Abstract
To date, measurements of the activity of aldehyde dehydrogenase-2 (ALDH2), a critical mitochondrial enzyme for eliminating certain cytotoxic aldehydes in the body and a promising target for drug development, have been largely limited to in vitro methods. Recent advancements in magnetic resonance spectroscopy (MRS) of hyperpolarized 13C-labeled substrates now provide a method to detect and image in vivo metabolic pathways with signal-to-noise ratio gains greater than 10,000 fold over conventional MRS techniques. However aldehydes, due to their toxicity and short T1 relaxation times, are generally poor targets for such 13C-labeled studies. In this work, we show that dynamic magnetic resonance spectroscopic imaging of hyperpolarized [1-13C]pyruvate and its conversion to [1-13C]lactate can provide an indirect in vivo measurement of ALDH2 activity via the concentration of NADH, a co-factor common to both the reduction of pyruvate to lactate and the oxidation of acetaldehyde to acetate. Results from a rat liver ethanol model (n = 9) show that changes in 13C-lactate labeling following the bolus injection of hyperpolarized pyruvate are highly correlated with changes in ALDH2 activity (R2=0.76).
Keywords: hyperpolarized 13C, ALDH2 activity, liver, ethanol, pyruvate, lactate, NADH
Introduction
The mitochondrial enzyme aldehyde dehydrogenase-2 (ALDH2), which plays a critical role in the detoxification of reactive aldehydes, such as acetaldehyde and 4-hydroxy-2-nonenal (4-HNE), has been identified as a promising drug development target for multiple pathologies including alcoholism [1-6], cardiac ischemia [7-16], and cancer [17-20]. To date, direct measurements of ALDH2 activity have largely been limited to in vitro methods, and the goal of this work was to investigate using dynamic magnetic resonance spectroscopic imaging (MRSI) of hyperpolarized [1-13C]pyruvate (Pyr) and its conversion to [1-13C]lactate (Lac) [21-24] as a method for in vivo measurement of ALDH2 activity.
Specifically, in vivo ethanol metabolism in the rat liver, which occurs via the breakdown of ethanol to acetaldehyde and acetaldehyde to acetate as catalyzed by the enzymes alcohol dehydrogenase (ADH) and acetaldehyde dehydrogenase (ALDH2) respectively, was used as a mechanism to generate acetaldehyde for ALDH2 activity investigation. Both steps of ethanol metabolism also reduce the coenzyme nicotinamide adenine dinucleotide (NAD+) to NADH, leading to the accumulation of NADH in the liver [25, 26]. Furthermore, the conversion of pyruvate to lactate takes place via the enzyme lactate dehydrogenase (LDH) and the concomitant oxidation of NADH to NAD+ [25]. At sufficiently high levels of pyruvate, this reaction is limited by the lactate pool size and NADH availability [27]. Hence, ethanol metabolism provides a mechanism by which NADH availability can be altered and used to modulate the NADH-dependent pyruvate-to-lactate metabolic pathway. ALDH2 activity, as reflected by NADH accumulation, can thus be measured via the increased maximum rate of [1-13C]Lac labeling following a bolus injection of hyperpolarized [1-13C]Pyr.
Previous studies have reported MRS and MRSI measurements of hyperpolarized [1-13C]Pyr to study rat liver metabolism modulated by ethanol [28, 29]. Changes in lactate production in the absence vs. presence of ethanol, as moderated by NADH accumulation, were presumed to be correlated with ALDH2 activity. Here we quantitatively measured both pyruvate-to-lactate 13C-labeling and ALDH2 activity in the same rat liver model using the ALDH2 inhibitor disulfiram to modulate ALDH2 activity.
Methods
Each polarized sample consisted of 40 μL of a mixture of 14-M [1-13C] pyruvic acid and 15-mM Ox063 trityl radical, to which 3 μL of a 1:50 dilution of Dotarem (Guerbet, France) was added prior to polarization. The sample was polarized using a HyperSense system (Oxford Instruments Molecular Biotools, Oxford, UK) to achieve approximately 20-25% liquid-state polarization at dissolution. The polarized sample was dissolved with a solution of 125-mM NaOH mixed with 40-mM Tris buffer, 50-mM NaCl and 0.1-g/L EDTA-Na2, leading to a 125-mM solution of hyperpolarized pyruvate with a pH of approximately 7.5. A volume of approximately 3.2 mL of the hyperpolarized pyruvate solution was injected into the tail vein at a rate of about 0.25 mL/s.
