deviations (n = 3).
According to sequence data, the central part of scs contains two polyadenylation signals that match potential transcription terminators operating in direct orientation (Figure 4B). To test for transcription terminator activity in the scs insulator, we used a bicistronic reporter based on two luciferase coding sequences driven by a single Drosophila actin 5C promoter. The IRES sequence from the Drosophila reaper gene [55] was inserted between Renilla luciferase (Rluc) and firefly luciferase (Fluc) (Figure 4B).
It was expected that if poly(A) signal was functional, a monocistronic Rluc mRNA would be produced; if poly(A) signal was non-functional or weakly functional, a longer mRNA would be generated, reaching the SV40 poly(A) signal located downstream of the Fluc. Thus, the Fluc-to-Rluc ratio would allow us to estimate the amount of long bicistronic mRNA relative to the total mRNA transcribed from construct.
In the bicistronic reporter, we inserted either SV40 terminator as a control or the central 392-bp HpaI–NdeI part of scs (scsm) that contains two polyadenylation signals (Figure 4B). As a result, we observed that the test scs fragment had a strong transcription terminator activity only in the direct orientation, corresponding to the presence of two polyadenylation signals. Thus, the scs insulator can function as a transcription terminator.
Testing the scs Insulator in the Promoterless white Assay
Our results suggested that the scs insulator contained two functional promoters at the ends and terminators in the middle.
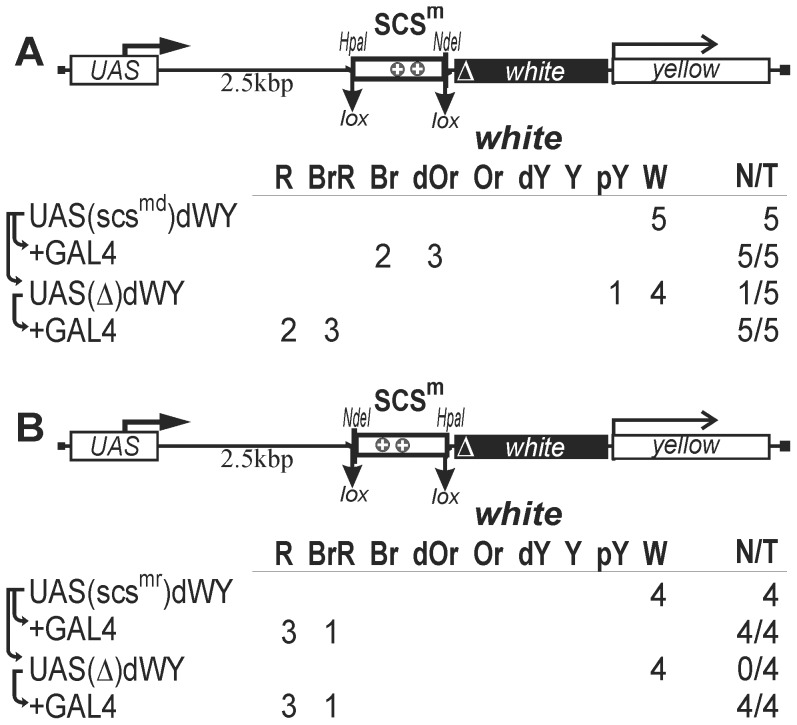
To test the terminators in scs for the ability to arrest transcription elongation in the eyes, we used a model system that contained the UAS promoter, a 2-kb spacer from the lacZ gene, and the promoterless mini-white gene with deleted Wari insulator (Figure 5). The white gene also contains an internal ribosome entry site that helps to translate mRNAs from the internal sites [26]. The yellow gene was used as a marker for selecting transgenic lines. The central 392-bp HpaI–NdeI part of scs (scsm) containing two polyadenylation signals flanked by lox sites was inserted into the spacer in either the direct (Figure 5A) or reverse orientation (Figure 5B). To express the GAL4 protein, we used the transgenic line carrying the GAL4 gene under control of the ubiquitous tubulin promoter (tubGAL4). The transgenic flies carrying the fragment of scs in either orientation had white eyes. Upon induction of the UAS promoter by crossing with the tubGAL4 line, flies carrying the scs fragment in direct orientation acquired eye pigmentation from dark orange to brown (Figure 5A). In derivative transgenic lines obtained by deleting the scs fragment, induction of the UAS promoter by GAL4 expression resulted in eye pigmentation ranging from brown-red to red, which was indicative of strong mini-white activation in transgenic flies. In contrast, GAL4 stimulated mini-white expression to the same level (brown-red eye pigmentation) in the presence or absence of the scs fragment inserted in reverse orientation (Figure 5B). These results suggest that the terminators contained in scs are functional in the transgenic lines.
