Abstract
We present the first example of chemo-selective site-specific spin labeling of a monomeric protein with two spectroscopically orthogonal spin labels: a Gadolinium (III) chelate complex and a nitroxide radical. A detailed analysis of the performance of two commercially available Gd(III) ligands in the Gd(III)-nitroxide pulse double electron-electron resonance (DEER or PELDOR) experiment is reported. A modification of the flip angle of the pump pulse in the Gd(III)-nitroxide DEER experiment is proposed to optimize sensitivity.
Keywords: EPR/ESR, DEER/PELDOR, lanthanide ions, nitroxide radicals, chemo-selective labeling, unnatural amino acid
Introduction
DEER (or PELDOR) spectroscopy1–3 in combination with site-directed spin labeling (SDSL)4–8 is a useful tool in structural studies of biomacromolecules. By attaching pairs of nitroxide spin labels to specific sites in biomolecules and measuring interspin distances in the DEER experiment one can obtain long-range distance constraints for structure determination or for monitoring the conformational changes of biomolecules or biomolecular complexes.9–14 To further develop the technique, attempts have been made to use spin labels different from conventional nitroxide radicals15–18 or non-identical (spectroscopically orthogonal) spin label pairs.19 An approach with pairs of non-identical spin labels requires, first, a clear spectroscopic separation of the two paramagnetic probes. In addition, the sensitivity, the depth of the DEER modulations, and the possibility to avoid orientation selection20,21 in distance measurement applications are primary factors that determine the choice of a particular spin pair.
We recently proposed a version of the DEER/SDSL approach based on the use of spectroscopically orthogonal Gadolinium (III) and nitroxide spin labels.22 This approach demonstrates good performance in distance measurements, and possesses a sensitivity that is comparable to that of the nitroxide-nitroxide DEER.22–24 In solution, the dynamics (in terms of rotational motion, bonds vibrations, shuffling of the coordinated groups)25 of the chelator wrapping the Gd(III) ion leads to the transient zero-field splitting (ZFS) interaction, which has the same order of magnitude as the static ZFS.26 The nearly featureless EPR spectra of Gd(III) complexes in frozen glassy state provide a snapshot of such transient ZFS. Importantly for distance measurements with Gd(III) labels, this strongly reduces the orientation selection even at high detection fields/frequencies. It has been shown that the use of high spin Gd(III) centers (spin S=7/2) in most cases does not introduce measurable ZFS-induced distortions to the obtained distance distributions.23 In the Gd(III)-nitroxide orthogonal spin pair each type of paramagnetic center can be addressed independently, which opens the possibility to characterize the local environment of each spin probe without the need of additional singly-labeled samples.24 Moreover, independent checks of macromolecular aggregation became possible with this technique.24 Importantly, several different distances can be measured in a single sample, thus increasing the information content available from DEER measurements.23,27
The orthogonal labeling of different subunits in a complex of non-identical biomacromolecules is rather straightforward, if each subunit can be labeled prior to complex formation. Two different paramagnetic centers can also be synthetically added to relatively small peptides.24,28 An orthogonal labeling approach is, however, also attractive for monomeric biomacromolecules, where it can potentially separate distance peaks due to protein aggregation from peaks associated with distances within the same protein molecule. For such studies on monomeric biomacromolecules or for studies of orthogonally labeled protomers in larger protein complexes appropriate orthogonal labeling schemes must be developed. In lactose permease, for example, site-directed labeling of a single-chain protein with a nitroxide spin label and a lanthanide ligand could be achieved by exploiting substrate protection of one of the two cysteine sites.29
Here, we present an alternative approach that does not rely on such fortuitous protection, but is rather based on the use of an unnatural amino acid for the nitroxide labeling30 and of an engineered cysteine for the attachment of a lanthanide chelator functionalized with a thiol-specific group. We also demonstrate that non-specific binding can be an important issue for Gd(III) labeling, most probably due to the large electric dipolar moment of many Gd(III) complexes. We evaluate the performance of two commercially available Gd(III) labels in terms of labeling efficiency, DEER modulation depth and sensitivity. In addition, we discuss optimum measurement conditions with respect to the interplay between the DEER echo reduction for a hard pumping pulse,23,27 and the reduction of the DEER modulation depth for a soft pumping pulse, thus proposing a modified DEER pulse sequence for obtaining optimum signal-to-noise ratio in Gd(III)-nitroxide distance measurements.
