Abstract
Pancreatic ductal adenocarcinoma is a disease of extremely poor prognosis for which there are no reliable markers of asymptomatic disease. To identify pancreatic cancer biomarkers, we focused on a genomic interval proximal to the most common fragile site in the human genome, chromosome 3p12, which undergoes smoking-related breakage, loss of heterozygosity, and homozygous deletion as an early event in many epithelial tumors, including pancreatic cancers. Using a functional genomic approach, we identified a seven-gene panel (TNC, TFPI, TGFBI, SEL-1L, L1CAM, WWTR1, and CDC42BPA) that was differentially expressed across three different expression platforms, including pancreatic tumor/normal samples. In addition, Ingenuity Pathways Analysis (IPA) and literature searches indicated that this seven-gene panel functions in one network associated with cellular movement/morphology/development, indicative of a “migration signature” of the 3p pathway. We tested whether two secreted proteins from this panel, tenascin C (TNC) and tissue factor pathway inhibitor (TFPI), could serve as plasma biomarkers. Plasma ELISA assays for TFPI/TNC resulted in a combined area under the curve (AUC) of 0.88 and, with addition of CA19-9, a combined AUC for the three-gene panel (TNC/TFPI/CA19-9), of 0.99 with 100% specificity at 90% sensitivity and 97.22% sensitivity at 90% specificity. Validation studies using TFPI only in a blinded sample set increased the performance of CA19-9 from an AUC of 0.84 to 0.94 with the two-gene panel. Results identify a novel 3p pathway–associated migration signature and plasma biomarker panel that has utility for discrimination of pancreatic cancer from normal controls and promise for clinical application.
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related mortality in both men and women in the United States. Estimates suggest that virtually 83% of the more than 42,470 cases in the United States diagnosed with the disease will also die from it, making pancreatic cancer the most deadly of cancers when grouped by organ site (1). Biomarkers for the early detection of pancreatic cancer are urgently needed. However, individual molecular biomarkers with high sensitivity and specificity needed for population-based screening have not been discovered. CA19-9 has been studied extensively and yet has failed to show the desired predictive value necessary for early detection and diagnosis (2). Although many high-throughput discovery platforms including proteomic, genomic, and transcriptomic approaches have been utilized and biomarker candidates identified, no one platform or molecule has been successfully validated in large population screens. As a part of the National Cancer Institute (NCI) Early Detection Research Network (EDRN), our goal is to assemble a panel of blood-based biomarker candidates, given that no one biomarker has yet shown promise for early detection. We hypothesized that a panel of early detection biomarkers for pancreatic cancer could be discovered by the identification of the earliest genetic pathways aberrant in smoking-related cancers such as lung, renal, and pancreatic cancers. Evidence that the driver events in smoking-related cancers remain to be discovered comes from the sequencing of the cancer genomes of 24 cases of pancreatic adenocarcinoma (3). Results indicated that although smokers have a significantly higher number of genetic alterations than do nonsmokers, including mutations, homozygous deletions, and amplifications, smoking-related genetic alterations did not seem to correlate with known driver genes mutated in pancreatic cancer, including KRAS, p53, CDKN2A/p16, etc., suggesting that the genetic determinants of smoking-related tumors are not driven by these major genes (3). Thus, we furthermore hypothesized that, to identify the earliest genetic alterations associated with pancreatic cancer, we must target the most common intervals of cytogenetic deletion associated with tobacco exposure, given that these alterations might be shared by both smoking-related and non–smoking-related pancreatic cancer. Loss of chromosome 3p has been documented as an initiating event in a cytogenetic pathway involved in smoking-related cancers; thus, we chose a functional genomic approach to target the genetic pathway deregulated by the deletion of the chromosome 3p12 region. This genomic interval is known to undergo loss of heterozygosity (LOH) and homozygous deletion in smoking-related tumors and is proximal to the most common fragile site in the human genome, FRA3B, which has been shown to be expressed in active smokers and thought to facilitate chromosome breakage following carcinogenic exposure from tobacco (4). High-frequency LOH has been observed in the FRA3B region encompassing 3p13–3p21 in a variety of epithelial cancers with homozygous deletions found proximal to the fragile site in lung, breast, and pancreatic tumors, suggestive that deletion of this genetic interval could be an early event in the genesis of smoking-related cancers (5). Our previous physical and functional mapping experiments showed that the introduction of a normal copy of chromosome 3p into renal cell carcinoma cell lines via microcell fusion suppressed tumorigenicity in nude mice in both orthotropic and subcutaneous injections (6–8). Fine mapping of suppressed and unsuppressed hybrids localized the NRC-1 tumor suppressor locus to a 4.75-Mb interval within chromosome 3p12 (9). In addition, we previously demonstrated high-frequency LOH in distinct intervals along 3p in malignant pancreatic islet cell tumors but not in precursor cystic lesions in a kindred with von Hippel–Lindau disease, an autosomal dominant cancer syndrome characterized by the development of multiple tumor types, two of which progress to malignancy (renal and pancreas), suggestive that loss of 3p proximal to VHL is requisite for malignant conversion (7). Experiments reported herein document the utility of a functional genomic approach not only to identify 3p12 pathway genes differentially expressed in pancreatic tumor/normal samples, but also to determine their relevance as blood-based pancreatic cancer biomarkers.
Materials and Methods
Patients and clinical samples
EDTA plasma samples from pancreatic adenocarcinoma cases were obtained from the TEXGEN repository, a Texas Medical Center consortium that houses sera and plasma from MD Anderson Cancer Center, Baylor, and Texas Children’s Hospital. Control plasma was obtained from individuals who were screened for different cancers and were free of malignancy or any benign condition. For validation purposes, a blinded set of EDTA plasma samples from controls and pancreatic adenocarcinoma patients were also obtained from The University of Alabama (UAB; W.E.G.).
RNA extraction, microarray, and quantitative real-time PCR analyses
Frozen pancreatic tumors and adjacent macroscopically and microscopically normal appearing pancreas from the same patients (matched) were obtained from untreated, retrospective pancreatic adenocarcinoma samples available from the M. D. Anderson Cancer Center tumor bank and our collaborator (MLF). Total RNA was extracted from these samples using a miRNeasy Mini Kit (Qiagen). Micro-array hybridization and scanning was done according to Affymetrix protocols. Quantitative real-time PCR (RT-PCR) analysis was carried out as per manufacturer’s protocol (Applied Biosystems), using specific primers. Data were analyzed according to the comparative Ct method and were normalized by glyceraldehyde-3-phosphate dehydrogenase expression.
Suppression subtractive hybridization library
The suppression subtractive hybridization (SSH) library was previously constructed using as starting materials for library construction microcell hybrids formed by the introduction of defined fragments of a normal chromosome 3p into a renal cell carcinoma cell line and subsequent assay of those microcell hybrids for tumor formation in athymic nude mice (10). We hypothesized that the resultant differentially expressed cDNAs obtained from the SSH library should represent genes up- or downregulated by the introduction of the NRC-1 tumor suppressor locus and could represent genes in a functional chromosome 3p12 tumor suppression pathway. Conversely, identification of this pathway could elucidate how loss of this genomic region and deregulation of this pathway could be involved in the early stages of pancreatic cancer. We furthermore hypothesized that characterization of this library could define genetic networks in pancreatic cancer that could serve as a source for biomarkers for early detection.