Figure 5. Testing the central 392-bp HpaI–NdeI part of scs (scsm) inserted in either (A) direct or (B) reverse orientation for the ability to terminate transcription in the eyes.
The UAS promoter is shown as the white rectangle marked “UAS.” “+GAL4” indicates that eye phenotypes in transgenic lines were examined after induction of GAL4 expression. In this case, N is the number of lines in which flies acquired a new w phenotype upon induction of GAL4. For other designations, see Figures 1 and 2.
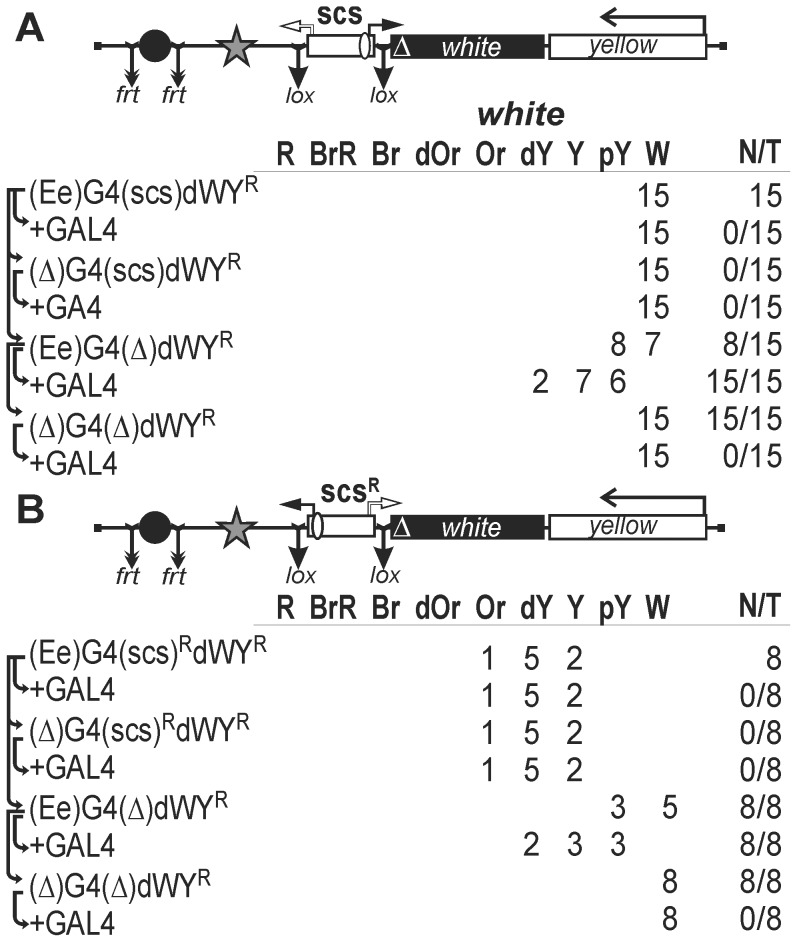
To find out if the promoters of scs are active in the eyes, we inserted the lox-flanked scs insulator upstream of the promoterless white gene in either direct (Figure 6A) or reverse orientation (Figure 6B). The eye enhancer flanked by frt sites and five GAL4-binding sites was inserted upstream of the scs insulator. As shown previously [53], the eye enhancer can substitute the promoter and drive transcription of the white gene in the eyes.
Figure 6. Testing (A) CG31211 or (B) Cad87A promoter in the scs insulator.
Index “R” indicates that the scs is inserted in the reverse orientation. Delta sign (Δ) indicates deletion of the white promoter. The star indicates four GAL4 binding sites inserted near the eye enhancer. “+GAL4” indicates that eye phenotypes were examined in transgenic lines after induction of GAL4 expression. Other designations are as in Figures 1 and 2.
We obtained 15 transgenic lines carrying the scs insulator inserted in direct orientation (Figure 6A). Flies of all these lines had white eye color, indicating that part of the CG31211 promoter included in scs was inactive in the eyes. Induction of GAL4 produced no change in eye pigmentation. When the scs insulator was deleted, flies with pale yellow eyes appeared in half of transgenic lines. Moreover, eye pigmentation further increased after GAL4 induction, suggesting that GAL4 stimulated transcription from the eye enhancer. The lack of white expression in the presence of the scs insulator could be explained by its function as a terminator of transcription initiated at the eye enhancer.
In eight transgenic lines carrying the construct with the scs insulator inserted in reverse orientation, flies had eye pigmentation ranging from yellow to orange (Figure 6B). Deletion of the scs insulator significantly reduced eye pigmentation, suggesting the main role for the Cad87A promoter in white expression. Induction of GAL4 or deletion of the eye enhancer had no effect on eye pigmentation, indicating that the eye enhancer failed to stimulate the Cad87A promoter. However, the Cad87A promoter could affect the activity of the eye enhancer by transcription interference in transgenic lines described in Figure 6A.
