Abstract
Methylated lysine and arginine residues on histones represent a crucial part of the histone code and their recognition by protein interaction domains modulates transcription. While some methylating enzymes appear to be specific for histones, many can modify both histone and non-histone substrates, and an increasing number are specific for non-histone substrates. Although some of the non-histone substrates can also be involved in transcription, a distinct subset of protein methylation reactions occurs at residues deeply buried in ribosomal proteins that may function in protein-RNA interactions rather than protein-protein interactions. Additionally, recent work has identified enzymes catalyzing protein methylation reactions on new sites in ribosomal and other proteins, including modifications of histidine and cysteine residues as well as the N terminus.
Key terms: histone methylation, ribosomal protein methylation, PRMTs, SET-domain methyltransferases
Protein methyltransferases: a brief overview
About 1–2% of genes in a variety of prokaryotic and eukaryotic organisms encode methyltransferases and a large fraction of these are specific for modifying protein substrates [1–4]. Most of these enzymes are in the seven-beta-strand [5], SET-domain [6], or SPOUT [7] structural families. While seven-beta-strand methyltransferases catalyze a wide variety of methylation reactions at many different types of residues, the SET-domain enzymes characterized to date are all protein lysine methyltransferases. With the recent exception of one protein arginine methyltransferase [8], SPOUT family enzymes appear to catalyze only RNA methylation reactions [7]. In humans, the largest group of methyltransferases encompasses some 56 SET-domain species [4,9]. Although the majority of these human SET-domain proteins appear to be histone methyltransferases, we are learning that more and more of them can modify other types of proteins. Protein methylation is perhaps most common at lysine and arginine residues (at least in eukaryotic cells). However, there are many other sites for such modification in proteins including histidine, glutamate, glutamine, asparagine, Daspartatel/L-isoaspartate, cysteine, N-terminal, and C-terminal residues [10,11]. Recent studies have now identified the first enzymes specific for catalyzing the methylation of three of these residues: N-terminal [12–13], histidines [14], and cysteines [15]. This review focuses on current advances in protein methyltransferase enzymology in yeast and mammalian systems, with particular emphasis on reactions in ribosomal systems.
Protein lysine methylation: not just for histones
Although most protein lysine methyltransferases are SET-domain family members [4,9], there are an increasing number of examples of seven-beta strand enzymes that catalyze similar reactions [16–19]. These enzymes result in the formation of monomethyl, dimethyl, and trimethyllysine residues. Some enzymes are specific for one or two of these modifications and some result in the formation of all three derivatives [20]. In histone tails, these modified residues are found at the surface of the nucleosome and are recognized by protein interaction domain species that lead to transcriptional activation or repression [17,21–24]. However, in ribosomal proteins these modifications can occur at buried residues that interact directly with rRNA species in the ribosomal interior [25].
The first methyltransferase identified with what would later be designated the SET-domain was the enzyme responsible for the trimethylation of lysine-14 of the large subunit of RUBISCO, the plant enzyme essential for fixing much of the carbon dioxide in the biosphere [26]. At that time, the amino acid sequence of this enzyme showed no similarity with other methyltransferases. Independently, the SET-domain had been previously established from the encoded amino acid sequences of three Drosophila genes associated with development: Su(var)3–9, enhancer of zeste, and trithorax. The realization that the sequences of these domains are similar to those of the plant methyltransferase opened the door to the discovery that the SET-domain represents a catalytic core of protein lysine methyltransferases and that each of these Drosophila proteins catalyze histone lysine methylation (reviewed in [21]). Interestingly, while the function of RUBISCO methylation is still unclear [27], we now know a tremendous amount about the enzymes that methylate histones and their biological role in maintaining and altering the histone code [22–24,28]. Indeed, for many scientists, SET-domain proteins and histone methyltransferases are almost interchangeable terms.
Analyses of the SET-domain proteins in the yeast Saccharomyces cerevisiae, however, revealed that at least half of these methyltransferases recognize non-histone substrates. The yeast genome encodes twelve SET-domain proteins that can be divided in two sequence-related subfamilies of six members each by iterative PSI- and PHI-BLAST searches [29] (Table 1). The first subfamily encodes three proteins that catalyze the methylation of histone proteins: Set1, which modifies lysine 4 of histone H3 [30]; Set2, which modifies lysine 36 of histone H3 [31]; and Set5, which modifies lysines 5, 8, and 12 of histone H4 [32]. No methyltransferase function has yet been assigned for the remaining three members of the group, Set3, Set4, and Set6. Surprisingly, none of the members of the second subfamily of SET-domain proteins in yeast modify histones. This family consists of four enzymes that modify ribosomal large subunit proteins, one enzyme that modifies elongation factor 1A, which brings amino acyl-tRNAs into the ribosome, and one enzyme that modifies cytochrome c (Table 1). These results suggest that the biological role of SET-domain methyltransferases includes important translational as well as transcriptional components.
Table 1.
