Abstract
The environmental heavy metal toxicant, lead (Pb), has been shown to be more harmful to the central nervous system (CNS) of children than to adults, given that Pb exposure affects the neural system during development. Because growth factors and cytokines play very important roles in development of the CNS, we have examined the impact of Pb exposure on the expression of cytokines during CNS development. Cytokine expression was studied in post-natal-day 21(pnd21) mice by microarray, real-time RT-PCR, Luminex, and ELISA methodologies. BALB/c mouse pups were exposed to Pb through the dam’s drinking water (0.1 mM Pb acetate), from gestation-day 8 (gd8) to pnd21. Two cytokines, interleukin-6 (IL-6) and transforming growth factor-β1 (TGF-β1), displayed significantly changed transcript levels in the presence of Pb. IL-6 and TGF-β1 both have signal transduction cascades that can cooperatively turn on the gene for the astrocyte marker glial-fibrillary acidic protein (GFAP). Microarray results indicated that Pb exposure significantly increased expression of GFAP. Pb also modulated IL-6, TGF-β1, and IL-18 protein expression in select brain regions. The deleterious effects of Pb on learning and long-term memory are posited to result from excessive astrocyte growth and/or activation with concomitant interference with neural connections. Differential neural expression of cytokines in brain regions needs to be further investigated to mechanistically associate Pb and neuroinflammation with behavioral and cognitive changes.
Keywords: astrocyte, brain, central nervous system, cytokine, lead, neuron, signal transduction, transcription
INTRODUCTION
The molecular mechanisms involved in lead (Pb)-induced neurotoxicity have not been fully elucidated. However, it is evident that Pb exposure during development can have serious, long-lasting, deleterious effects on the CNS [1]. The detrimental neurobehavioral outcomes due to Pb (blood level <10 µg/dl) exposure are more substantial during development, since similar detriments are not observed after adult exposures. Pb exposure during development lowers IQ, causes aggressive behavior, and impairs neuromuscular function [2]. Studies in mice and rats have indicated that absorption of Pb is greater in pups than in adult animals [3]. Furthermore, the absorption rate of Pb in pups was highest while they suckled milk from dams drinking Pb-containing water [4,5]. Young animals are likely to be more vulnerable to the toxic effects of Pb, because the blood brain barrier is not fully formed until later in postnatal development; as a result, Pb can easily pass into developing neural tissue [3] and directly affect neuronal synapses or indirectly affect neuronal connections via glial modulations.
Experiments performed in animals and in cell culture have demonstrated that Pb can have dramatic effects on growth and development of CNS cells. Studies with young rats have shown that Pb-exposed pups had fewer neurons, glial cells, and synapses per neuron than did their untreated counterparts [6]. Pb exposure has been reported to degenerate the myelin sheath [7], enhance basal release of neurotransmitter from presynaptic nerve endings [8], induce aberrant neural growth [9], increase proliferation and enlargement of astrocytes, and enhance breakdown of the blood-brain barrier [8,10].
Neonatal Pb exposure exacerbated sickness behavior in pups infected with Listeria monocytogenes; such sickness behavior was detected as loss of appetite and drinking, decreased body-weight gain, and lack of mobility [11]. Effects of Pb on sickness behavior suggested that Pb interactions within the CNS can influence peripheral immune responses [12]. Cytokines, such as IL-1, IL-6, and tumor necrosis factor -α (TNF-α), are the primary mediators of sickness behavior and are able to enter the brain via transport through the blood-brain barrier [13]. These proinflammatory cytokines can activate the hypothalamic-pituitary-adrenal axis, resulting in elevation of adrenocorticotropin hormone and glucocorticoids [14–16]. Increased levels of IL-6 correlate with fever induction after infection by a variety of pathogens [17,18]. Pb exposure increased lipopolysaccaride (LPS) induced IL-6 mRNA and protein levels in mouse brains [19].
It is now known that a number of cytokines are synthesized directly by cells within the CNS [20–23]. For instance, the pyrogenic cytokine IL-6 appears to be synthesized by CNS cells that include: astrocytes, oligodendrocytes, microglia, and several types of neuronal cells [24,25]. Most of the evidence for this conclusion originates from experiments involving cultured CNS cells. Interleukin-11 (IL-11) was detected in mouse brain by a variety of methods including: ribonuclease protection assay, in situ hybridization, RT-PCR, and western blot analysis [26]. IL-11 was found in hippocampus and spinal cord neurons consisting of motor neurons and preganglionic cells of the autonomic nervous system. Many of the cytokines expressed by cells in the brain are produced by microglial cells [22,24], such as IL-15 [27]. The IL-15 receptor complex has been detected on brain microglial cells, indicating that IL-15 has autocrine activity [28,29]. Expression of the proinflammatory cytokine, IL-18, its receptor IL-18R (IL-1R5/IL-1R7), and regulatory molecules, AcPL and IL-18BP, have been detected, by means of RT-PCR, in rat hypothalami [30–32]. Interestingly, in murine primary mixed glial cultures, IL-18 induced intracellular expression of IL-1α and IL-1β, as well as the release of IL-6 [33]. It has been observed that normal brain levels of IL-1β are very low, whereas IL-18 is expressed constitutively at relatively high levels [34]. Microglia, astrocytes, and oligodendrocytes express IL-18 [35,36]. Some cytokines also have neuronal forms; for example IL-16 has an elongated neuronal form (nIL-16), which interacts selectively with neuronal ion channels [37]. NIL-16 mRNA has been detected only in neurons within the cerebellum and hippocampus [37]. Cytokines of the TGF-β family (TGF-β1, -β2, and -β3) also are synthesized by cells of the CNS. Experiments performed in culture and by mRNA analysis of CNS-derived cells have indicated that TGF-βs are synthesized by glial (glioblastoma cells, astrocytes, and microglia) and neuronal cells [38,39]. IL-10 and IL-13 are synthesized at detectable levels in the brain only in disease states or as a result of injury; the known effects of these cytokines are reduction of inflammation and promotion of cell protection, via a shift toward a T-cell helper type 2 (Th2) immune response [22,23]. The cytokine IL-12 (p70) is also present at very low levels in the CNS, except during infection by various pathogens, or in autoimmune disease.
To address Pb effects on cytokine gene expression, we developmentally exposed BALB/c mouse pups to 0.1 mM Pb acetate in the drinking water of their dams, from gestational-day 8 (gd8) to post-natal day 21 (pnd21). The time frame of gd8 to pd21 and the 0.1 mM Pb concentration were chosen because previous studies from our laboratory indicated that a 0.1 mM Pb acetate exposure during gestational and lactational periods resulted in blood Pb levels of 15–20 µg/dl [5]. This blood Pb level range is comparable to levels found in environmentally Pb exposed children [1]. Our previous studies used gd15 as the initiation of Pb exposure, however, in order to obtain more of an impact of Pb exposure on development we chose gd8 for the present study. Pb exposure from day 0–8 of gestation was avoided in order to allow for embryo implantation and early neuronal and immune development in the absence of Pb. Cytokine expression was assessed for male and female mouse pups separately. Cytokine expression was differentially altered by Pb.
The objectives of this communication are two-fold, first to present new evidence pertinent to the field of Pb toxicology, and secondly, to review current literature regarding cytokines in the CNS and Pb effects on neuronal development.
MATERIALS AND METHODS
Animals and Treatments
BALB/c mice were obtained from Taconic Farms (Germantown, NY). Mice were housed two females with one male. Once mice with vaginal plugs were identified (checked daily), they were housed separately and were considered to be pregnant (gestational day 0, gd0). At gd8, these dams were placed on drinking water with or without 0.1 mM Pb acetate (3 litters per condition). Each litter was designated as N=1. Pregnant females were housed one per standard cage and allowed free access to food and water in our specialized pathogen-free, AAALAC-approved animal facility. All mice were maintained on a 12-h light/dark cycle and were seronegative for known background viral agents. All mouse pups were euthanized by CO2 asphyxiation at 21 days of age (pd21), and brain tissue from male and female pups were harvested and processed separately. All animals were treated humanely and all procedures were IACUC-approved.
Total RNA Isolation
We isolated total RNA using a Lipid Tissue RNA isolation kit and protocol from Qiagen, Inc. (Valencia, CA). Before removal from the cranium, the mouse pup brains were transcardially perfused with 30 ml of sterile, diethyl pyrocarbonate (DEPC)-treated PBS, and were immediately homogenized with the QIAzol reagent. We pooled brain tissue on the basis of litter and gender of the mouse pups. RNA was eluted from the midi columns with two aliquots of 250 µl each of RNase free water. Concentration and purity of the RNA were measured using a Beckman DU 640 spectrophotometer (Fullerton, CA). We checked RNA integrity both by a 1% agarose mini-gel using 1X TBE buffer and by a bioanalyzer. Mouse brains to be used for RNA isolation from specific brain regions were frozen at −80°C immediately after perfusion and removal. Cortex, striatum, hypothalamus, hippocampus, cerebellum, and substantia nigra were dissected from frozen brains and were not allowed to thaw before homogenization in the QIAzol reagent. Again, brain region tissues within a litter were pooled for RNA isolation. We assessed purity and RNA integrity as described for the whole-brain RNA.
Microarray Gene Expression Analysis
Affymetrix GeneChip hybridization and processing was performed by the Wadsworth Center, Microarray Core Facility (Albany, NY). Quality of the RNA was assessed by means of a bioanalyzer to check the ribosomal RNA S28/S18 ratio. RNA had to have a ratio of 1.8 or higher, if it was to be processed on the GeneChips. Six Affymetrix MG-430A GeneChips were hybridized, three with total RNA from one litter each of female distilled water controls, and three with total RNA from one litter each of female 0.1 mM Pb treated experimental pups. We analyzed the GeneChip results using the Affymetrix MAS 5.0 software, as well as GeneTraffic and PathwayAssist softwares.
Absolute Quantitative Real-Time RT-PCR
A two-step process was employed for cytokine mRNA quantification. First, cDNA was prepared from 1µg of total RNA using a 1st strand cDNA synthesis kit from Roche Applied Science (Indianapolis, IN). Following the synthesis reaction, a 200 µl aliquot of PCR-grade water was added to each tube. The second step involved separate amplification for each cytokine gene sequence using primer kits from Search-LC (Heidelberg, Germany). We did amplifications in duplicate with an Applied Biosystems 7500 real-time PCR instrument under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 68°C for 40 sec, and 72°C for 50 sec. Readings were taken at the third step of each cycle. Following amplification, melting curves were generated with 1 cycle of 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 min. Standard curves were generated for each run using standards of known copy number supplied by Search-LC. Quantification was performed for the following cytokine mRNAs: IL-5, IL-15, IL-16, IL-18, LT-β, TNF-α, TGF-β1, and IFN-γ. Quantitation results were recorded as copy number per microliter (CN), and all cytokine results were normalized to the copy number (CN) obtained for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from the same cDNA synthesis sample, according to the formula (cytokine CN/GAPDH CN) × 100.
