Summary
Establishment of germline sexual identity is critical for production of male and female germline stem cells, and sperm vs. eggs. Here we identify PHD Finger Protein 7 (PHF7) as an important factor for male germline sexual identity in Drosophila. PHF7 exhibits male-specific expression in early germ cells, germline stem cells and spermatogonia. It is required for germline stem cell maintenance and gametogenesis in males, whereas ectopic expression in female germ cells ablates the germline. Strikingly, expression of PHF7 promotes spermatogenesis in XX germ cells when they are present in a male soma. PHF7 homologs are also specifically expressed in the mammalian testis, and human PHF7 rescues Drosophila Phf7 mutants. PHF7 associates with chromatin and both the human and fly proteins bind histone H3 N-terminal tails with a preference for dimethyl lysine 4 (H3K4me2). We propose that PHF7 acts as a conserved epigenetic “reader” that activates the male germline sexual program.
Introduction
Sex determination is key to sexual reproduction, and both somatic cells and germ cells need to establish sex-specific developmental fates. Germline sexual development is essential for the production of two distinct gametes, and underlies important differences in the regulation of male vs. female fertility. In some species, germline stem cells are present in both males and females to sustain constant gamete production, but are regulated differently throughout development. In other species such as humans, sex-specific germ cell development produces a germline stem cell population only in males, while females have a much more limited capacity in making eggs. Defects in germline sexual development lead to a failure in gametogenesis, thus the study of germline sex determination is essential for understanding normal reproductive potential and treating infertility.
In some animals, such as mammals and Drosophila, the sex chromosome compositions of the soma and germline are interpreted independently, and the “sex” of the germline must match that of the soma for proper germ cell development to occur. For example, patients with Klinefelter’s Syndrome have an XXY sex chromosome constitution and are almost always infertile (Jacobs and Strong, 1959; Klinefelter et al., 1942). These individuals develop somatically as males due to the presence of a Y chromosome but the germline suffers from severe atrophy, including the loss of pre-meiotic germline and germline stem cells (Wikstrom and Dunkel, 2008). This is due to the presence of two X chromosomes in the germ cells, as the limited spermatogenesis in these patients is from germ cells that have lost one of the X chromosomes (Bergere et al., 2002; Sciurano et al., 2009). In Drosophila, XX females that are somatically transformed into males exhibit a similar germline loss due to a conflict in sexual identity between the masculinized soma and XX germline (Nothiger et al., 1989). Thus, fruitflies are a valuable model organism for studying how germ cells establish a proper sexual identity by coordinating intrinsic signals and those coming from the soma.
In Drosophila, the presence of two X chromosomes promotes female somatic identity by activating an alternative splicing cascade that acts through Sex lethal (SXL) and Transformer (TRA), and ultimately leads to production of either the male or female forms of the transcription factors Doublesex (DSX) and Fruitless (FRU) (reviewed in Camara et al., 2008; Siwicki and Kravitz, 2009). DSX and FRU are responsible for virtually all sexually dimorphic somatic traits in Drosophila, with DSX being the key factor in the somatic gonad. In contrast, the germline does not determine its sex with this cascade and factors like TRA and DSX are not required in germ cells (Marsh and Wieschaus, 1978; Schupbach, 1982). While SXL is required to promote female germ cell identity, its targets and mechanism of action in the germline are not known (Hashiyama et al., 2011; Nothiger et al., 1989; Schupbach, 1985; Steinmann-Zwicky et al., 1989). The transcription factor OVO and the ubiquitin protease Ovarian Tumor (OTU) are also required in the female germline and thought to function upstream of SXL (Oliver et al., 1990; Pauli et al., 1993). Even less is known about how sexual identity is specified in male germ cells. Male germ cells receive a signal through the JAK/STAT pathway that promotes their sexual identity (Wawersik et al., 2005), but the downstream factors that are subsequently activated are not known. Similarly, how male germ cells respond to their own sex chromosome constitution is also not known.
In this study, we report a histone code reader, Plant Homeodomain (PHD) Finger 7 (PHF7), that acts in the Drosophila germline to promote male sexual identity. PHF7 is specifically expressed in male germ cells from early stages of development and is restricted to male germline stem cells (GSCs) and spermatogonia. Phf7 is required for GSC maintenance and proper entry into spermatogenesis. Interestingly, expression of Phf7 in female germ cells causes ablation of the female germline. Moreover, Phf7 affects sexual compatibility between germline and soma. Loss of Phf7 in XY germ cells alleviates the germline loss typically observed when XY germ cells are surrounded by a female soma, and expression of Phf7 can induce spermatogenesis in XX germ cells nurtured by male soma. These findings indicate that Phf7 is an essential factor in determining sexual development in the Drosophila germline, and suggest that activation of the male identity occurs through interaction with the germline epigenome.
Results
Phf7 expression is specific to the male germline
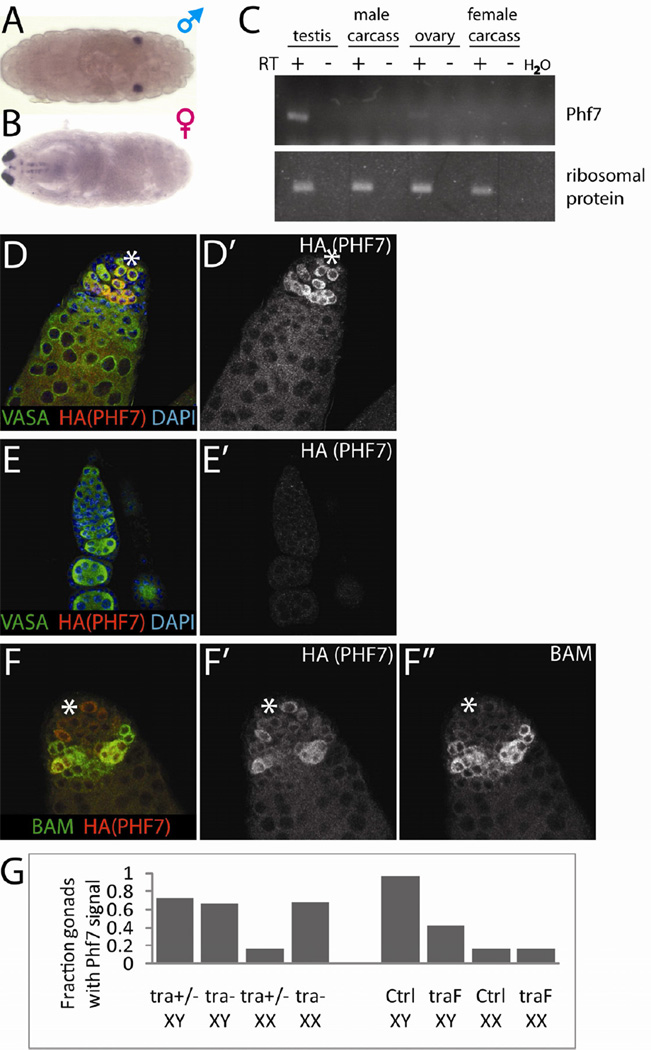
In an in situ screen for genes differentially expressed between the male and female embryonic gonad (Casper and Van Doren, 2009), we identified CG9576 as a male-specific gene (Figure 1A, B). Due to the extensive homology and likely orthology between CG9576 and mammalian Phf7 (see below), we will refer to the Drosophila gene as Phf7. RT-PCR on adult tissues indicated that Phf7 expression is exclusive to the gonads and strongly biased towards the testis (Figure 1C). To analyze expression at the protein level, we used recombineering to generate epitope-tagged Phf7 transgenes within the context of a large region of the genomic DNA surrounding the Phf7 locus. These transgenes fully rescue Phf7 loss-of-function mutants (Figure S1A, S3B). Western blot analyses of adult tissues confirmed that PHF7 expression is gonad-specific and strongly male-biased (Figure S1B). Immunofluorescence analyses of adult ovaries indicated that expression was below the limit of detection. In contrast, the testes exhibited substantial PHF7 expression in GSCs and spermatogonia (Figure 1D, E, Figure S1C, D). The peak of expression was seen in 4–8-cell spermatogonial cysts and declined sharply afterwards, as indicated by co-localization with Bag of Marbles (BAM), which is known to be present in 4–8 cell cysts (Figure 1F, McKearin and Ohlstein, 1995). The restriction of Phf7 expression to undifferentiated male germline suggests a possible role in early development of the male germline lineage.
Figure 1.