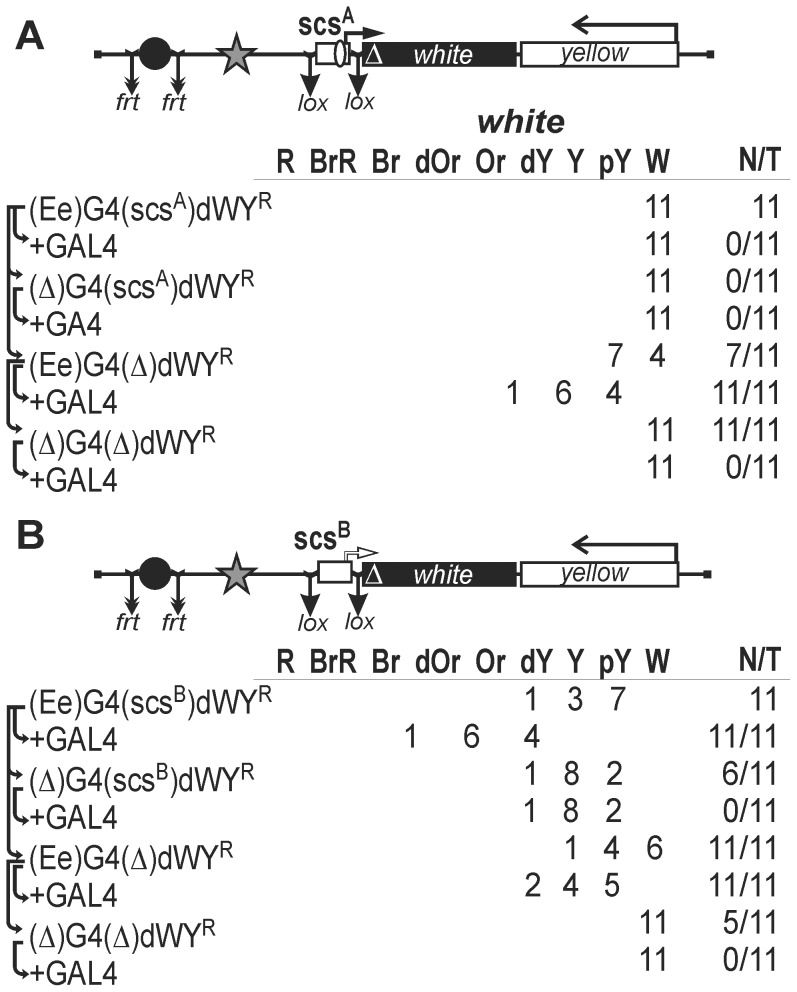
These results suggested that the eye enhancer failed to stimulate promoters contained in scs. However, it was possible that a certain region of the whole element blocked the interaction of the eye enhancer with the scs promoter. To test such a possibility, we inserted the lox-flanked parts of scs, including the CG31211 promoter (516-bp scsA, Figure 7A) and the Cad87A promoter (477-bp scsB, Figure 7B), into the promoterless white gene.
Figure 7. Testing (A) CG31211 or (B) Cad87A promoter in the scs insulator.
Other designations are as in Figures 1, 2, and 5. The scsA is 516-bp scs part including the CG31211 promoter. The scsB is 477-bp scs part including the Cad87A promoter. Other designations are as in Figures 1, 2 and 5.
Transgenic flies carrying scsA had white eye color, indicating that the CG31211 promoter was inactive in the eyes. Induction of the eye enhancer by GAL4 provided for an increase in eye pigmentation only after scsA was deleted (Figure 7A), which was evidence for the role of the transcription terminator in interrupting transcription initiated from the eye enhancer.
Next, we examined 11 transgenic lines carrying the scsB part of scs (Figure 7B). In all transgenic lines, flies had pigmented eyes, indicating the ability of the Cad87A promoter to drive the white expression in eyes. Deletion of the eye enhancer reduced eye pigmentation in most of transgenic lines, which might be explained either by the ability of the eye enhancer to weakly stimulate the Cad87A promoter or by the additive effect of transcription from the eye enhancer and the promoter. In any case, neither GAL4 activator nor the eye enhancer could effectively stimulate the Cad87A promoter.