Experimental details
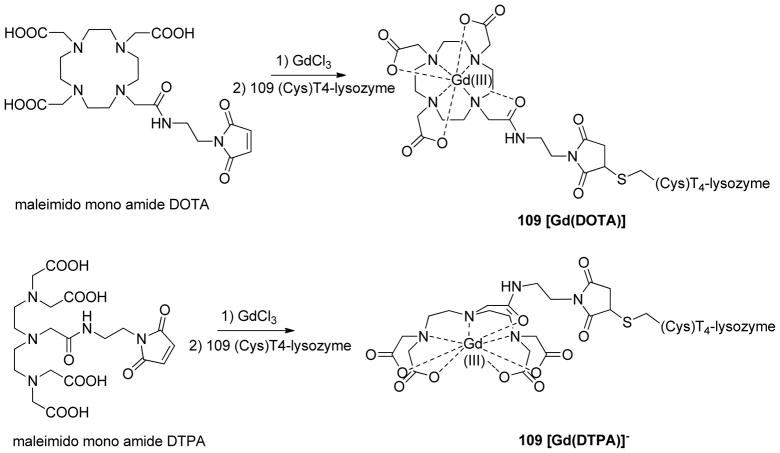
The Gd(III)-nitroxide orthogonal labeling approach was tested on two double mutants of bacteriophage T4-lysozyme with a genetically encoded unnatural amino acid p-acetyl-L-phenylalanine (p-AcPhe) either at site 68 or at site 131 and a cysteine at position 109 (Figure 1). The acetyl group of the residue p-AcPhe was reacted with the hydroxylamine pendant of a nitroxide radical to give rise to an ketoxime-linked nitroxide side chain K1 (Scheme 1).30 In each mutant the cysteine at site 109 was used for binding of the Gd(III)-based spin label. Two different commercially available (Macrocyclics, Inc., USA) functionalized chelators, maleimido-monoamide DOTA (1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid) and maleimido-monoamide DTPA (diethylene triamine pentaacetic acid), were used to efficiently trap Gd(III) ions. The cyclene derivative DOTA and the acyclic ligand DTPA are known to chelate lanthanide ions with high thermodynamic stability and kinetic inertness.31 The resulting chelate complexes, [Gd(DOTA)] and [Gd(DTPA)]− were then covalently attached to the protein through the chemo-specific reaction between the maleimido moiety and the cysteine sulfhydryl (Scheme 2). As a reference, two additional T4-lysozyme mutants with two cysteines at positions 68–109 and 109–131 were labeled with MTSSL to form the conventional R1 side chains (Scheme 1). The obtained Gd(III)–K1 DEER data were compared to the conventional R1–R1 DEER. Further sample preparation details can be found in the electronic supplementary information.
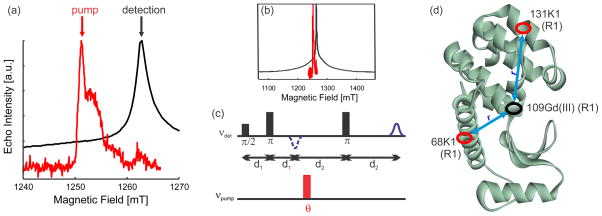
Figure 1.
Selective measurements of field swept ED EPR spectra, pulse sequence and investigated mutants in Gd(III) –nitroxide DEER measurements in T4-lysozyme. (a) Selective measurements of ED EPR spectra of [Gd(DOTA)] (black) and nitroxide radicals (red). Arrows mark positions of pump and detection pulses. The difference in microwave power setting between nitroxide and Gd(III) is 12 dB. Shot repetition times of 100 ms for the nitroxide and 1 ms for Gd(III) were employed. Spectra were normalized to their maximum intensity. (b) Corresponding ED EPR spectra in a broader field range. (c) Sequence of microwave pulses used in the DEER experiment to measure Gd(III)–nitroxide distances. Standard settings (θ = π) as well as settings with smaller flip angles θ for the pumping pulse were applied. (d) Ribbon model of wild-type T4L (PDB ID code 1L63) where the spin labeled sites have been marked with ellipses.
Scheme 1.
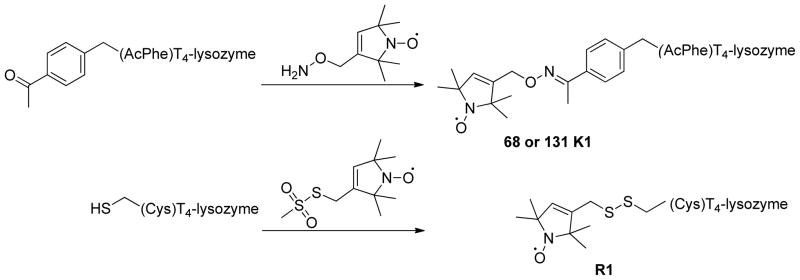
Site-directed labeling with nitroxide radicals.
On the top: p-acetyl-L-phenyl-alanine incorporated in T4-lysozyme. According to described procedure,27 the acetyl group was reacted with the hydroxylamine pendant of the nitroxide derivative to give rise to the oxime-linked nitroxide-side chain K1 located at sites 68 or 131 of T4-lysozyme. On the bottom: MTSSL used to label cysteines at sites 68 / 109 and 131 / 109 of T4-lysozyme in order to obtain 68R1 – 109R1 and 131R1 – 109R1 labeled mutants.
Scheme 2.
Site-directed labeling with lanthanide ions.
(Top) Maleimido mono amide DOTA chelating agent for Gd(III). After incubation with GdCl3 the resulting complex was reacted with the side chain of the cysteine at site 109. (Bottom) Maleimido mono amide DTPA chelating agent for Gd(III). After incubation with GdCl3 the resulting complex was reacted with the side chain of the cysteine at site 109.
The pulse settings for the Gd(III)-nitroxide DEER experiment are presented in Figure 1 together with the pulse sequence for the DEER experiment. The main contribution to the spectroscopic selectivity of nitroxide and Gd(III) species is the 12 dB difference in the optimum microwave power for the corresponding echo detection.22 An additional effect arises from the approximately two orders of magnitude difference in the longitudinal relaxation rates for the two types of paramagnetic species. For nitroxide and Gd(III) detection the limiting repetition rates were 10 and 1000–2000 repetitions per second, respectively. As in previous works22–24 we set the frequency of the pumping pulse at the maximum intensity of the nitroxide absorption, and the frequency of the detection pulses to the maximum of the Gd(III) absorption. Both for DOTA- and DTPA-based labels this corresponded to approximately 300 MHz lower frequency as compared to the frequency of the pump pulse. In addition to the DEER experiments with standard settings of the pump pulse (flip angle θ =π) we performed additional measurements decreasing the flip angle θ for the pump pulse, in order to verify the joint effect of the DEER modulation depth reduction and the echo reduction23,27 on the obtained overall signal-to-noise ratios. Particular pulse settings for DEER and relaxation measurements are given in the electronic supplementary information.
