Abstract
Objective:
Epithelial ovarian cancer (EOC) cells with CD44 and CK19 coexpression may represent a subset of ovarian cancer stem cells (OCSCs). This study was conducted to evaluate the correlation of the frequency of putative OCSCs (CD44 + CK19 + OCSCs) with the clinicopathologic features and the prognostic value in patients with recurrent advanced stage EOC.
Methods:
A retrospective study was carried out on 33 patients with EOC and a uniformly treated tissue microarray was constructed. A multiplexed, immunofluorescence-based method of automated in situ quantitative measurement of protein analysis was used for evaluation of the frequency or density of CD44 + CK19 + OCSCs in EOC.
Results:
The mean follow-up time was 42.8 ± 27.1 months. High frequency of EOC cells with CD44+ or CD44+/CK19+ was associated with chemoresistance (P = .033 and P = .02, respectively). Using K-M analysis with log-rank test, a high frequency of putative OCSCs was associated with short disease-free interval (7.9 months vs 20.9 months, P = .019). In univariable analysis, the frequency of OCSCs, International Federation of Gynecology and Obstetrics stage and residual tumor volume were significant predictor variables and were entered into multivariable analysis (P = .019, .037, and .005, respectively). Although no independent significant predictor was found, the frequency of putative OCSCs was the most promising predictor variable compared with the other 2 variables (hazard ratio = 2.344, P = .052).
Conclusion:
Our findings suggest that high frequency of OCSCs (CD44+ and CK19+) in epithelial ovarian tumors correlates with short progression-free intervals.
Keywords: ovarian cancer stem cells, CD44, CK19, AQUA
Introduction
Ovarian cancer varies widely in frequency among different geographic regions and ethnic groups, with a high incidence in Northern Europe and the United States, and a low incidence in Japan. The majority of cases are sporadic, and only 5% to 10% of ovarian cancers are familial.1
Approximately 75% of patients present with metastatic disease. While a large proportion have an excellent response initially when treated with optimal debulking and platinum-based chemotherapy, 80% of patients invariably will experience relapse of the disease and chemoresistance. The 5-year survival rate of patients with advanced stage disease is between 15% and 45%.2,3 The overall survival has modestly improved over the past decades.3
A better understanding of the biology of ovarian cancer is the cornerstone of defining relevant targets and developing novel approaches for therapy. More recently a series of studies have demonstrated the presence of a unique type of cells called cancer stem cells (CSCs). The concept of CSCs was first brought forward almost three decades ago, when a population of cancer cells with tumor-initiating properties was identified in hematologic cancers and some solid tumors.4,5
Cancer stem cells are a subgroup of malignant cells, which can give rise to a hierarchy of proliferating and progressively differentiating cells. Additionally, this subpopulation of tumor cells is resistant to conventional therapies and it has been suggested that these cells are the source of chemoresistance.6–9
We have recently identified a subgroup of ovarian cancer cells with stem cell (OCSC)-like properties. These cells are characterized by high tumorigenic capacity, resistance to chemotherapy, and differentiation potential both in vitro and in vivo.8–10 Several markers have been described for the identification of epithelial ovarian cancer (EOC) stem cells including CD44, CD133, CD24, ALDH1, MyD88, and CD117.11–14 Of these markers, the cell surface protein CD44 has been the most extensively studied.15 CD44+ OCSCs express pluripotency markers such as β-catenin, Oct-4, and SSEA-4 and have been demonstrated to be the chemoresistant progenitors in vivo and able to differentiate into the heterogeneous cell types comprising the tumor.8,16–18
Gene expression microarray analysis of CD44+ OCSCs revealed numerous differentially expressed genes including the intracellular protein cytokeratin 19 (CK19).8,19 Cytokeratin 19 has been found in a number of normal epithelial cells and was also described as a marker for mammary carcinoma cells.19 Most recently, a comparative proteomic study showed that CK19 were upregulated in poorly differentiated ovarian tumors.20
Evaluation and quantification of protein expression in paraffin section are limited because of lack of standardized methods. An immunofluorescence-based method of automated quantitative measurement of protein analysis (AQUA) is one of the most advanced pathological approaches for the quantification of proteins in tissue sections. Combined with tissue microarrays (TMAs), AQUA provides scoring, which is precise, reproducible, and free of the subjectivity associated with the pathologists’ evaluation.21
The objectives of this study were (1) to determine the value of CK19 alone or in combination with CD44 for the identification of OCSC in tissue sections and (2) to evaluate the potential use of these markers to predict the response to therapy. We hypothesized that patients whose tumors had a high frequency of OCSCs were more likely to present with chemoresistance and shorter progression-free interval. By using the AQUA system on TMAs, we demonstrated that the frequency of CD44 and CK19 coexpression as putative bimolecular markers of OCSCs was strongly associated with the response to chemotherapy and progression-free interval in patients with recurrent EOC.
Materials and Methods
Patients
A retrospective chart review from 2006 identified 33 consecutive patients diagnosed with first recurrence of epithelial ovarian carcinoma (EOC). These patients were treated with surgery and pre- or postoperative platinum-paclitaxel chemotherapy at Yale-New Haven Hospital. The stages were determined according to the International Federation of Gynecology and Obstetrics (FIGO) staging system. Histological grading evaluated the tumor architecture, the amount of solid neoplastic areas, the nucleus–cytoplasm ratio, and the nuclear polymorphism. The tumors were categorized as well (G1), moderately (G2), and poorly (G3) differentiated. The surgical approach consisted of total abdominal hysterectomy, bilateral salpingo-oophorectomy, total omentectomy, pelvic and paraaortic lymph node dissection, and maximum resection of all gross diseases.
Chemoresistance was defined as disease recurrence after intervals of <6 months following a platinum-based regimen, while platinum refractory patients had stable or progressive disease while being treated with chemotherapy. If the disease-free intervals were 6 months or longer, the tumors were defined as chemosensitive. All patients signed consent forms and the use of patient samples was approved by Yale University’s Human Investigations Committee.
Tissue Microarrays and Immunohistochemistry
A TMA consisting of tumors from all 33 patients was constructed at the Yale University Tissue Microarray Facility. Tissue cylinders with a diameter of 0.6 mm were punched from morphologically representative tissue areas of each “donor” tissue block and were placed in a recipient block.22 Tissue microarrays slide preparation including deparaffinization and staining was previously described.23 Briefly, slides were deparaffinized with xylene, washed, and rehydrated with alcohol of anticoncentration gradient. Antigen retrieval was accomplished by pretreatment module heating at 97°C for 20 minutes. Endogenous peroxidase activity was blocked by incubating in 0.3% hydrogen peroxide in methanol for 30 minutes at room temperature (RT). Nonspecific antibody binding was then blocked with 0.3% bovine serum albumin (BSA) in 0.1 mol/L of Tris-buffered saline (TBS) with 0.05% Tween for 30 minutes at RT. Following these steps, slides were incubated with the primary antibodies CD44, CK19, or a CK19 + CD44 cocktail by diluting at the desired concentration at 4°C overnight. The primary antibodies were rabbit monoclonal CD44 antibody (EPR1013Y, Abcam Corporation, Cambridge, Massachusetts, dilution 1:1000) and CK19 antibody (A53-B/A2, Abcam Corporation, dilution 1:50). For single target slides, the antibodies were incubated with goat anti-mouse (for CK19) or anti-rabbit (for CD44) secondary antibodies conjugated separately to a horseradish peroxidase–decorated polymer backbone (Envision, Dako North America, Inc, Carpinteria, California). For multiplexed immunohistochemistry, the slides were first incubated with anti-mouse antibody for 1 hour at RT. Cy5-tyramide (1:50) was added to each slide and incubated for 10 minutes and then benzoic hydrazide was used to block peroxidases. Anti-rabbit secondary antibodies were added to the multiplex immunohistochemistry (IHC) slide and followed with Alexa-750. Peroxidases were blocked again. Each slide was incubated with chicken monoclonal pan-cytokeratin (1:100, lab made, affinity purified) in 0.3% BSA/TBS at 4°C overnight. Goat anti-chicken secondary antibodies conjugated with Alexa 546 were diluted at 1:200 and used to stain each slide. Tissue nuclei were stained with 4′,6-diamidino-2-phynilindol (DAPI).
Automated Quantitative Analysis
A rigorous quantitative immunofluorescent-based method, the AQUA technology, was used to assess the protein expression. The details of AQUA have been described elsewhere.24–28 Briefly, the AQUA technology is a fluorescence immunohistochemistry–based method that generates objective and continuous protein expression scores in tissue by using automated fluorescence microscopy and advanced image analysis algorithms. The AQUA scores are directly proportional to molecules per unit area or protein concentration. Unlike traditional immunohistochemistry, the AQUA system produces strictly quantitative in situ protein expression data on a continuous scale rather than subjective, categorical data. Tissue samples are stained with markers that define the subcellular compartments of interest and the specific target (or targets) being studied. To measure the target gene expression, 4′-6-diamidino-2-phenylindole is used for the nuclear mask, anti-cytokeratin antibodies are used to identify and differentiate epithelium from stroma (tumor mask) and establish the cytoplasmic mask and an antibody directed against the target is used to visualize the target gene protein expression of interest. A high-resolution automated image acquisition is utilized after the TMA is stained. These images are individually thresholded to remove nonspecific signal then combined to produce a virtual image that represents pixels that are not only epithelial-specific but also represent cytoplasm and nuclear-specific pixels. Pixel intensities from a specific target that has been labeled for readout in a third fluorescent channel can subsequently be quantified within this ‘‘PLACEd’’ image.
The foundation of the AQUA score is the pixel-based locale assignment for compartmentalization of expression (PLACE) image analysis algorithm. Tissue is a complex mixture of various tissue components (ie, epithelium, stroma, and blood vessels) and subcellular components (ie, cytoplasm, nuclei, and membrane). The PLACE enables differential localization of image pixel intensities associated with target gene expression in these different components or masks. A critical step in the AQUA software is the setting of intensity thresholds that are used to delineate background or nonspecific pixels from signal-specific pixels. The AQUA analysis begins by generating a tumor mask through pixel intensity thresholding (binary gating) and spatial image analysis procedures. Images that have been masked in this way are subsequently combined in a mutually exclusive fashion such that pixels above the thresholds are assigned to specific subcellular compartments. Once pixels have been assigned to each compartment, the signal for the target biomarker can then be averaged over all of the pixels assigned to a given compartment, which is the AQUA score for that sample.28
For this study, serial monochromatic high-resolution image acquisitions were performed from each histospot by HistoRX PM-2000 platform (HistoFx, New Haven, Connecticut). For each histospot, in- and out-of-focus images were obtained using the signals from the DAPI channel, from the cytokeratin–Alexa 546 channel, from the CD44-Cy5 channel and from the CK19-Alexa 750 channel. A tumor mask, defined as the region of cytokeratin signal, was used to distinguish tumor cells from stromal and lymphocytic elements. Only expression within the tumor mask was calculated as a positive score. To measure the frequency of CK19 and CD44 coexpression, we binarized the CK19 signal within the tumor mask and created a CK19 compartment, which allowed us to measure the CD44 pixel intensity within the CK19 compartment. The AQUA score of a given target within the tumor mask or the CK19 compartment were calculated by dividing the signal intensity of the tumor mask area or CK19 compartment within the histospot. Histospots containing 5% tumor or less, as assessed by the percentage positive for cytokeratin, were excluded from further analysis.