SET-domain protein lysine methyltransferases in the yeast Saccharomyces cerevisiae
| Protein | Substrate(s) (position given from the mature N- terminal residue unless otherwise indicated) |
Product(s) | Methylated residue surface exposed or buried |
References |
|---|---|---|---|---|
| Subfamily one | ||||
| Set1 (as the catalytic component of the COMPASS complex) | histone H3 Lys-4 Dam1 Lys-233 (from initiator methionine) |
trimethyl dimethyl (possible trimethyl) |
surface unknown |
[30] |
| Set2 | histone H3 Lys-36 | trimethyl | surface | [31] |
| Set3 | no known methyltransferase activity | |||
| Set4 | no known methyltransferase activity | |||
| Set5 | histone H4 Lys-5, Lys-8, Lys12 histone H2A Lys-4, Lys-7 |
monomethyl at all three sites unknown |
surface | [32] |
| Set6 | no known methyltransferase activity | |||
| Subfamily two | ||||
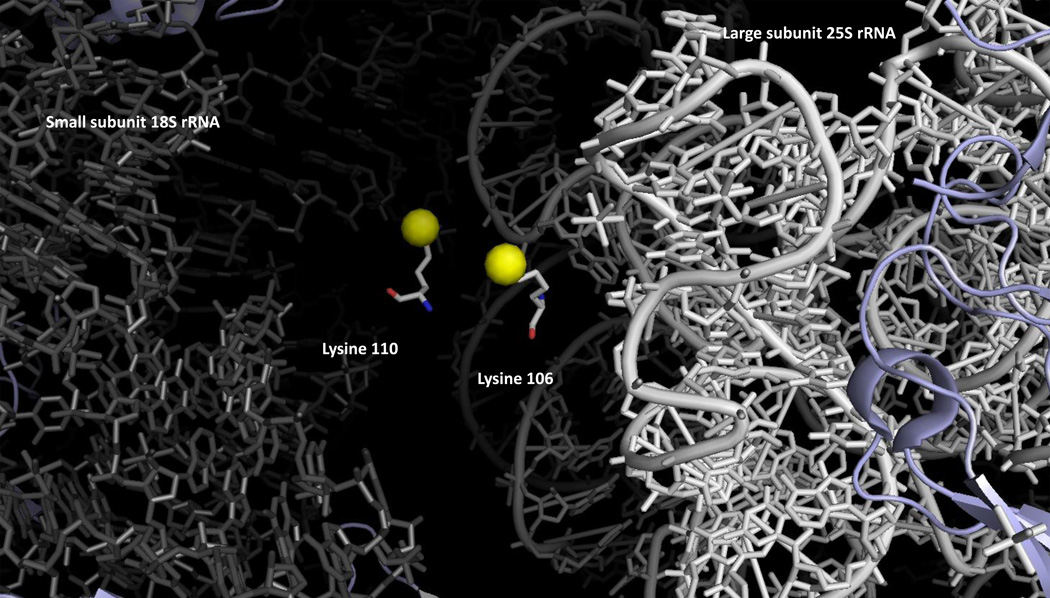
| Rkm1 | ribosomal protein Rpl23ab Lys-105, Lys-109 | dimethyl at both sites | surface (interface between small and large subunits) | [33] |
| Rkm2 | ribosomal protein Rpl12ab Lys-3 | trimethyl | surface | [25] |
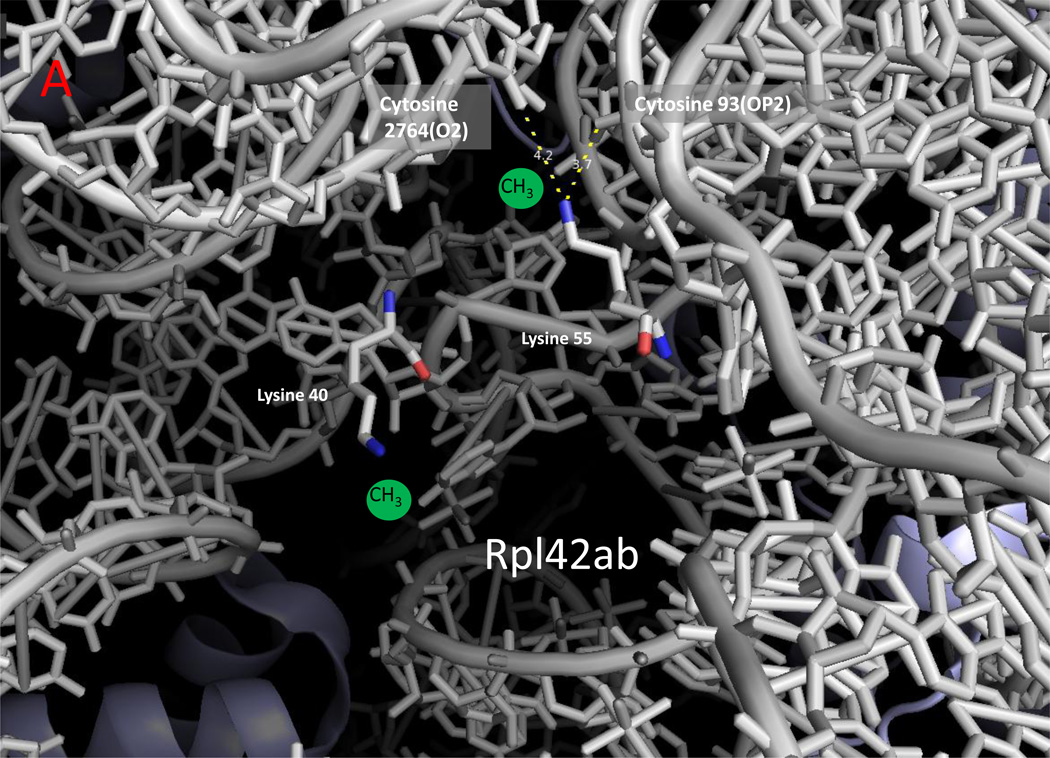
| Rkm3 | ribosomal protein Rpl42ab Lys-39 | monomethyl | buried | [25] |
| Rkm4 | ribosomal protein Rpl42ab Lys-54 | monomethyl | buried (close contacts to 25S rRNA O2 of cytosine 2764 and OP2 of cytosine-93) |
[25] |
| Efm1 | elongation factor eEF1A Lys-30 | monomethyl | surface | [34,35] |
| Ctm1 | iso-1-cytochrome c Lys-72 | trimethyl | surface | [36] |
Analyses of these yeast SET-domain enzymes have revealed that in most cases the enzyme appears to be specific for modifying a single protein substrate, at a single site in the protein sequence, and to a single degree of methylation (Table 1). However, there are exceptions. The Set5 enzyme monomethylates three nearby lysine residues in histone H4 [32] and the Rkm1 enzyme dimethylates two nearby lysine residues in the ribosomal protein Rpl23ab [33]. Perhaps more interestingly, the Set1 methyltransferase (as the catalytic unit of the COMPASS complex) has been shown to form a trimethyllysine residue on histone H3 and dimethyllysine or trimethyllysine residues on the kinetochore Dam1 protein [30]. In this case, it has been suggested these distinct modifications are linked in a regulatory crosstalk that relays changes in chromatin to the apparatus for chromosome segregation [30]. In any case, it is hard to rule out the possibility that other yeast SET-domain enzymes may also modify additional methyl-accepting substrates in the cell.
In humans, the SET-domain family is expanded to some 56 members [4]. Initial analyses indicated that most of these enzymes might be specific for histones, although the evidence was based largely on the comparison of sequence similarities. But current work, discussed later, has now established that many of these enzymes also catalyze protein lysine methylation of non-histone substrates and that some may not recognize histone substrates at all [37]. These results suggest a much greater impact of this class of enzymes on mammalian cellular physiology.
Additionally, other recent studies have demonstrated that an increasing number of protein lysine methyltransferases are non-SET domain enzymes. In yeast, four seven-beta-strand enzymes have been identified, including the Dot1 histone methyltransferase as well as enzymes that modify ribosomal proteins and translational elongation factors (Table 2). In fact, of the twenty-three protein methyltransferases with defined functions in yeast, sixteen modify proteins of the translational apparatus (ribosomal proteins, elongation and release factors) highlighting the broad importance of protein methylation in translation [35,44]. In mammalian cells, additional seven-beta strand protein lysine methyltransferases modify calmodulin [16] and VCP (valosin-containing protein), an ATP-dependent chaperone [19]. Sequence analysis has identified eight additional human gene products as potential protein lysine methyltransferases of this type [19]. It will be interesting to see whether these additional enzymes also have substrates associated with the translational apparatus.