Animal Model
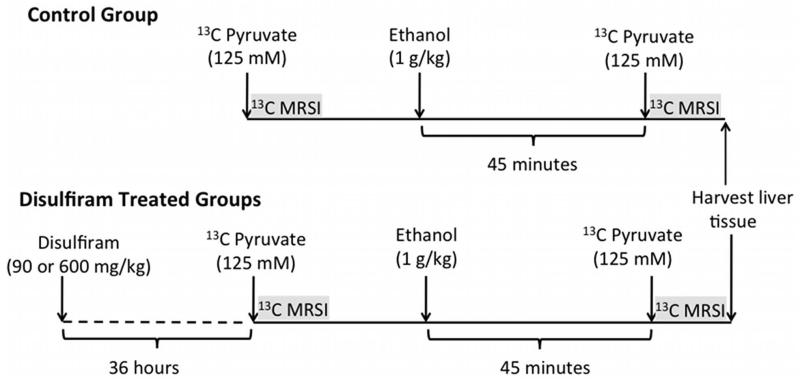
ALDH2 activity was manipulated using the FDA-approved ALDH2 inhibitor disulfiram. Disulfiram reduces ALDH2 activity in rat liver to approximately 60% of the normal level with a 90 mg/kg dose, and 25% of the normal level with a 600 mg/kg dose, at 24-48 hours after oral delivery [30]. Disulfiram was suspended in 3 mL of 5% weight/volume gum arabicum and delivered through oral gavage approximately 36 hours before the imaging experiments. Healthy male Wistar rats (n=9, 424±25 g body weight) were divided into three groups (n=3 each): control group which received no treatment; disulfiram-treated group receiving a 90-mg/kg dose; and disulfiram-treated group with a 600-mg/kg dose.
The rats were anesthetized with 1-3 % isoflurane in oxygen (~1.5 L/min) and a catheter was inserted in a tail vein. Respiration, temperature, heart rate, and oxygen saturation were monitored throughout the experiment session, with temperature regulated using a warm water blanket placed underneath the animals. Each rat received two injections of the hyperpolarized pyruvate solution, each followed by a 13C MRSI acquisition: one baseline measurement and another post-ethanol. Approximately 45 min before the second pyruvate injection, a 1.0-g/kg dose of a 20% ethanol solution was injected into the tail vein at the rate of 1 mL/min to achieve a targeted steady-state blood alcohol level (BAL) of approximately 100 mg/dL at the time of the second acquisition. The study followed the experimental design as shown in Fig. 1. At the end of the exam, liver tissue was harvested for ALDH2 enzyme activity assay. Immediately after euthanasia, an approximately 4 g sample of the liver was harvested, rapidly frozen in liquid nitrogen and stored in a −80° C freezer for subsequent ALDH2 enzymatic activity analysis using a spectrophotometrical assay described below. All animal procedures were approved by the local Institutional Animal Care and Use Committee.
Figure 1.
Experimental design with timing of injections and MRSI acquisitions. 13C MRSI measurements of hyperpolarized [1-13C]Pyr were performed twice on each animal, before and after ethanol infusion. The animals were divided into three groups (3 animals/group): controls, those receiving 90-mg/kg disulfiram dose, and those receiving 600-mg/kg disulfiram.
MR protocol
All experiments were performed on a clinical 3T Signa MR scanner (GE Healthcare, Waukesha, WI) equipped with self-shielded gradients (40 mT/m, 150 mT/m/ms). A custom-built dual-tuned (1H/13C) quadrature rat coil (inner diameter=80 mm, length=90 mm), operating at 127.9 MHz and 32.2 MHz, respectively, was used for both RF excitation and signal reception. A reference phantom containing an 8-M solution of 13C-urea placed on top of the animal was used for calibration of the transmit 13C RF power, and also for concentration quantification. Single-shot fast spin-echo 1H MR images with nominal in-plane resolution of 0.47 mm and 2-mm slice thickness were acquired in the axial, sagittal, and coronal planes throughout the scan session as anatomical references for prescribing the 13C-MRSI acquisitions.
Dynamic 13C MRSI data were acquired from a 10-mm slice through the liver using the 2D 3-shot spiral spectroscopic imaging sequence described in [31,32]. The sequence included a spatially non-selective lactate saturation pulse at the beginning of each sampling interval to saturate signal from in-flowing lactate generated in other organs, particularly the heart. Imaging parameters were: FOV=80 mm, nominal resolution=5×5 mm2, spectral width=276 Hz, variable flip angle scheme (35.3°, 45° 90° for the 3 interleaves) to effectively excite all the longitudinal magnetization at each temporal sampling point, echo time TE=3 ms. The acquisition time for the 3-shot spiral MRSI was 375 ms, and the sampling interval was 5 s, allowing approximately 4.55 s dead time between the lactate saturation pulse and the imaging for inflow of fresh pyruvate spins into the slice and 13C label exchange with lactate. The time from dissolution to start of pyruvate injection was approximately 20 s, and the scan was started coincident with the injection. The MRSI data were reconstructed similarly as described in [32], and metabolic maps for pyruvate and lactate were calculated by integrating the signal within±20 Hz around each peak in absorption mode. The mean time-resolved signal intensities for pyruvate and lactate were calculated in an ROI in the liver.