Expression profiles of Phf7. A, B) in situ hybridization of stage 17 male and female embryos for Phf7. The signal at the anterior (left) of the female embryo comes from hybridization of the Dfd-lacZ transgene used for sexing. C) RT-PCR of Phf7 in adult tissues. Carcass refers to whole adult flies with gonads removed. Ribosomal protein RpLP0 was used as internal control. D–F) Immunofluorescence of the C-terminally HA-tagged PHF7 BAC transgene, co-labeled as indicated on the panels. The ' panels display the PHF7 channel alone. Anti-VASA labels the germline and DAPI labels the DNA. D, D') Adult testis. E, E') Adult ovary. F, F', F") Colocalization of BAM with HA-tagged PHF7 in adult testis. The hub is indicated with a white asterisk. F" displays only the BAM channel. G) Fractions of stage 17 embryonic gonads expressing Phf7 by in situ hybridization of stage 17 mutant embryonic gonads. Genotypes are as indicated. Note that Phf7 is induced in XX germ cells in a male soma (tra-) and repressed in XY germ cells in a female soma (traF). See also Figure S1.
There are two known contributors to male germline gene expression: the signals from the somatic gonad and the germ cell's own sex chromosome constitution (Casper and Van Doren, 2009). To test if Phf7 expression is induced by the male soma, we asked if XX germ cells surrounded by a male somatic environment would upregulate Phf7 expression by examining masculinized (tra mutant) XX gonads. By in situ hybridization, such XX germline was found to express Phf7 at levels similar to control XY germline (Figure 1G). Conversely, we tested if XY germ cells surrounded by female soma can still express Phf7 by feminizing the somatic gonad of XY embryos through general expression of traF (U2AF-traF, Evans and Cline, 2007). We observed that Phf7 expression in XY germ cells is repressed when in contact with female soma, but the expression level is still higher than XX germ cells in contact with female soma (Figure 1G), suggesting that XY germ cells are able to upregulate Phf7 expression intrinsically. Taken together, these results indicate that Phf7 expression is regulated both extrinsically by the somatic gonad and intrinsically, by the germline sex chromosome constitution.
Phf7 is important for fertility in males but not females
To elucidate the role of Phf7 in gametogenesis, we generated loss of function alleles of Phf7 by P-element excision of the EP line EY03023. Three deletion mutants were obtained: ΔN2, ΔN18, and Δ88 (Figure S3A). In ΔN2 and ΔN18, the N-terminal portions of the Phf7 ORF have been deleted and are likely null alleles. Phf7 is completely removed in Δ88, but part of the downstream gene Rab35 is also deleted. Both males and females mutant for these Phf7 alleles are adult viable.
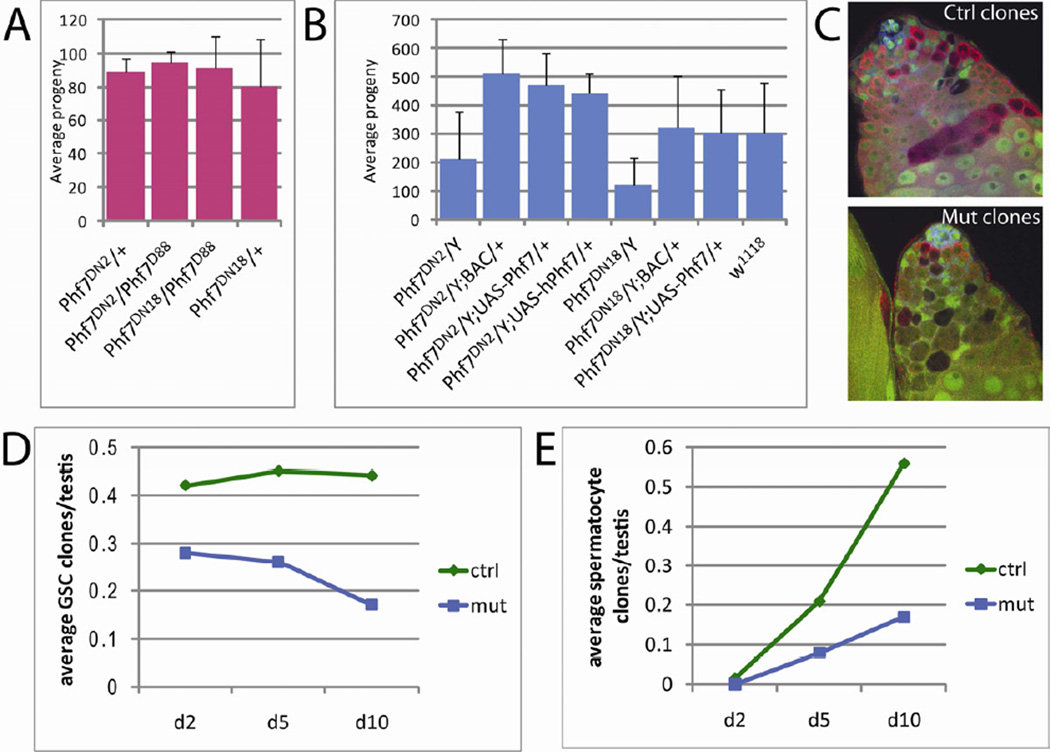
Fecundity tests were performed to test if loss of Phf7 affected spermatogenesis or oogenesis. We found that mutant females (Phf7ΔN2/Δ88 and Phf7ΔN18/Δ88) retained full fecundity, indicating that Phf7 does not contribute to oogenesis (Figure 2A). In contrast, males mutant for Phf7 (Phf7ΔN2/Y and Phf7ΔN18/Y) showed substantially impaired fecundity compared to genotype-matched controls (Phf7ΔN2/Y; BAC/+ and Phf7ΔN18/Y; BAC/+, Figure 2B). The defect in male fecundity can also be restored with the basal activity of a UAS-Phf7 cDNA construct, confirming that effects on fecundity in mutant males were indeed due to the loss of Phf7 (Figure 2B). There are single, clear homologs for Phf7 in the mouse and human genomes, and expression of these genes appears to be strongly biased toward the testis (Figure S2). Importantly, human PHF7 was able to rescue the fecundity defect in Phf7 mutants, indicating that the human and Drosophila PHF7 proteins are functionally orthologous (Figure 2B).
Figure 2.
Loss of Phf7 causes male specific developmental defects in the germline. A) Average fecundity of single female flies mutant for Phf7 compared to heterozygous controls. B) Average fecundity of single male flies mutant for Phf7 compared to mutant males rescued with either the BAC, UAS-Phf7 transgene, or UAS-human PHF7 (hPhf7) construct. Error bars in A and B represent standard deviations. C) Representative images of control or Phf7-mutant genetic mosaic clones at 10 days post clone induction. Clones are indicated by loss of GFP (green); VASA (germline, red); N-CADHERIN (hub, blue). Genotypes are as indicated in Experimental Procedures. D) Average numbers of GSC control (green diamonds) and mutant (blue squares) clones plotted for 2, 5, and 10 days after clonal induction. E) Average numbers of control (green diamonds) or mutant (blue squares) spermatocyte clones at 2, 5, and 10 days after clonal induction. See also Table S1.
Phf7 is required for male germline stem cell and spermatogonial development
The male-specific reduction in fecundity is consistent with the male-biased expression of Phf7, and prompted us to characterize the cellular basis of this defect. GSCs and spermatogonia represent the main cell types that express Phf7, thus we investigated the role of Phf7 in these populations using genetic mosaic clonal analysis. Since Phf7 is located on the X chromosome, it was not possible to utilize the standard approach for mitotic recombination (Golic, 1991). Instead, we used Phf7 mutant males carrying a Phf7 BAC rescue construct on a marked chromosome that could be eliminated by mitotic recombination (Experimental Procedures). Upon expression of FLP recombinase, clones of unmarked cells are generated that have lost the rescuing BAC and are therefore mutant for Phf7 (Figure 2C).
Using this technique, the maintenance of mutant germline clones over time was examined. We found that the number of Phf7-mutant GSCs declined steadily over time whereas those of control genotype were maintained (Figure 2D, Table S1), indicating that Phf7 is essential for GSC maintenance. The numbers of mutant vs. control spermatocyte clones were also recorded to monitor the role of Phf7 in spermatogonial development. While some mutant spermatocyte clones were observed, the numbers were substantially lower than those of controls (Figure 2E, Table S1). Since the decrease in Phf7-mutant spermatocytes compared to controls is far greater than the difference in initial GSC clones produced, we conclude that Phf7-mutant germ cells are also defective in spermatogonial development. In summary, mosaic clonal analyses indicate that Phf7 acts germline-autonomously to regulate maintenance of male GSCs and development of spermatogonia.