An experiment with detection on nitroxide radicals and pumping on Gd(III) ions is in principle possible as well, but would have a very low modulation depth of about the values obtained in Gd(III)-Gd(III) DEER measurements.16 Taking into account incomplete Gd(III) labeling (see below in the Results and discussion section), which further reduces the modulation depth in such an experiment, we did not test this inverse detection scheme on our T4 lysozyme samples.
Results and discussion
The T4-lysozyme mutants containing one nitroxide side chain K1 and one exposed cysteine residue have a strong tendency to cross-link. As a result, before Gd(III)-labeling the samples consisted to a large extent of cross-linked protein dimers, which could be monitored in the nitroxide-nitroxide DEER experiment (see electronic supplementary information for further details). Before Gd(III) labeling the T4-lysozyme samples were treated with DTT in order to cleave disulfide bridges between cross-linked proteins. DTT was removed from the protein solution shortly before Gd(III) labeling. After Gd(III) labeling (30 μM protein concentration, 3 equivalents of Gd(III) label), the modulation depth in the nitroxide-nitroxide DEER experiment was reduced by about 1/3. This indicated that about 30% of protein molecules were labeled with Gd(III) complexes, and thus unavailable for the cross-linking. Considering that samples were prepared under the same conditions, we assumed that the degree of dimerization is the same in all four samples. With these conditions we estimate the concentration of the orthogonally Gd(III)- and nitroxide-labeled protein to be about 30–40% of the concentration of all nitroxide-labeled protein molecules. The CW EPR measurements revealed nitroxide radical concentrations in the range 30–100 μM. The concentrations of orthogonally Gd(III)-nitroxide labeled T4-lysozyme should thus be 10–40 μM. In this concentration range good S/N was achieved in Gd(III)-nitroxide DEER, thus further reduction of the labeled protein concentrations is still possible. We estimate the limiting concentrations for this type of distance measurement to be 5–10 μM on our current high-power Q-band setup.32
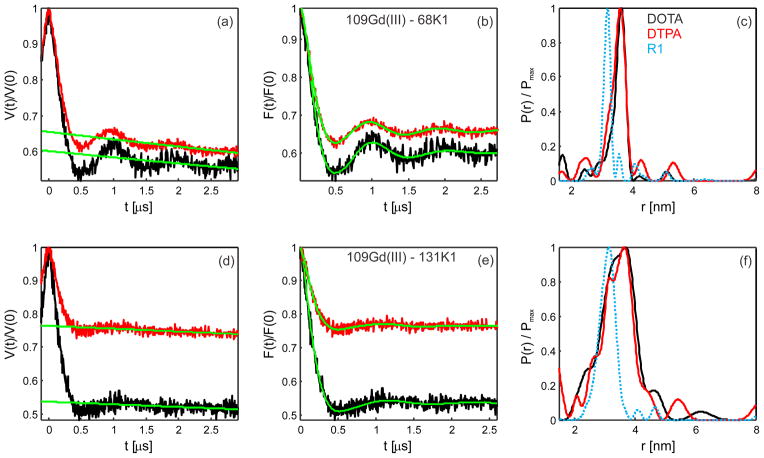
The samples for pulse EPR measurements were shock-frozen in a 1/1 v/v buffer/glycerol mixture, whereas spin labeling and sample preparation were performed with a MOPS buffer (pH 6.8), with low (25 mM NaCl) or high (500 mM) ionic strength, the latter one with additional 10% v/v glycerol. We found that, despite good labeling efficiencies for K1 nitroxide labels (>90%), the modulation depth in the Gd(III) nitroxide DEER experiment (with θ = π) was rather low for the samples prepared with a low ionic strength buffer (Figure 2). By washing the samples using the high ionic strength buffer with 10% v/v glycerol, the modulation depth could be dramatically increased for all four studied Gd(III)-nitroxide-labeled samples (Figure 2). To rationalize these observations, note that the modulation depth in the Gd(III)-nitroxide DEER experiment is determined by the labeling efficiency of the pumped spin labels (in our case, nitroxide radicals).24 The labeling efficiency for Gd(III) labels would only affect the obtained signal-to-noise ratios but not the DEER modulation depth. Since the oxime bond of the K1 side chain30 and the thioether bond4,14 linking the Gd(III)-complex to the engineered cysteine are stable under our experimental conditions, the washing procedure used could not change the fractions of nitroxide-only labeled and orthogonally Gd(III)-nitroxide labeled proteins. Therefore, the change of the DEER modulation depth has to be attributed to the presence of Gd(III) complexes detached from the protein in the sample prepared with the buffer of weaker ionic strength. We thus propose that in such a preparation there exists some fraction of Gd(III)-based spin labels that is non-specifically and non-covalently bound to the surface of T4-lysozyme. Upon admixing glycerol before shock-freezing with liquid nitrogen, these Gd(III) complexes were detached, thus participating in the formation of the Gd(III) spin echo signal but not in the DEER modulation depth. By washing the samples in a higher ionic strength buffer and with additional admixed glycerol, it was possible to strongly reduce such non-specific binding. Interestingly, for both mutants the DEER modulation depth obtained was lower for samples labeled with [Gd(DTPA)]− than for the samples labeled with neutral [Gd(DOTA)] (Figure 3). The difference might be explained by a higher degree of non-specific binding in the [Gd(DTPA)]− samples due to a stronger electrostatic interaction of the net negatively charged complex with the protein surface. Although the observed non-covalent labeling with Gd(III) chelates may be protein-specific, the electrostatic nature of the Gd(III) chelate complexes suggests a broader relevance of the observed effect.