The dynamic data were analyzed using the inflow-based single-slice quantification method described in [31]. The method is briefly described here. It exploits the inherently time-varying pyruvate concentration during a bolus injection to obtain independent estimates of apparent reaction velocity of lactate 13C label in each sampling interval. The injected pyruvate is assumed to be considerably greater than the endogenous pyruvate. Then, to estimate the in vivo pyruvate concentration at each time-point, the pyruvate signal is corrected for the polarization and T1 relaxation. The pyruvate percentage polarization at dissolution was estimated from the solid-state polarization value and the amount of the pyruvic acid sample based on independent calibration experiments. After dissolution, the pyruvate first experiences an in vitro T1 (approximately 60 s as measured in separate experiments) decay before injection and then an in vivo T1 (approximately 40 s in our study) decay after injection until the readout time. Finally, the corrected pyruvate signal is referenced to signal from the external 8-M 13C enriched urea phantom at thermal polarization to obtain the in vivo concentration. The influence of T1 decay for lactate is minimal given the 90° flip angle on lactate along with the spatially non-selective saturation of lactate signal every TR. The relationship between the estimated pyruvate concentration and apparent reaction velocity of lactate 13C labeling was mathematically approximated using a Michaelis-Menten-like formulation for saturatable kinetics with parameters corresponding to the apparent maximal reaction velocity Vmax and apparent Michaelis constant KM. While accurate quantification of the enzyme kinetics might need to account for the two-substrate – two-product sequential BiBi reaction, Zierhut et al. [33] demonstrated that the nonlinear relationship between the in vivo exchange rate constants and [1-13C]pyruvate dose was mathematically modeled well using a Michaelis–Menten-like formulation. Xu et al [31] also employed a Michaelis-Menten-like framework to model the saturatable kinetics between apparent reaction velocity and pyruvate concentration by taking advantage of pyruvate inflow. That model is used in this study, and the apparent reaction velocity of the 13C labeling estimated from the model is a combination of the net chemical conversion of pyruvate to lactate and isotopic exchange between pyruvate and lactate pools and also includes contributions from other factors such as organ perfusion and substrate transport kinetics. The estimated apparent Vmax values are unbiased with respect to experimental parameters including substrate dose, bolus shape and duration. The apparent Vmax estimates of the pyruvate-to-lactate 13C labeling process pre- and post-ethanol infusion were compared and the relative change in Vmax with ethanol, i.e. ΔVmax, was correlated with ALDH2 enzyme activity. Statistical significance was assessed using Student’s unpaired t-test between the control group and disulfiram-treated group.
ALDH2 enzyme assay
For the in vitro tissue assays, each 4-g liver sample was kept in 15 mL of a buffer solution (pH 7.4) containing 210 mM mannitol, 70 mM sucrose, 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS), and 1 mM EDTA. The protease inhibitor tablet (Sigma-Aldrich, St. Louis, MO) was also added to the buffer solution to prevent protein degradation of the liver samples. The liver samples were first homogenized with a Teflon-Glass Potter-Elvehjem homogenizer and the resulting homogenate was then centrifuged for 15 min at 3000 g in a Beckman L8-80M ultracentrifuge. After centrifugation, the supernatant on the top was a mixture of mitochondria and cytosol. Approximately 1 mL supernatant was carefully transported to a separate tube and about 1% triton solution was added to break down the cell membranes to assay the entire mitochondrial ALDH2 activity. Protein content in the mitochondria and cytosol mixture was quantitatively determined with Coomassie protein assay reagent (Pierce, Rockford, IL) with bovine serum albumin as the standard [34].
The ALDH2 enzymatic activity was determined spectrophotometrically using a kit (Trinity Biotech, NY, USA) to monitor the reductive reaction of NAD+ to NADH at λ = 340 nm. The assays were carried out at 25° C in a 96-well microplate. To start the reaction, 150 μL of 50 mM sodium pyrophosphate buffer (pH=9.5), 75 μL of 2.5 mM coenzyme NAD+, the sample solution with 94 μg of protein and 3 μL of 10 uM acetaldehyde were added. The accumulation of NADH was monitored for 10 min with measurements being taken every 30 s. The ALDH2 enzymatic activity A was proportional to the slope S of the recorded NADH accumulation curve. The slope was estimated by a linear fit for the data acquired over time t = 0 to 10 min. The empirical formula to calculate ALDH2 activity in units of μmole NADH formed/min/mg protein was: A=S×1000/(6.22×0.094), where 6.22 is the millimolar extinction coefficient and 0.094 is the target protein mass (mg) in the assay. For each sample, the assay was performed twice and the mean value used here.
