Abstract
The vertebrate multi-aminoacyl tRNA synthetase complex (MARS) is an assemblage of nine aminoacyl tRNA synthetases (ARSs) and three non-synthetase scaffold proteins, AIMP1 (aminoacyl tRNA synthetase complex-interacting multifunctional protein-1), AIMP2, and AIMP3. The evolutionary origin of the MARS is unclear, as is the significance of the inclusion of only nine of twenty tRNA synthetases. Eight of the nine amino acids corresponding to ARSs of the MARS are derived from two citric acid cycle intermediates, α-ketoglutatrate and oxaloacetate. We propose that the metabolic link with the citric acid cycle, the appearance of scaffolding proteins AIMP2 and AIMP3, and the subsequent disappearance of the glyoxylate cycle, together facilitated the origin of the MARS in a common ancestor of metazoans and choanoflagellates.
Keywords: tRNA synthetase, multi-aminoacyl tRNA synthetase complex, citric acid cycle, evolution
The multi-aminoacyl tRNA synthetase complex (MARS)
Many biological macromolecules exert their functions via formation of multi-component complexes. For example, the eukaryotic ribosome is made up of two large complexes comprising a total of about eighty proteins and four RNAs. Macromolecular complexes provide several functional advantages including increased reaction efficiency due to direct, vectorial transfer of substrates between components, minimization of the diffusion of substrates and intermediates, coordinated control of reaction rates, and compartmentalization of enzymatic reactions. Thus, when feasible, natural selection would favor the formation of macromolecular complexes that enable speed as well as parsimonious use of cellular energy resources. In support of this concept, a global analysis of the Saccharomyces cerevisiae (baker’s yeast) proteome has revealed more than five hundred protein complexes, with an average of about five proteins per complex [1].
Aminoacyl tRNA synthetases (ARSs) catalyze the ligation of specific amino acids to their cognate tRNAs. ARSs are classified into two groups (I and II) based primarily on the structural topologies of their catalytic domains. In vertebrates, nine ARSs along with three non-synthetase proteins form a stable macromolecular multiaminoacyl-tRNA synthetase complex (MARS, also abbreviated MSC) localized primarily in the cytoplasm [2-6]. The highly conserved components of this complex are LysRS, LeuRS, IleRS, GluProRS, MetRS, GlnRS, ArgRS, AspRS, and aminoacyl tRNA synthetase complex-interacting multifunctional proteins (AIMP)-1, -2, and -3, also known as p43, p38, and p18, respectively. The latter three proteins constitute non-synthetase, scaffolding components of the complex. GluProRS is a fused protein containing two distinct catalytic structures and activities, and is considered here as two synthetases.
Multiple functions have been proposed for the MARS including channeling of aminoacyl tRNAs for improving the efficiency of translation [7], serving as a depot to permit the stimulus-dependent release of constituents with non-canonical functions [8], and facilitating the transport of tRNAs from the nucleus to the cytoplasm [9]. Mutation of AIMP-2, a non-synthetase component of the MARS, results in loss of structural integrity of the complex and neonatal death in mice suggesting a critically important physiological function of the complex [10, 11].
Several smaller complexes containing ARSs (and non-ARS proteins) have been identified and shown to enhance translation by multiple mechanisms. For example, a complex of ValRS and EF-1A increases the rate of aminoacylation by ValRS [12]. In S. cerevisiae a complex of MetRS, GluRS, and Arc1p enhances the binding of the cognate tRNAs to the constituent ARSs [13, 14]. Finally, a complex of LysRS, LeuRS, ProRS, and EF-1A enhances aminoacylation by LysRS and ProRS in Methanothermobacter thermoautotrophicus [15]. These findings suggest that complexes comprising ARSs are favored during evolution because they enhance the efficiency of translation or its regulation. Thus, it is likely that a major force driving the origin and evolution of the MARS is its positive influence on translation efficiency. Indeed, the MARS increases the efficiency of translation by channeling charged tRNAs directly to the ribosome [16].
