Abstract
One primary goal in nanobiotechnology is designing new methodologies for molecular biomedical diagnosis at stages much earlier than currently possible and without use of expensive reagents and sophisticated equipment. In this work, we show the proof of principle for single-molecule detection of the nucleocapsid protein 7 (NCp7), a protein biomarker of the HIV-1 virus, using synthetic nanopores and the resistive-pulse technique. The biosensing mechanism relied upon specific interactions between NCp7 and aptamers of stem-loop 3 (SL3) in the packaging domain of the retroviral RNA genome. One critical step of this study was the choice of the optimal size of the nanopores for accurate, label-free determinations of the dissociation constant of the NCp7 protein - SL3 RNA aptamer complex. Therefore, we systematically investigated the NCp7 protein - SL3 RNA aptamer complex employing two categories of nanopores in a silicon nitride membrane: (i) small, whose internal diameter was smaller than 6 nm, and (ii) large, whose internal diameter was in the range of 7 through 15 nm. Here, we demonstrate that only the use of nanopores with an internal diameter that is smaller than or comparable with the largest cross-sectional size of the NCp7-SL3 aptamer complex enables accurate measurement of the dissociation constant between the two interacting partners. Notably, this determination can be accomplished without the need for prior nanopore functionalization. Moreover, using small solid-state nanopores, we demonstrate the ability to detect drug candidates that inhibit the binding interactions between NCp7 and SL3 RNA by using a test case of N-ethylmaleimide.
Keywords: Nanobiotechnology, Biosensors, Single-molecule detection, Single-channel electrical recordings, Chemical kinetics, Protein-RNA interactions
Nanopore-based detection has proven to be a successful method for probing a variety of molecules of biological interest, such as DNA, RNA and proteins.1–8 Of particular appeal is this technique’s ability to probe these molecules without the need for chemical modification or labeling, to do so at physiological conditions, and to examine single molecules at a time; this allows the possibility for results to come to light that would otherwise be masked in bulk measurements. Recent work in the field has exploited these properties in order to probe dynamic bimolecular interactions in real time.9–21 In addition to these studies, an emerging interest in adapting DNA and RNA aptamers for use with nanopores has arisen.7,22–28
Herein, we exploit the nanopore-probe technique to examine the single-molecule detection of the nucleocapsid protein (NCp7),29–31 a biomarker of the human immunodeficiency virus 1 (HIV-1). Single solid-state nanopores were created in a silicon nitride membrane using electron-beam ablation (Fig. 1).32 NCp7 is a 55-residue domain of the gag and gag-pol polyproteins in HIV-1 (Fig. 2A). It plays an important role in the selection of viral genomic RNA for packaging during the HIV-1 infection cycle. NCp7 contains two zinc knuckles that bind specifically to the exposed guanosines of RNA stem-loops in the packaging domain of genomic RNA at physiological salt concentrations.33–35 In this work, the detection scheme was based upon the specific interactions of NCp7 with three 20-nucleotide RNA aptamers of varying binding affinity, which were derived from the stem-loop 3 (SL3, also known as Ψ) from the packaging domain of the retroviral RNA (Fig. 2B–D).36 Thus, SL3 is a naturally occurring aptamer that binds NCp7 with high affinity (Fig. 2E). The results obtained from the nanopore measurements were compared to those derived previously from a well-established titration technique based on quenching the fluorescence of Trp37 in NCp7 by loop bases of bound SL3.37–40 In addition, N-ethylmaleimide (NEM), which is known to inhibit the binding interactions of NCp7 to DNA and SL3 RNA 41 was added to the solution to test whether inhibition of the binding could be monitored in real time. Beyond the potential medical interest of this system, aptamers represent a highly versatile class of biosensing components, since they can be targeted to a wide variety of analytes. 42–44 Aptamers are chemically stable and easily produced single-stranded nucleic acid molecules, representing a promising alternative to traditional antibody-based approaches used in molecular biomedical diagnosis and other biotechnological applications.45 By using modern screening techniques, they allow for the design of high specificity to numerous substrates, including peptides and proteins. Therefore, the methodology established in this paper is applicable to a wide range of systems.
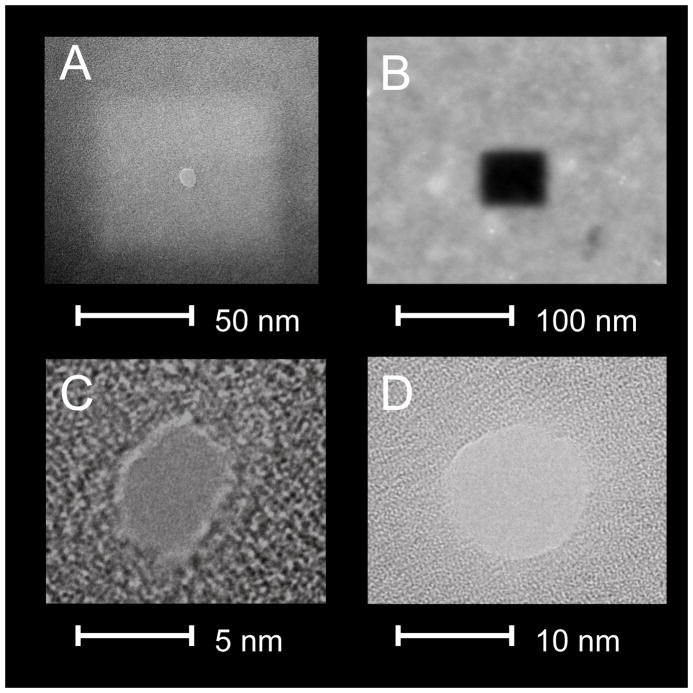
Figure 1. TEM images of nanopores.
(A) A bright-field TEM image of a thinned silicon nitride region with a small nanopore; (B) A high-angle annular dark field scanning TEM (HAADF-STEM) image of a thinned nitride region; (C) A TEM bright-field image of an ~3 nm-diameter nanopore in a silicon-nitride membrane; (D) A TEM bright-field image of an ~10 nm diameter nanopore in a silicon nitride membrane. All images were taken using a Technia F-20 S/TEM instrument.