Statistical Analysis
Biomarkers’ expression and their correlation with the following clinicopathologic features were evaluated by 1-way analysis of variance (ANOVA) test: age, histological type, FIGO stage, degree of tumor differentiation, and optimal (≤1 cm) versus suboptimal (>1 cm) debulking.
The X-tile program generated an optimal cutoff point of a marker’s AQUA score that best separated the cohort by the largest outcome difference, that is, time to disease progression.29 However, the cutoff point in one particular cohort can be cohort specific, that is, the same cutoff point may not be able to separate another cohort by the same outcome, and therefore X-tile generated cutoff points usually need to be validated in a secondary cohort. Nevertheless, X-tile generated cutoff points provide preliminary trend information when studying biomarkers. We have found in some biomarker studies that X-tile cannot generate an optimal cutoff point. In these cases, the distribution of biomarker expression in fast-progression tumors has a similar shape to the distribution in slow-progression tumors. Therefore, if X-tile is unable to generate an optimal cutoff point for a biomarker, at least it indicates that there is a difference in the biomarkers’ distribution between fast- and slow-progressing tumors.
The primary end point of this study was progression-free interval, defined as the time from when the first-line treatment was completed to the date of recurrence. Disease progression was diagnosed by rising cancer antigen-125 levels as recommended by the Gynecologic Cancer Intergroup in 2005 and/or imaging (http://gcig.igcs.org/CA125/respdef_nov2005.pdf).30,31 Overall survival was defined as the time from diagnosis to the date of death from any cause or the censoring date. Survival status was confirmed by medical chart review as of March 1, 2011. This date served as the censoring date. Kaplan-Meier curve analyses with log-rank tests were used to determine statistical significance. The univariate Cox regression model was applied to evaluate the hazard ratio (HR) of biomarkers on progression-free interval and overall survival. Only significant factors from univariate Cox regression analysis were entered into multivariate Cox regression analysis. Hazard ratios were calculated on log-transformed biomarkers and represented with their 95% confidence interval and 2-sided P value. P value <.05 was considered statistically significant.
All analyses were done using SPSS 15.0 for windows (SPSS Inc, Chicago, Illinois) and X-Tile 3.61 (Yale University, New Haven, Connecticut).
Results
The Study Population
Totally, 33 tumor samples were analyzed and the clinical course of these patients were followed retrospectively. The patients’ median age at diagnosis was 62.6 ± 9.8 years (range 44-86 years). By FIGO stage, 26 were stage III (26 of 33, 78.8%), 3 were stage IV (3 of 33, 9.1%), and 4 were stage X (4 of 33, 12.1%). The histological types were serous papillary carcinoma in 30 (90%) of 33 patients, clear cell carcinoma in 1 patient, mixed carcinoma (serous papillary and endometrioid with squamous differentiation) in 1 patient, and serotransitional carcinoma in 1 patient. The tumors were poorly differentiated in 26 (78.8%) of 33 cases, moderately differentiated in 6 (18.2%) of 33 cases, and well differentiated in 1 (3%) case. Of the 33 patients, 26 (78.8%) underwent optimal tumor debulking, whereas 7 (21.2%) of 33 patients underwent suboptimal debulking. All patients were treated with intravenous chemotherapy with paclitaxel and carboplatin. According to their response to chemotherapy, the patients were classified as chemosensitive (15 of 33, 45.5%) or chemoresistant (18 of 33, 54.5%). The follow-up time for these patients ranges from 4.3 months to 113 months, with a mean follow-up of 42.8 ± 27.1 months. Table 1 summarizes patients’ information.
Table 1.
Summary of the Patients’ Clinical and Pathologic Features.
| Variable | Frequency | Percentage |
|---|---|---|
| Age | ||
| ≤62 | 19 | 57.60 |
| >62 | 14 | 42.40 |
| Histology | ||
| Serous | 29 | 87.90 |
| Others | 4 | 12.10 |
| FIGO stage | ||
| III | 26 | 78.79 |
| IV | 3 | 9.09 |
| X | 4 | 12.12 |
| Differentiation | ||
| Well-moderate | 7 | 21.20 |
| Poor | 26 | 78.80 |
| Debulking | ||
| Optimal | 26 | 78.80 |
| Suboptimal | 7 | 21.20 |
| Lymph node | ||
| Negative | 13 | 39.40 |
| Positive | 14 | 42.40 |
| Unconfirmed | 6 | 18.20 |
| Chemoresponse | ||
| Sensitive | 15 | 45.50 |
| Resistance | 18 | 54.50 |
Abbreviation: FIGO, International Federation of Gynecology and Obstetrics.
During the follow-up interval, 12 patients died, 13 patients were followed-up to the censoring date, and 8 patients were lost to follow-up (19.3-63.6 months).