Bioinformatic analyses
To generate the highest quality expression data, the PDNN (positional-dependent-nearest-neighbor) model was chosen to account for existing probe variation in specific binding with the labeled target material (11). Existing algorithms, such as MAS 5.0, do not take into account probe-specific variation in binding efficiency and can result in variation in probe signal that vary more than 2 orders of magnitude within a single probe set. Probe normalization and summarization was done using the PerfectMatch software suite utilizing the PDNN algorithm (http://bioinformatics.mdanderson.org/software.html). For each probe set, the software outputs both the natural logarithm transformed expression level and correlation coefficient between the observed and modeled data. Data was filtered to identify genes exhibiting 2-fold or greater changes in gene expression.
Network and gene ontology analysis
Selected genes were investigated for network and gene functional interrelation by Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, www.ingenuity.com; ref. 12). IPA scans the set of input genes to identify networks byusing Ingenuity Pathways Knowledge Base for interactions between identified “Focus Genes,” in our case, the common genes identified from our pathways approach and known and hypothetical interacting genes stored in the knowledge base in IPA software, to generate a set of networks with a maximum network size of 35 genes/proteins. Networks are displayed graphically as genes/gene products (“nodes”) with the biological relationships between the nodes (“edges”) identified. All edges are from canonical information stored in the Ingenuity Pathways Knowledge Base. In addition, IPA computes a score for each network according to the fit of the user’s set of significant genes. The score indicates the likelihood of the Focus Genes in a network from the Ingenuity knowledge base being found together due to random chance. A score of 3, as the cutoff for identifying gene networks, indicates that there is only a 1/1,000 chance that the focus genes shown in a network are due to random chance. Therefore, a score of 3 or higher indicates a 99.9% confidence level to exclude random chance.
TNC-L ELISA
Plasma levels of the predominant isoform of tenascin C (TNC), TNC-large variant (TNC-L), were determined using a Human Tenascin-C Large (HMV; FNIII-B) ELISA kit (IBL-America), which detects the human TNC high-molecular-weight variant by sandwich ELISA. Samples were diluted 100-fold and then incubated in 96-well ELISA plates pre-coated with anti-TNC (4C8MS) antibody (Ab) for 1 hour at 37°C. After washing the wells 7 times with wash buffer, a horseradish peroxidase–conjugated anti-TNC (4F10TT) Ab was added and incubated for 30 minutes at 4°C. After washing the wells 9 times with wash buffer, chromogen was added and incubated at room temperature in dark for 30 minutes. The reaction was stopped by the addition of stop solution and the absorbance at 450 nm was determined using an ELISA plate reader (Spectramax Plus384 Microplate Reader; Molecular Devices) within 30 minutes of addition of stop solution, with the correction wavelength set at 540 nm. Results were mean absorbance of duplicate wells.
TFPI ELISA
Plasma tissue factor pathway inhibitor (TFPI) levels were determined using a commercially available Quantikine Human TFPI ELISA kit (DTFP10; R&D Systems, Inc.), which detects predominantly free TFPI and a very small percentage of LDL and HDL (low- and high-density lipoprotein)–bound TFPI by sandwich ELISA. The samples were diluted 200-fold and then incubated along with an assay diluent in 96-well ELISA plates precoated with anti-human TFPI monoclonal Ab for 2 hours at room temperature. After washing the wells 4 times with wash buffer, a horseradish peroxidase–conjugated anti-human TFPI was added and incubated for 1 hour at room temperature. After washing the wells 4 times with wash buffer, chromogen with substrate was added and incubated at room temperature in dark for 30 minutes. The reaction was stopped by the addition of stop solution and the absorbance at 450 nm was determined using an ELISA plate reader (Spectramax Plus384 Microplate Reader; Molecular Devices) within 30 minutes of addition of stop solution, with the correction wavelength set at 540 nm. Results were mean absorbance of duplicate wells.
CA19-9 ELISA
CA19-9 levels were measured in plasma samples (10 μL) using a commercially available ELISA kit (DRG International Inc.) according to manufacturer’s instructions. Ten microliters of each sample was incubated along with an assay buffer in 96-well ELISA plates precoated with murine monoclonal anti-CA19-9 Ab for 90 minutes at 37°C. After washing the wells 5 times with wash buffer, a horseradish peroxidase–conjugated anti-CA19-9 was added and incubated for 90 minutes at 37°C. After washing the wells 5 times with wash buffer, chromogen with substrate was added and incubated at room temperature in dark for 20 minutes. The reaction was stopped by the addition of stop solution and the absorbance at 450 nm was determined using an ELISA plate reader (Spectramax Plus384 Microplate Reader; Molecular Devices) within 15 minutes of addition of stop solution. Results were mean absorbance of duplicate wells.
For these experiments, we did not assign predetermined cutoff values to assess the specificity and sensitivity.
Statistics
Differences in plasma levels between normal and pancreatic adenocarcinoma were analyzed using the Student’s t test. To provide additional statistical rigor, the Mann–Whitney U test was also used to analyze the difference between normal and pancreatic adenocarcinoma samples. Two-sided P < 0.05 values were considered statistically significant. We constructed receiver operating characteristic (ROC) curves and calculated the area under the curve (AUC) to evaluate the specificity and sensitivity of predicting cases and controls by each protein and by the combination of these proteins. All statistical analyses were done using the Stata 10.1 (Stata Corporation).
Results
SSH library identifies candidate chromosome 3p12 pathway genes
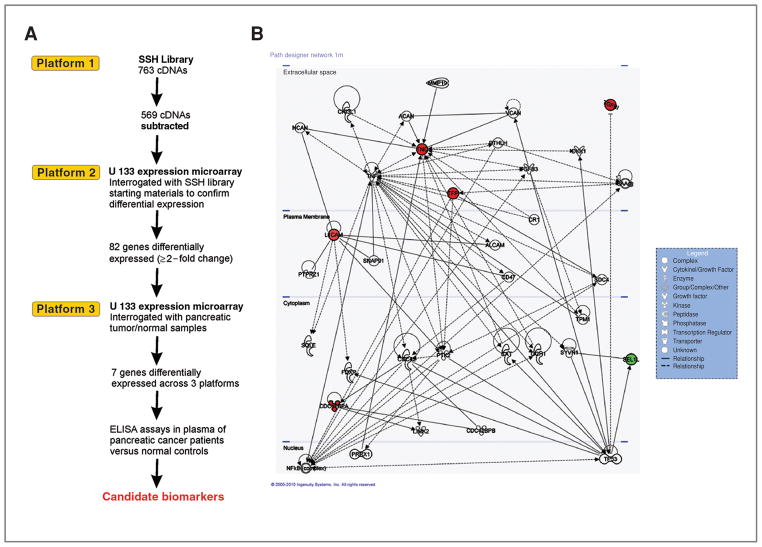
Three different expression platforms were utilized to identify genes in the chromosome 3p12 pathway to tumorigenesis in pancreatic cancer (schematically illustrated in Fig. 1A). We previously constructed an SSH library using as starting materials for library construction, microcell hybrid clones containing defined fragments of chromosome 3p12 that were either suppressed or unsuppressed for tumorigencity following injection of microcell hybrid clones into athymic nude mice (10). cDNAs differentially expressed from this SSH library should represent genes up- or down-regulated by the 3p12 tumor suppressor locus and, therefore, represent a 3p12 downstream pathway for biomarker discovery. From the SSH library, 880 partial cDNAs were obtained that were differentially expressed between suppressed and unsuppressed hybrids. PCR products were obtained from 763 clones (87% of the library) and used as templates for sequencing. A total of 569 of 763 clones (75%) were identified as subtracted because of the presence of appropriate adaptors and BLAST searches against the RefSeq database indicated 117 clones had no matches, short or poor quality sequence, or chimerism. The remainder of the 452 of 763 clones produced 2,297 sequences; however, a number of duplications were present in the library. After filtering for redundancy and for only those sequences with Entrez Gene matches, 507 Entrez Gene matches were obtained (Supplementary Table S1, Supplementary Appendix).