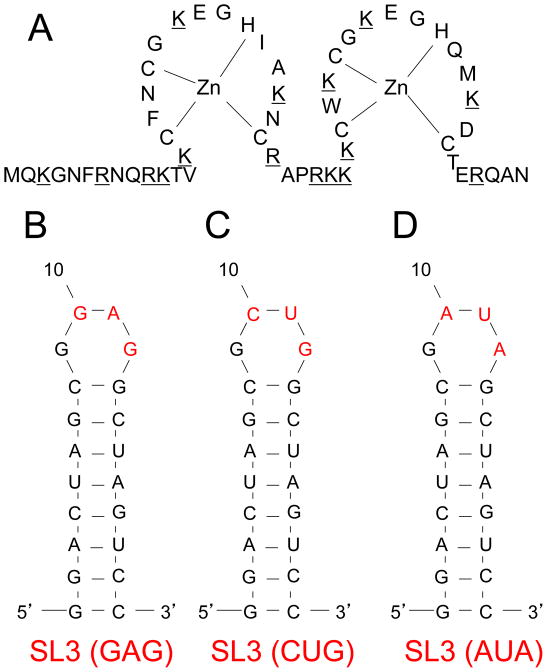
Figure 2. The primary sequence of the HIV-1 NCp7 protein and three variants of the SL3 RNA aptamers.
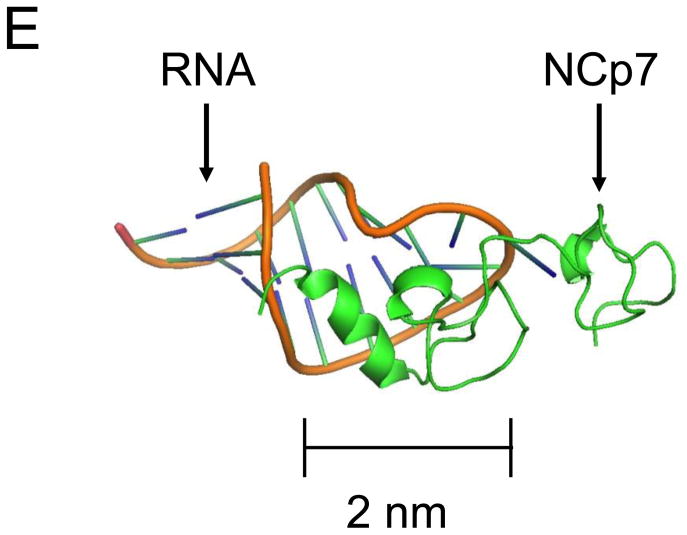
(A) The diagram displays the amino acid residues of the Zn2:NCp7 (1–55) protein. Positively charged amino acids are underlined; (B) The diagram indicates the nucleotide sequence of the high-affinity SL3 (GAG) aptamer; (C) The low-affinity SL3 (CUG) aptamer; (D) No-affinity SL3 (AUA) aptamer; (E) The panel shows the three-dimensional structure of the NCp7 protein bound to the SL3 stem-loop recognition element of the genomic Ψ RNA packaging signal, as determined by heteronuclear magnetic resonance spectroscopy (adapted from PDB 1A1T).33 The scale bar indicates the approximate dimension of the complex, with the largest cross-sectional dimension of approximately 5.5 nm.
In this work, the binding affinities of NCp7 with SL3 and two related RNA aptamers were extensively studied using two types of nanopores: (i) small nanopores, whose internal diameter was smaller than 6 nm, with a thickness of the silicon nitride membrane less than 15 nm, and (ii) large nanopores, whose internal diameter was in the range 7 through 15 nm, with a thickness greater than 20 nm. The translocation of the aptamers though small nanopores in ultrathin membranes was examined in detail. We present the titration experiments by adding increasing concentrations of NCp7 to the solution. The events observed when the aptamers were added to large nanopores in thicker membranes are also shown. Next, the events attributed to complexes of the SL3 RNA aptamers with the NCp7 protein are described. Finally, we employed a single small-diameter nanopore and N-ethylmaleimide (NEM) to perform real-time sampling of the inhibition of specific interactions between NCp7 and the high-affinity SL3 RNA aptamer. The ability of our system to detect the efficacy of NEM without labeling suggests that nanopores may be used to study drug inhibitors of binding protein-aptamer interactions.
Results and Discussion
Small nanopores
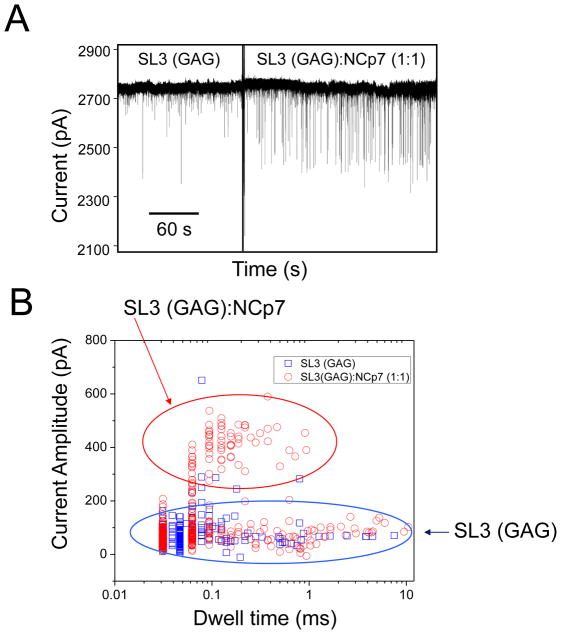
Nanopores with diameters less than 6 nm were formed in thin nitride membranes (~8–15 nm thick). Experiments were performed with 200 mM NaCl on the cis side of the chamber and 1 M NaCl on the trans side. The buffer on both sides was 5 mM NaH2PO4, at pH 7.0. The cis solution matched the salt and pH conditions used in previously published fluorescence-based titration experiments.35 The higher molarity of NaCl salt on the trans side was advantageously employed for both the drastic improvement in the signal-to-noise ratio of the acquired data as well as the substantial increase in the capture rate of the RNA aptamer.46 Conductance values for small nanopores under these conditions ranged from 4.5 through 13.3 nS (n=41). Small nanopores used in these experiments exhibited stable single-channel current signatures at voltages up to 400 mV. At positive applied voltages, NCp7 did not alter the single-channel current signature of small nanopores (Supplementary Information, Fig. S1), confirming its net positive charge. However, at negative voltages, it caused rapid fouling of the nanopore (Supplementary Information, Fig. S2) owing to protein adsorption on the inorganic surface of silicon nitride. 32 In contrast, SL3 RNA aptamers did not change the single-channel electrical signature of small nanopores at negative voltages (Supplementary Information, Fig. S3), confirming their net negative charge. However, when the applied voltage was positive, the SL3 RNA aptamers produced distinguishable current blockades. Fig. 3A shows a typical signature of the single-channel electrical trace after the addition of 500 nM SL3 (GAG) aptamer at a positive transmembrane potential of +200 mV.