Results and Discussion
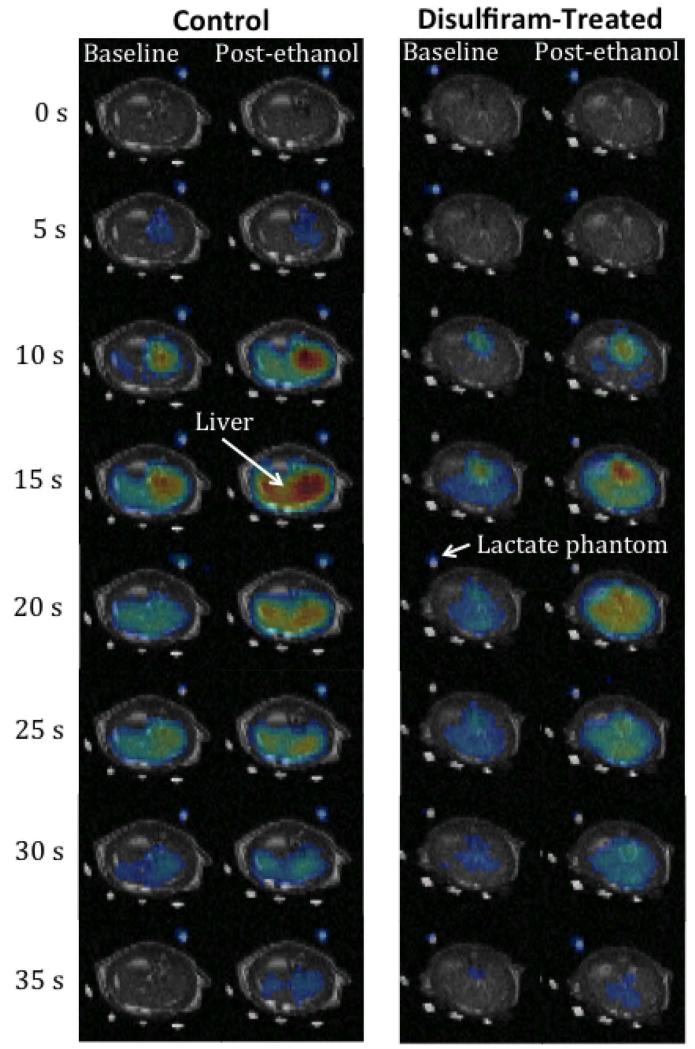
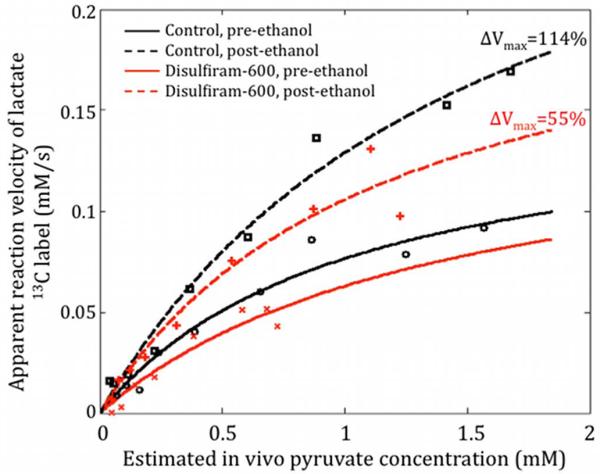
Figure 2 shows representative time-series of pre- and post-ethanol 13C lactate maps superimposed onto corresponding 1H MR images to illustrate the temporal dynamics and spatial distribution of the metabolite. Figure 3 plots the saturable kinetics in the liver ROI pre- and post-ethanol from two rats, one control and one treated with disulfiram, estimated using the inflow-based quantification method. Both, the 13C maps in Fig. 2 and the reaction velocity curves in Fig. 3, clearly show increased lactate signal post-ethanol compared to pre-ethanol. The relative change of the lactate apparent Vmax for the disulfiram-treated rat was smaller than that for the control rat, due to the lower ethanol-generated NADH from the partially inhibited ALDH2 activity.
Figure 2.