Figure 1.
A, schematic flow diagram of functional genomic approach to identify chromosome 3p12 pathway genes. Three expression platforms are denoted that were screened to identify candidates for validation in plasma as potential pancreatic cancer biomarkers. B, putative gene network derived from IPA software. Edges are displayed with labels that describe the nature of the relationship between the nodes. The lines between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect or hypothetical interactions. Color highlighting (red, upregulation; green, downregulation) indicates pathway-associated genes discovered by this study and nonhighlighted genes are those identified by IPA.
Expression profiling validates SSH library
To validate differential expression observed in the sequenced cDNA clones obtained from the SSH library, we utilized a second expression platform. Following analysis using the PerfectMatch software, probe sets overlapping the 507 Entrez Gene sequences from the SSH library were identified on a GeneChip U133 plus 2.0 array (Affymetix, Inc.) and examined for differential expression by interrogating the array with the starting materials for construction of the SSH library, i.e., microcell hybrids containing fragments of 3p used to construct the SSH library. By screening the same cDNAs identified from the SSH library on a commercial array, we were able to directly compare expression profiles across platforms to identify differentially expressed sequences, which we subsequently filtered by bioinformatics analyses to identify those genes with expression differences of 2-fold or greater. Supplementary Table S2 illustrates the 82-gene list of chromosome 3p12 pathways genes identified by this approach.
Expression analyses of pancreatic tumor/normal samples provide a third platform to stratify data
To identify those genes of the 82-gene list that were relevant to pancreatic cancer, we identified probe sets on a U133A plus 2.0 array corresponding to the 82-gene set and then interrogated the array with pancreatic tumor/normal samples. Frozen tumor and adjacent macroscopically and microscopically normal appearing pancreatic tissues from the same patient (matched) were obtained from untreated, retrospective pancreatic adenocarcinoma samples available from the M. D. Anderson Cancer Center tumor bank and our collaborator (MLF). The characteristics of the patients used for this study are presented in Supplementary Table S3. Total RNA from matched tumor/adjacent normal samples (8 paired samples) was utilized to interrogate the array and resultant bioinformatics analysis done to identify differentially expressed sequences across all 3 platforms. Bioinformatics analysis involved the stratification of the data set by selecting only those genes from the 82 gene list that were significantly differentially expressed across all 3 expression platforms resulting in a 7 gene set (P < 0.05; Table 1). The 7-gene panel [WWTR1 (13, 14), TGFBI (15, 16), TFPI (17, 18), CDC42BPA (19, 20), L1CAM (21, 22), TNC (23, 24), and SEL-1L (25, 26)] showed at least 2-fold or greater difference in gene expression and consistently remained as top candidates that were differentially expressed across 3 platforms. One gene, SEL-1L, was significantly downregulated in 8 of 8 tumors as compared with normal adjacent tissue, although the other 6 genes were upregulated. Importantly, 5 of 7 genes had also been previously published as being differentially expressed by immunohistochemistry (IHC) in pancreatic tumor samples. In addition, SEL-1L and TNC were differentially expressed by quantitative RT-PCR in 26 matched pancreatic tumor/normal samples (P = 0.002 and P = 0.038, respectively; Supplementary Table S4).
Table 1.
Seven-gene panel of candidate biomarkers identified from screening across 3 expression platforms
| Symbol | Title | Major biological functions | References |
|---|---|---|---|
| WWTR1/TAZ | WW domain containing transcription regulator 1 | Cofactor of transcription, cell migration, EMT | 13, 14 |
| TGFBI | Transforming growth factor, beta-induced, 68 kDa | Cell adhesion, migration, cell–matrix interaction | 15, 16 |
| TFPI | Tissue factor pathway inhibitor | Cell adhesion, migration, and proliferation | 17, 18 |
| CDC42BPA | CDC42 binding protein kinase alpha (DMPK-like) | Cell morphogenesis, cell signaling | 19, 20 |
| L1CAM | L1 cell adhesion molecule | Cell morphogenesis, migration, and cell survival | 21, 22 |
| TNC | Tenascin C (hexabrachion) | Cell adhesion, migration, proliferation, and angiogenesis | 23, 24 |
| SEL-1L | Sel-1 suppressor of lin-12-like | Negative regulation of colony formation, growth, and invasion; cell–matrix interaction | 25, 26 |
As another validation of our 7-gene biomarker panel, we conducted in silico analyses of publicly available microarray data sets from Oncomine (http://www.oncomine.org) to determine whether the 7 genes identified by our functional genomic screen were also found as differentially expressed in published pancreatic tumor/normal expression screens. Representative results of an individual data set (27), in which all our gene biomarkers of the 7-gene panel are expressed, are present in Supplementary Figure S1. Analysis of the biomarker panel showed that the expression of 6 genes (TNC, TFPI, TGFBI, LICAM, CDC42BPA, and WWTR1) was found to be significantly upregulated in pancreatic cancer when compared with normal pancreatic tissue (Supplementary Fig. S1). In contrast, SEL-1L was found to be significantly downregulated in pancreatic adenocarcinomas compared with the normal pancreas (Supplementary Fig. S1). Furthermore, we also analyzed the expression of our panel in 3 different expression data sets obtained from microdissected pancreatic tumor/normal samples (28–30). Importantly, although none of the 3 databases listed all 7 genes as differentially expressed, subsets of each gene biomarker of the 7-gene panel were represented in these data sets as well (Supplementary Figs. S2 and S3). Thus, in silico analyses confirmed our studies using cross-platform functional approaches, although none of the previously published compendiums of expression profiles identified these genes as a panel or studied their potential as blood-based pancreatic cancer biomarkers. Thus, our functional approach identified a novel panel differentially expressed in multiple data sets that hitherto had not been studied for blood-based biomarker development.
IPA identifies a single network and migration signature for 3p pathway genes
To determine the functional relationships among the 7 genes confirmed by our functional genomic pathway studies, IPA was queried for known or hypothetical interactions among the 7 genes in the panel and also all other genes in the Ingenuity database. With the exception of WWTR1/TAZ, which was not present in the IPA database, all the other 6 genes were used as focus genes for IPA. Unsupervised IPA network analysis identified a network of 35 genes that included all 6 focus genes and 29 additional genes (score = 16; Table 2). The interactive relationship between the genes in the network is shown in Figure 1B. Importantly, all 6 genes were classified into a single network related to cellular movement, cell morphology, and cellular development (Table 2). Of the 6 genes, 5 were also a part of a network involving cell signaling and cell interaction (P = 7.07E-06 to 3.07E-02) and cell movement (P = 1.38E-04 to 3.51E-02). The extremely low probability of obtaining this number of differentially expressed molecules in one network by chance alone is reflected by the P value for the network (P = 1.0E-16), indicating that this network is deregulated in a highly significant, nonrandom manner in pancreatic cancer cells. In addition, WWTR1/TAZ has also been reported to function in the regulation of cell migration in breast cancer (13). Therefore, we conclude that our functional genomic pathway approach has identified a gene signature related to cell movement, morphology, and organization, suggestive that the loss of the 3p12 locus in pancreatic cancer could be related to change in cell morphology and aberrant migration associated with early events in malignant transformation of pancreatic ductal epithelial cells (Table 2).
Table 2.
