Abstract
To form a gonad, germ cells (GCs) and somatic gonadal precursor cells (SGPs) must migrate to the correct location in the developing embryo and establish the cell-cell interactions necessary to create proper gonad architecture. During gonad morphogenesis, SGPs send out cellular extensions to ensheath the individual GCs and promote their development. We have identified mutations in the raw gene that result in a failure of the SGPs to ensheath the GCs, leading to defects in GC development. Using genetic analysis and gene expression studies, we find that Raw negatively regulates JNK signaling during gonad morphogenesis, and increased JNK signaling is sufficient to cause ensheathment defects. In particular, Raw functions upstream of the Drosophila Jun-related transcription factor to regulate its subcellular localization. Since JNK signaling regulates cell adhesion during the morphogenesis of many tissues, we examined the relationship between raw and the genes encoding Drosophila E-cadherin and β-catenin, which function together in cell adhesion. We find that loss of DE-cadherin strongly enhances the raw mutant gonad phenotype, while increasing DE-cadherin function rescues this phenotype. Further, loss of raw results in mislocalization of ß-catenin away from the cell surface. Therefore, cadherin-based cell adhesion, likely at the level of ß-catenin, is a primary mechanism by which Raw regulates germline-soma interaction.
Keywords: Drosophila, gonad formation, germ cells, raw, JNK signaling, DE-cadherin, shotgun
Introduction
During organogenesis, different cell types must recognize one another and control their relative positions and their shapes to create an organ with the proper cell-cell interactions and architecture. The failure of cells to receive the proper cues from each other and their surrounding microenvironment can affect cell differentiation and proliferation, and may lead to cell death or tumorigenesis. Defects in any of these steps may not only result in the failure to form a cohesive and functional tissue, but are likely to affect the viability and/or fertility of the organism.
One organ that is vital to the propagation of the species is the gonad. In most animals, the gonad is formed early in embryogenesis from two cell types: the somatic gonadal precursor cells (SGPs) and the germ cells (GCs) (reviewed in Richardson and Lehmann, 2010). SGPs and GCs are usually specified in different regions of the developing embryo and must find each other to create a functional gonad. In mammals, GCs require the somatic Sertoli cells in males, or the follicle cells in female, for the production of sperm and eggs (Liu et al., 2010; Wainwright and Wilhelm, 2010). Similarly in Drosophila, defects in soma-germline interactions can result in a failure to produce functional gametes (reviewed in Jemc, 2011). Studies using the Drosophila gonad as a model have provided a number of insights into the mechanisms that regulate the specification and migration of SGPs and GCs, while less is known about the molecular mechanisms that regulate gonad morphogenesis (reviewed in Jemc, 2011; Richardson and Lehmann, 2010).
In Drosophila, GCs are specified at the posterior pole of the embryo, and migrate through the gut to reach the mesoderm, where the SGPs are specified (reviewed in Jemc, 2011). Three clusters of SGPs are specified on each side of the embryo within parasegments (PS) 10, 11 and 12 of the mesoderm (Brookman et al., 1992). An additional cluster of SGPs is specified in PS 13 of males and females, but is only maintained in males (DeFalco et al., 2003). As the GCs contact the SGP clusters, these cells come together to form a cohesive tissue of intermingled GCs and SGPs (Boyle and DiNardo, 1995). At this time, the SGPs also begin to surround the GCs with cellular extensions in a process known as GC ensheathment (Jenkins et al., 2003). By stage 15 of embryogenesis, the gonad has compacted into its final rounded structure and the GCs are almost completely surrounded by SGPs, such that they make very little contact with one another (Boyle and DiNardo, 1995; Jenkins et al., 2003). In males, as the GCs contact the SGPs, they receive a signal from the SGPs that acts through the JAK/STAT pathway to regulate male-specific GC proliferation and male GC identity (Wawersik et al., 2005). During later stages of development, the GCs and SGPs will form either a testis or an ovary and begin to produce gametes.
Previously, a number of genes have been identified that are required for gonad morphogenesis, including those required for proper GC-SGP interaction and GC ensheathment (Jenkins et al., 2003; Li et al., 2003; Mathews et al., 2006; Van Doren et al., 2003; Weyers et al., 2011). In particular, Drosophila E-cadherin (DE-cad), which is encoded by the shotgun (shg) locus, is required at multiple steps of gonad formation, including GC ensheathment (Jenkins et al., 2003; Van Doren et al., 2003). shg mutants exhibit clumping of the GCs, and a failure of SGPs to intermingle with, and send out extensions around, the GCs (Jenkins et al., 2003). Interestingly, overexpression of DE-cad specifically in the GCs results in a similar phenotype, suggesting that the proper balance of adhesion protein levels between the SGPs and GCs is critical for GC ensheathment (Jenkins et al., 2003). Two other genes have also been demonstrated to function in GC ensheathment, and may function through DE-cad. The zinc ion transporter, Fear of intimacy (FOI), regulates DE-cad post-transcriptionally to promote GC ensheathment (Mathews et al., 2006; Van Doren et al., 2003). In addition, mutants for traffic jam (tj), which encodes a MAF family transcription factor, exhibit defects in the ability of the SGPs to properly intermingle with the GCs (Li et al., 2003). Analysis of tj mutant clones in adults revealed increased levels of a number of cell adhesion proteins, including DE-cad, upon tj mutation (Li et al., 2003), although it is unclear if tj regulates embryonic gonad formation by affecting DE-cad. Together, these results demonstrate that proper regulation of adhesion proteins is critical for establishment of germline-soma interactions.
In a genetic screen for mutations that affect gonad morphogenesis, mutations in the gene raw were found to exhibit GC ensheathment defects (Weyers et al., 2011). Similar to the mutants described above, raw mutants exhibit GC clumping in the gonad, and the SGPs fail to send out extensions to surround each GC. Previous studies of raw have found that it is required for dorsal closure, CNS retraction, and the morphogenesis of multiple tissues, including the salivary gland and malpighian tubules (Jack and Myette, 1997). It appears to negatively regulate JNK signaling during dorsal closure (Byars et al., 1999), although the mechanism for this regulation is unclear. In this paper, we examine the gonad phenotype exhibited by raw mutants, and explore the molecular mechanisms by which it functions to regulate germline-soma interactions.
Materials and Methods
Fly Stocks
The following fly stocks were used in this work: raw155.27 and raw134.47 (Weyers et al., 2011), shg g317 (Tepass et al., 1996), tub-e-cadWT (Pacquelet et al., 2003), tubulin-GAL4 (Lee and Luo, 1999), P{Dfd-lacZ-HZ2.7} on X (Bergson and McGinnis, 1990), and pucE69 (obtained from D. Bohmann). The 68-77 enhancer trap (Simon et al., 1990) expresses lacZ in the SGPs and was obtained from D. Godt and manipulated as previously described (Weyers et al., 2011). bsk1, jraIA109, UAS-bskDN, w1118, and Oregon R, were obtained from the Bloomington Stock Center. The TRE-GFP reporter, which contains four AP-1 binding sites downstream of an hsp70 promoter (Chatterjee and Bohmann, submitted), was a gift from D. Bohmann. Mutations were maintained over lacZ or GFP marked balancer chromosomes to allow for selection of mutants of the correct genotype.