In our previous study, transcription through the mini-white gene was found to result in a high level of its expression (from orange to red) in 38 (25%) out of 154 transgenic lines tested [26]. Deletion of the white promoter in these lines had no effect on eye pigmentation because of transgene insertion into the transcribed regions of genes that were active in the eye imaginal disks. Here, 23 derivative transgenic lines were obtained after deletion of the scs and the eye enhancer, and flies in all these lines had white eyes (Figure 6). In all 22 transgenic lines carrying scsA and scsB (Figure 7), the transgene was also inserted into genome regions that failed to support expression of promoterless mini-white gene. Thus, the transgenes carrying the scs insulator are rarely inserted into the genes expressed in the eye imaginal discs.
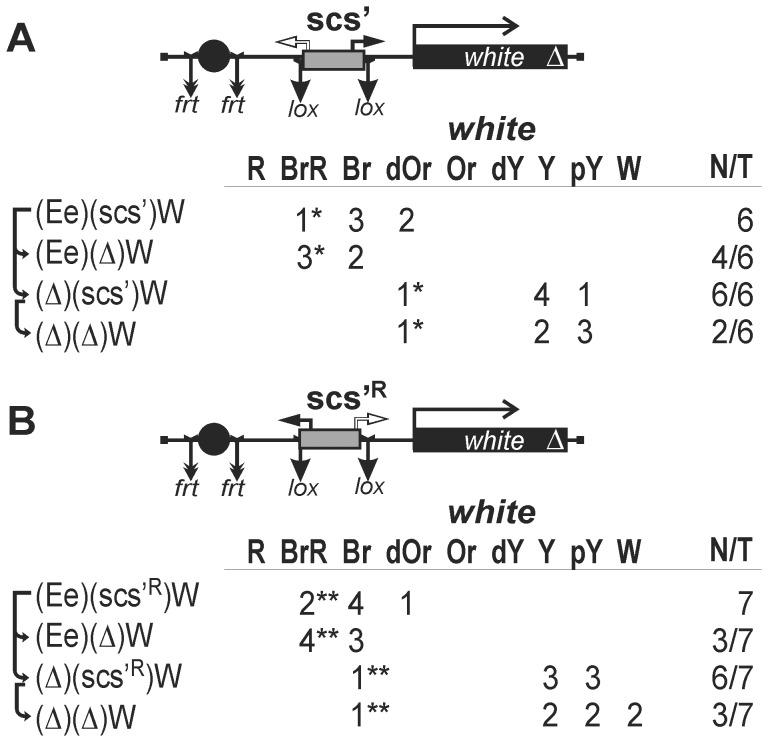
Testing the scs' Insulator in the promoterless white Assay
According to the results obtained in S2 cells, the scs' insulator contains two functional promoters. To determine the activity of these promoters in eye imaginal disks, we inserted the scs' insulator flanked by lox sites into the promoterless mini-white gene in either direct (Figure 8A) or reverse orientation (Figure 8B). The eye enhancer flanked by frt sites and five GAL4 binding sites were inserted upstream of the scs' insulator. Deletion of the eye enhancer in the transgenic lines carrying the scs' insulator inserted in the direct orientation (Figure 8A) did not change eye pigmentation, indicating that the aur promoter was functional in the eye imaginal disks. In contrast, the CG3281 promoter failed to drive white transcription (Figure 8B). Irrespective of scs' orientation, deletion of the eye enhancer had no significant influence on white expression, suggesting that the promoters in scs' are not sensitive to the white enhancer. GAL4 could weakly stimulate white expression only in the presence of the eye enhancer. This result confirms that the scs' insulator does not terminate transcription initiated in the eye enhancer.
Figure 8. Testing (A) aurora or (B) CG3281 promoter in the scs' insulator.
Asterisks indicate that the transgene was codirectionally inserted (*) in the first intron of the effet gene (3R:10565091) transcribed in the same direction as the mini-white gene or (**) in the first intron of the Dek gene (2R:12744143). Other designations are as in Figures 1, 2 and 5.
In two transgenic lines, flies had strongly pigmented eyes, with the pigmentation level remaining unchanged after deletion of the scs' insulator and the eye enhancer. The localization of insertion sites in these transgenic lines showed that the transgene was inserted into the genes whose transcription direction coincided with that of the mini-white gene (Figure 8B). Thus, the scs' insulator fails to protect the mini-white gene from transcription starting upstream of the transgene integration site.