Relative Quantitative RT-PCR
Again, a two-step process was employed to quantitate the cytokine mRNA. First, cDNA was synthesized from 2 µg of total RNA by use of the cDNA archive kit from Applied Biosystems, Inc. (Foster City, CA) according to the manufacturer’s directions. After the 2-hr incubation at 37°C the 100 µl reaction mixtures were diluted with an equal volume of PCR-grade water. Relative quantitation was then carried out by use of a TaqMan method with primers, probes, and master mixes from Applied Biosystems. Gene expression TaqMan kits used for these experiments were for mouse IL-6 (Mm00446190_m1), IL-11 (Mm00434162_m1), IL-13 (Mm00434204_m1), IL-18 (Mm00434225_m1), TGF-β1 (Mm00441724_m1) and GADPH (Mm99999915_g1). We determined expression of IL-18 and TGF-β1 by both the TaqMan method and the absolute quantification method as a control to verify that the two methodologies gave the same outcome. The house-keeping gene gapdh (GenBank: NM_008084) was again used as the endogenous control for normalization of the cytokine mRNA quantities. We performed amplification in an Applied Biosystems 7500 real-time PCR instrument under the following conditions: 50°C for 2 min, then 95°C for 10 min followed by 40 cycles of 95°C for 30 sec and 60°C for 1 min. Readings were taken during the second step of each cycle. We evaluated cytokine expression using the Applied Biosystems SDS software, with one of the control samples set as a reference, i.e., set at 1.0 and by the equation, cytokine expression = 2−(ExpCt-GAPDH Ct) × 1000. Verification of gender for each of the RNA samples was carried out by the TaqMan method using primers and probes from Applied Biosystems for a Y chromosome-specific gene transcript, ubiquitously transcribed tetratricopeptide repeat, Uty (Mm00447710_ml).
Cytokine Protein Detection
Brain regions were dissected from frozen perfused brains of 21 day old female mouse pups. Dissected tissue was immediately homogenized in an appropriate volume of M-PER protein extraction reagent plus the protease cocktail, HALT, from Pierce (Rockford, IL). Homogenates were spun at 13,000 rpm in a microcentrifuge at 4°C for 20 min to pellet non-solubilized material. Tissues from 2 mouse pups were pooled from each litter (3 control and 3 Pb treated litters). Total protein concentrations were determined by means of a kit from Pierce (Rockford, IL). IL-18 concentrations were determined by means of an ELISA kit from BD Biosciences (San Jose, CA). Brain region homogenates were diluted 20 fold and the manufacturer’s protocol was closely followed. The cytokine, TGF-β1, was detected by using an ELISA kit from R & D System (Minneapolis, MN). The manufacturer's protocol was followed. Briefly, the plate was coated with anti-mouse TGF-β1 monoclonal antibody (mAb). After washing, serially diluted standards and samples were loaded on the plate. After incubation, biotin-labeled anti-mouse TGF-β1 mAb was applied, followed by avidin-peroxidase and finally the substrate. The plates were read using an ELISA plate reader (EL310; Bio-Tek, Burlington, VT) at 450 nm. The cytokine IL-6 was detected by using a Fluorokine Map Mouse IL-6 Kit (R & D System, Minneapolis, MN). The assay protocol was provided by manufacturer. Briefly, a microplate was washed and loaded with diluted microparticle mixture, which contained the beads coated with capture antibody to target cytokine. Then, the serial diluted standards and brain homogenate were added. After incubation for three hours at room temperature on a horizontal microplate shaker, the microplate was washed and biotin antibody cocktail was added. Finally, Streptavidin-PE was used as the fluorescent signal. The assay was run on a Luminex 100 instrument. The results were analyzed by using the software provided by Upstate Biologicals (Lake Placid, NY).
Statistical Analysis
Student’s t-test was used to check for significance of difference between male and female and between Pb and control, gene expression levels. For all experiments, statistical significance was assumed at P ≤ 0.05.
RESULTS
Cytokine RNA Quantification in the Whole Brain
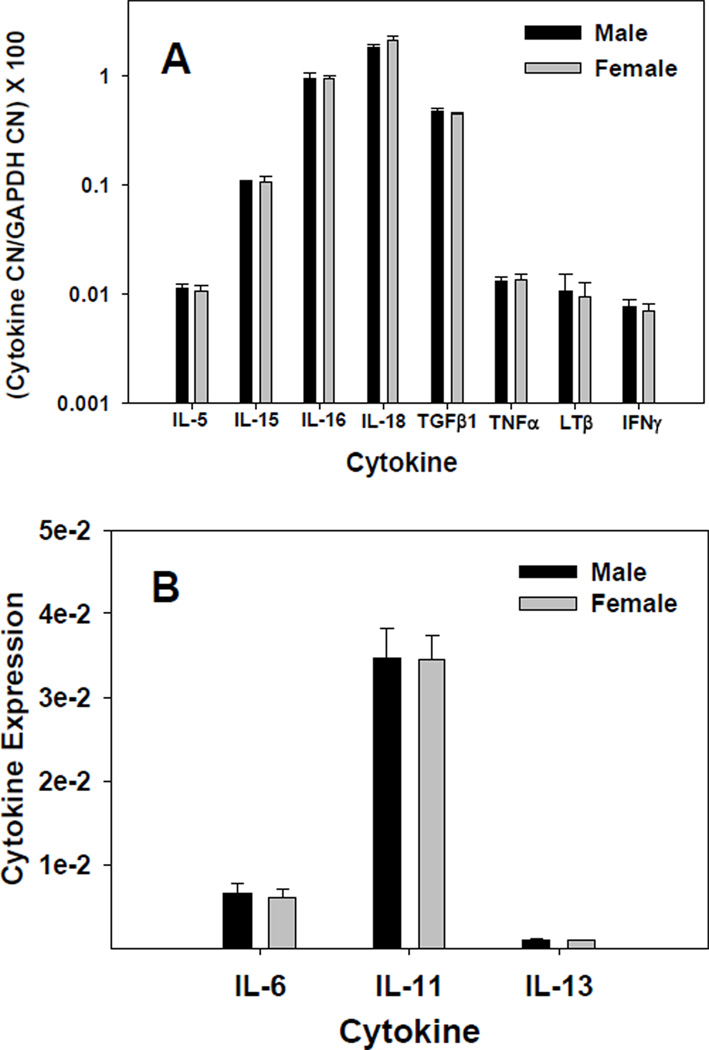
To assess CNS cytokine gene expression during development, we analyzed the relative expression of a selected group of common cytokines by quantitative RT-PCR and GeneChip protocols. Since there is some evidence now that neuroimmunological differences may exist based on sex and gender we analyzed male and female mouse perfused brain tissue separately [40]. Figures 1A & 1B show the relative levels of detectable cytokine mRNAs, between males and females, as measured by real-time RT-PCR. The most highly expressed cytokine was IL-18, followed by IL-16, TGF-β1, and IL-15 (Figure 1A). Interleukin-11 was expressed at a higher level, in both males and females, than IL-6 (P≤0.002), while IL-13 levels were very low; being close to the limits of detection (Figure 1B). There were no significant differences in the expression levels between males and females for any of the cytokines shown in Figure 1. We also attempted to measure transcript levels for IL-4 and IL-10, but these were found to be below our limit of detection in mouse brains at pnd21 (data not shown).
Figure 1.
Constitutive expression of CNS cytokines. RNA from brains of male and female BALB/c mouse pups at 21 days of age was quantified by real-time RT-PCR, either by absolute quantitation using a standard curve (A) or by relative quantitation via the TaqMan method (B). Whole brain RNA was isolated using the Qiagen Lipid Tissue Midi RNA isolation kit. Brains were pooled by gender within each litter. Cytokine mRNA quantity was normalized to endogenous control GAPDH. Each bar represents mean ± SE for an N of 3 litters. Cytokine levels that did not significantly differ (P≤0.05) from each other were TNFα from IFNγ, and LTβ from IL-5, TNFα and IFNγ.
Quantification of IL-16, IL-18, TGF-β1, IL-11, and IL-6 in Brain Regions
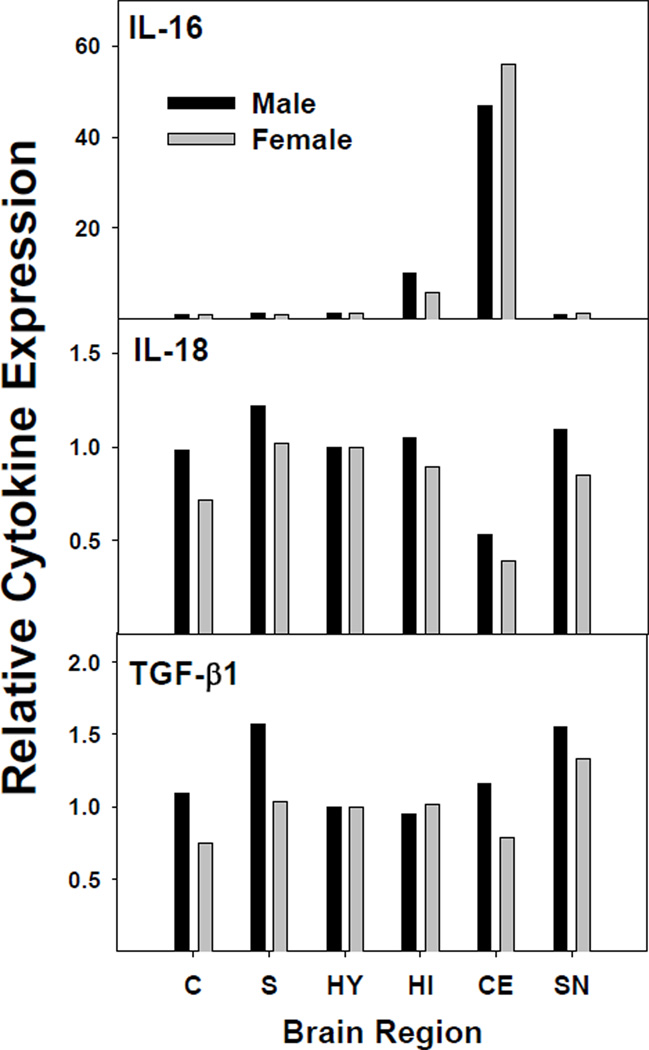
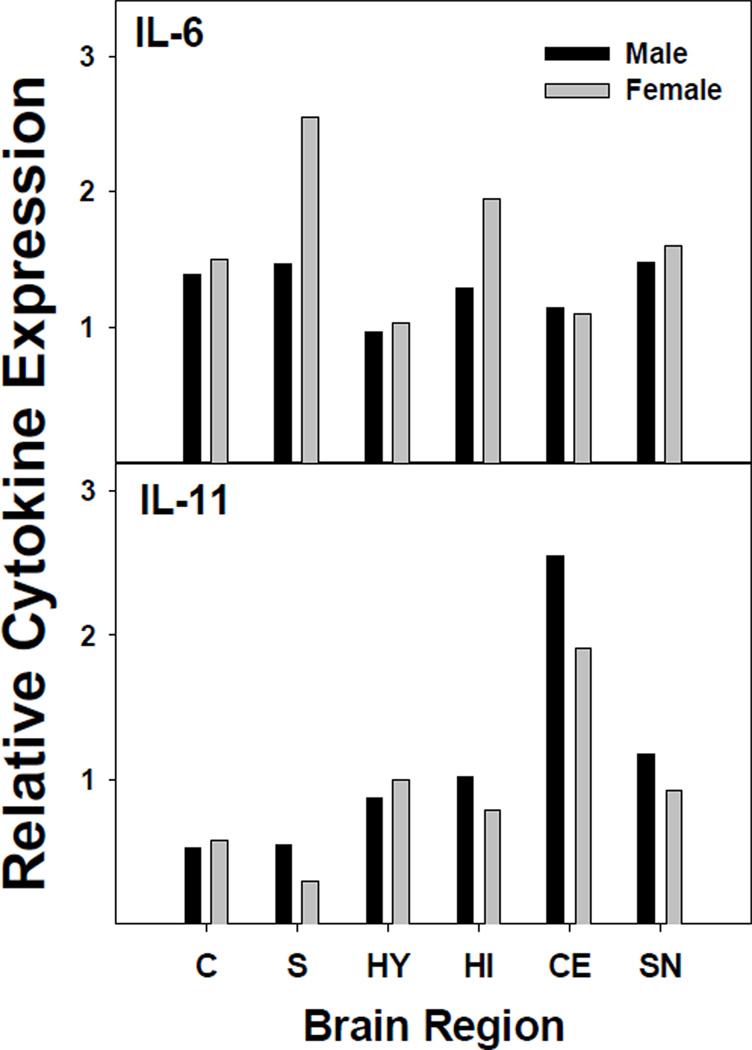
We used absolute quantification of cytokine mRNA to measure cytokine gene expression in a number of specific brain regions, namely; cortex, striatum, hypothalamus, hippocampus, cerebellum, and substantia nigra. Figure 2 shows the pattern of expression for each of the more highly expressed cytokines (IL-16, IL-18, and TGF-β1). As indicated, the pattern of expression was similar between male and female mouse pups. IL-18 and TGF-β1 were expressed across the brain regions studied, while IL-16 was most highly expressed in the cerebellum and hippocampus, as reported previously [37]. The primary form of detectable IL-16 was likely the neuronal form of IL-16 (nIL-16), given that nIL-16 has been reported to be expressed primarily by neurons in the hippocampus and cerebellum. However, our data suggested that low IL-16 expression is observable in other regions. Interestingly, the expression profiles of IL-18 and TGF-β1 were similar. In general, IL-18 and TGF-β1 had slightly greater expression in the striatum and the substantia nigra of males. The different expression levels could be a reflection of the number of microglia cells present in the designated brain regions, in that substantia nigra is known to have the greatest number of microglia cells [41,42]. Figure 3 shows the relative expression levels of IL-6 and IL-11 in the brain regions. IL-6 expression was evenly distributed among the brain regions, but the profile for IL-11 was different from IL-6, in that IL-11 expression was highest in the cerebellum for males and females. A notable difference was that females had relatively lower expression of IL-11 than males in striatum, hippocampus, and cerebellum.
Figure 2.
Relative cytokine mRNA concentration in brain regions. Cytokines measured were IL-16, IL-18, and TGF-β1. Total RNA was isolated, using the Qiagen Lipid Tissue Mini kit, from mouse brain regions of two litters. Cytokine mRNA was quantified by real-time RT-PCR using an absolute quantification method. Copy number results were normalized to the endogenous control GAPDH. The values for cortex (C), striatum (S), hypothalamus (HY), hippocampus (HI), cerebellum (CE), substantia nigra (SN) from males or females from two litters are presented as a proportion of the hypothalamic value set at 1.
Figure 3.
Relative cytokine mRNA concentration for IL-6 and IL-11 in brain regions. Total RNA was isolated and quantified using the TaqMan method from Applied Biosystems, Inc. Cytokine expression levels in the various brain regions of male and female mice are presented as described in Figure 2.
Pb Effect on Cytokine Expression in the Whole Brain
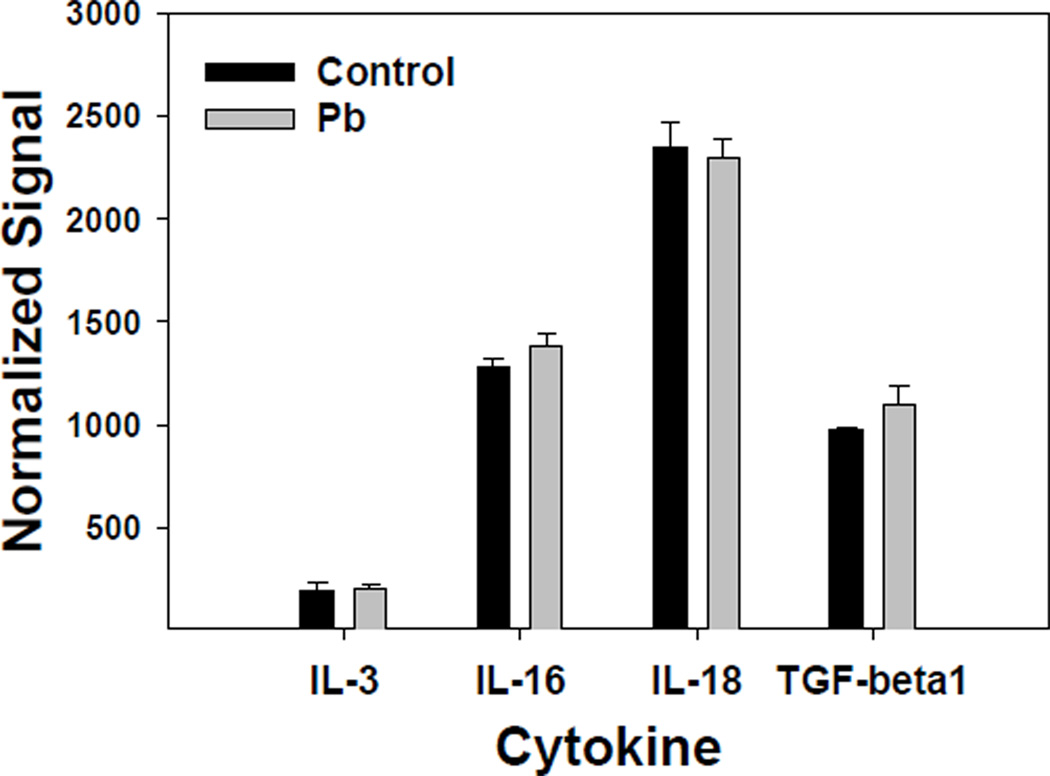
Affymetrix GeneChips were employed both to provide a more complete picture of general CNS cytokine gene expression and to provide information about the effect of low-level Pb exposure on the developing CNS. GeneChip results substantiated the identification of the cytokines that were most highly expressed and it provided information about the effects of Pb on cytokine gene expression. Figure 4 shows the relative expression profile of cytokines that were given a “call” of present (P) by the Affymetrix MAS 5.0 software, as normalized to GADPH. These cytokines were IL-3, IL-16, IL-18, and TGF-β1. The GeneChip results indicated no significant Pb effect on the expression of TGF-β1, which had the lowest P-value of the group at P=0.24. All of the other common cytokines were given a call of absent (A) by the software. Real-time RT-PCR was performed on IL-16, IL-18, and TGF-β1, and in agreement with the GeneChip data, the results indicated high expression of these cytokines. Furthermore, quantitative RT-PCR results for IL-16 and IL-18 did not show a significant difference with the 0.1 mM Pb exposure (Table 1). However, when mouse pups were exposed to 0.5 mM Pb and sacrificed at pnd21, exposed female mouse pups had increased IL-18 levels (data not shown). Real-time RT-PCR analysis of TGF-β1 gene expression indicated a significant increase in females exposed to 0.1 mM Pb (Table 1).
Figure 4.
Affymetrix GeneChip analysis of CNS cytokine gene expression in brains from Pb-treated and control female mouse pups. Pb-treated female mouse pups were exposed from gd8-pd21 to 0.1 mM Pb acetate via the dam’s drinking water. RNA was isolated, using the Qiagen Lipid Tissue Midi kit, from brain tissue pooled within each litter; 3 litters per treatment. The GeneChip used for the analysis was MG 430A from Affymetrix. Cytokine RNA expression was normalized to endogenous expression levels for GAPDH and results represent mean ± SD.
Table 1.
Effect of Pb on Gene Expression of Cytokines in the Brain
| Femalea | Malea | |||||
|---|---|---|---|---|---|---|
| Cytokineb | Control Cytokine expression ± SEM |
Control Cytokine expression ± SEM |
P-value | Control Cytokine expression ± SEM |
Pb Cytokine expression ± SEM |
P-value |
| IL-5 | 0.010 ±0.002 | 0.011 ±0.002 | 0.933 | 0.011 ±0.001 | 0.011 ±0.002 | 0.813 |
| IL-15 | 0.108 ±0.01 | 0.098 ±0.007 | 0.468 | 0.108 ±0.003 | 0.098 ±0.006 | 0.204 |
| IL-16 | 0.951 ±0.05 | 1.117 ±0.06 | 0.097 | 0.963 ±0.099 | 1.078 ±0.073 | 0.402 |
| IL-18 | 2.125 ±0.18 | 1.964 ±0.137 | 0.515 | 1.827 ±0.086 | 2.093 ±0.116 | 0.14 |
| TNF-α | 0.013 ±0.002 | 0.014 ±0.002 | 0.949 | 0.013 ±0.001 | 0.013 ±0.001 | 0.929 |
| LT-β | 0.010 ±0.003 | 0.010 ±0.003 | 0.857 | 0.011 ±0.004 | 0.012 ±0.005 | 0.832 |
| IFN-γ | 0.007 ±0.001 | 0.007 ±0.001 | 0.837 | 0.008 ±0.0001 | 0.006 ±0.001 | 0.287 |
| TGF-β1 | 0.440 ±0.016 | 0.528 ±0.025 | 0.039c | 0.470 ±0.03 | 0.540 ±0.036 | 0.206 |
N = 3 litters
Cytokine gene expression was determined by quantitative real-time RT-PCR and the absolute CN (copy number/µl) was normalized to the endogenous control, GAPDH, CN using the formula (cytokine CN/GAPDH CN) × 100.
Significant at P≤0.05 by a two-tailed Student’s t-test.
Although the GeneChip data indicated that expression was absent for other cytokines that are known to be present in the CNS, (both from studies reported in the literature and from experiments performed in our laboratory), we performed quantitative RT-PCR for several of the “absent” cytokines, since real-time RT-PCR is a more quantitative and sensitive procedure. One of the cytokines that is often mentioned as exerting an influence on CNS after infection by various pathogens is TNF-α. This cytokine plays a major role in the display of sickness behavior and is known to stimulate expression of IL-6 [43]. Pb exposure did not change gene expression of TNF-α in either male or female mice (Table 1). Cytokines that had been detected in the CNS by other investigators include IL-15 and LT-β [44]. Real-time RT-PCR results for IL-15 and LT-β also indicated that Pb had no significant effect on the expression of these cytokines (Table 1). Two other cytokines, IL-5 and IFN-γ, were of interest, since Pb has been observed to skew the immune system toward a Th2 response [45]. IFN-γ expression is representative of a Th1 response, and IL-5 is indicative of a Th2 response [46]. Comparison of the expression of IL-5 and IFN-γ, between Pb-treated and untreated male and female mice, showed that Pb did not significantly perturb expression of either cytokine, in either gender (Table 1).
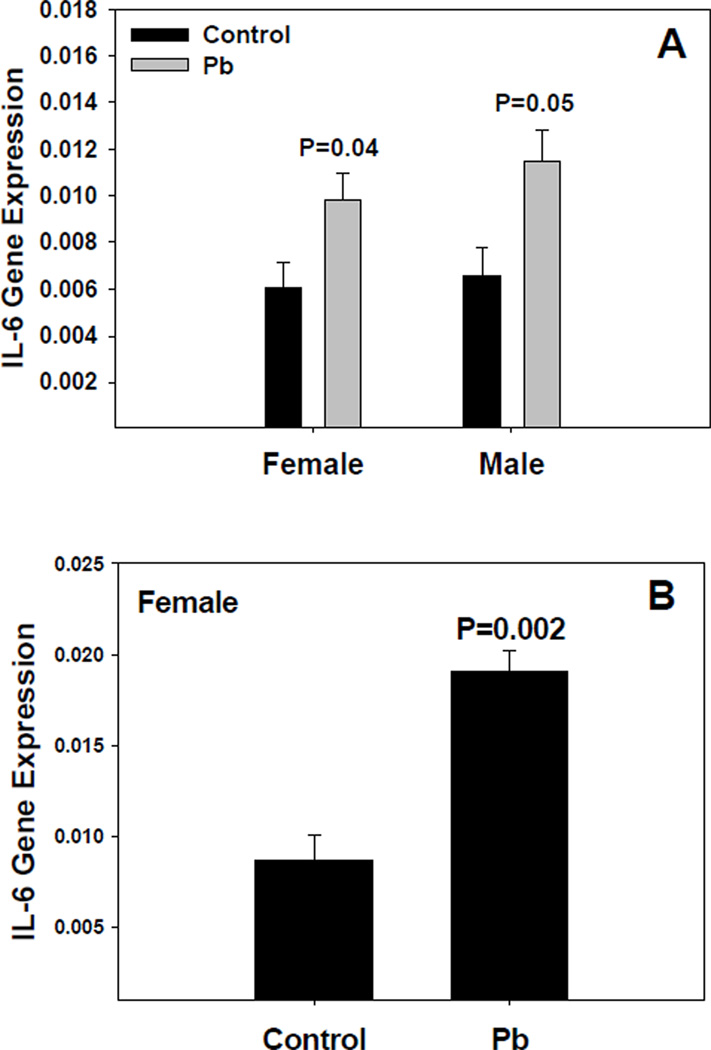
IL-6 was of particular interest, given that Pb has been found to enhance IL-6 expression in the CNS of adult mice treated with LPS [19]. Relative quantitative analysis was carried out for IL-6 mRNA. The results indicate that IL-6 expression was increased, in both females and males, by 0.1 mM Pb (Figure 5A). This increase in IL-6 gene expression was even more pronounced in females when the Pb exposure was increased to 0.5 mM (Figure 5B). Preliminary results for IL-11, an IL-6 family member, indicated some alteration of expression by Pb, but further data is needed to verify these results.
Figure 5.
Pb exposure modifies IL-6 gene expression in the brain as measured by real-time TaqMan RT-PCR. Male and female mice were exposed to 0.1 mM Pb acetate from gd8 to pnd21 (A), or 0.5 mM Pb from gd0 to pnd21 (B) through the dam’s drinking water. All were assayed on pnd21. Cytokine mRNA was quantified and normalized to that of GAPDH within each sample. The data represent an N of 3 litters for male and female mice Pb-treated or untreated, i.e., 6 litters total. Results represent mean ± SE; the * indicates a P≤0.05.
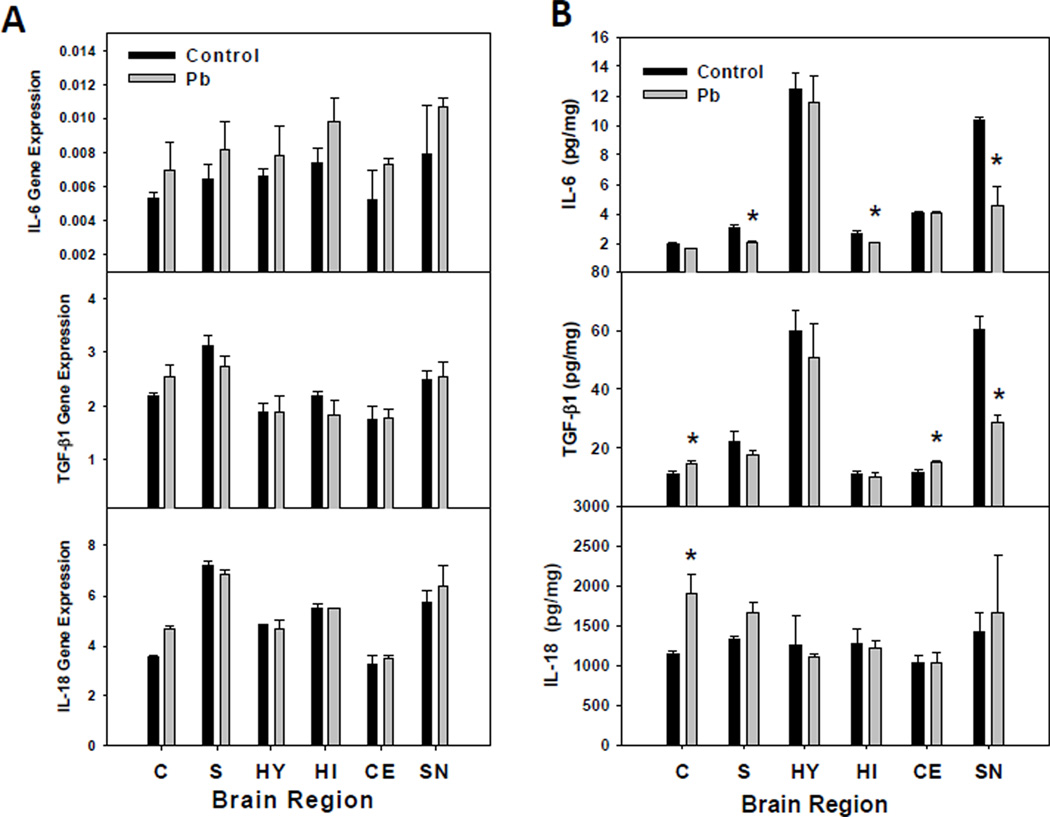
Pb Effect on Expression of IL-6, IL-18, and TGF-β1 in Brain Regions
Among all of the cytokines examined in this study, only members of the IL-6 and TGF-β1 families had their expression significantly altered by exposure to Pb under the conditions described here for this study. Thus, regional expression levels of these cytokines were measured along with that of IL-18, the most highly expressed pro-inflammatory cytokine in the brain. Gene expression changes after exposure to Pb are presented in Figure 6A. For both IL-18 and TGF-β1 the cytokine expression was most increased by Pb in the frontal cortex. IL-6 message levels were generally higher than control in each region studied, but these differences were highly variable and were not significant. Perhaps, this is due to the low amount of IL-6 mRNA present in each region, which results in a higher standard deviation (Figure 6A). Protein levels of IL-6, TGF-β1, and IL-18 corresponded to their overall mRNA levels for each, since IL-18>TGF-β1>IL-6 (Figure 6B). Pb had no effect on the protein levels of IL-6 in the frontal cortex, hypothalamus or the cerebellum, but decreased protein levels were observed in the striatum, hippocampus, and the substantia nigra. TGF-β1 protein concentration was increased by Pb in the frontal cortex and cerebellum, but was decreased by Pb in the substantia nigra. Slightly decreased concentrations of TGF-β1 were observed in the striatum, hippocampus and hypothalamus, but these changes were not significantly different from the control. Pb increased gene and protein expression in the frontal cortex, but had no effect on expression in other brain regions (Figure 6A/B).
Figure 6.
Regional CNS cytokine expression after Pb exposure. (A) IL-6, TGF-β1, and IL-18 gene expression in different female brain regions were assayed by real-time TaqMan RT-PCR as described in the methods. Relative expression was calculated using the formula (2−(ExpCT-GAPDH CT) × 1000), where CT is the threshold cycle number. (B) Protein concentrations for IL-6, TGF-β1, and IL-18 were measured by ELISA (IL-18 and TGF-β1) or Luminex (IL-6). The protein data represent an N of 3 litters each for control and Pb treated female pups. The bars represent mean ±SD; the * indicates a P≤0.05.
Pb Effect on the Signal Transduction Pathways of IL-6 and TGF-β1
The expression of genes known to influence IL-6 and TGF-β expression were evaluated; c-jun and c-fos were increased by Pb (Table 2), and c-jun and c-fos bind to the activator protein 1 (AP-1) promoter site for IL-6 and TGF-β [47–50]. Administration of Pb also slightly enhanced the transcription factors MAD homolog (Smad) and signal transducer and activator of transcription 3 (STAT3) as well as multiple mitogen activated protein kinases (Mapks) [51–54] (Table 2). Table 2 presents GeneChip data only for the Map kinase genes (of the 42 that were represented on the GeneChip) that had a “call” of present (P). Map kinases whose expression was significantly increased by Pb included Mapk6, Mapk11(p38), Map4k5, Map4k6, and Mapkapk2.
Table 2.
Effect of Pb on Signal Transduction and Transcription Factorsa
| Signalb ± S.D. | ||||
|---|---|---|---|---|
| GeneBank ID | Gene | Control | Pb | P-valuec |
| NM_010591 | c-jun | 1904 ± 70 | 2072 ± 106 | 0.04 |
| AV026617 | fos | 7313 ±106 | 8106 ± 681 | 0.06 |
| AB008192 | Smad3 | 82 ± 38 | 288 ± 144 | 0.08 |
| AI325183 | Stat3 | 3288 ±160 | 3539 ± 313 | 0.14 |
| D76446.1 | Tak1 | 1052 ± 91 | 990 ± 130 | 0.32 |
| NM_011951 | Mapk1 | 12119 ±719 | 12998 ± 1083 | 0.15 |
| NM_015806 | Mapk6 | 1485 ± 107 | 1655 ± 73 | 0.04 |
| BC024684 | Mapk11(p38) | 256 ± 32 | 358 ± 52 | 0.02 |
| AF128892 | Mapk14 | 3350 ± 173 | 3638 ± 171 | 0.06 |
| BM240207 | Map2k4 | 5412 ± 244 | 5894 ± 358 | 0.06 |
| AW541674 | Map2k7 | 1157 ± 77 | 1232 ± 69 | 0.14 |
| AA929089 | Map3k7 | 3832 ± 167 | 4152 ± 401 | 0.14 |
| BF166991 | Map4k2 | 500 ± 29 | 583 ± 80 | 0.08 |
| BB734681 | Map4k5 | 363 ± 50 | 490 ± 90 | 0.05 |
| NM_016713 | Map4k6 | 2589 ± 64 | 3086± 86 | 0.005 |
| BG918951 | Mapkapk2 | 783 ± 20 | 994 ± 91 | 0.009 |
Affymetrix MG430A GeneChip Data. The data represent mouse brain total RNA from female mouse pups from 3 litters of untreated control mice and 3 litters of 0.1 mM Pb acetate treated experimental mice.
Signal was normalized to the GAPDH signal on each respective GeneChip
Statistics were performed by means of a one-tailed Student’s t-test, significant at P≤0.05.
Since IL-6 and TGF-β play an active role in neural stem cell differentiation and can act in synergy to promote astrocyte differentiation from neuronal cell precursors [55], an IL-6 and TGF-β signaling pathway was examined. An affected gene, which includes binding sites for both STAT3 and Smad, is gfap (GenBank:BB183081, X78141, X02801), coding for the astrocyte marker protein, GFAP, [39,55–57]. GeneChip results for gfap, recorded in signal units (SU), were normalized to gapdh signal units from the same chip. Pb significantly increased transcription of the gfap gene in female mouse pups (control n=3, 6,237.2 ± 84.5 SU; Pb n=3, 6,823.3 ± 50.1 SU, P ≤ 0.004); males were not assayed.
DISCUSSION
Quantification of the constitutive expression of common cytokine mRNAs in the brain of BALB/c mouse pups at pd21 revealed that transcripts were most abundant for IL-18 >IL-16 >TGF-β1 >IL-15. Previous studies on expression of IL-18 suggested that this cytokine is constitutively expressed by astrocytes and microglial cells [36]. Although CNS IL-18 expression did not differ significantly by gender, females consistently showed higher expression, suggesting that there may be hormonal regulation of this cytokine. Both the mRNA and the protein results indicated that IL-18 was increased by Pb in the frontal cortex. Increases in IL-18 expression within the frontal lobe area of the brain have been reported in certain neurodegenerative disorders [58]. This cytokine has been shown to impair long-term potentiation and N-methyl-D-aspartate (NMDA) receptor mediated transmission in vitro [59]. It was implied that this occurs by IL-18 activation of the p38 MAPK cascade which includes Map4k6 and Mapkapk2, all of which are up-regulated by Pb exposure according to our GeneChip results (Table 2). Stimulation by Pb of the p38 MAPK cascade in CNS tissue cultures of hippocampal and cerebellum sections has been observed [60,61]. Previous studies have linked p38 MAPK cascade signaling to changes in synaptic plasticity [62]. Our, in vivo, results regarding IL-18 expression, suggest that Pb might have a potentially strong effect in the frontal cortex with regard to synaptic plasticity. Another highly expressed cytokine was IL-16. Interleukin-16 transcript levels were found to be very high in the cerebellum and the hippocampus; however, we were unable to differentiate nIL-16 from its truncated form, which functions as a chemokine for CD4+ T-cells. Since lymphocytes have been suggested to constitutively traffic into the CNS [63], it is possible that chemokines for lymphocytes are produced within the brain. IL-16 immunoreactivity has been observed in microglial nodules, along with infiltrating lymphocytes [64].
Significant modulation of cytokine gene expression in whole brain by Pb was observed for TGF-β1 (Table 1) and IL-6 (Figure 5). A possible explanation for this finding is that Pb affects DNA-binding proteins that can regulate transcription of IL-6 and TGF-β1 genes. Pb has been reported to activate protein kinase C (PKC) [65–68]; this activation in turn induces genes that are regulated by AP-1 transcription factors [69–71]. These AP-1 factors are protein dimers of the jun family or heterodimers of the jun and fos family of genes, termed immediate early response genes (IERG) [69]. IERG genes are transcribed within minutes after stimulation. Since jun and fos must be phosphorylated before they can dimerize, Mapks also play a role in this transduction pathway. It has been reported that Pb, via its activation of PKC, can perturb the Map kinase system in some way [72]. As indicated, Pb increased expression of jun and fos and certain Map kinases. Pb also has been reported to affect the transcription factor Sp1, in a process involving PKC alpha and Map kinases [73,74]. Thus, if the cytokine genes for il6 and tgfb1 have a site for either of these regulatory elements, Pb could have an effect on increasing the cytokine’s gene transcription. The genes for il6 and tgfb1 each exhibit the presence of at least one AP-1 binding site, the conserved sequence TGACTCA, [47–50,75–77]. Also, promoter regions for il6 and tgfb1 appear to contain Sp1 sites [78–80], the conserved sequence GGGCGG.
Increased gene expression of IL-6 and TGF-β1 during development may adversely affect neuronal growth and differentiation [25,81–83]. Over-expression of IL-6 has been shown to result in reactive gliosis, as evidenced by increased size and numbers of astrocytes and ramified microglia. Over-expression of IL-6 also has an adverse effect on NMDA glutamate receptor neurons; it causes excessive activation of these neurons, leading to cell death by necrosis. The effects of TGF-β are more difficult to predict than those of IL-6 in that TGF-β is known to have opposite effects (inflammatory and anti-inflammatory), which seems to be dependent on multiple additional immunoregulatory factors [39,84,85].
IL-6 and TGF-β1 play active roles in neural stem cell differentiation [55,86,87]. They act in synergy to promote astrocyte differentiation from neuronal cell precursors. The IL-6 family of cytokines function via membrane-bound gp130, to activate the JAK/STAT pathway [82,88,89]. Specifically, the IL-6 pathway leads to tyrosine-phosphorylation of STAT3, and that process promotes STAT3 dimerization, followed by translocation to the nucleus, where the pSTAT3 dimers promote gene transcription. Likewise, the TGF-β cytokines bind to membrane-bound receptors, leading to activation of Smad molecules. Like STAT3, the Smad proteins dimerize and translocate to the nucleus, where they promote gene transcription [55,90]. Again, Map kinases play roles in these transduction pathways [51,54]. Pb enhanced expression of STAT3 and Smad, as well as GFAP, an astrocytic protein induced by STAT3 and Smad. Interestingly, binding of STAT3 or Smad alone does not induce transcription of gfap. Transcription of the gfap gene occurs only if both STAT3 and Smad are bound, along with a connecting molecule, p300, to bridge the space between STAT3 and Smad [55]. Interestingly, GFAP gene expression was found significantly increased at pnd20 in the cortical region of the brain during studies performed on rats exposed to Pb during various stages of development, but in the hippocampus the GFAP expression peaked at pnd12 in Pb exposed rats and had fallen below control (unexposed) levels by pnd20 [91]. These GFAP gene expression results are in agreement with our proposed mechanism, since our brain region cytokine gene expression results (Figure 6) show that both IL-6 and TGF-β1 mRNAs are elevated at pnd21 in the cortex but not in the hippocampus at this time. In the hippocampus gene expression of TGF-β1 at pnd21 was slightly decreased in the presence of Pb (Figure 6). GFAP gene expression in the cerebellum of developmentally Pb exposed rats at pnd20 was similar to untreated controls, but later significantly decreased to below control levels at pd30 and pd50 [92]. Our cytokine gene expression results for the cerebellum at pnd21 indicate that IL-6 and TGF-β1 mRNA levels in Pb-treated mouse pups were similar to those detected in controls. The discrepancy between message and protein levels for IL-6 and TGF-β1 in the different brain regions is likely due to kinetic differences in binding to their receptors and uptake/utilization. This is especially apparent for IL-6, in that the protein levels were lower with Pb, whereas the mRNA levels were increased. The reason for this may be that we are not detecting IL-6 bound to its receptor. Our detection method for IL-6 protein may only measure free IL-6 and not IL-6 bound to its receptor [19]. Therefore the discrepancy between IL-6 protein and mRNA levels may be due to more IL-6 bound to receptors in Pb treated animals. For this reason it may be important to determine the Pb effect on expression of the IL-6 receptor. Expression of this receptor may be up-regulated by Pb. Since IL-6 and TGF-β are needed to induce GFAP and GFAP expression was increased by Pb, it is likely that IL-6 protein was made and utilized. With regard to brain regions, future studies also may need to focus more on the frontal cortex. Earlier reports on Pb in the CNS suggested that the hippocampus was a major target for Pb effects; however, these cytokine expression studies suggest that the cortex also may be a target for the toxic effects of Pb.
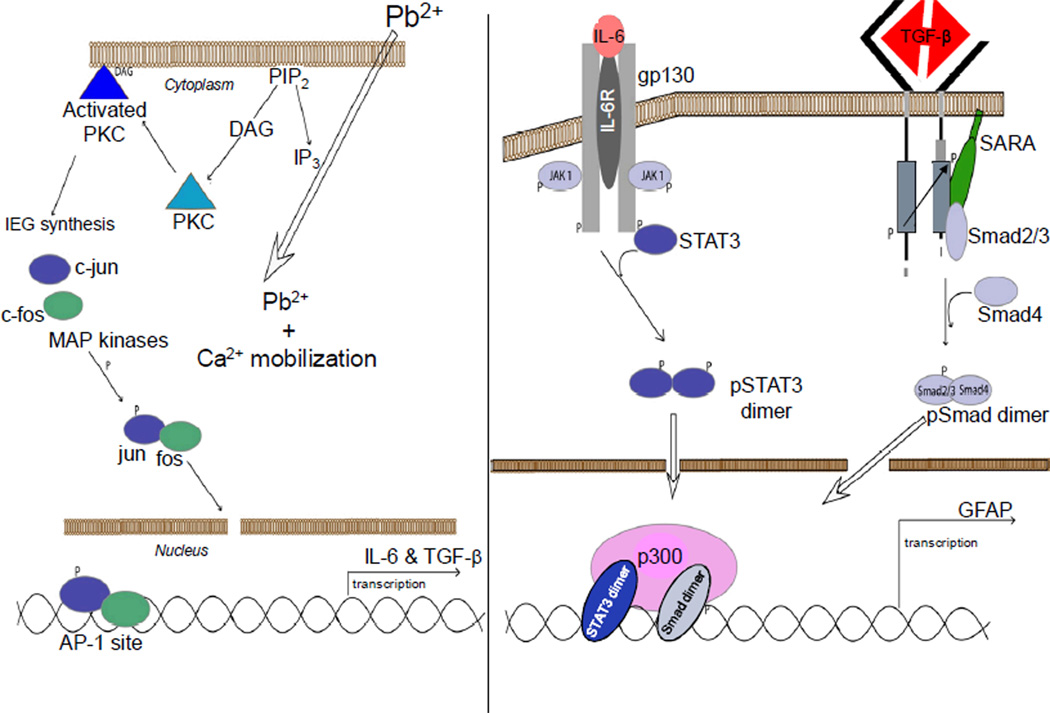
It has been noted that differentiation of neurons occurs early (gd12) during development, whereas differentiation of astrocytes occurs at a later time (gd14) [93]. This developmental difference has been proposed to be due to methylation of the STAT3 site during the early stages of development, given that STAT3 cannot bind methylated DNA [55]. Methylated DNA would then promote neuronal differentiation and block differentiation of astrocytes. However, in the study presented here, the Pb treatment was continued to pd21, a timepoint at which the STAT3 site should no longer be methylated. Lack of DNA methylation would allow gfap transcription and astrocyte differentiation to proceed [55]. The Pb treatment did increase gfap transcription in female mice. Therefore, Pb could be producing a mild gliosis, such that astrocyte growth is favored over neuronal growth. In support of this possibility, Pb-induced PKC activation has been observed to inhibit proliferation and differentiation of oligodendroglial progenitor cells in culture [94]. Effects on IL-6 and TGF-β by environmental toxicants, such as Pb, could differentially affect particular brain regions and induce abnormal behavior. It has been demonstrated in rats that developmental Pb exposure produced lasting changes in sustained attention, response initiation, and reactivity to errors [95]. Ultimately, the behavioral feature for Pb exposure could be a decrease in long-term memory [96]. The present data in conjunction with that already reported in the literature suggests the mechanism diagrammed (Figure 7). Pb enters the cell and mobilizes calcium which leads to cleavage of phosphatidylinositol bisphospate (PIP)2 into inositol triphosphate (IP)3 and diacylglycerol (DAG). The DAG then binds to PKC which becomes activated and migrates to the cytoplasmic membrane. Activation of PKC leads to transcription of immediate early response genes, c-jun and c-fos, whose products can be phosphorylated by Map kinases, dimerize, and migrate to the nucleus where they initiate transcription of genes, such as il6 and tgfb1, that have AP-1 sites present in their promoter regions. These cytokines are then synthesized and secreted by the cell. The newly made cytokines then bind to their respective receptors and promote the transcription of genes, such as gfap, via their respective signaling cascades. In astrocytes, increased transcription of gfap and other genes could enhance astrocyte numbers and functions, which could upset the developmental balance between astrocytes and neurons and interfere with neuronal connections.
Figure 7.
Posited Pb-induced disruption of CNS development. Pb enters the cell and mobilizes calcium setting in motion a chain of events that culminate in the increased expression of IL-6 and TGF-β1 as described in the discussion (A). The newly synthesized cytokines then bind to their respective receptors and via their individual signaling cascades, promote the transcription of specific genes, such as GFAP (B), suggesting astrocyte activation and proliferation.
ACKNOWLEDGMENTS
This work was supported by NIH grants ES011135 and ES013857. We thank Michael Ryan and the late Robin Pietropaolo from the Wadsworth Center Microarray Core facility for their assistance in performing the microarray analysis, Nina G. Pabello and Greg Lyng for assistance in obtaining regional brain tissue for isolation of RNA.
REFERENCES
- 1.Needleman H. Lead poisoning. Annu Rev Med. 2004;55:209–222. doi: 10.1146/annurev.med.55.091902.103653. [DOI] [PubMed] [Google Scholar]
- 2.Lanphear BP, Hornung R, Khoury J, Yolton K, Baghurst P, Bellinger DC, et al. Low-level environmental lead exposure and children’s intellectual function: An international pooled analysis. Environ Health Perspect. 2005;113(7):894–899. doi: 10.1289/ehp.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein GW. Developmental neurobiology of lead toxicity. In: Needleman HL, editor. Human Lead Exposure. Boca Raton, FL: CRC; 1992. pp. 137–154. [Google Scholar]
- 4.Bradbury MWB, Deane R. Permeability of the blood-brain barrier to lead. Neurotoxicology. 1993;14(2–3):131–136. [PubMed] [Google Scholar]
- 5.Snyder JE, Filipov NM, Parsons PJ, Lawrence DA. The efficiency of maternal transfer of lead and its influence on plasma IgE and splenic cellularity of mice. Toxicol Sci. 2000;57(1):87–94. doi: 10.1093/toxsci/57.1.87. [DOI] [PubMed] [Google Scholar]
- 6.Krigman MR. Neuropathology of heavy metal intoxication. Environ Health Perspect. 1978;26:117–120. doi: 10.1289/ehp.7826117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toews AD, Krigman MR, Thomas DJ, Morell P. Effect of inorganic lead exposure on myelination in the rat. Neurochem Res. 1980;5(6):605–616. doi: 10.1007/BF00964782. [DOI] [PubMed] [Google Scholar]
- 8.Bressler JP, Goldstein GW. Mechanisms of lead neurotoxicity. Biochem Pharmacol. 1991;41(4):479–484. doi: 10.1016/0006-2952(91)90617-e. [DOI] [PubMed] [Google Scholar]
- 9.Kern M, Audesirk G. Inorganic lead may inhibit neurite development in cultured rat hippocampal neurons through hyperphosphorylation. Toxicol Appl Pharmacol. 1995;134(1):111–123. doi: 10.1006/taap.1995.1174. [DOI] [PubMed] [Google Scholar]
- 10.Qian Y, Tiffany-Castiglioni E. Lead-induced endoplasmic reticulum (ER) stress responses in the nervous system. Neurochem Res. 2003;28(1):153–162. doi: 10.1023/a:1021664632393. [DOI] [PubMed] [Google Scholar]
- 11.Dyatlov VA, Lawrence DA. Neonatal lead exposure potentiates sickness behavior induced by Listeria monocytogenes infection of mice. Brain Behav Immun. 2002;16(4):477–492. doi: 10.1006/brbi.2001.0641. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence DA. Immuntoxicity of heavy metals. In: Dean JH, Luster MJ, Munson R, et al., editors. Immunotoxicology and Immunopharmacology. New York: Raven Press; 1985. pp. 341–350. [Google Scholar]
- 13.Banks WA. Blood-brain barrier transport of cytokines: a mechanism for neuropathology. Curr Pharm Des. 2005;11:973–984. doi: 10.2174/1381612053381684. [DOI] [PubMed] [Google Scholar]
- 14.Dunn AJ, Wang J. Cytokine effects on CNS biogenic amines. Neuroimmunomodulation. 1995;2:319–328. doi: 10.1159/000097211. [DOI] [PubMed] [Google Scholar]
- 15.Harbuz MS, Stephanou A, Sarlis N, Lightman SL. The effects of recombinant human interleukin (IL)-1 alpha, IL-1 beta or IL-6 on hypothalamo-pituitary-adrenal axis activation. J Endocrinol. 1992;133(3):349–355. doi: 10.1677/joe.0.1330349. [DOI] [PubMed] [Google Scholar]
- 16.Stouthard JM, Romijn JA, van der Poll T, Endert E, Klein S, Bakker PJ, et al. Endocrinologic and metabolic effects of interleukin-6 in humans. Am J Physiol. 1995;268:E813–E819. doi: 10.1152/ajpendo.1995.268.5.E813. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins SJ, Rothwell NJ. Cytokines and the nervous system I: expression and recognition. Trends Neurosci. 1995;18(2):83–88. [PubMed] [Google Scholar]
- 18.Luheshi G, Rothwell N. Cytokines and fever. Int Arch Allergy Immunol. 1996;109:301–307. doi: 10.1159/000237256. [DOI] [PubMed] [Google Scholar]
- 19.Kishikawa H, Lawrence DA. Differential production of interleukin-6 in the brain and spleen of mice treated with lipopolysaccharide in the presence and absence of lead. J Toxicol Environ Health A. 1998;53:357–373. doi: 10.1080/009841098159222. [DOI] [PubMed] [Google Scholar]
- 20.Allan SM, Rothwell NJ. Cytokines and acute neurodegeneration. Nat Rev Neurosci. 2001;2(10):734–744. doi: 10.1038/35094583. [DOI] [PubMed] [Google Scholar]
- 21.Ransohoff RM, Benveniste EN, editors. Cytokines and the CNS. New York: Taylor & Francis Group; 2006. [Google Scholar]
- 22.Strle K, Zhou J-H, Shen W-H, Broussard SR, Johnson RW, Freund GG, et al. Interleukin-10 in the brain. Crit Rev Immunol. 2001;21:427–449. [PubMed] [Google Scholar]
- 23.Szelenyi J. Cytokines and the central nervous system. Brain Res Bull. 2001;54(4):329–338. doi: 10.1016/s0361-9230(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 24.Gadient RA, Otten U. Expression of interleukin-6 (IL-6) and interleukin-6 receptor (IL-6R) mRNAs in rat brain during postnatal development. Brain Res. 1994;637(1):10–14. doi: 10.1016/0006-8993(94)91211-4. [DOI] [PubMed] [Google Scholar]
- 25.Gruol DL, Nelson TE. Physiological and pathological roles of interleukin-6 in the central nervous system. Mol Neurobiol. 1997;15:307–339. doi: 10.1007/BF02740665. [DOI] [PubMed] [Google Scholar]
