Abstract
Organogenesis is a complex process requiring multiple cell types to associate with one another through correct cell contacts and in the correct location to achieve proper organ morphology and function. To better understand the mechanisms underlying gonad formation, we performed a mutagenesis screen in Drosophila and identified twenty-four genes required for gonadogenesis. These genes affect all different aspects of gonad formation and provide a framework for understanding the molecular mechanisms that control these processes. We find that gonad formation is regulated by multiple, independent pathways; some of these regulate the key cell adhesion molecule DE-cadherin, while others act through distinct mechanisms. In addition, we discover that the Slit/Roundabout pathway, best known for its role in regulating axonal guidance, is essential for proper gonad formation. Our findings shed light on the complexities of gonadogenesis and the genetic regulation required for proper organ formation.
Keywords: Drosophila, Gonad formation, Morphogenesis, slit, robo, Organogenesis
Introduction
A fundamental question in developmental biology is how different cell types work together to form coherent tissues and organs with the proper cellular architecture. Many of the cellular processes underlying organogenesis, such as cell migration, cell-cell recognition and contact, and the control of cell shape, are shared among different organs. However, the molecular mechanisms regulating these processes are poorly understood. Development of the Drosophila embryonic gonad involves many of these shared cellular events, yet this organ is formed from relatively few, well-defined cell types. Therefore, the Drosophila embryonic gonad provides an elegant system in which to study the various cellular processes that are required for organogenesis.
The Drosophila embryonic gonad is formed from two primary cell types: somatic cells, known as somatic gonadal precursors (SGPs), and germ cells (GCs). The GCs form at the posterior pole of the embryo early in embryogenesis and later migrate to make contacts with the SGPs (Santos and Lehmann, 2004; Starz-Gaiano et al., 2001). The SGPs are specified as three distinct bilateral clusters in the dorsolateral domain of the mesoderm in parasegments (PS) 10-12 (Boyle and DiNardo, 1995; Brookman et al., 1992; Cumberledge et al., 1992; Greig and Akam, 1995; Moore et al., 1998a; Riechmann et al., 1997; Riechmann et al., 1998; Warrior, 1994), and have distinct identities depending on from which PS they arise (Boyle and DiNardo, 1995; DeFalco et al., 2004). A fourth cluster of SGPs, the male-specific SGPs (msSGPs) is specified in PS13, but only survives to join the embryonic gonad in males (DeFalco et al., 2003).
After SGP clusters are specified, the gonad forms in several steps (Fig. 1A,B). At embryonic stage 12, individual clusters of SGPs, along with arriving GCs, move toward each other and join to form a contiguous band of cells, a process termed SGP cluster fusion. As GCs join the gonad, they are individually surrounded by SGP cellular extensions in a process called ensheathment, which separates the GCs from each other (Jenkins et al., 2003). Ensheathment continues during compaction, when the SGPs and their ensheathed GCs coalesce into a spherical structure. By embryonic stage 15, both compaction and ensheathment are complete and the embryonic gonad is fully formed (Fig. 1A,B). At this stage, both SGPs and GCs have a sex-specific identity (Camara et al., 2008) but apart from the msSGPs, the initial formation of the embryonic gonad is similar in males and females.
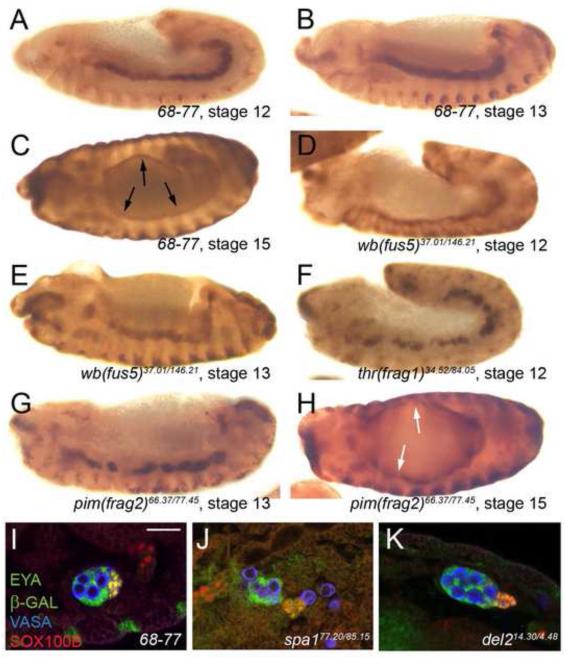
Figure 1. Gonad formation and phenotypes observed in the screen.
(A) Steps of embryonic gonad formation. SGPs, red; GCs, blue. (B) Antibody labeling of SGPs (68-77 enhancer trap, anti-β-GAL, red) and GCs (anti-VASA, blue) at indicated stages. (C-E) Embryos carrying the 68-77 enhancer trap treated by X-Galactosidase reaction. White arrows indicate β-GAL expression within SGPs. Stage as indicated. (F-N) Phenotypes observed in the screen. Stage 15 embryos immunolabeled for the 68-77 enhancer trap (anti-β-Galactosidase). Bar = 20μm. (F) Wild type embryo (68-77). Small holes in the immunohistochemical staining represent GCs. (G) Sparse gonad (en(spa2)). (H) Fragmented gonad (pim(frag2)). (I) Unfused two-clustered gonad (aop/yan(fus6)). Note the anterior cluster has no GCs incorporated. (J) Unfused SGP clusters that are contacting but not integrating together into a coherent tissue (robo2(fus4)). (K) Elongated gonad (com1). (L) Gonad with incomplete compaction (ptc(com4)). (M) Gonad with an ensheathment defect (raw(ensh)). Large holes in the immunohistochemical staining indicate location of clumped GCs, as compared to the smaller holes in panel A. (N) Embryo with misplaced gonads (LanB1(mspl)). Though both gonads are morphologically wild type, they are in different parasegments, rather than both in parasegment 10, as in wild type.
Although the steps of gonad formation are morphologically defined, the mechanisms behind these cellular movements are not well understood. Only a few genes have been implicated, including traffic jam (tj), shotgun (shg, encoding DE-cadherin, DE-CAD), and fear of intimacy (foi). TJ, a transcription factor, is expressed in the SGPs and tj mutants show disruptions in ensheathment, causing GCs to cluster together inside the gonad (Kawashima et al., 2003; Li et al., 2003). While TJ regulates levels of adhesion molecules in the adult ovary (Li et al., 2003), its mechanistic role in the embryonic gonad is undefined. DE-CAD, an adhesion molecule, is expressed in both SGPs and GCs (Jenkins et al., 2003). shg (DE-cad) mutants exhibit ensheathment defects as well as incomplete gonad compaction (Jenkins et al., 2003). Mutations in foi also result in compaction and ensheathment defects in the gonad (Van Doren et al., 2003). FOI, a zinc transporter, affects DE-CAD levels post-transcriptionally within the gonad (Mathews et al., 2006; Mathews et al., 2005). Together, these results suggest that proper regulation of cell adhesion is vital for gonad formation.
Although tj, shg (DE-cad), and foi are clearly important for gonad formation, many of the mechanisms necessary for gonadogenesis remain unclear. To identify additional regulators of gonad formation we performed a mutagenesis screen of the second chromosome, assaying gonadal cell position using a marker for SGPs. We identified twenty-four genes involved in gonad formation. Further analysis of these genes indicates that there are several unforeseen points of regulation within gonad formation, including differences between SGPs and msSGPs, DE-CAD dependent and independent mechanisms, and the unexpected involvement of the Slit/Robo pathway. These findings provide valuable insights into the formation of the Drosophila embryonic gonad and the cellular processes regulating organogenesis in general.
Materials and Methods
Fly Stocks
The following fly stocks were used: eyacli-IID, shgR64a, tjPL3, en54, thr1, Egfr2, scw5, pimIL, slit2, Df(2R)Jp5 (a slit deficiency), P[PZ]slit05248, robo1and 2, robo24 and 8 (B. Dickson; (Rajagopalan et al., 2000b), leak2, robo31 (B. Dickson; (Rajagopalan et al., 2000b), P[HS-hid] (Grether et al., 1995), and Df(2L)Exel7006 (a robo3 deficiency). Balancer chromosomes carrying a P[Kr-Gal4, UAS-GFP], P[ftz-LacZ], or P[Dfd-YFP] transgene were used to identify homozygous mutant embryos. The Bloomington deficiency kit and the Baylor P-element mapping kit (Zhai et al., 2003) were used for mapping purposes. Mapped mutations were confirmed with embryos transheterozygous for a screen allele and a previously characterized allele of the gene in question. All such combinations showed phenotypes similar to those seen with screen alleles alone. Unspecified stocks are from the Bloomington Stock Center (Indiana University, Bloomington, IN, USA).
The fly stock referred to as “68-77”, originally made by Simon (D. Godt; #FBtp0001579; (Simon et al., 1990), was manipulated to add the genetic markers aristaeless (al) and speck (sp). This was then isogenized and confirmed to exhibit no background defects in gonad formation. The resulting stock was the starting stock for the screen (Fig. S1) and was used as the wild type control unless stated otherwise.
Mutagenesis and Screening
The al 68-77 sp starting stock was mutagenized using ethane methyl sulfonate (EMS) and homozygous mutant embryos were generated using the scheme outlined in figure S1. Gonad formation was analyzed by X-galactosidase (X-Gal) staining to reveal the ß-galactosidase activity present in the SGPs. Mutagenesis, use of heat-shock-head involution defective (HS-hid) transgene, embryo collection, embryo fixation, X-Gal reactions, and cuticle preparations were all carried out as previously published (Moore et al., 1998b). During the screening process, an average lethality rate of 89.4% was observed. This corresponds to, via Poisson distribution, an average “hit rate” of 2.24 lethals per fly line, and over seven-fold coverage of the second chromosome. Our screen therefore achieved theoretical saturation. Mutant lines were placed into complementation groups by testing for trans-heterozygous lethality and confirming each group’s gonad defect through X-Gal staining.
Immunohistochemistry and Microscopy
All analyses were carried out with embryos trans-heterozygous for two of the strongest alleles available from the screen (Table S1), however, since screen alleles are uncharacterized, the observed phenotypes may not represent the null condition in all cases. Anterior is left in all figures.
Embryo fixation and immunostaining against SLIT, ROBO, ROBO2, and ROBO3 were as in Simpson et al. (Simpson et al., 2000b). All other fluorescent antibody labeling was as previously described (Le Bras and Van Doren, 2006). Non-fluorescent immunolabelings, done as in Eldon and Pirrotta (Eldon and Pirrotta, 1991), were visualized by horseradish peroxidase reactions via a biotinylated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) and the ABC Elite Kit (Vector Labs, Burlingame, CA, USA) using DAB (3′3′-diaminobenzidine) as a substrate (Vector Labs).
Fluorescent immunolabelings were mounted in 70% glycerol with 2.5% DABCO (Sigma-Aldrich, St. Louis, MO, USA) and analyzed with Zeiss LSM 510 Meta or Confocor systems. Non-fluorescent immunolabelings were mounted in 70% glycerol or Aquatex (EMD Chemicals, Gibbstown, NJ, USA) and viewed on a Zeiss Axioskop using a Coolsnap camera with RS image software (Roper Scientific (now Photometrics), Tucson, AZ, USA). Embryos were staged by gut morphology, according to Campos-Ortega and Hartenstein (Campos-Ortega and Hartenstein, 1997).
Antibodies used were as follows (dilution, source): mouse anti-β-Galactosidase (GAL) (1:10000, Promega, Madison, WI, USA); rabbit anti-β-GAL (1:10000, Cappel); mouse anti-EYA 10H6 (1:25, Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA, USA), S. Benzer/N.M. Bonini); rabbit anti-SOX100B (1:1000, 1:2000 for histochemical labelings, S. Russell); rat anti-DE-CAD DCAD2 (1:20, DSHB, T. Uemura); mouse anti-FASIII 7G10 (1:30, DSHB, C. Goodman); chick anti-VASA (1:10000, K. Howard); rabbit anti-VASA (1:10000, R. Lehmann); guinea pig anti-TJ (1:5000, D. Godt); mouse anti-SLIT C555.6D (1:10, DSHB, S. Artavanis-Tsakonas); mouse anti-ROBO 13C9 (1:10, DSHB, C. Goodman); mouse anti-ROBO2 (1:200, B. Dickson; (Rajagopalan et al., 2000b); mouse anti-ROBO3 extracellular 14C9 (1:10, DSHB, C. Goodman); mouse anti-GFP (1:50, Santa Cruz); rabbit anti-GFP (1:2000 for fluorescent labelings, 1:20000 for histochemical labelings, Torrey Pines, East Orange, NJ, USA); rabbit anti-Zfh-1 (1:5000, R. Lehmann); and mouse anti-Neurotactin BP106 (1:10, DSHB, C. Goodman). Biotin conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA), as well as Alexa 488, 546, or 633 (Molecular Probes (Invitrogen), Carlsbad, CA USA) conjugated secondary antibodies were used at 1:500.
Results
Identification of new regulators of Drosophila embryonic gonad formation
To discover new genes involved in early gonad formation we performed a large-scale EMS mutagenesis screen of the second chromosome. We used an enhancer trap line (68-77) that expresses lacZ in SGPs (Boyle and DiNardo, 1995; Simon et al., 1990; Warrior, 1994), enabling efficient visualization of the gonad (Fig. 1B, C-E, S2), and allowing us to bias the screen towards genes that affect gonad morphogenesis and against mutations that block SGP specification. Embryos from over 7,000 uniquely mutagenized fly lines (Fig. S1) were analyzed by ß-Galactosidase staining, and 269 lines exhibiting perturbations in gonad morphology were maintained. Of these lines, 131 were found to be linked to the second chromosome and exhibited their respective phenotypes with at least 30% penetrance. Complementation testing placed 80 of the 131 mutant lines into 24 complementation groups, each named according to its predominant phenotype (Table S1). Thus, we identified 24 different second chromosome genes with roles in gonad formation. The remaining 51 lines have phenotypes either caused by synthetic effects or are the only identified allele of that gene. Due to complications related to working with possible synthetic effects, no further analysis was performed on these lines.
Mutations affect different stages of gonad formation
A wide range of gonad phenotypes was observed in the lines established in the screen, however, most resulted from disruptions in the known steps of gonad formation. The following phenotypes were found:
Sparse SGPs
(Groups spa1, engrailed (en, spa2), spa3) These embryos had fewer SGPs than normal. Their gonads were often fragmented, perhaps because there were too few SGPs present to form a coherent cluster (Fig. 1G). Low numbers of SGPs could indicate a problem with the initial specification of SGPs or with survival of the SGPs after their specification.
Fragmented gonads
(Groups three rows (thr, frag1), pimples (pim, frag2)) Fragmented gonads had loosely gathered SGPs (Fig. 1H). This could result from either a loss of cell adhesion within individual SGP clusters or a disruption in the initial location of individual cells.
SGP cluster fusion defects
(Groups ribbon (rib, fus1), fus2, slit(fus3), roundabout2 (robo2/leak, fus4), wing blister (wb, fus5), anterior open (aop/yan, fus6), longitudinals lacking (lola, fus7)) Gonads showing this phenotype had at least one SGP cluster that did not fuse with other SGPs. Clusters were either fully separated (Unfused, Fig. 1I) or in contact with one another but not fully integrated together (Contacting, Fig. 1J).
Compaction defects
(Groups com1, com2, com3, patched (ptc, com4), shg/DE-cad(com5)) These gonads had completed SGP cluster fusion, but failed to condense completely into the round shape characteristic of the mature embryonic gonad. There were two types: elongated gonads, which remained as the original band of tissue that formed after SGP cluster fusion (Elongated, Fig. 1K), and those which had condensed slightly but were misshapen (Uncompacted, Fig. 1L). Misshapen gonads ranged from having skinny or bulbous extensions, to simply being too oblong.
Ensheathment defects
(raw(ensh)) In gonads with ensheathment defects, SGPs failed to wrap individual germ cells. Consequently, the unensheathed germ cells were associated in larger clusters, rather than being separated by the SGPs (Unensheathed, Fig. 1M).
Misplaced gonads
(Laminin B1 (LanB1, mspl)) Gonads showing this defect were often morphologically indistinguishable from wild type gonads, but they were not located in PS10, as in wild type (Fig. 1N). Interestingly, bilateral gonads were not necessarily in the same segment with respect to each other, indicating that gonad positioning was independently established for each gonad.
Delayed gonad formation
(eyes absent (del1, eya), del2, del3) Gonads in embryos from these groups demonstrated formation defects at the earlier time points of gonad formation, but were able to fully form by later stages of embryogenesis, giving the appearance of a delayed time frame of gonad formation.
Morphological Defects
(Epidermal growth factor receptor (Egfr, mor1), screw (scw, mor2)) Embryos in these groups demonstrated gonad formation defects, but also exhibited severe morphological defects that precluded staging of embryos.
Analysis of gonad defects immediately revealed that many complementation groups exhibited more than one phenotype. In order to characterize this surprising phenotypic range and determine the predominant phenotype present in each of the twenty-four complementation groups, the occurrence of gonad phenotypes was scored at embryonic stages 13/14, 15, and 16/17 (Fig. 2). Results showed that each complementation group exhibited a primary defect affecting one aspect of gonad formation, such as SGP cluster fusion (Unfused and Contacting) or gonad compaction (Elongated and Uncompacted). For comparison, shgR64a/2 (DE-cad) embryos were also quantitated, with results as follows: for stages 13/14 (n=114), 15 (n=41), and 16/17 (n=49), respectively, 39%, 58.5%, and 44.9% of mutant embryos displayed abnormal gonads (38.6%, 4.9%, and 2.0% fusion defects, and 0.9%, 53.6%, and 42.9% compaction defects at each stage).
Figure 2. Graph of phenotypic penetrances for each group.
The phenotypic range of each group at stages 13/14, 15, and 16/17. Each bar represents the percentage of gonads within that group showing the indicated phenotype. Every group shows a wide variety of phenotypes, though each was named according to its predominant defect. Groups denoted by a single asterisk were unable to be staged during late embryogenesis due to defects in embryonic morphology. Those denoted by a double asterisk were unable to be staged during all stages of embryogenesis pertinent to gonad formation, and the results were grouped together. Gonads cannot be considered to have compaction defects until stage 15 when a wild type gonad would be fully compacted, however, some younger gonads may be classified as having compaction defects if their gonads appear to be compacting incorrectly. Unstg = Unstageable
Ensheathment defects can occur independently of other gonad phenotypes (Jenkins et al., 2003; Li et al., 2003; Mathews et al., 2006) and thus were independently assessed in all groups. Ensheathment was the primary defect in one group (raw(ensh), Fig. 1M), and strong ensheathment defects were seen in five additional groups (Table S2). Twelve groups showed partial ensheathment defects but exhibited them transiently or had other gonad defects that made it difficult to assess ensheathment properly, while six groups exhibited no signs of ensheathment problems (Table S2).
Overall, each phenotype observed in the screen was found to be predominate in at least one mutant group (Table S2), indicating we have identified regulators of each step of gonad formation. Some groups also exhibited altered embryo morphology severe enough to interfere with staging. Two of the most severely effected, mor1 and 2, were mapped to Egfr and screw, respectively. All groups except one were embryonic lethal, but despite any defects in embryo morphology, all groups survived long enough to produce an external cuticle which forms after gonad formation is complete. In most cases the cuticles revealed only subtle or no patterning defects (data not shown). Thus, for most of our complementation groups, the gonad defects were not due to early lethality or gross defects in embryonic patterning.
Surprisingly, many complementation groups exhibited a more severe phenotype at stage 14, when the gonad initially forms, than at later stages (Fig. 2). This compensation is most strongly seen in three groups, eya(del1), del2, and del3, where malformed gonads “rescue” to a nearly wild type appearance by later stages, causing gonad formation to appear delayed (Fig. 2). This may indicate the existence of parallel pathways regulating gonad morphogenesis, such that when one pathway is debilitated another pathway can ultimately compensate.
SGP specification is unaffected in most groups
To determine if our screen mutants exhibited gonad phenotypes due to defects in SGP identity, we assayed EYA, a transcription factor essential for SGP specification (Boyle et al., 1997). EYA expression normally begins in all SGPs at embryonic stage 12 and is later restricted to the posterior of the gonad (Boyle et al., 1997; Boyle and DiNardo, 1995). Four complementation groups exhibited defects in EYA expression. Further analysis revealed that one of these groups (del1) represents mutations in eya itself. Though eya is required for SGP identity, previous work has shown that in the absence of eya, some SGPs still initially express some SGP markers and interact with GCs, but proper gonads do not form in strong eya mutants (Boyle et al., 1997). Our eya alleles likely represent hypomorphic mutants that delay but do not prevent gonad formation. Another group, spa2, represents mutations in engrailed, which is known to influence SGP specification (Riechmann et al., 1998). In the other two groups (thr(frag1) and spa3) EYA was expressed normally at stage 13, but was no longer detected at stage 15 (data not shown), indicating that these genes may be required for maintenance of SGP identity or SGP survival. In all remaining groups, EYA expression was normal (Table S2), indicating that improper SGP identity was not the primary cause of most phenotypes revealed in the screen.
Most gonad defects are not due to disruptions in mesodermal patterning
Since SGPs are specified from the mesoderm, we investigated whether the gonad phenotypes we observed reflected general defects in mesodermal patterning and morphogenesis (Azpiazu and Frasch, 1993; Patel et al., 1987). For this we used the marker FasIII to examine the visceral mesoderm which, like the SGPs, is specified as individual clusters of mesodermal cells that must join together to form a contiguous tissue (Azpiazu and Frasch, 1993; Campos-Ortega and Hartenstein, 1997). Embryos in seven groups exhibited visceral mesoderm defects; slit(fus3) and wb(fus5) mutants had minor morphological defects (Fig. 3D,E and not shown), com3 mutants lost FASIII expression (data not shown) and thr(frag1), pim(frag2), screw(mor2), and spa3 mutants all had breaks in their FASIII expression (Fig. 3F-H and not shown), most likely the result of incomplete visceral mesoderm cluster fusion. These seven mutations, therefore, cause defects within at least two mesodermal tissues, the visceral mesoderm and the gonad. Embryos in the seventeen remaining groups, however, exhibited normal FASIII expression in the visceral mesoderm (Table S2). Thus, most gonad defects are not caused by broad disruptions in the entire mesoderm. This also suggests that even though both the gonadal mesoderm and the visceral mesoderm undergo a process of cluster fusion, the cellular mechanisms involved likely require different genetic regulation.
Figure 3. Visceral mesoderm and msSGP phenotypes observed in gonad mutants.
(A-H) Visceral mesoderm phenotypes. Embryos immunolabeled for the visceral mesoderm marker FASIII (anti-FASIII), stages and genotypes indicated. (A-C) In wild type the visceral mesoderm is a smooth, coherent tissue at stages 12 (A) and 13 (B), and has surrounded the gut by stage 15 (black arrows, C). (D, E) The visceral mesoderm in wb(fus5) mutants is fused into a coherent tissue at both stage 12 (D) and 13 (E), but is thinner than normal, and jagged in appearance. (F-H) The visceral mesoderm in thr(frag1) (F) and pim(frag2) (G, H) mutants does not completely fuse, leaving breaks in the tissue. At younger stages (F, G), some visceral mesoderm clusters appear to be contacting each other but have not integrated together. (H) The visceral mesoderm is able to surround the gut by stage 15 in pim(frag2) mutants, but several thin spots or even holes are seen in the tissue (white arrows). (I-K) Examples of msSGP phenotypes. Embryos immunolabeled for SGPs (68-77 enhancer trap, anti-β-Galactosidase, and anti-EYA, both green), msSGPs (anti-SOX100B, red and anti-EYA, green – appear yellow) and GCs (anti-VASA, blue). Stage 15 embryos, all male. Bar = 20μm. (I) Wild type embryo. msSGPs have joined and integrated into the gonad. (J) A gonad whose msSGPs failed to join (spa1). (K) A gonad with msSGPs that have joined but are loosely gathered (del2).
msSGPs and SGPs exhibit parallel as well as distinct regulatory mechanisms
msSGPs are specified independently of the SGPs (DeFalco et al., 2004; DeFalco et al., 2003) and therefore may respond to genetic disruptions in unique ways. We analyzed the msSGPs in all complementation groups using an msSGP marker, SOX100B, and identified msSGP defects in fourteen groups. Six groups had msSGPs that often failed to join the gonad and remained as an independent cluster of cells within the mesoderm, similar to a cluster fusion defect (Fig. 3J). Five groups had msSGPs that consistently joined the SGPs but were loosely packed and failed to condense into the tissue, similar to a compaction defect (Fig. 3K). Finally, three groups, rib(fus1), aop/yan(fus6), and spa3, appeared to not specify msSGPs (based on co-expression of EYA and SOX100B, >50 embryos scored per genotype). In the ten remaining groups, however, msSGPs joined the gonad normally despite obvious defects in other aspects of gonad formation. Thus, the mechanisms regulating msSGP development and morphogenesis can clearly be separated from those regulating the SGPs.
DE-cadherin-independent regulation of gonad formation
The cell adhesion molecule DE-CAD has an important role in gonad formation. shg (DE-cad) mutants have defects in gonad compaction and germ cell ensheathment (Jenkins et al., 2003; Van Doren et al., 2003), and indeed, one of the complementation groups we identified in our screen (com5) represents new alleles of shg (DE-cad). Therefore, we wanted to determine if DE-CAD expression and localization were affected in any other mutants identified in the screen. We found that many of our groups exhibited defects in DE-CAD levels or localization. Seven groups showed low levels, or in some cases a complete absence of DE-CAD expression in the gonad (Fig. 4B). In six groups, DE-CAD protein was localized in punctae that often appeared to be intracellular, as opposed to the uniform cell surface labeling characteristic of wild type (Fig. 4C). And four groups showed disorganized DE-CAD (Fig. 4D), where the protein also appeared more intracellular than wild type, but was not as punctate as in Fig. 4C.
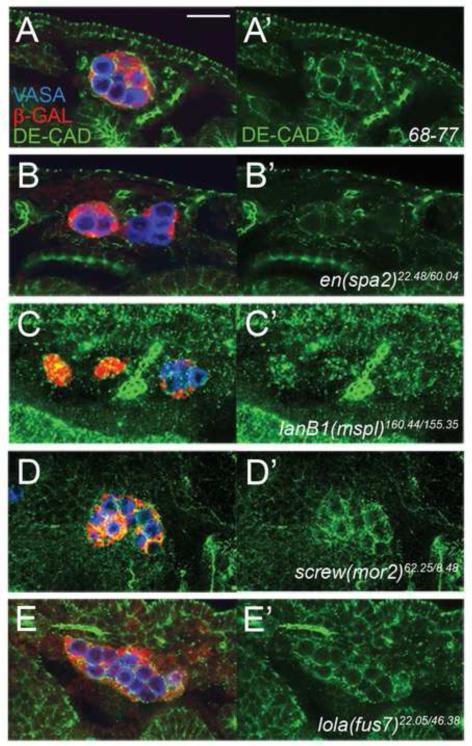
Figure 4. DE-CAD expression and localization in screen mutants.
Examples of disruptions in DE-CAD localization and/or expression. Stage 15 embryos immunolabeled for DE-CAD (green), the 68-77 enhancer trap (anti-β-Galactosidase, red), and GCs (anti-VASA, blue). Bar = 20μm. (A) In wild type, DE-CAD is localized to the plasmalemma of gonad cells. (B) Gonad with greatly reduced DE-CAD expression (en(spa2)). (C) Gonad where DE-CAD is more punctate than normal (LanB1(mspl)). (D) Gonad where DE-CAD is present but disorganized (screw(mor2)). Note the co-localization of DE-CAD and β-GAL labeling in panels C and D, indicating that some DE-CAD is intracellular. (E) Some mutations cause defects in gonad formation but do not disrupt DE-CAD expression and localization (lola(fus7)).
For some of the groups in which the pattern of DE-CAD localization was affected, the mutant gonad phenotype resembled partial or complete loss of zygotic shg (DE-cad) function (e.g. com1, com2, del2, Egfr(mor1)), suggesting that the affects on DE-CAD may be the primary cause of the gonad defect. However, some of these groups exhibited a mutant phenotype that is different or more severe than that of shg (DE-cad) mutants (e.g. rib(fus1), thr(frag1), spa1, screw(mor2)). These mutants may affect the function of maternally contributed DE-CAD protein (e.g. by regulating cell surface levels), or affect other aspects of gonad formation in addition to DE-CAD.
Interestingly, we also found that seven of our groups exhibited normal DE-CAD expression and localization despite having mutant gonads (Fig. 4E). Thus, DE-CAD expression is not necessarily disrupted simply as a consequence of the failure to form a gonad. Further, while previous work clearly indicated the importance of DE-CAD in gonad formation, these groups indicate that there are pathways regulating this process independently of DE-CAD.
The Slit/Robo pathway plays a role in gonad formation
The screen also revealed the surprising involvement of the Slit/Robo pathway in gonadogenesis; the complementation groups fus3 and fus4 carry mutations in the genes slit and roundabout2 (robo2, also known as leak), respectively. (Thus, the fus3 group alleles become slit5.05 and slit58.08, and the fus4 alleles leak58.22 and leak112.03. See Supplemental Table 1.) SLIT, a secreted protein (Rothberg and Artavanis-Tsakonas, 1992; Rothberg et al., 1990), regulates cell migration and axonal pathfinding, and can act as either an attractive or repellent signal depending on the cell type (Englund et al., 2002; Kramer et al., 2001). SLIT guides and positions tissues from both short and long distances (Kolesnikov and Beckendorf, 2005; Rajagopalan et al., 2000a; Rajagopalan et al., 2000b; Simpson et al., 2000a; Simpson et al., 2000b), including axons in the developing CNS (Kidd et al., 1999), the trachea (Englund et al., 2002), salivary glands (Kolesnikov and Beckendorf, 2005), and heart tube (MacMullin and Jacobs, 2006; Qian et al., 2005; Santiago-Martinez et al., 2006). SLIT is a ligand for the Roundabout (Robo) family of receptors (Brose et al., 1999; Kidd et al., 1999). In Drosophila, there are three Robos: ROBO, ROBO2 (or LEAK), and ROBO3 (Kidd et al., 1999; Rajagopalan et al., 2000b; Simpson et al., 2000b).
We observed that mutants for slit or robo2 exhibited unfused SGP clusters at stage 13 (Figs 5C,G, 6), perhaps indicating a role in guiding SGP cluster movement. By stage 15, SGP clusters had fused, but gonads then failed to compact properly (Figs 5D,H, 6). Although robo and robo3 were not identified in our screen, we found that mutants for these genes exhibited gonad compaction defects (Figs 5E,F,I,J, 6). Additionally, mutants for any of these four genes exhibited ensheathment defects (Fig. 5D,F,H,J and Table 1).
Figure 5. slit, robo, robo2, and robo3 mutants exhibit gonad malformations.
Embryos immunolabeled for the 68-77 enhancer trap (anti-β-Galactosidase, red), Neurotactin (NRT, anti-NRT, green), and GCs (anti-VASA, blue). NRT, a cell surface protein expressed strongly in SGPs but only weakly in GCs, provides an indication of the extent of ensheathment (Jenkins et al., 2003). Embryonic stages and genotypes are indicated, except that one copy of the 68-77 enhancer trap was also present in each genotype. Bar = 20μm. Insets are NRT only from within boxed area. Both slit and robo2 mutants exhibit cluster fusion defects at stage 13 (C, G). All four mutant genotypes display compaction defects and ensheathment defects at stage 15 (D, F, H, J).
Figure 6. Graph of phenotypic penetrance of gonad phenotypes in slit/robo mutants.
Phenotypic variability of each group at stages 13/14, 15, and 16/17. Bars represent the percentage of gonads with each given genotype showing indicated phenotype.
Table 1.
Ensheathment penetrance of slit/Robo mutants
| Genotype |
Percentage of gonads exhibiting
unensheathed GCs (n) |
|---|---|
| 68-77 | 14.1 (220) |
| raw(ensh)134.47/ raw(ensh)155.27 | 56.4 (110) |
| slit2/ Df(2R)Jp5 | 20.1 (249) |
| robo1/ robo2 | 29.6 (243) |
| robo24/ robo28 | 28.0 (168) |
| robo31/ Df(2L)Exel7006 | 31.8 (214) |
The penetrance of cluster fusion or compaction defects in slit mutants was higher than that for mutations in any individual robo (Fig. 6), consistent with the multiple Robo receptors acting in a partially redundant manner in response to the SLIT ligand. While double mutants between Robo and either Robo2 or Robo3 did not exhibit an increased penetrance of gonad formation defects (Fig. 6), the proximity of Robo2 and Robo3 precluded analysis of that double mutant or the triple mutant. Interestingly, the penetrance of ensheathment defects in robo, robo2, and robo3 mutants individually was higher than that of slit mutants (Table 1) suggesting that Robo proteins may have some function that is independent of SLIT for this process. It has been previously demonstrated that the Robos can act independently of SLIT as adhesion molecules (Hivert et al., 2002). Again, the possible double mutant combinations among the Robo’s and between the Robos and slit did not exhibit a significant change in penetrance over the single mutants.
To further explore the role of the Slit/Robo system in gonad formation, we determined the expression pattern of the Robos within the gonad. ROBO was observed in the gonad at stage 13 (Fig. 7A), and accumulated to higher levels within the gonad as coalescence occurred (Fig. 7B). It was found between SGPs, but was most strongly localized around individual GCs. Since GCs are ensheathed by SGPs (Jenkins et al., 2003), this immunofluorescence may represent the interface between SGPs and GCs (Fig. 7B) and suggests that ROBO functions in cell contacts between SGPs and GCs. ROBO2 was expressed in the gonad as well as in several immediately surrounding tissues including the trachea (as previously reported (Parsons et al., 2003)). Its expression levels within the gonad increased as the gonad formed (Fig. 7C,D). ROBO2 was observed at contact sites between SGPs as well as between SGPs and GCs (Fig. 7D). This suggests it is important for both types of cell-cell contacts, and likely explains the greater phenotypic penetrance and severity of robo2 mutants than that of robo and robo3 mutants (Figs 5F,H,J, 6). Anti-ROBO3 labeling was also observed within the gonad, however, the immunoreactivity did not appear localized to the cell surface, and so was inconclusive (data not shown).
Figure 7. ROBO, ROBO2, and ROBO3 are found in the gonad, while SLIT is not zygotically expressed in the gonad.
Embryonic stages and genotypes are indicated. Bar = 20μm. Insets represent green channel only from within boxed area. Antibodies were previously shown to identify their respective antigens, and immunoreactivity disappeared in mutants for robo (Kidd et al., 1998), robo2 (Rajagopalan et al., 2000b), or slit (Rothberg et al., 1988), respectively. (A-F) Embryos immunolabeled for SGPs (68-77 enhancer trap, anti-β-Galactosidase, red), GCs (anti-VASA, blue), and either anti-ROBO (A, B) or anti-ROBO2 (C, D) (green). (A, B) ROBO is present in the gonad at low levels at stage 13 (A), but accumulates to higher levels as the gonad compacts (B). It appears most strongly localized between SGP and GCs. (C, D) ROBO2 is found in the gonad at stage 13 (C), and it is present at slightly higher levels at stage 15 (D). ROBO2 localizes to interfaces between SGPs and SGPs (arrows) as well as SGPs and GCs (D). (E, F) slit enhancer trap expression. Embryos labeled for the enhancer trap (anti-β-Galactosidase, green), SGPs (anti-TJ, red), and GCs (anti-VASA, blue). The lack of β-GAL in or near the gonad indicates that slit is not expressed by gonadal cells. (G, H) SLIT localization. Embryos immunolabeled for SGPs (anti-ZFH-1, red), GCs (anti-VASA, blue), and SLIT (green). SLIT is found in punctae in the wall of the gut as well as at muscle attachment sites in the ectoderm (as in (Rothberg et al., 1990). Immunoreactivity is observed in the GC nuclei but does not represent zygotic expression of slit, as this labeling was still present in embryos homozygous for a slit deletion. It is unclear whether this nuclear staining represents maternal protein or cross-reactivity.
SLIT, being a secreted protein, can act locally or from a distance (Kolesnikov and Beckendorf, 2005; Rajagopalan et al., 2000a; Rajagopalan et al., 2000b; Simpson et al., 2000a; Simpson et al., 2000b). To determine which occurs here, we examined slit expression in the context of the gonad. A slit enhancer trap, P[PZ]slit05248, showed expression within the CNS midline, ectoderm, and gut as previously reported (Rothberg et al., 1990), but no expression was observed within the gonad or any immediately surrounding tissue (Fig. 7E,F). Similarly, an anti-SLIT immunoserum showed the presence of SLIT at the midline, ectoderm and gut, but not within the SGPs or in tissues surrounding the gonad (Fig. 7G,H). While SLIT is not expressed within the gonad, all three Robos are. Thus, SLIT likely acts on the gonad from a distance to help drive SGP cluster fusion and gonad compaction.
Discussion
The formation of the Drosophila embryonic gonad relies upon the interaction of multiple cell types to establish a functional organ. Though the morphogenetic changes involved in gonadogenesis have been previously described (Jenkins et al., 2003), the regulation of these interactions is poorly understood; only a few genes have been implicated in the process. Thus, to increase our knowledge of the mechanisms underlying gonad formation, we performed an EMS mutagenesis screen, revealing 24 genes involved in regulating gonadogenesis. Analysis of these genes enhances our understanding of gonad formation and provides new insights into the cellular mechanisms and genetic interactions required for the development of this organ.
Of the genes we identified, only three, shg (DE-cad), eya, and en had previously known roles in gonad formation or SGP specification. While their identification was not unexpected, the discovery of new mutations in these genes confirmed the screen was aptly designed to isolate mutations affecting gonad development. Missing from this list, however, are some additional genes on the second chromosome which exhibit defects in gonad formation, including tj (Li et al., 2003), robo and robo3.
Previously, all known genes with roles in gonadogenesis either had disrupted DE-CAD within the gonad (foi (Jenkins et al., 2003; Mathews et al., 2006)) or were known to regulate DE-CAD in other contexts (tj (Li et al., 2003)). Thus, DE-CAD is clearly an important player in gonad development. This is supported by our identification of several new mutants affecting DE-CAD expression or localization within the gonad. However, we also identified a number of new mutants that affect gonad formation without affecting DE-CAD expression. Thus, there are likely to be DE-CAD-independent mechanisms regulating gonad formation as well.
Analysis of mutant phenotypes
The genes we have discovered are likely to play a direct role in gonad formation, and the phenotypes observed are unlikely to be due to secondary defects in other aspects of development. Our mutant phenotypes are not due to early embryonic lethality, and most mutants did not disrupt SGP identity or development of a neighboring mesodermal tissue, the visceral mesoderm. While we might have expected to find some genes that are important for more general processes such as mesoderm development or cell migration, most of our mutants do not have global defects in other tissues.
The range of gonad phenotypes observed within each group was rather wide (Fig. 2). This pleiotropy suggests that some steps of gonad formation are not as genetically separable as was originally expected. Although every complementation group exhibits one predominant gonad defect, no single mutation shows only SGP cluster fusion defects or only compaction defects. One explanation is that the disruption of a cellular function, such as adhesion between SGPs, may affect multiple aspects of gonad morphogenesis. Thus, a mutation in a gene acting in SGP adhesion might exhibit defects in both SGP cluster fusion and gonad compaction. Another potential reason for the pleiotropy found in the screen may be that later steps of gonad formation are dependent upon the timely completion of earlier steps. This would explain why mutations primarily affecting SGP cluster fusion often also exhibit defects in compaction, while mutations primarily affecting compaction have fewer defects in SGP cluster fusion (Fig 2).
On the other hand, it is possible to identify mutations that affect ensheathment without affecting other aspects of gonad formation (e.g. tj (Li et al., 2003) and raw(ensh) (this work)). This may be because ensheathment involves a unique cell shape change within SGPs that is not required for other steps of gonad formation. Also, it represents a specific interaction between GCs and SGPs, whereas other steps in gonad formation are primarily dependent on SGPs and can occur in the absence of GCs (Brookman et al., 1992).
Lastly, our data indicate there are parallel genetic pathways affecting gonad formation. In most groups, the percentage of mutant gonads decreases in late stages of embryogenesis (Fig. 2), demonstrating that even when a gonad has failed to form within the normal time frame, it can often form properly at a later stage. It is possible that some aspects of this delay/rescue phenotype are because some of the alleles we have identified may not be null alleles. Overall, however, it suggests that there are redundant pathways at work, such that debilitation of one pathway leads to defects in the timing and fidelity of gonad formation that can be compensated for by another pathway. Consistent with the idea of multiple pathways, we find that some mutations act by affecting DE-cadherin expression or localization, while others do not.
msSGPs mimic SGP behaviors, but are regulated independently
The mutations identified in our screen also provide further support for the unique nature of the msSGPs. SGP and msSGP identity appear similar in some ways, in that they express some of the same molecular markers and exhibit a similar process of cluster fusion to join the gonad (DeFalco et al., 2003). However, msSGPs are distinct from SGPs, as they are specified from a different mesodermal location and do not require some of the genes required for SGP identity, such as eya (DeFalco et al., 2003). Thus, it was unclear whether the morphogenesis of the msSGPs would be regulated in the same manner as the SGPs.
Indeed, we find that there are clear differences in the genetic regulation of how the SGPs and msSGPs join the gonad. On one hand, the msSGPs sometimes exhibit defects similar to the SGPs for a particular mutation. For example, some groups with SGP cluster fusion defects have msSGPs that fail to join the gonad, and some groups with compaction defects also have msSGPs that loosely attach to the gonad (e.g. ptc(com4), fus2). Thus, some genes may regulate SGPs and msSGPs in a coordinated manner. On the other hand, the msSGPs sometimes exhibit defects different than the SGPs. For example, some groups with compaction defects have msSGPs that fail to join the gonad (e.g. com1, com2). There are also clear examples of mutations that strongly affect the SGPs, but have no apparent affect on the msSGPs (e.g. com3, raw(ensh), lola(fus7)). Thus, the regulation of msSGP morphogenesis can be distinct from that of the SGPs, strengthening the conclusion that these two cell types have distinct identities and developmental programs.
The involvement of the Slit/Robo pathway in gonad formation
Unexpectedly, the screen revealed the role of the Slit/Robo pathway in gonadogenesis. Although all three Robos function in gonad formation, their phenotypes and localization patterns suggest that each one has a slightly different role in the process. ROBO2 is the only Robo expressed at detectable levels as SGP cluster fusion occurs, as well as the only robo gene that causes a substantial cluster fusion defect when mutated (Figs 5G, 6, 7C). Therefore, ROBO2 appears to be the principal Robo protein mediating cluster fusion. Once the SGP clusters have merged, all three Robos contribute to gonad compaction and ensheathment, as all exhibit defects in these processes when mutant (Figs 5, 6, 7, Table 1). Though their mutant phenotypes are similar, ROBO and ROBO2 have slightly different localizations, with ROBO more prominent between SGPs and GCs, and ROBO2 between SGPs as well as between SGPs and GCs. This suggests potentially distinct functions, and would be consistent with other reports that the Robos perform separate functions (Englund et al., 2002; Parsons et al., 2003; Rajagopalan et al., 2000a; Rajagopalan et al., 2000b; Simpson et al., 2000a; Simpson et al., 2000b). On the other hand, slit mutants have stronger compaction and SGP cluster fusion defects than mutants for any one robo (Fig. 6), suggesting SLIT is the required ligand for Robo function, and that there is also some functional redundancy between the different Robo receptors during these aspects of gonad formation.
Though slit mutants demonstrate a gonad phenotype, no zygotic SLIT expression was detected within or immediately surrounding the embryonic gonad. Therefore, SLIT could be acting upon the gonad from a distance. Potential sources of SLIT include the ectoderm at muscle attachment sites and the walls of the gut (Fig. 7E-H, and (Rothberg et al., 1990)), both of which are adjacent to the mesoderm where the gonad is located. These locations suggest that SLIT repels migrating SGPs away from these surrounding tissues, preventing SGPs from exploring too far medially or laterally, and in effect guiding SGP clusters together and helping them to condense together during compaction. In an effort to explore this model further, we expressed SLIT in various tissues surrounding the gonad, however, these experiments gave ambiguous results which neither refuted nor supported our model of SLIT as a guidance factor in gonadogenesis. Rather, these results suggested that the amount of SLIT present was more important than the location. Thus, it is also possible that SLIT provides a permissive signal to gonadal cells, rather than a directionally instructive one.
The Robos may also contribute to gonad formation through adhesive mechanisms; ROBO and ROBO2 exhibit both homophilic and heterophilic adhesion properties (Hivert et al., 2002; Simpson et al., 2000b). Studies with shg (DE-cad) or foi mutants indicate that a loss of adhesion in the gonad can account for incomplete compaction and ensheathment (Jenkins et al., 2003; Mathews et al., 2006). Therefore, the defects observed in mutants for the robo genes may be due to a disruption in Robo-mediated adhesion within the gonad. Robo adhesion can occur independently of SLIT (Hivert et al., 2002) and would therefore account for the SLIT independent nature of ensheathment.
The Slit/Robo pathway may also influence cadherin-mediated adhesion in the gonad. While some studies have demonstrated a negative relationship between cadherin-based adhesion and Slit/Robo signaling (Rhee et al., 2007; Rhee et al., 2002; Santiago-Martinez et al., 2008), Slit/Robo have also been observed to upregulate cadherins (Prasad et al., 2008; Shiau and Bronner-Fraser, 2009). While we have not observed a significant change in DE-CAD expression in Slit/Robo pathway mutants, it remains possible that these two important mechanisms for regulating gonad formation are cooperating in this process.
Supplementary Material
Supplemental Figure 1 – Screen Genetics. Mutant lines from our screen were created using the genetic scheme outlined here. See materials and methods for more details.
Supplemental Figure 2 – Balancer chromosome ftz-lacZ expression. Sample embryo from our screening process treated by X-Galactosidase reaction. This embryo carries both the 68-77 enhancer trap (gonad expression denoted by the white arrow) and the ftz-lacZ construct (denoting the presence of the balancer chromosome, black arrow). Compare with embryos in Fig. 1C-E which all lack the ftz-lacZ balancer construct.
Research Highlights.
A mutagenesis screen identifies 24 genes involved in gonad formation
These genes preferentially affect the behavior of the somatic gonad rather than the germ cells
Gonad formation is regulated by both DE-cadherin-dependent and independent pathways
The slit/robo pathway is essential for organogenesis of the gonad
Acknowledgments
We are grateful to the numerous colleagues who have generously supplied us with fly stocks and reagents for this work, and we have specifically acknowledged them within the Materials and Methods. We also acknowledge the Bloomington Stock Center (Indiana University) and the Developmental Studies Hybridoma Bank (University of Iowa) for providing reagents, and FlyBase (Tweedie et al., 2009) for providing essential information. We thank the JHU Integrated Imaging Center for assistance in confocal microscopy, the Kramer and Dickson labs for assistance and advice on Slit/Robo, and Marnie Halpern and members of the Van Doren Lab for helpful discussions and critical reading of the manuscript. This work was supported by the National Institutes of Health (HD46619) to M.V.D. and (5F32GM085925) to J.C.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arora K, Levine MS, O’Connor MB. The screw gene encodes a ubiquitously expressed member of the TGF-beta family required for specification of dorsal cell fates in the Drosophila embryo. Genes Dev. 1994;8:2588–2601. doi: 10.1101/gad.8.21.2588. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Boyle M, Bonini N, DiNardo S. Expression and function of clift in the development of somatic gonadal precursors within the Drosophila mesoderm. Development. 1997;124:971–982. doi: 10.1242/dev.124.5.971. [DOI] [PubMed] [Google Scholar]
- Boyle M, DiNardo S. Specification, migration and assembly of the somatic cells of the Drosophila gonad. Development. 1995;121:1815–1825. doi: 10.1242/dev.121.6.1815. [DOI] [PubMed] [Google Scholar]
- Bradley PL, Andrew DJ. ribbon encodes a novel BTB/POZ protein required for directed cell migration in Drosophila melanogaster. Development. 2001;128:3001–3015. doi: 10.1242/dev.128.15.3001. [DOI] [PubMed] [Google Scholar]
- Brookman JJ, Toosy AT, Shashidhara LS, White RA. The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development. 1992;116:1185–1192. doi: 10.1242/dev.116.4.1185. [DOI] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Byars CL, Bates KL, Letsou A. The dorsal-open group gene raw is required for restricted DJNK signaling during closure. Development. 1999;126:4913–4923. doi: 10.1242/dev.126.21.4913. [DOI] [PubMed] [Google Scholar]
- Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr. Top. Dev. Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. 2nd ed Springer; Berlin ; New York: 1997. [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Cumberledge S, Szabad J, Sakonju S. Gonad formation and development requires the abd-A domain of the bithorax complex in Drosophila melanogaster. Development. 1992;115:395–402. doi: 10.1242/dev.115.2.395. [DOI] [PubMed] [Google Scholar]
- D’Andrea RJ, Stratmann R, Lehner CF, John UP, Saint R. The three rows gene of Drosophila melanogaster encodes a novel protein that is required for chromosome disjunction during mitosis. Mol. Biol. Cell. 1993;4:1161–1174. doi: 10.1091/mbc.4.11.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T, Le Bras S, Van Doren M. Abdominal-B is essential for proper sexually dimorphic development of the Drosophila gonad. Mech. Dev. 2004;121:1323–1333. doi: 10.1016/j.mod.2004.07.001. [DOI] [PubMed] [Google Scholar]
- DeFalco TJ, Verney G, Jenkins AB, McCaffery JM, Russell S, Van Doren M. Sex-specific apoptosis regulates sexual dimorphism in the Drosophila embryonic gonad. Dev. Cell. 2003;5:205–216. doi: 10.1016/s1534-5807(03)00204-1. [DOI] [PubMed] [Google Scholar]
- Eldon ED, Pirrotta V. Interactions of the Drosophila gap gene giant with maternal and zygotic pattern-forming genes. Development. 1991;111:367–378. doi: 10.1242/dev.111.2.367. [DOI] [PubMed] [Google Scholar]
- Englund C, Steneberg P, Falileeva L, Xylourgidis N, Samakovlis C. Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development. 2002;129:4941–4951. doi: 10.1242/dev.129.21.4941. [DOI] [PubMed] [Google Scholar]
- Fessler LI, Campbell AG, Duncan KG, Fessler JH. Drosophila laminin: characterization and localization. J. Cell Biol. 1987;105:2383–2391. doi: 10.1083/jcb.105.5.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E, Tietje K, Jan LY, Jan YN. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development. 1994;120:1385–1398. doi: 10.1242/dev.120.6.1385. [DOI] [PubMed] [Google Scholar]
- Graner MW, Bunch TA, Baumgartner S, Kerschen A, Brower DL. Splice variants of the Drosophila PS2 integrins differentially interact with RGD-containing fragments of the extracellular proteins tiggrin, ten-m, and D-laminin 2. J. Biol. Chem. 1998;273:18235–18241. doi: 10.1074/jbc.273.29.18235. [DOI] [PubMed] [Google Scholar]
- Greig S, Akam M. The role of homeotic genes in the specification of the Drosophila gonad. Curr. Biol. 1995;5:1057–1062. doi: 10.1016/s0960-9822(95)00210-7. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V. Robo1 and Robo2 are homophilic binding molecules that promote axonal growth. Mol. Cell. Neurosci. 2002;21:534–545. doi: 10.1006/mcne.2002.1193. [DOI] [PubMed] [Google Scholar]
- Jenkins AB, McCaffery JM, Van Doren M. Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development. 2003;130:4417–4426. doi: 10.1242/dev.00639. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Nakamura A, Yasuda K, Kageyama Y. Dmaf, a novel member of Maf transcription factor family is expressed in somatic gonadal cells during embryonic development and gametogenesis in Drosophila. Gene Expr Patterns. 2003;3:663–667. doi: 10.1016/s1567-133x(03)00093-0. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK. NETRIN and SLIT guide salivary gland migration. Dev. Biol. 2005;284:102–111. doi: 10.1016/j.ydbio.2005.04.037. [DOI] [PubMed] [Google Scholar]
- Kornberg T. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc Natl Acad Sci U S A. 1981;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer SG, Kidd T, Simpson JH, Goodman CS. Switching repulsion to attraction: changing responses to slit during transition in mesoderm migration. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–620. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M. Development of the male germline stem cell niche in Drosophila. Dev. Biol. 2006;294:92–103. doi: 10.1016/j.ydbio.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Livneh E, Glazer L, Segal D, Schlessinger J, Shilo BZ. The Drosophila EGF receptor gene homolog: conservation of both hormone binding and kinase domains. Cell. 1985;40:599–607. doi: 10.1016/0092-8674(85)90208-9. [DOI] [PubMed] [Google Scholar]
- MacMullin A, Jacobs JR. Slit coordinates cardiac morphogenesis in Drosophila. Dev. Biol. 2006;293:154–164. doi: 10.1016/j.ydbio.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Mathews WR, Ong D, Milutinovich AB, Van Doren M. Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development. 2006;133:1143–1153. doi: 10.1242/dev.02256. [DOI] [PubMed] [Google Scholar]
- