Abstract
Purpose
Several studies have examined the prognostic value of the codon 72 polymorphism of the p53 gene in colorectal adenocarcinoma, but none have addressed patient race/ethnicity. Therefore, this study assessed the prognostic value of this polymorphism in African American and Caucasian colorectal adenocarcinoma patients separately.
Experimental Design
Colorectal adenocarcinomas from137 African Americans and 236 non-Hispanic Caucasians were assessed for p53 mutations and genotyped for the codon 72 polymorphism. The phenotypes were correlated with p53 mutational status, clinicopathologic features, and patient survival using the χ2 test and Kaplan-Meier and Cox regression models.
Results
The incidence of p53 mutations was similar in African American and Caucasian patients (50% versus 54%, respectively); however, the homozygous Pro72 allele frequency was higher in African Americans (17%) as compared with Caucasians (7%). In contrast, the homozygous Arg72 allele frequency was higher in Caucasians (36%) than in African Americans (19%). In African Americans but not Caucasians, the Pro/Pro phenotype significantly correlated with a higher incidence of missense p53 mutations and with nodal metastasis. African Americans, but not Caucasians, with the Pro/Pro phenotype had significantly higher mortality (log-rank P = 0.005 versus. P = 0.886) and risk of death due to colorectal adenocarcinoma (hazard ratio, 2.15; 95% confidence interval, 1.02-4.53 versus hazard ratio, 1.60; 95% confidence interval, 0.69-3.18) than those with the phenotype Arg/Arg orArg/Pro.
Conclusions
The higher frequency of the Pro/Pro phenotype of p53 in African American patients with colorectal adenocarcinoma is associated with an increased incidence of p53 mutations, with advanced tumor stage, and with short survival.
In the United States, there are racial differences in colorectal cancer incidence, mortality, and survival; the highest colorectal cancer incidence and mortality rates and the lowest survival rates occur among African Americans (1). Most studies have attributed this disparity principally to socioeconomic differences (2, 3); however, variability in clinicopathologic characteristics (4, 5) have also been suggested as primary factors contributing to survival differences between African American and Caucasian patients. Previous studies from our laboratory suggested that more clinicopathologically aggressive cancers account for the poor survival in African American patients compared with non-Hispanic Caucasian patients (4, 6). Nevertheless, limited attention has been focused on understanding the genetic and molecular basis for the discrepancy in clinical outcomes among colorectal cancer patients with different ethnic backgrounds.
In about 50% of human cancers, including colorectal cancer, the p53 gene is mutated; missense point mutations that disrupt p53 or its functional domains or protein sequestration result in its inactivation, which in turn promotes tumor progression and leads to poor patient survival (7, 8). The mutations that affect structural or functional domains and those in evolutionarily conserved regions are associated with aggressive tumors (9) and chemoresistance (10). p53 mutations within certain domains (the L2 loop, the L3 loop, and the Loop Sheet Helix motif) and mutations in the evolutionarily nonconserved region of p53 (11) are associated with aggressive phenotypes (12). For various malignancies, the heterogeneity of p53 mutant phenotypes apparently leads to heterogeneous clinical outcomes (13, 14).
p53 is a highly conserved gene. To date, only five polymorphisms have been reported in the coding region; four in exon 4 at codons 34, 36 (15), 47 (16), and 72 (17); and one in exon 6 at codon 213 (18). However, p53 polymorphisms are also found in the intronic regions, two in intron 1 (19, 20), one in intron 2 (21), one in intron 3 (22), two in intron 6 (23, 24), five in intron 7 (25, 26), and one in intron 9 (27). Among all these, codon 47 and 72 polymorphisms of p53 have been functionally well characterized. The codon 72 polymorphism is a common alteration in the general population that results in either an arginine or a proline residue at position 72 in the proline-rich domain (residues 64-92) of the p53 protein, resulting in a marked change in its protein structure (17). Several studies, in vivo and in vitro, have highlighted the functional difference between the Pro/Pro and Arg/Arg variants, with the Arg/Arg form of wild-type p53 harboring a greater apoptosis-inducing potential than the Pro/Pro variant (28, 29). In contrast, there is an increased capability for cancer cell proliferation for the Pro/Pro phenotype (28). Furthermore, when compared with other known p53 polymorphisms, the codon 72 polymorphism exhibits a higher level of frequency variation among different racial/ethnic groups (30) and correlates with cancer progression (31). Thus, it is important to understand the effect of the codon 72 polymorphism of p53 on disease progression and clinical outcomes based on race/ethnicity.
Previous studies have examined the role of the p53 codon 72 polymorphism in causing other mutations within the p53 gene. These efforts have identified a higher incidence of missense p53 mutations in breast cancer, but not in colorectal or bladder cancer, when the tissues exhibit Arg/Arg homozygotes compared with Arg/Pro heterozygotes or Pro/Pro homozygotes (32). In non-small cell lung cancers, the p53 codon 72 Pro allele is preferentially mutated (33) and is associated with an increased risk of tumor progression (34) and poor patient survival (35). Thus, the p53 codon 72 polymorphism could contribute to the differences between individuals or racial groups in susceptibility and severity of disease. The incidence and the prognostic importance of the codon 72 polymorphism in African American and Caucasian patients with colorectal cancer, however, have not been investigated separately. Therefore, in this study, we examined, for colorectal cancers, associations between the codon 72 polymorphism and the type of p53 mutations, aggressive tumor pathologic features, and patient survival based on patient race/ethnicity.
Materials and Methods
Patient population
Approval for these studies was obtained from the Institutional Review Board of the University of Alabama at Birmingham (UAB). All patients included in this study had undergone surgery for primary colorectal cancer at the UAB hospital.
Retrospective colorectal cancer samples
We selected all eligible 137 African American and 236 non-Hispanic Caucasian patients from a total of 268 African American and 492 non-Hispanic Caucasian colorectal cancer patients who had undergone surgical resection for “first primary” colorectal cancer from 1985 to 1995 at UAB. The 760 retrospective samples were an “unselected” patient population. The intent of using patients from this time period was to maximize postsurgery follow-up. Formalin-fixed, paraffin-embedded tissue blocks from these patients were obtained from the Anatomic Pathology Division at UAB. These histologically confirmed colorectal cancers and the corresponding normal (benign colonic epithelial tissues 8 cm away from colorectal cancer) tissues were analyzed for the p53 gene status and the codon 72 polymorphism of p53.
During our initial selection process, those patients who died within a week of their surgery; those with surgical margin-involvement, unspecified tumor location, multiple primaries within the colorectum, or multiple malignancies; or those patients with family of hereditary nonpolyposis colorectal cancer, familial adenomatous polyposis, or personal histories of colorectal cancer were excluded from our study population. Because, based on the information in the patient charts, we recognized it would be difficult to identify the familial versus sporadic nature of colorectal cancers, this retrospective cohort can be described as a “consecutive” patient population. We included patients who had undergone surgery alone as a therapeutic intervention, but we excluded patients who received presurgical or postsurgical chemotherapy or radiation therapy to control for treatment bias. This study included patients with stages I through IV; however, patients with stage III or IV did not receive any adjuvant therapy for various reasons but were subjected to surgery with a palliative intent.
Pathologic features
In our study, two pathologists (CKS and WEG) individually reviewed H&E-stained slides of all cases for the degree of histologic differentiation and regraded all lesions as well, moderate, poor, or undifferentiated. Cases with disagreement were resolved by reevaluating the slides together to reach a consensus. Subsequently, we pooled well and moderately differentiated tumors into a low-grade group and poor and undifferentiated tumors into a high-grade group (36). The pathologic staging was done according to the criteria of the American Joint Commission on Cancer (37). The International Classification of Diseases for Oncology (ICD-O) codes was used to specify the anatomic location of the tumor (38). The anatomic subsites were the proximal colon, the distal colon, and the rectum. Three-dimensional tumor size (length, breadth, and depth) was taken into consideration; the largest of the three dimensions was used for statistical purposes.