Table 2.
Non-SET domain protein methyltransferases and their established substrates in the yeast Saccharomyces cerevisiae.
| Protein (unless otherwise indicated, all are seven-beta strand methyltransferases) | Substrate and position (given from the mature N-terminal residue, unless otherwise designated) | Product(s) | Methylated residue surface exposed or buried | References |
|---|---|---|---|---|
| Protein lysine methyltransferases | ||||
| Dot1 | histone H3 Lys-79 | mono, di, and trimethyl | surface | [17] |
| See1 | elongation factor eEF1A (Tef1/Tef2) Lys-316 | dimethyl | surface | [34] |
| Efm2 | elongation factor EF3A (Yef3) Lys-186 | trimethyl | unknown | [35] |
| Rkm5 | ribosomal protein Rpl1 Lys-46 | monomethyl | surface | [18] |
| Protein arginine methyltransferases | ||||
| Rmt1 (Hmt1) | many | omega-monomethyl; omega-asymmetric dimethyl | [38] | |
| Rmt2 | ribosomal protein Rpl12ab Arg-66 | delta-monomethyl | protein surface | [39] |
| Hsl7 | unknown | omega-monomethyl; omega-symmetric dimethyl | [40] | |
| Sfm1 (SPOUT family methyltransferase) | ribosomal protein Rps3 Arg-145 | omega-monomethyl | buried - close contacts with 18S rRNA adenine-1427 | [8] |
| N-terminal protein methyltransferase | ||||
| Nmt1 | ribosomal protein Rpl12ab Pro-1 ribosomal protein Rps25ab Pro-1 several others? |
N-dimethyl N-dimethy1 |
surface surface |
[12] |
| Protein histidine methyltransferase | ||||
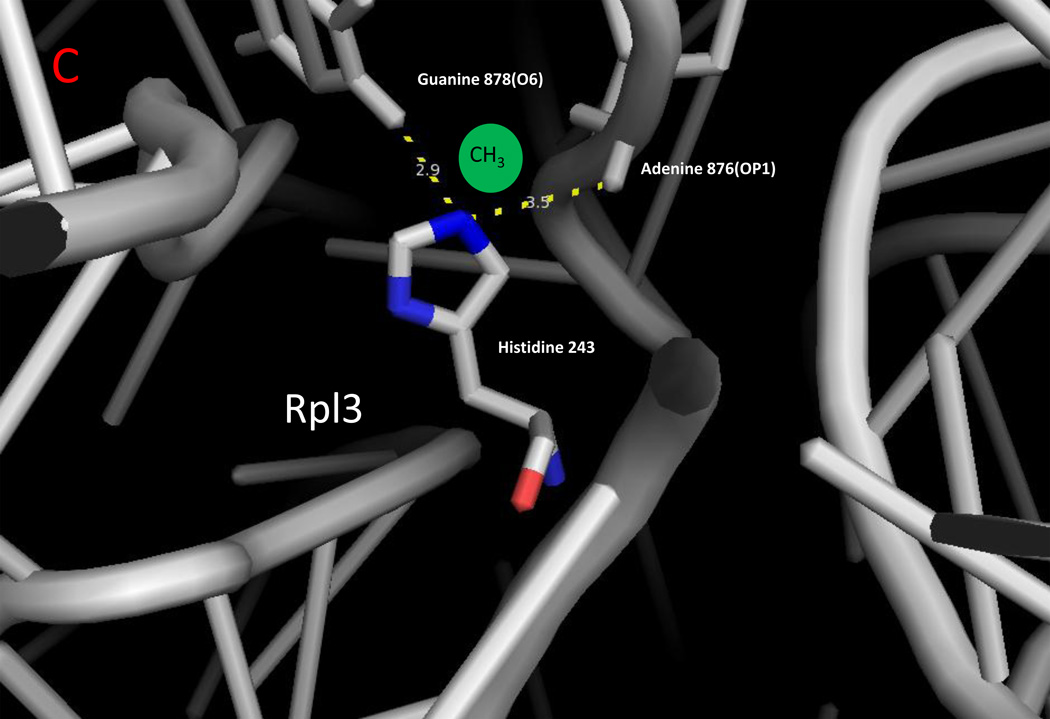
| Hpm1 | Ribosomal protein Rpl3 His-242 | 3-(tau)methyl | buried - contacts with 25S rRNA O6 guranine-878 and OP1 adenine-876 | [14] |
| Protein glutamine methyltransferases | ||||
| Mtq1 | mitochondrial translational release factor Mrf1 Gln-287 (from initiator Met) | N-5-monomethyl amide | surface | [41] |
| Mtq2 | cytoplasmic translational release factor Sup45 Gln-182 | N-5-monomethyl amide | surface | [41] |
| C-terminal protein methyltransferase | ||||
| Ppm1 | Protein phosphatase 2A catalytic subunit C-terminal Leu | methyl ester | surface | [42] |
| C-terminal protein isoprenylcysteine methyltransferase | ||||
| Ste14 (membrane-bound methyltransferase) | Many | C-terminal isoprenylated cysteine methyl ester | surface | [43] |
The diversity of SET-domain enzymes and seven-beta strand methyltransferases specific for protein lysine residues suggest a wide range of physiological roles for the modification reactions that they catalyze. Recent work, described below, has provided evidence that protein lysine methylation may be particularly important not only in the function of proteins involved in translation but in that of non-histone proteins associated with transcriptional processes.
Methylation of ribosomal proteins, transcription factors, and other non-histone proteins at lysine residues
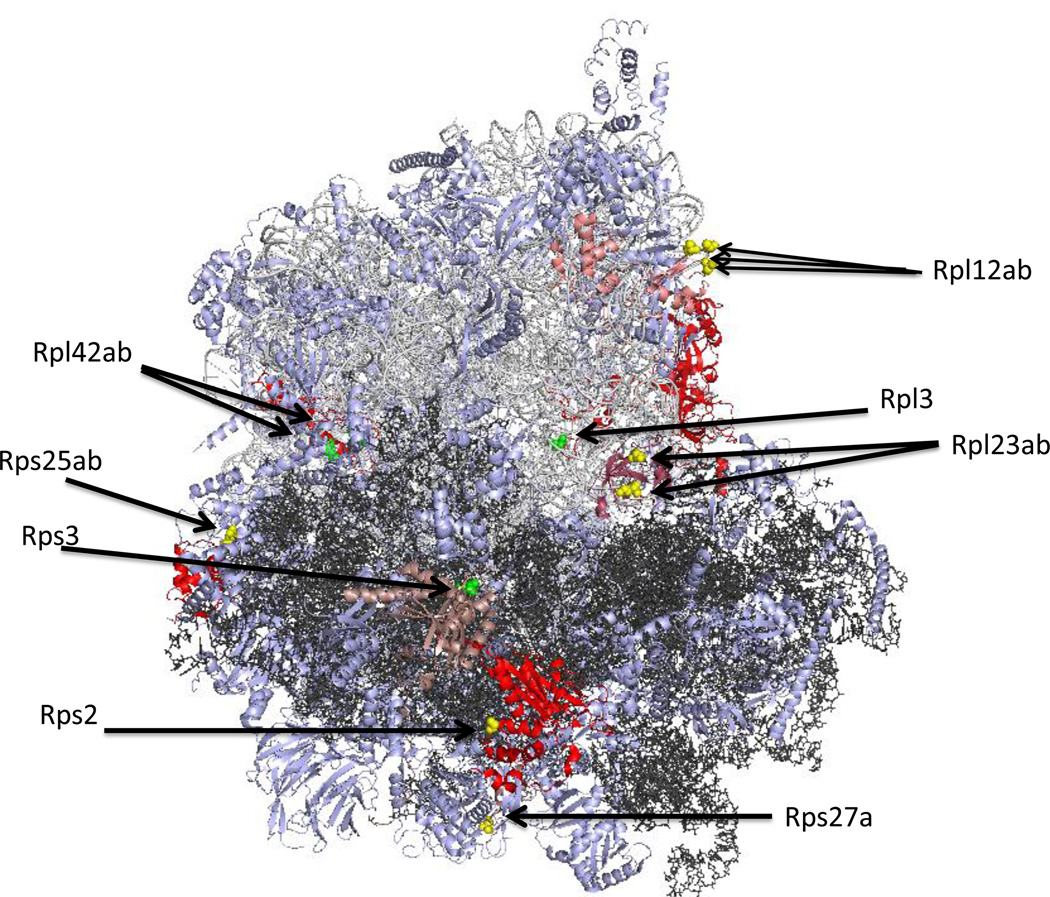
In Saccharomyces cerevisiae, five non-histone proteins are modified at lysine residues by six SET-domain methyltransferases (Table 1) and three non-histone proteins are modified at lysine residues by three seven-beta-strand enzymes (Table 2). Interestingly, with the exception of cytochrome c, all of these proteins are involved in translation, either as ribosomal proteins or elongation factors. The availability of an atomic-resolution structure of the yeast ribosome at 3-Å resolution [45] allows us to map the position of most of the modified residues (Figure 1). While methyl groups were not modeled into this structure, it is clear that some of the methylated sites are exposed to the surface and some of the sites are buried deeply within the ribosome. For example, in this structure, Rpl1 density was not found, presumably because it was easily detached from the ribosomal surface, and the N-terminal tail of Rpl12ab containing the trimethylated lysine-3 appeared to be disordered at the ribosomal surface. Thus, the methylated lysine residues on Rpl1 and Rpl12ab would be expected to be exposed. By contrast, it is apparent that the two methylated residues on Rpl42ab are localized deeply within the ribosomal structure (Figure 2a). Here, the monomethylated Lys-55 residue is modeled with van der Waals contacts with two cytidine residues on the 25S rRNA. For the two dimethyllysine residues on Rpl23ab, it appears that these residues form part of the interface with the small ribosomal subunit, perhaps in a position to interact with the incoming mRNA (Figure 3). These interactions of protein lysine methyl groups with RNA appear to be novel; all other interactions described to date involve proteins [46]. Modification of Rpl42ab presumably occurs before final ribosomal assembly to allow the methyltransferase access to the lysine side chain, although it is possible that dynamic flexibility in ribosomes may allow for the residue to flip out of the ribosomal structure for the methylation reaction.
Figure 1.
Surface and buried sites of methylation on cytoplasmic ribosomal proteins in the yeast Saccharomyces cerevisiae. The 25S ribosomal RNA of the large subunit is shown in light gray; the 18S ribosomal RNA of the small subunit is shown in dark gray. Non-methylated proteins are shown in light blue; methylated proteins (Tables 1 and 2) are shown in pink (Rpl12ab, Rpl23ab, Rps27a, Rps3) and red (Rpl3, Rps2, Rps25ab, Rpl42ab). The approximate positions of surface-exposed methyl groups are shown as yellow spheres; buried methyl groups are represented as green spheres. The illustration was made using PyMOL from the PDB structures 3U5F, 3U5G, 3U5H, and 3U5I [45].
Figure 2.
Zoomed in view of methylated lysine, arginine, and histidine residues with ribosomal RNA in yeast cytoplasmic ribosomes. The monomethylated residues of Rpl42ab ((a); Lys-39; Lys-54), Rps3 ((b); Arg-145), and Rpl3 ((c); His-242) are shown with nitrogen atoms in blue and oxygen atoms in red [8,14,25]. Ribosomal 25S (Panels A and C) and 18S (Panel B) RNA are shown in gray. Methyl groups were not modeled into these structures; possible locations are suggested by the green spheres. Examples of close contact distances (less than 5 Å) are shown between the methylated atom in the protein and the RNA to emphasize the apposition of the methylated residue to the RNA, although these interactions may differ in a refined structure that includes the methyl groups. The illustration was made using PyMOL and the PDB structures 3U5F, 3U5G, 3U5H, and 3U5I [45].
Figure 3.
Intrasubunit localization of the methylated lysine residues of Rpl23ab in yeast cytoplasmic ribosomes. Dimethyllysine residues 105 and 109 are shown with the epsilon amino group as a yellow sphere. These residues are positioned at the interface of the small and large ribosomal subunits; 18S rRNA is shown in gray on the left and 25S rRNA in white on the right. The illustration was made using PyMOL from the PDB structures 3U5F, 3U5G, 3U5H, and 3U5I [45].
It can be speculated that the methyl groups may guide productive interactions between the ribosomal RNA and ribosomal proteins. This may occur by methyl groups blocking unfavorable interactions by steric exclusion or disruption of hydrogen bonding patterns. Additionally, it is now clear that hydrogen atoms on methyl groups attached to nitrogen atoms can themselves be hydrogen bond donors [47] and may thus contribute to new hydrogen bonding networks. For example, such interactions are central to the recognition of trimethyllysine residues by a novel histone H3 binding module [48].
