Abstract
The nuclear factor Y (NF-Y), which is a ubiquitous transcription factor found in eukaryotes, is composed of three distinct subunits, namely, NF-YA, NF-YB, and NF-YC. Here, we firstly characterized the detailed function of the Arabidopsis NFYA1 factor. It is found that the 35S::AtNFYA1-overexpressed lines were hypersensitive to salt stress and Abscisic acid (ABA) during the early-postgermination growth stages. The transgenic lines exhibited a severe postgermination growth arrest compared with the wild-type (WT) under salt stress and ABA treatment. Interestingly, sodium tungstate, which is an ABA synthesis inhibitor, restored the salt-sensitive phenotype of the 35S::AtNFYA1 lines. Results of the qRT-PCR analysis showed that the mRNA levels of ABI3 and ABI5, as well as their downstream genes AtEM1 and AtEM6, were more greatly upregulated under salt stress during seed germination in the transgenic lines compared with those in WT. On the other hand, the NFYA1-RNAi lines were found to be insensitive to salt stress and exhibited decreased levels of ABI3, ABI5, EM1, and EM6 transcripts. Our results provide clear evidence supporting a role of AtNFYA1 in regulating postgermination growth arrest under salt stress.
Introduction
Seed germination is a critical process in the plant life cycle, which is determined by many environmental factors, such as temperature, light, oxygen, and water status, among others. Seeds often fail to germinate and develop under unfavorable conditions even after the break of dormancy. The Abscisic acid (ABA) is an important phytohormone that regulates the responses of plants to various environmental stresses, such as high salinity and drought [1]. ABA also plays a significant role in the mediation of seed dormancy, seed germination, and postgermination growth [2], [3]. ABA levels decrease upon seed inhibition to allow the seeds to germinate and develop into seedlings under normal conditions. However, ABA levels remain high under abiotic stress conditions, arresting plant growth and development [4]. The exposure of seeds to exogenous ABA during germination leads to a rapid but reversible arrest in postgermination growth. This ABA-mediated growth arrest of germinated seeds serves as a protective mechanism for the survival of young seedlings under stress conditions [2].
ABI3 and ABI5 are important ABA-insensitive genes that regulate the mediation of ABA-dependent growth arrest during germination [5], [6], [7], [8]. ABI5 encodes a member of basic leucine zipper transcription factors, which can be induced by ABA only within a short poststratification interval, and thus defines a narrow developmental checkpoint following germination [8]. ABI3 encodes a putative acidic domain transcription factor that is necessary for the expression of a large number of late embryogenesis genes, which are supposedly required for desiccation tolerance [7], [9]. The accumulation of ABI3 is also triggered by ABA within a short developmental window. ABI5 mRNA levels were found to be greatly reduced in abi3-1 mutant. Additionally, ABI5 can rescue the ABA-insensitivity of abi3-1. This phenomenon shows that ABI3 acts upstream of ABI5, and therefore indicates the dependency of growth arrest efficiency on ABI5 levels [8]. AtEM1 and AtEM6 are two important Group 1 Late Embryogenesis Abundant (LEA) genes that are expressed during the late period of embryo maturation and during the developmentally regulated period of dehydration at the end of seed development [10]. In the presence of ABA, ABI5 can regulate the expression of AtEM1 and AtEM6 by binding to their promoters [8], [11]. Moreover, AtEM1 and AtEM6 can also be re-induced during the postgermination arrest process under water deficit while the ABI5 expression is upregulated, which might be responsible for the acquired stress tolerance of germinated seeds [8].
Several studies revealed that CPR5, WRKY2 and HYL1 genes also mediate postgermination growth arrest. The phenotypes of cpr5, wrky2 and hyl1 mutants exhibited ABA-hypersensitivity during germination and displayed different degrees of postgermination growth arrest [12], [13], [14]. Moreover, these mutants showed significantly altered expression levels of ABI3, ABI5, EM1, and EM6, implying their direct or indirect involvement in the ABA-regulated postgermination pathways.
Nuclear factor Y (NF-Y) is a kind of ubiquitous transcription factor present in nearly all eukaryotes, which includes three subunits: NFYA, NFYB and NFYC [15], [16]. In animals, the NF-YB and NF-YC subunits firstly form a heterdimer in the cytoplasm, and then it is transported into the nucleus and recruits the NF-YA subunit to generate the mature, heterotrimeric NF-Y transcription factor [17], [18], [19]. This complex has high affinity and sequence specificity for the CCAAT box, which is a cis-element that exists in approximately 25% of eukaryotic gene promoters [18], [20], [21].
Arabidopsis has multiple genes encoding the three subunits, namely, 10 NF-YA, 13 NF-YB, and 13 NF-YC homologs [19]. Due to the potential combinatorial diversity among numerous NFY factors, as well as the high affinity and sequence specificity for the extensive CCAAT box in the eukaryotic genomes, these members play significant roles in the mediation of diverse genes and involve in regulating various processes. For instance, the Leafy Cotyledon1, (LEC1 or NFYB9) which was the first cloned and described plant NF-Y, is essential in both early and late embryo development and necessary for controlling the transition from embryo to adult status [22], [23], [24]. Altered expression of NFYB1 (HAP3a) and NFYB2 (HAP3b) in Arabidopsis could greatly affect the flowering time [21], [25], [26], Several NF-Y members also appear to be important in regulating drought responses, such as the maize NFYB1 and the Arabidopsis NFYA5, both of which could confer drought stress tolerance in the transgenic plants [27], [28],. Additionally, some NFYA subunits such as Arabidopsis NFYA2, NFYA3, NFYA5 were reported to participate in nitrogen nutrition [29], and broader functions of NFY factors like regulation of light signaling, ER stress, Chloroplast biogenesis etc were also revealed [29], [30].
Regarding Arabidopsis NFYA1, its possible role in delaying flowering was reported earlier under the control of a tissue-specific promoter [26]. Recently, Mu etal reported the function of NFYA1 in reproductive development and seed germination [31]. In this study, we demonstrate novel roles for NFYA1 in salt stress. Transgenic lines overexpressing NFYA1 were shown to be hypersensitive to salt stress during post-germination growth. Moreover, genes with known involvement in post-germination development, such as ABI3, ABI5, EM1, and EM6, were greatly upregulated in the transgenic lines. The results of this study show that NFYA1 can be an important regulator in the mediation of post-germination growth arrest under salt stress and ABA treatment.
Materials and Methods
Plant Material
Arabidopsis thaliana ecotype Columbia was used in this study. Surface sterilized seeds were sown on full strength MS medium supplemented with 3% sucrose and stratified at 4°C for 2 d in the dark prior to germination. Seedlings were grown on MS plates or soil under LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions at 22°C. Seeds of abi3, abi5 were obtained from the Arabidopsis Biological Resource Center (ABRC).