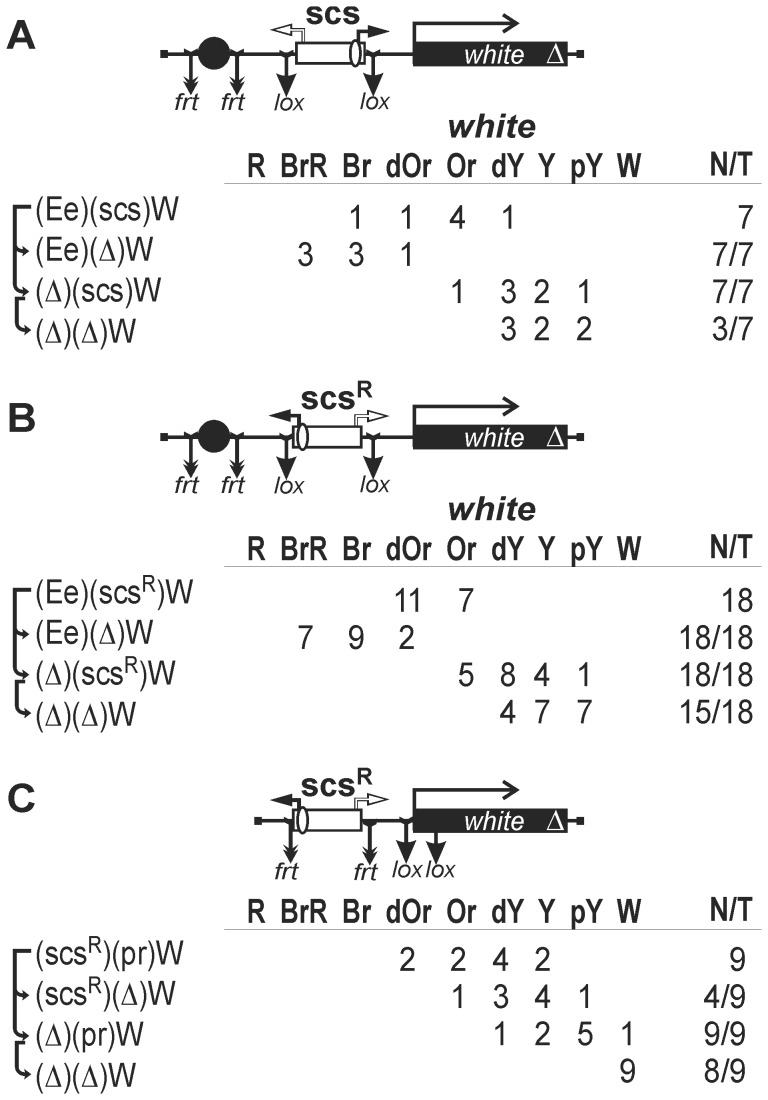
Testing the scs and scs' Insulators in Enhancer-blocking and CPE Assays
In the transgenic lines described in Figures 2 and 3, the eye enhancer was partially attenuated by the yellow promoter. To check whether the scs insulator could block the strong enhancer–promoter communication, we used the eye enhancer–white gene system with the deleted Wari insulator. The scs insulator flanked by lox sites was inserted in either direct (Figure 9A) or reverse orientation (Figure 9B) between the frt-flanked eye enhancer and the white promoter. In the resultant 25 transgenic lines, the scs insulator only partially blocked the eye enhancer activity. This was confirmed by the fact that deletion of the eye enhancer resulted in further reduction of eye pigmentation: in all 25 derivative transgenic lines with the deleted eye enhancer, flies had eye color phenotypes ranging from pale yellow to orange. Additional deletion of scs in any of the lines did not provide for an increase in eye pigmentation. This observation confirms our finding that the scs insulator directs integration of the transgenic construct into the genome regions that do not stimulate mini-white expression.
Figure 9. Testing the scs insulator inserted in (A) direct or (B) reverse orientation in enhancer-blocking and CPE assays and (C) testing for interference between the white promoter and the Cad87A promoter in the scs insulator.
We noticed that eye pigmentation was darker in flies from transgenic lines carrying the scs insulator inserted in reverse orientation (Figures 9A, 9B), which could be explained by the activity of the Cad87A promoter. To test for cooperation between the white and Cad87A promoters, we made the construct in which the frt-flanked scs was inserted in reverse orientation upstream of the lox-flanked white promoter (Figure 9C). Transgenic flies had eye color phenotypes in the range from dark orange to yellow, and deletion of either scs or the white promoter reduced eye pigmentation, suggesting that the Cad87A and white promoters cooperate in the mini-white gene expression.
Next, we tested the scs' insulator in the enhancer blocking assay (Figure 10). We found that scs' failed to effectively block the eye enhancer: deletion of scs' led to a slight enhancement of eye pigmentation in only 7 out of 13 transgenic lines. At the same time, flies from two transgenic lines had a relatively high level of eye pigmentation after deletion of scs' and the eye enhancer. In both these lines, the transgene was inserted into genes whose transcription direction coincided with that of the mini-white gene (Figure 10). These results support our conclusion that scs' does not protect white expression from transcription initiated upstream of the transgene.
Figure 10. Testing the scs' insulator inserted in (A) direct or (B) reverse orientation in enhancer-blocking and CPE assays.