Immunofluorescence Staining of CD44, CK19, or CD44/CK19 Coexpression in Tumor Mask
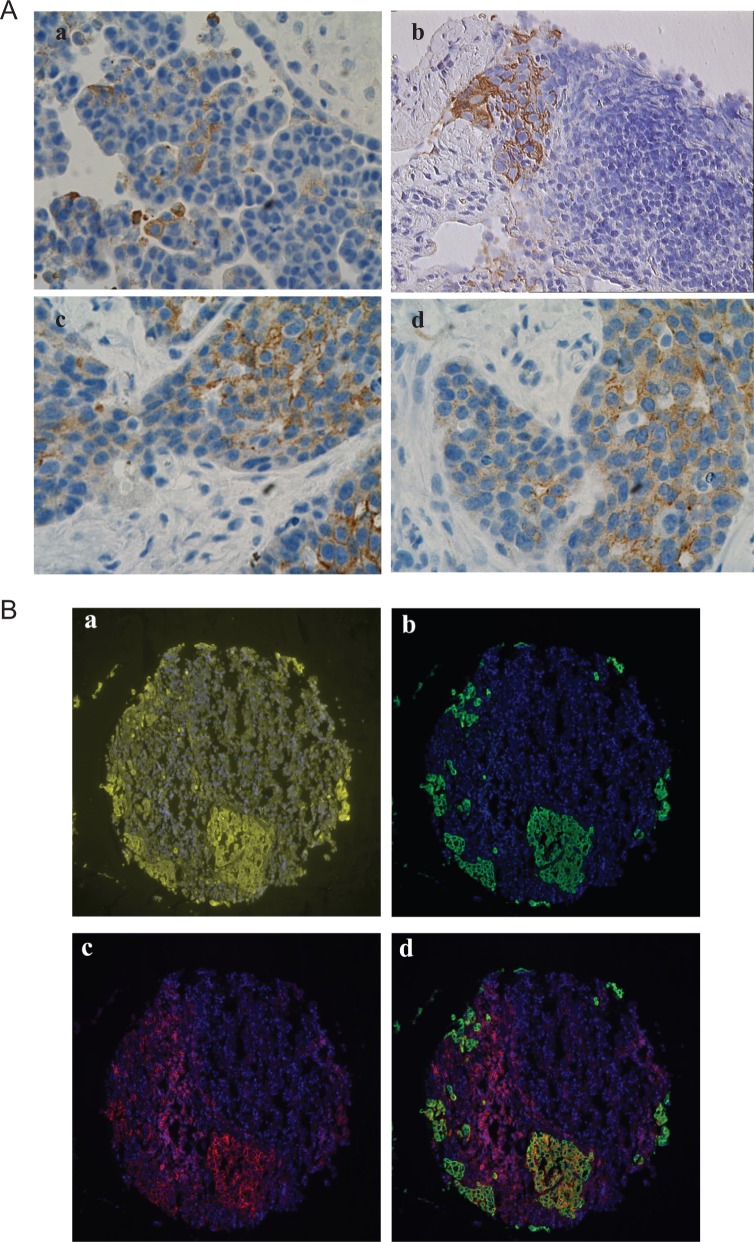
CK19 was predominantly expressed in the epithelial cells although its expression was heterogeneous. The percentage of CK19+ epithelial cells varied between samples (Figure 1A). Some tumors presented clusters of CK19+ cells surrounded by CK19− cancer cells (Figure 1B, a and b), while others had a higher percentage of CK19+ cells with only few CK19− cancer cells (Figure 1B, c and d). These findings suggest that the expression of CK19 is associated with a specific group of cancer cells and its quantification could provide important clinical information. Expression of CD44 was found not only in epithelial cells but also in various stroma and/or immune cells; which makes difficult to quantify the expression levels of CD44 by cancer cells. The heterogeneity of CD44 was previously studied.19 We were able to overcome this problem by using the AQUA system. This allows removal of noncancer cells by application of the masking system using multiple antigens.32 The exclusion of stroma and immune cells was done through defined regions of the epithelial cells mask created by pan-cytokeratin. Using this approach, we could identify the CD44+ and CK19 ovarian cancer cells. Most CD44+ cells were found either as small clusters or as single cells. CK19+ cells commonly appeared as clusters or small islands. These expressions were widely heterogeneous. Coexpression of CD44 and CK19 varied between tumor cells and tumor samples (Figure 1).
Figure 1.
Expression of CK-10 and CD44 in ovarian cancer. A, Representative immunostaining for CK19 in ovarian cancer samples. B, Representative examples for CD44 and CD44/CK19 staining patterns in Yale ovarian cancer 203 TMA. 4′,6-Diamidino-2-phenylindole ([DAPI] nucleus) is blue, pan-cytokeratin is yellow, CK19 is green, and CD44 is red. (a) Pan-cytokeratin and DAPI staining. (b) CD44 and DAPI staining. (c) CK19 and DAPI staining. (d) CD44, CK19, and DAPI staining merged.
Correlation of CD44, CK19, and CD44/CK19 Expression to Clinicopathologic Features and Progression-Free Interval
Next, we quantified the expression of CD44+ and CK9+ cells. The expression of CD44 in tumor mask, CK19 in tumor mask, and CD44 in the CK19 compartment were measured by AQUA analysis in 2-fold redundancy. As described in the Materials and Methods section, AQUA scores for duplicate tissue cores were averaged to obtain a mean AQUA score for each tumor. Of the 33 patients, 28 were scoreable in 2-fold redundancy with greater than 5% tumor mask within the histospot. Five patients had only 1 histospot that reached the inclusion criteria, so the single AQUA score characterized these patients.
Using ANOVA test, we found that the mean level of CD44 expression in tumor cells correlates with the response to initial chemotherapy (P = .033), that is, tumors with high CD44 expression are likely to be chemoresistant. An even stronger positive association with primary chemoresistance was found in the patients with tumors coexpressing CD44/CK19 (P = .02). No significant correlations were found between CK19 expression and other clinicopathologic features such as age, stage, debulking, histology, grade, and chemoresponse (Table 2).
Table 2.