Immunohistochemistry
Antibody stains were performed as previously described (Jenkins et al., 2003; Moore et al., 1998), with the exception of stage 17 embryos stained for STAT, which were sonicated to increase antibody penetrance (Le Bras and Van Doren, 2006). The following primary antibodies were used (dilutions; source): mouse α-β-GAL (1:10,000; Promega), rabbit α-β-GAL (1:10,000; Cappel), rabbit α-GFP (1:2,000; Torrey Pines Biolabs), mouse α-GFP (1:50; Santa Cruz), rabbit α-pH3 (1:1000; Millipore), mouse α-EYA (1:25; DSHB), mouse α-NRT BP106 (1:10; Developmental Studies Hybridoma Bank (DSHB)), rat α-DE-cadherin (DCAD2, 1:20; DSHB), mouse α-ARM (N27A1; 1:100; DSHB), chick a-VASA (1:10,000; K. Howard), rat a-VASA (1:50; DSHB), guinea pig α -TJ (1:1,000; generated using the same epitope as previously described (Li et al., 2003)), rabbit α-JRA (1:1,500; D. Bohmann), rabbit α-JUN (1:100; Santa Cruz), rabbit α -STAT (1:50; S. Hou). Alexa 488, 546, and 633 conjugated secondary antibodies used at 1:500 from (Molecular Probes, Invitrogen). For diaminobenzidine (DAB) detection, biotin conjugated secondaries (Jackson ImmunoResearch) were used, and the stain was developed using the ABC Elite kit (Vector Labs) using DAB as a substrate (Vector Labs). Nuclei were stained by incubating embryos for 10 minutes in 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS with 0.1% Tween. Embryos were staged using gut morphology according to Campos-Ortega and Hartenstein (Campos-Ortega and Hartenstein, 1985). Sex of embryos was determined when necessary using P{Dfd-lacZ-HZ2.7} (Bergson and McGinnis, 1990) crossed in from the paternal X chromosome.. Fluorescently-stained embryos were mounted in 70% glycerol containing 2.5% DABCO (Sigma) and visualized using a Zeiss LSM 510 Meta or Confocor confocal microscope. Non-fluorescent immunostainings were mounted in Aquatex (EMD Chemicals) and visualized using a Zeiss Axioskop with Coolsnap camera and RS software (Photemetrics).
Quantitation of Protein levels
JRA and TJ levels were compared in raw mutant (raw134.47/raw155.27) and control (raw155.27/+ or raw134.47/+) SGPs. The pixel intensity of JRA was measured using the LSM 510 software across the region of high TJ expression in each SGP (assumed to be the nucleus) as well as 1-2 μm flanking each side of the TJ staining. The average intensity of JRA or TJ staining across 1 μm of the flanking region and nuclear region was calculated for each SGP. The values for each cell were averaged together, and the standard deviation was calculated.
As it is difficult to quantitate ARM expression in comparable regions across multiple samples, ARM levels were ranked subjectively on a scale of 1-3 and the average expression was calculated.
Western Blot Analysis
For Western blot analysis, embryos were collected from the following cross: raw134.47/Cyo,twist-GAL4,UAS-GFP × raw155.27/CyO, twist-GAL4, UAS-GFP, and sorted into GFP positive and GFP negative populations using a COPAS Select embryo sorter (Union Biometrica). For analysis of JRA and DE-cad levels, embryos were lysed in RIPA buffer containing protease inhibitors (Roche), sodium orthovanadate and Ser/Thr phosphatase inhibitors (Sigma). Samples were run on 10% SDS-PAGE and protein expression was compared in control embryos (GFP positive population) and raw mutant embryos (GFP negative population). For analysis of phospho-ARM levels, embryos were lysed in NP-40 lysis buffer containing protease inhibitors (Roche) and Ser/Thr phosphatase inhibitors (Sigma). Samples were run on 8% SDS-PAGE and protein expression was compared in control embryos (Oregon R) and raw mutant embryos (GFP negative population). Immunoblotting was performed with the following antibodies according to standard methods: rabbit α-JUN was used at (1:1,000; Santa Cruz), mouse α-tubulin (DM1A; 1:1,000; Sigma), rat α-DE-cad (DCAD2; 1:500; DSHB), mouse α-ARM (N27A1; 1:1,000; DSHB), and phospho-β-catenin (1:1,000; Cell Signaling). HRP conjugated secondaries (Jackson Immunoresearch) were used at 1:1,000-2,000 and detected used the Amersharm ECL or ECL-plus kit (GE Healthcare). Relative levels of Jun and DE-cad were determined using ImageJ software.
Generation of Rescue Construct
UAS-rawRA was generated by inserting the rawRA transcript into pUASpB (a modified version of pUASP (Rorth, 1998) with an attB site for phiC31 mediated integration). The 5′ fragment of rawRA was amplified by PCR from RE05256 (Berkeley Drosophila Genome Project) using the following primers: 5′-CATGAGCGGCCGCGCCGCCACCATGAAAACTGAAAGCAGCAGTTATC-3′ and 5′-CGGGCACCACATTGCATCTGGCCTTTGCTCGCCATGCATTCAGAACTTCT-3′. The 3′ fragment was derived from GH23250 (Berkeley Drosophila Genome Project) that was used to generate a pUASpB-rawRB transgene. This fragment was isolated by digestion using XcmI and XbaI. Both fragments were ligated into pUASpB that was digested with NotI and XbaI, and the construct was confirmed by sequencing. This construct was inserted into flies using phiC31 integration into P{CARYP}attP40 (Groth et al., 2004; Markstein et al., 2008) by Genetics Services (Cambridge, Massachusetts).
Results
raw is required for GC ensheathment by SGPs
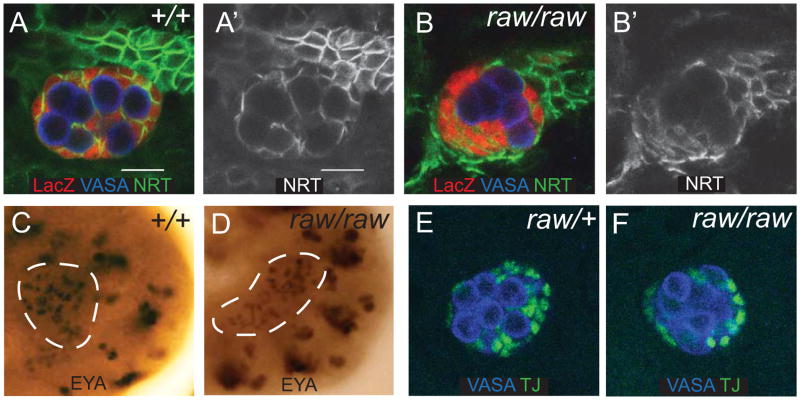
Previously, we identified two mutant alleles of a gene that exhibited defects in the ability of the SGPs to ensheath the GCs and mapped this gene to the raw locus (Weyers et al., 2011). In order to further examine the raw mutant phenotype, we scored the frequency of ensheathment defects in raw mutants using a lacZ enhancer trap, called 68-77, which expresses lacZ cytoplasmically in the SGPs (Boyle and DiNardo, 1995; Simon et al., 1990; Warrior, 1994). While only ∼5% of gonads exhibited defects in GC ensheathment in heterozygous controls, raw mutants exhibited ensheathment defects in ∼45% of gonads. Antibody staining using the SGP cell surface marker Neurotactin (NRT), revealed clumps of GCs surrounded by SGPs at the periphery of the gonad (Fig. 1A, B), demonstrating that raw is required for GC ensheathment by the SGPs and formation of a gonad with proper architecture. Given the 45% frequency of ensheathment, it appeared possible that the phenotype might be sex-specific. However, examination of sexed male and female embryos revealed that this was not the case (data not shown).
Figure 1. raw mutants exhibit ensheathment defects.
(A,B) Stage 15 gonads stained to examine germ cell ensheathment. (A, A′) 68-77/+. (B, B′) raw134.47, 68-77/raw155.27, 68-77. Embryos are immunostained for VASA (blue, germ cells), 68-77-lacZ (red, SGPs), and Neurotactin (NRT) (green, SGP cell surface). (A′, B′) NRT alone. (C, D) Stage 12 gonads stained to examine EYA expression as an indicator of SGP specification. SGPs are outlined with a white dashed line. (C) Control gonad: 68-77/68-77. (D) raw134.47, 68-77/raw155.27, 68-77 gonad. (E, F) Expression of TJ in stage 15 embryos to assess SGP identity. Traffic jam (green, SGPs) and VASA (blue, germ cells). (E) Control gonad: raw134.47, 68-77/+ or raw155.27, 68-77/+. (F) raw134.47, 68-77/raw155.27, 68-77 gonad. Scale bar: 10 μm for all immunofluorescence images. Posterior right.
Since defects in gonad formation could arise from defects in cell fate specification, we wanted to confirm that the SGPs were specified properly. SGPs are specified from the mesoderm, and we previously showed that other mesodermally derived tissues, specifically the visceral mesoderm, formed normally in raw mutants (Weyers et al., 2011). To examine SGP identity, we analyzed expression of two markers expressed in SGPs, Eyes absent/Clift (EYA/CLI) and TJ. EYA is required for SGP specification (Boyle et al., 1997; Boyle and DiNardo, 1995), while TJ is expressed in SGPs from stage 13 onward (Li et al., 2003). In raw mutants, EYA expression was observed in stage 12 SGPs, following SGP specification (Fig. 1C,D and Weyers et al., 2011), and TJ was observed in SGPs from stage 13 onward (Fig. 1 E, F; data not shown). Therefore, the defects in GC ensheathment do not appear to be a result of improper SGP specification or identity, but rather defects in the communication or interaction between the germline and soma.