The mechanistic basis for the inclusion of nine specific ARSs in the MARS remains unclear. The nine MARS-bound ARSs belong to both class I and class II aminoacyl-tRNA synthetases, thereby excluding class as an important criterion for inclusion. Likewise, the substrates of the nine resident ARSs do not fall into a single class, for example, they include both polar and hydrophobic amino acids. A relationship between MARS-bound ARSs and the size of their corresponding amino acids has been suggested, namely, only ARSs corresponding to intermediate-sized amino acids are in the MARS [17]. According to this hypothesis, the binding of aminoacyl-tRNAs corresponding to intermediate-size amino acids to eEF-1A is disfavored thermodynamically, but balanced by MARS-facilitated transfer of aminoacyl-tRNAs to eEF-1A. However, this intriguing hypothesis does not explain the absence of MARS in single-celled organisms such as bacteria and yeast.
The MARS and the citric acid cycle
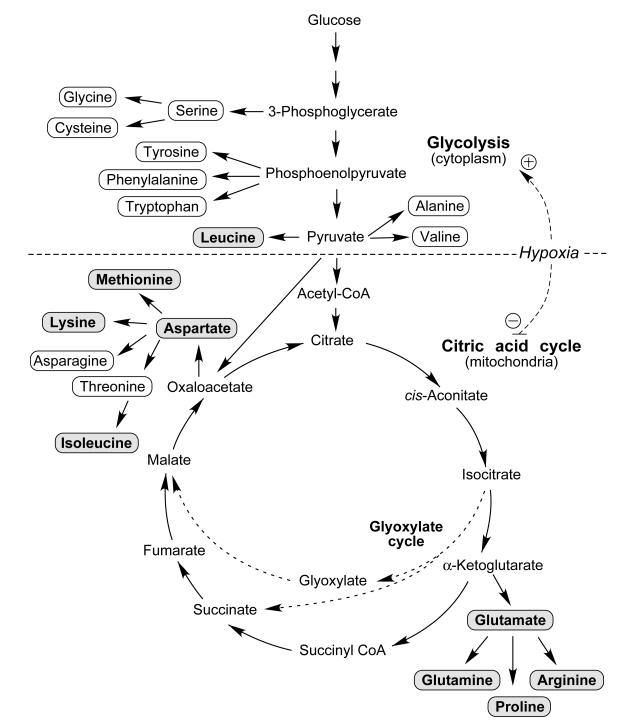
Remarkably, there appears to be a previously unrecognized connection between MARS and the citric acid (also known as Krebs or tricarboxylic acid) cycle. Ten amino acids are derived from intermediates of the citric acid cycle that takes place in mitochondria (Figure 1). Eight of the nine amino acids whose corresponding ARSs constitute the MARS are generated from two intermediates of the citric acid cycle: Glu, Gln, Pro, and Arg are derived from α-ketoglutarate, whereas Lys, Ile, Met, and Asp are derived from oxaloacetate (along with Asn and Thr) (Figure 1 and Table 1) [18]. Leu is an exception as it is a component of the MARS but is not directly generated from intermediates of the citric acid cycle.
Figure 1.
Amino acid biosynthetic pathways. Nineteen of the twenty amino acids (rounded rectangles) are synthesized directly or indirectly from the metabolites of glycolysis (nine) and the citric acid cycle (ten) (pathways separated by dashed horizontal line). Amino acids whose corresponding ARSs are components of the MARS are highlighted (gray background). Key components of glyoxylate cycle are shown (arrows with dashed lines). The citric acid cycle takes place in the mitochondria, whereas glycolysis and MARS are localized in the cytoplasm.
Table 1.