Association Between Clinicopathologic Features and Expression of CD44 in Tumor Mask, CK19 in Tumor Mask, or CD44 in CK19 Compartment.
| Variables | n | CD44Ck19 (Range) | P Value | CD44 (Range) | P Value | CK19 (Range) | P Value |
|---|---|---|---|---|---|---|---|
| Age | .101 | .05 | .316 | ||||
| <62 | 19 | 55.52 (13.46-163.94) | 51.43 (19.07-130.68) | 120.29 (17.40-240.36) | |||
| >62 | 14 | 88.63 (21.07-283.47) | 90.96 (23.23-279.88) | 143.76 (21.17-281.51) | |||
| Histology | .388 | .534 | .314 | ||||
| Serous | 29 | 72.81 (13.46-283.47) | 70.56 (19.07-279.88) | 134.57 (17.40-281.51) | |||
| Others | 4 | 46.03 (21.07-65.52) | 51.05 (23.23-85.35) | 98.88 (34.93-146.10) | |||
| Debulk | .469 | .678 | .721 | ||||
| Optimal | 26 | 65.75 (13.46-283.47) | 65.99 (19.07-279.88) | 132.40 (21.17-281.51) | |||
| Suboptimal | 7 | 83.71 (39.34-163.94) | 76.41 (39.34-130.68) | 122.23 (17.40-173.80) | |||
| Stage | .876 | .922 | .515 | ||||
| III | 26 | 71.06 (13.46-283.47) | 69.72 (19.07-279.88) | 128.15 (21.17-281.51) | |||
| IV | 3 | 75.09 (37.84-135.34) | 69.95 (36.06-113.57) | 170.03 (123.63-240.36) | |||
| X | 4 | 55.70 (26.88-102.16) | 60.00 (27.31-93.87) | 113.99 (17.40-173.80) | |||
| Grade | .92 | .772 | .079 | ||||
| Well-moderate | 7 | 71.53 (13.46-283.47) | 73.92 (23.23-279.88) | 91.79 (34.93-143.99) | |||
| Poor | 26 | 69.04 (16.56-170.18) | 66.65 (19.07-233.32) | 140.60 (17.40-281.51) | |||
| Chemo | .02a | .033a | .833 | ||||
| Sensitive | 15 | 44.68 (13.46-127.31) | 45.1 (19.07-97.42) | 132.94 (44.71-281.51) | |||
| Resistant | 18 | 90.30 (21.07-283.47) | 87.45 (23.23-279.88) | 128.00 (17.40-211.08) |
a P < .05.
Correlation of CD44, CK19, and CD44/CK19 Expression With Progression-Free Interval and Overall Survival
We sought to further evaluate the prognostic value of these biomarkers for putative OCSCs as a predictive factor for progression-free interval and overall survival. For this purpose, an optimal cutoff point was generated by the X-Tile software. The cutoff AQUA score for CD44, CK19, and CD44/CK19 compartment was determined at 38, 112, and 38, respectively. The classification was high expression above the cutoff value and low expression below the cutoff value. Table 3 lists the number of patients with high expression and low expression.
Table 3.
AQUA Cutoff Values for Each Marker and Combination.
| Marker | AQUA Cutoff Value | High Expression, N | Low Expression, N |
|---|---|---|---|
| CD44 | 38 | 21 | 12 |
| CK19 | 112 | 19 | 14 |
| CD44/CK19 | 38 | 22 | 11 |
Abbreviations: AQUA, automated quantitative analysis; N, number of patients.
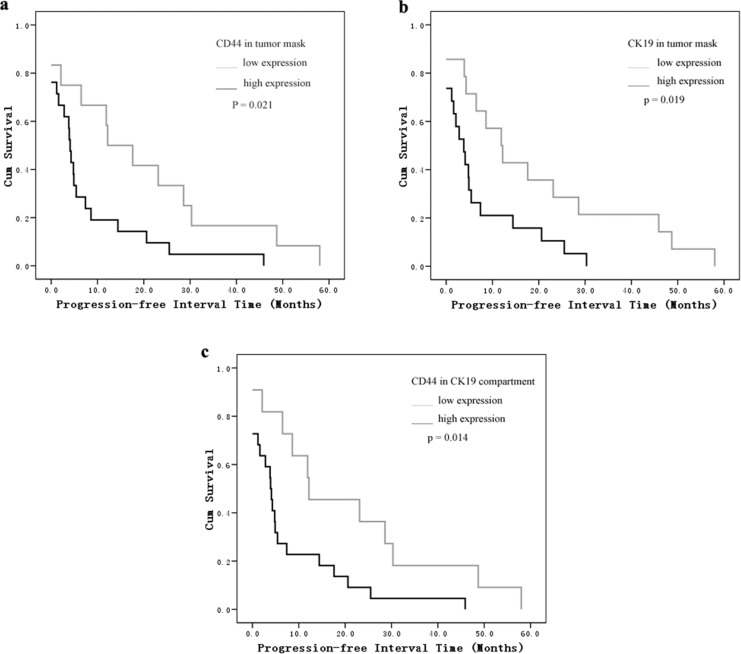
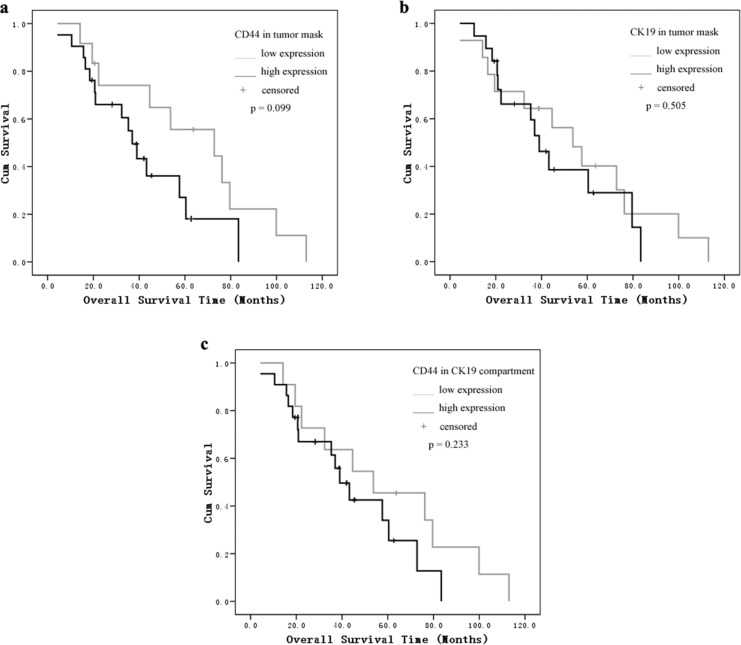
As shown in the Kaplan-Meier analysis and log-rank test, the most significant difference was found in the CD44/CK19 compartment group. The mean progression-free interval in patients with high CD44/CK19 coexpression in tumor cells was 7.6 months versus 20.9 months in the low CD44/CK19 coexpression groups (P = .014). Patients whose tumors showed high CD44 or CK19 expression had a shorter progression-free interval than patients with low expression tumors (CD44: 7.6 months [high] vs 19.9 months [low]; CK19: 6.8 months [high] vs 19.2 months [low], P = .021, P = .019, respectively, Figure 2). We did not find a correlation between CD44 and CK19 expression and overall survival (Figure 3).