Ensheathment is required for proper germline development
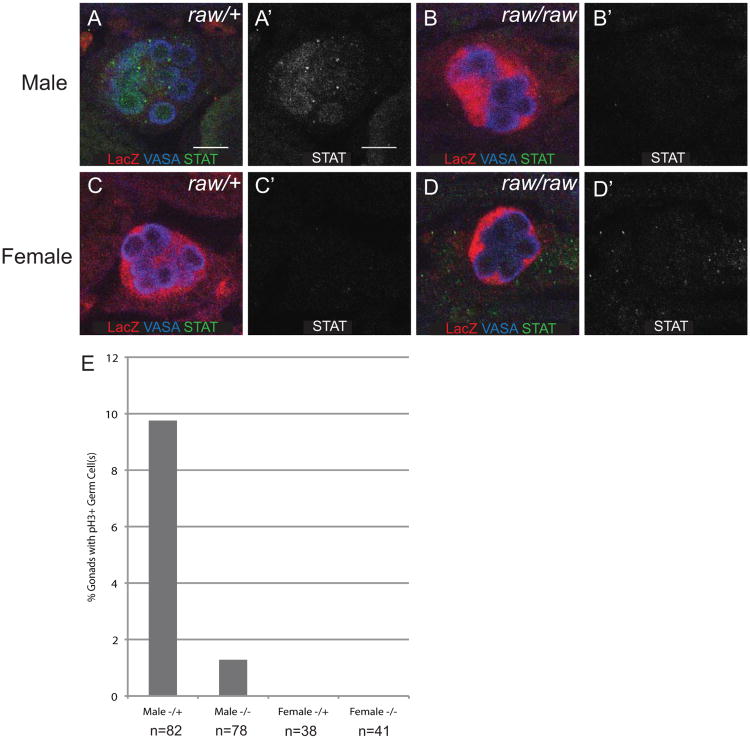
We next wanted to test whether the defects in germ cell ensheathment caused by raw mutants had effects on proper soma-germline communication and germline development. In males, GCs begin to proliferate at stage 15 of embryogenesis, while they remain quiescent in females until late embryonic stages (Sato et al., 2008; Wawersik et al., 2005). We examined GC proliferation in raw mutants by scoring the number of gonads that contained GCs positive for the mitotic marker phospho-histone 3 (pH3). In control males, approximately 9% of gonads contained at least one mitotic GC (Fig. 2E). In contrast, in raw mutant males, only ∼1% of gonads contained a mitotic GC (Fig. 2E). In female controls and female raw mutants, pH3 positive GCs were not observed, consistent with previous results (Fig. 2E). These results suggest that defects in GC ensheathment affect the sexually dimorphic development of GCs.
Figure 2. Ensheathment regulates germ cell development.
(A-D) STAT expression in stage 17 embryonic gonads. Embryos immunostained for VASA (blue, germ cells), 68-77-lacZ (red, SGPs), and STAT (green) (A′-D′) STAT alone. Posterior is to the right. Scale bar: 10μm. (A, A′) Male raw heterozygous control gonads express STAT in germ cells. (B, B′) Male raw134.47/raw155.27 gonads rarely express STAT in germ cells. (C, C′) Female raw heterozygous controls. (D, D′) Female raw134.47/raw155.27. (E) Stage 15 gonads scored for the presence of pH3 positive germ cells to assess germ cell proliferation.
Sex-specific signaling from the male SGPs to the GCs through the JAK/STAT pathway regulates the proliferation of male GCs as well as other sex-specific GC characteristics (Wawersik et al., 2005). Male-specific activation of the JAK/STAT pathway can be observed as an increase in STAT92E immunoreactivity in male GCs (Wawersik et al., 2005), and we used this assay to test whether the ensheathment defects in raw mutants interrupted soma-GC signaling. While STAT92E immunoreactivity was observed in the germline of raw heterozygous male control embryos, raw mutant males failed to exhibit STAT92E expression in their germline (Fig. 2A, B). The loss of STAT92E did not correlate with the strength of the ensheathment defect in raw mutant males. While STAT92E was reduced in all male gonads examined, not all gonads exhibited strong ensheathment defects (data not shown). In females, STAT92E staining was not observed in either controls or raw mutants (Fig. 2C, D), as expected. We conclude that the failure of male GC proliferation observed in raw mutants is due to defects in JAK/STAT signaling between the soma and the germline, and therefore that the ensheathment of the GCs by the SGPs is likely important for soma-germline communication.
raw negatively regulates JNK signaling
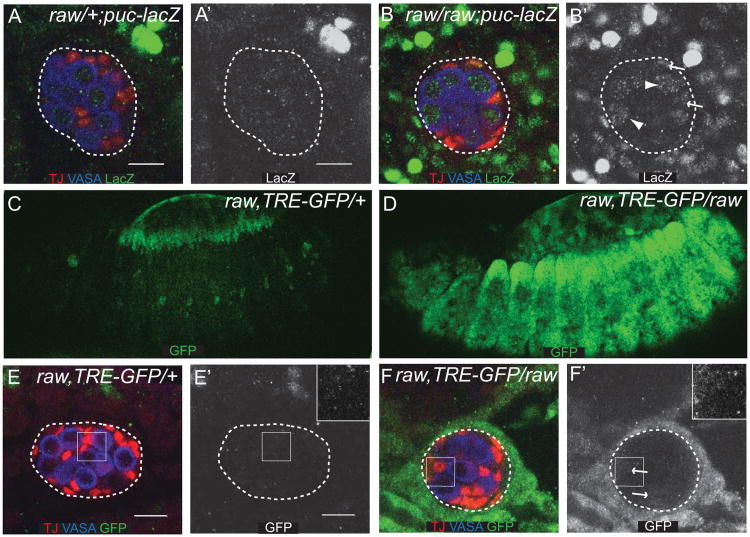
While raw is required for the morphogenesis of a number of tissues during development, how it regulates these processes is not well understood. Previous studies have shown that raw acts as a negative regulator of JNK signaling during dorsal closure in Drosophila (Bates et al., 2008; Byars et al., 1999), suggesting that raw could function in a similar manner during gonad morphogenesis. In order to determine if JNK signaling is upregulated in raw mutants, we examined expression of targets of the JNK pathway. First, we examined expression of the previously identified JNK pathway target puckered (puc) to determine if its expression was upregulated in or around the embryonic gonad. To do this, we used an enhancer trap that had been shown to reflect increased levels of JNK pathway activity in the dorsal ectoderm (Ring and Martinez Arias, 1993). We found that this reporter exhibited increased activity in raw mutants relative to controls (Fig. 3A, B). Increased activity was observed in many different cell types in the embryo, as previously reported (Byars et al., 1999). Higher levels of activity were observed outside the gonad, compared to inside the gonad (Fig. 3A, B); however, some activity was observed within the gonad. puc-lacZ was increased in both GCs and SGPs (Fig. 3A, B). To examine further whether expression of puc truly reflects increased JNK pathway activity mediated by the downstream AP-1 transcription factor, consisting of FOS and JUN (Jun-related antigen or JRA, in Drosophila), we utilized a synthetic GFP reporter that contains a multimerized AP-1 binding site downstream of an hsp70 promoter (Chatterjee and Bohmann, submitted). Expression of this reporter was observed in the leading edge of the dorsal ectoderm in wild-type embryos (Fig 3C), a place that is well known to be active for JNK pathway activity (Glise et al., 1995; Hou et al., 1997; Sluss et al., 1996). In raw mutants, broad upregulation of this reporter was observed throughout the embryo (Fig. 3D). In the region of the embryonic gonad, we observed no reporter expression in wild-type embryos (Fig. 3E), while in raw mutants, the reporter was strongly upregulated in cells surrounding the gonad, weakly activated in the SGPs (Fig. 3F), and undetectable in the GCs. We conclude that mutations in raw lead to a dramatically increased JUN transcriptional activity both in the region of the embryonic gonad and in many other tissues.
Figure 3. JNK signaling is increased in raw mutants.