Metabolic links between multi-ARS complexes and the citric acid cycle
| Domain of life | Complex componentsa |
Metabolic pathway connectingb complex to citric acid cycle |
Glyoxylate cycle |
|---|---|---|---|
|
M. janaschii
c (Archaea) |
ProRS AspRS LysRS Mj1338 |
α-Ketoglutarate → → Pro Oxaloacetate → Asp Oxaloacetate → → Lys |
absent |
|
M. thermautotrophicus (Archaea) |
LysRS LeuRS ProRS EF-1A |
Oxaloacetate → → Lys Pyruvate → → Leu α-Ketoglutarate → → Pro |
absent |
| Mammals and insects (Eukarya) |
LysRS LeuRS IleRS GluProRS MetRS GlnRS ArgRS AspRS AIMP1 AIMP2 AIMP3 |
Oxaloacetate → → Lys Pyruvate → → Leu Oxaloacetate → → Ile α-Ketoglutarate → Glu α-Ketoglutarate → → Pro Oxaloacetate → → Met α-Ketoglutarate → → Gln α-Ketoglutarate → → Arg Oxaloacetate → Asp |
absent (except in nematodes) |
Only complexes with three or more ARSs are shown
Single arrow indicates single-step pathway, double arrow indicates multi-step pathway
Proposed complex
The observation that eight of the nine ARSs in the MARS are related by their ligand to the citric acid cycle, but only two of eleven non-MARS ARSs have this relationship, is unlikely to be coincidental. In fact, a chi-square test with Yates’ correction for low expected frequencies indicates a p-value of <0.01. We hypothesize that the citric acid cycle has had a positive influence on the emergence of the MARS during evolution via amino acids synthesized from its intermediary metabolites. The mechanism by which the citric acid cycle could have facilitated the origin and evolution of the MARS is likely to be related to its positive influence on the rate of protein synthesis. In organisms as diverse as bacteria, yeast, and mammals, under conditions of limitation of specific amino acids the corresponding ARS is induced, suggesting that the ARS is rate-limiting for protein synthesis under this condition [19-23]. During early evolution there might have been an extended period, or possibly episodic intervals, where the flux through the citric acid cycle was diminished and insufficient to meet the cellular demand for amino acids derived from its intermediates. This condition favored the evolution of novel mechanisms to more efficiently utilize the limited amino acid pool, for example, by amalgamating specific ARSs into complexes, which by interaction with the protein synthesis machinery can directly channel charged tRNAs to the elongating polypeptide [16, 24].
Asn and Thr are two exceptions to this principle because both are derived from the oxaloacetate intermediate in the citric acid cycle, but their synthetases are not present in the MARS. The reason for these exceptions is not certain. It is possible, if not likely, that factors unrelated to the citric acid cycle also contributed to the selection of specific synthetases residing in the MARS. For example, free AsnRS and ThrRS might have been required for non-canonical activities that precluded their residence in the MARS. In fact, several MARS-localized ARSs exhibit such noncanonical functions, but have evolved specific stimulus-dependent mechanisms to induce their release [8, 25-27]. Also, formation and maintenance of such a large complex needs proper scaffolding that might have precluded inclusion of some synthetases.
In summary, there is an association between the citric acid cycle and the MARS. We propose that the former has influenced the evolution of the latter. However, the citric acid cycle is present in all aerobic organisms. Then, what prevented its emergence earlier in evolution, for example in bacteria and fungi?
The MARS and the glyoxylate cycle
A comparative analysis of intermediary metabolism provides a potentially important insight. The glyoxylate cycle is a citric acid cycle variant present in bacteria, some archaeal species, and fungi, but absent in nearly all animals [28]. Importantly, bacteria and fungi, which possess the glyoxylate cycle, lack the MARS. The citric acid cycle intermediary metabolite α-ketoglutarate is absent in the glyoxylate cycle, possibly decoupling the regulation of synthesis of α-ketoglutarate-derived amino acids from oxaloacetate-derived amino acids (Figure 1). Remarkably, specific loss of the glyoxylate cycle in the archaea Methanocaldococcus jannaschii and Methanothermobacter thermoautotrophicus was accompanied by the appearance of small, multi-ARS complexes (Table 1) [29]. The components of these archaeal complexes, like the MARS, show relationships to the citric acid cycle; ProRS, LysRS, and LeuRS are constituents of a complex in M. thermoautotrophicus, and ProRS, LysRS, and AspRS are predicted to form a complex in M. janaschii [15, 29]. Interestingly, the amino acids corresponding to these ARSs, again with the exception of Leu, all are derived from the citric acid cycle (Table 1). These observations suggest that bypassing α-ketoglutarate by the glyoxylate cycle severs the stoichiometric relationship beween amino acids derived from oxaloacetate and α-ketoglutarate, thereby negatively influencing the formation of complexes containing ARSs derived from these branch points, including the MARS. Thus, we hypothesize that the disappearance of the glyoxylate cycle during evolution was a major metabolic factor contributing to the origin and evolution of the MARS.