Figure 2.
Kaplan-Meier survival analysis and log-rank test for progression-free intervals by (A) CD44 in tumor mask, (B) CK19 in tumor mask, and (C) CD44 in CK19 compartment expression as determined by automated quantitative analysis (AQUA) score.
Figure 3.
Kaplan-Meier survival analysis and log-rank test for overall survival by (A) CD44 in tumor mask, (B) CK19 in tumor mask, and (C) CD44 in CK19 compartment expression as determined by automated quantitative analysis (AQUA) score.
Univariate and Multivariate Cox Regression Analysis for Progression-Free Interval
Using the Cox proportional hazards model, we carried out univariable analysis to assess the predictive value of these markers for the patients’ progression-free intervals. The patients with high expression of CD44, CK19, or the combination CD44/CK19 had a shorter disease-free interval, with P values of .027, .025, and .019, respectively. In addition, the residual postoperative tumor volume and the clinical stage were also significant prognostic factors for decreased progression-free intervals (P = .005 and .037, respectively; Table 4). The 3 sets of markers described above were not completely independent of each other. The CD44/CK19 coexpression was the most significant factor in the Kaplan-Meier survival analysis. For this reason, only the markers of CD44/CK19 coexpression as well as the postoperative tumor residual and the clinical stage were entered into the multivariate Cox analysis. None of the 3 factors was significant. However, the set of CD44/CK19 coexpression was the nearest independent poor prognostic factor, with a calculated hazard ratio of 6.499 (P = .052). In this cohort, the well-recognized prognostic factors’ residual tumor volume and tumor stage were not statistically significant (P = .222 and .31; Table 5).
Table 4.
Univariate Cox Regression Analyses for Progression-Free Intervals.
| Variable | HR | 95% CI | P Value |
|---|---|---|---|
| Age | .211 | ||
| <62 | 1 | ||
| >62 | 1.585 | 0.770-3.260 | |
| Histology | .705 | ||
| Serous | 1 | ||
| Others | 0.815 | 0.283-2.348 | |
| Debulking | .005a | ||
| Optimal | 1 | ||
| Suboptimal | 4.047 | 1.522-10.759 | |
| Stage | .037a | ||
| III | 1 | ||
| IV | 0.978 | 0.291-3.29 | |
| X | 5.085 | 1.457-17.744 | |
| Grade | .285 | ||
| 1-2 | 1 | ||
| 3 | 1.649 | 0.659-4.123 | |
| Lymph node | .366 | ||
| Negative | 1 | ||
| Positive | 1.433 | 0.657-3.125 | |
| CD44/CK19 | .019a | ||
| Low | 1 | ||
| High | 2.612 | 1.17-5.834 | |
| CD44 | .027a | ||
| Low | 1 | ||
| High | 2.411 | 1.104-5.263 | |
| CK19 | .025a | ||
| Low | 1 | ||
| High | 2.381 | 1.113-5.095 | |
Abbreviations: CI, confidence interval; HR, hazard ratio.
a P < .05.
Table 5.
Multivariable Cox Regression Analysis for Progression-Free Intervals.
| Variables | HR | 95% CI | P Value |
|---|---|---|---|
| CD44/CK19 | .052 | ||
| Low | 1 | ||
| High | 2.344 | 0.993-5.531 | |
| Debulk | .222 | ||
| Optimal | 1 | ||
| Sub | 2.136 | 0.632-7.221 | |
| Stage | .31 | ||
| III | 1 | ||
| IV | 1.291 | 0.371-4.491 | |
| X | 3.034 | 0.705-13.055 | |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Discussion
In this study, we demonstrate the correlation between the number of ovarian CSCs, as determined by the expression of CD44 and CK19, and clinical outcome. We demonstrate that the use of AQUA system allows the quantification of CD44+/CK19+ EOC cells within tumor tissue and its potential association with patients’ response to therapy. Our findings suggest that a high ratio of CD44+/CK19+ cells within tumor tissue is associated with chemoresistance and poor clinical outcome.