(A, B) Stage 15 embryos immunostained for a puc-lacZ enhancer trap (green), TJ (red, SGPs), and VASA (blue, germ cells). (A′, B′) puc-lacZ alone. (A, A′) raw heterozygous controls. (B, B′) raw134.47/raw155.27. Arrows indicate pucE69-lacZ expression in SGPs; arrowheads indicate pucE69-lacZ expression in GCs. The gonad has been outlined with a dashed line. (C-F) Embryos immunostained to label the TRE-GFP AP-1 reporter. (C, D) Stage 13 embryos. TRE-GFP AP-1 reporter (green). (E, F) Stage 15 embryos. TRE-GFP AP-1 reporter. (green), VASA (blue, germ cells), and TJ (red, SGPs). (E′, F′) GFP alone. (C, E, E′) raw134.47 heterozygous controls. Inset in E′ indicated by solid square. (D, F, F′) raw134.47/raw155.27. Arrows indicate TRE-GFP expression in the SGPs. Inset in F′ indicated by solid square. The gonad has been outlined with a dashed line. Scale bar: 10 μm. Posterior is to the right.
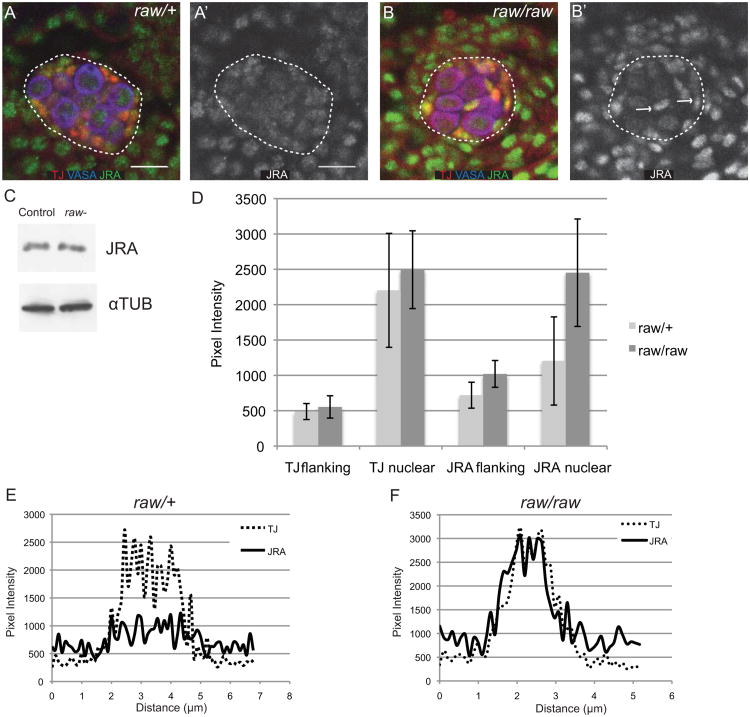
We next wanted to examine how raw influences JNK pathway activity. The most downstream component of the JNK pathway is transcription factor AP-1, and so we examined expression of JRA and FOS in raw mutants. While FOS expression was not changed significantly in raw mutants relative to controls (data not shown), JRA expression appeared strikingly different in raw mutants, both in the gonad and in the surrounding tissue (Fig. 4A, B). In raw mutants, JRA immunoreactivity appeared more intense and concentrated in a smaller subcellular region relative to controls (Fig. 4A, B).
Figure 4. raw regulates JRA subcellular localization.
(A, B) Stage 15 embryos immunostained stained for JRA (green), TJ (red, SGPs), VASA (blue, germ cells). (A′, B′) JRA alone. (A, A′) raw heterozygous controls. (B, B′) raw134.47/raw155.27. The gonad has been outlined with a dashed line. Scale bar: 10 μm. Posterior is to the right. Arrows indicate SGPs with altered JRA expression. (C) Western blot to analyze JRA levels in raw134.47/raw155.27 mutant embryos relative to controls. Lane 1 is control embryos (+/+ or raw155.27/+ or raw134.47/+). Lane 2 is raw-(raw134.47/raw155.27embryos). (D) Quantitation of TJ and JRA immunofluorescence intensity within the nucleus and in flanking regions. See methods for details. (E, F) Representative plots of JRA and TJ immunofluoresence intensity in individual SGPs in raw heterozygous controls (E) or raw134.47/raw155.27 (F). Note that JRA is more uniform across the cell in controls (E) but co-localizes more closely with TJ (nucleus) in raw mutants (F).
When we restored raw expression to a subset of cells in raw-mutant embryos, using a driver that is expressed in a pair-rule pattern (prd-Gal4), we observed a decreased intensity of JRA nuclear staining in regions where raw was expressed (Fig. S1). In order to examine overall JRA protein levels, we performed Western blot analysis on lysates from control and raw mutant embryos. Surprisingly, we did not observe a significant increase in total JRA protein in raw mutants relative to controls (Fig. 4C; quantification of JRA relative to alpha-tubulin loading control reveals the ratio of raw/wt = 0.85, std. dev.=0.12, n=3), despite the apparent increase in JRA levels upon immunostaining. Therefore, we next asked if the more concentrated immunoreactivity of JRA in raw mutants reflects increased localization to the nucleus relative to the cytoplasm. To do this, we compared JRA immunostaining to that of another SGP transcription factor, TJ, and quantified the fluorescence intensity in different regions. TJ was used, as it colocalized with DAPI, but had a more uniform distribution in the nucleus than DAPI, making quantification of pixel intensity across the nucleus more reliable (Fig. S2). In wild-type embryos, JRA staining was relatively uniform across the cell, whereas TJ staining was increased in the nuclear region of the cell (Fig. 4D-E). In contrast, in raw mutants, JRA immunostaining across a cell more closely resembled that of TJ (Fig. 4D, F). To compare nuclear levels vs. cytoplasmic levels across many individual SGPs, we defined the TJ “bright” region as the nucleus, and compared JRA fluorescence intensity relative to TJ intensity in the TJ “bright” vs. flanking region of the cell (Fig. 4D-F). We found that the levels of nuclear JRA increased dramatically in raw mutants relative to controls (Fig. 4D). Based on this analysis, we conclude that loss of raw function results in increased nuclear localization of JRA, and that raw may normally act as a negative regulator of the JNK pathway by restricting access of JRA to the nucleus.
Increased JNK signaling causes ensheathment defects
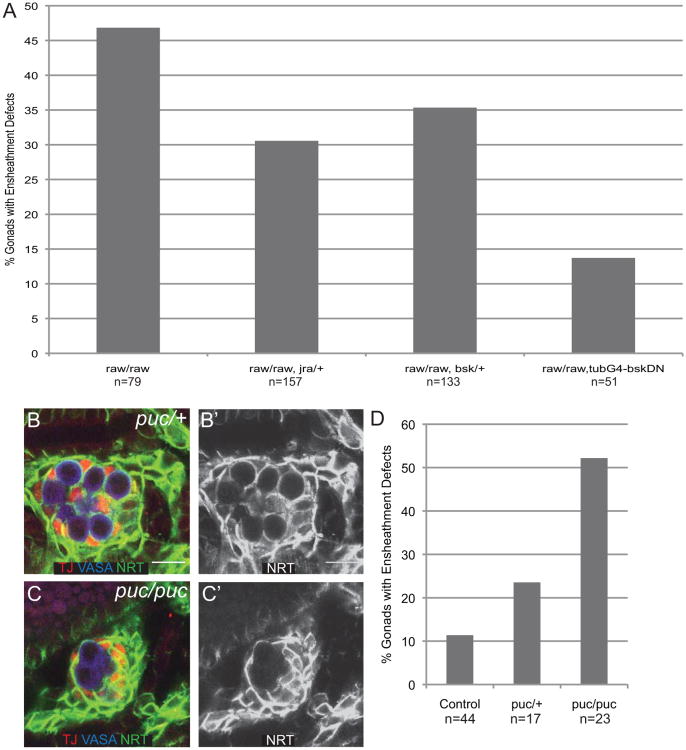
The above results suggest that the ensheathment defects observed in raw mutants occurred as a result of increased JNK signaling. If this is the case, we expected that mutating members of the JNK signaling pathway would suppress the ensheathment defects observed in raw mutants. To test this hypothesis, we examined embryos that were homozygous mutant for raw and also heterozygous for mutations in the Drosophila JNK, basket (bsk), or jra. In both cases we observed a suppression of the raw ensheathment defect (Fig. 5A). To further decrease the activity of the JNK pathway, we expressed a dominant negative form of UAS-bsk in raw mutants. In this case, we observed a stronger suppression of the raw mutant phenotype (Fig. 5A). These results suggest that the raw phenotype is caused by the increase in JNK signaling. If this is the case, we expected that directly increasing JNK signaling would be sufficient to cause defects in GC ensheathment independent of raw. To test this, we examined the effect of removing the negative feedback regulator of JNK signaling, puc. We found that puc heterozygotes exhibited a 25% frequency of ensheathment defects compared to only 11% in controls, while puc homozygous mutants exhibited a 50% frequency of ensheathment defects (Fig. 5B-D), which is a similar penetrance to raw mutants. These results demonstrate that increasing JNK signaling is sufficient to cause ensheathment defects.
Figure 5. raw interacts genetically with JNK signaling pathway members.
(A) Quantitation of % gonads with ensheathment defects. Ensheathment scored using anti-ß-gal immunostaining of 68-77-lacZ in stage 14/15 embryos. Genotypes: raw/raw (raw134.47, 68-77/raw155.27, 68-77); raw/raw, jra (raw155.27, 68-77/raw134.47, jraIA109, 68-77); raw/raw, bsk (raw134.47, 68-77/raw155.27, bsk1) ; raw/raw, tubG4-bskDN (UAS-bskDN; raw134.47, 68-77/raw155.27, 68-77; tubG4). (B, C) Stage 15 gonads immunostained stained to examine germ cell ensheathment. VASA (blue, germ cells), TJ (red, SGPs), Neurotactin (NRT) (green, SGP cell surface). (B′, C′) NRT alone. (B, B′) pucE69/+ controls. (C, C) pucE69/pucE69. (D) Quantitation of ensheathment defects in control (w1118; +/+), pucE69/+ and pucE69/pucE69 gonads using anti-NRT immunostaining.
raw regulates cadherin-based adhesion
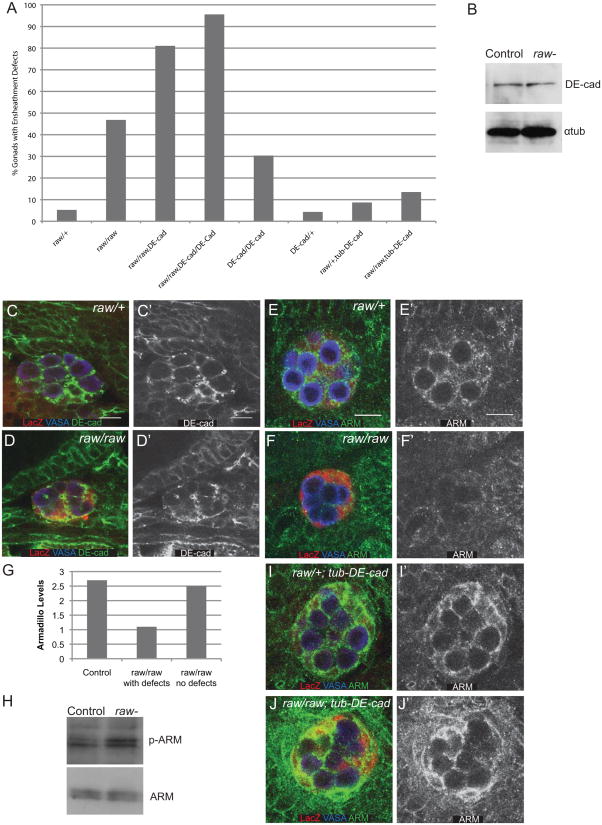
JNK signaling has been demonstrated to affect a number of cellular processes including cell adhesion, leading us to ask if the function of cell adhesion proteins is affected in raw mutants. The ensheathment defects observed in raw mutants are very similar to those previously seen in mutants for Drosophila E-cadherin (DE-cad) (Jenkins et al., 2003), which is encoded by the shotgun (shg) locus (Tepass et al., 1996). Therefore, we examined the possibility that decreased DE-cad function might be responsible for the raw mutant phenotype. If this is the case, we expect that mutating a single copy of shg in a raw mutant should enhance frequency of ensheathment defects. Indeed, loss of a single copy of shg enhanced the frequency of ensheathment defects in raw mutants (Fig. 6A). Embryos homozygous mutant for both raw and shg exhibited an even greater frequency of ensheathment defects (Fig. 6A). This is still consistent with the genes being in the same pathway, rather than parallel pathways, as both genes are known to be contributed maternally. To determine whether defects in DE-cad function are the primary defect in raw mutant gonads, we determined whether forced expression of DE-cad paternally could rescue the raw mutant phenotype. Surprisingly, expression of additional DE-cad using the alpha-tubulin promoter (Pacquelet et al., 2003) was sufficient to strongly rescue the ensheathment defects in raw mutant gonads. Since the alpha-tubulin promoter is expressed in the soma but not the germline (Fig. S3 and Mathews et al., 2006), it is likely that raw causes defects in GC ensheathment primarily by affecting DE-cad function in the soma.
Figure 6. raw regulates germline-soma interactions through cadherin-based adhesion.
(A) Quantitation of ensheathment defects using anti-ß-gal immunostaining of 68-77-lacZ. Note that loss of DE-cadherin (shotgun) enhances the ensheathment defects observed in raw mutants while increasing DE-cadherin expression with tub-DE-cad rescues these defects. Genotypes: raw/+ (raw134.47, 68-77/+ or raw155.27, 68-77/+); raw/raw (raw134.47, 68-77/raw155.27, 68-77); raw/raw, DE-cad (raw134.47, shgg317, 68-77/raw155.27, 68-77); raw/raw, DE-cad/DE-cad (raw134.47, shgg317, 68-77/raw134.47, shgg317, 68-77); DE-cad/DE-cad (shgg317, 68-77/shgg317, 68-77); DE-cad/+ (shgg317, 68-77/+) ; raw/+; tub-DE-cad (raw15527, 68-77/+; tub-DE-cadWT) ; raw/raw; tub-DE-cad (raw134.47, 68-77/raw155.27, 68-77; tub-DE-cadWT). (B) Western blot analysis of DE-cad protein levels in raw mutants (raw134.47/raw155.27) relative to controls. (C, D) DE-Cad expression in stage 15 embryos. VASA (blue, germ cells), 68-77-lacZ (red, SGPs), DE-cad (green). (C′, D′) DE-cad alone. (C, C′) raw heterozygous control, (D, D′) raw134.47/raw155.27. (E, F) ARM expression in stage 15 embryos. VASA (blue, germ cells), 68-77-lacZ (red, SGPs), ARM (green). (E′, F′) ARM alone. (E, E′) raw heterozygous control, (F, F′) raw134.47/raw155.27. (G) ARM levels at sites of SGP-GC contact in raw/+ (raw134.47, 68-77/+ or raw155.27, 68-77/+), and raw/raw (raw134.47, 68-77/raw155.27, 68-77) with or without ensheathment defects in stage 14/15 embryos. (H) Western blot analysis of phospho-ARM protein levels relative to total ARM in raw mutants (raw134.47raw155.27) relative to controls (Oregon R). (I, J) ARM expression in stage 15 embryos overexpressing DE-cad. VASA (blue, germ cells), 68-77-lacZ (red, SGPs), ARM (green). (I′, J′) ARM alone. (I, I′) raw/+; tub-DE-cad (raw155.27or raw134.47, 68-77/+;tub-DE-cadWT). (J, J′) raw/raw; tub-DE-cad (raw134.47, 68-77/raw155.27, 68-77; tub-DE-cadWT). Scale bars: 10 μm. Posterior right.
Given that raw and DE-cad appear to function cooperatively to mediate germline-soma interactions, we next wanted to examine DE-cad protein in raw mutants. We examined DE-cad protein levels using Western blot analysis on raw mutant embryos compared to controls and found only a small reduction in DE-cad levels in raw mutants (Fig. 6B; raw/wt=0.82, Std. Dev.=0.03, n=2). Though DE-cad levels were not strongly affected in raw mutants, it remained possible that its localization or function could be affected, and so we examined DE-cad localization by immunostaining. We found that DE-cad is still localized to sites of SGP-GC interaction in raw mutants similar to controls (Fig.6C, D). To further address DE-cad function, we examined the localization of another member of the cadherin adhesion complex, β-catenin, which is encoded by armadillo (arm) in Drosophila. In contrast to what we observed with DE-cad, we find that ARM localization to the membrane is strongly decreased in raw mutants (Fig. 6E, F). The decrease in ARM immunostaining correlated with germ cell ensheathment defects; those embryos with ensheathment defects were also the embryos that exhibited the clearest decreases in ARM staining (Fig. 6G). One way in which ß-catenin interaction with cadherins is known to be regulated is through phosphorylation (Aberle et al., 1997; Lee et al., 2009; Lilien and Balsamo, 2005). Indeed, a slight increase in the S44P/S48P phosphoform of ARM is observed in whole-embryo extracts from raw mutants relative to controls, while total ARM levels appear unchanged (Fig. 6H). Finally, since the raw mutant ensheathment defect can be rescued by overexpression of DE-cad, we examined whether ARM immunolocalization was also rescued in these embryos. Indeed, in raw mutants overexpressing DE-cad we observed an increase in ARM immunostaining that was now comparable to controls overexpressing DE-cad (Fig. 6I, J). Based on these results, we propose a model in which Raw acts as a negative regulator of the JNK pathway, which, in turn acts to negatively regulate DE-cad-based adhesion, at least in part by influencing the localization of ARM/ß-catenin.
An alternative model, however, is that raw primarily acts by affecting cadherin-based adhesion, and that changes in cell-cell adhesion lead to the observed changes in JNK pathway activity and JRA localization. To distinguish between these models, we tested whether DE-cad acts upstream of downstream of the JNK pathway in germ cell ensheathment. To do this, we examined JRA localization in raw mutants that have been rescued by overexpression of DE-cad. If the JNK pathway acts upstream of DE-cad, we expect that expression of DE-cad would rescue the raw mutant defects but would not affect JRA localization. However, if it is the changes in DE-cad function that lead to the changes in JRA localization, then we would expect DE-cad expression should rescue both the ensheathment defects and the changes in JRA localization. We found that, in raw mutants in which DE-cad is overexpressed, we do not observe a change in JRA localization (Fig. S4), even though the ensheathment defects were rescued (Fig. 6A). This supports the model that JNK signaling acts upstream of DE-cad/ARM and that raw mutants affect DE-cad/ARM function by causing an upregulation of the JNK pathway.
Discussion
Here we have shown that raw is an important regulator of embryonic gonad morphogenesis and the establishment of proper gonad architecture. raw mutants exhibit a failure of SGPs to ensheath GCs in the gonad, resulting in defects in GC development. We have also found that raw affects gonad morphogenesis primarily by acting as a negative regulator of the JNK signaling pathway. Finally, we find that raw mutants exhibit defects in cadherin-based cell adhesion, and that this is the primary cause of the failure of gonad morphogenesis. These results have clear implications for our understanding of how important cell signaling pathways are regulated to control normal organogenesis and may be misregulated to cause disease.
raw is a negative regulator of the JNK pathway
Previously, raw has been proposed to be a negative regulator of the JNK pathway during closure of the dorsal epidermis, based on changes in JNK-dependent expression of target genes such as dpp and puc (Bates et al., 2008; Byars et al., 1999). Indeed, we have also seen an increase in puc expression in the region of the embryonic gonad, and more broadly throughout the embryo (Figs. 3). Further, we observed upregulation of a dedicated AP-1 reporter construct (Fig. 3), indicating that the changes in target gene expression are directly due to changes in AP-1 transcriptional activity regulated by the JNK pathway. When we upregulated the JNK pathway via independent means, we observed similar defects in gonad morphogenesis, indicating that the changes in the JNK pathway were the primary mechanism by which raw mutants cause gonad defects. Therefore, our results support and extend the previous observations that raw acts as a negative regulator of JNK pathway, both in the gonad and in other tissues in which raw mutants exhibit defects in morphogenesis.
How might raw be regulating the JNK pathway? Our evidence indicates that raw regulates the JNK pathway at the level of transcription factor JRA. We found that the nuclear localization of JRA, but not FOS, was altered in raw mutants. JRA was more strongly concentrated in the nucleus in a variety of cell types in raw mutants, whereas we did not observe changes in the global levels of JRA protein. These observations are consistent with previous genetic epistasis experiments that indicated that raw acts at the level of JRA, rather than further upstream in the pathway (Bates et al., 2008). It has been proposed that raw acts as a general negative regulator of the JNK pathway to suppress basal activity and perhaps establish a threshold for pathway activation (Bates et al., 2008; Byars et al., 1999). Our data are consistent with this hypothesis, as we saw a general nuclear accumulation of JRA in a variety of cell types in the embryo (Fig. 4), along with generalized activation of the transcriptional reporter for AP-1 activity (Fig. 3). Presumably, not all of these different cells are normally exposed to activators of the JNK pathway at this time, indicating that the pathway may be activated in cells in which the pathway would normally be turned off. Thus, rather than being just a modulator of the level of signal a cell might receive under conditions of JNK pathway activation, raw is likely also responsible for ensuring that the pathway remains inactive in cells that are not experiencing pathway activation.
It is difficult to predict exactly how RAW may be regulating JNK signaling, as the Raw protein has no readily identifiable protein domains and exhibits only limited homology to proteins of other species. It may be the case that similar JNK pathway regulators are present in other species and have structural and/or functional conservation with Raw, but are difficult to identify based on primary sequence homology. Studies examining the subcellular localization of Raw in cultured mammalian (Bates et al., 2008) or Drosophila (J. Jemc and M. Van Doren, unpublished data) cells indicate that it is primarily found in the cytoplasm. One attractive hypothesis is that Raw directly binds to JRA to block its nuclear translocation and sequester JRA in the cytoplasm. Unfortunately, our efforts to identify a direct, physical interaction between Raw and JRA have so far been unsuccessful.
The JNK pathway is subject to negative regulation at several levels. Most familiar are the MAP kinase phosphatases (MKPs, a subfamily of Dual-specificity phosphatases), like Drosophila Puckered (Martin-Blanco et al., 1998), that provide negative feedback by dephosphorylating activated MAP kinases such as JNK (Bermudez et al., 2010). Additional modes of regulation include nuclear repressors of AP-1 target genes (e.g. Anterior open, (Riesgo-Escovar and Hafen, 1997)) and a secreted protease that acts in negative feedback on the JNK pathway (Scarface, (Rousset et al., 2010; Sorrosal et al., 2010)). Raw appears to represent a distinct mode of regulation, acting on the ability of JRA/JUN to translocate to the nucleus. Regulation of the subcellular localization of transcription factors and cofactors is a strategy that is commonly deployed to regulate signaling pathway activity, and many transcription factors are sequestered in the cytoplasm as a mechanism for negatively regulating their activity. We propose that JRA is subject to such regulation as a means to repress its activity in cells that are not experiencing sufficient levels of JNK pathway activity. Further studies of Raw are necessary to determine how Raw functions at a molecular level to regulate JRA subcellular localization.
raw regulates cadherin-based adhesion in the gonad
We have found that raw mutants also exhibit defects in cadherin-based cell adhesion, which is known to be important for proper gonad morphogenesis and GC ensheathment by SGPs (Jenkins et al., 2003; Van Doren et al., 2003). Loss of DE-cad function exacerbates the gonad defects observed in raw mutants while increasing DE-cad function strikingly rescues these defects (Fig. 6). It is likely that the increase in JNK pathway activity in raw mutants leads to defects in DE-cad-based adhesion and that this is the primary cause of the gonad morphogenesis defects. This is in contrast to the role of raw and the JNK pathway in the closure of the dorsal epidermis, which is largely thought to be due to regulation of dpp expression (Byars et al., 1999). Consistent with this, we observed less up-regulation of dpp in the region of the gonad, relative to the overall activation of the AP-1 transcriptional reporter (Figs. 3 and data not shown).
Previous studies in mammalian cells have implicated JNK signaling in negative regulation of cadherin-based cell adhesion (Lee et al., 2009), while in other contexts the JNK pathway has also been observed to upregulate DE-cad (Dobens et al., 2001). Our results favor a repressive role for the JNK pathway on DE-cad in the gonad. It is also known that cadherins can act upstream of the JNK pathway, and that loss of cadherin can lead to an increase in c-Jun protein levels (Knirsh et al., 2009). However, our results are consistent with DE-cad acting downstream of the JNK pathway, since DE-cadherin expression could rescue gonad morphogenesis independently of rescuing JRA localization (Figs. 6 and S4). We conclude that during gonad morphogenesis, raw acts as a negative regulator of the JNK pathway, and increased JNK pathway activity observed in raw mutants leads to a downregulation of DE-cadherin based cell adhesion and a failure of proper ensheathment of the GCs by the somatic gonad.
While we did not observe a change in DE-cad localization in the gonad, the localization of ARM/ß-catenin was dramatically altered. Since ARM is essential for proper DE-cad function in cell adhesion (Gorfinkiel and Arias, 2007; Pacquelet et al., 2003), this indicates that DE-cadherin-based adhesion is strongly affected in raw mutants. It has been shown that JNK can directly phosphorylate ß-catenin and negatively regulate its activity (Lee et al., 2009). Consistent with this, we observed a modest increase in the relevant phospho-form of ARM/ß-catenin in raw mutants (Fig. 6). Thus, this may represent one aspect of how the JNK pathway regulates DE-cad based adhesion in the gonad. However, the change in ARM/ß-catenin phosphorylation observed is unlikely to account for the more dramatic change in ARM/ß-catenin immunostaining observed in the gonad. Considering that we also observe a strong increase in transcriptional activation by AP-1 in raw mutants, and that mutations in the JRA transcription factor can partially suppress the gonad morphogenesis defects observed in raw mutants, we conclude that at least some of the JNK pathway effect on DE-cad function and ß-catenin localization is likely to depend on changes in gene expression mediated by AP-1. Since we did not observe overall changes in protein levels for DE-cad or ARM/ß-catenin in raw mutants, the changes in gene expression may reflect changes in other regulators of DE-cad based cell adhesion. Interestingly, our previous work identified a zinc transporter, Fear of intimacy, that also affects gonad morphogenesis and GC ensheathment by regulating DE-cad (Mathews et al., 2006; Mathews et al., 2005; Van Doren et al., 2003). Regardless of whether there is an interesting connection between zinc transport and the JNK pathway, or these represent independent pathways, they highlight the importance of careful regulation of cadherin-based cell adhesion in controlling morphogenesis.
Our previous work has indicated that GC ensheathment requires preferential adhesion between SGPs and GCs, such that SGP-GC adhesion is favored over GC-GC or SGP-SGP adhesion (Jenkins et al., 2003). Indeed, just increasing the adhesion between GCs via DE-cadherin expression in these cells is sufficient to prevent ensheathment of the GCs by SGPs (Jenkins et al., 2003). In raw mutants, we primarily observe changes in the JNK pathway in SGPs and surrounding somatic cells. In addition, expression of DE-cad in the soma, but not the germline, is sufficient to rescue the ensheathment defects in raw mutants. Together, these data indicate that raw mutants likely affect gonad ensheathment by decreasing DE-cad function in the SGPs, which decreases SGP-GC adhesion relative to GC-GC adhesion. While it is possible that effects of raw on somatic cells outside of the gonad affect ensheathment within the gonad, it is less easy to imagine how decreasing DE-cad activity in these cells would influence ensheathment.
Regulation of JNK in development and disease
The JNK pathway has been implicated in many diseases, including birth defects, neurodegeneration, inflammatory diseases, and cancer, to name a few (Johnson and Nakamura, 2007). Signaling pathways must be tightly regulated both positively, to ensure rapid and robust signaling responses, and negatively, to terminate signaling events and prevent inappropriate signaling. As a negative regulator of JNK pathway signaling, raw represents the type of gene that might be mutated or misregulated in diseases caused by altered JNK pathway activity. This idea is supported by the strong developmental phenotypes associated with mutations in negative regulators of the JNK pathway in Drosophila (Bates et al., 2008; Byars et al., 1999; Rousset et al., 2010; Sorrosal et al., 2010) and mice (Bermudez et al., 2010).
One disease where the JNK pathway has been particularly well studied is cancer. The JNK pathway's role in cancer is complex, however, and the pathway can act in tumor suppression or oncogenesis, depending on the context (Shaulian, 2010). In mouse and Drosophila models of cancer due to activated Ras, upregulation of the JNK pathway is required for tumor formation and disease progression (Cellurale et al., 2011; Igaki et al., 2006; Uhlirova and Bohmann, 2006). Interestingly, downregulation of E-cadherin is also associated with cancer progression (Berx and van Roy, 2009), including in the models of activated Ras where the JNK pathway is involved (Cellurale et al., 2011; Igaki et al., 2006). Thus, a similar link between the JNK pathway and cadherin regulation that we observed in morphogenesis of the gonad during development may play a role in oncogenesis. Since upregulation of the JNK pathway promotes cancer in these examples, negative regulators of the pathway such as the MAPK phosphatases or Raw would act as tumor suppressors whose mutation could contribute to disease progression. A better understanding of how the JNK pathway is regulated, and how Raw contributes to this regulation, is essential for understanding the normal roles of the JNK pathway in development and homeostasis, and how it is misregulated to cause disease.
Supplementary Material
Stage 15 embryos immunostained for JRA. (A) raw155.27/raw134.47, UAS-rawRA. (B) raw155.27/raw134.47, UAS-rawRA; paired-G4. Scale bar: 50μm. Posterior right.
(A) Stage 15 raw/+ (raw134.47, 68-77/+ or raw155.27, 68-77/+) immunostained for DAPI (blue) and TJ (red; SGPs). (A′) DAPI alone. (A′′) TJ alone. Two SGPs have been outlined.
(A) tubulin-G4, UAS-GFP embryos immunostained for GFP (green), SO×100B (male-specific SGPs; red), and germ cells (blue). (A′) GFP alone. White arrowhead indicates germ cells; black arrow indicates SGPs. The GFP channel was saturated to emphasize the absence of GFP expression in the germ cells. Posterior right.
(A,B) Stage 15 embryos immunostained to label JRA (red), VASA (blue, germ cells), and 68-77 lacZ (Green, SGPs). (A) raw134.47/raw155.27. (B) raw134.47/raw155.27; tubulin-DE-cad. Scale Bar: 10μm. Posterior right. Note that increased expression of DE-cad does not rescue the increased JRA nuclear localization observed in raw mutants, even though it rescues the ensheathment defects (Fig 6A). Thus, DE-cad does not act upstream of JRA.
Research Highlights.
Raw regulates ensheathment of germ cells by somatic cells during gonad formation.
Raw negatively regulates JNK signaling during gonad formation.
Raw regulates JUN subcellular localization.
Raw modulates germline-soma interactions via E-cadherin and ARM.
Acknowledgments
Special thanks to members of the fly community that supplied us with fly stocks and reagents for this work, which have been cited in Materials and Methods. In particular, we thank Nirmalya Chatterjee and Dirk Bohmann for generously supplying us with their AP-1 transcriptional reporter prior to publication. We also thank the Bloomington Stock Center (Indiana University), the Developmental Studies Hybridoma Bank (University of Iowa), and the Drosophila Genome Resource Center for reagents. Special thanks to the Integrated Imaging Center at Johns Hopkins for assistance with confocal imaging, and members of the Van Doren and Chen labs for helpful discussions. This work was supported by the National Institutes of Health Grants F32GM085925 to J. C. J. and R01GM084356 to M. V. D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jennifer C. Jemc, Email: jcjm127@gmail.com.
Alison B. Milutinovich, Email: abjenk@hotmail.com.
Jill J. Weyers, Email: jillweyers@gmail.com.