- 26.Du X, Everett ET, Wang G, Lee W-H, Yang Z, Williams D. Murine interleukin-11 (IL-11) is expressed at high levels in the hippocampus and expression is developmentally regulated in the testis. J Cell Physiol. 1996;168(2):362–372. doi: 10.1002/(SICI)1097-4652(199608)168:2<362::AID-JCP15>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 27.Maslinska D. The cytokine network and interleukin-15 (IL-15) in brain development. Folia Neuropathol. 2001;39:43–47. [PubMed] [Google Scholar]
- 28.Hanisch U-K, Lyons SA, Prinz M, Nolte C, Weber JR, Kettenmann H, et al. Mouse brain microglia express interleukin-15 and its multimeric receptor complex functionally coupled to Janus Kinase activity. J Biol Chem. 1997;272(46):28853–28860. doi: 10.1074/jbc.272.46.28853. [DOI] [PubMed] [Google Scholar]
- 29.Kurowska M, Rudnicka W, Maslinska D, Maslinska W. Expression of IL-15 and IL-15 receptor isoforms in select structures of human fetal brain. Ann NY Acad Sci. 2002;966:441–445. doi: 10.1111/j.1749-6632.2002.tb04245.x. [DOI] [PubMed] [Google Scholar]
- 30.Andre R, Wheeler RD, Collins PD, Luheshi GN, Pickering-Brown S, Kimber I, et al. Identification of a truncated IL-18Rβ mRNA: a putative regulator of IL-18 expressed in rat brain. J Neuroimmunol. 2003;145(1):40–45. doi: 10.1016/j.jneuroim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both TH1 and TH2 responses. Annu Rev Immunol. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler RD, Culhane AC, Hall MD, Pickering-Brown S, Rothwell NJ, Luheshi GN. Detection of the interleukin 18 family in rat brain by RT-PCR. Mol Brain Res. 2000;77(2):290–293. doi: 10.1016/s0169-328x(00)00069-3. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler RD, Brough D, Le Feuvre RA, Takeda K, Iwakura Y, Luheshi GN, et al. Interleukin-18 induces expression and release of cytokines from murine glial cells: interactions with interleukin-1β. J Neurochem. 2003;85(6):1412–1420. doi: 10.1046/j.1471-4159.2003.01787.x. [DOI] [PubMed] [Google Scholar]
- 34.Culhane AC, Hall MD, Rothwell NJ, Luheshi GN. Cloning of rat brain interleukin-18 cDNA. Mol Psychiatry. 1998;3:362–366. doi: 10.1038/sj.mp.4000389. [DOI] [PubMed] [Google Scholar]
- 35.Cannella B, Raine CS. Multiple sclerosis: cytokine receptors on oligodendrocytes predict innate regulation. Ann Neurol. 2004;55(1):46–57. doi: 10.1002/ana.10764. [DOI] [PubMed] [Google Scholar]
- 36.Conti B, Park LC, Calingasan NY, Kim Y, Kim H, Bae Y, et al. Cultures of astrocytes and microglia express interleukin 18. Brain Res Mol Brain Res. 1999;67(1):46–52. doi: 10.1016/s0169-328x(99)00034-0. [DOI] [PubMed] [Google Scholar]
- 37.Kurschner C, Yuzaki M. Neuronal interleukin-16 (NIL-16): A dual function PDZ domain protein. J Neurosci. 1999;19(18):7770–7780. doi: 10.1523/JNEUROSCI.19-18-07770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Constam DB, Philipp J, Malipiero UV, ten Dijke P, Schachner M, Fontana A. Differential expression of transforming growth factor-β1, β2, and β3 by glioblastoma cells, astrocytes, and microglia. J Immunol. 1992;148(5):1404–1410. [PubMed] [Google Scholar]
- 39.Gomes FCA, Sousa VdeO, Romao L. Emerging roles for TGF-β1 in nervous system development. Int J Dev Neurosci. 2005;23(5):413–424. doi: 10.1016/j.ijdevneu.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Darnall BD, Suarez EC. Sex and gender in psychoneuroimmunology research: past, present, and future. Brain, Behavior, and Immunity. 2009;23(5):595–604. doi: 10.1016/j.bbi.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim WG, Mohney RP, Wilson B, Jeohn GH, Liu B, Hong JS. Regional difference in susceptibility to LPS-induced neurotoxicity in the rat brain: role of microglia. J Neurosci. 2000;20(16):6309–6316. doi: 10.1523/JNEUROSCI.20-16-06309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39(1):151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 43.Aloisi F, Care A, Borsellino G, Gallo P, Rosa S, Bassani A, et al. Production of hemolymphopoietic cytokines (IL-6, IL-8, colony-stimulating factor) by normal human astrocytes in response to IL-1β and tumer necrosis factor-α. J Immunol. 1992;149(7):2358–2366. [PubMed] [Google Scholar]
- 44.Cannella B, Sizing ID, Benjamin CD, Browning JL, Raine CS. Antibodies to lymphotoxin α (LTα) and LTβ recognize different glial cell types in the central nervous system. J Neuroimmunol. 1997;78(2):172–179. doi: 10.1016/s0165-5728(97)00098-2. [DOI] [PubMed] [Google Scholar]
- 45.Heo Y, Lee WT, Lawrence DA. Differential effects of lead and cAMP on development and activities of Th1- and Th2-lymphocytes. Toxicol Sci. 1998;43(1):172–185. doi: 10.1006/toxs.1998.2457. [DOI] [PubMed] [Google Scholar]
- 46.Wanf J, Shannon MF, Young IG. A role for Ets1, synergizing with AP-1 and GATA-3 in the regulation of IL-5 transcription in mouse Th2 lymphocytes. Int Immunol. 2006;18(2):313–323. doi: 10.1093/intimm/dxh370. [DOI] [PubMed] [Google Scholar]
- 47.Dai J, Huang C, Wu J, Yang C, Frenkel K, Huang X. Iron-induced interleukin-6 gene expression: possible mediation through the extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways. Toxicology. 2004;203(3):199–209. doi: 10.1016/j.tox.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Liu G, Ding W, Liu X, Mulder K. c-Fos is required for TGFβ1 production and the associated paracrine migratory effects of human colon carcinoma cells. Mol Carcinog. 2006;45(8):582–593. doi: 10.1002/mc.20189. [DOI] [PubMed] [Google Scholar]
- 49.Persson E, Voznesensky OS, Huang Y-F, Lerner UH. Increased expression of interleukin-6 by vasoactive intestinal peptide is associated with regulation of CREB, AP-1 and C/EBP, but not NF-κB, in mouse calvarial osteoblasts. Bone. 2005;37(4):513–529. doi: 10.1016/j.bone.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 50.Xie J, Pan H, Yoo S, Gao S-J. Kaposi’s Sarcoma-associated herpesvirus induction of AP-1 and interleukin-6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J Virol. 2005;79(24):15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abecassis L, Rogier E, Vazquez A, Atfi A, Bourgeade M-F. Evidence for a role of MSK1 in transforming growth factor-β-mediated responses through p38α and Smad signaling pathways. J Biol Chem. 2004;279(29):30474–30479. doi: 10.1074/jbc.M403294200. [DOI] [PubMed] [Google Scholar]
- 52.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 53.ten Dijke P, Hill CS. New insights into TGF-β- Smad signaling. Trends Biochem Sci. 2004;29(5):265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF–β–induced transcription. Nature. 1998;394(6696):909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 55.Taga T, Fukuda S. Role of IL-6 in the neural stem cell differentiation. Clin Rev Allergy Immunol. 2005;28:249–256. doi: 10.1385/CRIAI:28:3:249. [DOI] [PubMed] [Google Scholar]
- 56.de Sampaio e Spohr TCL, Martinex R, da Silva EF, Neto VM, Gomes FCA. Neuro-glia interaction effects on GFAP gene: a novel role for transforming growth factor-β1. Eur J Neurosci. 2002;16(11):2059–2069. doi: 10.1046/j.1460-9568.2002.02283.x. [DOI] [PubMed] [Google Scholar]
- 57.Sousa VdeO, Romao L, Neto VM, Gomes FCA. Glial fibrillary acidic protein gene promoter is differently modulated by transforming growth factor beta-1 in astrocytes from distinct brain regions. Eur J Neurosci. 2004;19(7):1721–1730. doi: 10.1111/j.1460-9568.2004.03249.x. [DOI] [PubMed] [Google Scholar]
- 58.Ojala J, Alafuzoff I, Herukka S-K, van Groen T, Tanila H, Pirttila T. Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients. Neurobiology of Aging. 2009;30(2):198–209. doi: 10.1016/j.neurobiolaging.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Curran B, O’Conner JJ. The pro-inflammatory cytokine interleukin-18 impairs long-term-potentiation and NMDA receptor mediated transmission in the rat hippocampus in vitro. Neuroscience. 2001;108(1):83–90. doi: 10.1016/s0306-4522(01)00405-5. [DOI] [PubMed] [Google Scholar]
- 60.Cordova FM, Rodrigues ALS, Giacomelli MBO, Oliveira CS, Posser T, Dunkley PR, Leal RB. Lead stimulate ERK1/2 and p38MAPK phosphorylation in the hippocampus of immature rats. Br Res. 2004;998(1):65–72. doi: 10.1016/j.brainres.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 61.Leal RB, Ribeiro SJ, Posser T, Cordova FM, Rigon AP, Filho EZ. Modulation of ERK1/2 and p38MAPK by lead in the cerebellum of Brazilian catfish Rhamdia quelen. Aq Toxicol. 2006;77(1):98–104. doi: 10.1016/j.aquatox.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Thomas GM, Huganir RL. MAPK cascade signaling and synaptic plasticity Nature Rev. Neuroscience. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 63.Schwab JM, Nguyen TD, Meyermann R, Schluesener HJ. Human focal cerebral infarctions induce differential lesional interleukin-16 (IL-16) expression confined to infiltrating granulocytes, CD8+ T-lymphocytes and activated microglia/macrophages. J Neuroimmunol. 2001;114(2):232–241. doi: 10.1016/s0165-5728(00)00433-1. [DOI] [PubMed] [Google Scholar]