Networks identified from IPA
| Network | Genes in Ingenuity networka | Functions | Scoreb |
|---|---|---|---|
| 1 | ACAN, ALCAM, CAT, CD47, CDC42, CDC42BPA, CDC42BPB, CHI3L1, CR1, DKK1, FDXR, GCH1, L1CAM, LIMK2, MMP19, NCAN, NFkB (complex), PRRX1, PTHLH, PTK2, PTPRZ1, SAA@, SDC4, SEL-1L, SNAP91, SQLE, SYVN1, TFPI, TGFB3, TGFBI, TNC, TNF, TP53, TPM1, VCAN | Cellular movement, Cell morphology, Cellular development | 16 |
Genes in boldface were identified from our functional genomic pathway approach as differentially expressed across 3 platforms as focus genes; additional genes listed were identified by IPA.
A score >3 is considered significant.
TNC and TFPI are candidate plasma biomarkers that distinguish pancreatic cancer from normal screening controls
Four of the genes of the 7-gene panel (TGFBI, TFPI, LICAM, and TNC) were also secreted proteins. Sandwich ELISA assays were then done on 2 of the 4 secreted proteins, TFPI and TNC, for which commercial ELISAs were available to determine their ability to function as plasma biomarkers. The patient population characteristics used in the present study with respect to age, sex, alcohol intake history, smoking history, diabetic history, site of the disease, staging, and survival data are presented in Table 3. Results indicated that individual plasma TNC-L levels, presented in the form of a scatter plot (Fig. 2A), were significantly different between pancreatic cancer patients and normal screening controls in that the median plasma TNC-L levels was 342.6 pg/mL in patients with pancreatic adenocarcinoma (n = 36) as compared with levels in normal subjects (243.3; n = 19; Student’s t test, P = 0.0006; Mann–Whitney’s U test, P = 0.0004). Figure 2B illustrates the ROC curve for TNC. The AUC was 0.79, with a specificity of 47% at 90% sensitivity and sensitivity of 25% at 90% specificity.
Table 3.
Characteristics of samples used in the study
| Characteristics | Test set (TEXGEN)
|
Validation set (UAB)
|
||
|---|---|---|---|---|
| No. of patients (n = 36) | No. of controls (n = 19) | No. of patients (n = 37) | No. of controls (n = 15) | |
| Sex | ||||
| Male | 20 | 11 | 22 | 4 |
| Female | 16 | 8 | 15 | 11 |
| Age, y | ||||
| <50 | 2 | 3 | 2 | 4 |
| 50–60 | 12 | 12 | 6 | 3 |
| 61–70 | 15 | 3 | 12 | 3 |
| 71–80 | 7 | 1 | 13 | 5 |
| 81–90 | 0 | 0 | 4 | 0 |
| Race | ||||
| White | 36 | 19 | 33 | 13 |
| Black | 0 | 0 | 4 | 2 |
| Histology | ||||
| Adenocarcinoma | 36 | 0 | 37 | 0 |
| Alcohol history | ||||
| Current | 10 | N/A | N/A | N/A |
| Former | 14 | N/A | N/A | N/A |
| Never | 12 | N/A | N/A | N/A |
| Smoking history | ||||
| Current | 10 | N/A | N/A | N/A |
| Former | 13 | N/A | N/A | N/A |
| Never | 13 | N/A | N/A | N/A |
| Diabetes history | ||||
| Yes | 7 | N/A | N/A | N/A |
| No | 29 | N/A | N/A | N/A |
| Stage | ||||
| I | 0 | – | – | – |
| II | 6 | – | 1 | – |
| III | 5 | – | 2 | – |
| IV | 8 | – | 9 | – |
| N/A | 17 | – | 25 | – |
| Site | ||||
| Body | 4 | – | N/A | – |
| Head | 21 | – | N/A | – |
| Pancreas overlapping lesion | 7 | – | N/A | – |
| Tail | 4 | – | N/A | – |
| Stage | ||||
| Direct extension | 9 | – | N/A | – |
| Direct extension + lymph node | 3 | – | N/A | – |
| Distant | 17 | – | N/A | – |
| Localized | 2 | – | N/A | – |
| Regional lymph node involvement | 1 | – | N/A | – |
| N/A | 4 | – | N/A | – |
Abbreviation: N/A, not available.
Figure 2.
Candidate biomarkers from functional genomic approach validated in plasma of pancreatic cancer patients versus controls. A, plasma TNC-L concentrations of pancreatic carcinoma patients and normal subjects. Line, median plasma TNC level. The difference between normal and pancreatic adenocarcinoma samples is statistically significant (Mann–Whitney’s U test, P = 0.0004). B, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of the plasma TNC ELISA assay. The AUC was 0.79. The specificity was 47% given 90% sensitivity and the sensitivity was 25% given 90% specificity; C, plasma TFPI concentrations in pancreatic carcinoma patients and normal subjects. Line, median plasma TFPI level. The difference between normal and pancreatic adenocarcinoma samples is statistically significant (Mann–Whitney’s U test, P < 0.0001). D, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of the plasma TFPI ELISA assay. The AUC was 0.87. The specificity was 63% given 90% sensitivity and the sensitivity was 64% given 90% specificity; E, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of the combinations of 2 markers, plasma TNC and TFPI ELISA. The combined AUC is 0.88. F, plasma CA19-9 concentrations of pancreatic carcinoma patients and normal subjects. Line, median plasma CA19-9 level. The difference between normal and pancreatic adenocarcinoma samples is statistically significant (Mann–Whitney’s U test, P < 0.00001). G, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of plasma CA19-9 ELISA assay. The AUC was 0.93, with a specificity of 94.74% at 90% sensitivity and sensitivity of 91.67% at 90% specificity; H, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of combinations of 2 markers, plasma CA19-9 and TFPI ELISA. The combined AUC was 0.99.
We next extended our study to further validate the significance of TFPI as a potential plasma biomarker by ELISA. Figure 2C depicts the individual plasma TFPI levels of normal and pancreatic cancer patients. Plasma TFPI levels of patients with pancreatic adenocarcinoma were significantly higher than normal subjects (Student’s t test, P = 0.0004; Mann–Whitney’s U test, P< 0.0001), with the median plasma TFPI level of 27.0 ng/mL in pancreatic adeonocarcinoma patients (n = 36) compared with normal subjects (15.3; n = 19). Figure 2D illustrates the ROC curve which compares the ability of plasma TFPI to distinguish between patients with pancreatic cancer and normal subjects. The AUC for TFPI was 0.87, with a specificity of 63% given 90% sensitivity and a sensitivity of 64% given 90% specificity.
We next tested whether the combination of the 2 markers TNC and TFPI could increase sensitivity and specificity for discrimination between cancer and normal plasma samples. The combined AUC for both markers was 0.88 (Fig. 2E). The combination of markers resulted in a specificity of 63% given 90% sensitivity and a sensitivity of 67% given 90% specificity. These combined results, then, suggest that the 2-gene panel identified through our functional genomic studies has high sensitivity and specificity to discriminate tumor and normal samples in the plasma and indicate that these genes are candidate blood-based biomarkers for pancreatic cancer. The diagnostic potential of candidate biomarkers TNC and TFPI, relative to and in combination with CA19-9, the standard serum biomarker for pancreatic cancer, was determined. Results indicated that individual plasma CA19-9 levels, presented in the form of a scatter plot (Fig. 2F), were significantly different between pancreatic cancer patients and normal screening controls in that the median plasma CA19-9 levels were 173 U/mL in patients with pancreatic adenocarcinoma (n = 36) as compared with levels in normal subjects (11.9; n = 19; Student’s t test, P < 0.00001; Mann–Whitney’s U test, P < 0.00001). Figure 2G illustrates the ROC curve for CA19-9, and the AUC was 0.93, with a specificity of 94.74% at 90% sensitivity and sensitivity of 91.67% at 90% specificity. Figure 2H illustrates the ROC curve for CA19-9 and TFPI combined. The AUC was 0.99, with a specificity of 100% at 90% sensitivity and sensitivity of 97.22% at 90% specificity.