Figure 2.
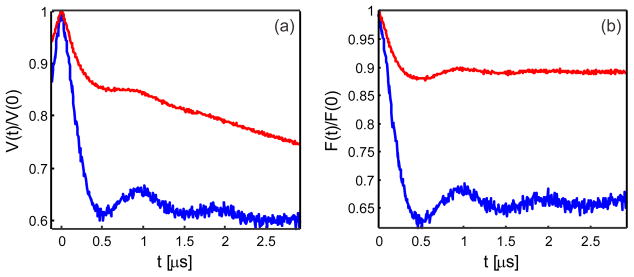
Effect of the buffer ionic strength on modulation depths in the Gd(III)–nitroxide DEER experiment. (a) Primary DEER data and (b) background-corrected form factor traces as calculated by DeerAnalysis 2011 for 109 [Gd(DTPA)]− –68 K1. Results obtained using a 25 mM NaCl-containing buffer and a 500 mM NaCl plus 10% v/v glycerol-containing buffer are displayed in red and blue respectively. A significant increase of the Gd(III)-nitroxide DEER modulation depth is observed. Q-band DEER measurements were recorded at 10 K with a pump pulse flip angle θ = π.
Figure 3.
Q-band DEER measurements of [Gd(DOTA)]-nitroxide (black) and [Gd(DTPA)]− -nitroxide (red) labeled T4-lysozyme. On the top, (a), (b) and (c) results for 109 Gd(III) – 68 K1. On the bottom, (d), (e) and (f) results for 109 Gd(III) –131 K1. (a, d) Primary DEER time traces with background functions (green), (b, e) background-corrected form factors traces and (c, f) the obtained distance distributions as calculated by DeerAnalysis 2011. Best fits (b, e, in green) were obtained with a Tikhonov regularization parameter of 10 with a model-free fitting. The nominal flip angle for the pump pulse was set to π. The dotted-cyan distance distribution (c, f) was obtained in samples doubly-labeled with the R1 side chain. DEER experiments were performed at 10K with d1 = 400 ns and d2 = 3200 ns.
The set of Gd(III)-nitroxide DEER measurements performed for a standard pump flip angle θ = π on the samples treated with a 10% v/v glycerol, 500 mM NaCl buffer is presented in Figure 3 together with the data analysis. The experimental data were processed with use of the DeerAnalysis 2011 package33 with model free fits, supplemented by the Tikhonov regularization with L-curve analysis34–36 for obtaining the optimum smoothness distance distributions (Figure 3). In addition, model fits with a single Gaussian distribution and with a linear combination of two Gaussian distributions were also performed (see electronic supplementary information for further details). Due to the low protein concentrations, the intermolecular background was nearly linear for all four studied samples. The background artifacts were, thus, minimized.
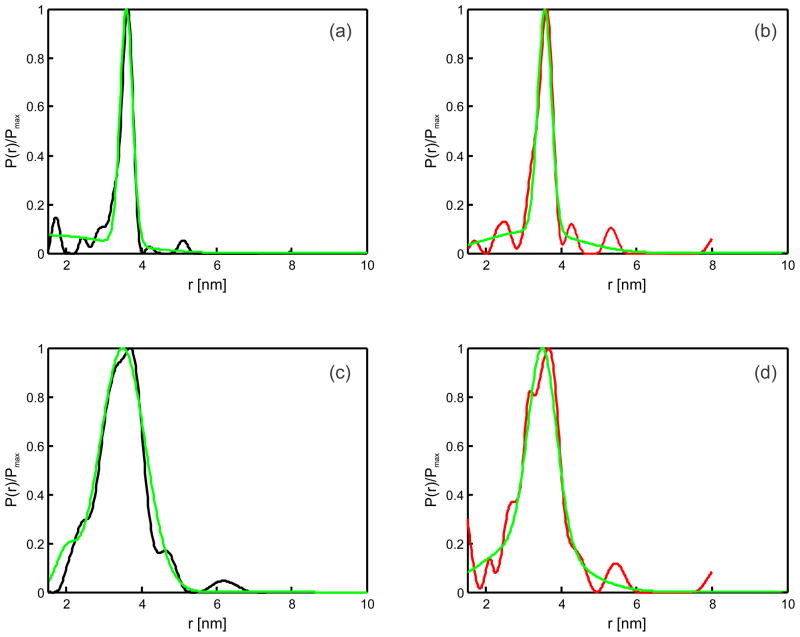
The distance distributions obtained from the Gd(III)-nitroxide DEER experiment showed a main peak together with a small fraction of broadly distributed distances, which are best visible upon fitting the data with a linear combination of two Gaussian distributions (Figure 4 and electronic supplementary information). The position and the width of the main peak was nearly identical in the model free fit and in the two Gaussians fit (Figure 4). In the model free fit the broad component is broken into a collection of individual small peaks, due to the property of the Tikhonov regularization to provide a distance distribution with similar peak widths throughout the whole populated distance range. The two Gaussians fit demonstrates that this broad component is well fitted by a broad Gaussian distribution as well. Note that a single Gaussian fit resulted in a much worse correspondence between the simulated time trace and the experimental data (see electronic supplementary information). In that fit the width of the peak became artificially increased in order to partially incorporate the broad component.
Figure 4.
Comparison of distance distributions obtained with model free fits, based on Tikhonov regularization, for [Gd(DOTA)] (black, (a, c)) and [Gd(DTPA)]− (red, (b, d)) with the corresponding distance distributions obtained with a two Gaussian model fit (green). Data are shown for 109 Gd(III) – 68 K1 (a, b) and for 109 Gd(III) – 131 K1 (c, d). Further Gaussian model fit details can be found in the electronic supplementary information.