- 64.Zhao M-L, Si Q, Lee SC. IL-16 expression in lymphocytes and microglia in HIV-1 encephalitis. Neuropathol Appl Neurobiol. 2003;30(3):233–242. doi: 10.1046/j.0305-1846.2003.00527.x. [DOI] [PubMed] [Google Scholar]
- 65.Chen H-H, Ma T, Ho IK. Protein kinase C in rat brain is altered by developmental lead exposure. Neurochem Res. 1999;24(3):415–421. doi: 10.1023/a:1020993802239. [DOI] [PubMed] [Google Scholar]
- 66.Costa LG, Guizzetti M, Lu H, Bordi F, Vitalone A, Tita B, et al. Intracellular signal transduction pathways as targets for neurotoxicants. Toxicology. 2001;160(1):19–26. doi: 10.1016/s0300-483x(00)00435-2. [DOI] [PubMed] [Google Scholar]
- 67.Cremin JD, Jr, Smith DR. In vitro and in vivo Pb effects on brain protein kinase C activity. Environ Res. 2002;90(3):191–199. doi: 10.1016/s0013-9351(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 68.Long GJ, Rosen JF, Schanne FAX. Lead activation of protein kinase C from rat brain. J Biol Chem. 1994;269(2):834–837. [PubMed] [Google Scholar]
- 69.Bressler J, Kim K, Chakraborti T, Goldstein G. Molecular mechanisms of lead neurotoxicity. Neurochem Res. 1999;24(4):595–600. doi: 10.1023/a:1022596115897. [DOI] [PubMed] [Google Scholar]
- 70.Kim K-A, Chakraborti T, Goldstein GW, Bressler JP. Immediate early gene expression in P12 cells exposed to lead: Requirement for protein kinase C. J Neurochem. 2000;74(3):1140–1146. doi: 10.1046/j.1471-4159.2000.741140.x. [DOI] [PubMed] [Google Scholar]
- 71.Ramesh GT, Manna SK, Aggarwal BB, Jadhav AL. Lead exposure activates nuclear factor kappa B, activator protein-1, c-jun N-terminal kinase and caspases in the rat brain. Toxicol Lett. 2001;123(3):195–207. doi: 10.1016/s0378-4274(01)00395-2. [DOI] [PubMed] [Google Scholar]
- 72.Lu H, Guizzetti M, Costa L. Inorganic lead activates the mitogen-activated protein kinase kinase-mitogen-activated protein kinase-p90RSK signaling pathway in human astrocytoma cells via a protein kinase C-dependent mechanism. J Pharmacol Exp Ther. 2002;300(3):818–823. doi: 10.1124/jpet.300.3.818. [DOI] [PubMed] [Google Scholar]
- 73.Atkins DS, Basha MR, Zawia NH. Intracellular signaling pathways involved in mediating the effects of lead on the transcription factor Sp1. Int J Dev Neurosci. 2003;21(5):235–244. doi: 10.1016/s0736-5748(03)00067-4. [DOI] [PubMed] [Google Scholar]
- 74.Crumpton T, Atkins DS, Zawia NH, Barone S. Lead exposure in pheochromocytoma (PC12) cells alters neural differentiation and Sp1 DNA-binding. Neurotoxicology. 2001;22(1):49–62. doi: 10.1016/s0161-813x(00)00008-5. [DOI] [PubMed] [Google Scholar]
- 75.Bamba S, Andoh A, Yasui H, Makino J, Kim S, Fujiyama Y. Regulation of IL-11 expression in intestinal myofibroblasts: role of c-Jun, AP-1-and MAPK-dependent pathways. Am J Physiol Gastrointest Liver Physiol. 2003;285(3):529–538. doi: 10.1152/ajpgi.00050.2003. [DOI] [PubMed] [Google Scholar]
- 76.NCBI (National Center for Biotechnology Information) Nucleotide. [accessed 1 September 2006];2006 http://www.ncbi.nlm.nih.gov Available:
- 77.Yang L, Yang Y-C. Regulation of interleukin (IL)-11 gene expression in IL-1 induced primate bone marrow stromal cells. J Biol Chem. 1994;269(52):32732–32739. [PubMed] [Google Scholar]
- 78.Kang S-H, Brown DA, Kitajima I, Xu X, Heidenreich O, Gryaznov S, et al. Binding and functional effects of transcriptional factor Sp1 on the murine interleukin-6 promotor. J Biol Chem. 1996;271(13):7330–7335. doi: 10.1074/jbc.271.13.7330. [DOI] [PubMed] [Google Scholar]
- 79.Kim Y, Ratziu V, Choi S-G, Lalazar A, Theiss G, Dang Q, et al. Transcriptional activation of transforming growth factor β1 and its receptors by the Kruppel-like Zf9/core promoter-binding protein and Sp1. J Biol Chem. 1998;273(50):33750–33758. doi: 10.1074/jbc.273.50.33750. [DOI] [PubMed] [Google Scholar]
- 80.Sanceau J, Kaisho T, Hirano T, Wietzerbin J. Triggering of the human interleukin-6 gene by interferon-γ and tumor necrosis factor-α in monocytic cells involves cooperation between interferon regulatory factor-1, NFκB, and Sp1. J Biol Chem. 1995;270(46):27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- 81.Klasen HJ, Imfeld KL, Kirov II, Tai L, Gage FH, Young MJ, et al. Expression of cytokines by multipotent neural progenitor cells. Cytokine. 2003;22(4):101–106. doi: 10.1016/s1043-4666(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 82.Van Wagoner NJ, Benveniste EN. Interleukin-6 expression and regulation in astrocytes. J Neuroimmunol. 1999;100(1):124–139. doi: 10.1016/s0165-5728(99)00187-3. [DOI] [PubMed] [Google Scholar]
- 83.Wyss-Coray T, Borrow P, Brooker MJ, Mucke L. Astroglial overproduction of TGF-β1 enhances inflammatory central nervous system disease in transgenic mice. J Neuroimmunol. 1997;77(1):45–50. doi: 10.1016/s0165-5728(97)00049-0. [DOI] [PubMed] [Google Scholar]
- 84.Flanders KC, Ren RF, Lippa CF. Transforming growth factor-βs in neurodegenerative disease. Prog Neurobiol. 1998;54(1):71–85. doi: 10.1016/s0301-0082(97)00066-x. [DOI] [PubMed] [Google Scholar]
- 85.Wahl SM, Allen JB, McCartney-Francis N, Marganti-Kossmann MC, Kossmann T, Ellingsworth L, et al. Macrophage- and astrocyte-derived transforming growth factor β as a mediator of central nervous system dysfunction in acquired immune deficiency syndrome. J Exp Med. 1991;173(4):981–991. doi: 10.1084/jem.173.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buisson A, Lesne S, Docagne F, Ali C, Nicole O, MacKenzie ET, et al. Transforming growth factor-β and ischemic brain injury. Cell Mol Neurobiol. 2003;23(4):539–549. doi: 10.1023/A:1025072013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cafferty WBJ, Gardiner NJ, Das P, Qiu J, McMahon SB, Thompson SWN. Conditioning injury-induced spinal axon regeneration fails in interleukin-6 knock-out mice. J Neurosci. 2004;24(18):4432–4443. doi: 10.1523/JNEUROSCI.2245-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bravo J, Heath JK. Receptor recognition by gp130 cytokines. EMBO J. 2000;19(11):2399–2411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teng FYH, Tang BL. Axonal regeneration in adult CNS neurons-signaling molecules and pathways. J Neurochem. 2006;96(6):1501–1508. doi: 10.1111/j.1471-4159.2006.03663.x. [DOI] [PubMed] [Google Scholar]
- 90.Sometani A, Kataoka H, Nitta A, Fukumitsu H, Nomoto H, Furukawa S. Transforming growth factor-b1 enhances expression of brain-derived neurotrophic factor and its receptor, TrkB, in neurons cultured from rat cerebral cortex. J Neurosci Res. 2001;66(3):369–376. doi: 10.1002/jnr.1229. [DOI] [PubMed] [Google Scholar]
- 91.Harry GJ, Schmitt TJ, Gong Z, Brown H, Zawia N, Evans HL. Lead-induced alterations of glial fibrillary acidic protein (GFAP) in the developing rat brain. Toxicol Appl Pharmacol. 1996;139(1):84–93. doi: 10.1006/taap.1996.0145. [DOI] [PubMed] [Google Scholar]
- 92.Zawia NH, Harry GJ. Developmental exposure to lead interferes with glial and neuronal differential gene expression in the rat cerebellum. Toxicol Appl Pharmacol. 1996;138(1):43–47. doi: 10.1006/taap.1996.0095. [DOI] [PubMed] [Google Scholar]
- 93.Zhu G, Mehler MF, Mabie PC, Kessler JA. Developmental changes in progenitor cell responsiveness to cytokines. J Neurosci Res. 1999;56(2):131–145. doi: 10.1002/(sici)1097-4547(19990415)56:2<131::aid-jnr3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 94.Deng W, Poretz RD. Protein kinase C activation is required for the lead-induced inhibition of proliferation and differentiation of cultured oligodendroglial progenitor cells. Brain Res. 2002;929(1):87–95. doi: 10.1016/s0006-8993(01)03385-6. [DOI] [PubMed] [Google Scholar]
- 95.Morgan RE, Garavan H, Smith EG, Driscoll LL, Levitsky DA, Strupp BJ. Early lead exposure produces lasting changes in sustained attention, response initiation, and reactivity to errors. Neurotoxicol Teratol. 2001;23(6):519–531. doi: 10.1016/s0892-0362(01)00171-4. [DOI] [PubMed] [Google Scholar]
- 96.Vazquez A, Pena de Ortiz S. Lead (Pb+2) impairs long-term memory and blocks learning-induced increases in hippocampal protein kinase C activity. Toxicol Appl Pharmacol. 2004;200(1):27–39. doi: 10.1016/j.taap.2004.03.011. [DOI] [PubMed] [Google Scholar]