Time series of baseline and post-ethanol 13C lactate maps superimposed onto 1H images from a control rat and a disulfiram-treated rat (600-mg/kg dose). The ethanol infusion led to greater lactate signal post-ethanol compared to baseline. The increase in 13C lactate label generated was lower for a disulfiram-treated rat than the control. All images are displayed on the same scale.
Figure 3.
Apparent reaction velocity estimates in the liver pre- and post-ethanol from two rats: one treated with disulfiram and one untreated. The change in apparent Vmax pre-to-post ethanol was lower when ALDH2 activity was inhibited via disulfiram.
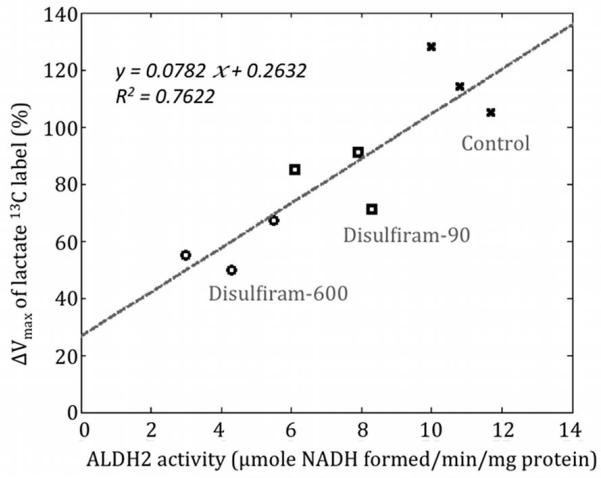
Table 1 summarizes the estimated apparent Vmax values of the pyruvate-to-lactate 13C labeling process pre- and post-ethanol infusion for all rats and the ALDH2 enzyme activities. The relative change of the lactate apparent Vmax with ethanol (ΔVmax) was 116% ± 12% (mean ± std, n=3) for the control group, 82% ± 10% for the disulfiram-90 group (p=0.029, unpaired t-test with control group) and 57% ± 9% for the disulfiram-600 group (p=0.028, unpaired t-test with control group). Compared to the mean ALDH2 activity of the control group, the ALDH2 activity of the disulfiram-90 group was 68.2% ± 10.8% and of the disulfiram-600 group was 39.3% ± 11.5%. Thus, the higher disulfiram dose led to a larger reduction in the Pyr-to-Lac ΔVmax with ethanol, consistent with a greater inhibition of ALDH2 activity. Figure 4 plots the measured ΔVmax as a function of ALDH2 activity for all rats, showing that the ΔVmax of the pyruvate-to-lactate 13C labeling process correlates well with the ALDH2 activity assay results (linear fit R2=0.76).
Table 1.
The estimated apparent Vmax values of the pyruvate-to-lactate 13C labeling process pre and post-ethanol infusion for all rats. The inhibition of ALDH2 activity by disulfiram led to reduced the ΔVmax. The average apparent ΔVmax with ethanol was 116% for the control group, 82% for the disulfiram-90 group and 57% for the disulfiram-600 group and correlated with ALDH2 enzyme activity. The unit of ALDH2 enzymatic activity is μmole NADH formed/min/mg protein.
| Control group | Disulfiram-90 | Disulfiram-600 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Rat ID | H211 | H212 | H233 | H239 | H240 | H243 | H228 | H229 | H232 |
| Vmax pre-ethanol (mM/s) | 0.10 | 0.13 | 0.16 | 0.17 | 0.22 | 0.16 | 0.28 | 0.15 | 0.27 |
| Vmax post-ethanol (mM/s) | 0.23 | 0.26 | 0.33 | 0.31 | 0.37 | 0.31 | 0.42 | 0.22 | 0.46 |
| ΔVmax (post-pre)/pre | 128% | 105% | 114% | 85% | 71% | 91% | 50% | 55% | 67% |
| ALDH2 enzyme activity | 10.0 | 11.7 | 10.8 | 6.1 | 8.3 | 7.9 | 4.3 | 3.0 | 5.5 |
Figure 4.
ΔVmax of the pyruvate-to-lactate 13C labeling process correlates well with the ALDH2 enzyme activity of the liver tissue. The y-axis of the plot shows the percent change in Vmax from pre-to-post ethanol.
The y-axis intercept of the fitted line in Fig. 4 is approximately 26% and this intercept corresponds to the case in which the ALDH2 activity is completely inhibited (i.e., the second reaction of the ethanol metabolism is completely blocked). Because the first step of the ethanol metabolism (ethanol to acetaldehyde) still generates the coenzyme NADH, there still remains some increase in lactate production after ethanol infusion. However, since acetaldehyde quickly accumulates in rat liver due to the complete ALDH2 inhibition and the equilibrium point of the first reaction of the ethanol metabolism is consequently shifted to the ethanol side, there is less than half of NADH produced compared with the control scenario. Therefore, ΔVmax should be less than half of ΔVmax in the control group (i.e, ΔVmax < 0.5*116% = 58%).