Given the presumed absence of multi-ARS complexes in most archaeal species and in bacteria, the archaeal and eukaryotic complexes most likely originated independently, supporting a propensity for ARSs corresponding to citric acid cycle-derived amino acids to form complexes. In a less parsimonious alternative scenario, formation of the earliest multi-ARS complex might pre-date the divergence of the archaea and eukaryota branches of life. This would require subsequent losses in most archaea and in animals (and subsequent reappearance in animals regained hundreds of millions of years later), possibly due to the continued presence of the glyoxylate cycle. In this case, the archaeal complexes might represent early precursors to MARS formed by gradual accretion. The notable presence of ProRS and LysRS in all three complexes might be due to preferred interactions between these ARSs; alternatively, the order of selection for early inclusion might be stochastic.
The nematode C. elegans is a unique case. Nematodes are the only metazoans known to possess the glyoxylate cycle. The C. elegans genome harbors a horizontally acquired gene that encodes a bifunctional enzyme with isocitrate lyase and malate synthase catalytic activities, the unique enzymes of the glyoxylate shunt [30]. Interestingly, C. elegans has a multi-ARS complex with a composition different from that of the common MARS; the complex lacks AIMP3, ProRS, and AspRS, whereas ValRS is included [31]. Moreover, GluRS and ProRS are not fused and are encoded by separate genes, unlike in other bilaterians. Possibly, acquisition of the glyoxylate cycle by horizontal transfer in C. elegans induced transformation of the common MARS into a novel, reduced complex, consistent with the concept of an inverse relationship between the glyoxylate cycle and the multisynthetase complex. However, in this case the gain of the glyoxylate cycle induced a reduction in size of the multi-synthetase complex, not a complete loss (further discussed below). The presence of small multi-ARS complexes in some archaea and a reduced complex in C. elegans is consistent with a negative influence of the glyoxylate cycle on the origin and evolution of MARS.
Evidence for the origin of MARS in a common ancestor of metazoans and choanoflagellates
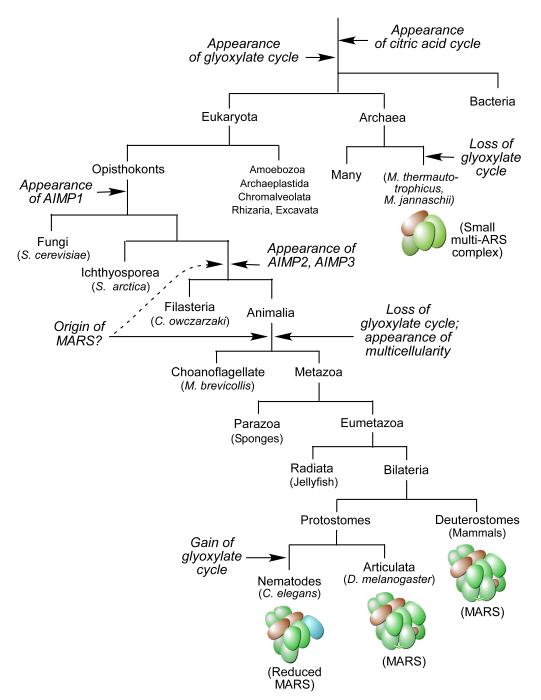
Experimental evidence for the MARS has been reported in multiple mammals as well as in insects [3, 32], indicating it appeared before the division of bilateria into protostome and deuterostome clades. The MARS has not been detected in prokaryotes and yeast, and its presence in lower metazoans such as sponges has not been reported. The genes encoding malate synthase and isocitrate lyase are not present in the genomes of choanoflagellates and metazoans (with the exception of nematodes) indicating the glyoxylate cycle is likewise absent (Figure 2). Given the hypothesis that the glyoxylate cycle has had a negative influence on the formation of the MARS in evolution, we propose that the MARS first appeared in a common ancestor of choanoflagellates and metazoans (Figure 2). Thus, the appearance of the MARS might have approximately coincided with the root of the basal metazoan tree. Although organisms belonging to filasteria are unicellular, choanoflagellates exhibit both unicellular and colonial growth and metazoans are strictly multicellular [33]. Thus, the appearance of the MARS might have coincided with the appearance of multicellularity.
Figure 2.
The origin of the MARS and other multi-ARS complexes. Tree of life with key branch points is shown (with example organisms in parentheses). The appearance and disappearance of AIMP1, 2, and 3, the glyoxylate cycle, and multi-ARS complexes are indicated (arrows); possible early origin of MARS is shown by dashed line. The presence or absence of AIMP2, AIMP3, isocitrate lyase, malate synthase (the latter two are glyoxylate cycle-specific enzymes) was determined using NCBI database and Origins of Multicellularity database of the Broad institute.