Before the tissues were utilized for DNA extraction and subsequent mutational and genotyping analyses, a section was cut from each block and stained with H&E to assess the proportion of tumor versus uninvolved tissue in the sample and to permit macrodissection (using a simple microscope) to separate tumor from uninvolved tissue.
Patient demographics and follow-up information
This study includes both African American and non-Hispanic Caucasian patients. Information on their race/ethnicity background was obtained from the patient charts and assignment was self-described or self-identified. For example, blacks of the metro-Birmingham area are overwhelmingly classified as African Americans, because migration of African blacks from other continents was rare during this study period. However, we recognize there is some diversity in identification within any race/ethnic group.
Patient demographics along with clinical and follow-up information were retrieved retrospectively from medical records, physician charts, and pathology reports as well as from the UAB Tumor Registry. Patients were followed either by the patients’ physician or by the UAB tumor registry until their death or the date of the last documented contact within the study time frame. The Tumor Registry ascertained outcome (mortality) information directly from the patients (or living relatives) and from the physicians of the patients through telephone and mail contacts. This information was further validated against the state death registry. Demographic data, including patient age at diagnosis, gender, race/ethnicity, date of surgery, date of the last follow-up (if alive), date of recurrence (if any), and date of death, were collected. The Tumor Registry updated follow-up information every 6 mo, and follow-up of our retrospective cohort ended in May 2008. The laboratory investigators (VRK and XJ) were blinded to the outcome information until the completion of the assays. The mean follow-up periods for African Americans and Caucasian patients are 10.9 y (range <1 to < 22 y) and 10.7 y (range <1 to < 21 y), respectively.
p53 mutational analysis and genotyping
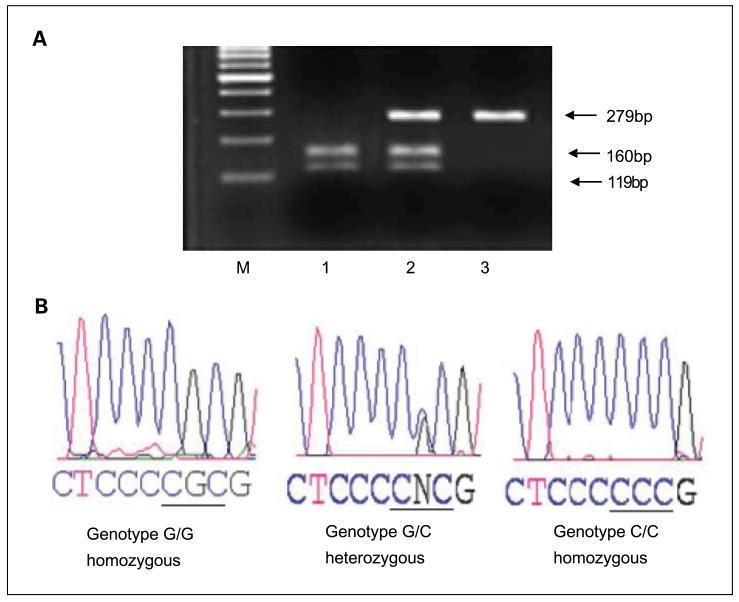
DNA extraction from formalin-fixed, paraffin-embedded archival tissues of 373 colorectal cancers and matching normals was done following a modified deparaffinization protocol (39). The status of the p53 gene was assessed by PCR and direct sequencing of exons 4 through 9, by use of exon-specific primers (Table 1). The genotype of p53 for the codon 72 polymorphism at exon 4 was determined by PCR-restriction fragment length polymorphism (Fig. 1; ref. 30). Exons 4, 5, 6, 7, and 8-9 of the p53 gene were amplified separately by incubating on a Robocycler (Strategene) for 10 min at 94°C for initial denaturation followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The final extension step was 72°C for 7 min. The standard reaction mixture (25 μL) contained 100 ng of genomic DNA, 0.25 μmol/L of each primer, 0.2 mmol/L of each dNTP, 10 × PCR buffer (Invitrogen), 2 mmol/L MgCl2, and 0.5 units of platinum Taq DNA polymerase (Invitrogen). Electrophoresis was done for PCR products in 3% agarose gel prepared in 0.4 × tris-borate-EDTA buffer. The purified PCR product was directly sequenced on an ABI 3100 sequencer. Compilation and sequence analysis was done with LASER-GENE (DNA STAR, Inc.) sequence analysis software, which allows for direct analysis of sequencing electrophoretograms for the detection of duplex sequence signals at each position to identify mutation/polymorphisms. Nucleotide changes in each exon sequence were confirmed by sequencing the opposite strand.
Table 1.
Oligonucleotide primer sequences for the p53 gene amplification and sequencing
| Exon | Primers for gene amplification | Size (bp) | Primers for sequencing |
|---|---|---|---|
| Exon-4 | S 5′TCCCCCTTGCCGTCCCAA3′ | 279 | S 5′TCCCCCTTGCCGTCCCAA3′ |
| A 5′CGTGCAAGTCACAGACTT3′ | A 5′CGTGCAAGTCACAGACTT3′ | ||
| Exon-5 | S 5′TTTCAACTCTGTCTCCTTCCT3′ | 229 | S 5′CCTTCCTCTTCCTGGAGTAC3′ |
| A 5′GCCCCCAGCTGCTCACCATC3′ | A 5′AGCTGCTCACCATCGCTATC3′ | ||
| Exon-6 | S 5′CACTGATTGCTCTTAGGTCTG3′ | 144 | S 5′TCTTAGGTCTGGCCCCTCCT3′ |
| A 5′AGTTGCAAACCAGACCTCAG3′ | A 5′ACCAGACCTCAGGCGGCTCA3′ | ||
| Exon-7 | S 5′GTGTTGTCTCCTAGGTTGGC3′ | 150 | S 5′CCTAGGTTGGCTCTGACTGT3′ |
| A 5′TGTGCAGGGTGGCAAGTGGC3′ | A 5′GGGTGGCAAGTGGCTCCTGA3′ | ||
| Exons 8-9 | S 5′CCTATCCTGAGTAGTGGTAA3′ | 346 | S 5′TGGTAATCTACTGGGAGCAG3′ |
| A 5′ACTTGATAAGAGGTCCCAAG3′ | A 5′CCCAAGACTTAGTACCTGAA3′ |
Abbreviations: S, sense primer; A, antisense primer.
Fig. 1.
PCR with PCR-restriction fragment length polymorphism (PCR-RFLP) and sequence analysis. Genotyping of p53 for the codon 72 polymorphism by PCR-RFLP. A, the 279-bp target DNA fragment, containing the CGC/CCC site of the p53 gene codon 72 located in exon 4, was digested with Bstu 1. The digested product was separated on a 3% agarose gel with ethidium bromide and photographed with an UltraViolet Product Image Store system.The C/C genotype (Pro/Pro phenotype) produced a single 299-bp band due to loss of the Bstu1restriction site; the wild-type G/G genotype (Arg/Arg phenotype) produced two bands (119-bp and 160-bp); and the G/C genotype (Arg/Pro phenotype) produced three bands (119-bp, 160-bp, and 279-bp). B, direct sequencing analysis of DNA fragments confirmed by PCR-RFLP.
Classification of p53 mutations
Mutations of the p53 gene were classified into two categories, disruptive and nondisruptive, based on their locations within p53 (40); mutations at these sites disrupt important functional domains of p53 and predict amino acid alterations (http://www.bioscience.org/urllists/aminacid.htm). Disruptive mutations are DNA sequence alterations that (a) introduce a STOP sequence resulting in disruption of p53 protein production or (b) any DNA sequence alteration that occurs within DNA contact surface regions or functional regions L2 (codons 163-195), L3 (codons 236-251), and LSH (codons 273-286) that results in the replacement of an amino acid from one polarity/charge category with an amino acid from another category. Nondisruptive mutations are those occurring outside the L2, L3, or LSH regions (except stop codons) and mutations within the L2, L3, or LSH domains of DNA contact surface regions that result in the replacement of an amino acid with another from the same polarity/charge category.