The disulfiram-600 ALDH2 activity was somewhat higher than the 25% of normal reported in literature, and may be due to the presence of high-KM ALDH1 in the mitochondria and cytosol mixture of the liver tissue assayed. Disulfiram does not affect the ALDH1 activity [4].
An alternate pathway for ethanol metabolism is the microsomal ethanol-oxidizing system (MEOS), which depends on cytochrome P-450 and generates acetaldehyde by the oxidation of NADPH to NADP+ resulting in free radical release. Ethanol oxidation via MEOS is considerably lower than ADH and this pathway probably only accounts for about 10% of total alcohol metabolism [35], and likely has minimal effect in this study as reflected by the good correlation obtained between ALDH2 activity and pyruvate-to-lactate apparent reaction velocity. However, it may need to be considered in future quantitative studies. Ethanol-derived acetate can also enter the citric acid cycle via conversion to acetyl-CoA, for instance as a fuel for brain astroglia [36], producing additional NADH. However, in the liver, processing by the citric acid cycle of the acetyl-CoA from ethanol is blocked as the excess NADH from ethanol metabolism inhibits the enzymes isocitrate dehydrogenase and α-ketoglutarate dehydrogenase. The accumulation of acetyl-CoA can then lead to formation of ketone bodies and fatty acid synthesis (leading to the “fatty liver” condition) [25]. All of these factors, including altered redox state (NAD+/NADH) from ethanol metabolism, reactive oxygen species production and oxidative stress contribute to alcohol-induced hepatic fibrosis [37]. While the pyruvate-to-lactate 13C labeling process reflects the increased NADH/NAD+ due to ethanol metabolism, it does not completely represent the redox state. For example, ethanol metabolism via MEOS could alter the redox state (NADP+/NADPH) without the same impact on 13C labeling. Methods for assessing the redox state using hyperpolarized 13C have been reported in other studies [38,39].
This study demonstrates an application of hyperpolarized 13C MRSI for indirect detection of a metabolic process occurring over timescales greater than the short T1 relaxation time constraint of hyperpolarized 13C studies. The manipulation of ALDH2 activity by disulfiram occurred over 36 hours and the buildup of NADH from ethanol took place over 45 min prior to the bolus injection of Pyr. The changes in the enzyme activity were then detected via the conversion of Pyr to Lac using the coenzyme NADH linking the two processes.
Conclusion
This work demonstrates that dynamic in vivo MRSI measurements of the relative change of [1-13C]Lac labeling before and after ethanol infusion, or ΔVmax, following a bolus injection of hyperpolarized [1-13C]Pyr can potentially serve as a non-invasive indicator of ALDH2 activity in this ethanol-treated rat model. Future work will involve extending these results to measure ALDH2 activity in the heart following ischemia/reperfusion injury [7].
Acknowledgments
Funded by
NIH grants AA018681, AA05965, AA13521-INIA, EB009070, P41 EB015891l; GE Healthcare
Abbreviations used
- ALDH2
aldehyde dehydrogenase-2
- BAL
blood alcohol level
- EDTA
ethylene-diaminetetraacetic acid
- MRSI
magnetic resonance spectroscopic imaging
- MEOS
microsomal ethanol-oxidizing system
- NAD+
nicotinamide adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide, reduced form
- NADP+
nicotinamide adenine dinucleotide phosphate
- ROI
region of interest
References
- 1.O’Keefe JH, Bybee KA, Lavie CJ. Alcohol and cardiovascular health: the razor-sharp double-edged sword. J Am Coll Cardiol. 2007;50(11):1009–14. doi: 10.1016/j.jacc.2007.04.089. [DOI] [PubMed] [Google Scholar]
- 2.Peng GS, Wang MF, Chen CY, Luu SU, Chou HC, Li TK, Yin SJ. Involvement of acetaldehyde for full protection against alcoholism by homozygosity of the variant allele of mitochondrial aldehyde dehydrogenase gene in Asians. Pharmacogenetics. 1999;9(4):463–76. [PubMed] [Google Scholar]