Constructs and Generation of Transgenic Plants
To generate 35S::NFYA1 construct, NFYA1 sequence without 3′UTR was PCR amplified from the Arabidopsis cDNA and inserted downstream of 35S promoter in the binary vector pBI121 after confirmed by sequencing. To generate 35S::NFYA1-GFP construct, stop codon removed NFYA1 was ligated in frame to GFP and confirmed by sequencing. To generate AtNFYA1-RNAi construct, the Arabidopsis pFGC5941 vector (ordered from ABRC) for dsRNA production was used. A 427 bp fragment of NFYA1 cDNA was amplified by PCR using the forward primer 5′-TCTAGAGGCGCGCCGCTCTGCTGTGAATTTCCACT-3′ and the reverse primer 5′-GGATCCATTTAAATCCTGATATGGGTTTGGGACAC-3′. The fragment was first cloned between the AscI and SwaI sites of pFGC5941 before an inverted repeat of the same fragment was inserted into the BamHI and XbaI sites of pFGC5941 already containing the sense repeat. All the recombinant binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 and the WT Arabidopsis were transformed by the floral dip method [32]. All the sequence information is from TAIR database (http://www.arabidopsis.org/). Details about other primer sequences are shown in Table S1.
Microscopy Analysis
For DAPI staining, seedlings were stained in 0.2 mg/L DAPI for 15 min, washed three times in PBS solution. For GFP analysis, root tips from 3-day-old transgenic seedlings or onion epidermal cells carrying 35S::NFYA1-GFP expression vector were mounted on slides, and then DAPI or GFP signal was observed with a fluorescence microscope (BX51, model 7.3; Olympus, Japan).
Stress Assays
For germination assay under stresses, seeds were surface sterilized with 70% ethanol for 5 min, with 2.6% hypochlorite for 10 min, and then rinsed with sterile deionized water. Germination assays were carried out with three replicates of 50 seeds. Seeds were sown on MS medium supplemented with 3% sucrose, and the plates were placed at 4°C for 3d in the dark and then transferred to the growth chamber (16 h light/8 h dark) at 22°C. The seeds were regarded as germinated when the radicles protrude from the seed coat. For direct comparison of germination rates, each plate was subdivided, and seeds of all genotypes were put on the same plate.
Statistical Analysis
Data were subjected to Data Processing System (DPS) and significant differences between individual means established using a Student’s t test. Differences at the 1% level were considered significant and denoted by the lowercase letters among different groups.
Quantitative RT-PCR
For qRT-PCR, Reverse transcription reactions were performed using 5 µg RNA by M-MLV reverse transcriptase (Transgene, CHINA) according to the supplier’s manual after incubation with RNase-free DNase I. QRT-PCR reactions were performed with a Bio-Rad real-time thermal cycling system using SYBR-Green to detect gene expression abundances. The reaction mixture (25 µl) contained 0.5 µM of each primer and appropriate amounts of enzymes, cDNA and fluorescent dyes. All runs used a negative control without adding target cDNA, resulting in no detectable fluorescence signal from the reaction. A range of five dilutions of the total cDNA was tested in the same conditions as the samples. Amplification reactions were initiated with a pre-denaturing step at 95°C for 30s and followed by denaturing (95°C for 5s), annealing (60°C for 10s) and extension (72°C for 15s) steps for 49 cycles during the second stage, and a final stage of 55°C to 95°C to determine dissociation curves of the amplified products. All reactions were done in at least three replicates. Data were analyzed using Bio-Rad CFX Manager software. Primer information for qRT-PCR assay is included in Table S1.
Results
Characterization of the AtNFYA1 Transcription Factor Expression
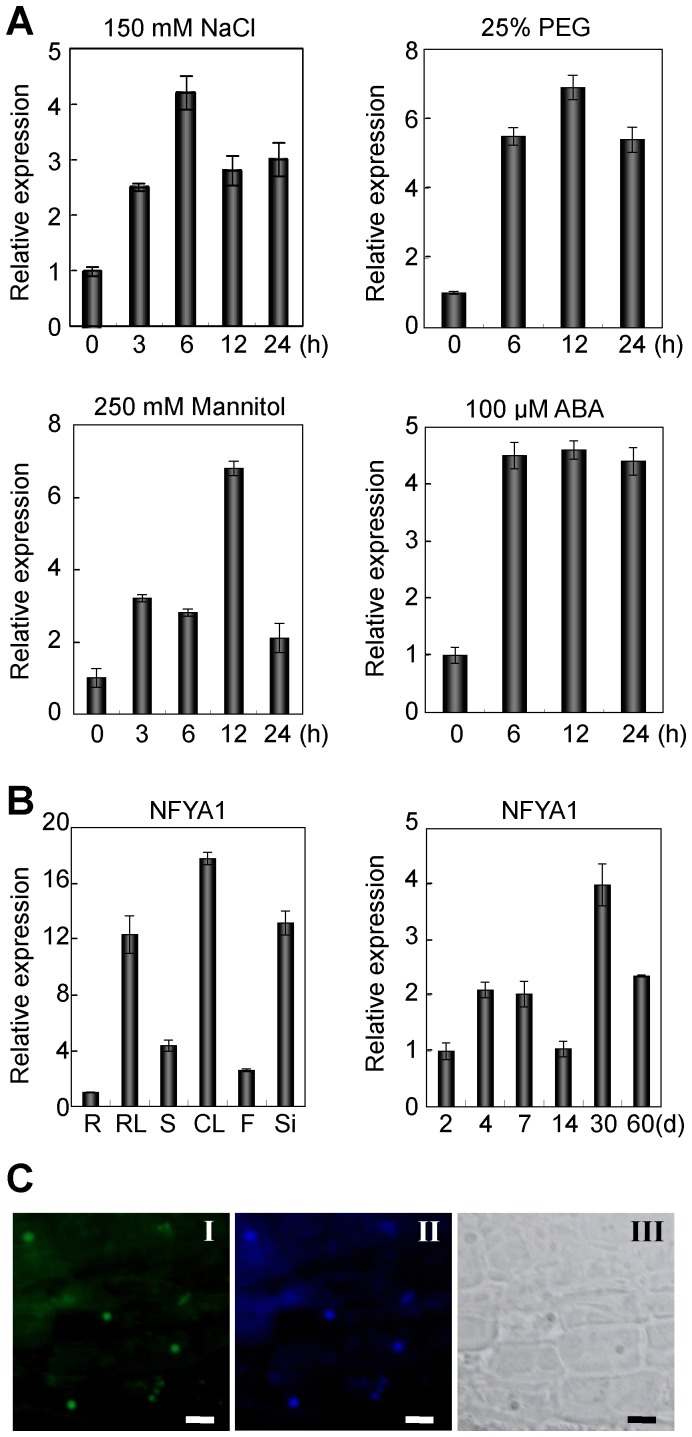
To explore the possible biological roles of the NFYA family under abiotic stress, gene analysis was firstly performed by Genevestigator database (www.genevestigator.com), and the NFYA1 (At5g12840) was found to respond to various environmental stimuli. Subsequently, qRT-PCR analysis was performed, and the results indicated that the expression of NFYA1 was obviously induced by NaCl, mannitol, PEG, and ABA treatments (Fig. 1A), suggesting that NFYA1 is involved in plant responses to environmental stimuli. In addition, the spatio-temporal expression pattern shows that the Arabidopsis NFYA1 can be detected in all developmental stages from 2-day-old to 60-day-old plants, and it is preferentially expressed in leaves and siliques, while weakly expressed in roots and flowers (Fig. 1B).