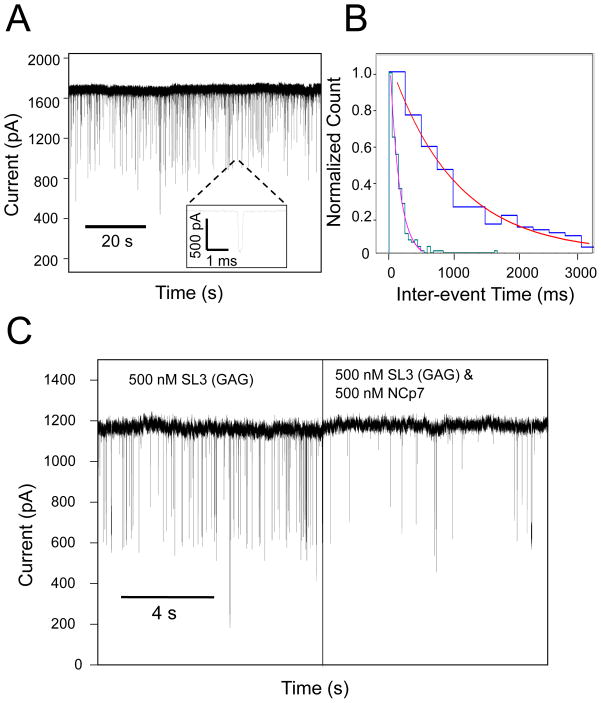
Figure 3. Sampling SL3 RNA aptamer - NCp7 interactions using a small nanopore.
(A) A representative single-channel electrical trace demonstrating SL3 RNA aptamer–produced current blockades. The chamber contained 500 nM high-affinity SL3 (GAG) aptamer. The internal diameter of the nanopore was smaller than 6 nm, whereas the thickness of the silicon nitride membrane was smaller than 10 nm; (B) Histogram showing the change in inter-event time when 500 nM NCp7 was added to 500 nM SL3 (GAG) aptamer. A fitting of the histogram for 500 nM SL3 (GAG) alone (green) gives an inter-event time of τon = 149 ± 15 ms, while the fitting of the same data after addition of 500 nM NCp7 protein (blue) shows an increased inter-event time of τon = 1030 ± 60 ms; (C) Trace showing a drastic reduction of event frequency corresponding to the above histograms. The applied voltage was +200 mV. The buffer solution was 200 mM NaCl, 5mM NaH2PO4, pH 7.0 on the cis side, and 1 M NaCl, 5 mM NaH2PO4, pH 7.0 on the trans side.
The frequency of the current blockades produced by SL3 RNA aptamers can be determined by τon histograms, which can be fit to a single exponential using a log likelihood ratio test protocol (Fig. 3B).47 Thus, the inverse of τon gives the event frequency.48 Addition of NCp7 did not alter the dwell time or amplitude of the current blockades (Supplementary Information, Fig. S4). Instead, it changed their event frequency, as measured by a τon histogram. Fig. 3C shows a representative single-channel electrical trace demonstrating the drastic reduction in the event frequency of current blockades when 500 nM NCp7 is added to 500 nM SL3 (GAG) aptamer. This reduction suggests that titration experiments can be performed using these small nanopores.26 Voltage dependence tests of the dwell time of the SL3 RNA aptamer-produced current blockades displayed a biphasic signature, featuring a peak of the dwell time of the SL3 (GAG) aptamer between 200 mV and 250 mV (Supplementary Information, Fig. S5). This sort of biphasic signature of the dwell time has also been observed in the past with other charged polymers, such as polypeptides interacting with protein nanopores.48–50 Experiments with SL3 (GAG) (number of distinct experiments was n=5), SL3 (CUG) (n=3) and SL3 (AUA) (n=3) were performed such that the SL3 RNA concentration was fixed at 500 nM and the NCp7 concentration progressively increased from 0 nM to 125 nM, 250 nM, 375 nM, 500 nM, 750 nM, 1000 nM, and finally 1250 nM.
Titration curves using small nanopores
The binding affinities of NCp7 to each of the three SL3 RNA aptamers may be calculated using a titration curve.26 In previous fluorescence experiments, 36,40 this was achieved by assuming the fluorescence intensity to be directly proportional to the free NCp7 concentration. For the small nanopores used in this study, a titration may be analyzed by assuming that the frequency of events is directly proportional to the concentration of free SL3 RNA aptamer. The curves may then be fit using the equation:35,36,40
| (2) |
where Pt and Rt are the total NCp7 protein and SL3 RNA concentrations, respectively, f is the frequency of low-amplitude current blockades, f0 and f∞ are the event frequency at the initial time and at saturation with NCp7, respectively, and Kd is the dissociation constant. This equation assumes that Pt = Pf + Pbound Rt = Rf + Rbound, and Kd =(Rf Pf)/(RP). Here, Rf and Pf are the concentrations of free SL3 RNA aptamer and NCp7 protein in aqueous phase, respectively. Rbound and Pbound denote the concentrations of bound SL3 RNA aptamer and NCp7 protein in aqueous phase, respectively. RP is the concentration of the NCp7-SL3 aptamer complex in solution.
Here, we assume that all events observed with the small nanopores are due to the SL3 RNA passing into the nanopore and that the NCp7-SL3 complexes are completely excluded, despite their net negative charge, so that the events sample the concentration in the chamber well. Two considerations justify this assumption: (i) there is a lack of change observed in the current amplitude or dwell time of the blockade events after NCp7 is added to the cis chamber, and (ii) in work with larger nanopores (see next section) there is a significant alteration in the amplitude of current blockades that is probably due to blockage by the complex.
The Kd values determined by the small nanopores may differ somewhat from values derived using the fluorescence data, because the protein’s fluorescence is quenched most efficiently when the NCp7 protein binds to the loop of the SL3 RNA aptamer. In contrast, additional reduction of current blockades may occur in the nanopores due to non-specific interactions of NCp7 with the SL3 stem bases in solution; there may also be SL3 stems bound weakly to the substrate surface, and those bound near a nanopore could bind NCp7 and reduce current flow. Surface-bound RNA could also form complexes that reduce the solution concentrations of RNA and protein, and the effective event frequency. The effects of non-specific binding of RNA and RNA-protein complexes to the surface, and of RNA stem bases to NCp7 are likely to be small at the 200 mM NaCl used here, as it has been shown that non-specific SL3-NCp7 interactions are largely suppressed at monovalent cation concentrations above 150 mM. 35
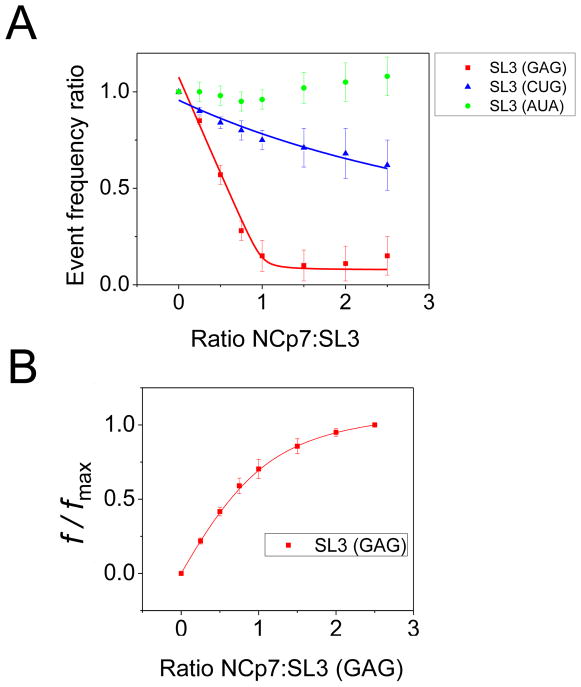
Fig. 5A shows the data obtained for all three SL3 RNA aptamers by using small nanopores. In this plot, the event frequency ratio is the frequency of the SL3 RNA aptamer-produced current blockades after addition of NCp7 to the cis side normalized to the initial event frequency (eq. 2). The Kd values calculated by fitting to the titration curves given in Fig. 5A are provided in Table 1. These Kd values show the same trends as in the previously published fluorescence data.36
Figure 5. Titration curves of the SL3 RNA variants interacting with NCp7.