- 3.Umulis DM, Gurmen NM, Singh P, Fogler HS. A physiologically based model for ethanol and acetaldehyde metabolism in human beings. Alcohol. 2005;35:3–12. doi: 10.1016/j.alcohol.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Tottmar O, Marchner H. Disulfiram as a tool in the studies of metabolism of acetaldehyde in rats. Acta Pharmacol Toxicol. 1976;38:366–375. doi: 10.1111/j.1600-0773.1976.tb03132.x. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson CJP. The Role of Acetaldehyde in the Actions of Alcohol. Alcohol Clin Exp Res. 2001;25(5 Suppl ISBRA):15S–32S. doi: 10.1097/00000374-200105051-00005. [DOI] [PubMed] [Google Scholar]
- 6.Zakhari S. In: Alcohol Metabolism, Tobacco and Cancer. Cho C, Purohit V, editors. Basel; Karger: 2006. pp. 29–47. [Google Scholar]
- 7.Chen CH, Budas GR, Churchill EN, Disatnik M-H, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–5. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchill E, Budas G, Vallentin A, Koyanagi T, Mochly-Rosen D. PKC isozymes in chronic cardiac disease: possible therapeutic targets? Annu Rev Pharmacol Toxicol. 2008;48:569–99. doi: 10.1146/annurev.pharmtox.48.121806.154902. [DOI] [PubMed] [Google Scholar]
- 9.Aberle NS, 2nd, Burd L, Zhao BH, Ren J. Acetaldehyde-induced cardiac contractile dysfunction may be alleviated by vitamin B1 but not by vitamins B6 or B12. Alcohol Alcohol. 2004;39(5):450–4. doi: 10.1093/alcalc/agh085. [DOI] [PubMed] [Google Scholar]
- 10.Budas GR, Mochly-Rosen D. Mitochondrial protein kinase Cepsilon (PKCepsilon): emerging role in cardiac protection from ischaemic damage. Biochem Soc Trans. 2007;35(Pt 5):1052–4. doi: 10.1042/BST0351052. [DOI] [PubMed] [Google Scholar]
- 11.Buja LM. Myocardial ischemia and reperfusion injury. Cardiovasc Pathol. 2005;14(4):170–5. doi: 10.1016/j.carpath.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Collins MA, Neafsey EJ, Mukamal KJ, Gray MO, Parks DA, Das DK, Korthuis RJ. Alcohol in Moderation, Cardioprotection, and Neuroprotection: Epidemiological Considerations and Mechanistic Studies. Alcohol Clin Exp Res. 2009;33(2):206–19. doi: 10.1111/j.1530-0277.2008.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings RB, Steenbergen C, Jr, Reimer KA. Myocardial ischemia and reperfusion. Monogr Pathol. 1995;37:47–80. [PubMed] [Google Scholar]
- 14.Mochly-Rosen D, Wu G, Hahn N, Osinska H, Liron T, Lorenz JN, Yatani A, Robbins J, Dorn GW., 2nd Cardiotrophic effects of protein kinase C epsilon: analysis by in vivo modulation of PKCepsilon translocation. Circ Res. 2000;86(11):1173–9. doi: 10.1161/01.res.86.11.1173. [DOI] [PubMed] [Google Scholar]
- 15.Sydow K, Daiber A, Oelze M, Chen Z, August M, Wendt M, Ulrich V, Mülsch A, Schulz E, Keaney JF, Jr, Stamler JS, Münzel T. Central role of mitochondrial aldehyde dehydrogenase and reactive oxygen species in nitroglycerin tolerance and cross-tolerance. J Clin Invest. 2004;113(3):482–9. doi: 10.1172/JCI19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagi S, Iwai N, Yamauchi R, Kojima S, Yasuno S, Baba T, Terashima M, Tsutsumi Y, Suzuki S, Morii I, Hanai S, Ono K, Baba S, Tomoike H, Kawamura A, Miyazaki S, Nonogi H, Goto Y. Aldehyde dehydrogenase 2 gene is a risk factor for myocardial infarction in Japanese men. Hypertens Res. 2002;25(5):677–81. doi: 10.1291/hypres.25.677. [DOI] [PubMed] [Google Scholar]
- 17.Jones DE, Jr, Evces S, Lindahl R. Expression of tumor-associated aldehyde dehydrogenase during rat hepatocarcinogenesis using the resistant hepatocyte model. Carcinogenesis. 1984;5(12):1679–87. doi: 10.1093/carcin/5.12.1679. [DOI] [PubMed] [Google Scholar]
- 18.Randi G, Altieri A, Gallus S, Francheschi S, Negri E, Talamini R, La Vecchia C. History of cirrhosis and risk of digestive tract neoplasms. Ann Oncol. 2005;16(9):1551–5. doi: 10.1093/annonc/mdi266. [DOI] [PubMed] [Google Scholar]
- 19.Seitz HK, Matsuzaki S, Yokoyama A, Homann N, Väkeväinen S, Wang XD. Alcohol and cancer. Alcohol Clin Exp Res. 2001;25(5 Suppl ISBRA):137S–143S. doi: 10.1097/00000374-200105051-00024. [DOI] [PubMed] [Google Scholar]