We next tested whether the combination of the 2 markers, TNC and TFPI, with CA19-9 could further increase sensitivity and specificity for discrimination between cancer and normal plasma samples. The combined AUC for all the markers was 0.99 (Supplementary Fig. S4). The combination of markers resulted in a specificity of 100% given 90% sensitivity and a sensitivity of 97.22% given 90% specificity.
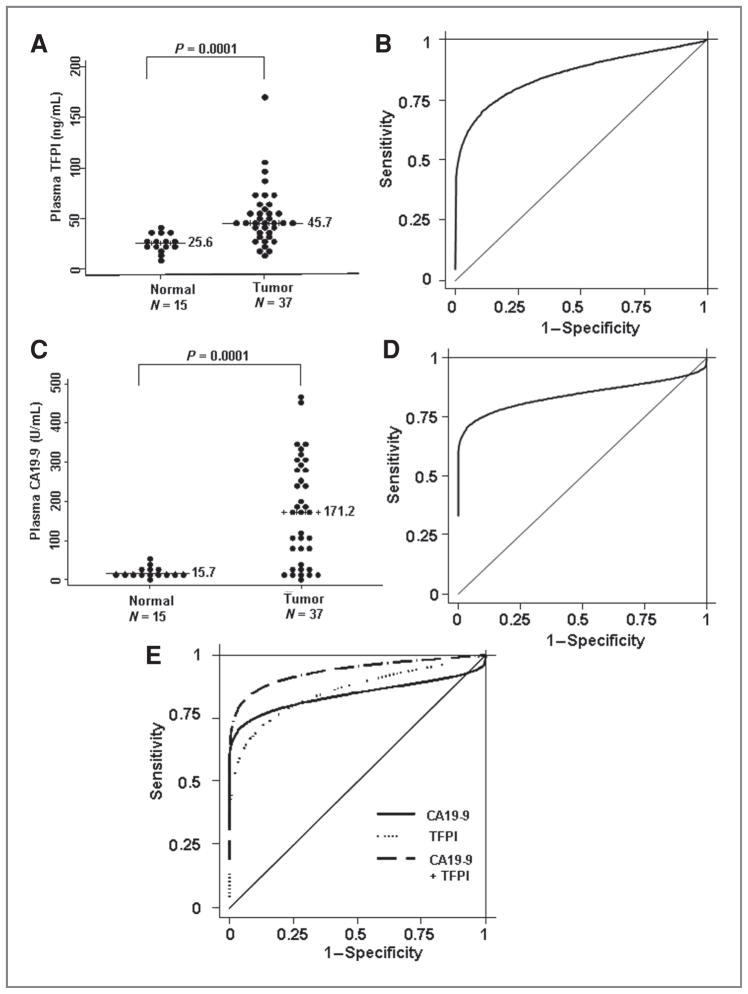
To further strengthen our findings and to validate our results in a different sample set, we tested TFPI levels by ELISA in plasma samples from normal and pancreatic adenocarcinoma patients collected at the UAB. A description of the UAB sample set is given in Table 3. Because the addition of TNC did not add significantly to the sensitivity and specificity (Supplementary Fig. S4), we analyzed only TFPI levels in these samples. Figure 3A depicts the individual plasma TFPI levels of normal and pancreatic cancer patients. Plasma TFPI levels of patients with pancreatic adenocarcinoma were significantly higher than normal subjects (Student’s t test, P = 0.000014; Mann–Whitney’s U test, P = 0.0001), with the median plasma TFPI level of 45.7 ng/mL in pancreatic adeonocarcinoma patients (n = 37) compared with normal subjects (25.6; n = 15). Figure 3B illustrates the ROC curve to compare the ability of plasma TFPI in distinguishing between patients with pancreatic cancer and normal subjects. The AUC for TFPI was 0.87 with a specificity of 46.67% at 90% sensitivity and 70.27% sensitivity at 90% specificity.
Figure 3.
A, plasma TFPI concentrations of pancreatic carcinoma patients and normal subjects. Line, median plasma TFPI level. The difference between normal and pancreatic adenocarcinoma samples is statistically significant (Mann–Whitney’s U test, P= 0.0001). B, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of plasma TFPI ELISA assay. The AUC was 0.87 with a specificity of 46.67% at 90% sensitivity and 70.27% sensitivity at 90% specificity. C, plasma CA19-9 concentrations of pancreatic carcinoma patients and normal subjects. Line, median plasma CA19-9 level. The difference between normal and pancreatic adenocarcinoma samples is statistically significant (Mann–Whitney’s U test, P= 0.0001). D, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of plasma CA19-9 ELISA assay. The AUC was 0.84, with a specificity of 13.33% at 90% sensitivity and 75.68% sensitivity at 90% specificity. E, ROC curve for differentiating normal and pancreatic carcinoma patients on the basis of combinations of 2 markers, plasma CA19-9 and TFPI ELISA. The combined AUC was 0.94 and resulted in a specificity of 86.67% at 90% sensitivity and 83.78% sensitivity at 90% specificity.
Next, we analyzed CA19-9 by ELISA in these samples. Results indicated that individual plasma CA19-9 levels, presented in the form of a scatter plot (Fig. 3C), were significantly different between pancreatic cancer patients and normal screening controls in that the median plasma CA19-9 levels was 171.2 U/mL in patients with pancreatic adenocarcinoma (n = 37) as compared with levels in normal subjects (15.7; n = 15; Student’s t test, P = 0.0000000400227; Mann–Whitney’s U test, P = 0.0001). Figure 3D illustrates the ROC curve for CA19-9, in which the AUC was 0.84, with a specificity of 13.33% at 90% sensitivity and 75.68% sensitivity at 90% specificity. The combined AUC for TFPI and CA19-9 was 0.94 (Fig. 3E). The combination of markers resulted in a specificity of 86.67% at 90% sensitivity and 83.78% sensitivity at 90% specificity.
Discussion
Attempts to identify pancreatic cancer biomarkers have failed to produce a single marker with the sensitivity and specificity necessary for population screening. We reasoned that a targeted strategy to identify differentially expressed genes related to the earliest cytogenetic aberrations might be more successful in developing such biomarkers because we would be able to focus on those aberrantly expressed genes that may be involved in initiating the pathways that ultimately lead to tumorigenesis, invasion, and metastasis. Using 3 different expression platforms, we identified a 7-gene set as being differentially expressed between pancreatic cancer and normal samples and from which we have validated a subset of markers for differential expression by ELISA assays. Our results indicate that we have been able to identify 2 relevant blood-based biomarker candidates for pancreatic cancer. Several published reports document the potential of using immunohistochemical staining of TNC, an extracellular matrix protein, as a potential marker of early disease as well as a predictor of poor prognosis in several tumor types, including colon, bladder, and pancreas (31–34). TNC has been shown to be overexpressed in the stroma by IHC in a variety of different cancers, including pancreatic cancer (31–33, 35). In addition to stromal expression, TNC expression increases from low-grade PanIN-1A and -1B intraductal precursor lesions to high-grade PanIN-2 and -3 lesions to invasive lesions, suggestive that TNC could be an early immunohistochemical marker of disease (31). However, reports of the utility of TNC as a blood-based biomarker have been limited to colorectal cancer (36), in which TNC spliced variant overexpression in plasma was observed and is considered a potential biomarker for colorectal cancer. Our data implicate TNC as a potential plasma marker for pancreatic cancer, which based on its expression in precursor lesions and upregulation with increasing stage, suggests that it might be a marker of early disease.