Yas Takeda, Email: yas.takeda@gmail.com.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates KL, Higley M, Letsou A. Raw mediates antagonism of AP-1 activity in Drosophila. Genetics. 2008;178:1989–2002. doi: 10.1534/genetics.107.086298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergson C, McGinnis W. An autoregulatory enhancer element of the Drosophila homeotic gene Deformed. EMBO J. 1990;9:4287–4297. doi: 10.1002/j.1460-2075.1990.tb07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez O, Pages G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:C189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M, Bonini N, DiNardo S. Expression and function of clift in the development of somatic gonadal precursors within the Drosophila mesoderm. Development. 1997;124:971–982. doi: 10.1242/dev.124.5.971. [DOI] [PubMed] [Google Scholar]
- Boyle M, DiNardo S. Specification, migration and assembly of the somatic cells of the Drosophila gonad. Development. 1995;121:1815–1825. doi: 10.1242/dev.121.6.1815. [DOI] [PubMed] [Google Scholar]
- Brookman JJ, Toosy AT, Shashidhara LS, White RA. The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development. 1992;116:1185–1192. doi: 10.1242/dev.116.4.1185. [DOI] [PubMed] [Google Scholar]
- Byars CL, Bates KL, Letsou A. The dorsal-open group gene raw is required for restricted DJNK signaling during closure. Development. 1999;126:4913–4923. doi: 10.1242/dev.126.21.4913. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer-Verlag; New York: 1985. [Google Scholar]
- Cellurale C, Sabio G, Kennedy NJ, Das M, Barlow M, Sandy P, Jacks T, Davis RJ. Requirement of c-Jun NH(2)-terminal kinase for Ras-initiated tumor formation. Mol Cell Biol. 2011;31:1565–1576. doi: 10.1128/MCB.01122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco TJ, Verney G, Jenkins AB, McCaffery JM, Russell S, Van Doren M. Sex-specific apoptosis regulates sexual dimorphism in the Drosophila embryonic gonad. Dev Cell. 2003;5:205–216. doi: 10.1016/s1534-5807(03)00204-1. [DOI] [PubMed] [Google Scholar]
- Dobens LL, Martin-Blanco E, Martinez-Arias A, Kafatos FC, Raftery LA. Drosophila puckered regulates Fos/Jun levels during follicle cell morphogenesis. Development. 2001;128:1845–1856. doi: 10.1242/dev.128.10.1845. [DOI] [PubMed] [Google Scholar]
- Glise B, Bourbon H, Noselli S. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N, Arias AM. Requirements for adherens junction components in the interaction between epithelial tissues during dorsal closure in Drosophila. J Cell Sci. 2007;120:3289–3298. doi: 10.1242/jcs.010850. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Goldstein ES, Perrimon N. Drosophila Jun relays the Jun amino-terminal kinase signal transduction pathway to the Decapentaplegic signal transduction pathway in regulating epithelial cell sheet movement. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Jack J, Myette G. The genes raw and ribbon are required for proper shape of tubular epithelial tissues in Drosophila. Genetics. 1997;147:243–253. doi: 10.1093/genetics/147.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemc JC. Somatic gonadal cells: The supporting cast for the germline. Genesis. 2011 doi: 10.1002/dvg.20784. [DOI] [PubMed] [Google Scholar]
- Jenkins AB, McCaffery JM, Van Doren Mx. Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development. 2003;130:4417–4426. doi: 10.1242/dev.00639. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim Biophys Acta. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirsh R, Ben-Dror I, Spangler B, Matthews GD, Kuphal S, Bosserhoff AK, Vardimon L. Loss of E-cadherin-mediated cell-cell contacts activates a novel mechanism for up-regulation of the proto-oncogene c-Jun. Mol Biol Cell. 2009;20:2121–2129. doi: 10.1091/mbc.E08-12-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M. Development of the male germline stem cell niche in Drosophila. Dev Biol. 2006;294:92–103. doi: 10.1016/j.ydbio.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Lee MH, Koria P, Qu J, Andreadis ST. JNK phosphorylates beta-catenin and regulates adherens junctions. FASEB J. 2009;23:3874–3883. doi: 10.1096/fj.08-117804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr Opin Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Liu CF, Liu C, Yao HH. Building pathways for ovary organogenesis in the mouse embryo. Curr Top Dev Biol. 2010;90:263–290. doi: 10.1016/S0070-2153(10)90007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews WR, Ong D, Milutinovich AB, Van Doren M. Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development. 2006;133:1143–1153. doi: 10.1242/dev.02256. [DOI] [PubMed] [Google Scholar]
- Mathews WR, Wang F, Eide DJ, Van Doren M. Drosophila fear of intimacy encodes a Zrt/IRT-like protein (ZIP) family zinc transporter functionally related to mammalian ZIP proteins. J Biol Chem. 2005;280:787–795. doi: 10.1074/jbc.M411308200. [DOI] [PubMed] [Google Scholar]
- Moore LA, Broihier HT, Van Doren M, Lunsford LB, Lehmann R. Identification of genes controlling germ cell migration and embryonic gonad formation in Drosophila. Development. 1998;125:667–678. doi: 10.1242/dev.125.4.667. [DOI] [PubMed] [Google Scholar]
- Pacquelet A, Lin L, Rorth P. Binding site for p120/delta-catenin is not required for Drosophila E-cadherin function in vivo. J Cell Biol. 2003;160:313–319. doi: 10.1083/jcb.200207160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo-Escovar JR, Hafen E. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- Ring JM, Martinez Arias A. puckered, a gene involved in position-specific cell differentiation in the dorsal epidermis of the Drosophila larva. Dev Suppl. 1993:251–259. [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Rousset R, Bono-Lauriol S, Gettings M, Suzanne M, Speder P, Noselli S. The Drosophila serine protease homologue Scarface regulates JNK signalling in a negative-feedback loop during epithelial morphogenesis. Development. 2010;137:2177–2186. doi: 10.1242/dev.050781. [DOI] [PubMed] [Google Scholar]
- Sato T, Ueda S, Niki Y. Wingless signaling initiates mitosis of primordial germ cells during development in Drosophila. Mech Dev. 2008;125:498–507. doi: 10.1016/j.mod.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Shaulian E. AP-1--The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O'Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluss HK, Han Z, Barrett T, Goberdhan DC, Wilson C, Davis RJ, Ip YT. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- Sorrosal G, Perez L, Herranz H, Milan M. Scarface, a secreted serine protease-like protein, regulates polarized localization of laminin A at the basement membrane of the Drosophila embryo. EMBO Rep. 2010;11:373–379. doi: 10.1038/embor.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. Embo J. 2006;25:5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Mathews WR, Samuels M, Moore LA, Broihier HT, Lehmann R. fear of intimacy encodes a novel transmembrane protein required for gonad morphogenesis in Drosophila. Development. 2003;130:2355–2364. doi: 10.1242/dev.00454. [DOI] [PubMed] [Google Scholar]
- Wainwright EN, Wilhelm D. The game plan: cellular and molecular mechanisms of mammalian testis development. Curr Top Dev Biol. 2010;90:231–262. doi: 10.1016/S0070-2153(10)90006-9. [DOI] [PubMed] [Google Scholar]
- Warrior R. Primordial germ cell migration and the assembly of the Drosophila embryonic gonad. Dev Biol. 1994;166:180–194. doi: 10.1006/dbio.1994.1306. [DOI] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–567. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers JJ, Milutinovich AB, Takeda Y, Jemc JC, Van Doren M. A genetic screen for mutations affecting gonad formation in Drosophila reveals a role for the slit/robo pathway. Dev Biol. 2011;353:217–228. doi: 10.1016/j.ydbio.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stage 15 embryos immunostained for JRA. (A) raw155.27/raw134.47, UAS-rawRA. (B) raw155.27/raw134.47, UAS-rawRA; paired-G4. Scale bar: 50μm. Posterior right.
(A) Stage 15 raw/+ (raw134.47, 68-77/+ or raw155.27, 68-77/+) immunostained for DAPI (blue) and TJ (red; SGPs). (A′) DAPI alone. (A′′) TJ alone. Two SGPs have been outlined.
(A) tubulin-G4, UAS-GFP embryos immunostained for GFP (green), SO×100B (male-specific SGPs; red), and germ cells (blue). (A′) GFP alone. White arrowhead indicates germ cells; black arrow indicates SGPs. The GFP channel was saturated to emphasize the absence of GFP expression in the germ cells. Posterior right.
(A,B) Stage 15 embryos immunostained to label JRA (red), VASA (blue, germ cells), and 68-77 lacZ (Green, SGPs). (A) raw134.47/raw155.27. (B) raw134.47/raw155.27; tubulin-DE-cad. Scale Bar: 10μm. Posterior right. Note that increased expression of DE-cad does not rescue the increased JRA nuclear localization observed in raw mutants, even though it rescues the ensheathment defects (Fig 6A). Thus, DE-cad does not act upstream of JRA.