(A) Titration curves derived with small nanopores using a concentration of 500 nM for different SL3 RNA aptamer variants. The event frequency ratio is the frequency of the SL3 RNA aptamer-produced current blockades after addition of NCp7 to the chamber normalized to the initial event frequency; (B) Curve derived from large nanopores using a concentration of 1000 nM for SL3 RNA (GAG). Curves are fit as described in the text. Kd values for the 1:1 complexes are derived from best fittings and are given in Table 1.
Table 1. The dissociation constant, Kd, of the NCp7 protein-SL3 RNA aptamer complexes.
Values for small and large nanopore data were arrived at using best-fit curves to titrations shown in Fig. 5.
| SL3 RNA Aptamer | Small nanopore | Large nanopore | Fluorescenceb |
|---|---|---|---|
| SL3 (GAG) | 2 ± 4 nM | 278 ± 166 nM | 28 ± 2 nM |
| SL3 (CUG) | 1960 ± 300 nM | NDa | 850 ± 250 nM |
| SL3 (AUA) | NDa | NDa | 20000 nM |
ND stands for not determined data. For large nanopores, we were unable to fit the low-affinity aptamer data to eqn. (3). The binding affinity of the SL3 (AUA) to NCp7, which was obtained from fluorescence measurements, is rounded up to the nearest significant digit.
Fluorescence values are from the literature.36
We observed a somewhat greater binding affinity of NCp7 to SL3 RNA (GAG) when using the nanopore-probe technique as compared to the fluorescence approach (Table 1). While this may reflect non-specific interactions discussed in the previous paragraph, it is also difficult to accurately determine Kd values by fluorescence when Kd < 20 nM or Kd > 300 nM, where small measurement errors exert dramatic effects on the derived Kd.35 The same error considerations apply to the nanopore data. Briefly, at Kd < 20 nM, there are few measured data points that differ from a binding isotherm that corresponds to Kd → 0 (affinity → ∞), and the results of the fitting algorithm are largely dependent on the one or two points at the curved elbow of the titration curves (e.g., the curve for SL3(GAG) in Fig 5A; a Kd → 0 titration would consist of two straight lines intersecting at a 1:1 ratio of [NCp7]:[SL3]). The experimental error for Kd > 300 nM is dominated by uncertainty in the extrapolation to a saturating concentration of NCp7 (e.g., the curve for SL3 (CUG) in Fig. 5A). For low-affinity complexes it is not practical to continue the titrations to very large [NCp7] to determine the saturation limit, as such high concentrations would favor stoichiometries where more than one protein is bound per RNA.31,35,51
Recently, we derived the affinities of NCp7 to 24-nucleotide long DNA aptamers of varying affinity using an engineered protein nanopore derived from ferric hydroxamate uptake component A (FhuA) of Escherichia coli. 26 Since, this engineered protein nanopore is cation selective, it was more convenient to use a detection mechanism based upon the partitioning of the positively-charged NCp7 protein into the nanopore lumen, which contrasts to the biosensing approach presented in this work. Several other studies have used smaller-sized nanopores for force-spectroscopy.10,13,52,53 In that technique, the electric field produced in the nanopore was used to directly dissociate molecules from each other. The time to dissociation was measured and interpreted to derive affinity data. In these experiments, we were not able to produce current blockades with lifetimes long enough to perform such an analysis. However, the force of the electric field at or near the nanopore orifice could conceivably act upon an NCp7-SL3 aptamer complex, catalyzing the dissociation process. This may explain why the event frequencies of the high-affinity aptamers, shown in Fig. 5A, do not go to zero at saturating concentrations of NCp7. While there are differences in the Kd values determined using fluorescence, the silicon nitride nanopores, and the engineered FhuA nanopores, the most significant result is that the overall trends are similar.
Large nanopores
Nanopores with diameters greater than 10 nm enlarged significantly with time when drilled into ultrathin silicon nitride membranes, whose thickness was less than 10 nm. Therefore, thicker silicon nitride membranes, ranging from 20 through 30 nm, were used. A variety of nanopore diameters was tested with data collected on nanopores with diameters ranging from 7 nm to 15 nm, as measured by bright-field TEM. We were not able to isolate systematic trends based solely on the nanopore diameter due to our use of different thicknesses of nitride in these experiments. Experiments were performed with 200 mM NaCl on the cis side of the chamber and 1 M NaCl on the trans side. The buffer on both sides was 5 mM NaH2PO4, at pH 7.0. Under these experimental circumstances, large nanopores showed stable current versus voltage profiles when only buffer solution was in the chamber. The addition of NCp7 to the cis chamber did not produce any alteration in the current signature at a transmembrane potential of +200 mV. In contrast, the addition of the SL3 RNA aptamers created very short-lived current spikes of non-uniform, low amplitude, many of which exhibited dwell times near the rise-time of the filter (Fig. 4A).47 The addition of the NCp7 protein changed both the dwell time and current amplitude of the blockades (Fig. 4B). As expected, dwell time alterations were not reproducible in different-size nanopores. Importantly, the proportion of current blockades exhibiting a greater current amplitude increased with increasing concentrations of NCp7, suggesting that these events can be attributed to the NCp7 protein-SL3 RNA aptamer complex. Experiments with SL3 (GAG) (n=3) were performed such that the SL3 RNA aptamer concentration was fixed at 1000 nM and the NCp7 concentration progressively increased from 0 to 250 nM, 500 nM, 750 nM, 1000 nM, 1500 nM, 2000 nM, and finally 2500 nM. We also performed similar single-channel electrical recordings using SL3 (CUG) (n=3) and SL3 (AUA) (n=3) aptamers (Supplementary Materials, Fig. S6). One would expect a decrease in the frequency of low-amplitude, SL3 RNA aptamer-induced current blockades to be close to that observed with the small nanopores; however, this was not the case. The event frequency decreased, yet not in the same manner as that recorded with small-diameter nanopores and not reproducibly for each large nanopore (Supplementary Materials, Fig. S7). Therefore, we speculate that some of the low-amplitude current blockades recorded with large nanopores are due to “bumping” events of the larger NCp7-SL3 RNA complex that cannot be separated by the current amplitude alone. Alternatively, there could be NCp7-SL3 RNA complexes that have small amplitude blockades due to short residence time and attenuation by the filter.