Our results also identify TFPI as a novel pancreatic plasma biomarker candidate. TFPI is a major inhibitor of the tissue factor pathway of blood coagulation in vivo. Plasma TFPI levels have been shown to increase significantly at the time of diagnosis compared with controls and reach near normal levels following surgical removal of pancreatic tumor (37). Plasma TFPI has also been reported to be increased in acute pancreatitis compared with normal subjects (38). Plasma levels of total TFPI has been found to be upregulated in a number of solid tumors involving colon, pancreas, and stomach (39). Recently, increased TFPI expression levels have also been reported in colon and breast tumor tissues (40). Our study suggests that plasma TFPI levels may be a potential biomarker for pancreatic cancer and can also serve as a prognostic marker owing to its modulation following surgery.
Our results indicate that the combined analysis of TFPI and CA19-9 in plasma can discriminate pancreatic adenocarcinoma patients from normal screening controls with better sensitivity and specificity than CA19-9 alone, at least in our pilot studies. These results could have significance for the early screening of pancreatic cancer given that CA19-9 alone fails to have adequate predictive value. Owing to CA19-9 being typically a serum marker, these results would furthermore suggest that future analyses of the value of CA19-9 screening in plasma as well as its use in combination with TFPI are warranted to determine whether the combined panel could result in a viable test for general population screening.
Furthermore, using a functional genomic approach, we have identified a cancer-associated network associated with the differential expression of the chromosome 3p12 locus implicated in smoking-related malignancies. IPA identified cell movement/cell morphology/cell differentiation as the single critical network associated with the 3p12 pathway, with 7 of 7 genes in the panel functionally implicated previously in the regulation of cell movement and migration. Thus, although the importance of migration has been thoroughly characterized in relationship to metastasis, the role of cell movement and migration in early stages of cancer initiation is not well understood. Because the chromosome 3p12 region has been shown to undergo LOH and homozygous deletion as an early event in smoking-related cancers, potentially, loss of this region could play a role in the loss of polarity, cell migration, and movement in the earliest stages of malignant transformation and epithelial-mesenchymal transition (EMT) related to invasive disease. We have earlier characterized one novel gene DEAR1 (annotated as TRIM62) in detail from our SSH library. Genetic complementation of DEAR1 in a breast cancer cell line carrying a DEAR1 mutation resulted in restoration of acinar morphogenesis whereas knockdown resulted in loss of polarity and tissue architecture in 3-dimensional (3D) culture (10). Thus, DEAR1 falls within this 3p pathway and functions in the regulation of cellular morphology and differentiation, as it relates to changes in 3D acinar morphogenesis. In addition, other members of the 7-gene panel have also been documented to play a role in cell migration and early stages of pancreatic cancer, suggestive that this network may be critically deregulated in early pancreatic cancer. TNC has been shown to mediate proliferation and migration of astrocytes in a wound assay (41). Tumor-associated isoforms of TNC has been shown to promote breast cancer cell invasion and growth (42). TNC signaling has been reported to play an important role in mammary tumor growth and metastasis, and knockdown of TNC exhibited significant impairment in cell migration and anchorage-dependent cell proliferation in breast cancer cell line (43). In addition, TNC has also been shown to stimulate glioma cell migration (44). TFPI has been shown to control migration of endothelial cells (45). Extracellular matrix bound TFPI through an interaction with tissue factor/VIIa complex localized on cancer cells has also been shown to facilitate cancer cell migration and adhesion (18).
SEL-1L is the only member of the 7-gene panel shown to be downregulated in pancreatic tumors versus normal samples in this study. It is expressed abundantly only in the normal pancreas and is downregulated in pancreatic cancers. SEL-1L was first reported as a pancreas-specific transcript but later found to be highly expressed in normal pancreas and present at very low levels in several other adult tissues (25, 46, 47). Its loss of expression has been observed in 17% of pancreatic adenocarcinomas. Thus, SEL-1L represents a gene that shows a tissue-restricted pattern of expression, with the only tissue showing abundant expression being the pancreas. In addition, SEL-1L maps into a genomic interval for the insulin-dependent diabetes mellitus locus at chromosome 14q24.3-q31 but was later excluded as a candidate gene for diabetes (48). Induced expression of SEL-1L in pancreatic cancer cells has been shown to decrease the clonogenity and anchorage-independent growth, and it also delayed tumor growth in immunodeficient mice (49). In addition, SEL-1L has been reported to affect pancreatic cancer cell cycle and invasiveness by modulating the expression of PTEN and genes involved in cell–matrix interactions (26). SEL-1L has been shown to be a negative regulator of Notch signaling. The Notch pathway has been extensively studied and regulates cell fate decisions in a large number of adult and embryonic tissues. Components of the Notch signaling pathway have been shown to be overexpressed in pancreatic adenocarcinomas (50), with activation of Notch signaling observed in PanIN lesions. Thus, SEL-1L loss of expression could represent a very early marker of pancreatic cancer.
L1CAM has been reported to be overexpressed in a number of different tumors types including colon, breast, and ovarian tumors, melanoma, gliomas, neuroblastomas, and pancreatic neuroendocrine tumors (21). Immunohistochemical staining of L1CAM was observed in chronic pancreatitis tissues and was absent in normal pancreatic tissues (51). Importantly, L1CAM has been shown to play a role both in migration and in the malignant transformation of pancreatic adenocarcinoma (51). Importantly, upregulation of L1CAM expression by IHC has been observed in later stage, high-grade PanIN lesions as compared with PanIN-1A/1B lesions that are not thought to have a high risk for progression to pancreatic cancer, suggestive of the role of L1CAM early in transition to pancreatic adenocarcinoma (52). Interestingly, WWTR1/TAZ, a transcription cofactor, was also found to regulate cell migration and invasion (13, 14). WWTR1 has also been reported to be amplified in pancreatic cancer cell lines and in pancreatic cancer (53). It was found to play a role in the migration, invasion, and tumorigenesis of breast cancer cells (13). TGFBI has been reported to be overexpressed in colon and pancreatic cancer (16, 54). TGFBI is an excreted extracellular matrix protein reported to play a role in cell–matrix regulation as well as cell migration in bone (55). It has recently been found as one of a gene panel upregulated during hematopoietic stem cell lineages as they differentiated and became migratory, suggesting a role for TGFBI in stem cell migration between niches (56). CDC42BPA, a protein kinase, has also been implicated in tumor cell invasion (20) and forms a complex with a leucine-rich adaptor protein, LRAP35a, and MYO18A, shown to play a crucial role in cell protrusion and migration (57). Therefore, all 7 genes in our panel have been closely linked functionally to the control of cell migration in cancer and potentially in the early stages of pancreatic tumorigenesis.
In addition, 3 of the 7 genes, L1CAM, TGFBI, and CDC42BPA, identified as most differentially expressed, are mutated in the germline in genetic disorders including CRASH syndrome, Thiel–Behnke corneal dystrophy, and Crohn’s disease, respectively (58, 59). Given that germline mutations underlying genetic disorders are very rare, the finding that our study identified 3 of 7 genes as being mutated in hereditary diseases indicates the functional significance of this migration pathway in early development, the deregulation of which could be of critical importance in pancreatic cancer initiation and progression.