CK19 is a filamentous protein and belongs to the type I group of cytokeratins.33 CK19 expression has been associated with cancer, and it has been used for the detection of circulating breast cancer tumor cells.34 It has also been reported that CK19 expression is increased in a number of different solid tumors such as hepatocellular carcinoma, renal cell carcinoma, and cervical squamous cell carcinoma.33,35 Most recently, it was reported that CK19 expression was upregulated in ovarian cancer samples compared with normal ovarian tissue by a comparative proteomic study.20 In the present study, we demonstrated that CK19 is highly expressed in a subgroup of ovarian cancer cells, suggesting its potential association with ovarian CSCs.
CD44 belongs to a multifunctional family of type I transmembrane proteins and is expressed as a wide variety of isoforms in many cells.15,36 A weight of evidence demonstrated that CD44 plays crucial roles in cancer biology, such as the adhesion and migration of tumor cells, niche generation and modulation, epithelial–mesenchymal transformation, and resistance to drugs.18,37
CD44 has been identified as a marker for CSCs including OCSCs.18 We have shown recently that ovarian cancer cells expressing CD44 are highly tumorigenic, have capabilities of self-renewal, and recapitulate the heterogeneous phenotype of the tumor.8,10,38 In addition, CD44+ cells are resistant to chemotherapy and are able to survive treatment with conventional drugs such as carboplatin and paclitaxel.39
The findings described in this study are in support of the hypothesis that the CSCs are significant players in chemoresistance. In this model, chemotherapy only targets ovarian cancer cells but has no effect on the CSCs.
Several studies have reported an association between CD44 expression and clinical outcomes of patients with ovarian cancer. Patients with CD44+ tumors had a mean survival of 25 months compared with patients with CD44− tumors whose mean survival was 52 months.40,41 However, none of these studies have been able to elucidate the biological and cellular meaning of these differences. Our findings suggest that the expression of CD44 may be related to this unique cell population, the ovarian CSCs.
While other studies found no correlation between CD44 expression and clinical outcome, Sillanpaa et al found that CD44 overexpression was a good predictor of clinical outcome.42,43 An explanation for the differences in results is the complexity of CD44, which exist in a wide variety of isoforms. Many of these studies used antibodies for specific variants of CD44 but not pan-CD44. In addition, the method used to quantify the expression of the proteins in paraffin section such as IHC has some inherent limitations. CD44 is extensively expressed on stromal and immune cells and this creates a problem for semiquantitative methods, such as the H score. The application of the TMA and AQUA system for the quantification and identification of CD44+ cells improves the reliability and reproducibility of the results. It also decreases the methodological bias, since it allows the simultaneous studying of multiple specimens, the uniform handling of all specimens, and a far more efficient workflow. The AQUA system can easily preclude the nonepithelial cells from tissue removing potential contaminants. In addition, based on the high specificity and sensitivity of fluorescent techniques, it can target multiple antigens at the same time and on the same slide. The yield of these methods has been demonstrated in several studies.44
Limitations of this study are the relatively small cohort and need of data validation for the X-tile generated cutoff points. Although the use of TMA has numerous advantages, especially since it allows the evaluation of multiple patients’ samples under the same conditions, one of its limitations is the fact that it only reflects the condition of a small area of the tumor. A clinical application of our findings will require the quantification of multiple sections of the tumor. Therefore, the results of this study can be characterized as hypothesis generating and need to be validated in a larger sample size. The small sample size limited the statistical power and did not allow adjustment for all potential confounding factors.
In conclusion, the frequency of CD44+/CK19+ EOC cells was associated with chemoresistance and decreased progression-free interval in patients with ovarian cancer. Use of the AQUA system may allow the appropriate quantification of these cells in tissue samples and therefore its potential clinical application.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: in part by grants from NCI/NIH (grants numbers RO1CA127913; RO1CA118678); The Sands Family Foundation; and the Discovery To Cure Research Program.
References
- 1.Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19(1):3–10. [DOI] [PubMed] [Google Scholar]
- 2.Nossov V, Amneus M, Su F, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol. 2008;199(3):215–223. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. [DOI] [PubMed] [Google Scholar]
- 4.Garvalov BK, Acker T. Cancer stem cells: a new framework for the design of tumor therapies. J Mol Med (Berl). 2011;89(2):95–107. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien CA, Kreso A, Dick JE. Cancer stem cells in solid tumors: an overview. Semin Radiat Oncol. 2009;19(2):71–77. [DOI] [PubMed] [Google Scholar]
- 6.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 7.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. [DOI] [PubMed] [Google Scholar]
- 8.Alvero AB, Chen R, Fu HH, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8(1):158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mor G, Yin G, Chefetz I, Yang Y, Alvero A. Ovarian cancer stem cells and inflammation. Cancer Biol Ther. 2011;11(8):708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvero AB, Fu HH, Holmberg J, et al. Stem-like ovarian cancer cells can serve as tumor vascular progenitors. Stem Cells. 2009;27(10):2405–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curley MD, Therrien VA, Cummings CL, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27(12):2875–2883. [DOI] [PubMed] [Google Scholar]