Figure 4. Representative single-channel electrical trace showing the NCp7-SL3 RNA aptamer interactions using a large nanopore.
(A) The addition of 1 μM NCp7 protein increases the frequency of large amplitude current blockades observed with a large-diameter (~7nm) nanopore in a thick (~30 nm) silicon nitride membrane when added to a solution containing 1 μM high-affinity SL3 (GAG) aptamer; (B) A scatter plot of current amplitude versus dwell time showing distinct event types. Low-amplitude current blockades are attributed to the SL3 RNA aptamer alone. Large-amplitude events are interpreted as current blockades produced by the NCp7-SL3 RNA aptamer complex. Square events represent measurements taken with SL3 (GAG) solution in the chamber, while circle events indicate data taken with both SL3 (GAG) and NCp7 added to the chamber. The applied transmembrane potential was +200 mV. The buffer solution contained 200 mM NaCl, 5mM NaH2PO4, pH 7.0 on the cis side, and 1 M NaCl, 5 mM NaH2PO4, pH 7.0 on the trans side.
Titration curves using large nanopores
While smaller nanopores offer a straightforward method of binding affinity analysis, results with larger nanopores are more complicated to interpret. In large nanopores, events due to both the SL3 RNA and the NCp7-SL3 RNA aptamer complex are observed. In a previous study by Wanunu and coworkers, 18 the binding of various aminoglycosides to an A-site RNA have been detected and discriminated by differences in current amplitude. In that study, an affinity curve was constructed, enabling the determination of reasonable binding affinity values matching bulk measurements. The major assumption was that the proportion of a drug molecule:A-site RNA complex and an A-site RNA entered the nanopore at the same rate, which is reasonable given their similar size. In contrast, using such an analysis in this work did not return reasonable values (Supplementary Material, Fig. S6) leading us to reject such an assumption for further data analysis. A likely explanation is that the hydrodynamic radius of the NCp7-SL3 RNA complex is greater than that of the SL3 RNA aptamer alone, leading to different diffusion dynamics in solution and therefore a distinction in the capture rate between the two partitioning molecules. Major determinants of the difference in the capture rates between the SL3 RNA aptamer and the NCp7-SL3 RNA complex include different nature of the energetic barriers54,55 for the translocation of the aptamer and aptamer-target complex as well as a possible electro-osmotic effect.56,57
An alternative method for calculating the Kds with large nanopores is to measure the increase and saturation in large-amplitude current blockades with progressively higher concentrations of NCp7. Assuming 1:1 binding, the concentration of the NCp7-SL3 aptamer complex in solution can be expressed as:
| (3) |
where [Complex] is the concentration of the NCp7-SL3 complex in solution and [Complex]max is the maximum concentration of the complex observed after saturation with NCp7. Here, x denotes the added concentration of NCp7 divided by the added concentration of the SL3 RNA aptamer. xmax indicates the highest added concentration of NCp7 divided by the added concentration of the SL3 RNA aptamer. C is the dissociation constant (Kd) divided by the added concentration of the SL3 RNA aptamer. Equation (3) was derived by the relation Kd =(Rf Pf)/(RP), as in equation (2), with the assumption that the concentration of the NCp7-SL3 aptamer complex is directly proportional to the frequency of the large-amplitude current blockades. Fig. 5B shows a plot of the ratio of the frequency of the large-amplitude current blockades to the frequency of large-amplitude events at the maximum used NCp7-to-SL3 (GAG) ratio. Fitting using equation (3) for C and multiplying by the concentration of SL3 (GAG) gives the value for Kd of 278 ± 166 nM (Table 1). We were not able to obtain satisfactory fits to this equation for data acquired with the low-binding affinity SL3 RNA variants.
N-ethylmaleimide addition to small nanopores
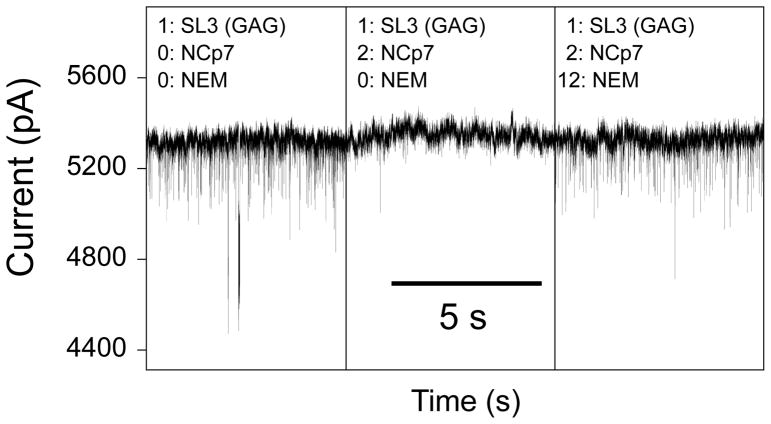
Given that the binding affinities arrived at using small nanopores are in reasonable accord with those obtained by fluorescence data, the possibility of probing a drug candidate against NCp7 becomes feasible. NEM reacts covalently with the cysteine residues in the zinc-fingers of NCp7, unfolding the fingers and preventing the protein from making specific interactions with the SL3 RNA.58,59 We added a 6-fold molar excess of NEM to the test solution after suppressing events by the addition of the NCp7 protein to the SL3 (GAG) aptamer at an applied transmembrane potential of +400 mV. This experiment was repeated three times on the same nanopore of ~6 nm diameter. Addition of the NEM rapidly brought the event frequency to near its original value (Table 2), demonstrating that NEM suppresses the formation of the NCp7-SL3 complex (Fig. 6).
Table 2. Normalized capture frequency of the SL3 (GAG) aptamer-produced current blockades in the presence of NCp7 and NEM.
Event frequency of current blockades produced by the SL3 (GAG) aptamer was measured before and after addition of NCp7 and then NEM. The capture frequency was normalized to the value determined using solution that contained only the SL3 (GAG) aptamer. The initial concentration of the SL3 (GAG) aptamer was 5 μM. Error bars represent standard deviations for 3 separate single-channel experiments.
| Concentration ratio SL3(GAG):NCp7:NEM | Normalized capture frequency |
|---|---|
| 1:0:0 | 1 |
| 1:1:0 | 0.19 ± 0.10 |
| 1:2:0 | 0.03 ± 0.10 |
| 1:2:12 | 0.85 ± 0.20 |
Figure 6. Effect of N-ethylmaleimide on formation of the NCp7-SL3 RNA complex.