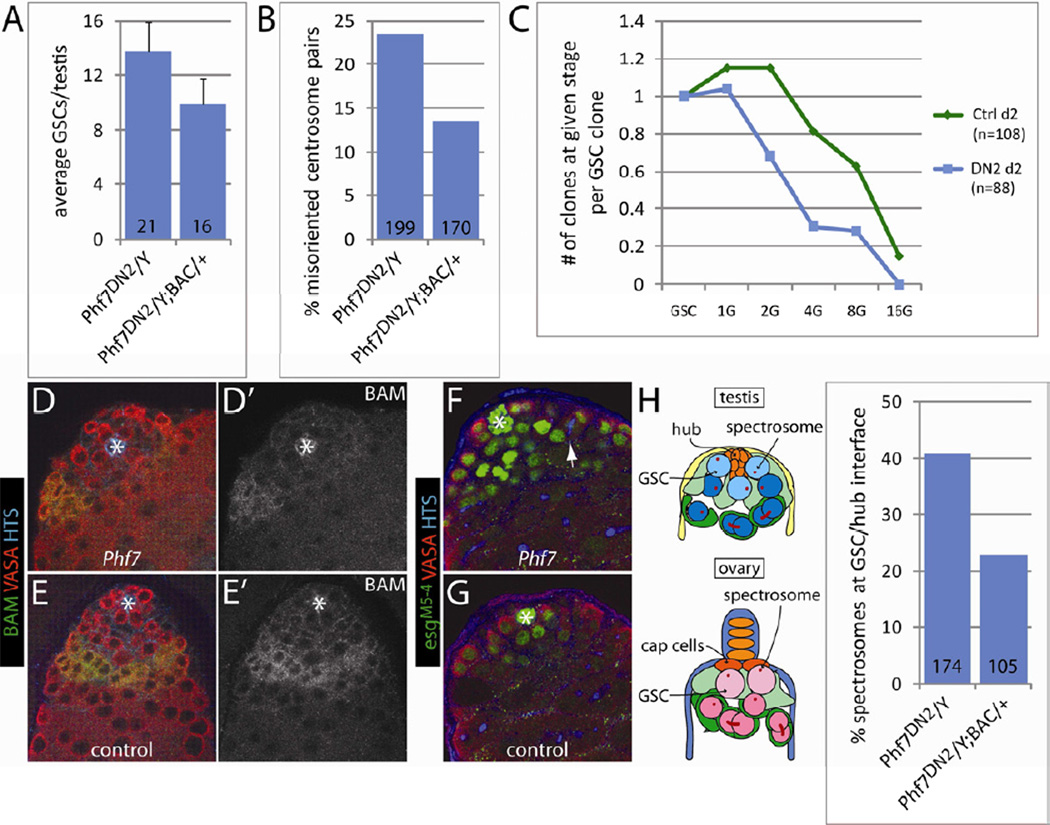
In addition to the genetic mosaic clonal analyses, we also wanted to establish the effects of Phf7 in male GSCs and spermatogonia in the context of whole mutant testes. Interestingly, we did not observe a decrease in GSC number in Phf7 mutants and, in fact, GSC number was moderately elevated (Figure 3A), a trend that persisted even with aging (data not shown). It is intriguing that mutant GSCs are retained when present with other mutant GSCs, but not when they are in competition with wild-type GSCs, as was the case in our mosaic clonal analysis. However, this phenomenon has been observed previously, for example with Stat92E-mutant GSCs, which are lost rapidly if they are in competition with wild-type GSCs (Kiger et al., 2001; Tulina and Matunis, 2001), but are retained when all GSCs are mutant (Leatherman and Dinardo, 2008). Thus, placing GSCs in competition with wild-type GSCs is likely a more sensitive assay for defects in GSC maintenance. Activation of the JAK-STAT pathway was not affected in mutant GSCs (data not shown). Mutant GSCs were also examined for their ability to carry out oriented divisions as is the case in wild-type GSCs (Yamashita et al., 2003). In Phf7 mutants, GSCs were at least as mitotically active as control GSCs (Figure S3C), but their centrosomes were not as reliably oriented toward the hub-GSC interface as in wild-type (Figure 3B), though this did not result in an increase in misoriented GSC divisions (Figure S3D). In contrast, we found that the spectrosomes, a germline-specific organelle, were more likely to be localized to the hub-GSC interface (Figure 3H). Interestingly, the localization of the spectrosomes, but not the centrosomes, to the niche-GSC interface is a characteristic of female GSCs rather than male GSCs (de Cuevas and Spradling, 1998; Lin et al., 1994), indicating that Phf7 mutant GSCs are feminized (see below).
Figure 3.
Profiles of GSCs and spermatogonia in hemizygous Phf7-mutant testes compared to controls. A) Average numbers of GSCs in mutant and BAC-rescued testes. Error bars represent standard deviations. B) Percentages of centrosome pairs in GSCs that did not exhibit hub-proximal orientation in mutant and BAC-rescued testes. C) FLP-mediated lineage analysis of GSCs and developing spermatogonia in hemizygous Phf7-mutant (blue squares) and control testes (green diamonds). Genotypes are as in Experimental Procedures. The number of spermatogonial clones at 1-, 2-, 4-, 8-, and 16-cell stage per GSC clone is plotted at 2 days post clonal induction. D–G) Adult testes are immunolabeled as indicated. D' and E' show the BAM staining alone. Hubs are marked with white asterisks. Note the reduced population of BAM-positive spermatogonial cysts in Phf7-mutant (D, Phf7ΔN2/Y) compared to BAC-rescued testes (E, Phf7ΔN2/Y; BAC/+). F, G) Expanded population of esgM5-4-expressing germline in Phf7-mutant (F, Phf7ΔN2/Y) testes compared to controls (G, Phf7ΔN2/Y; BAC/+). Arrow points to a branched fusome connecting a 4-cell spermatogonial cyst. H) Localization of spectrosomes is altered in Phf7-mutant GSCs compared to BAC-rescued counterparts. Cartoons on the left depict the typical spectrosome localization patterns that are different in male and female GSCs. The percentages of GSCs exhibiting hub-proximal spectrosomes are plotted on the graph to the right for both Phf7-mutants and BAC-rescued samples. For all bar graphs, the numbers at the base of the bars indicate the sample size for each genotype. See also Figure S3.
To better examine the dynamics of spermatogonial development in hemizygous Phf7-mutant vs. control testes, we carried out a lineage tracing experiment. Using standard FLP/FRT techniques, we labeled a fraction of GSCs in either mutant or control testes and tracked the numbers of spermatogonial cysts at various stages at 2 days post labeling. By calculating the number of spermatogonia produced by each labeled GSC, it was possible to determine the developmental capacity of spermatogonia in mutant testes compared to controls. We found that the number of spermatogonia generated per GSC was far lower in mutants than that of controls (Figure 3C), with less than half of the 4-cell and 8-cell cysts produced in Phf7 mutants, indicating that loss of Phf7 impairs the ability of male germline to transit through spermatogonial stages.
To address defects in spermatogonial development, we first examined BAM, a pro-differentiation factor expressed in 4–8 cell germline cysts (Gonczy et al., 1997; McKearin and Spradling, 1990). In Phf7 mutants, we still observe BAM expression in 4–8 cell cysts, but the region of BAM staining is diminished compared to that in controls (Figure 3D), indicating that fewer BAM-positive cysts are present in mutant testes. Another marker examined was esgM5-4, an enhancer trap that is expressed in the hub, GSCs, and gonialblasts. Interestingly, in Phf7 mutants, a larger domain of esgM5-4 expression was observed that frequently persisted until the 4-and occasionally 8-cell stages (Figure 3F). The failure to downregulate esgM5-4 and the decrease in BAM staining are consistent with defects in transitioning from undifferentiated GSC and early spermatogonia to later stages of cyst development and suggest that Phf7 is required for male germline differentiation.