Asterisks indicate that the transgene was codirectionally inserted (*) in an intron of the capr gene (3L:18665421) or (**) in an intron of the CG7950 gene (3R:25882796). Other designations are as in Figures 1 and 2.
Discussion
Our experiments with transgenic lines support previous observations [28], [48]–[51], [56] that scs is one of the strongest Drosophila insulators, while scs' has only a weak insulator activity. Here, we provide the first experimental evidence that pairing between scs and scs', described previously [33], has a functional outcome in improving the scs' insulator activity. The scs and scs' insulators in transgenic lines were located at a distance of about 9 kb, which is similar to the 14-kb distance between them in the endogenous genome region. As expected, scs is much more effective in reinforcing insulation mediated by Zw5 binding sites, which supports our previous observation that Zw5 can facilitate long-distance interactions [42]. The improvement of enhancer blocking by the interaction of scs and scs' may be explained by the formation of a chromatin loop between these insulators, which interferes with enhancer–promoter communication. However, it is also possible that the interaction between these insulators facilitates cooperative binding of insulator proteins to their sites, with consequent reinforcement of their enhancer-blocking activity.
The scs and scs' insulators contain promoters that are active in S2 cells, suggesting that both insulators may block enhancers according to the promoter competition model [40]. However, only the Cad87A promoter of scs and the aur promoter of scs' can drive white expression in the eyes. Since the CG31211 gene is strongly transcribed in eyes (FlyBase database), we suggest that the scs insulator lacks certain regulatory elements that are important for the activity of this promoter. Unexpectedly, we have found that the eye enhancer or GAL4 fails to effectively stimulate the Cad87A and aur promoters. This may be explained either by the specificity of the eye enhancer to stimulate only the white promoter or by the inability of the promoters in scs and scs' to be stimulated by the activators bound to the enhancer or GAL4.
In transgenic lines, the scs insulator located on the 3' side of the yellow gene interacts with the promoter [54]. The scs insulator blocks the enhancers to the same extent in all transgenic lines, indicating that chromatin loop formation with insulators located outside the transgene is not required for enhancer blocking. Our results are in accordance with the previous observation that one copy of the scs insulator can block the eye enhancer on an episome, out of the chromatin context [56]. Taken together, these observations suggest that direct interactions of proteins bound to the scs modules and the white enhancer and/or promoter are responsible for effective blocking of the eye enhancer. Interestingly, even eight binding sites for the Zw5 protein block the enhancers to a much lesser extent than does the scs insulator that has only one Zw5 binding site [31]. This is evidence that additional, as yet unidentified proteins are required for insulation mediated by scs.
As shown previously, transcription induced by the Cad87A promoter of scs inserted into the regulatory region of the bithorax complex can affect the activity of the enhancers that stimulate Abd-a and Abd-B genes [35]. In contrast to our previous observation that transcription through the transgene inactivates the mini-white promoter [26], it has been found that transcription from the Cad87A promoter does not interfere with activity of the white promoter. When the white and Cad87A promoters have the same direction in transgenic lines, they function additively in stimulating white expression.
A number of experiments performed to date indicate that a major portion of the genome is being transcribed and that a large percentage of the transcripts is accounted for by long non-protein-coding sequences (lncRNA) [57]; [58]. Recent data suggests that many of lncRNA have important roles in regulation of transcription [59]. Therefore, to functionally separate two adjacent chromatin domains, the boundaries should contain transcription terminators. Here, we have found that the scs insulator contains terminators that stop transcription. Interestingly, SF1, a chromatin boundary located in the Drosophila Antennapedia complex (ANT-C) [60], also contains a functional transcription terminator (DL and OM, unpublished). Thus, the presence of transcription terminators may well be a common feature of chromatin boundaries.