The width of the broad component present in the distance distributions cannot be explained by the entire conformational flexibility of the two spin labels and is thus attributed to sample imperfections. This broad component could be tentatively attributed to a certain degree of protein misfolding. Note that in the R1-R1 nitroxide-nitroxide DEER measurements a small broader component was present as well.30 The R1-R1 DEER experiments reproduced in the present work at Q band confirm this feature (Figure 3), which is thus specific to the protein rather than to the type of label.
The Gd(III)-nitroxide DEER traces of the samples with K1 at site 68 revealed several full oscillations. This resulted in rather narrow distribution of distances for both [Gd(DTPA)]− - and [Gd(DOTA)]-labeled samples. For the K1 side chain placed at site 131 broader distance distributions were obtained. The distance distributions obtained with [Gd(DTPA)] − and [Gd(DOTA)] were nearly identical as well. In both cases a lower DEER modulation depth was obtained for [Gd(DTPA)] −-labeled samples as compared to the [Gd(DOTA)]-labeled ones. Both for K1 side chain at position 68 and at position 131, the distance distributions obtained from Gd(III)-nitroxide DEER experiment are similar to the R1-K1 nitroxide-nitroxide distance distributions measured between the same pairs of sites in terms of mean distances and distribution widths.30 In contrast, the corresponding K1-K1 nitroxide-nitroxide distance distributions were significantly broader.
The interspin distance is sensitive to the displacement of one of the spin centers along the interspin vector direction significantly more than to the displacement in the direction perpendicular to the interspin vector. A distance distribution obtained from the DEER experiment allows for estimating the sum of projection widths for the distributions of positions of a pair of spin labels, with the projection direction being along the average interspin vector. The choice of the two pairs of label positions used in this work allows for roughly estimating the width of the distribution of Gd(III) centers in two very different directions30 (Figure 1). This is done by comparing the Gd(III)-K1 distance distributions to the R1-K1 and K1-K1 nitroxide-nitroxide distance distributions reported previously.30 From such a comparison we estimate that the conformational freedom of the Gd(III)-labels is comparable to that of the R1 side chain. It is possible that the Gd(III) complex electrostatically sticks to the surface of protein, analogously to what was observed for a conformationally constrained DOTA-derivative attached to ubiquitin.37
At 10 K DEER traces with a length of at least 4 μs could be measured at Q band for all four samples (5 μs for 68 K1 – 109 [Gd(DOTA)] sample). Due to the fast longitudinal relaxation of Gd(III) (see electronic supplementary information), the repetition rates up to 2000 shots per second could be used in Gd(III)-nitroxide DEER experiments. We estimated the characteristic D-values of the Zero Field Splitting (ZFS) term in the spin Hamiltonian of Gd(III) centers to be about 600 MHz for [Gd(DOTA)] and about 1500 MHz for [Gd(DTPA)] − (see electronic supplementary information). These D-values are in agreement with previously reported values for corresponding complexes without maleimido linker.38 The lower D-value for [Gd(DOTA)] results in a narrower central transition of the corresponding echo-detected (ED) EPR spectrum. This is advantageous regarding signal-to-noise ratios obtained in the DEER experiment, as for a narrower EPR line more spins can be excited by a microwave pulse with a given excitation bandwidth. The transverse relaxation rate of [Gd(DTPA)] − has a stronger temperature dependence than that for [Gd(DOTA)] (see electronic supplementary information). In both cases measurement of 3–4 μs DEER traces should be feasible at 20 K. The Gd(III)-nitroxide DEER measurements in the temperature range 5–20 K are thus possible for both types of Gd(III) labels. By increasing the temperature, the accessible distance range would, of course, be reduced due to the shortening of the characteristic transverse relaxation times.
An important factor influencing the sensitivity of the Gd(III)-nitroxide DEER experiment is the reduction of the amplitude of the refocused echo upon applying the pump pulse.23,27 Due to the difference in spin operator transition moments for nitroxide radicals (S=1/2) and Gd(III) centers (S=7/2), the nominal nitroxide flip angle of π for the pump pulse corresponds to the flip angle of 4π for the most intense |−1/2〉↔|+1/2〉 transition of Gd(III) species (and of 2π for the nitroxide flip angle of π/2). In this range the excitation bandwidth of the pump pulse, with respect to unwanted excitation of Gd(III) species, changes significantly with the change of the flip angle. Reducing the excitation bandwidth of the pump pulse leads to a smaller DEER echo reduction, which is favorable for better signal-to-noise ratios. On the other hand, reducing the flip angle of the pump pulse causes lower DEER modulation depth.
The described effects depend on the strengths of the pulses used in the Gd(III)-nitroxide DEER experiment. In the following we restricted the duration of all pulses on the detection frequency (Gd(III) species) to 12 ns, which is the shortest pulse length at the 300 MHz frequency offset reproducibly achievable on our high-power Q-band setup. The sensitivity of refocused echo detection reduces rather strongly with reduction of flip angles or excitation bandwidths of the corresponding pulses, and we thus assume that settings with all 12 ns pulses are close to the optimal ones.