Event frequency modulation caused by the addition of 5 μM high-affinity SL3 (GAG) aptamer, with 10 μM NCp7, causing reduction in the event frequency. Introduction of 60 μM N-ethylmaleimide returns the event frequency to near its pre-NCp7 level. The applied transmembrane potential was +400 mV. The buffer solution contained 200 mM NaCl, 5mM NaH2PO4, pH 7.0 on the cis side, and 1 M NaCl, 5 mM NaH2PO4, pH 7.0 on the trans side. The nanopore was ~6 nm in diameter in a 15-nm thick nitride.
Concluding remarks
In summary, our studies have established that solid-state nanopores can be used to perform real-time measurements of the affinity of a small protein with short RNA aptamers, even when the overall charge of the protein analyte-aptamer complex is negative. The results obtained with small nanopores are a satisfactory match to the binding affinity values obtained in previously published fluorescence studies, 36 suggesting that affinity can be monitored with nanopores, whose internal diameter is comparable or smaller than the largest cross-sectional diameter of the binding protein-RNA aptamer complex. This confirms that sensitive quantification of disease biomarkers, such as the HIV-1 nucleocapsid protein, can be accomplished using stable nanopores. The methodology can be expanded to other protein-RNA ligand complexes 60 for a rapid, label-free determination of the dissociation constants with no further requirement for functionalization of the silicon nitride surface. Additionally, the demonstration of the inhibition of binding by NEM suggests that nanopores could be used for screening potential drug targets, especially as massively parallel nanopore devices become available.61–63
Experimental Section
Fabrication of solid-state nanopores
First, 40-nm thick membranes of freestanding low-stress silicon nitride were created using standard photolithography techniques. Next, a 50 μm square section of nitride was thinned to either ~10 nm or ~20 nm using a FEI Strata 400 STEM focused ion beam (Fig. 1A and Fig. 1B). Nanopores were then created directly using a FEI Tecnai F20 S/TEM in a STEM mode, as described previously.32 Nanopore diameter was measured in bright-field TEM mode (Fig. 1C and Fig. 1D). Nanopores were wetted using isopropanol and flushed with deionized water before being loaded into a custom-built Teflon chamber. The silicon-supporting chip was coated with a fast curing silicone elastomer (Kwik-Cast, World Precision Instruments, Sarasota, FL) to reduce the current noise.64 Large nanopores were treated for 10 minutes in piranha solution (a 3:1 ratio mixture of H2SO4:H2O2) before the isopropanol wash. Initial bath solution was 1 M NaCl, 5 mM NaH2PO4, pH 7.0 on both sides of the chamber. Current measurements were performed using Ag/AgCl electrodes connected to an Axon 200B amplifier (Axon Instruments, Foster City, CA) and filtered at 10 kHz with an 8-pole, low-pass Bessel filter, Model 900 (Frequency Devices, Ottawa, IL) before being digitized by an Axon 1400A digitizer (Axon). Only nanopores displaying low 1/f noise and stable current signatures were used in these experiments.65 In general, we found that the noise depends on two parameters: nanopore thickness and nanopore diameter. For thick nanopores, current noise was more pronounced. In thinner nanopores, less noise was detected.66 As was discussed by Smeets and coworkers (2008),65 it is thought that “wetting” of nanopores is closely related to the current noise, and large-diameter nanopores are generally easier to “wet” than small-diameter nanopores. A broad range of both nanopore diameter and thickness was used in these experiments. Thin nanopores, with a thickness smaller than 10 nm, had an excellent success rate and almost always displayed low current noise, while large-diameter, thicker nanopores have a success rate closer to 70% after piranha treatment.
Current versus voltage curves were constructed to confirm that nanopores matched the expected size according to the formula66:
| (1) |
where I is the open-state current of the nanopore and V is the applied transmembrane voltage. Here, μNa+ and μCl− are the electrophoretic mobilites of the Na+ ions and Cl− ions, respectively. e, heff and d denote the elementary charge, the effective length of the nanopore and its diameter, respectively. nNaCl is the number density of NaCl. Following confirmation of size, solution on the cis (grounded) side of the chamber was changed to 200 mM NaCl, 5 mM NaH2PO4, pH 7.0. All analytes were added to the cis side of the chamber.
Preparation of NCp7 and SL3 RNA aptamers
The NCp7 protein (Fig. 2A) was expressed, purified and had its concentration measured as described previously.40 The high-affinity SL3 (GAG) aptamer (Fig. 2B) used in this study is a mimic of the sequence that occurs in the packaging domain of genomic RNA, while the low-affinity SL3 (CUG) (Fig. 2C) and no-affinity SL3 (AUA) (Fig. 2D) aptamers represent mimics lacking one and both exposed guanosine(s), respectively, reducing the binding affinity of the NCp7-aptamer complex.36 The RNA stem-loops used in this study were purchased from IDT (Integrated DNA Technologies, Coralville, IA). SL3 RNA aptamer samples were dissolved in Milli-Q purified water (EMD Millipore Corporation, Billerica, MA). Prior to use in RNA experiments, samples were heated briefly to 90°C, then cooled on ice for 15 minutes to form hairpins.
Given the highly specific interactions between NCp7 and SL3 genomic RNA region and that SL3 packing is highly conserved in different mutations of HIV, the zinc knuckles have become an attractive target for drugs seeking to treat HIV. 39,67 NEM is an alkylating agent that covalently reacts with cysteine thiols; this causes irreversible ejection of zinc and the inability of the protein to adopt a biologically active conformation.58,59 In this work, its ability to inactivate the binding mechanism of NCp7 to SL3 allowed it to be used as a test molecule for assessing whether nanopores can perform real-time detection of drug candidates. At pH 7.0, NCp7 has a formal positive charge of +9e and the SL3 aptamers a charge of -19e each. The complex will therefore have a net charge of -10e.35 The solution structure of NCp7 bound to the SL3 RNA (GAG) aptamer33 was used to show that the volume is approximately 4 times that of the RNA aptamer alone, with the SL3 RNA aptamer approximated by a cylinder with a diameter of ~2 nm and a length of ~3 nm (Fig. 2E). It is difficult to obtain an accurate volume for the NCp7 protein alone. For example, in the NCp7-SL3 RNA complex, NCp7 is rather compact, while the unbound form is largely a random coil except for the two zinc fingers. The largest cross-sectional size of the NCp7-SL3 complex is ~5.5 nm.