- 20.Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res. 2007;149(6):293–7. doi: 10.1016/j.trsl.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Golman K, Petersson JS. Metabolic imaging and other applications of hyperpolarized 13C1. Acad Radiol. 2006;13(8):932–42. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Kurhanewicz J, Bok R, Nelson SJ, Vigneron DB. Current and potential applications of clinical 13C MR spectroscopy. J Nucl Med. 2008;49(3):341–4. doi: 10.2967/jnumed.107.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malloy CR, Merritt ME, Sherry AD. Could 13C MRI assist clinical decision-making for patients with heart disease? NMR Biomed. 2011;24:973–979. doi: 10.1002/nbm.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg J, Tymoczko J, Stryer L. Biochemistry. W.H. Freeman; 2006. [Google Scholar]
- 26.Deitrich R, Petersen D, Vasiliou V. Removal of acetaldehyde from the body. Novartis Found Symp. 2007;285:23–40. doi: 10.1002/9780470511848.ch3. [DOI] [PubMed] [Google Scholar]
- 27.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13(11):1382–7. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 28.Spielman DM, Mayer D, Yen YF, Tropp J, Hurd RE, Pfefferbaum A. In vivo measurement of ethanol metaboism in the rat liver using magnetic resonance spectroscopy of hyperpolarized [1-13C] pyruvate. Magn Reson Med. 2009;62:307–313. doi: 10.1002/mrm.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josan S, Spielman D, Yen YF, Hurd R, Pfefferbaum A, Mayer D. Fast volumetric imaging of ethanol metaboism in rat liver with hyperpolarized [1-13C] pyruvate. NMR Biomed. 2012;25(8):993–9. doi: 10.1002/nbm.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersson H, Tottmar O. Inhibition of Aldehyde Dehydrogenases in Rat Brain and Liver by Disulfiram and Coprine. Journal of Neurochemistry. 1978;39:628–634. doi: 10.1111/j.1471-4159.1982.tb07939.x. [DOI] [PubMed] [Google Scholar]
- 31.Xu T, Mayer D, Gu M, Yen YF, Josan S, Tropp J, Pfefferbaum A, Hurd R, Spielman D. Quantitation of in vivo metabolic kinetics of hyperpolarized pyruvate in rat kidneys using dynamic 13C1 MRSI. NMR Biomed. 2011;24(8):997–1005. doi: 10.1002/nbm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer D, Yen YF, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. Application of subsecond spiral chemical shift imaging to real-time multislice imaging of rat in vivo after injection of hyperpolarized 13C1 pyruvate. Magn Reson Med. 2009;62:557–564. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zierhut ML, Yen Y-F, Chen AP, Bok R, Albers M, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE, Nelson SJ. Kinetic modeling of hyperpolarized 13C1 pyruvate metabolism in normal rats and TRAMP mice. J. Magn Reson. 2010;202:85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Lieber CS. Microsomal Ethanol-Oxidizing System (MEOS): The First 30 Years (1968-1998)-A review. Alcohol: Clin Exp Res. 1999;23:991–1007. [PubMed] [Google Scholar]
- 36.Patel AB, de Graaf RA, Rothman DL, Behar KL, Mason GF. Evaluation of cerebral acetate transport and metabolic rates in the rat brain in vivo using 1H-[13C]-NMR. J Cereb Blood Flow Metab. 2010;30:1200–1213. doi: 10.1038/jcbfm.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegmund SV, Brenner DA. Molecular pathogenesis of alcohol-induced hepatic fibrosis. Alcohol Clin Exp Res. 2005;29:102S–109S. doi: 10.1097/01.alc.0000189275.97419.58. [DOI] [PubMed] [Google Scholar]
- 38.Keshari KR, Kurhanewicz J, Bok R, Larson PEZ, Vigneron DB, Wilson DM. Hyperpolarized 13C dehydroascorbate as an endogenous redox sensor for in vivo metabolic imaging. PNAS. 2011;108:18606–18611. doi: 10.1073/pnas.1106920108. doi:10.1073/pnas.1106920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohndiek SE, Kettunen MI, Hu D-E, Brett WC, Boren J, Gallagher FA, Brindle K. Hyperpolarized [1-13C]-Ascorbic and Dehydroascorbic Acid: Vitamin C as a Probe for Imaging Redox Status in Vivo. JACS. 2011;133:11795–11801. doi: 10.1021/ja2045925. [DOI] [PMC free article] [PubMed] [Google Scholar]