The 56 members of the SET-domain family encoded by the human genome can be divided into ten classes by their amino acid sequences [4]. Nine of these classes, including 53 members, initially were surmised to catalyze histone methylation based on sequence similarity to known protein lysine methyltransferases. Only one class (VII) consisting of 3 members was initially associated with non-histone substrates by their sequence similarity to the RUBISCO methyltransferase [4]. Recent work has shown that the situation is increasingly more complex. In many cases, enzymes have been found to have activity on both histone and non-histone substrates (Table 3). This development is an important one because so many of the initially characterized protein methyltransferases displayed the "tripartite" specificity seen in most of the yeast enzymes in Tables 1 and 2: one protein substrate, one site within that protein, and one level of modification of that site for each enzyme. This is clearly not the case for the nine mammalian SET-domain methyltransferases described in Table 3. Eight of these enzymes (EZH2, SETD1A, EHMT1/2, SETDB1, SMYD3, SETD6, SETD8, and SETD7) clearly methylate both histone and non-histone substrates. An additional enzyme, SMYD2, modifies multiple non-histone substrates. The rapid pace of discovery of novel non-histone protein substrates for these enzymes suggests that we may only be at the beginning of understanding the true range of their methyl-accepting substrates. The recognition of multiple substrates for protein lysine methyltransferases has certainly opened a Pandora's box. How many additional substrates have been missed in characterizing these protein lysine methyltransferases? May other SET-domain enzymes thought to only modify a single substrate and site in fact be capable of modifying additional proteins or additional sites in the protein in the cell?
Table 3.
Mammalian SET-domain protein methyltransferases that have been shown to modify non-histone substrates
| Protein | SET-domain family [4] |
Substrate and methylation position (from the mature N- terminus unless otherwise indicated) |
References |
|---|---|---|---|
|
Enzyme(s) that methylate histones and non-histone substrates |
|||
| EZH2 (KMT6) | Class I - EZ | histone H3 Lys-27 retinoic acid-related orphan nuclear receptor α (RORα) Lys-38 transcription factor GATA4 Lys-299 |
[49,50] [49] [50] |
| SETD1A (KMT2F) | Class II - SET2 | histone H3 Lys-4 heat-shock protein 70 (HSP70; HSPA1A) Lys-560 (dimethyl) |
[51] [52] |
| EHMT1 (KMT1D)(GLP)/ EHMT2 (KMT1C)(G9A) |
Class V - Suvar-3-9 - histone H3 Lys-9 |
histone H1 histone H3 Lys-9 (mono and dimethyl) histone H3 Lys-27 chromodomain Y-like protein (CDYL1) Lys- 135 (mono, di, and trimethyl) widely-interspaced zinc finger-containing protein (WIZ) Lys- 1162 (di and trimethyl) apoptotic chromatin condensation inducer in the nucleus (ACINUS) Lys-654 (di and trimethyl) p53 transcription factor Lys-373 (dimethyl) MyoD transcription factor Lys-104 |
[53] [53] [53] [54] [54] [54] [55] [56] |
| SETDB1 | Class V - Suvar-3–9 - histone H3 Lys-9 |
histone H3 Lys-9 HIV1 Tat transcriptional activator Lys-50, Lys- 51 |
[57] |
| SMYD3 | Class VI – SMYD | histone H4 Lys-5 Vascular endothelial growth factor receptor 1 tyrosine kinase Lys- 831 |
[58] [59] |
| SETD6 | Class VII - SET6 | histone H2AZ Lys-7 RelA subunit of NF- kappa B transcription factor Lys-310 (monomethyl) serine/threonine protein kinase PLK1 serine/threonine protein kinase PAK4 |
[60] [61] [61] [61] |
| SETD8 (SET8) (KMT5A)(PrR/SET7) |
PR/SET | histone H4 Lys-20 (monomethyl) p53 transcription factor Lys-382 |
[62] [62,63] |
| SETD7 (KMT7) (SET7/9)(SET7)(SET9) |
SET7 | A wide variety of substrates including: histone H2A histone H2B DNMT1 DNA methyltransferase Lys-142 estrogen receptor-α p53 transcription factor Lys-372 TFIID protein TAF10 Lys-189 FOXO3 Lys-270 |
[37,64,65] |
|
Enzymes that methylate only non- histone substrates |
|||
| SMYD2 (KMT3C) | Class VI - SMYD | monomethylation at: p53 Lys-370 Hsp90 Lys-209, Lys- 615 RB1 retinoblastoma- associated protein Lys-850 |
[66] [67] [68] |
What can we conclude at this point on the nature of the identified non-histone substrates compiled in Table 3? It appears that the apple does not fall far from the tree here; many of these non-histone substrates are also involved in transcriptional control. Significantly, thirteen of these lysine methyl-acceptors are transcription factors or closely associated with transcription factors: RORα, GATA4, CDYL1, WIZ, p53, myoD, HIV Tat protein, the RelA subunit of NF-κB, DNMT1, estrogen receptor-α, TFIID protein 10, FOXO3, and RB1 protein (Table 3). Of the remaining six non-histone substrates, one is involved in apoptosis, two are heat shock proteins, one is a receptor tyrosine protein kinase, and two are serine/threonine protein kinases involved in cell cycle control (Table 3). Certainly more substrates remain to be identified and it will be of interest to see if the majority of them are also associated with transcriptional regulation. Interestingly, as of yet no mammalian ribosomal proteins have been found to be modified by SET-domain methyltransferases, suggesting that different organisms may use protein lysine methyltransferases to different ends. However, less is known about mammalian ribosomal methylation and it will be instructive to identify the enzymes responsible for the known sites of methylation on lysine residues, including Lys-4 of RL29 and Lys-22 of RL40 in rat ribosomes [69].
How substrate specificity is determined in SET-domain enzymes has been explored in several recent papers. A study with peptide substrate arrays for the human SETD8 protein that methylates both histones and p53 (Table 3) revealed a 7-residue consensus sequence R-H-R/K/Y-K-V/I/L/F/Y-R [62]. Searching human sequences for this consensus sequence identified a number of new candidate substrate proteins, but interestingly none of these appeared to be recognized by SETD8 [62]. A similar approach for the SETD7 methyltransferase was more successful, leading to the identification of nine new non-histone substrates [64]. An alternative approach to identifying new substrates involves the utilization of protein arrays. This approach was used with success to identify new substrates for SETD7 and SETD6 [70] in arrays containing over 9,500 human proteins. Finally, it is possible to test banks of SET-domain enzymes on specific substrates to identify which enzyme is responsible for a specific modification [61].