In conclusion, we have taken a pathways approach to biomarker discovery by utilizing 3 different expression-based platforms to identify chromosome 3p12 pathway genes differentially expressed between pancreatic tumor/normal samples, which could serve as candidate biomarkers for the early detection of pancreatic cancer. Biomarker panels described herein will be further validated in larger case–control studies with the EDRN of the NCI. Additional candidates from the 7-gene list and associated IPA network members should also be investigated for their ability to improve performance of current panels. Future studies are also warranted to investigate the role of cell polarity and migration in the initiation of pancreatic cancer and the potential for biomarker discovery by a targeted pathway approach.
Acknowledgments
We gratefully acknowledge the NCI Early Detection Research Network for support and guidance in our ongoing biomarker discovery projects. We are also grateful to Drs. David Gold and David Stivers, who provided bioinformatics support for this project.
Grant Support
This research was supported by grant UO1CA111302 to A.M. Killary from the National Cancer Institute Early Detection Research Network.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerprevres.aacrjournals.org/).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CACancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kim HR, Lee CH, Kim YW, Han SK, Shim YS, Yim JJ. Increased CA 19–9 level in patients without malignant disease. Clin Chem Lab Med. 2009;47:750–4. doi: 10.1515/CCLM.2009.152. [DOI] [PubMed] [Google Scholar]
- 3.Blackford A, Parmigiani G, Kensler TW, Wolfgang C, Jones S, Zhang X, et al. Genetic mutations associated with cigarette smoking in pancreatic cancer. Cancer Res. 2009;69:3681–8. doi: 10.1158/0008-5472.CAN-09-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein CK, Glover TW, Palmer JL, Glisson BS. Direct correlation between FRA3B expression and cigarette smoking. Genes Chromosomes Cancer. 2002;34:333–40. doi: 10.1002/gcc.10061. [DOI] [PubMed] [Google Scholar]
- 5.Shridhar R, Shridhar V, Wang X, Paradee W, Dugan M, Sarkar F, et al. Frequent breakpoints in the 3p14. 2 fragile site, FRA3B, in pancreatic tumors. Cancer Res. 1996;56:4347–50. [PubMed] [Google Scholar]
- 6.Sanchez Y, El-Naggar A, Pathak S, Killary AM. A tumor suppressor locus within 3p14-p12 mediates rapid cell death of renal cell carcinoma in vivo. Proc Natl Acad Sci USA. 1994;91:3383–7. doi: 10.1073/pnas.91.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lott ST, Lovell M, Naylor SL, Killary AM. Physical and functional mapping of a tumor suppressor locus for renal cell carcinoma within chromosome 3p12. Cancer Res. 1998;58:3533–7. [PubMed] [Google Scholar]
- 8.Lovell M, Lott ST, Wong P, El-Naggar A, Tucker S, Killary AM. Thegenetic locus NRC-1 within chromosome 3p12 mediates tumor suppression in renal cell carcinoma independently of histological type, tumor microenvironment, and VHL mutation. Cancer Res. 1999;59:2182–9. [PubMed] [Google Scholar]
- 9.Zhang K, Lott ST, Jin L, Killary AM. Fine mapping of the NRC-1 tumor suppressor locus within chromosome 3p12. Biochem Biophys Res Commun. 2007;360:531–8. doi: 10.1016/j.bbrc.2007.06.066. [DOI] [PubMed] [Google Scholar]
- 10.Lott ST, Chen N, Chandler DS, Yang Q, Wang L, Rodriguez M, et al. DEAR1 is a dominant regulator of acinar morphogenesis and an independent predictor of local recurrence-free survival in early-onset breast cancer. PLoS Med. 2009;6:e1000068. doi: 10.1371/journal.pmed.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21:818–21. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 12.Nomura H, Uzawa K, Yamano Y, Fushimi K, Ishigami T, Kato Y, et al. Network-based analysis of calcium-binding protein genes identifies Grp94 as a target in human oral carcinogenesis. Br J Cancer. 2007;97:792–801. doi: 10.1038/sj.bjc.6603948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–8. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 14.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11:511–22. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- 16.Ma C, Rong Y, Radiloff DR, Datto MB, Centeno B, Bao S, et al. Extra-cellular matrix protein betaig-h3/TGFBI promotes metastasis of colon cancer by enhancing cell extravasation. Genes Dev. 2008;22:308–21. doi: 10.1101/gad.1632008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broze GJ, Jr, Warren LA, Novotny WF, Higuchi DA, Girard JJ, Miletich JP. The lipoprotein-associated coagulation inhibitor that inhibits the factor VII-tissue factor complex also inhibits factor Xa: insight into its possible mechanism of action. Blood. 1988;71:335–43. [PubMed] [Google Scholar]
- 18.Fischer EG, Riewald M, Huang HY, Miyagi Y, Kubota Y, Mueller BM, et al. Tumor cell adhesion and migration supported by interaction of a receptor-protease complex with its inhibitor. J Clin Invest. 1999;104:1213–21. doi: 10.1172/JCI7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Loyer P, Li H, Valentine V, Kidd V, Kraft AS. Cloning and chromosomal location of a novel member of the myotonic dystrophy family of protein kinases. J Biol Chem. 1997;272:10013–20. doi: 10.1074/jbc.272.15.10013. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson S, Paterson HF, Marshall CJ. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat Cell Biol. 2005;7:255–61. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- 21.Raveh S, Gavert N, Ben-Ze’ev A. L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 2009;282:137–45. doi: 10.1016/j.canlet.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 22.Rathjen FG, Schachner M. Immunocytological and biochemical characterization of a new neuronal cell surface component (L1 antigen) which is involved in cell adhesion. EMBO J. 1984;3:1–10. doi: 10.1002/j.1460-2075.1984.tb01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiquet-Ehrismann R, Mackie EJ, Pearson CA, Sakakura T. Tenascin: an extracellular matrix protein involved in tissue interactions during fetal development and oncogenesis. Cell. 1986;47:131–9. doi: 10.1016/0092-8674(86)90374-0. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar S, Nuttall RK, Liu S, Edwards DR, Yong VW. Tenascin-C stimulates glioma cell invasion through matrix metalloproteinase-12. Cancer Res. 2006;66:11771–80. doi: 10.1158/0008-5472.CAN-05-0470. [DOI] [PubMed] [Google Scholar]
- 25.Biunno I, Appierto V, Cattaneo M, Leone BE, Balzano G, Socci C, et al. Isolation of a pancreas-specific gene located on human chromosome 14q31: expression analysis in human pancreatic ductal carcinomas. Genomics. 1997;46:284–6. doi: 10.1006/geno.1997.5018. [DOI] [PubMed] [Google Scholar]
- 26.Cattaneo M, Fontanella E, Canton C, Delia D, Biunno I. SEL1L affects human pancreatic cancer cell cycle and invasiveness through modulation of PTEN and genes related to cell-matrix interactions. Neoplasia. 2005;7:1030–8. doi: 10.1593/neo.05451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segara D, Biankin AV, Kench JG, Langusch CC, Dawson AC, Skalicky DA, et al. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin Cancer Res. 2005;11:3587–96. doi: 10.1158/1078-0432.CCR-04-1813. [DOI] [PubMed] [Google Scholar]
- 28.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–57. [PubMed] [Google Scholar]