Supplementary Material
Acknowledgments
We are grateful to J. Grazul, M. Thomas and J. Treichler for their help during the early stages of this work. We also thank colleagues in the Movileanu and Borer laboratories for stimulating discussions. Nanopores were made at the Electron Microscopy Facility of the Cornell Center for Materials Research (CCMR) with support from the National Science Foundation - Materials Research Science and Engineering Centers (MRSEC) program (DMR-0520404). The preparation of the silicon nitride membranes was performed at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation (Grant ECS-0335765). This work is funded in part by grants from the National Science Foundation (DMR-1006332) and the National Institutes of Health (R01 GM088403) to L.M.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: (i) control experiments with NCp7 and SL3 RNA aptamers using small nanopores, (ii) comparison of the values of the dwell time and amplitude of the current blockades recorded with a small-diameter nanopore before and after addition of NCp7 to solution, (iii) voltage dependence of the dwell time of the current blockades produced by the SL3 RNA aptamers; (iv) large event ratio with smaller events for all three SL3 RNA aptamers; (v) frequency of low-amplitude, SL3 RNA aptamer-induced current blockades observed with large nanopores. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Bezrukov SM. Ion Channels As Molecular Coulter Counters to Probe Metabolite Transport. J Membr Biol. 2000;174:1–13. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]
- 2.Bayley H, Cremer PS. Stochastic Sensors Inspired by Biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 3.Dekker C. Solid-State Nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 4.Movileanu L. Squeezing a Single Polypeptide Through a Nanopore. Soft Matter. 2008;4:925–931. doi: 10.1039/b719850g. [DOI] [PubMed] [Google Scholar]
- 5.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The Potential and Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Movileanu L. Interrogating Single Proteins Through Nanopores: Challenges and Opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Gu LQ, Shim JW. Single Molecule Sensing by Nanopores and Nanopore Devices. Analyst. 2010;135:441–451. doi: 10.1039/b907735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of Biological Pores in Nanomedicine, Sensing, and Nanoelectronics. Curr Opin Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benner S, Chen RJ, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Sequence-Specific Detection of Individual DNA Polymerase Complexes in Real Time Using a Nanopore. Nat Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-Molecule Analysis of DNA-Protein Complexes Using Nanopores. Nat Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 11.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a Single Protein in a Nanopore Through Electrostatic Traps. J Am Chem Soc. 2008;130:4081–4088. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 12.Mohammad MM, Movileanu L. Excursion of a Single Polypeptide into a Protein Pore: Simple Physics, But Complicated Biology. Eur Biophys J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 13.Dorvel B, Sigalov G, Zhao Q, Comer J, Dimitrov V, Mirsaidov U, Aksimentiev A, Timp G. Analyzing the Forces Binding a Restriction Endonuclease to DNA Using a Synthetic Nanopore. Nucleic Acids Res. 2009;37:4170–4179. doi: 10.1093/nar/gkp317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nanopore Based Sequence Specific Detection of Duplex DNA for Genomic Profiling. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczyk SW, Tuijtel MW, Donkers SP, Dekker C. Unraveling Single-stranded DNA in a Solid-state Nanopore. Nano Lett. 2010;10:1414–1420. doi: 10.1021/nl100271c. [DOI] [PubMed] [Google Scholar]
- 16.Spiering A, Getfert S, Sischka A, Reimann P, Anselmetti D. Nanopore Translocation Dynamics of a Single DNA-bound Protein. Nano Lett. 2011;11:2978–2982. doi: 10.1021/nl201541y. [DOI] [PubMed] [Google Scholar]
- 17.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Controlling Protein Translocation Through Nanopores with Bio-Inspired Fluid Walls. Nat Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wanunu M, Bhattacharya S, Xie Y, Tor Y, Aksimentiev A, Drndic M. Nanopore Analysis of Individual RNA/Antibiotic Complexes. ACS Nano. 2011;5:9345–9353. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Reading DNA at Single-nucleotide Resolution with a Mutant MspA Nanopore and phi29 DNA Polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-A Precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic Sensing of Proteins with Receptor-modified Solid-state Nanopores. Nat Nanotechnol. 2012;7:257–263. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- 22.Shim JW, Gu LQ. Encapsulating a Single g-quadruplex Aptamer in a Protein Nanocavity. J Phys Chem B. 2008;112:8354–8360. doi: 10.1021/jp0775911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim JW, Tan Q, Gu LQ. Single-molecule Detection of Folding and Unfolding of the G-quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2009;37:972–982. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding S, Gao C, Gu LQ. Capturing Single Molecules of Immunoglobulin and Ricin with an Aptamer-Encoded Glass Nanopore. Anal Chem. 2009;81:6649–6655. doi: 10.1021/ac9006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawano R, Osaki T, Sasaki H, Takinoue M, Yoshizawa S, Takeuchi S. Rapid Detection of a Cocaine-binding Aptamer Using Biological Nanopores on a Chip. J Am Chem Soc. 2011;133:8474–8477. doi: 10.1021/ja2026085. [DOI] [PubMed] [Google Scholar]
- 26.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J Am Chem Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotem D, Jayasinghe L, Salichou M, Bayley H. Protein Detection by Nanopores Equipped with Aptamers. J Am Chem Soc. 2012;134:2781–2787. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012;12:4895–4900. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosa G, Harbron EJ, Zeng Y, Liu HW, O’Connor DB, Eta-Hosokawa C, Musier-Forsyth K, Barbara PF. Secondary Structure and Secondary Structure Dynamics of DNA Hairpins Complexed with HIV-1 NC Protein. Biophys J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. Nucleic Acid Binding and Chaperone Properties of HIV-1 Gag and Nucleocapsid Proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, et al. Complex Interactions of HIV-1 Nucleocapsid Protein with Oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedzwiecki DJ, Grazul J, Movileanu L. Single-molecule Observation of Protein Adsorption onto an Inorganic Surface. J Am Chem Soc. 2010;132:10816–10822. doi: 10.1021/ja1026858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 Nucleocapsid Protein Bound to the SL3 Psi-RNA Recognition Element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 34.Linial ML, Miller AD. Retroviral RNA Packaging: Sequence Requirements and Implications. Curr Top Microbiol Immunol. 1990;157:125–152. doi: 10.1007/978-3-642-75218-6_5. [DOI] [PubMed] [Google Scholar]