In the transgenic assay used to test insulators for protection from chromosomal position effects (CPE), transcription terminators contained in the insulators could be partially responsible for CPE suppression. For example, we have shown previously that the SV40 transcriptional terminator was efficient in protecting mini-white expression from positive position effects [26]. In 4 out of the total 33 transgenic lines carrying the construct with scs' (12%), transcription through the transgene led to white expression in eyes. The scs' insulator failed to terminate such transcription, indicating that this insulator could not effectively protect from CPE. In addition to the ability of scs to terminate transcription, we found that this insulator reduced the frequency of insertion of the transgene into the regions that stimulate white expression in the eyes. It is possible that proteins bound to the scs insulator interact with chromatin proteins that recruit the transgene to certain genomic regions. Our results contradict the data by Cuvier et al. [21] who obtained flies with strongly pigmented eyes in 6 out of 19 transgenic lines (32%) carrying the mini-white gene flanked on the 3' side by the scs insulator. To explain the difference, we hypothesize that the white promoter and the Wari insulator also determine the sites of transgene insertion. We have previously found that the Wari insulator interacts with the white promoter and potentiates its activity [54]. In the experiments by Cuvier et al. [21], the transgene contained both regulatory elements of the white gene, while in our constructs either the promoter or insulator was deleted and, consequently, only the scs insulator was involved in determining the transgene insertion site. It is noteworthy that, as shown previously, human matrix attachment regions (MARs) can insulate transgene expression from CPE in Drosophila melanogaster [22]. However, excision of MARs from the transgenes has proved to have no effect on white expression [23]. It has been suggested that MAR sequences can cause transposon to home to the nuclear matrix prior to integration, thereby targeting transposon integration to specific kinds of sites within the Drosophila genome. Such genome region-specific targeting of transgenes carrying regulatory elements such as scs and MARs may reflect global organization of the genome [38], [61], [62]. Further study is required to understand the mechanisms of this phenomenon.
Acknowledgments
We are grateful to N.A. Gorgolyuk for his help in preparing the manuscript.
Funding Statement
This study was supported by the Russian Foundation for Basic Research (project no. 12-04-92423-EMBL-a) and grants from the Molecular and Cellular Biology Program of the Russian Academy of Sciences (to OK), the Ministry of Science and Education of the Russian Federation (Project no. 8103), the Government of the Russian Federation (order #220) (to VS), and the President's Stipendy (no. SP-1960.2012.4) (to OM). Experiments were performed using the equipment of IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation (grant no. 16.552.11.7067). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dean A (2011) In the loop: Long range chromatin interactions and gene regulation. Brief Funct. Genomics 10: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuhn EJ, Geyer PK (2003) Genomic insulators: Connecting properties to mechanism. Curr Opin Cell Biol 15: 259–265. [DOI] [PubMed] [Google Scholar]
- 4. Brasset E, Vaury C (2005) Insulators are fundamental components of the eukaryotic genomes. Heredity 94: 571–576. [DOI] [PubMed] [Google Scholar]
- 5. Maksimenko OG, Chetverina DA, Georgiev PG (2006) Insulators of higher eukaryotes: Properties, mechanisms of action, and role in transcription regulation. Russ J Genet 42: 845–857. [PubMed] [Google Scholar]
- 6. Maeda RK, Karch F (2007) Making connections: Boundaries and insulators in Drosophila . Curr Opin Genet Dev 17: 394–399. [DOI] [PubMed] [Google Scholar]
- 7. Wallace JA, Felsenfeld G (2007) We gather together: Insulators and genome organization. Curr Opin Genet Dev 17: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raab JR, Kamakaka RT (2010) Insulators and promoters: Closer than we think. Nature Rev Genet 11: 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mukhopadhyay S, Schedl P, Studitsky VM, Sengupta AM (2011) Theoretical analysis of the role of chromatin interactions in long-range action of enhancers and insulators. Proc Natl Acad Sci U S A 108: 19919–19924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herold M, Bartkuhn M, Renkawitz R (2012) CTCF: Insights into insulator function during development. Development 139: 1045–1057. [DOI] [PubMed] [Google Scholar]
- 11. Holdridge C, Dorsett D (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. . Mol Cell Biol 11: 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geyer PK, Corces VG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- 13. Cai H, Levine M (1995) Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. [DOI] [PubMed] [Google Scholar]
- 14. Sigrist CJ, Pirrotta V (1997) Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mallin DR, Myung JS, Patton JS, Geyer PK (1998) Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Comet I, Savitskaya E, Schuettengruber B, Nègre N, Lavrov S, et al. (2006) PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell 11: 117–124. [DOI] [PubMed] [Google Scholar]
- 17. Kellum R, Schedl P (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. [DOI] [PubMed] [Google Scholar]
- 18. Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, et al. (1995) A P-element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. . Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Majumder P, Roy S, Belozerov VE, Bosu D, Puppali M, et al. (2009) Diverse transcription influences can be insulated by the Drosophila SF1 chromatin boundary. Nucleic Acids Res 37: 4227–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chung JH, Whiteley M, Felsenfeld G (1993) A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila . Cell 74: 505–514. [DOI] [PubMed] [Google Scholar]