This review has concentrated on enzymes involved in lysine methylation in yeast and mammalian cells. However, one interesting enzyme from a prokaryote is worthy of mention. The most promiscuous protein lysine methyltransferase may be an enzyme found in the hyperthermophilic archaeal species Sulfolubus islandicus [71]. This enzyme is a seven-beta strand enzyme that appears to modify numerous proteins at multiple sites within each protein species. The large-scale conversion of lysine residues to dimethyllysine appears to be associated with the resistance of a protein to heat denaturation [71]. Whether such stabilization occurs for the eukaryotic proteins described here, including the proteins of the translational apparatus, remains to be seen.
At this point, our knowledge of the importance of protein lysine methylation is best established for the regulation of chromatin function although we are beginning to understand better the role of this modification in translation. However, protein methylation occurs at many other residues and may play similar or distinct roles in a wide variety of systems. For all of the modifications described below, recent evidence has pointed to roles in ribosomal structure and function, including additional examples suggesting direct interactions of methylated residues and RNA.
Non-histone protein methylation at arginine residues
Protein arginine methylation has been well studied in both yeast and mammalian systems (for recent reviews, see Refs. [72–74]). In histones and in non-histone proteins, ω-monomethyl, ω-asymmetric, and ω-symmetric dimethylated residues are recognized by tudor protein interaction domains [74], much in the same way as methylated lysine residues are recognized. In mammalian cells, these methylation reactions are catalyzed by a sequence-related family of nine seven-beta-strand methyltransferases designated protein arginine methyltransferase 1 (PRMT1) to PRMT9. Six of these enzymes (PRMT1, 2, 3, 4, 6, and 8) have been shown to catalyze asymmetric dimethylation while one enzyme (PRMT5) has been shown to catalyze symmetric dimethylation [72,74]. One enzyme (PRMT7) appears unique in that it may only catalyze ω-monomethylation [75], while the specificity of PRMT9 (4q31) [76] (erroneously designated as PRMT10 in UniProt) has not been established. The specificity of these enzymes for protein substrates is generally much broader than that of the protein lysine methyltransferases. For all of the enzymes for which activity has been shown, they can modify multiple substrates, often at multiple sites within a given protein [72]. In yeast, a smaller family of three seven-beta-strand enzymes includes Rmt1, the homolog of PRMT1/2/3/4/6/8, Hsl7, the homolog of PRMT5, and a distinct enzyme (Rmt2) that catalyzes the specific modification of the bridge, or δ-guanidino nitrogen atom, in an arginine residue of the large subunit ribosomal protein Rpl12ab (Table 2; [39,73]). Interestingly Sfm1, an enzyme of the SPOUT family, whose members generally modify RNA species [7], has been recently shown to catalyze the ω-monomethylation of an arginine residue in the ribosomal small subunit protein Rps3 [8]. Although the methylated arginine residue in Rps3 is on the surface of this protein, it does not contact the surface of the ribosome or other ribosomal proteins. Rather, the methylated site is buried within the ribosomal RNA and makes close contact with the nitrogen atoms on adenine-1427 [8] (Figure 2b). Genes encoding orthologs of the Rmt2 and Sfm1 enzymes do not appear to be found in animal species. However, the discovery of these proteins does suggest that the family of protein arginine methyltransferases may be broader than previously imagined.
Unlike protein lysine methylation, for which much of the interest and work has centered on histone substrates, protein arginine methylation has been studied extensively not only with histone substrates and transcriptional control, but with substrates involved in signal transduction, DNA repair, and RNA splicing [72–74].
Present challenges in protein arginine methylation include better defining the substrate specificity of mammalian PRMT7 and PRMT9 (4q31) enzymes and determining whether additional enzymes are encoded by mammalian genomes. As described above, mammals lack genes encoding proteins with amino acid sequences similar to the S. cerevisiae Rtm2 and Sfm1 protein arginine methyltransferases [8,39]. It was previously suggested that the mammalian FBXO10 and FBXO11 proteins had PRMT activity [77], but these claims have not been supported by further work [72]. Finally, it is clear that PRMTs function in the cytoplasm and in the nucleus [78]. However, strong evidence for methylation of rat luminal Golgi proteins [79] suggests that one or more of the existing mammalian PRMTs can localize to the Golgi or that at least one novel enzyme is present.
Histidine methylation
The modification of protein histidine residues by methylation of the N-1(π) or N-3(τ) atoms of the imidazole ring has been established for a small group of proteins in prokaryotic and eukaryotic cells. These proteins include mammalian actin, myosin heavy chains, myosin light chain kinase, a migration inhibitor factor related protein, and the alpha chain of methyl-coenzyme M reductase (see [14] for a review). Much of the attention has been focused on the role of the widely-conserved N-3 methylation of histidine-73 of actin [80]. The recent discovery that histidine-242 in the "tryptophan finger" of the cytoplasmic yeast ribosomal protein Rpl3 is methylated on the N-3 position allowed for the identification of the first enzyme catalyzing this process [14]. Significantly, this residue is buried deep within the ribosomal 25S RNA; the methylated N-3 atom has close contacts with the O6 atom of guanine-878 and the OP1 atom of adenine-876 (Figure 2c). This site is near the ribosomal A-site and the peptidyltransferase center and may be important in the "rocker switch" coordinating the binding and dissociation of the incoming aminoacyl-tRNA-elongation factor 1A complex [81]. The human ortholog of this enzyme is C1orf156; it is presently unknown whether this enzyme is responsible for the modification of any of the known mammalian proteins methylated at the N-1 or N-3 positions of histidine residues.
Cysteine methylation
Methylation of cysteine residues can occur in enzymes that transfer methyl groups from alkylated DNA in a repair reaction [82], in intermediate steps of catalysis [83] and in automethylation reactions [84]. However, the first example of a protein cysteine methyltransferase with a separate methyl-accepting substrate was the NleE protein of a pathogenic strain of E. coli [15]. This enzyme is secreted into the mammalian host and modifies a cysteine residue in a four-cysteine zinc cluster in the TAB2 and TAB3 adapter proteins involved in NF-κB signaling. Methylation destabilizes this zinc-cysteine cluster and disrupts the signaling pathway that would normally lead to the inflammatory response against the bacterium.