Expression of Phf7 leads to ablation of female germ cells
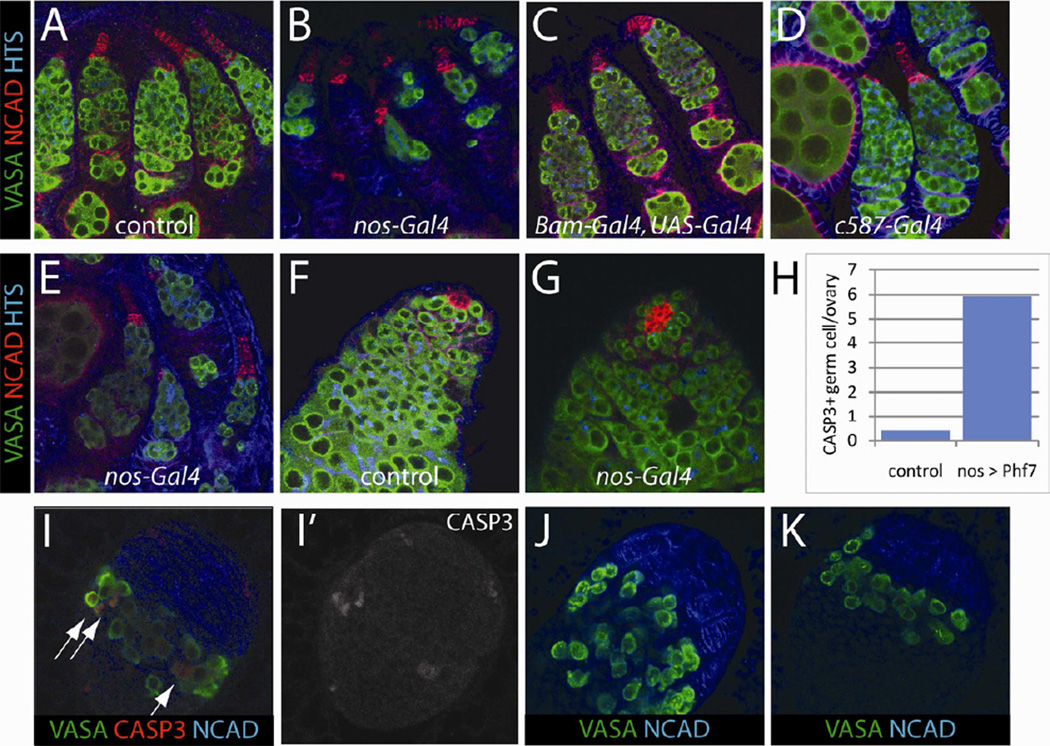
In addition to the male-specific loss of function phentoype, another dramatic sex-specific effect of Phf7 occured when it was expressed in female germ cells. The female germline suffered severe loss when Phf7 was expressed in the germline using nos-Gal4 (VASA, Figure 4B). In newly eclosed adults, a majority of ovarioles exhibited severe germline depletion, and by 5 days after eclosion few ovarioles contained any germline. In contrast, the male germline was unaffected morphologically and those males were fully fertile (VASA, Figure 4G, data not shown). This effect was observed when using either an EP line containing a UAS sequence inserted ~500bp upstream of the transcription start site of Phf7 (EY03023) or a UAS-Phf7 cDNA construct. While both fly lines were able to cause female germline loss when driven with nos-Gal4, the effect with the EP line was stronger, possibly due to higher expression levels (Figure 4B, E). Intriguingly, only undifferentiated female germline was sensitive to Phf7 overexpression. When Phf7 was expressed in differentiating germ cells starting at the 2-cell cyst stage (Bam-Gal4, UAS-Gal4), no germline loss was observed (Figure 4C). Similarly, no effect was observed when Phf7 was expressed in the somatic cells of the gonad (Figure 4D).
Figure 4.
Overexpression of Phf7 induces apoptosis only in the undifferentiated female germline. A–E) Ovaries from freshly eclosed females are immunolabeled as indicated; VASA (green, germline), N-CADHERIN (red, terminal filaments in female, hub in male), HTS (blue, fusome). A, control ovary (nos-Gal4). B, EY03023/w; nos-Gal4/+. C, EY03023/w; UAS-Gal4/+; Bam-Gal4/+. D, EY03023/c587-Gal4. E, UAS-Phf7/nos-Gal4. F, G) Testes from newly eclosed males are immunolabeled as in A-E. F, control testis (nos-gal4). G, EY03023/Y; nos-Gal4/+. H) Average numbers of activated Capase-3 positive germ cells in control (nos-Gal4) and UASPhf7/nos-Gal4 3rd instar ovaries. I–K) 3rd instar larval ovaries are immunolabeled as indicated. I) UAS-Phf7/nos-Gal4 3rd instar ovaries highlighting germline undergoing Capase-3-dependent apoptosis. I' shows the activated Caspase-3 signal alone. J, K) Loss of germline observed in 3rd instar ovaries of control (J, nos-Gal4) and UAS-Phf7/nos-Gal4 (K).
To understand the nature of the germline loss, females expressing Phf7 were examined as third instar larvae when ovarian differentiation normally begins (Godt and Laski, 1995; Zhu and Xie, 2003). We found that significant loss of germline had already occurred at the time the female germline stem cell niche (terminal filaments, NCAD) is forming (Figure 4K). Moreover, female germ cells expressing Phf7 at this stage were positive for activated Caspase-3 at a frequency ~10-fold higher than in controls (Figure 4H, I). Thus the female germline appears to become sensitive to Phf7 expression as germ cells are becoming GSCs, suggesting that PHF7 interferes with an early stage of the female germ cell developmental program. Interestingly, this is the same stage at which male (XY) germ cells begin to atrophy when present in a female soma (unpublished data, S. Murray and M. Van Doren).
Phf7 promotes the male program of germ cell development
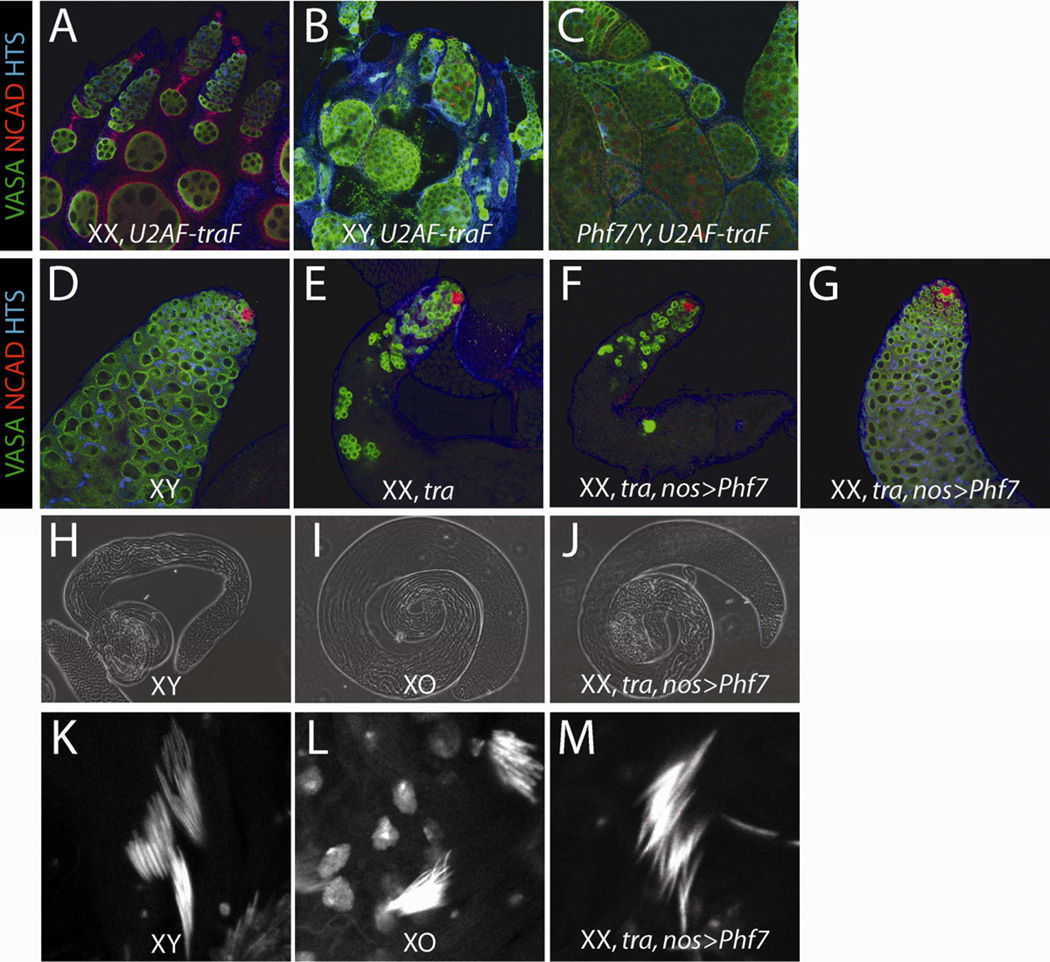
The phenotypes of Phf7 mutant males and PHF7-expressing females suggested that PHF7 is involved in establishing the male germline sexual program. Consistent with this, we observed that Phf7 mutant male GSCs were more likely to have their spectrosomes adjacent to the hub-GSC interface, and their centrosomes displaced from this interface, which is more characteristic of female GSCs. To investigate further if XY germline is feminized in the absence of Phf7, we examined XY flies that have been transformed into females. Normally, an XY germ cell present in a female soma fails to develop properly due to the sexual incompatibility between the germline and the soma (Steinmann-Zwicky et al., 1989; Van Deusen, 1977). We reasoned that if Phf7 promotes a male germline identity, then the loss of Phf7 function from the germ cells in these animals would make the XY germ cells more compatible with a female soma. To feminize the soma, we expressed traF ubiquitously (U2AF-traF) in an otherwise XY animal, which causes the XY germline to degenerate (VASA, Figure 5B). When Phf7 function was removed from these XY females, we observed much improved germline retention and organization in the XY pseudo-ovaries (VASA, Figure 5C), suggesting that loss of Phf7 has indeed shifted XY germ cells towards a partially feminized identity.