- 29.Buchholz M, Braun M, Heidenblut A, Kestler HA, Klöppel G, Schmiegel W, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–36. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 30.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–27. [PubMed] [Google Scholar]
- 31.Esposito I, Penzel R, Chaib-Harrireche M, Barcena U, Bergmann F, Riedl S, et al. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J Pathol. 2006;208:673–85. doi: 10.1002/path.1935. [DOI] [PubMed] [Google Scholar]
- 32.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juuti A, Nordling S, Louhimo J, Lundin J, Haglund C. Tenascin C expression is upregulated in pancreatic cancer and correlates with differentiation. J Clin Pathol. 2004;57:1151–5. doi: 10.1136/jcp.2003.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kressner U, Lindmark G, Tomasini-Johansson B, Bergström R, Gerdin B, Påhlman L, et al. Stromal tenascin distribution as a prognostic marker in colorectal cancer. Br J Cancer. 1997;76:526–30. doi: 10.1038/bjc.1997.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunner A, Mayerl C, Tzankov A, Verdorfer I, Tschörner I, Rogatsch H, et al. Prognostic significance of tenascin-C expression in superficial and invasive bladder cancer. J Clin Pathol. 2004;57:927–31. doi: 10.1136/jcp.2004.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda A, Otani Y, Iseki H, Takeuchi H, Aikawa K, Tabuchi S, et al. Clinical significance of large tenascin-C spliced variant as a potential biomarker for colorectal cancer. World J Surg. 2007;31:388–94. doi: 10.1007/s00268-006-0328-6. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl AK, Odegaard OR, Sandset PM, Harbitz TB. Coagulation inhibition and activation in pancreatic cancer. Changes during progress of disease. Cancer. 1992;70:2067–72. doi: 10.1002/1097-0142(19921015)70:8<2067::aid-cncr2820700809>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 38.Yasuda T, Ueda T, Kamei K, Shinzaki W, Sawa H, Shinzeki M, et al. Plasma tissue factor pathway inhibitor levels in patients with acute pancreatitis. J Gastroenterol. 2009;44:1071–9. doi: 10.1007/s00535-009-0096-9. [DOI] [PubMed] [Google Scholar]
- 39.Iversen N, Lindahl AK, Abildgaard U. Elevated plasma levels of the factor Xa-TFPI complex in cancer patients. Thromb Res. 2002;105:33–6. doi: 10.1016/s0049-3848(01)00404-2. [DOI] [PubMed] [Google Scholar]
- 40.Sierko E, Wojtukiewicz MZ, Zimnoch L, Kisiel W. Expression of tissue factor pathway inhibitor (TFPI) in human breast and colon cancer tissue. Thromb Haemost. 2010;103:198–204. doi: 10.1160/TH09-06-0416. [DOI] [PubMed] [Google Scholar]
- 41.Nishio T, Kawaguchi S, Yamamoto M, Iseda T, Kawasaki T, Hase T. Tenascin-C regulates proliferation and migration of cultured astrocytes in a scratch wound assay. Neuroscience. 2005;132:87–102. doi: 10.1016/j.neuroscience.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 42.Hancox RA, Allen MD, Holliday DL, Edwards DR, Pennington CJ, Guttery DS, et al. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res. 2009;11:R24. doi: 10.1186/bcr2251. [Epub 2009 Apr 30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvo A, Catena R, Noble MS, Carbott D, Gil-Bazo I, Gonzalez-Moreno O, et al. Identification of VEGF-regulated genes associated with increased lung metastatic potential: functional involvement of tenascin-C in tumor growth and lung metastasis. Oncogene. 2008;27:5373–84. doi: 10.1038/onc.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivasankaran B, Degen M, Ghaffari A, Hegi ME, Hamou MF, Ionescu MC, et al. Tenascin-C is a novel RBPJkappa-induced target gene for Notch signaling in gliomas. Cancer Res. 2009;69:458–65. doi: 10.1158/0008-5472.CAN-08-2610. [DOI] [PubMed] [Google Scholar]
- 45.Provençal M, Michaud M, Beaulieu E, Ratel D, Rivard GE, Gingras D, et al. Tissue factor pathway inhibitor (TFPI) interferes with endothelial cell migration by inhibition of both the Erk pathway and focal adhesion proteins. Thromb Haemost. 2008;99:576–85. doi: 10.1160/TH07-10-0623. [DOI] [PubMed] [Google Scholar]
- 46.Harada Y, Ozaki K, Suzuki M, Fujiwara T, Takahashi E, Nakamura Y, et al. Complete cDNA sequence and genomic organization of a human pancreas-specific gene homologous to Caenorhabditis elegans sel-1. J Hum Genet. 1999;44:330–6. doi: 10.1007/s100380050171. [DOI] [PubMed] [Google Scholar]
- 47.Donoviel DB, Donoviel MS, Fan E, Hadjantonakis A, Bernstein A. Cloning and characterization of Sel-1l, a murine homolog of the C. elegans sel-1 gene. Mech Dev. 1998;78:203–7. doi: 10.1016/s0925-4773(98)00146-4. [DOI] [PubMed] [Google Scholar]
- 48.Donoviel DB, Bernstein A. SEL-1L maps to human chromosome 14, near the insulin-dependent diabetes mellitus locus 11. Genomics. 1999;56:232–3. doi: 10.1006/geno.1998.5534. [DOI] [PubMed] [Google Scholar]
- 49.Cattaneo M, Orlandini S, Beghelli S, Moore PS, Sorio C, Bonora A, et al. SEL1L expression in pancreatic adenocarcinoma parallels SMAD4 expression and delays tumor growth in vitro and in vivo. Oncogene. 2003;22:6359–68. doi: 10.1038/sj.onc.1206665. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 51.Geismann C, Morscheck M, Koch D, Bergmann F, Ungefroren H, Arlt A, et al. Up-regulation of L1CAM in pancreatic duct cells is transforming growth factor beta1- and slug-dependent: role in malignant transformation of pancreatic cancer. Cancer Res. 2009;69:4517–26. doi: 10.1158/0008-5472.CAN-08-3493. [DOI] [PubMed] [Google Scholar]
- 52.Bergmann F, Wandschneider F, Sipos B, Moldenhauer G, Schniewind B, Welsch T, et al. Elevated L1CAM expression in precursor lesions and primary and metastastic tissues of pancreatic ductal adenocarcinoma. Oncol Rep. 2010;24:909–15. doi: 10.3892/or.2010.909. [DOI] [PubMed] [Google Scholar]
- 53.Lin L-J, Asaoka Y, Tada M, Sanada M, Nannya Y, Tanaka Y, et al. Integrated analysis of copy number alterations and loss of hetero-zygosity in human pancreatic cancer using a high-resolution, single nucleotide polymorphism array. Oncology. 2008;75:102–12. doi: 10.1159/000155813. [DOI] [PubMed] [Google Scholar]
- 54.Schneider D, Kleeff J, Berberat PO, Zhu Z, Korc M, Friess H, et al. Induction and expression of betaig-h3 in pancreatic cancer cells. Biochim Biophys Acta. 2002;1588:1–6. doi: 10.1016/s0925-4439(02)00052-2. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto K, Noshiro M, Ohno S, Kawamoto T, Satakeda H, Akagawa Y, et al. Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochim Biophys Acta. 1997;1355:303–14. doi: 10.1016/s0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- 56.Ciriza J, García-Ojeda ME. Expression of migration-related genes is progressively upregulated in murine lineage-Sca-1+c-Kit+ population from the fetal to adult stages of development. Stem Cell Res Ther. 2010;1:14. doi: 10.1186/scrt14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan I, Yong J, Dong JM, Lim L, Leung T. A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell. 2008;135:123–36. doi: 10.1016/j.cell.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Yamasaki M, Thompson P, Lemmon V. CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics. 1997;28:175–8. doi: 10.1055/s-2007-973696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pampukha VM, Kravchenko SA, Tereshchenko FA, Livshits LA, Drozhyna GI. Novel L558P mutation of the TGFBI gene found in Ukrainian families with atypical corneal dystrophy. Ophthalmologica. 2009;223:207–14. doi: 10.1159/000202645. [DOI] [PubMed] [Google Scholar]