Statistical methods
Data management for statistical analyses
Deaths due to colorectal cancer were the outcomes (events) of interest. Patients who died within 1 mo after surgery were excluded from these analyses. Nine (7%) African American and 16 (7%) Caucasian patients were lost to follow-up and 5 (4%) African Americans and 8 (3%) Caucasians died within 1 mo after surgery; therefore, the survival analyses were done on the remaining 123 African American and 212 Caucasian patients. Because the Arg allele is preferentially retained in patients heterozygous for this polymorphism, the Arg/Arg and Arg/Pro phenotypes were pooled for survival analyses (41), and the data were compared with that for Pro/Pro phenotypes. The time at risk was measured by calculating the number of months from date of surgery to time of event or date of last contact.
Statistical analyses
Comparison of baseline characteristics in each ethnic group was done with χ2 tests (42). These analyses were also used to examine univariate associations with covariates and potential confounders. The baseline characteristics included demographic variables (age and gender), pathologic variables (tumor location, size, histologic type, differentiation, and stage), and p53 mutational status. The type I error rate of each test was controlled at <0.05. All analyses were done with SAS statistical software version 9.0 (43).
Survival analysis was used to model time from date of surgery until death due to colorectal cancer. Those patients who died of any cause other than colorectal cancer and those who were alive at the end of the study were considered to be censored. A log-rank test and Kaplan-Meier survival curves (44) were used to compare the Pro/Pro phenotype with the phenotype Arg/Arg or Arg/Pro in terms of survival in each ethnic group. The type I error rate of each test is controlled at <0.05.
Besides the primary analysis determining the effect of the codon 72 polymorphism described above, secondary analyses were done to consider covariates known to be potential confounders or independent risk factors for death. These included age, gender, tumor location, tumor stage, tumor size, and p53 mutation status. For these analyses, Cox regression models (45) were used within each ethnic group, with a final Cox model including those covariates for which P < 0.05.
The bootstrap method was used to show the robustness of our results. By resampling the rows of the data matrix with replacement, we constructed 200 separate datasets following the methods described earlier (46). The full and final models were fit to each of the 200 data sets. The median P value for each variable obtained from the 200 separate models was selected as an estimate of the statistical significance of each variable in the full (final) model.
Results
Study cohort characteristics
Demographic and tumor characteristics for African American and Caucasian patients with colorectal cancers are given in Table 2. There were no significant differences by race/ethnicity with regard to age at diagnosis (χ2 P = 0.66), gender (χ2 P = 0.16), depth of tumor invasion (χ2 P = 0.79), tumor stage (χ2 P = 0.96), tumor grade (χ2 P = 0.65), or tumor size (χ2 P = 0.48), but there was a significant difference in anatomic location of tumor within the colorectum (χ2 χ = 0.005). African American patients were more likely to present with proximal tumors (58%) as compared with Caucasian patients (43%).
Table 2.
Clinicopathologic and molecular features
| Variable | African Americans,* n = 137 (%) | Caucasians,† n= 236 (%) | χ2 P |
|---|---|---|---|
| Age group (y) | |||
| <65 | 56 (41) | 102 (43) | 0.66 |
| ≥65 | 81 (59) | 134 (57) | |
| Gender | |||
| Female | 60 (44) | 86 (36) | 0.16 |
| Male | 77 (56) | 150 (64) | |
| Tumor location | |||
| Proximal colon | 80 (58) | 101 (43) | 0.005 |
| Distal colon | 41 (30) | 79 (34) | |
| Rectum | 16 (12) | 55 (23) | |
| Depth of tumor invasion | |||
| pTx | 0 | 1 (<1) | 0.79 |
| pT1 | 4 (2) | 6 (3) | |
| pT2 | 20 (15) | 42 (18) | |
| pT3 | 86 (63) | 135 (58) | |
| pT4 | 27(20) | 50 (21) | |
| Nodal status | |||
| Nx | 0 | 1 (<1) | 0.64 |
| N0 | 73 (53) | 131 (56) | |
| N1-3 | 64 (47) | 102 (44) | |
| Distant metastasis | |||
| M0 | 118 (86) | 203 (87) | 0.86 |
| M1 | 19 (14) | 31 (13) | |
| Tumor stage | |||
| I | 17(12) | 33 (14) | 0.96 |
| II | 53 (39) | 92 (39) | |
| III | 48 (35) | 78 (34) | |
| IV | 19 (14) | 31 (13) | |
| Tumor grade | |||
| Low | 103 (76) | 179 (78) | 0.65 |
| High | 33 (24) | 51 (22) | |
| Tumor size (cm) | |||
| ≤5 | 73 (53) | 132 (59) | 0.48 |
| >5 | 62 (47) | 96 (41) | |
| p53 status | |||
| Wild-type | 69 (50) | 109 (46) | 0.44 |
| Mutated | 68 (50) | 127(54) | |
| p53 mutation type‡ | |||
| Disruptive | 31 (46) | 51 (40) | 0.46 |
| Nondisruptive | 37(54) | 76 (60) | |
| p53 codon 72 phenotypes | |||
| Arg/Arg | 26 (19) | 85 (36) | 0.002 |
| Arg/Pro | 87(64) | 134 (57) | |
| Pro/Pro | 24 (17) | 17 (7) | |
| Vital status at the end of follow-up§ | |||
| Alive | 36 (28) | 64 (29) | 0.98 |
| Died due to CRC | 61 (48) | 104 (47) | |
| Died due to other causes | 31 (24) | 52 (24) |
Abbreviations: n, total number of cases; CRC, colorectal cancer.
Information on tumor grade for one case and tumor size for two cases was not available for African Americans.
Information on tumor location for one case; pT, pN and M for two cases; tumor stage for two cases; tumor grade for six cases; and tumor size for eight cases was not available for Caucasians.
p53 mutation that introduces an amino acid that disrupts functional domains of p53.
As described in Materials & Methods, 9 African American and 16 Caucasian patients were lost to follow-up.
Frequencies of p53 mutations and codon 72 polymorphism
The incidence of all and disruptive mutations were similar for African American patients (50% and 46%, respectively) and Caucasian patients (54% and 40%, respectively; Table 2). These mutations were clustered in exons 4 through 8. Analysis of p53 at codon 72 for the status of variant forms by genotyping (Fig. 1) in these colorectal cancers revealed a higher frequency of Arg/Pro phenotypes than Arg/Arg or Pro/Pro in both racial groups; however, homozygous Pro/Pro was more predominant in African Americans (24 of 137; 17%) than Caucasians (17 of 236; 7%; Tables 2 and 3). The correlation of p53 codon 72 polymorphism status based on race, along with demographic, tumor, and other molecular features, is shown in Table 3.
Table 3.