Another example of methylation of cysteine residues in a zinc-cysteine cluster was recently found in the Rps27a protein of the small subunit of the yeast cytoplasmic ribosome [8]. No evidence has been found yet for a methyltransferase activity that might be responsible for this modification. Indeed, based on the similarity of the methylated zinc-cysteine cluster in Rps27a to one in the N-terminal domain of the E. coli Ada protein involved in repair of DNA phosphotriester damage, it has been speculated that the unmodified form of Rps27a may also be involved in DNA repair by serving as an acceptor site for an unwanted DNA methyl group in a non-enzymatic scavenging reaction [8].
Ribosomal protein methylation has now been shown to occur at lysine, arginine, histidine, and cysteine residues. Recent work has demonstrated enzymes catalyzing one additional site of methylation, at the N terminus, that appears to be more widely dispersed in nature.
Protein modification at the N terminus in eukaryotes
N-terminal methylation has long been established for a small group of prokaryotic and eukaryotic proteins, and it was predicted over 25 years ago that the eukaryotic methyltransferase would recognize Xxx-Pro-Lys sequences [85]. This alpha N-terminal protein methyltransferase, designated Ntm1 in yeast and NTMT1 in humans, was finally identified in two recent studies: an analysis of large subunit ribosomal proteins in yeast [12] and of the regulator of chromatin condensation 1 (RCC1) protein in humans [13]. Both studies confirmed the Xxx-Pro-Lys specificity and pointed to the large range of possible new substrates. A recent study has shown even more relaxed substrate specificity for this methyltransferase, but confirmed that the initially identified motif is the preferred substrate [86]. The specificity of this enzyme suggests that a large group of proteins may be methylated by this enzyme [12,13,86]. It has been hypothesized that the quaternization of the N-terminal alpha nitrogen atom by methylation results in a fixed positive charge that would allow the N terminus to maintain its charge even in hydrophobic environments. The methylated N terminus may be recognized by specific protein binding domains. Alternatively, such methylation might interfere with the recognition of other modified residues near the N terminus by protein binding domains.
Conclusions and future directions
While it appears that we may have identified already many (and perhaps most) of the genes encoding protein methyltransferases, we are still near the beginning of establishing the range of their physiological targets. Newer approaches using peptide arrays and protein arrays appear to be powerful, especially when combined with the validation of sites in vivo. It has also become clear that "crosstalk" between modification pathways could be a general phenomenon, so that it might be essential to understand the pathways not only leading to methylation of specific sites on a "naked" protein but also those leading to methylation of substrates that have been previously modified by acetylation, phosphorylation, or other posttranslational modifications. It has also been of interest to see the examples described here where methylated sites on lysine, arginine, and histidine residues of yeast ribosomal proteins are poised to interact with rRNA. While there are many cases where protein methylation interactions have been shown to facilitate or block protein-protein interactions [46], the knowledge that methylated residues interact with RNA opens new windows to understanding the functional range of protein methylation.
Acknowledgments
This work was supported by US Public Health grant GM026020. I thank Qais Al-Hadid for his help in the preparation of the figures, and to my colleagues for their expert advice, including Albert Courey, Alexander Patananan, Jonathan Lowenson, and Lauren Budenholzer. I am especially indebted to Brian Young, Anne McBride, David Weiss, Cecilia Zurita-Lopez, Kristofor Webb, and Tanya Petrossian for their contributions to the studies from our laboratory reported here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katz JE, et al. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics. 2003;2:525–540. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Petrossian TC, Clarke S. Bioinformatic identification of novel methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wlodarski T, et al. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS ONE. 2011 doi: 10.1371/journal.pone.0023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol. Cell. Proteomics. 2011 doi: 10.1074/mcp.M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubert HL, et al. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tkaczuk KL, et al. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007 doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young BD, et al. Identification of methylated proteins in the yeast small ribosomal subunit: A role for SPOUT methyltransferases in protein arginine methylation. Biochemistry. 2012;51:5091–5104. doi: 10.1021/bi300186g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richon VM, et al. Chemogenetic analysis of human protein methyltransferases. Chem. Biol. Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 10.Clarke SG, Tamanoi F, editors. The Enzymes Volume 24: Protein Methyltransferases. Amsterdam: Academic Press; 2006. pp. 3–570. [Google Scholar]
- 11.Walsh CT. Posttranslational Modifications of Proteins: Expanding Nature's Inventory. Englewood Colorado: Roberts and Company; 2006. Protein Methylation; pp. 121–149. [Google Scholar]
- 12.Webb KJ, et al. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry. 2010;49:5225–5235. doi: 10.1021/bi100428x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tooley CE, et al. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466:1125–1128. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb KJ, et al. A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J. Biol. Chem. 2010;285:37598–37606. doi: 10.1074/jbc.M110.170787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature. 2012;481:204–210. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 16.Magnani R, et al. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nature Commun. 2010 doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Develop. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webb KJ, et al. The ribosomal L1 protuberance in yeast is methylated on a lysine residue catalyzed by a seven-β-strand methyltransferase. J. Biol. Chem. 2011;286:18405–18413. doi: 10.1074/jbc.M110.200410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kernstock S, et al. Lysine methylation of VCP by a member of a novel protein methyltransferase family. Nature Commun. 2012 doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 20.Couture JR, et al. Structural origins for the product specificity of SET domain protein methyltransferases. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 22.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black JC, et al. Histone lysine methylation dynamics; establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb KJ, et al. Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J. Biol. Chem. 2008;283:35561–35568. doi: 10.1074/jbc.M806006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein RR, Houtz RL. Cloning and developmental expression of pea ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol. Biol. 1995;27:249–261. doi: 10.1007/BF00020181. [DOI] [PubMed] [Google Scholar]
- 27.Houtz RL, et al. Co- and post-translational modifications in Rubisco: unanswered questions. J. Exp. Bot. 2008;59:1635–1645. doi: 10.1093/jxb/erm360. [DOI] [PubMed] [Google Scholar]
- 28.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Porras-Yakushi TR, et al. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at the N-terminus. J. Biol. Chem. 2006;281:35835–35845. doi: 10.1074/jbc.M606578200. [DOI] [PubMed] [Google Scholar]