Because primitive multicellular organisms did not possess a circulatory system, cells in central, protected regions of the organism, distant from the external atmosphere and environment, might be subjected to a relatively hypoxic condition. Importantly, hypoxia has a marked inhibitory effect on the citric acid cycle, but it enhances glycolysis [34, 35]. We propose that the exposure of certain centrally important cells to hypoxia in primitive multicellular organisms reduced flux through the citric acid cycle, thereby reducing the generation of amino acids derived from cycle intermediates, as well as reducing the relative level of charged tRNA for these amino acids. The deficit of these amino acids and charged tRNAs stimulated the development of mechanisms to more effectively utilize the limited pools. One such mechanism is the assembly of the relevant synthetases into a ribosome-interacting complex, such as the MARS, that more efficiently channels amino acids from charged tRNAs to the elongating polypeptide chain [16, 24].
Intriguingly, the proposed influence of hypoxia on the evolution of the MARS might also clarify the inclusion of LeuRS, despite the fact that leucine is not derived from a citric acid cycle intermediate. A recent metabolite profiling experiment in yeast has shown that hypoxia markedly and specifically reduces intracellular leucine, while elevating valine and alanine (all three amino acids are derived from pyruvate, Figure 1) [36]. Thus, hypoxia not only reduces the amount of amino acids derived from the citric acid cycle, but also the availability of leucine, providing a plausible and mechanistic rationale for the inclusion of LeuRS in the MARS.
Role of the AIMP scaffold proteins in MARS structure and evolution
The three non-synthetase proteins in the vertebrate MARS are critical for complex assembly. AIMP2 connects the two major sub-complexes that constitute the holo-MARS [37]. Sub-complex I consists of MetRS, GluProRS, IleRS, LeuRS, and AIMP3, and sub-complex II contains GlnRS, ArgRS and AIMP1. AIMP3 anchors MetRS to MARS, and AIMP1 anchors GlnRS and ArgRS to the complex via its interaction with AIMP2 [37]. siRNA-mediated knockdown of any of the AIMPs disrupts MARS assembly [4, 37]. Thus, appearance of the accessory proteins during evolution might also have contributed to the origin of the MARS by solving scaffolding problems associated with assembly of a large multi-protein complex. In fact, the important role of scaffolding and nucleating factors in the formation of large macromolecular complexes has been reported [38]. Although AIMP1 is present in all opisthokonts (i.e., fungi and animals), AIMP2 and AIMP3 made their first appearance in the common ancestor of filasteria and animalia that predates the loss of glyoxylate cycle in animalia. It is possible that the three AIMP proteins first appeared for activities unrelated to protein synthesis (as has been described for the modern AIMPs [39-41]), and later evolved scaffolding functions to facilitate MARS assembly in a common ancestor of metazoans and choanoflagellates (Figure 2).
Constructive neutral evolution in MARS of higher organisms
Higher organisms cannot synthesize certain amino acids. For example, Homo sapiens cannot produce the essential amino acids His, Ile, Leu, Lys, Met, Phe, Thr, Trp, and Val. Therefore, the availability of certain amino acids, including some whose corresponding ARSs are in the MARS, does not depend on intermediary metabolism in “higher” organisms such as Homo sapiens. This raises the question as to why the MARS is maintained in extant higher organisms. We propose a constructive neutral evolution model for the MARS to explain this apparent inconsistency with our model. This model has been applied to explain complex molecular phenomena; for example, RNA editing and splicing [42, 43]. According to this model, biological complexity arises out of neutrally-fixed features that are not readily unfixed because they are unidirectional [44, 45]. In this case, once a stable complex such as MARS is formed, the function of the components becomes dependent on one another due to fixation of neutral mutations. Therefore, they partially lose the independence that their ancestors enjoyed and cannot function as effectively outside the complex. This compromise in independence of constituent ARSs favors the maintenance of the MARS during evolution despite the release from the metabolic dependence on the citric acid cycle. Furthermore, this model also explains why a partial, MARS-like complex was retained in C. elegans even after re-acquisition of the glyoxylate cycle.
Several studies support the constructive neutral evolution model for the MARS. ArgRS has two isoforms in mammals derived from the same gene; one is a part of the MARS and the other is free. However, the free ArgRS isoform is less efficient than the complex-bound isoform [16]. siRNA-mediated knockdown experiments have shown a physical dependence of MARS components on the intact holo-complex [4]. These studies suggest that complex-bound ARSs have partially lost both functional and physical independence thereby providing support for the constructive neutral evolution model.