Association between p53 codon 72 phenotypes and clinic pathologic characteristics
| Variable | African Americans (n = 137) |
Caucasians (n = 236) |
||||||
|---|---|---|---|---|---|---|---|---|
| Arg/Arg |
Arg/Pro |
Pro/Pro |
χ2 P | Arg/Arg |
Arg/Pro |
Pro/Pro |
χ2 P | |
| 26 (19%) | 87 (64%) | 24 (17%) | 85 (36%) | 134 (57%) | 17 (7%) | |||
| Age group (y) | ||||||||
| <65 | 9 (35) | 35 (40) | 12 (50) | 0.531 | 35 (40) | 65 (49) | 2 (12) | 0.014 |
| ≥65 | 17(65) | 52 (60) | 12 (50) | 50 (60) | 69 (51) | 15 (88) | ||
| Gender | ||||||||
| Female | 14 (53) | 37(43) | 9 (38) | 0.570 | 37 (44) | 39 (29) | 10 (59) | 0.013 |
| Male | 12 (47) | 50 (57) | 15 (62) | 48 (56) | 95 (71) | 7 (41) | ||
| Tumor location* | ||||||||
| Proximal colon | 17(65) | 46 (53) | 17(7 1) | 0.342 | 41 (49) | 50 (37) | 2 (12) | 0.012 |
| Distal colon | 5 (19) | 31 (36) | 5 (20) | 27(32) | 48 (36) | 12 (71) | ||
| Rectum | 4 (16) | 10 (12) | 2 (11) | 16 (19) | 36 (27) | 3 (18) | ||
| Nodal status* | ||||||||
| N0 | 11 (44) | 54 (62) | 8 (32) | 0.017 | 43 (52) | 76 (57) | 12 (70) | 0.354 |
| N1-3 | 14 (56) | 33 (38) | 17(68) | 40 (48) | 57 (43) | 5 (30) | ||
| Tumor grade* | ||||||||
| Low | 17 (68) | 71 (82) | 15 (63) | 0.093 | 56 (68) | 112 (85) | 11 (73) | 0.012 |
| High | 8 (32) | 16 (18) | 9 (37) | 27 (32) | 20 (15) | 4 (27) | ||
| p53 status | ||||||||
| Wild-type | 11 (42) | 51 (59) | 7(29) | 0.025 | 33 (34) | 65 (49) | 11 (65) | 0.112 |
| Mutated | 15 (58) | 36 (41) | 17(7 1) | 52 (66) | 69 (51) | 6 (35) | ||
| p53 mutation type† | ||||||||
| Disruptive | 5 (33) | 16 (44) | 10 (59) | 0.345 | 25 (48) | 24 (35) | 2 (33) | 0.317 |
| Nondisruptive | 10 (67) | 20 (56) | 7 (31) | 27 (52) | 45 (65) | 4 (67) | ||
Information on some cases was not available.
p53 mutation that introduces an amino acid that disrupts functional domains of p53.
In African Americans, the colorectal cancers homozygous for the Pro/Pro (17 of 24; 71%) or Arg/Arg (15 of 26; 58%) phenotypes had a higher incidence of p53 missense mutations compared with the heterozygous Arg/Pro phenotype (36 of 87; 41%; Table 3). In contrast, in Caucasian patients, the homozygous Arg/Arg (52 of 85; 66%) and heterozygous Arg/Pro (69 of 134; 51%) phenotypes exhibited a higher frequency of p53 mutations than the homozygous Pro/Pro phenotype (6 of 17; 35%; Table 3). In addition to codon 72 polymorphisms, we also observed polymorphisms in exon-6 at codon 213 (CGA>CGG, Arg>Arg, silent alteration) in two Caucasian patients (data not shown).
Differential p53 mutational spectra at different structural and functional regions were observed among African American and Caucasian patients in relation to the mutant Pro/Pro phenotype (Table 4). In African Americans, most of these mutations were the disruptive type, clustered in the S10 β sheet, an important structural motif of p53 with a DNA-binding function (Table 4). In relation to the Pro/Pro phenotype, mutations in the S10 β sheet domain (24%; 4 of 17), in the evolutionary nonconserved region (35%; 6 of 17), in the DNA contact region (24%; 4 of 17), and in exon 6 (35%; 6 of 17) of p53 were higher in African American patients; however, no Caucasian patient with the Pro allele exhibited mutations in these regions. Point mutations leading to a high degree of disruption and truncation of the p53 protein were 59% (10 of 17) in African American patients compared with 33% (2 of 6) in Caucasian patients. Mutations in the LSH motif (codon 273-286) were different in these two ethnic groups. In colorectal cancers with advanced tumor stage, the point mutations at different structural and functional regions of the p53 gene were higher in African American patients (13 of 17; 75%) in relation to the Pro/Pro phenotype compared with Caucasian patients (1 of 6; 17%; Table 4). The key observation was that the association between p53 mutation and codon 72 phenotypes was different between these racial groups. Among the colorectal cancers with the Pro/Pro phenotype in African Americans (n = 24), 17 had mutations (71%) of which 10 were disruptive (42%) type, compared with the colorectal cancers with Pro/Pro phenotype in Caucasians (n = 17) with 6 having mutations (35%) of which 2 were disruptive type (12%; Fisher’s exact test, P = 0.046; Table 4).
Table 4.
Mutational spectra of p53 in African American and Caucasian patients in relation to Pro/Pro phenotype
| Case No. |
p53 codon |
Nucleotide change |
Amino acid change |
Structural element affected* |
Amino acid contact† |
Mutation type |
Exon-conserved area affected |
Tumors stage at diagnosis |
|---|---|---|---|---|---|---|---|---|
| African American patients (17of 24; 71% of cases with p53 mutations) | ||||||||
| p47 | 270 | TTT>TGT | Phe>Cys | S10 β sheet | Buried | Disruptive | 8-Cons V | III C |
| p106 | 152 | CCG>CTG | Pro>Leu | - | PE | Nondisruptive | 5-Outside Cons | III C |
| p110 | 153 | CCC>CTC | Pro>Leu | - | PE | Nondisruptive | 5-Outside Cons | III C |
| p132 | 196 | CGA>TGA | STOP | Truncated | - | Disruptive | 6-Cons III | IV D |
| p138 | 176 | TGC>TAC | Cys>Tyr | L2 loop | Zn binding | Nondisruptive | 5-Cons III | II B |
| p146 | 220 | TAT>CAT | Tyr>His | - | Buried | Nondisruptive | 6-Outside Cons | III C |
| p168 | 282 | CGG>TGG | Arg>Trp | H2 helix | Buried | Disruptive | 8-Cons V | IV D |
| p187 | 213 | CGA>TGA | STOP | Truncated | - | Disruptive | 6-Outside Cons | I A |
| p198 | 196 | CGA>TGA | STOP | Truncated | - | Disruptive | 6-Outside Cons | III C |
| p222 | 248 | CGG>CAG | Arg>Gln | L3 loop | DNA contact | Disruptive | 7-Cons IV | III C |
| p251 | 258 | GAA>GAC | Glu>Asp | S9 β sheet | Buried | Nondisruptive | 7-Cons IV | III C |
| p262 | 273 | CGT>CAT | Arg>His | S10 β sheet | DNA contact | Nondisruptive | 8-Cons V | II B |
| p276 | 194 | CTT>CGT | Leu>Arg | L2 loop | Buried | Disruptive | 6-Outside Cons | III C |
| p283 | 273 | CGT>TGT | Arg>Cys | S10 β sheet | DNA contact | Disruptive | 8-Cond V | IV D |
| p285 | 175 | CGC>CAC | Arg>His | L2 loop | Buried | Nondisruptive | 5-Cons III | IV D |
| p291 | 282 | CGG>TGG | Arg>Trp | H2 helix | Buried | Disruptive | 8-Cons V | II B |
| p401 | 273 | CGT>CTT | Arg>Leu | S10 β sheet | DNA contact | Disruptive | 8-Cons V | III C |
| Caucasian patients (6 of 17; 35% of cases with p53 mutations) | ||||||||
| P195 | 175 | CGC>CAC | Arg>His | L2 loop | Buried | Nondisruptive | 5-Cons III | II B |
| P207 | 242 | TGC>TAC | Cys>Tyr | L3 loop | Zn binding | Nondisruptive | 7-Cons IV | II B |
| P243 | 286 | GAA>AAA | Glu>Lys | H2 helix | PE | Disruptive | 8-Cons V | II B |
| P252 | 282 | CGG>TGG | Arg>Trp | H2 helix | Buried | Disruptive | 8-Cons V | IVD |
| P281 | 245 | GGC>AGC | Gly>Ser | L3 loop | Buried | Nondisruptive | 7-Cons IV | II B |
| P302 | 236 | TAC>TGC | Tyr>Cys | S8 β sheet | Buried | Nondisruptive | 7-Cons IV | II B |
Abbreviations: Cons, evolutionarily conserved area; Outside cons, outside evolutionarily conserved area. Buried, <15% of DNA binding site surface area exposed. PE, partially exposed (>15 or <50% of DNA binding site surface area exposed); Disruptive, mutation that introduces an amino acid that disrupts the biding site; Nondisruptive, mutation that introduces an amino acid that does not disrupt the binding site.
Structural element feature as described in Cho et al. (1994).
Contact features of amino acids with DNA.
As shown in Table 3, in African-American patients the higher incidence of the Pro/Pro phenotype was significantly associated with nodal metastasis (17 of 24; 68%; χ2 P = 0.017). In African American patients, these tumors exhibited disruptive mutations (10 of 24; 59%) and high-grade differentiation. In Caucasian patients, the higher incidence of the mutant Pro/Pro phenotype significantly correlated with old age, female gender, distal colon location, and high-grade differentiation.
Survival analyses
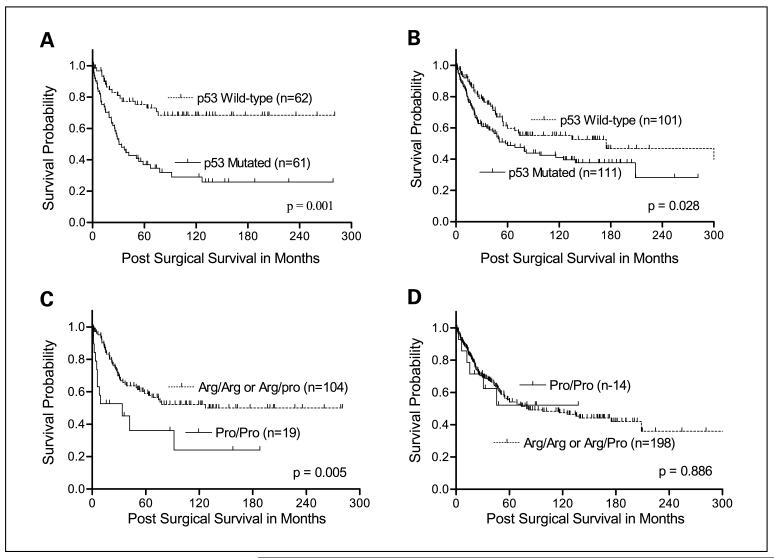
Univariate Kaplan-Meier survival analyses based on race/ethnicity showed that colorectal cancers with p53 mutations were significantly associated with short overall survival as compared with those with wild-type p53 in both African American (log rank, P = 0.001; Fig. 2A) and Caucasian (log rank, P = 0.028; Fig. 2B) patients. The Pro/Pro phenotype of colorectal cancers was significantly associated with poor survival of African American patients (log-rank, P = 0.005; Fig. 2C), but not Caucasian patients (log-rank, P = 0.886; Fig. 2D).
Fig. 2.
Kaplan-Meier survival curves for impact of p53 mutations and the codon 72 polymorphism based on race. For both African Americans (A, log-rank, P = 0.001) and Caucasians (B, log-rank, P = 0.028) colorectal cancers with p53 mutations had significantly poor survival compared with those with wild-type p53. C, African American patients with colorectal cancers having the mutant Pro/Pro phenotype of p53 had significantly poorer survival compared with those with the wild-type Arg/Arg phenotype or those heterozygous for the Arg/Pro phenotypes (log-rank, P = 0.005). D, for Caucasian patients, there was no survival difference between the codon 72 phenotypes of p53 (log-rank, P = 0.866).
The independent prognostic significance of the Pro/Pro phenotype on colorectal cancer-specific survival based on race was evaluated with a Cox regression model. These multivariate survival analyses confirmed the independent effect of the Pro/Pro phenotype on colorectal cancer-specific survival only in African American patients (Table 5). African American patients with the Pro/Pro phenotype of p53 were 2.15 times (hazard ratio, 2.15; 95% confidence interval, 1.02-4.53) more likely to die of colorectal cancer compared with African Americans with the Arg/Arg or Arg/Pro phenotype. The point estimate of risk in the Caucasian patients was smaller (hazard ratio, 1.60; 95% confidence interval, 0.69-3.18; Table 5). In our analyses, tumor stage and p53 mutations were also independent prognostic indicators in both African American and Caucasian patients. However, tumor location (proximal colon) and tumor size were identified as independent prognostic indicators only for African American patients (Table 5).
Table 5.
Cox regression analysis to determine prognostic significance of p53 codon 72 phenotypes
| Prognostic variables | Indicator of poor prognosis | Hazard ratio*(95% confidence intervals) | P |
|---|---|---|---|
| African American patients Codon 72 polymorphism |
|||
| Pro/Pro vs. Arg/Arg or Arg/Pro | Pro/Pro | 2.15 (1.02-4.53) | 0.045 |
| p53 status | |||
| Mutated vs. wild-type | Mutated | 2.30 (1.19-4.45) | 0.014 |
| Tumor location | |||
| Proximal colon vs. Rectum | Proximal colon | 2.18 (1.24-3.82) | 0.007 |
| Distal colon vs. Rectum | Distal colon | 1.27(0.4 2-3.92) | 0.674 |
| Tumor stage | |||
| II vs. I | II | 3.77 (0.85-16.79) | 0.081 |
| III vs. I | III | 17.17 (1.65-31.21) | 0.008 |
| IV vs. I | IV | 27.46 (5.88-128.28) | <0.0001 |
| Tumor size | |||
| >5 cm vs. ≥5 cm | >5 cm | 0.41 (0.23-0.74) | 0.003 |
| Caucasian patients Codon 72 polymorphism |
|||
| Pro/Pro vs. Arg/Arg or Arg/Pro | Pro/Pro | 1.60 (0.69-3.18) | 0.277 |
| p53 status | |||
| Mutated vs. wild-type | Mutated | 1.55 (1.04-2.32) | 0.032 |
| Tumor stage | |||
| II vs. I | II | 1.63 (0.74-3.59) | 0.229 |
| III vs. I | III | 4.10 (1.92- 8.77) | 0.0003 |
| IV vs. I | IV | 10.71 (4.72-24.29) | <0.0001 |
Adjusted for the p53 codon 72 polymorphism, p53 gene mutations, age, gender, and TNM tumor stage, tumor location within the colorectum, tumor grade, and tumor size.
As described above, bootstrapped data sets were used to validate the robustness of the results. The median P value for each variable obtained from the bootstrapping of the Cox regression models was selected as an estimate of the statistical significance of each variable. The median P value for the p53 codon 72 Pro/Pro phenotype (versus Arg/Pro and Arg/Arg) for African Americans was 0.041, and that for the p53 mutation (versus wild-type) was 0.012. The corresponding median P values for Caucasian patients were 0.297 and 0.027, respectively (data not shown). Because the median P values of these variables obtained from bootstrap were consistent with the final models generated from each racial group (as shown in Table 5), the robustness of our findings is apparent.
Discussion
In this study, for the first time, we assessed the prognostic value of the codon 72 polymorphism of p53 in African American and Caucasian patients with colorectal cancers and showed that this polymorphism, specifically the Pro/Pro phenotype, is an independent factor of prognosis for African American patients. Similar frequencies of p53 mutations were observed in both African American and Caucasian patients. However, the incidence of the homozygous mutant variant Pro/Pro was higher in African American patients as compared with Caucasians; in contrast, the wild-type Arg/Arg variants were more common in Caucasians than in African Americans. In African American patients, colorectal cancers with Pro/Pro mutant phenotypes exhibited a higher incidence of p53 mutations, specifically of the disruptive type, and were associated with nodal metastasis and short overall survival. These aggressive phenotypic features of the Pro/Pro mutant phenotype were not observed in Caucasian patients. Thus, these findings suggest that the Pro/Pro mutant phenotype is associated with advanced tumor stage and short survival of African American patients.
In the United States, African Americans have the highest incidence of colorectal cancer, a younger mean age at colorectal cancer diagnosis (47), a higher proportion of proximal colon cancers (48), and a lower survival rate compared with Caucasian patients (49, 50). Consistent with these studies, our present study also showed that there was a preponderance of proximal tumors in African Americans and that the proximal tumor site is an independent prognostic indicator for this group. Relating to the differences in the distribution of adenocarcinomas within the colorectum in these two racial groups, there are various hypotheses relating to differences in diet, alcohol consumption, hormone status, socioeconomic status, and physical activity (51-53). It is likely that there are differences throughout the colon/rectum regarding sensitivity to dietary carcinogen exposure. Indeed, in experimental studies in vivo, a diet high in fat increased the incidence of tumor development in the proximal colon compared with the distal colon (54). A subsequent study showed that the difference in the incidence of tumors between the proximal and distal colon was attributable to the greater capacity of the distal colon to cope with initial damage to DNA caused by carcinogens (55). These findings suggest the importance of studies focused on understanding the underlying mechanisms involved in tumor development in relation to anatomic tumor site and the consideration of tumor location in assessing the aggressiveness and the outcome of patients based on race/ethnicity.
Although the differences in colorectal cancer incidence and survival between Caucasian and African American patients might be due to contributions of various factors, including dietary habits, physical activities, access to screening/health care, or distinct genetic or molecular features, there are no established biological explanations for these differences. The differences in frequency or in mutational spectra at different structural and functional domains of oncogenes/tumor suppressor genes or differences in incidence of genetic or epigenetic events among African American and Caucasian patients suggest that genetic susceptibility, in terms of allelic loss, methylation, and mutagenic events or environmental exposure, could be different. Alternatively, genetic traits that protect against mutations in vital genes in different pathways could be present. Indeed, in endometrial cancers, the incidence of p53 overexpression as a result of point mutations is higher in African American than in Caucasian patients, suggesting that alterations in p53 might be one of the key contributing factors to the racial disparity in disease outcome (56). Further, such alterations, including the codon 72 polymorphism of p53 as observed in this study, might be helpful in understanding the biological differences related to tumor aggressiveness between races. Other factors may, of course, contribute to the racial disparity in patient clinical outcome.
More than 85% of known, cancer-related p53 mutations are missense mutations. For several cancers, including colorectal cancers, mutations of p53 and overexpression of mutant p53 protein (nuclear accumulation) are associated with advanced tumor stage and poor survival (57, 58). Missense mutations occurring in vivo lead to single amino acid substitutions, which result in the disruption of p53 conformation and in loss of function (59). The consequences of these changes might contribute to tumor progression and to a poor prognosis. In agreement with these results, there was, in the present study, no racial disparity in the missense mutation frequency or prognostic importance of p53 mutations in African American and Caucasian patients; in both racial groups, p53 missense mutations were associated with poor patient survival.
Several single-nucleotide polymorphisms have been identified in the coding region of the p53 gene. Because of the silent nature of codons 34 and 36 in exon 4, and codon 213 in exon 6 polymorphisms of p53, their functional characteristics were not studied. However, an additional single-nucleotide polymorphism at codon 47 in exon 4 is a rare germ-line polymorphism that results in an amino acid change, proline > serine (16). The functional characterization studies show that the mutant p53Ser47 phenotype has decreased capacity to induce apoptosis; to transactivate two p53 target genes, p53AIP1 and PUMA; and to bind the MAPK1 protein as compared with the wild-type p53Pro47 phenotype (60). Additionally, a recent report indicated that the p53Pro47Ser polymorphism was neither involved in susceptibility to developing malignancy (glioma) nor helpful in predicting patient survival (61). Nevertheless, future studies on larger populations of different ethnic groups are essential to determine a definitive role of the p53Pro47Ser polymorphism, alone or combination with other alterations within the p53 gene, in tumor development and clinical outcomes.
In addition to missense mutations and other known polymorphisms within the coding region of the p53 gene, polymorphism at codon 72 located in a proline-rich region (residues 64-92) of the p53 protein are functionally important in the growth suppression and apoptosis mediated by p53 but not for cell cycle arrest (30). Indeed, the Arg72 form of p53 has 15-fold enhanced capacity to induce apoptosis, compared with Pro72, and this is due to more efficient localization of the Arg72 form to mitochondria than the Pro72 form (28). Moreover, several earlier studies have reported a significant association between Pro72 and the risk of developing colorectal cancers (31, 62). Other studies have noted different biological and biochemical activity between the p53 mutant protein containing arginine at codon 72 and mutant p53 protein containing proline at this position (63). However, a recent study with lung cancer has shown that polymorphism at codon 72 (CGC to CCC; substitution of an arginine residue with a proline residue) is a risk factor for lung cancer development and that tumors with the Pro/Pro phenotype have an increased incidence of missense point mutations in the p53 gene, indicating a poor prognosis (33). Consistent with these studies, our results show a higher incidence of p53 mutations associated with the Pro/Pro phenotype. This phenotype correlates with aggressive tumor behavior and a poor prognosis in colorectal cancers compared with the Arg/Arg or Arg/Pro phenotype, specifically in African American patients. As outlined above, this study shows that differences in p53 mutation rates and differential p53 mutational spectra in relation to important structural elements (h sheets and helix motifs) and functional domains (L2, L3, and LSH) relate to linkage disequilibrium among codon 72 phenotypes. Such disequilibrium could be a basis for differences among Arg and Pro alleles in relation to tumor aggressiveness and poor patient survival.
DNA polymorphisms and their incidences, susceptibility for occurrence of genetic alterations, and the risk of tumor progression for patients with cancer can vary substantially between different racial groups (30, 64). Although most polymorphisms are functionally neutral, some affect regulation of gene expression or the function of the coded protein. These functional polymorphisms, including the codon 72 polymorphisms of p53, despite being of low occurrence, could contribute to the differences between individuals or races in susceptibility and severity of disease (30, 64). Thus, it is important to determine if common structural polymorphisms are in linkage disequilibrium with genetic (mutations and allele loss) and epigenetic events (methylation and acetylation) and their differential profiles between races. Indeed, the effects of these genetic alterations, alone or in combination, or through interaction with environmental factors, have been implicated in angiogenic pathways and in lung cancer susceptibility and/or severity of disease in African Americans and Caucasians (30, 64). In another study, a higher prevalence of the Pro/Pro phenotype of p53 and its association with high risk of developing lung cancer in African American patients was reported, suggesting that there is substantial interindividual variation in susceptibility to genetic events and to tumor development (30). It is evident from our data that an increased rate of p53 mutations, specifically disruptive mutations or mutations of outside evolutionary conserved regions, and decreased survival were noted among African American patients who exhibit the Pro/Pro phenotype. These findings suggest that the Pro/Pro phenotype of p53 is a race-specific molecular prognostic marker for African American patients with colorectal cancer.
The findings of this study also have implications in the interpretation of molecular variability in colorectal cancer in relation to patient race/ethnicity. An earlier colorectal cancer study in a German patient population, which included all stages of tumor, reported a preferential loss of the Pro72 allele and an increased frequency of missense mutations of p53 in the retained Arg72 allele both in primary as well as liver metastatic lesions, and correlated with aggressive tumor behavior (65). A similar increase in Arg72 status of p53 with advanced disease was reported in urinary tract cancers (transitional cell carcinoma) in a Japanese patient population (66). These as well as several other population genetics studies suggest that race/ethnicity categories aid in identifying unique germline alleles and that allelic combinations can modify both cancer risk and/or progression in some molecular subsets of human malignancies. Furthermore, as the lifestyles and/or environmental exposures (e.g., dietary carcinogens or exposure to infections) vary according to race/ethnicity (51-53), studies based on race/ethnicity could be useful for understanding how differences among different populations affect the pathobiology of colorectal cancer.