- 30.Lantham JA, et al. Chromatin signaling to kinetochores: Transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs SM, et al. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J. Biol. Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green EM, et al. Methylation of H4 lysines 5, 8, and 12 by yeast Set5 calibrates chromatin stress responses. Nature Struct. Mol. Biol. 2012;19:361–363. doi: 10.1038/nsmb.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porras-Yakushi TR, et al. Yeast ribosomal/cytochrome c SET domain methyltransferase subfamily: Identification of Rpl23ab methylation sites and recognition motifs. J. Biol. Chem. 2007;283:12368–12376. doi: 10.1074/jbc.M611896200. [DOI] [PubMed] [Google Scholar]
- 34.Lipson RS, et al. Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2010;500:137–143. doi: 10.1016/j.abb.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couttas TA, et al. Methylation of translation-associated proteins in Saccharomyces cerevisiae: Identification of methylated lysines and their methyltransferases. Proteomics. 2012;12:960–972. doi: 10.1002/pmic.201100570. [DOI] [PubMed] [Google Scholar]
- 36.Polevoda B, et al. Cytochrome c methyltransferase, Ctm1p, of yeast. J. Biol. Chem. 2000;275:20508–20513. doi: 10.1074/jbc.M001891200. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genetics Develop. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Jackson CA, et al. Proteomic analysis of interactors for yeast protein arginine methyltransferase Hmt1 reveals novel substrate and insights into additional biological roles. Proteomics. 2012;12:3304–3314. doi: 10.1002/pmic.201200132. [DOI] [PubMed] [Google Scholar]
- 39.Chern MK, et al. Yeast ribosomal protein L12 is a substrate of protein-arginine methyltransferase 2. J. Biol. Chem. 2002;277:15343–15353. doi: 10.1074/jbc.M111379200. [DOI] [PubMed] [Google Scholar]
- 40.Sayegh J, Clarke SG. Hsl7 is a substrate-specific type II protein arginine methyltransferase in yeast. Biochem. Biophys. Res. Commun. 2008;372:811–815. doi: 10.1016/j.bbrc.2008.05.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polevoda B, et al. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006;281:2562–2571. doi: 10.1074/jbc.M507651200. [DOI] [PubMed] [Google Scholar]
- 42.Leulliott N, et al. Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity. J. Biol. Chem. 2004;279:8351–8358. doi: 10.1074/jbc.M311484200. [DOI] [PubMed] [Google Scholar]
- 43.Hahne K, et al. Evaluation of substrate and inhibitor binding to yeast and human isoprenylcysteine carboxyl methyltransferases (Icmts) using biotinylated benzophenone-containing photoaffinity probes. Biochem. Biophys. Res. Commun. 2012;423:98–103. doi: 10.1016/j.bbrc.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polevoda B, Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Shem A, et al. The structure of the eucaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 46.Erce MA, et al. The methylproteome and the intracellular methylation network. Proteomics. 2012;12:564–586. doi: 10.1002/pmic.201100397. [DOI] [PubMed] [Google Scholar]
- 47.Horowitz S, Trievel RC. Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem. 2012;287:41576–41582. doi: 10.1074/jbc.R112.418574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwase S, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nature Struct. Mol. Biol. 2011;18:769–777. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JM, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 50.He A, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Develop. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tate CM, et al. CXXC finger protein 1 restricts the Setd1a histone H3K4 methyltransferase complex to euchromatin. FEBS J. 2010;277:210–223. doi: 10.1111/j.1742-4658.2009.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho HS, et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nature Commun. 2012 doi: 10.1038/ncomms2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Develop. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rathert P, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nature Chem. Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ling BMT, et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pagans S, et al. Characterization of HIV Tat modifications using novel methyl-lysine-specific antibodies. Methods. 2010;53:91–96. doi: 10.1016/j.ymeth.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Aller GS. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunizaki M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 60.Binda A, et al. SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics. 2013;8:1–7. doi: 10.4161/epi.23416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy D, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nature Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kudithipudi S, et al. The SET8 H4K20 protein lysine methyltransferase has a long recognition sequence covering seven amino acid residues. Biochimie. 2012;94:2212–2218. doi: 10.1016/j.biochi.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 63.Shi X, et al. Modulation of p53 function by SET8-mediated p53 methylation at lysine 382. Mol. Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhayalan A, et al. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 65.Calnan DR, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging. 2012;4:462–479. doi: 10.18632/aging.100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferguson AD, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 67.Abu-Farna, et al. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J. Mol. Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 68.Cho HS, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williamson NA, et al. Post-translational processing of rat ribosomal proteins: Ubiquitious methylation of Lys22 within the zinc-finger motif of RL40 (carboxyl-terminal extension protein 52) and tissue-specific methylation of Lys4 in RL29. Eur. J. Biochem. 1997;246:786–793. doi: 10.1111/j.1432-1033.1997.00786.x. [DOI] [PubMed] [Google Scholar]
- 70.Levy D, et al. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin. 2011 doi: 10.1186/1756-8935-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chu Y, et al. Identification and characterization of a highly conserved crenarchael protein lysine methyltransferase with broad substrate specificity. J. Bacteriol. 2012;194:6917–6926. doi: 10.1128/JB.01535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Low JKK, Wilkins MR. Protein arginine methylation in Saccharomyces cerevisiae. FEBS J. 2012;279:4423–4443. doi: 10.1111/febs.12039. [DOI] [PubMed] [Google Scholar]
- 74.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nature Rev. Cancer. 2012;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 75.Zurita-Lopez CI, et al. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J. Biol. Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J, et al. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 77.Krause CD, et al. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Therapeut. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 78.Hermann, et al. Human protein arginine methyltransferases in vivo – distinct properties of eight canonical members of the PRMT family. J. Cell Sci. 2009;122:667–677. doi: 10.1242/jcs.039933. [DOI] [PubMed] [Google Scholar]
- 79.Wu CC, et al. Organellar proteomics reveals Golgi arginine dimethylation. Mol. Biol. Cell. 2004;15:2907–2919. doi: 10.1091/mbc.E04-02-0101. Correction Mol. Biol. Cell 15 (11) page 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nyman T, et al. The role of MeH73 in actin polymerization and ATP hydrolysis. J. Mol. Biol. 2002;317:577–589. doi: 10.1006/jmbi.2002.5436. [DOI] [PubMed] [Google Scholar]
- 81.Meskauskas A, Dinman JD. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nuc. Acids Res. 2010;38:7800–7813. doi: 10.1093/nar/gkq641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sedgwick B, et al. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 83.Boal AK, et al. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siddique AN, et al. Auto-methylation of the mouse DNA-(cytosine C5)-methyltransferase Dnmt3a at its active site cysteine residue. FEBS J. 2011;278:2055–2063. doi: 10.1111/j.1742-4658.2011.08121.x. [DOI] [PubMed] [Google Scholar]
- 85.Stock A, et al. N-terminal methylation of proteins: structure, function, and specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 86.Petkowski JJ, et al. Substrate specificity of mammalian N-terminal alpha-amino methyltransferase. Biochemistry. 2012;51:5942–5950. doi: 10.1021/bi300278f. [DOI] [PMC free article] [PubMed] [Google Scholar]