Concluding remarks
mRNA translation and intermediary metabolism are among the most ancient, fundamental, and widespread systems of life on earth. Thus, there was substantial time and opportunity for each system to influence the function and evolution of the other. Our hypothesis provides an example of functional co-evolution that links the origin of the MARS to the citric acid cycle. This model does not exclude the possibility that other factors also influenced the formation and evolution of the MARS [17]. Moreover, because ARS complexes enhance the efficiency of translation, we speculate that natural selection favors the inclusion of all twenty ARSs in a complex. Possibly, this accretion process is ongoing. Alternatively, issues of co-regulation and availability of sufficient scaffolding might not permit the assembly of such a large complex. Early studies suggested the presence of a complex containing all twenty ARSs; however, the evidence, based exclusively on size fractionation, was also consistent with transient and indirect interactions of all ARSs via translating ribosomes, rather than in a single pansynthetase complex [46-50]. Even if such a large, loosely assembled complex of all 20 ARSs does exist, the nine MARS synthetases might serve as the stable core complex, and the same evolutionary pressures described here would be responsible for selection of synthetases for the core complex. Recent whole-genome sequencing of a plethora of early-appearing organisms has enabled the development of plausible hypotheses linking the appearance of the MARS to intermediary metabolism and scaffold proteins. The continuing addition of genome sequences from other organisms will permit a still finer temporal analysis of these relationships. However, testing these hypotheses and developing a more complete portrait of the origin of the MARS is likely to require real biochemical analysis of the MARS in key branch-point organisms.
ACKNOWLEDGMENTS
This work was supported by NIH grants P01 HL029582, P01 HL076491, and R01 GM086430 (to P.L.F.). S.M.E. was supported by a fellowship from the American Heart Association, Great Rivers Affiliate. Neither of the authors have any financial conflict of interest with the information in this manuscript
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 2.Mirande M, et al. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur. J. Biochem. 1985;147:281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- 3.Kerjan P, et al. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim. Biophys. Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 4.Han JM, et al. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J. Biol. Chem. 2006;281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 5.Norcum MT. Isolation and electron microscopic characterization of the high molecular mass aminoacyl-tRNA synthetase complex from murine erythroleukemia cells. J. Biol. Chem. 1989;264:15043–15051. [PubMed] [Google Scholar]
- 6.Filonenko VV, Deutscher MP. Evidence for similar structural organization of the multienzyme aminoacyl-tRNA synthetase complex in vivo and in vitro. J. Biol. Chem. 1994;269:17375–17378. [PubMed] [Google Scholar]
- 7.Negrutskii BS, Deutscher MP. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray PS, et al. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Nathanson L, Deutscher MP. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 2000;275:31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- 10.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MJ, et al. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 12.Negrutskii BS, et al. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 13.Simos G, et al. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 14.Simos G, et al. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 15.Praetorius-Ibba M, et al. Functional association between three archaeal aminoacyl-tRNA synthetases. J. Biol. Chem. 2007;282:3680–3687. doi: 10.1074/jbc.M609988200. [DOI] [PubMed] [Google Scholar]
- 16.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol. Cell. 2008;29:419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfson A, Knight R. Occurrence of the aminoacyl-tRNA synthetases in high-molecular weight complexes correlates with the size of substrate amino acids. FEBS Lett. 2005;579:3467–3472. doi: 10.1016/j.febslet.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 18.Stryer L. Biochemistry. W.H. Freeman; 1995. [Google Scholar]
- 19.Luo D, et al. Structure and regulation of expression of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol. 1997;179:2472–2478. doi: 10.1128/jb.179.8.2472-2478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazard M, et al. Overexpression of mammalian phenylalanyl-tRNA synthetase upon phenylalanine restriction. FEBS Lett. 1987;216:27–30. doi: 10.1016/0014-5793(87)80750-0. [DOI] [PubMed] [Google Scholar]