Figure 5.
Phf7 controls the male germline sexual program. A–C) Adult ovaries are immunolabeled as indicated. A, control XX ovary (+/+; U2AF-traF/+). B, Feminized XY gonad overexpressing traF (+/Y; U2AF-traF/+). C, Phf7-mutant XY gonad overexpressing traF (Phf7ΔN2/Y; U2AF-traF/+). Note the significant rescue of germ cell number and organization in C compared to B. D–G) Adult testes are immunolabeled as indicated. D, control XY testis (nos-Gal4). E, masculinized (tra-mutant) XX pseudo-testis (+/+; nos-Gal4, tra1/Df(tra)). F and G, Phf7-overexpressing, tra mutant XX pseudo-testis (EY03023/+; nos-Gal4, tra1/Df(tra)). F is an example of an unrescued pseudo-testis, while G is an example of a fully-rescued, wild-type looking pseudo-testis. H–J) Phase contrast images of adult testes. H, XY control (Oregon-R, +/Y). I, XO males (yw/O; st1ncdD/+). J, Phf7-overexpressing, tra mutant XX pseudo-testis (EY03023/+; nos-Gal4, tra1/Df(tra)). K–M) Nuclear condensation of maturing sperm at the base of adult testis coils revealed by DAPI stains. K, XY control (Oregon-R, +/Y). L, XO males (yw/O; ncdD/+). M, Phf7-overexpressing, tra mutant XX pseudo-testis (EY03023/+; nos-Gal4, tra1/Df(tra)). J and M are from examples of Phf7-rescued pseudo-testes. See also Figure S4.
Next, we asked the reciprocal question of whether Phf7 expression can induce male germline development in XX germ cells. To address this, we first tested if the effects of Phf7 expression are synergistic with those thought to masculinize XX germline. Mutations in otu cause tumorous ovaries and, in the strongest cases, germline loss, similar to XY germ cells in a female soma (King et al., 1986). Interestingly, when Phf7 was expressed in the XX germline under conditions that exhibited only partial germline loss (UAS-Phf7, 1 day old adults), being heterozygous for mutations in otu caused a significant enhancement of the germline loss phenotype (Figure S4). This supports the view that Phf7 expression is indeed able to promote male identity in XX germ cells.
We next wanted to determine whether Phf7 expression was sufficient to induce XX germ cells to make sperm, the most stringent test of male germ cell identity. This was done by examining XX flies that have been transformed to a male somatic identity due to mutations in tra. XX germ cells in these animals cannot develop normally and give rise to atrophic gonads (Figure 5E), again because the XX germline is sexually incompatible with a male soma. When Phf7 was overexpressed in these XX germ cells (nos-Gal4, EY03023), a fraction (6%) of the gonads appeared completely rescued and were identical in size and morphology to wild-type testes (Figure 5G, J, Table 1). This rescue was not observed in any of our tra-mutant controls, including the Gal4 driver alone, the UAS element alone, or the Gal4 driver expressing a control construct (UAS-lacZ). In addition, these testes exhibited late-stage sperm, including sperm tails and the condensed chromatin characteristic of differentiated sperm heads (Figure 5M). Since these animals lack a Y chromosome, which contains genes required for sperm function, we could not test these animals for fertility. However, the pseudo-testes and sperm that developed resembled those produced by otherwise normal males that lack a Y chromosome (XO males, Figure 5I, J, L, M). The remaining, unrescued pseudo-testes appeared similar to XX tra-mutant pseudo-testes that lacked Phf7 expression (Figure 5F). The low percentage of rescue may be due to residual female character promoted by the presence of two X chromosomes, the lack of optimal expression pattern generated for Phf7, or the requirement of another male-promoting pathway. Regardless, the fact that Phf7 is able to induce spermatogenesis in XX germ cells is a strong indication that it can promote the male program of germ cell development.
Table 1.
Expression of Phf7 in XX germ cells can induce spermatogenesis with the support of a masculinized somatic environment.
| Genotype | Age (days) |
Rescued pseudo- testes |
Total pseudo- testes |
Percent rescuea (%) |
|---|---|---|---|---|
| +/+; nos-Gal4,tra1/Df(tra) | 1–3 | 0 | 86 | 0 |
| EY03023/+; tra1/Df(tra) | 1–3 | 0 | 210 | 0 |
| EY03023/+; nos-Gal4,tra1/Df(tra) | 1–3 | 10 | 292 | 3.4 |
| +/+; nos-Gal4,tra1/Df(tra) | 5 | 0 | 134 | 0 |
| +/+; nos-Gal4,tra1/UAS-lacZ,Df(tra) | 5 | 0 | 26 | 0 |
| EY03023/+; nos-Gal4,tra1/Df(tra) | 5 | 5 | 84 | 6.0 |
percent rescue is calculated by dividing the number of rescued pseudo-testes by the total number of pseudo-testes analyzed.
PHF7 specifically associates with chromatin
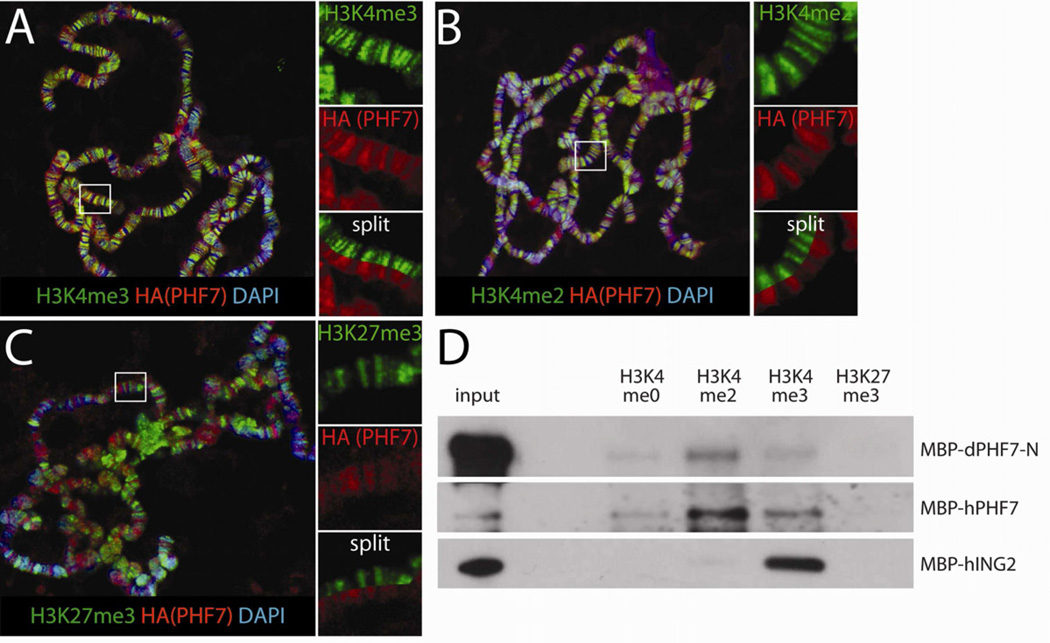
To understand how Phf7 is able to induce the male germline sexual program, we searched for predicted domains present in the protein. The N-terminal 300 amino acids of Drosophila PHF7 exhibits a high degree of homology to human PHF7 (32% identity, 52% similarity, Figure S5), and a lower degree of homology to other PHD finger-containing proteins. The remaining 220 amino acids of D. melanogaster PHF7 lack homology to other proteins, and are absent even in some Drosophila species. PHD fingers are frequently found in chromatin-associated proteins and most commonly interact with the N-terminal tail of histone H3, often specifically recognizing either the methylated or unmethylated form of H3 lysine 4 (H3K4me0 vs. H3K4me2/3, reviewed in Sanchez and Zhou, 2011). The two HA-tagged versions of our Phf7 genomic rescue construct (C- vs. N-terminus, Figure S1A) exhibited similar cell type immunoreactivity, but different degrees of nuclear localization (Figure 1D, Figure S1C). The observation that PHF7 can localize to the nucleus is consistent with it being a chromatin-associated molecule. PHF7 contains a putative nuclear export signal at its N-terminus (la Cour et al., 2004), suggesting that the protein may shuttle between the nucleus and cytoplasm as an added layer of regulation. This is supported by the fact that the N-terminal tagged version of PHF7 is more strongly localized to the nucleus, indicating that the tag may interfere with the nuclear export signal.