Figure 1. AtNFYA1 expression pattern and transcriptional regulation.
(A) Induction of AtNFYA1 by NaCl, PEG, Mannitol and ABA assessed by qRT-PCR. 2-week-old Arabidopsis wild type seedlings growing on agar medium under LD condition were treated with the hydroponic solution in petri dishes with filter paper. Expression levels of non-treated samples (0 h) were set to 1.0. (B) Spatio-temporal expression of AtNFYA1 assessed by qRT-PCR. NFYA1 accumulation in roots and 2-day-old seedlings was set to 1.0, respectively. R: roots, RL: rosette leaves, S: stems, CL: cauline leaves, F; flowers, Si: siliques. qRT-PCR quantifications were normalized to ACTIN. Error bars represent SE for three independent experiments. (C) Nuclear localization of NFYA1. The NFYA1-GFP fusion construct was expressed in transgenic Arabidopsis under the control of the CaMV 35S promoter, and GFP signal was observed in the nucleus of the root tip cells (I), with DAPI staining of the nucleus (II) for comparison and morphology of the cells under bright field (III). Bar, (I)–(III), 100 µm.
The NFYA factors in mammals are located in the nucleus and interact with the NFYB/NFYC dimers transported from the cytoplasm [16]. In this study, a C-terminal fusion to the green fluorescent protein (GFP) was generated under the control of the cauliflower mosaic virus 35S promoter to confirm the subcellular localization of the NFYA1 protein. The GFP signal in the 35S::NFYA1-GFP transgenic Arabidopsis was observed predominantly in the nucleus of the root tip cells (Fig. 1C). In addition, the nuclear localization was also confirmed in onion epidermal cells via transient expression (Fig. S1).
Overexpression of AtNFYA1 Leads to Postgermination Growth Arrest under Salt Stress
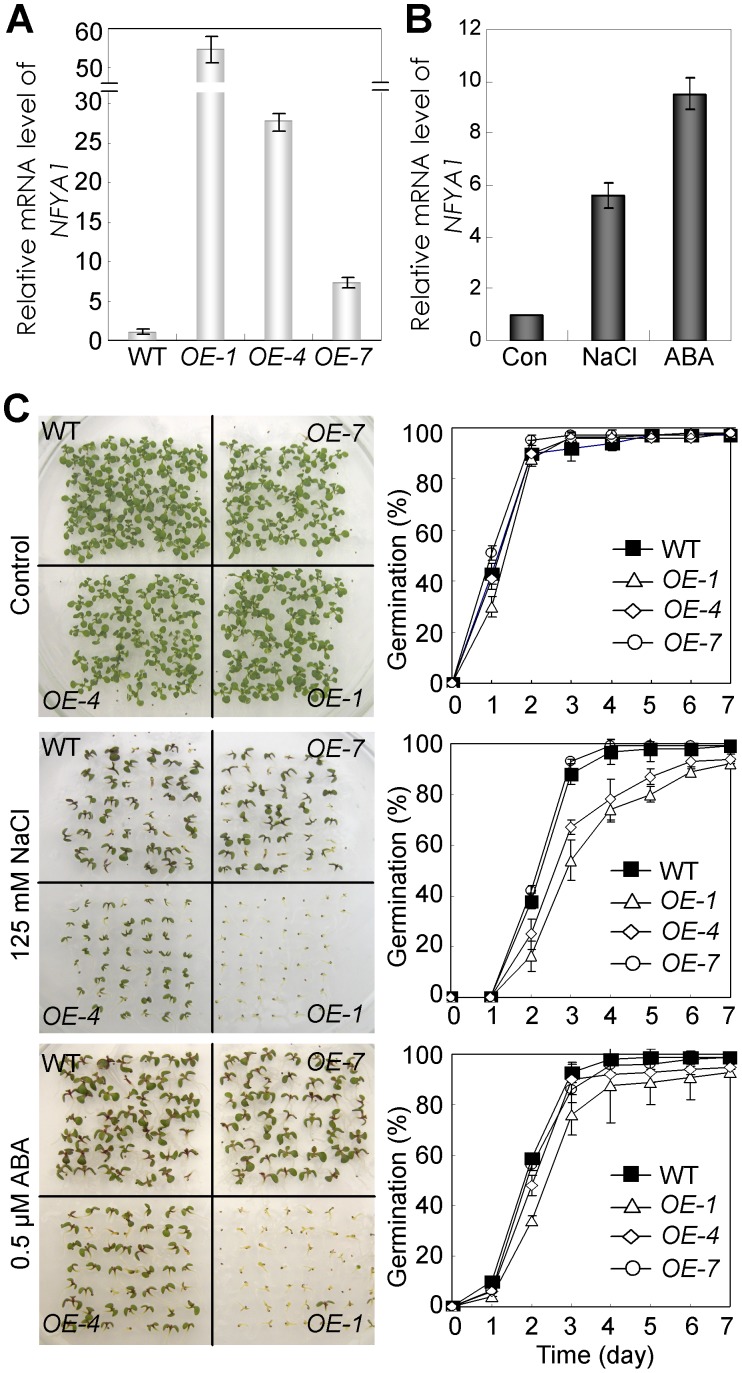
To demonstrate the function of NFYA1 in the regulation of plant responses to abiotic stresses, the transgenic plants overexpressing NFYA1 under the control of the CaMV 35S promoter were generated. Several independent lines that express various levels of NFYA1 were obtained. Line 1 (OE-1), Line 4 (OE-4), and Line 7 (OE-7), which exhibited high, moderate, and low NFYA1 mRNA levels, respectively, were chosen for further analysis (Fig. 2A). Although NFYA1 mRNA is obviously induced by salt and ABA in both seedlings (Fig. 1A ) and germinated seeds (Fig. 2B ), and the NFYA1 transgenic plants were observed to be clearly tolerant to salt stress (Fig. S2), when considering the seed germination and subsequent growth of WT and NFYA1 transgenic seedlings, a remarkable postgermination growth arrest of OE-1 was observed after seed germination (Fig. 2C). As shown in Fig. 2C, the WT and NFYA1 transgenic plants showed no difference after growing for 2 weeks on the medium without NaCl. However, on the medium supplemented with 125 mM NaCl, OE-1 germinated but failed to develop into seedlings, and OE-4 exhibited smaller seedlings as well as weaker growth compared with WT and OE-7. These observations indicate that the sensitivity of the germinated seeds to salt stress depends on the NFYA1 expression levels. Additionally, when we grew the same transgenic lines on medium supplemented with ABA, postgermination growth arrest was also observed, and a more severe phenotype was found in the OE-1 plants than the other two weaker lines OE-4 and OE-7.