- 12.Kryczek I, Liu S, Roh M, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130(1):29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29(18):2672–2680. [DOI] [PubMed] [Google Scholar]
- 14.Luo L, Zeng J, Liang B, et al. Ovarian cancer cells with the CD117 phenotype are highly tumorigenic and are related to chemotherapy outcome. Exp Mol Pathol. 2011;91(2):596–602. [DOI] [PubMed] [Google Scholar]
- 15.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. [DOI] [PubMed] [Google Scholar]
- 16.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. [DOI] [PubMed] [Google Scholar]
- 17.Yin G, Chen R, Alvero AB, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29(24):3545–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. [DOI] [PubMed] [Google Scholar]
- 19.Steffensen KD, Alvero AB, Yang Y, et al. Prevalence of epithelial ovarian cancer stem cells correlates with recurrence in early-stage ovarian cancer. J Oncol. 2011;2011:620523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim R, Lappas M, Ahmed N, Permezel M, Quinn MA, Rice GE. 2D-PAGE of ovarian cancer: analysis of soluble and insoluble fractions using medium-range immobilized pH gradients. Biochem Biophys Res Commun. 2011;406(3):408–413. [DOI] [PubMed] [Google Scholar]
- 21.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97(24):1808–1815. [DOI] [PubMed] [Google Scholar]
- 22.Kluger HM, Dolled-Filhart M, Rodov S, Kacinski BM, Camp RL, Rimm DL. Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin Cancer Res. 2004;10(1 pt 1):173–177. [DOI] [PubMed] [Google Scholar]
- 23.Psyrri A, Kountourakis P, Yu Z, et al. Analysis of p53 protein expression levels on ovarian cancer tissue microarray using automated quantitative analysis elucidates prognostic patient subsets. Ann Oncol. 2007;18(4):709–715. [DOI] [PubMed] [Google Scholar]
- 24.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1327. [DOI] [PubMed] [Google Scholar]
- 25.Gustavson MD, Bourke-Martin B, Reilly D, et al. Standardization of HER2 immunohistochemistry in breast cancer by automated quantitative analysis. Arch Pathol Lab Med. 2009;133(9):1413–1419. [DOI] [PubMed] [Google Scholar]
- 26.Gustavson MD, Bourke-Martin B, Reilly DM, et al. Development of an unsupervised pixel-based clustering algorithm for compartmentalization of immunohistochemical expression using Automated QUantitative Analysis. Appl Immunohistochem Mol Morphol. 2009;17(4):329–337. [DOI] [PubMed] [Google Scholar]
- 27.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1327. [DOI] [PubMed] [Google Scholar]
- 28.Gustavson MD, Molinaro AM, Tedeschi G, Camp RL, Rimm DL. AQUA analysis of thymidylate synthase reveals localization to be a key prognostic biomarker in 2 large cohorts of colorectal carcinoma. Arch Pathol Lab Med. 2008;132(11):1746–1752. [DOI] [PubMed] [Google Scholar]
- 29.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. [DOI] [PubMed] [Google Scholar]
- 30.Rustin GJ, Quinn M, Thigpen T, et al. Re: New guidelines to evaluate the response to treatment in solid tumors (ovarian cancer). J Natl Cancer Inst. 2004;96(6):487–488. [DOI] [PubMed] [Google Scholar]
- 31.Rustin GJ. Can we now agree to use the same definition to measure response according to CA-125? J Clin Oncol. 2004;22(20):4035–4036. [DOI] [PubMed] [Google Scholar]
- 32.Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol. 2010;176(5):2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain R, Fischer S, Serra S, Chetty R. The use of Cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol. 2010;18(1):9–15. [DOI] [PubMed] [Google Scholar]
- 34.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13(4):1033–1067. [DOI] [PubMed] [Google Scholar]
- 35.Mertz KD, Demichelis F, Sboner A, et al. Association of cytokeratin 7 and 19 expression with genomic stability and favorable prognosis in clear cell renal cell cancer. Int J Cancer. 2008;123(3):569–576. [DOI] [PubMed] [Google Scholar]
- 36.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. [DOI] [PubMed] [Google Scholar]
- 37.Marhaba R, Klingbeil P, Nuebel T, Nazarenko I, Buechler MW, Zoeller M. CD44 and EpCAM: cancer-initiating cell markers. Curr Mol Med. 2008;8(8):784–804. [DOI] [PubMed] [Google Scholar]
- 38.Yin G, Alvero AB, Craveiro V, et al. Constitutive proteasomal degradation of TWIST-1 in epithelial-ovarian cancer stem cells impacts differentiation and metastatic potential [published online February 20, 2012]. Oncogene. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casagrande F, Cocco E, Bellone S, et al. Eradication of chemotherapy-resistant CD44+ human ovarian cancer stem cells in mice by intraperitoneal administration of clostridium perfringens enterotoxin. Cancer. 2011;117(24):5519–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayastha S, Freedman AN, Piver MS, Mukkamalla J, Romero-Guittierez M, Werness BA. Expression of the hyaluronan receptor, CD44S, in epithelial ovarian cancer is an independent predictor of survival. Clin Cancer Res. 1999;5(5):1073–1076. [PubMed] [Google Scholar]
- 41.Rodriguez-Rodriguez L, Sancho-Torres I, Mesonero C, Gibbon DG, Shih WJ, Zotalis G. The CD44 receptor is a molecular predictor of survival in ovarian cancer. Med Oncol. 2003;20(3):255–263. [DOI] [PubMed] [Google Scholar]
- 42.Berner HS, Davidson B, Berner A, et al. Expression of CD44 in effusions of patients diagnosed with serous ovarian carcinoma—diagnostic and prognostic implications. Clin Exp Metastasis. 2000;18(2):197–202. [DOI] [PubMed] [Google Scholar]
- 43.Sillanpaa S, Anttila MA, Voutilainen K, et al. CD44 expression indicates favorable prognosis in epithelial ovarian cancer. Clin Cancer Res. 2003;9(14):5318–5324. [PubMed] [Google Scholar]
- 44.Dolled-Filhart M, Gustavson M, Camp RL, Rimm DL, Tonkinson JL, Christiansen J. Automated analysis of tissue microarrays. Methods Mol Biol. 2010;664:151–162. [DOI] [PubMed] [Google Scholar]