To investigate if PHF7 indeed associates with chromatin, we first examined its localization on polytene chromosomes in Drosophila larval salivary glands, which are composed of ~1000 aligned chromosomes allowing for direct visualization of proteins bound to specific regions of the genome. When Phf7 was expressed in the salivary glands, we observed extensive co-localization between PHF7 and polytene regions positive for histone H3 di- and trimethylated on lysine 4 (Figure 6A, B), marks of active transcription. This pattern was observed in polytene chromosomes from both male and female larvae (data not shown), and PHF7 also co-localized well with interbands (less intense DAPI staining), regions that are enriched for active genes. In contrast, PHF7 did not co-localize with the repressive chromatin mark of trimethylated histone H3K27 (H3K27me3, Figure 6C). To determine whether PHF7 is able to directly associate with modified histones, as has been observed for other PHD finger proteins, we used a binding assay between recombinant PHF7 and peptides representing the N-terminal tail of histone H3 that contain specific modifications. Interestingly, we found that both a fragment of Drosophila PHF7 covering all three putative PHD domains (aa 1–274) as well as full-length human PHF7 were able to associate with H3 N-terminal tail peptides, and exhibited a specificity for those modified by dimethylation of lysine 4 (H3K4me2, Figure 6D). In contrast, human ING2 exhibited a preference for H3K4me3, as previously reported (Shi et al., 2006). Together, these data indicate that PHF7 is a chromatin-associated factor found at sites of active transcription that can bind specifically to modified histone H3, specifically H3K4me2.
Figure 6.
PHF7 is a histone code reader. A–C) Polytene chromosomes isolated from salivary glands expressing UAS-Phf7-HA with fkh-Gal4 are co-labeled with antibodies for HA (PHF7) and various modifications on histone H3 tails as indicated. DAPI labels the DNA. A, H3K4me3. B, H3K4me2. C, H3K27me3. Smaller panels represent larger magnification of regions of the chromosomes (white rectangles) showing either the H3 mark (green) or HA-PHF7 (red) alone, or with the single channels split and aligned on the same image for direct comparison. D) α-MBP western blot analyses of MBP-tagged proteins precipitated with H3 N-terminal tail peptides modified as indicated. dPHF7-N is the N-terminal PHD finger domain of Drosophila PHF7, hPHF7 is full length human PHF7 and hING2 is the PHD finger of human ING2. Inputs are 1% for dPHF7 and hING2 and 10% for hPHF7. Note that the low specific activity of dPHF7-N in the binding assay was due to a tendency to aggregate in solution that was remarkably consistent with different protein fragments and purification conditions. See also Figure S5.
Discussion
Here we report the identification of PHF7, whichacts as a critical factor to trigger male germline sexual development. Phf7 is required for the proper male development in XY germ cells and can even induce spermatogenesis in XX germ cells when present in a male soma. Further, the presence of Phf7 orthologs that are expressed in the male germline in mammals indicates that this family of proteins represents conserved regulators of male germline sexual development.
PHF7 and germline sexual identity
Our data indicate that Phf7 acts to promote a male identity in the germline. Loss of Phf7 function affected male GSC maintenance and spermatogenesis, but had no effect in females (Figure 2, 3). Phf7-mutant GSCs exhibited a more female-like pattern of spectrosome localization (Figure 3H), and male (XY) germ cells mutant for Phf7 were more compatible with a female soma than were wild-type male germ cells (Figure 5B, C). Further, expression of PHF7 was able to masculinize the female germline: PHF7 expression induced apoptosis in developing XX germ cells and interacted with mutations in otu in a manner that indicates XX germ cells that express PHF7 are more male-like (Figure 4, Figure S4). Strikingly, PHF7 expression was able to induce spermatogenesis in XX germ cells when they are present in a male soma (Figure 5), something that XX germ cells are normally not able to do. Taken together, these results indicate that Phf7 promotes and is sufficient to induce male identity in the germline.
Sex determination is thought to be initiated early during development, and sex-specific differences in the male and female germline are first observed during embryogenesis (Casper and Van Doren, 2009; Hashiyama et al., 2011; Poirie et al., 1995; Staab et al., 1996; Wawersik et al., 2005). Our data indicate that Phf7 plays a role in early germline sexual development, rather than a late role to regulate germ cell differentiation and gametogenesis. First, PHF7 expression is observed in the embryonic gonad and, in the adult, PHF7 is found in the GSCs and early gonia and disappears dramatically as gonia transition to spermatocytes (Figure 1D, F). Further, forced PHF7 expression disrupts early female germ cell development, around the time when they are first forming GSCs (Figure 4). Expression of PHF7 after the early cystoblast stage (Bam-Gal4, UAS-Gal4) had no effect on the female germline, indicating that it can only affect early stages of female germ cell development. Phf7 mutants show defects in male GSC behavior and maintenance, and in the initial progression to form spermatocytes, but it is possible that these defects are due to even earlier problems in male sexual identity.
Germline sexual identity is determined by both the germ cell sex chromosome constitution and signals from the surrounding soma (Murray et al., 2010). Phf7 expression is activated in XX germ cells when in contact with a male soma and repressed in XY germ cells when contacting a female soma (Figure 1G). However, in a female somatic environment, XY germ cells are somewhat more likely than XX germ cells to express Phf7, indicating that Phf7 may also respond to the sex chromosome constitution of the germ cells in addition to being regulated by the soma. Further, exogenous expression of Phf7 is required to promote spermatogenesis in XX germ cells when in a male soma. Thus, the Phf7 expression that is normally initiated in such germ cells by the male soma must either not be maintained, or may be insufficient to overcome the influence of the XX sex chromosome genotype.
It is likely that Phf7 is not acting alone to control male sexual identity. Phf7 mutant males are still able to undergo spermatogenesis, but at a much reduced capacity. This appears to be the null phenotype for Phf7 as our mutants have lost significant portions of the coding sequence (see Experimental Procedures). Further, when PHF7 is expressed in XX germ cells present in a male soma, these germ cells can undergo spermatogenesis, but the penetrance of this phenotype is low. Interestingly, the rescue of spermatogenesis in these XX germ cells follows an “all or nothing” pattern; either the rescue is largely complete to give full testes and sperm production, or little rescue is observed. Therefore, there appears to be a threshold that must be crossed to promote male germline sexual identity, and that once this threshold is met, those germ cells either take over the testis, or induce other germ cells to also follow the male pathway. The simplest explanation for both the incomplete block to spermatogenesis in Phf7 mutants and the incomplete rescue of spermatogenesis by Phf7 in XX males is that an additional factor (or factors) exist which promotes male identity in addition to Phf7. Such a factor could function parallel to Phf7 in a single pathway, or represent independent input regarding germline sex determination (e.g. independent signals from the soma that influence germline sex).
PHF7 is a histone code reader
PHD fingers, such as those found in PHF7, are best known for their ability to specifically bind histones that have been modified on their N-terminal tails, in particular methylated H3K4 (Bienz, 2006). Here we show that both Drosophila and human PHF7 can directly associate with dimethylated H3K4, indicating that PHF7 is indeed a histone code reader. It is uncommon for PHD domains to associate preferentially with H3K4me2 over H3K4me3, but this specificity has been observed previously (Kim and Buratowski, 2009; Wilson et al., 2008), and is likely important for how PHF7 modulates expression of its targets. Both di- and trimethylated H3K4 are found at actively transcribed genes, but H3K4me2 is normally localized at the 5' end of coding sequences, downstream of H3K4me3 which is near promoters (Barrera and Ren, 2006). The two marks are also regulated by different demethylases (Eissenberg et al., 2007; Kavi and Birchler, 2009; Rudolph et al., 2007). A few recent studies have started to dissect unique effects of H3K4me2 on gene transcription (Kim and Buratowski, 2009; Pinskaya and Morillon, 2009), but the exact mechanisms are not well understood. Some PHD finger proteins also contain other domains, such as those that modify histones enzymatically. This does not appear to be the case for PHF7, and the region of homology between PHF7 homologs of different species is restricted to the PHD domains. However, individual PHD fingers can bind modified histone tails independently, and it is yet unclear which PHD finger in PHF7 contacts H3K4me2 and what activities the others might have. The logic of how PHF7 is recruited to specific loci and affects chromatin structure and gene activity are interesting questions for future work.