Despite the increased interest of oncology medical practitioners to use molecular biomarkers in predicting the aggressiveness of colorectal cancer and clinical outcomes, markers that are indicative of prognosis are still lacking. The significant correlations we report in this article between germline alleles of p53 leading to Pro/Pro phenotypes and increased incidence of p53 mutations, advanced tumor stage, and short survival are clear indications that the codon 72 polymorphism of the p53 gene is involved in colorectal cancer progression specifically in African American patients. Although these correlations need to be validated in future large prospective studies, our findings suggest that together with other confounding factors of disease progression, analysis of the codon 72 polymorphism of the p53 gene might aid in understanding racial differences in determining aggressiveness of colorectal cancer and in designing optimal treatment regimens.
Translational Relevance.
Understanding molecular defects and information on differences in colorectal cancer incidence, aggressiveness, and clinical outcomes in relation to different race/ethnic groups is important in the individualization of treatment and the elimination of race/ethnic disparities. However, race/ethnicity-specific markers indicative of predicting the aggressiveness of colorectal cancer and clinical outcomes are lacking. Therefore, for the first time, the current study has analyzed the prognostic value of the codon 72 polymorphism of p53 in African American and Caucasian patients with colorectal cancer separately, and suggested that colorectal cancers with Pro/Pro mutant phenotypes exhibited a higher incidence of p53 mutations and were associated with nodal metastasis, and short overall survival of African Americans only. These findings suggest that togetherwith other confounding factors of disease aggressiveness, analysis of the codon 72 polymorphism of the p53 gene might aid in understanding racial differences in aggressive progression and clinical outcomes of colorectal cancer, and suggest considering race/ethnicity in the design of treatment regimens.
Acknowledgments
Also, we thank Donald L. Hill, Ph.D., Department of Preventive Medicine, University of Alabama at Birmingham, for his critical review of this manuscript.
Grant support: NIH/National Cancer Institute (RO1-CA98932-01 and U54-CA118948) to Dr. U. Manne.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest with any of the material submitted in this article.
The amino acids database published in Frontiers in Biosciences (http://www.bioscience.org/urllists/aminacid.htm).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Mandelblatt J, Andrews H, Kao R, Wallace R, Kerner J. The late-stage diagnosis of colorectal cancer: demographic and socioeconomic factors. Am J Public Health. 1996;86:1794–7. doi: 10.2105/ajph.86.12.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcella S, Miller JE. Racial differences in colorectal cancer mortality. The importance of stage and socioeconomic status. J Clin Epidemiol. 2001;54:359–66. doi: 10.1016/s0895-4356(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 4.Alexander D, Jhala N, Chatla C, et al. High-grade tumor differentiation is an indicator of poor prognosis in African Americans with colonic adenocarcinomas. Cancer. 2005;103:2163–70. doi: 10.1002/cncr.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polite BN, Dignam JJ, Olopade OI. Colorectal cancer model of health disparities: understanding mortality differences in minority populations. J Clin Oncol. 2006;24:2179–87. doi: 10.1200/JCO.2005.05.4775. [DOI] [PubMed] [Google Scholar]
- 6.Alexander DD, Waterbor J, Hughes T, Funkhouser E, Grizzle W, Manne U. African-American and Caucasian disparities in colorectal cancer mortality and survival by data source: an epidemiologic review. Cancer Biomark. 2007;3:301–13. doi: 10.3233/cbm-2007-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- 8.Powell B, Soong R, Iacopetta B, Seshadri R, Smith DR. Prognostic significance of mutations to different structural and functional regions of the p53 gene in breast cancer. Clin Cancer Res. 2000;6:443–51. [PubMed] [Google Scholar]
- 9.Goh HS, Chan CS, Khine K, Smith DR. p53 and behaviour of colorectal cancer. Lancet. 1994;344:233–4. doi: 10.1016/s0140-6736(94)93000-7. [DOI] [PubMed] [Google Scholar]
- 10.Aas T, Borresen AL, Geisler S, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med. 1996;2:811–4. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 11.Kressner U, Inganas M, Byding S, et al. Prognostic value of p53 genetic changes in colorectal cancer. J Clin Oncol. 1999;17:593–9. doi: 10.1200/JCO.1999.17.2.593. [DOI] [PubMed] [Google Scholar]
- 12.Skaug V, Ryberg D, Kure EH, et al. p53 mutations in defined structural and functional domains are related to poor clinical outcome in non-small cell lung cancer patients. Clin Cancer Res. 2000;6:1031–7. [PubMed] [Google Scholar]
- 13.Borresen AL, Andersen TI, Eyfjord JE, et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995;14:71–5. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- 14.Erber R, Conradt C, Homann N, et al. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene. 1998;16:1671–9. doi: 10.1038/sj.onc.1201690. [DOI] [PubMed] [Google Scholar]
- 15.Vos M, Adams CH, Victor TC, van Helden PD. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of theTP53 gene in a population at high risk for esophageal cancer in South Africa. Cancer Genet Cytogenet. 2003;140:23–30. doi: 10.1016/s0165-4608(02)00638-6. [DOI] [PubMed] [Google Scholar]
- 16.Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet. 1993;53:752–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–3. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbone D, Chiba I, Mitsudomi T. Polymorphism at codon 213 within the p53 gene. Oncogene. 1991;6:1691–2. [PubMed] [Google Scholar]
- 19.Buchman VL, Chumakov PM, Ninkina NN, Samarina OP, Georgiev GP. A variation in the structure of the protein-coding region of the human p53 gene. Gene. 1988;70:245–52. doi: 10.1016/0378-1119(88)90196-5. [DOI] [PubMed] [Google Scholar]
- 20.Futreal PA, Barrett JC, Wiseman RW. An Alu polymorphism intragenic to theTP53 gene. Nucleic Acids Res. 1991;19:6977. doi: 10.1093/nar/19.24.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliva MR, Saez GT, Latres E, Cordon-Cardo C. A new polymorphic site in intron 2 ofTP53 characterizes LOH in human tumors by PCR-SSCP. Diagn Mol Pathol. 1995;4:54–8. doi: 10.1097/00019606-199503000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Lazar V, Hazard F, Bertin F, Janin N, Bellet D, Bressac B. Simple sequence repeat polymorphism within the p53 gene. Oncogene. 1993;8:1703–5. [PubMed] [Google Scholar]
- 23.Chumakov PM, Jenkins JR. BstNI/NciI polymorphism of the human p53 gene (TP53) Nucleic Acids Res. 1991;19:6969. doi: 10.1093/nar/19.24.6969-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniel T, Carbone D, Takahashi T, et al. The MspI polymorphism in intron 6 of p53 (TP53) detected by digestion of PCR products. Nucleic Acids Res. 1991;19:4796. doi: 10.1093/nar/19.17.4796-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser J, Condie A. Biallelic ApaI polymorphism of the human p53 gene (TP53) Nucleic Acids Res. 1991;19:4799. doi: 10.1093/nar/19.17.4799-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf J, Merk B, Maurer U, Muller E, Bergmann L. Identification of novel polymorphisms in intron 7 of the human p53 gene in acute myeloid leukemia and healthy donors. Leuk Lymphoma. 2001;41:655–8. doi: 10.3109/10428190109060356. [DOI] [PubMed] [Google Scholar]
- 27.Graziani D, Romagnoli S, Cassani B, Alfano RM, Roncalli M, Coggi G. An Ava I polymorphism in the TP53 gene. Mol Cell Probes. 1999;13:393–5. doi: 10.1006/mcpr.1999.0256. [DOI] [PubMed] [Google Scholar]
- 28.Dumont P, Leu JI, Della Pietra AC, III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 29.Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–9. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Zhao H, Amos CI, et al. p53 genotypes and haplotypes associated with lung cancer susceptibility and ethnicity. J Natl Cancer Inst. 2002;94:681–90. doi: 10.1093/jnci/94.9.681. [DOI] [PubMed] [Google Scholar]
- 31.Lung FW, Lee TM, Shu BC, Chang FH. p53 codon 72 polymorphism and susceptibility malignancy of colorectal cancer in Taiwan. J Cancer Res Clin Oncol. 2004;130:728–32. doi: 10.1007/s00432-004-0605-4. [DOI] [PubMed] [Google Scholar]
- 32.Langerod A, Bukholm IR, Bregard A, et al. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev. 2002;11:1684–8. [PubMed] [Google Scholar]
- 33.HuY McDermott, MP Ahrendt, SA The. p53 codon 72 proline allele is associated with p53 gene mutations in non-small cell lung cancer. Clin Cancer Res. 2005;11:2502–9. doi: 10.1158/1078-0432.CCR-04-1913. [DOI] [PubMed] [Google Scholar]
- 34.Shao Y, Tan W, Zhang S. p53 gene codon 72 polymorphism and risk of esophageal squamous cell carcinoma: a case/control study in a Chinese population. Dis Esophagus. 2008;21:139–43. doi: 10.1111/j.1442-2050.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang YC, Lee HS, Chen SK, Chang YY, Chen CY. Prognostic significance of p53 codon 72 polymorphism in lung carcinomas. Eur J Cancer. 1999;35:226–30. doi: 10.1016/s0959-8049(98)00369-4. [DOI] [PubMed] [Google Scholar]
- 36.Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–94. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 37.Green FL, Page DL, Fleming ID, et al. American Joint Committee on Cancer . Cancer staging handbook from the AJCC cancer staging manual. 6th ed. Springer-Verlag; New York: 2006. [Google Scholar]
- 38.WHO . International classification of diseases for oncology. World Health Organization; Geneva: 1990. [Google Scholar]
- 39.Fredricks DN, Relman DA. Paraffin removal from tissue sections for digestion and PCR analysis. Biotechniques. 1999;26:198–200. doi: 10.2144/99262bm04. [DOI] [PubMed] [Google Scholar]
- 40.Friend S. p53: a glimpse at the puppet behind the shadow play. Science. 1994;265:334–5. doi: 10.1126/science.8023155. [DOI] [PubMed] [Google Scholar]
- 41.Brooks LA, Tidy JA, Gusterson B, et al. Preferential retention of codon 72 arginine p53 in squamous cell carcinomas of the vulva occurs in cancers positive and negative for human papillomavirus. Cancer Res. 2000;60:6875–7. [PubMed] [Google Scholar]
- 42.Fleiss J. Statistical methods for rates and proportions. JohnWiley and Sons; New York (NY): 1981. [Google Scholar]
- 43.Allison P. Survival analysis using the SAS System: a practical guide. SAS Institute Inc; Cary (NC): 1995. [Google Scholar]
- 44.Kaplan E, Meier P. Non-parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 45.Cox DR. Regression models and life tables. J Roy Stat Soc. 1972;34:187–220. [Google Scholar]
- 46.Mooney CZ, Duval RD. Bootstrapping: a non-parametric approach to statistical inference. Sage; Newbury Park (CA): 1993. [Google Scholar]
- 47.Troisi RJ, Freedman AN, Devesa SS. Incidence of colorectal carcinoma in the U S.: an update of trends by gender, race, age, subsite, and stage, 1975-1994. Cancer. 1999;85:1670–6. [PubMed] [Google Scholar]
- 48.Ozick LA, Jacob L, Donelson SS, Agarwal SK, Freeman HP. Distribution of adenomatous polyps in African-Americans. Am J Gastroenterol. 1995;90:758–60. [PubMed] [Google Scholar]
- 49.Alexander D, Chatla C, Funkhouser E, Meleth S, Grizzle WE, Manne U. Postsurgical disparity in survival between African Americans and Caucasians with colonic adenocarcinoma. Cancer. 2004;101:66–76. doi: 10.1002/cncr.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayberry RM, Coates RJ, Hill HA, et al. Determinants of black/white differences in colon cancer survival. J Natl Cancer Inst. 1995;87:1686–93. doi: 10.1093/jnci/87.22.1686. [DOI] [PubMed] [Google Scholar]
- 51.Devesa SS, Chow WH. Variation in colorectal cancer incidence in the United States by subsite of origin. Cancer. 1993;71:3819–26. doi: 10.1002/1097-0142(19930615)71:12<3819::aid-cncr2820711206>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 52.Gapstur SM, Potter JD, Folsom AR. Alcohol consumption and colon and rectal cancer in post-menopausal women. Int J Epidemiol. 1994;23:50–7. doi: 10.1093/ije/23.1.50. [DOI] [PubMed] [Google Scholar]
- 53.Slattery ML, Schaffer D, Edwards SL, Ma KN, Potter JD. Are dietary factors involved in DNA methylation associated with colon cancer? Nutr Cancer. 1997;28:52–62. doi: 10.1080/01635589709514553. [DOI] [PubMed] [Google Scholar]
- 54.Holt PR, Mokuolu AO, Distler P, Liu T, Reddy BS. Regional distribution of carcinogen-induced colonic neoplasia in the rat. Nutr Cancer. 1996;25:129–35. doi: 10.1080/01635589609514435. [DOI] [PubMed] [Google Scholar]
- 55.Hong MY, Chapkin RS, Morris JS, et al. Anatomical site-specific response to DNA damage is related to later tumor development in the rat azoxymethane colon carcinogenesis model. Carcinogenesis. 2001;22:1831–5. doi: 10.1093/carcin/22.11.1831. [DOI] [PubMed] [Google Scholar]
- 56.Maxwell GL, Risinger JI, Hayes KA, et al. Racial disparity in the frequency of PTEN mutations, but not microsatellite instability, in advanced endometrial cancers. Clin Cancer Res. 2000;6:2999–3005. [PubMed] [Google Scholar]
- 57.Hamelin R, Laurent-Puig P, Olschwang S, et al. Association of p53 mutations with short survival in colorectal cancer. Gastroenterology. 1994;106:42–8. doi: 10.1016/s0016-5085(94)94217-x. [DOI] [PubMed] [Google Scholar]
- 58.Manne U, Myers RB, Moron C, et al. Prognostic significance of Bcl-2 expression and p53 nuclear accumulation in colorectal adenocarcinoma. Int J Cancer. 1997;74:346–58. doi: 10.1002/(sici)1097-0215(19970620)74:3<346::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 59.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–55. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280:24245–51. doi: 10.1074/jbc.M414637200. [DOI] [PubMed] [Google Scholar]
- 61.Pinto GR, Yoshioka FK, Silva RL, et al. Prognostic value of TP53 Pro47Ser and Arg72Pro single nucleotide polymorphisms and the susceptibility to gliomas in individuals from Southeast Brazil. Genet Mol Res. 2008;7:207–16. doi: 10.4238/vol7-1gmr415. [DOI] [PubMed] [Google Scholar]
- 62.Sjalander A, Birgander R, Athlin L, et al. P53 germ line haplotypes associated with increased risk for colorectal cancer. Carcinogenesis. 1995;16:1461–4. doi: 10.1093/carcin/16.7.1461. [DOI] [PubMed] [Google Scholar]
- 63.Marin MC, Jost CA, Brooks LA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 64.Jin X, Wu X, Roth JA, et al. Higher lung cancer risk for younger African-Americans with the Pro/Pro p53 genotype. Carcinogenesis. 1995;16:2205–8. doi: 10.1093/carcin/16.9.2205. [DOI] [PubMed] [Google Scholar]
- 65.Schneider-Stock R, Boltze C, Peters B, et al. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia. 2004;6:529–35. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furihata M, Takeuchi T, Matsumoto M, et al. p53 mutation arising in Arg72 allele in the tumorigenesis and development of carcinoma of the urinary tract. Clin Cancer Res. 2002;8:1192–5. [PubMed] [Google Scholar]