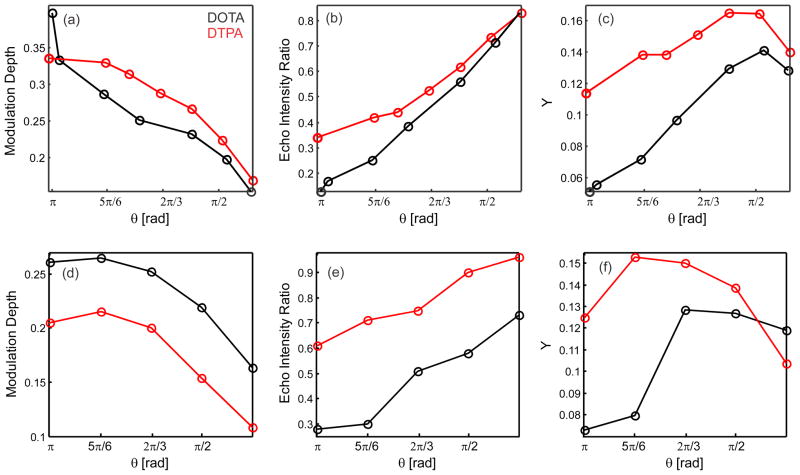
In order to find the optimum conditions for the interplay of the modulation depth reduction and the DEER echo amplitude reduction effects, we first performed a series of Gd(III)-nitroxide DEER measurements with different flip angles θ for a pump pulse of 12 ns duration (Figure 5(a–c) and electronic supplementary information). The shape of the obtained DEER traces did not depend on the strength of the pump pulse, thus identical distance distributions were obtained regardless of the flip angle θ. For a given spin pair concentration, the signal-to-noise ratio in the Gd(III)-nitroxide DEER experiment is proportional to the product of the echo reduction ratio and the modulation depth (Y parameter). Figure 5(c) shows that the Y parameter reaches its maximum at θ ~ π/2 independently of the type of Gd(III) chelate complex used.
Figure 5.
Variation of the Gd(III)-nitroxide DEER modulation depth (a, d) and of the echo intensity ratio (b, e) as a function of the flip angle θ for the pump pulse. The echo intensity ratios were calculated by measuring the DEER echo with and without pump pulse. The value Y (c, f) is the product between the DEER modulation depth and the echo intensity ratio. This value represents the relative S/N ratio for the corresponding DEER experiment. A first series with constant pump pulse length of 12 ns and with variation of microwave power for the pump pulse is presented in (a–c), a second series with constant microwave power for the pump pulse (optimal value from the first series) and with variation of the pump pulse duration is shown in (d–f). Data were obtained for 109 Gd(III) - 68 K1. Results for [Gd(DOTA)] and [Gd(DTPA)]− are shown in black and red respectively. Q-band DEER traces were obtained at 10 K.
In the second experiment, we fixed the microwave power to the optimum value found in the previous series (a power that corresponds to a π/2 pulse for the 12 ns pulse duration) and varied the length of the pump pulse. The corresponding dependencies of Gd(III)-nitroxide DEER modulation depth, echo reduction and cumulative parameter Y(θ) are shown in Figure 5(d–f). One can see that a pump pulse flip angle range between π/2 and 2π/3 corresponds to the best sensitivity conditions. Thus, a 12 ns pulse with θ = π/2 can be used to optimize sensitivity in the Gd(III)-nitroxide DEER experiment. The DEER modulation depth at these conditions is about 0.2 for both types of Gd(III) labels (compare to 0.35–0.4 for θ = π and tp=12 ns, Figure 5(a)). This value is still acceptable in most applications, unless samples with very low labeling efficiency are to be studied. Performing Gd(III)-nitroxide DEER experiments with reduced flip angle for the pump pulse is thus advantageous in terms of signal-to-noise ratio, especially if [Gd(DOTA)]-based labels are used.
Conclusions
We showed for the first time a site-specific orthogonal Gd(III)-nitroxide spin labeling on a model 19 kDa protein, using an unnatural amino acid approach for the attachment of a nitroxide label (K1 side chain) and thiol specific attachment of a Gd(III)-label to an engineered cysteine residue. This approach does not exploit any specific properties of the used model protein, and should thus be rather general. The use of high ionic strength, glycerol-containing buffer is recommended for Gd(III) labeling to avoid non-covalent, non-specific attachment of the ligand to the protein. The performance of commercially available [Gd(DOTA)]- and [Gd(DTPA)]− -based spin labels in Gd(III)-nitroxide DEER experiment was evaluated in detail. In particular, for the studied labeling site, the Gd(III)-based spin labels have a rather restricted conformational freedom, which makes the widths of the distance distributions comparable to those obtained with the conventional R1 nitroxide side chains. These widths are mainly determined by the conformational freedom of the K1 nitroxide side chain, located at the unnatural amino acid site. For pumping on a nitroxide and observing on a Gd(III) label, we propose a modified 4-pulse-DEER sequence with a π/2 flip angle and 12 ns duration for the pump pulse. This sequence optimizes the signal-to-noise ratios in the Gd(III)-nitroxide DEER experiment at a cost of about 50% reduction of the modulation depth as compared to the all-12-ns standard 4 pulse DEER sequence. The optimization was done for the 12 ns detection pulses and for pumping and detecting at the maxima of corresponding EPR spectra of nitroxide radicals and Gd(III) ions. These conditions may not be achievable with low power Q-band setups or with narrow bandwidth probe heads. Q-band DEER-based Gd(III)-nitroxide distance measurements should be possible down to 5–10 μM concentrations of orthogonally-labeled protein molecules. The proposed labeling approach opens the possibility to benefit from advantages of spectroscopically orthogonal spin pairs in EPR studies on proteins.
Supplementary Material
Acknowledgments
We acknowledge financial support by the SNF (Grant No. 200021_121579), NIH grant EY05216 and the Jules Stein Professorship (to WLH). We are grateful to Professors Kalman Hideg and Tamas Kalai for the generous gift of the hydroxylamine spin label used in these experiments.
Footnotes
Supporting Information: sample preparation; experimental details; data processing; Q-band Gd(III)-nitroxide DEER measurements; X-band Gd(III)-nitroxide DEER measurements; Gaussian fits of Gd(III)-nitroxide DEER measurements; nitroxide-nitroxide DEER measurements for the pair of R1 side chains; assessing the Gd(III) labeling efficiency; conditions for optimal signal-to-noise in Gd(III)-nitroxide DEER; effect of nuclear modulation on the quality of the DEER trace; echo detected EPR spectra of Gd(III) centers; relaxation properties of Gd(III) centers. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Milov AD, Salikhov KM, Shirov MD. Application of ELDOR in Electron-Spin Echo for Paramagnetic Center Space Distribution in Solids. Fiz Tverd Tela (Leningrad) 1981;23:957–982. [Google Scholar]; Milov AD, Ponomarev AB, Tsvetkov YuD. Electron Electron Double Resonance in Electron-Spin Echo-Model Biradical Systems and the Sensitized Photolysis of Decalin. Chem Phys Lett. 1984;110:67–72. [Google Scholar]
- 2.Martin RE, Pannier M, Diederich F, Gramlich V, Hubrich M, Spiess HW. Determination of End-to-End Distances in a Series of TEMPO Diradicals of up to 2.8 nm Length with a New Four-Pulse Double Electron Electron Resonance Experiment. Angew Chem Int Ed Engl. 1998;37:2834–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-Time Free Measurements of Dipole-Dipole Interactions between Electron Spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 4.Griffith OH, McConnell HM. A Nitroxide-Maleimide Spin Label. Proc Natl Acad Sci. 1966;55:8–11. doi: 10.1073/pnas.55.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altenbach C, Marti T, Khorana HG, Hubbell WL. Transmembrane Protein Structure: Spin Labeling of Bacteriorhodopsin Mutants. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 6.Hubbell WL, Cafiso DS, Altenbach C. Identifying Conformationl Changes with Site-Directed Spin Labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 7.Klare JP, Steinhoff HJ. Spin Labeling EPR. Photosynth Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 8.Bordignon E. Site-Directed Spin Labeling of Membrane Proteins. Curr Top Chem. 2012;321:121–157. doi: 10.1007/128_2011_243. [DOI] [PubMed] [Google Scholar]
- 9.Jeschke G, Polyhach Ye. Distance Measurements on Spin-Labelled Biomacromolecules by Pulsed Electron Paramagnetic Resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 10.Schiemann O, Prisner TF. Long-range Distance Determination in Biomacromolecules by EPR Spectroscopy. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 11.Tsvetkov YD, Milov AD, Maryasov AG. Pulsed Electron-Electron Double Resonance (PELDOR) as EPR Spectroscopy in Nanometer Range. Russ Chem Rev. 2008;77:487–520. [Google Scholar]
- 12.Krsti I, Edenward B, Margraf D, Marko A, Prisner T. Structure and Dynamic of Nucleic Acids. Curr Top Chem. 2012;321:159–198. doi: 10.1007/128_2011_300. [DOI] [PubMed] [Google Scholar]
- 13.Jeschke G. DEER Distance Measurements on Proteins. Ann Rev Phys Chem. 2012;63:1–28. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 14.Zhou A, Abu-Baker S, Sahu ID, Liu L, McCarrick RM, Dabney-Smith C, Lorigan GA. Determining α–Helical β-Sheet Secondary Structures via Pulsed Electron Spin Resonance Spectroscopy. Biochem. 2012;51:7417–7419. doi: 10.1021/bi3010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Amsterdam IM, Ubbink M, Canters GW, Huber M. Measurement of a Cu-Cu Distance of 26 A by Pulsed EPR Method. Angew Chem Int Ed. 2003;42:62–64. [PubMed] [Google Scholar]
- 16.Raitsimring AM, Gunanathan C, Potapov A, Efremenko I, Martin JML, et al. Gd3+ Complexes as Potential Spin Labels for High Field Pulsed EPR Distance Measurements. J Am Chem Soc. 2007;129:14138–14139. doi: 10.1021/ja075544g. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Kise D, Saxena S. An Approach Towards the Measurements of Nanometer Range Distances Based on Cu2+ Ions and ESR. J Phys Chem B. 2010;114:6165–6174. doi: 10.1021/jp911637s. [DOI] [PubMed] [Google Scholar]
- 18.Reginsson GW, Kunjir NJ, Sigurdsson NT, Schiemann O. Trityl Radicals: Spin-Label for Nanometer Distance Measurements. Chem Eur J. 2012;18:13580–13584. doi: 10.1002/chem.201203014. [DOI] [PubMed] [Google Scholar]
- 19.Narr E, Godt A, Jeschke G. Selective Measurements of a Nitroxide-Nitroxide Separation of 5 nm and a Nitroxide-Copper Separation of 2.5 nm in a Terpyridine-Based Copper(II) Complex by Pulse EPR Spectroscopy. Angew Chem Int Ed. 2002;41:3907–3910. doi: 10.1002/1521-3773(20021018)41:20<3907::AID-ANIE3907>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]; Jeschke G, Zimmermann H, Godt A. Isotope Selection in Distance Measurements between Nitroxides. J Magn Res. 2006;180:137–146. doi: 10.1016/j.jmr.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Polyhach Ye, Godt A, Bauer C, Jeschke G. Spin Pair Geometry Revealed by High-Field DEER in the Presence of Conformational Distribution. J Magn Reson. 2007;185:118–129. doi: 10.1016/j.jmr.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Bode BE, Plackmeyer J, Prisner TF, Schiemann O. PELDOR Measurements on a Nitroxide-Labeled Cu(II) Porphyrin: Orientation Selection, Spin-Density Distribution and Conformational Flexibility. J Phys Chem A. 2008;112:5064–5073. doi: 10.1021/jp710504k. [DOI] [PubMed] [Google Scholar]
- 22.Lüders P, Jeschke G, Yulikov M. Double Electron-Electron Resonance Measured between Gd3+ Ions and Nitroxide Radicals. J Phys Chem Lett. 2011;2:604–609. [Google Scholar]
- 23.Yulikov M, Lüders P, Warsi MF, Chechik V, Jeschke G. Distance Measurements in Au Nanoparticles Functionalized with Nitroxide Radicals and Gd3+-DTPA Chelate Complexes. Phys Chem Chem Phys. 2012;14:10732–10746. doi: 10.1039/c2cp40282c. [DOI] [PubMed] [Google Scholar]
- 24.Lüders P, Jäger H, Hemminga MA, Jeschke G, Yulikov M. Distance Measurements on Orthogonally Spin Labelled Membrane Spanning WALP23 Polypeptides. J Phys Chem B. 2013;117:2061–2068. doi: 10.1021/jp311287t. [DOI] [PubMed] [Google Scholar]
- 25.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) Chelates as MRI Contrast Agents: Structure, Dynamics and Applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]; Su X-C, Otting G. Paramagnetic Labeling of Proteins and Oligonucleotides for NMR. J Biomol NMR. 2010;46:101–112. doi: 10.1007/s10858-009-9331-1. [DOI] [PubMed] [Google Scholar]; Keizers PHJ, Ubbink M. Paramagnetic Tagging for Protein Structure and Dynamic Analysis. Progr NMR Spectrosc. 2011;58:88–96. doi: 10.1016/j.pnmrs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Raitsimring AM, Astashkin AV, Caravan; P. High-Frequency EPR and ENDOR Characterization of MRI Contrast Agent. Biological Magnetic Resonance. In: Berliner L, Hanson G, editors. Biological Magnetic Resonance. Vol. 28. Springer–Verlag; New York: 2009. pp. 581–621. [Google Scholar]
- 27.Kaminker I, Yagi H, Huber T, Feintuch A, Otting G, Goldfarb D. Spectroscopic Selection of Distance Measurements in a Protein Dimer with Mixed Nitroxide and Gd3+ Spin Labels. Phys Chem Chem Phys. 2012;14:4355–4358. doi: 10.1039/c2cp40219j. [DOI] [PubMed] [Google Scholar]
- 28.Lüders P, Jaeger H, Hemminga MA, Jeschke G, Yulikov M. Multiple Pathway Relaxation Enhancement in the System Composed of Three Paramagnetic Species: Nitroxide Radical-Ln3+-O2. J Phys Chem Lett. 2012;3:1336–1340. doi: 10.1021/jz300316q. [DOI] [PubMed] [Google Scholar]
- 29.Voss J, Wu J, Hubbell WL, Jacques V, Meares CF, Kaback HR. Helix Packing in Lactose Permease of Escherichia Coli: Distances between Site-Directed Nitroxides and a Lanthanide. Biochemistry. 2001;40:3184–3188. doi: 10.1021/bi002333e. [DOI] [PubMed] [Google Scholar]
- 30.Fleissner MR, Brustad EM, Tamas K, Altenbach C, Cascio D, Peters FB, Hideg K, Peukere S, Schultz PG, Hubbell WL. Site-Directed Spin Labeling of a Genetically Encoded Unnatural Amino Acid. Proc Natl Acad Sci. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rocklage SM, Watson AD. Chelates of Gadolinium and Dysprosium as Contrast Agents for MR Imaging. J Magn Res Imaging. 1993;3:167–178. doi: 10.1002/jmri.1880030129. [DOI] [PubMed] [Google Scholar]
- 32.Polyhach Y, Bordignon E, Tschaggelar R, Gandra S, Godt A, Jeschke G. High Sensitivity and Versatility of the DEER Experiment on Nitroxide Radical Pairs at Q-Band Frequencies. Phys Chem Chem Phys. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- 33.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DEER Analysis 2006 – A Comprehensive Software Package for Analyzing Pulsed ELDOR Data. Appl Magn Reson. 2006;30:473–498. http://www.epr.ethz.ch/software/index. [Google Scholar]
- 34.Tikhonov AN, Arsenin VY. Solutions of Ill-Posed Problems. John Wiley & Sons; New York: 1977. [Google Scholar]
- 35.Jeschke G, Panek G, Godt A, Bender A, Paulsen H. Data Analysis Procedures for Pulse ELDOR Measurements of Broad distance distributions. Appl Magn Reson. 2004;26:223–244. [Google Scholar]
- 36.Chiang YW, Borbat PP, Freed JH. The Determination of Pair Distance Distributions by Pulsed EPR Using Tikhonov Regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Häussinger D, Huang J, Grzesiek S. DOTA-M8: an Extremely Rigid, High-Affinity Lanthanide Chelating Tag for PCS NMR Spectroscopy. J Am Chem Soc. 2009;131:14761–14767. doi: 10.1021/ja903233w. [DOI] [PubMed] [Google Scholar]
- 38.Raitsimring AM, Astashkin AV, Poluektov OG, Caravan P. High-Field Pulsed EPR and ENDOR of Gd3+ Complexes in Glassy Solutions. Appl Magn Reson. 2005;28:281–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.