- 35.Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. Effects of the Nature and Concentration of Salt on the Interaction of the HIV-1 Nucleocapsid Protein with SL3 RNA. Biochemistry. 2010;49:3525–3533. doi: 10.1021/bi901279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paoletti AC, Shubsda MF, Hudson BS, Borer PN. Affinities of the Nucleocapsid Protein for Variants of SL3 RNA in HIV-1. Biochemistry. 2002;41:15423–15428. doi: 10.1021/bi026307n. [DOI] [PubMed] [Google Scholar]
- 37.Cornille F, Mely Y, Ficheux D, Savignol I, Gerard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Solid Phase Synthesis of the Retroviral Nucleocapsid Protein NCp10 of Moloney Murine Leukaemia Virus and Related “zinc-fingers” in Free SH forms. Influence of Zinc Chelation on Structural and Biochemical Properties. Int J Pept Protein Res. 1990;36:551–558. [PubMed] [Google Scholar]
- 38.Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mely Y. Nucleic Acid Sequence Discrimination by the HIV-1 Nucleocapsid Protein NCp7: a Fluorescence Study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 39.Summers MF, Henderson LE, Chance MR, Bess JW, Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, III, Hare DR. Nucleocapsid Zinc Fingers Detected in Retroviruses: EXAFS Studies of Intact Viruses the Solution-state Structure of the Nucleocapsid Protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of Packaging Domain Loops in HIV-1 RNA for the Nucleocapsid Protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 41.Yang L. PhD Dissertation. Syracuse University; 2008. NMR Analysis of Structural Features of the HIV-1 Nucleocapsid Protein in Response to Mutation and Interaction with RNA and Drug Candidates. [Google Scholar]
- 42.Famulok M, Hartig JS, Mayer G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 43.Song S, Wang L, Li J, Zhao J, Fan C. Aptamer-Based Biosensor. Trends Anal Chem. 2008;27:108–117. [Google Scholar]
- 44.Abelow AE, Schepelina O, White RJ, Vallee-Belisle A, Plaxco KW, Zharov I. Biomimetic Glass Nanopores Employing Aptamer Gates Responsive to a Small Molecule. Chem Commun (Camb ) 2010;46:7984–7986. doi: 10.1039/c0cc02649b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arrondo JL, Alonso A. Advanced Techniques in Biophysics. Springer-Verlag; Berlin-Heidelberg: 2006. [Google Scholar]
- 46.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Movileanu L, Cheley S, Bayley H. Partitioning of Individual Flexible Polymers into a Nanoscopic Protein Pore. Biophys J. 2003;85:897–910. doi: 10.1016/S0006-3495(03)74529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. Catalyzing the Translocation of Polypeptides Through Attractive Interactions. J Am Chem Soc. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 49.Movileanu L, Schmittschmitt JP, Scholtz JM, Bayley H. Interactions of the Peptides with a Protein Pore. Biophys J. 2005;89:1030–1045. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bikwemu R, Wolfe AJ, Xing X, Movileanu L. Facilitated Translocation of Polypeptides Through a Single Nanopore. J Phys :Condens Matter. 2010;22:454117. doi: 10.1088/0953-8984/22/45/454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagan N, Fabris D. Direct Mass Spectrometric Determination of the Stoichiometry and Binding Affinity of the Complexes between Nucleocapsid Protein and RNA Stem-loop Hairpins of the HIV-1 Psi-recognition Element. Biochemistry. 2003;42:10736–10745. doi: 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
- 52.Tabard-Cossa V, Wiggin M, Trivedi D, Jetha NN, Dwyer JR, Marziali A. Single-Molecule Bonds Characterized by Solid-state Nanopore Force Spectroscopy. ACS Nano. 2009;3:3009–3014. doi: 10.1021/nn900713a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudko OK, Mathe J, Szabo A, Meller A, Hummer G. Extracting Kinetics from Single-Molecule Force Spectroscopy: Nanopore Unzipping of DNA Hairpins. Biophys J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muthukumar M, Baumgartner A. Effects of Entropic Barriers on Polymer Dynamics. Macromolecules. 1989;22:1937–1941. [Google Scholar]
- 55.Niedzwiecki DJ, Mohammad MM, Movileanu L. Inspection of the Engineered FhuA ΔC/Δ4L Protein Nanopore by Polymer Exclusion. Biophys J. 2012;103:2115–2124. doi: 10.1016/j.bpj.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu LQ, Cheley S, Bayley H. Electroosmotic Enhancement of the Binding of a Neutral Molecule to a Transmembrane Pore. Proc Natl Acad Sci U S A. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong CT, Muthukumar M. Polymer Capture by Electro-osmotic Flow of Oppositely Charged Nanopores. J Chem Phys. 2007;126:164903. doi: 10.1063/1.2723088. [DOI] [PubMed] [Google Scholar]
- 58.Chertova EN, Kane BP, McGrath C, Johnson DG, Sowder RC, Arthur LO, Henderson LE. Probing the Topography of HIV-1 Nucleocapsid Protein with the Alkylating Agent N-ethylmaleimide. Biochemistry. 1998;37:17890–17897. doi: 10.1021/bi980907y. [DOI] [PubMed] [Google Scholar]
- 59.Jenkins LM, Byrd JC, Hara T, Srivastava P, Mazur SJ, Stahl SJ, Inman JK, Appella E, Omichinski JG, Legault P. Studies on the Mechanism of Inactivation of the HIV-1 Nucleocapsid Protein NCp7 with 2-mercaptobenzamide Thioesters. J Med Chem. 2005;48:2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- 60.Han A, Creus M, Schurmann G, Linder V, Ward TR, de Rooij NF, Staufer U. Label-free Detection of Single Protein Molecules and Protein-protein Interactions Using Synthetic Nanopores. Anal Chem. 2008;80:4651–4658. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- 61.Osaki T, Suzuki H, Le PB, Takeuchi S. Multichannel Simultaneous Measurements of Single-Molecule Translocation in Alpha-Hemolysin Nanopore Array. Anal Chem. 2009;81:9866–9870. doi: 10.1021/ac901732z. [DOI] [PubMed] [Google Scholar]
- 62.Kleefen A, Pedone D, Grunwald C, Wei R, Firnkes M, Abstreiter G, Rant U, Tampe R. Multiplexed Parallel Single Transport Recordings on Nanopore Arrays. Nano Lett. 2010;10:5080–5087. doi: 10.1021/nl1033528. [DOI] [PubMed] [Google Scholar]
- 63.Atas E, Singer A, Meller A. DNA Sequencing and Bar-Coding Using Solid-state Nanopores. Electrophoresis. 2012;33:3437–3447. doi: 10.1002/elps.201200266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabard-Cossa V, Trivedi D, Wiggin M, Jetha NN, Marziali A. Noise Analysis and Reduction in Solid-state Nanopores. Nanotechnology. 2007;18:305505. [Google Scholar]
- 65.Smeets RM, Keyser UF, Dekker NH, Dekker C. Noise in Solid-state Nanopores. Proc Natl Acad Sci U S A. 2008;105:417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid Electronic Detection of Probe-Specific MicroRNAs Using Thin Nanopore Sensors. Nat Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 67.Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, Henderson LE, Bess JW, Jr, Arthur LO, McDougal JS. Inhibition of HIV-1 Infectivity by Zinc-Ejecting Aromatic C-Nitroso Compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.