- 21.Meussdoerffer F, Fink GR. Structure and expression of two aminoacyl-tRNA synthetase genes from Saccharomyces cerevisiae. J. Biol. Chem. 1983;258:6293–6299. [PubMed] [Google Scholar]
- 22.Condon C, et al. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6992–6997. doi: 10.1073/pnas.93.14.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lofgren DJ, Thompson LH. Relationship between histidyl-tRNA level and protein synthesis rate in wild-type and mutant Chinese hamster ovary cells. J. Cell Physiol. 1979;99:303–312. doi: 10.1002/jcp.1040990304. [DOI] [PubMed] [Google Scholar]
- 24.Raina M, et al. Association of a multi-synthetase complex with translating ribosomes in the archaeon Thermococcus kodakarensis. FEBS Lett. 2012;586:2232–2238. doi: 10.1016/j.febslet.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo M, Schimmel P. Homeostatic mechanisms by alternative forms of tRNA synthetases. Trends Biochem. Sci. 2012;37:401–403. doi: 10.1016/j.tibs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang P, et al. Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8239–8244. doi: 10.1073/pnas.1100224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo M, et al. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashov FA, et al. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hausmann CD, Ibba M. Aminoacyl-tRNA synthetase complexes: molecular multitasking revealed. FEMS Microbiol. Rev. 2008;32:705–721. doi: 10.1111/j.1574-6976.2008.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, et al. Bifunctional glyoxylate cycle protein of Caenorhabditis elegans: a developmentally regulated protein of intestine and muscle. Dev. Biol. 1995;169:399–414. doi: 10.1006/dbio.1995.1156. [DOI] [PubMed] [Google Scholar]
- 31.Havrylenko S, et al. Caenorhabditis elegans evolves a new architecture for the multi-aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2011;286:28476–28487. doi: 10.1074/jbc.M111.254037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellermann O, et al. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. I. Species specificity of the polypeptide composition. J. Biol. Chem. 1982;257:11041–11048. [PubMed] [Google Scholar]
- 33.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster KA. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J. Exp. Biol. 2003;206:2911–2922. doi: 10.1242/jeb.00516. [DOI] [PubMed] [Google Scholar]
- 35.Nakano MM, et al. Anaerobic regulation of Bacillus subtilis Krebs cycle genes. J. Bacteriol. 1998;180:3304–3311. doi: 10.1128/jb.180.13.3304-3311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gleason JE, et al. Analysis of hypoxia and hypoxia-like states through metabolite profiling. PLoS One. 2011;6:e24741. doi: 10.1371/journal.pone.0024741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminska M, et al. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2009;284:6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- 38.Peterson-Kaufman KJ, et al. Nucleating the assembly of macromolecular complexes. Chembiochem. 2010;11:1955–1962. doi: 10.1002/cbic.201000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shalak V, et al. The EMAPII cytokine is released from the mammalian multisynthetase complex after cleavage of its p43/proEMAPII component. J. Biol. Chem. 2001;276:23769–23776. doi: 10.1074/jbc.M100489200. [DOI] [PubMed] [Google Scholar]
- 40.Han JM, et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh YS, et al. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell. 2010;9:810–822. doi: 10.1111/j.1474-9726.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 42.Stoltzfus A. On the possibility of constructive neutral evolution. J. Mol. Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 43.Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- 44.Gray MW, et al. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 45.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. U.S.A. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deutscher MP. The eucaryotic aminoacyl-tRNA synthetase complex: suggestions for its structure and function. J. Cell. Biol. 1984;99:373–377. doi: 10.1083/jcb.99.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandyopadhyay AK, Deutscher MP. Complex of aminoacyl-transfer RNA synthetases. J. Mol. Biol. 1971;60:113–122. doi: 10.1016/0022-2836(71)90451-7. [DOI] [PubMed] [Google Scholar]
- 48.Kaminska M, et al. Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J. Biol. Chem. 2009;284:13746–13754. doi: 10.1074/jbc.M900480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ussery MA, et al. Subcellular distribution of aminoacyl-tRNA synthetases in various eukaryotic cells. Eur. J. Biochem. 1977;72:491–500. doi: 10.1111/j.1432-1033.1977.tb11272.x. [DOI] [PubMed] [Google Scholar]
- 50.Turkovskaya HV, et al. Renaturation of rabbit liver aminoacyl-tRNA synthetases by 80S ribosomes. Int. J. Biochem. Cell Biol. 1999;31:759–768. doi: 10.1016/s1357-2725(99)00031-x. [DOI] [PubMed] [Google Scholar]