Another point of interest is how a reader of such a common epigenetic mark would have a sexspecific role in regulating male germline identity. It has been observed that mutation of an H3K4me2 demethylase in C. elegans, which leads to increased dimethylation at H3K4, results in ectopic activation of male-specific germline genes (Katz et al., 2009). A similar mutation in Drosophila causes female germline developmental defects (Szabad et al., 1988), which may be related to the germline atrophy we observed when PHF7 expression was up-regulated in female germ cells. These data are consistent with the hypothesis that H3K4me2 has a unique role in regulating the male germline genome. Interestingly, we have recently identified another germline chromatin factor, No child left behind (NCLB), that is expressed in germ cells of both sexes but required for GSC function only in males (Casper et al., 2011). Thus, NCLB may cooperate with PHF7 in regulating the male GSC transcriptional program.
Mammalian orthologs of Phf7 likely also function in male germline development
Based on sequence homology, orthologs of Phf7 are present in a wide range of mammalian species. Human and mouse PHF7 share extensive homology to Drosophila PHF7 throughout the N-terminus where the PHD fingers are present (Figure S5), and our results confirm that human PHF7 recognizes H3K4me2, similar to the fly protein. Interestingly, EST profiling (UniGene, Figure S2) indicates strong testis biases for Phf7 expression in many species, including humans, mice, rats and dogs. Moreover, several studies that performed genome-wide RNA profiling from purified mouse germline populations indicate that mouse Phf7 expression is present in spermatogonia and is further induced in spermatocytes (Fallahi et al., 2010; Rossi et al., 2008). Remarkably, human PHF7 was able to rescue fecundity defects in male flies mutant for Phf7 (Figure 2B). Thus, the sequence conservation observed between mammalian and Drosophila Phf7 represents true functional orthology.
As in Drosophila, germline sex determination in mouse is regulated at an early stage and is controlled by important signals from the soma, which for the mouse include retinoic acid and FGF9 (Bowles et al., 2006; Colvin et al., 2001; Koubova et al., 2006). Such signals regulate the timing of meiotic entry, which is different between the sexes, and also influence sex-specific programs of germline gene expression, such as expression of the key male-specific factor nanos2 (Tsuda et al., 2003). Significant changes in germ cell chromatin occur during this critical time in germ cell development, including changes in the H3K4 methylation state (Hajkova et al., 2008). Thus, Phf7 represents a prime candidate for interpreting these chromatin changes in a sex-specific manner to regulate male-specific gene expression. It will be of great interest to determine whether Phf7 plays a critical role in mouse and human male germ cell development, as we propose for Drosophila.
Experimental Procedures
Fly strains, mosaic analysis, and fecundity
The fly stocks used were obtained from the Bloomington Stock Center unless otherwise indicated and as following: Oregon-R, w1118, EY03023, P{Dfd-lacZ-HZ2.7} on X (W. McGinnis), nos-Gal4, c587-Gal4 (S. Selleck), Bam-Gal4 (D. McKearin), UAS-Gal4, otu1, U2AF-traF (T. Cline), fkh-Gal4 (D. Andrews). The combination of tra1 and Df(3L)st-j7 (denoted as Df(tra) in the Result section and Figure Legends) was used to generate tra mutants. XO males were made by crossing ncdD females to yw males.
Phf7ΔN2, Phf7ΔN18, Phf7Δ88 were generated by P-element excision of EY03023. Phf7ΔN2 lost ~2kb upstream of the annotated transcription start site and the first 595bp of the transcript (resulting in deletion of aa1–137 of 520). Phf7ΔN18 has its ~1.5kb upstream of the annotated transcription site deleted together with the first 811bp of the transcript (deleting aa1–211). Phf7Δ88 has not been mapped, but the entire open reading frame (ORF) as well as the 5' portion of the Rab35 ORF have been removed.
The genotypes for mosaic genetic clonal analysis were: hsFLP/Y; FRT80B, Ubi-GFP/FRT80B (for generating control clones) and Phf7ΔN2, hsFLP/Y; FRT80B, BAC, Ubi-GFP/FRT80B (for generating Phf7-mutant clones).
The genotypes for the lineage tracing experiment were: FRT40A, arm-lacZ/FRT40A (for marking of clones in control testes) and Phf7ΔN2/Y; FRT40A, arm-lacZ/FRT40A (for marking of clones in Phf7-mutant testes).
To induce unmarked clones in both the genetic mosaic clonal analysis and lineage tracing experiments, flies were heat-shocked in a circulating water bath at 37°C for 45 minutes, rested at room temperature for 1h, then treated at 37°C again for 45 minutes.
Fecundity tests for females were performed by mating single test virgin females to three newly-eclosed Oregon-R males for one week, and the resulting progeny were counted 14 days after the cross was set up. For males, newly-eclosed single test males were mated to 15 virgin Oregon-R females for one week, and the numbers of progeny produced were recorded 14 days after the day the cross started.
BAC tagging and cDNA constructs
The BAC construct CH322-177L19 (BACPAC Resources Center, Venken et al., 2009) was recombineered to generate three different epitope-tagged transgenes of Phf7: TAP tag (3xFlag-Strep-6xHis) at the N-terminus of Phf7 (Tiefenbach et al., 2010), 3xHA at the N-terminus, and 3xHA at the C-terminus. UAS-Phf7 was constructed from a full-length cDNA clone (LD43541, Drosophila Genome Resource Center) cloned into pUASPB, a modified version of pUASP (Rorth, 1998) containing an attB site for phiC31 mediated transgenesis. pUASP-Phf7-HA has 3xHA inserted into the C-terminus of the Phf7 ORF. pUASP-hPhf7 contains the human cDNA of PHF7 (BC022002, Open Biosystems) in pUASPB. For transgenesis, BAC constructs were inserted into PBac{yellow[+]-attP-3B}VK00033 (Venken et al., 2009) and cDNA constructs were inserted into P{CaryP}attP2 (Groth et al., 2004).
in situ hybridization, RT-PCR, western blotting, and histone tail binding assay
Embryonic in situ hybridization was carried out as previously described (Lehmann and Tautz, 1994). Antisense probe for Phf7 was generated by in vitro transcription of a Phf7 cDNA clone (LD43541). For RT-PCR, total RNA isolated from various tissues was converted to cDNA with Superscript II (Invitrogen) and PCR-amplified using Phf7 primers (9576cdsF: 5'-GAGCTGATCTTCGGCACTGT-3'; 9576cdsR: 5'-GCGTATAGGCAGGTCACCAC-3') and ribosomal protein (RpLP0) primers (RpLP0F: 5'-AACATCTCGCCCTTCTCGTA-3'; RpLP0R: 5'-CCACTCCCTGTTGGAACTTG-3') as controls. Western blots were conducted by separating homogenized adult tissue samples on acrylamide gels, transferred onto PVDF membranes (Millipore), and blotted with mouse-α-FLAG (M2, 1:1000, Sigma) and rabbit-α-histone H3 as loading control (1:5000, Abcam).
For histone tail binding assays, aa 1–274 of Drosophila PHF7 (dPHF7-N) and full-length human PHF7 were cloned into pMBP (S. Fugmann) to introduce the maltose-binding protein (MBP) as a N-terminal tag, expressed in bacteria, and purified using amylose resin (NEB). MBP-dPHF7-N, MBP-hPHF7, or MBP-ING2 (Wilson et al., 2008) was mixed with biotinylated histone H3 peptides carrying specific modifications (Millipore, Anaspec) in binding buffer (50mM Tris pH7.5, 150mM NaCl, 0.1% NP40, 100ng/µL BSA), pulled-down with streptavidin agarose (Sigma), and analyzed by western blotting with rabbit-α-MBP (1:10000, NEB).
Immunofluorescence
Immunofluorescence on adult gonads and polytene chromosomes were performed as previously described (Gonczy et al., 1997; Swaminathan et al., 2005). Confocal images were taken on ConfoCor3 (Zeiss).
Primary antibodies and the concentrations used are as following. Rabbit-α-vasa, 1:10000 (R. Lehmann); chicken-α-vasa, 1:10000 (K. Howard); rat-α-N-cadherin, 1:20 (DN-EX#8, DSHB); mouse-α-hts, 1:5 (1B1, DSHB); mouse-α-α-spectrin, 1:2 (DSHB); mouse-α-orb, 1:30 (4H8, DSHB); rabbit-α-GFP, 1:1000 (Torrey Pines Biolabs); rabbit-α-β-gal, 1:10000 (Cappel); mouse-α-α-tubulin, 1:500 (DM1a, Sigma); mouse-α-γ-tubulin, 1:100 (Sigma); rat-α-HA, 1:100 (3F10, Roche); mouse-α-BamC, 1:25 (DSHB); rabbit-α-trimethylated H3K4, 1:100 (Millipore), mouse-α-unmethylated H3K4, 1:100 (CMA301, Millipore); rabbit-α-trimethylated H3K27, 1:100 (Millipore); rabbit-α-activated Caspase-3, 1:100 (BD); rabbit-α-phospho-histone H3, 1:100 (Millipore).