- 21. Cuvier O, Hart CM, Laemmli UK (1998) Identification of a class of chromatin boundary elements. Mol Cell Biol 18: 7478–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Namciu SJ, Blochlinger KB, Fournier RE (1998) Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. . Mol Cell Biol 18: 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Namciu SJ, Fournier RE (2004) Human matrix attachment regions are necessary for the establishment but not the maintenance of transgene insulation in Drosophila melanogaster . Mol Cell Biol 24: 10236–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levis R, Hazelrigg T, Rubin GM (1985) Effects of genomic position on the expression of transduced copies of the white gene of Drosophila . Science 229: 558–561. [DOI] [PubMed] [Google Scholar]
- 25. Pirrotta V, Steller H, Bozzetti MP (1985) Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J 4: 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silicheva M, Golovnin A, Pomerantseva E, Parshikov A, Georgiev P, et al. (2010) Drosophila mini-white model system: New insights into positive position effects and the role of transcriptional terminators and gypsy insulator in transgene shielding. Nucleic Acids Res 38: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Udvardy A, Maine E, Schedl P (1985) The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341–358. [DOI] [PubMed] [Google Scholar]
- 28. Kellum R, Schedl P (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12: 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vazquez J, Schedl P (1994) Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J 13: 5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao K, Hart CM, Laemmli UK (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889. [DOI] [PubMed] [Google Scholar]
- 31. Gaszner M, Vazquez J, Schedl P (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev 13: 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hart CM, Zhao K, Laemmli UK (1997) The scs' boundary element: Characterization of boundary element-associated factors. Mol Cell Biol 17: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blanton J, Gaszner M, Schedl P (2003) Protein : protein interactions and the pairing of boundary elements in vivo. Genes Dev 17: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Avramova Z, Tikhonov A (1999) Are scs and scs' neutral' chromatin boundaries of the 87A7 locus in vivo? Trends Genet 15: 138–139. [DOI] [PubMed] [Google Scholar]
- 35. Hogga I, Karch F (2002) Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb mediated silencing. Development 129: 4915–4922. [DOI] [PubMed] [Google Scholar]
- 36. Jiang N, Emberly E, Cuvier O, Hart CM (2009) Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol 29: 3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, et al. (2010) A comprehensive map of insulator elements for the Drosophila genome. PloS Genet. 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Negre N, Brown CD, Ma L, Bristow CA, Miller SW, et al. (2011) A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chopra VS, Cande J, Hong JW, Levine M (2009) Stalled Hox promoters as chromosomal boundaries. Genes Dev 23: 1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, Zaytseva O, et al. (2008) Red flag on the white reporter: A versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res 36: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, Georgiev P (2008) Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res 36: 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pirrotta V (1988) Vectors for P-mediated transformation in Drosophila. Biotechnology 10: 437–456. [DOI] [PubMed] [Google Scholar]
- 44. Karess RE, Rubin GM (1984) Analysis of P transposable element functions in Drosophila. . Cell 38: 135–146. [DOI] [PubMed] [Google Scholar]
- 45. Siegal ML, Hartl DL (2000) Application of Cre/loxP in Drosophila. Site-specific recombination and transgene coplacement. Methods Mol Biol 136: 487–495. [DOI] [PubMed] [Google Scholar]
- 46. Golic KG, Lindquist S (1989) The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- 47. Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P (2007) Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer–promoter communication. Mol Cell Biol 27: 3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuhn EJ, Viering MM, Rhodes KM, Geyer PK (2003) A test of insulator interactions in Drosophila . EMBO J 22: 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuhn EJ, Hart CM, Geyer PK (2004) Studies of the role of the Drosophila scs and scs' insulators in defining boundaries of a chromosome puff. Mol Cell Biol 24: 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Majumder P, Cai HN (2003) The functional analysis of insulator interactions in the Drosophila embryo. Proc Natl Acad Sci U S A 100: 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gohl D, Aoki T, Blanton J, Shanower G, Kappes G, et al. (2011) Mechanism of chromosomal boundary action: Roadblock, sink, or loop? Genetics 187: 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geyer PK, Corces VG (1987) Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster . Genes Dev 1: 996–1004. [DOI] [PubMed] [Google Scholar]
- 53. Qian S, Varjavand B, Pirrotta V (1992) Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics 131: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Erokhin M, Davydova A, Kyrchanova O, Parshikov A, Georgiev P, et al. (2011) Insulators form gene loops by interacting with promoters in Drosophila . Development 138: 4097–4106. [DOI] [PubMed] [Google Scholar]
- 55. Hernandez G, Vazquez-Pianzola P, Sierra JM, Rivera-Pomar R (2004) Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parnell TJ, Geyer PK (2000) Differences in insulator properties revealed by enhancer blocking assays on episomes. EMBO J 19: 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Birney E, Stamatoyannopoulos JA, Dutta A, Guigό R, Gingeras TR, et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488. [DOI] [PubMed] [Google Scholar]
- 59. Wang K, Chang H (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belozerov VE, Majumder P, Shen P, Cai HN (2003) A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila . EMBO J 22: 3113–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Steensel B (2011) Chromatin: Constructing the big picture. EMBO J. 30: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]