Figure 2. 35S::NFYA1 transgenic lines are hypersensitive to salt stress and ABA during postgermination growth.
(A) Relative expression levels of different 35S::NFYA1 lines assessed by qRT-PCR. 2-week-old Arabidopsis seedlings on MS agar medium under LD condition were harvested and RNA was extracted. Expression levels of NFYA1 in wild type were set to 1.0. (B) Accumulation of NFYA1 mRNA in germinated seeds of under ABA and salt treatment. Wild type seeds were germinated on MS liquid medium moistened filter paper for 24 hours after stratification, and then were transferred onto filtered paper moistened with water (control), 125 mM NaCl and 0.5 µM ABA respectively. After treated for 8 hours, the samples were harvested and RNA was extracted. Here, three biological repeats were done and each qRT-PCR was also performed three times. (C) Wild type, NFYA1 transgenic lines OE-1, OE-4 and OE-7 growing on MS medium supplemented with 0 mM (control) and 125 mM NaCl for 14 days, and germination rate of WT, OE-1, OE-4 and OE-7 seeds on the above medium counted for 7 days after stratification. For these data, at least three independent biological repeats were done, and above 50 seeds were used for each line. Pictures were taken 14 d after stratification.
AtNFYA1 Functions in the Postgermination Checkpoint
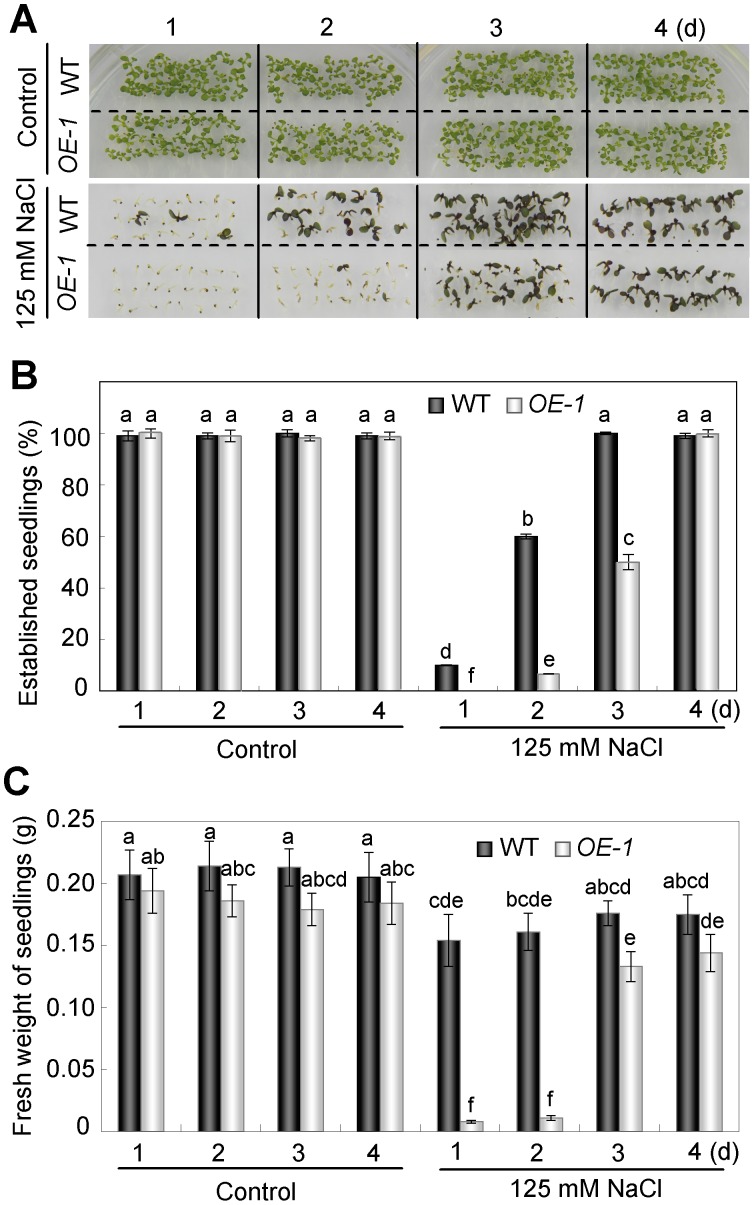
Since we saw clearly salt tolerance in the NFYA1-overexpressed seedlings and big plants (Fig. S2), whereas severe growth quiescence of the transgenic lines after seed germination, we assume that NFYA1 functions in regulating the postgermination growth arrest of the plants during a very short period after seed germination, To figure out the developmental stage that NFYA1 might involve in, the seeds of WT and OE-1 were geminated under normal conditions for 1, 2, 3, and 4 days, respectively, after 3-day stratification at 4°C. Then, the germinated seeds (or young seedlings) were transferred onto MS medium supplemented with 125 mM NaCl and exposed to salt stress for two weeks. As shown in Fig. 3, the proportion of green seedlings gradually increased as the delay of the transfer (Fig. 3B). The growth arrest happened to almost all transgenic seedlings transferred from normal condition to salt stress medium within 2 days after germination. They showed nearly no increase of fresh weight and remained white and quiescent after 2 weeks under salt stress condition. (Fig. 3C). However, the exposure of the 3- and 4-day-old young transgenic seedlings growing on MS medium to salt stress condition failed to arrest their subsequent growth (Fig. 3B,C). Therefore, NFYA1 is hypothesized to be involved in the mediation of postgermination growth arrest under salt stress during a limited developmental window before the seedlings turn green.
Figure 3. 35S::NFYA1 transgenic lines are hypersensitive to salt stress during a limited developmental window.
(A) Seeds of WT and OE-1 lines were sown on MS medium for germination, and after 1d, 2d, 3d and 4d, the germinated seeds were transferred respectively onto MS medium supplemented with 125 mM NaCl for postgermination growth for 14 days under LD condition. (B) The number of established seedlings under each condition was counted. (C) After 14 days, the fresh weight of 50 transferred seedlings under each condition was measured. For these data, at least three independent biological repeats were done. Samples with different letters are significantly different: P<0.01.
Involvement of AtNFYA1 in the ABA Signaling Pathway Instead of in the Synthesis Pathway
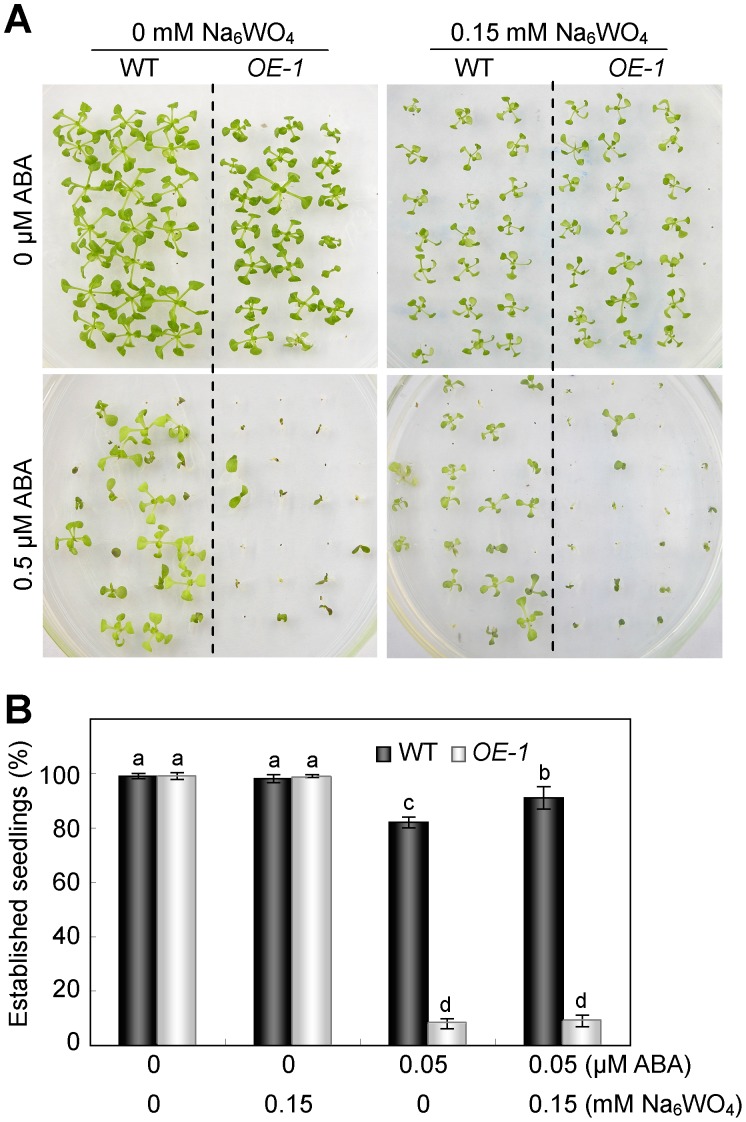
ABA is a critical signaling molecule involved in seed germination, postgermination growth, and abiotic stress resistance. In this study, to reveal whether the postgermination growth arrest under salt stress affected the ABA-related pathway. Sodium tungstate (Na6Wo4), which is one of the ABA synthesis inhibitors, was employed to block the endogenous ABA synthesis under salt treatment [33], [34], [35], [36]. As can be seen in Fig. 4, the application of sodium tungstate led to better growth of both the WT and OE-1 plants under salinity conditions and recovered the growth arrest of OE-1 seeds. Under more severe stress, higher concentration of sodium tungstate is required for the recovery of the salt sensitive phenotype (Fig. 4). For instance, the development of most OE-1 seeds remained quiescent on MS medium supplemented with 125 mM NaCl and 0.1 mM sodium tungstate. However, in the presence of 0.15 mM sodium tungstate, they began to turn green and exhibit normal growth, implying that sodium tungstate has a dose effect in recovering the arrested growth of the NFYA1 transgenic seedlings.
Figure 4. Sodium tungstate recovered the postgermination growth arrest of NFYA1 overexpression line under salt stress.
(A) WT and 35S::NFYA1 seeds were germinated on MS medium for 24 hours after stratification and transferred onto MS medium supplemented with NaCl and Na6Wo4 of different concentrations. (B) The rate of established seedlings was counted after growing for 14 days on NaCl and sodium tungstate medium. Samples with different letters are significantly different: P<0.01.
However, the concurrent application of exogenous ABA with sodium tungstate to the WT and OE-1 plants failed to recover the quiescent transgenic seedlings to normal development. In addition, sodium tungstate alone cannot lead to any difference in between the WT and 35S::NFYA1 transgenic lines, which could serve as a control showing that sodium tungstate contributes nothing to the arrested growth of the germinated seeds (Fig. 5). These findings reasonably demonstrate that the NFYA1 participates in regulating the ABA signaling pathway rather than the ABA synthesis pathway.
Figure 5. Sodium tungstate cannot recover the postgermination growth arrest of NFYA1 overexpression line under exogenous ABA treatment.
(A) WT and 35S::NFYA1 seeds were germinated on MS medium for 24 hours after stratification and transferred onto MS medium supplemented with ABA and Na6Wo4 of different concentrations. (B) The rate of established seedlings was counted after 14 days growing on the medium under LD condition. Samples with different letters are significantly different: P<0.01.
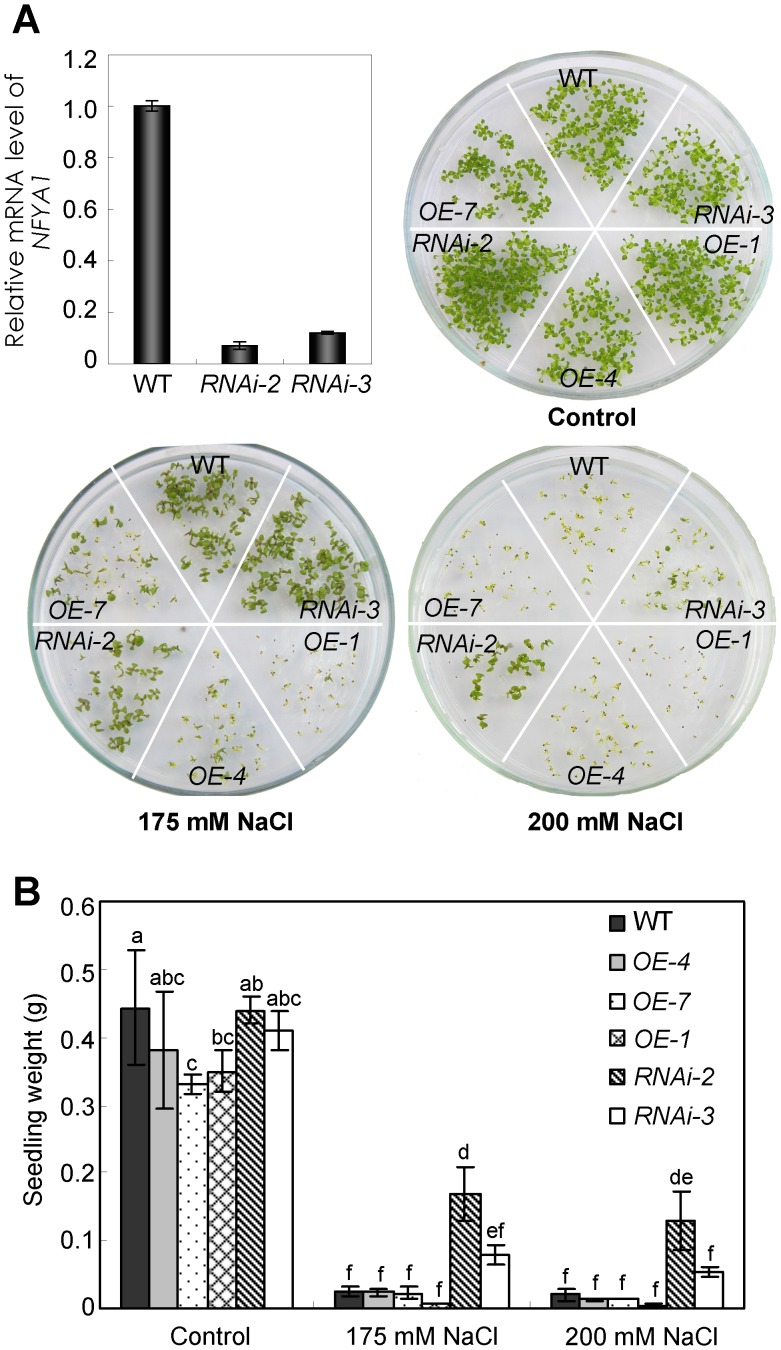
NFYA1-RNAi Line is Insensitive to Salt Stress during Postgermination Growth
To further demonstrate the role of NFYA1 in regulating postgermination growth, we generated NFYA1-RNAi lines driven by 35S promoter. RNAi-2 and RNAi-3, in which the NFYA1 mRNA level was dramatically downregulated, were chosen for further experiment (Fig. 6A). As shown in Fig. 6B, both RNAi-2 and RNAi-3 could develop into green seedlings under 175 mM NaCl like WT, while the NFYA1 overexpression lines failed. Under higher salt stress (200 mM NaCl), where the growth arrest also happened to WT, the RNAi lines remain growing. These results indicate that NFYA1 acts as a negative regulator in affecting the seed postgermination growth, which is consistent with our above results.
Figure 6. NFYA1-RNAi lines are tolerant to salt stress during germination and postgermination growth.
(A) Expression level of NFYA1 in NFYA1-RNAi lines assessed by qRT-PCR. Phenotype of wild type, NFYA1-RNAi lines and 35S::NFYA1 lines growing on MS medium supplemented with 0 mM (Control), 175 mM and 200 mM NaCl for 14 days under LD condition, and then weights of 25 seedlings were measured for each line (B). Three replicates were performed for each experiment. Samples with different letters are significantly different: P<0.01.
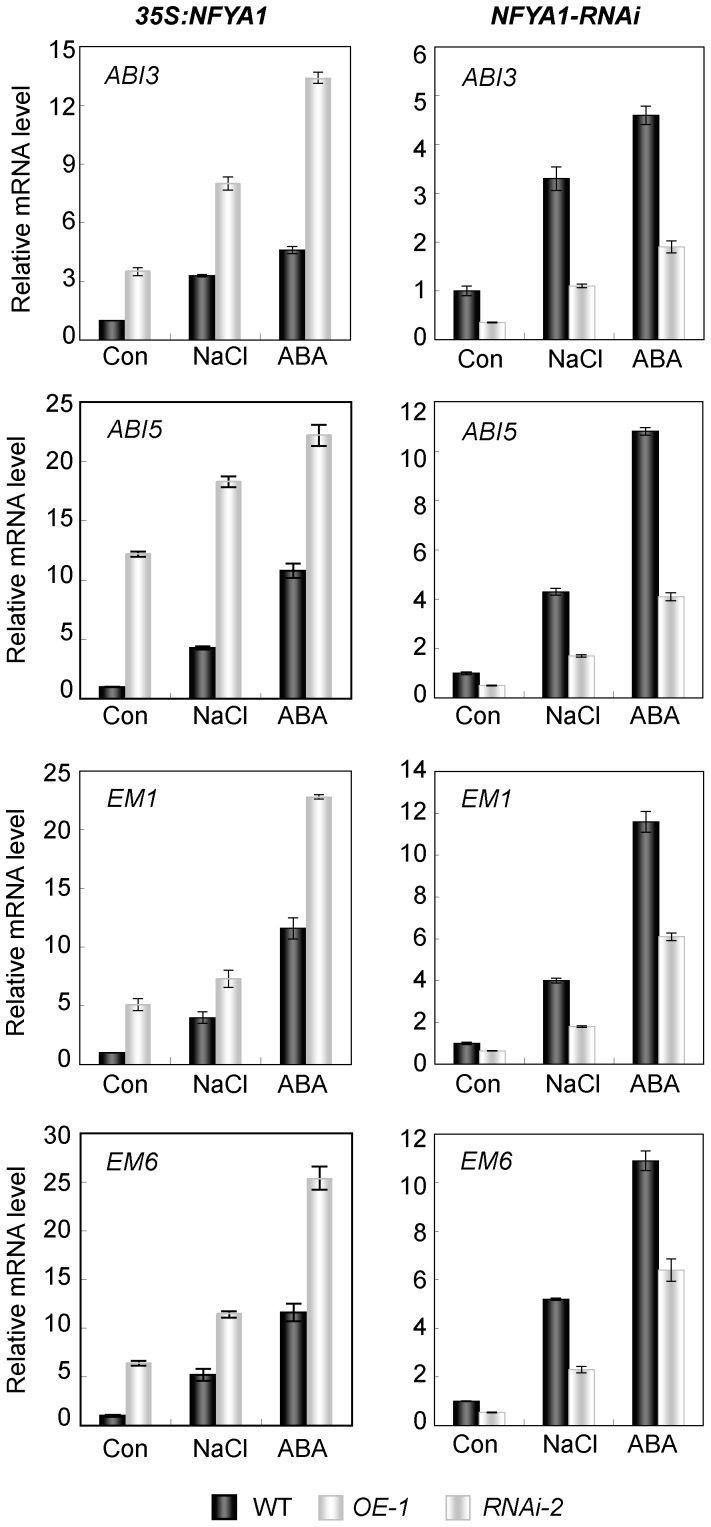
AtNFYA1 Affects the ABA Signaling Pathway as an Upstream Regulator
The aforementioned study indicated that the growth arrest of the germinated seeds under salt stress is an adaptive mechanism regulated by the ABA signaling pathway. To determine the variety of genes affected and which one(s) contributed to the growth arrest of the seeds within this short developmental period, the ABI3 and ABI5 expressions were detected using qRT-PCR in the germinated seeds of WT and NFYA1 transgenic lines treated with 125 mM NaCl and 0.5 µM ABA. Compared with WT, both ABI3 and ABI5 were elevated in the NFYA1 transgenic lines under normal conditions, demonstrating that NFYA1 acted upstream and positively regulated the ABI3 and ABI5 expressions. However, compared with normal condition (Control), higher expression levels of ABI3 and ABI5 were observed in the NFYA1 transgenic lines under NaCl and ABA treatments. In addition, EM1 and EM6, which act downstream of ABI5, exhibited similar expression patterns as those of ABI3 and ABI5 under these conditions (Fig. 7). The expressions of other genes involved in the seed germination and the ABA signaling pathway, such as ABI3-interacting protein 3 (AIP3), ABI4, and RAB18, were also assessed, but only minor expressional changes were observed in the WT and NFYA1 transgenic lines (data not shown). However, in the NFYA1-RNAi lines, the expression levels of ABI3, ABI5, EM1, and EM6 were dramatically downregulated (Fig. 7). The above data suggested that NFYA1 is involved in regulating ABI3-regulated cascades of the ABA signaling pathway.
Figure 7. Expression levels of ABA pathway-related genes in 35S::NFYA1 line (OE-1) and NFYA1-RNAi transgenic line (RNAi-2) detected by qRT-PCR.
Seeds were germinated on MS liquid medium moistened filter paper for 24 hours after stratification, and then were transferred onto filtered paper moistened with water (Con), 125 mM NaCl and 0.5 µM ABA respectively. After treated for 8 hours, the samples were harvested and RNA was extracted. Here, three biological repeats were done and each qRT-PCR was also performed three times.
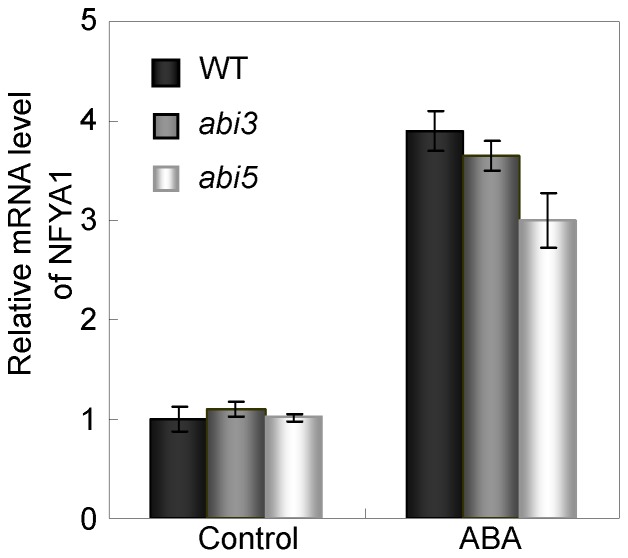
To further confirm the above results, we detected the expression of NFYA1 in the abi3 and abi5 knockout mutants. The qRT-PCR results revealed that the level of NFYA1 is not altered in both mutants under control conditions, and it is increased to similar levels in WT and the mutants under ABA treatments (Fig. 8). These findings demonstrate that ABA induced expression of NFYA1 is not dependent on ABI3 and ABI5, which is supportive to our conclusion that NFYA1 acts upstream of ABI3 and ABI5 to regulate postgermination growth.
Figure 8. Expression of NFYA1 in WT and abi3, abi5 mutants detected by qRT-PCR.
Seeds of WT, abi3, abi5 were germinated on MS liquid medium moistened filter paper for 24 hours after stratification, and then were transferred onto filtered paper moistened with water (control), 125 mM NaCl and 0.5 µM ABA, respectively. After treated for 8 hours, the samples were harvested and RNA was extracted. Here, three biological repeats were done and each qRT-PCR was also performed three times.
Discussion
Arabidopsis NFYA1 is a member of the NFY family, whose function is to be discovered. In our study, we reported a novel role of AtNFYA1 in mediating seed postgermination growth arrest under salt stress, which also affected the ABA signaling pathway. It is observed that the NFYA1-overexpressed lines are hypersensitive to salt treatment in a does-dependent manner during seed postgermination growth. Dramatic development quiescence of the young embryos was observed in the presence of NaCl (Fig. 2C). It is, however, inconsistent with our initial finding that NFYA1 is remarkably induced by several abiotic stresses such as salt, mannitol and PEG (Fig. 1), and the 35S::NFYA1 plants exhibit a salt resistant phenotype compared with that of wild type Arabidopsis (Fig. S2). These interesting observations demonstrate that NFYA1 is involved in different signaling pathways at different developmental stages.
Under salt stress, it is found that the hypersensitivity of NFYA1 overexpressers only confined in a limited developmental window during the postgermination growth, which is, before the cotyledons turn green and develop into young seedlings (Fig. 3). It is a very important checkpoint in this period under salt stress, because postgermination growth arrest can protect the germinated seeds from water deficit [2]. It is documented that the application of exogenous ABA can lead to arrested growth of the developing embryos after germination, and the formation of quiescent, germinated seeds is considered an adaptive mechanism to increase the survival rate of young seedlings under stresses [2], [37]. Genes involved in the regulation of postgermination growth arrest was also revealed in other studies by mutant screening, such as cpr5, wrky2 and hyl1, and most of the knockout lines were dependent on the ABA signaling pathways and hypersensitive to exogenous ABA [12], [13], [14]. Several abiotic stresses, particularly water stress, can result in the accumulation of endogenous ABA, which in turn, triggers the upregulation of the defense system in plants to abiotic stresses [35], [38]. Apart from NaCl (Fig. 2C), ABA was also identified as an efficient stimulus of postgermination growth arrest in this study (Fig. 2C). Further experiments revealed that the salt-sensitive phenotype of the 35S::NFYA1 lines can be relieved by the application of the ABA synthesis inhibitor sodium tungstate, suggesting that salt stress might lead to the overaccumulation of endogenous ABA in the NFYA1 transgenic seeds (Fig. 4). However, sodium tungstate failed to recover the arrested seeds into normal growth under exogenous ABA treatment (Fig. 5), implying that the growth arrest of the germinated seeds is not due to the increase of ABA but as a result of the perturbation of the downstream ABA signaling pathway. These findings further demonstrate that the Arabidopsis NFYA1 participates in regulating the ABA signaling pathway rather than the ABA synthesis pathway.
The transcription factors ABI3 and ABI5 are known to be important regulators of ABA-dependent growth arrest during seed germination and early growth, and their expressions define a narrow developmental checkpoint following germination. ABI5 acts downstream of ABI3, and ABA-induced ABI5 can activate the expression of two late embryogenesis-abundant genes EM1 and EM6 by occupying their promoters [8], [11]. To gain insight into the NFYA1 function during seed early growth after germination, we analyzed the expression levels of the above four genes in WT and 35S::NFYA1 transgenic lines. Interestingly, ABI3, ABI5, and their downstream genes EM1 and EM6 were all dramatically elevated in NFYA1-overexpressed lines and were downregulated in NFYA1-RNAi lines under ABA treatment and salt stress (Fig. 7). As compared with WT Arabidopsis, the higher expression levels of the four genes in NFYA1-overexpressed lines stimulated growth arrest. Here, our study reported that NFYA1 might represent another branch in regulating the seed postgermination growth arrest via the ABI3-controlled cascades.
Some individual NFY subunits were reported to play pleiotropic regulating roles during plant growth and development. In our study, we observed novel functions of NFYA1 in abiotic stress responses at different developmental stages. Recently, Mu etal. reported the involvement of NFYA1 in male gametogenesis, embryogenesis and seed development [31]. These pleiotropic functions of NFYA1 might be explained by the diverse combination of NFYA1 with other NFYB/NFYC factors, which thus affected different regulating pathways. In a word, it is interesting to gain insight into the connection of distinct NFYA1 functions, and further study on protein-protein or protein-DNA interactions is also needed to investigate the inner mechanisms.
Supporting Information
Specific localization of NFYA1 in the nucleus of onion epidermal cells. Agrobacterium tumefaciens strain LBA4404 carrying 35S::GFP and 35S::NFYA1-GFP expression vectors was transformed into the onion epidermal cells and then visualized with a fluorescence microscope (BX51, model 7.3; Olympus, Japan). Bar, 100 µm.
(TIF)
Enhanced salt stress tolerance of 35S:NFYA1 transgenic plants. (A) WT, OE-1 and OE-4 seeds were germinated on MS medium and the 5-day-old seedlings were transferred onto MS control medium without NaCl and MS medium supplemented with 125 mM NaCl for 10 days, (B) Statistical analysis of root elongation, (C) salt stress tolerance and (D) survival rate of 4-week-old WT and OE-1 plants under SD conditions after treated with 0 mM (Control) and 250 mM NaCl for 10 days. Samples with different letters are significantly different: P<0.01.
(TIF)
Primers used in the qRT-PCR assay.
(TIF)
Funding Statement
Funding was provided by National Natural Science Foundation (Grant No. 31000121), http://www.nsfc.gov.cn/Portal0/default166.htm; National Basic Research Program (Grant No. 2012CB114200), http://www.973.gov.cn/English/Index.aspx; and Genetically modified organisms breeding major projects of China (Grant No. 2011ZX08009-003-002), http://www.nmp.gov.cn/zxjs/zjy/201012/t20101208_2129.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hetherington AM (2001) Guard cell signaling. Cell 107: 711–714. [DOI] [PubMed] [Google Scholar]
- 2. Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nambara E, Marion-Poll A (2003) ABA action and interactions in seeds. Trends Plant Sci 8: 213–217. [DOI] [PubMed] [Google Scholar]
- 4. Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606. [DOI] [PubMed] [Google Scholar]
- 5. Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547. [DOI] [PubMed] [Google Scholar]
- 7. Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, et al. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328. [DOI] [PubMed] [Google Scholar]
- 9. Bies-Etheve N, da Silva Conceicao A, Giraudat J, Koornneef M, Leon-Kloosterziel K, et al. (1999) Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol 40: 1045–1054. [DOI] [PubMed] [Google Scholar]
- 10. Manfre AJ, Lanni LM, Marcotte WR Jr (2006) The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development. Plant Physiol 140: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, et al. (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao G, Zhang S, Wang C, Yang X, Wang Y, et al. (2011) Arabidopsis CPR5 independently regulates seed germination and postgermination arrest of development through LOX pathway and ABA signaling. PLoS One 6: e19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang W, Yu D (2009) Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu C, Han MH, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci U S A 99: 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frontini M, Imbriano C, Manni I, Mantovani R (2004) Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 3: 217–222. [PubMed] [Google Scholar]
- 16. Kahle J, Baake M, Doenecke D, Albig W (2005) Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol 25: 5339–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng Y, Jahroudi N (2003) The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J Biol Chem 278: 8385–8394. [DOI] [PubMed] [Google Scholar]
- 18. Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, et al. (2008) The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol 28: 2047–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siefers N, Dang KK, Kumimoto RW, Bynum WEt, Tayrose G, et al. (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maity SN, de Crombrugghe B (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci 23: 174–178. [DOI] [PubMed] [Google Scholar]
- 21. Cai X, Ballif J, Endo S, Davis E, Liang M, et al. (2007) A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol 145: 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, et al. (1994) LEAFY COTYLEDON1 Is an Essential Regulator of Late Embryogenesis and Cotyledon Identity in Arabidopsis. Plant Cell 6: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lotan T, Ohto M, Yee KM, West MA, Lo R, et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- 24. Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci U S A 100: 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, et al. (2007) AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol 40: 1083–1089. [DOI] [PubMed] [Google Scholar]
- 26. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li WX, Oono Y, Zhu J, He XJ, Wu JM, et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci U S A 104: 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2012) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. [DOI] [PubMed]
- 30. Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, et al. (2012) The Promiscuous Life of Plant NUCLEAR FACTOR Y Transcription Factors. Plant Cell 24: 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mu J, Tan H, Hong S, Liang Y, Zuo J (2013) Arabidopsis Transcription Factor Genes NF-YA1, 5, 6, and 9 Play Redundant Roles in Male Gametogenesis, Embryogenesis, and Seed Development. Mol Plant 6: 188–201. [DOI] [PubMed] [Google Scholar]
- 32. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 33. Hansen H, Grossmann K (2000) Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol 124: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu X, Zhang A, Zhang J, Jiang M (2006) Abscisic acid is a key inducer of hydrogen peroxide production in leaves of maize plants exposed to water stress. Plant Cell Physiol 47: 1484–1495. [DOI] [PubMed] [Google Scholar]
- 35. Jiang M, Zhang J (2002) Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J Exp Bot 53: 2401–2410. [DOI] [PubMed] [Google Scholar]
- 36. Nenghui Ye GZ, Yinggao Liu, Yingxuan Li, Jianhua Zhang (2011) ABA Controls H2O2 Accumulation Through the Induction of OsCATB in Rice Leaves Under Water Stress. Plant Cell Physiol 52: 689–698. [DOI] [PubMed] [Google Scholar]
- 37. Kinoshita N, Berr A, Belin C, Chappuis R, Nishizawa NK, et al. (2010) Identification of growth insensitive to ABA3 (gia3), a recessive mutation affecting ABA Signaling for the control of early post-germination growth in Arabidopsis thaliana. Plant Cell Physiol 51: 239–251. [DOI] [PubMed] [Google Scholar]
- 38. Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, et al. (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146: 1368–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specific localization of NFYA1 in the nucleus of onion epidermal cells. Agrobacterium tumefaciens strain LBA4404 carrying 35S::GFP and 35S::NFYA1-GFP expression vectors was transformed into the onion epidermal cells and then visualized with a fluorescence microscope (BX51, model 7.3; Olympus, Japan). Bar, 100 µm.
(TIF)
Enhanced salt stress tolerance of 35S:NFYA1 transgenic plants. (A) WT, OE-1 and OE-4 seeds were germinated on MS medium and the 5-day-old seedlings were transferred onto MS control medium without NaCl and MS medium supplemented with 125 mM NaCl for 10 days, (B) Statistical analysis of root elongation, (C) salt stress tolerance and (D) survival rate of 4-week-old WT and OE-1 plants under SD conditions after treated with 0 mM (Control) and 250 mM NaCl for 10 days. Samples with different letters are significantly different: P<0.01.
(TIF)
Primers used in the qRT-PCR assay.
(TIF)