To analyze oriented centrosome pairs in male GSCs, testes were dissected from freshly eclosed males, fixed, and stained with mouse-α-γ-tubulin along with other markers. Z-stack confocal images were taken to analyze whether centrosomes were at the interface between GSCs and the hub. Only GSCs containing a pair of replicated centrosomes were scored. The procedure for analyzing spindle orientiation was similar, except testes were stained with mouse-α-α-tubulin. Mitotic indices of GSCs and cyst stem cells (CySCs) were scored by calculating the number of either cell type positive for phospho-histone H3 (pH3) per testis. CySCs were defined as pH3+ nuclei not co-stained with the germline marker vasa.
Supplementary Material
Acknowledgements
We are very grateful to members of the fly community that supplied us with fly stocks and reagents for this work, as specifically cited in Materials and Methods. We also thank the Bloomington Stock Center (Indiana University), the Developmental Studies Hybridoma Bank (University of Iowa), and the Drosophila Genome Resource Center for reagents. In addition, we would like to thank Dr. Sebastian Fugmann for help on protein purification, and Drs. Geraldine Seydoux, Xin Chen, and the Van Doren Lab for helpful discussions. Imaging was performed at the Integrated Imaging Center at The Johns Hopkins University. This work was supported by the National Institutes of Health (GM084356).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells-- insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol. 2006;18:291–298. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Bergere M, Wainer R, Nataf V, Bailly M, Gombault M, Ville Y, Selva J. Biopsied testis cells of four 47,XXY patients: fluorescence in-situ hybridization and ICSI results. Hum Reprod. 2002;17:32–37. doi: 10.1093/humrep/17.1.32. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- Casper AL, Baxter K, Van Doren M. no child left behind encodes a novel chromatin factor required for germline stem cell maintenance in males but not females. Development. 2011;138:3357–3366. doi: 10.1242/dev.067942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AL, Van Doren M. The establishment of sexual identity in the Drosophila germline. Development. 2009;136:3821–3830. doi: 10.1242/dev.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- Evans DS, Cline TW. Drosophila melanogaster male somatic cells feminized solely by TraF can collaborate with female germ cells to make functional eggs. Genetics. 2007;175:631–642. doi: 10.1534/genetics.106.066332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallahi M, Getun IV, Wu ZK, Bois PRJ. A Global Expression Switch Marks Pachytene Initiation during Mouse Male Meiosis. Genes. 2010;1:469–483. doi: 10.3390/genes1030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D, Laski FA. Mechanisms of cell rearrangement and cell recruitment in Drosophila ovary morphogenesis and the requirement of bric a brac. Development. 1995;121:173–187. doi: 10.1242/dev.121.1.173. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Matunis E, DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development. 1997;124:4361–4371. doi: 10.1242/dev.124.21.4361. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiyama K, Hayashi Y, Kobayashi S. Drosophila Sex lethal gene initiates female development in germline progenitors. Science. 2011;333:885–888. doi: 10.1126/science.1208146. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Strong JA. A Case of Human Intersexuality Having a Possible XXY Sex-Determining Mechanism. Nature. 1959;183:302–303. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A. C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi HH, Birchler JA. Drosophila KDM2 is a H3K4me3 demethylase regulating nucleolar organization. BMC Res Notes. 2009;2:217. doi: 10.1186/1756-0500-2-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC, Mohler D, Riley SF, Storto PD, Nicolazzo PS. Complementation between alleles at the ovarian tumor locus of Drosophila melanogaster. Dev Genet. 1986;7:1–20. doi: 10.1002/dvg.1020070102. [DOI] [PubMed] [Google Scholar]
- Klinefelter HF, Reifenstein EC, Albright F. Syndrome Characterized by Gynecomastia, Aspermatogenesis without A-Leydigism, and Increased Excretion of Follicle-Stimulating Hormone. The Journal of Clinical Endocrinology. 1942;2:615–627. [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Marsh JL, Wieschaus E. Is sex determination in germ line and soma controlled by separate genetic mechanisms. Nature. 1978;272:249–251. doi: 10.1038/272249a0. [DOI] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- Murray SM, Yang SY, Van Doren M. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol. 2010;22:722–729. doi: 10.1016/j.ceb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothiger R, Jonglez M, Leuthold M, Meier-Gerschwiler P, Weber T. Sex determination in the germ line of Drosophila depends on genetic signals and inductive somatic factors. Development. 1989;107:505–518. doi: 10.1242/dev.107.3.505. [DOI] [PubMed] [Google Scholar]
- Oliver B, Pauli D, Mahowald AP. Genetic evidence that the ovo locus is involved in Drosophila germ line sex determination. Genetics. 1990;125:535–550. doi: 10.1093/genetics/125.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli D, Oliver B, Mahowald AP. The role of the ovarian tumor locus in Drosophila melanogaster germ line sex determination. Development. 1993;119:123–134. doi: 10.1242/dev.119.1.123. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Morillon A. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity. Epigenetics. 2009;4:302–306. doi: 10.4161/epi.4.5.9369. [DOI] [PubMed] [Google Scholar]
- Poirie M, Niederer E, Steinmann-Zwicky M. A sex-specific number of germ cells in embryonic gonads of Drosophila. Development. 1995;121:1867–1873. doi: 10.1242/dev.121.6.1867. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Rossi P, Lolicato F, Grimaldi P, Dolci S, Di Sauro A, Filipponi D, Geremia R. Transcriptome analysis of differentiating spermatogonia stimulated with kit ligand. Gene Expr Patterns. 2008;8:58–70. doi: 10.1016/j.modgep.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011 doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T. Autosomal mutations that interfere with sex determination in somatic cells of Drosophila have no direct effect on the germline. Dev Biol. 1982;89:117–127. doi: 10.1016/0012-1606(82)90300-1. [DOI] [PubMed] [Google Scholar]
- Schupbach T. Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex-lethal in Drosophila melanogaster. Genetics. 1985;109:529–548. doi: 10.1093/genetics/109.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciurano RB, Luna Hisano CV, Rahn MI, Brugo Olmedo S, Rey Valzacchi G, Coco R, Solari AJ. Focal spermatogenesis originates in euploid germ cells in classical Klinefelter patients. Hum Reprod. 2009;24:2353–2360. doi: 10.1093/humrep/dep180. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki KK, Kravitz EA. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr Opin Neurobiol. 2009;19:200–206. doi: 10.1016/j.conb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab S, Heller A, Steinmann-Zwicky M. Somatic sex-determining signals act on XX germ cells in Drosophila embryos. Development. 1996;122:4065–4071. doi: 10.1242/dev.122.12.4065. [DOI] [PubMed] [Google Scholar]
- Steinmann-Zwicky M, Schmid H, Nothiger R. Cell-autonomous and inductive signals can determine the sex of the germ line of drosophila by regulating the gene Sxl. Cell. 1989;57:157–166. doi: 10.1016/0092-8674(89)90181-5. [DOI] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabad J, Reuter G, Schroder MB. The effects of two mutations connected with chromatin functions on female germ-line cells of Drosophila. Mol Gen Genet. 1988;211:56–62. doi: 10.1007/BF00338393. [DOI] [PubMed] [Google Scholar]
- Tiefenbach J, Moll PR, Nelson MR, Hu C, Baev L, Kislinger T, Krause HM. A live zebrafish-based screening system for human nuclear receptor ligand and cofactor discovery. PLoS One. 2010;5:e9797. doi: 10.1371/journal.pone.0009797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Van Deusen EB. Sex determination in germ line chimeras of Drosophila melanogaster. J Embryol Exp Morphol. 1977;37:173–185. [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–567. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom AM, Dunkel L. Testicular function in Klinefelter syndrome. Horm Res. 2008;69:317–326. doi: 10.1159/000117387. [DOI] [PubMed] [Google Scholar]
- Wilson DR, Norton DD, Fugmann SD. The PHD domain of the sea urchin RAG2 homolog, SpRAG2L, recognizes dimethylated lysine 4 in histone H3 tails. Dev Comp Immunol. 2008;32:1221–1230. doi: 10.1016/j.dci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Zhu CH, Xie T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 2003;130:2579–2588. doi: 10.1242/dev.00499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.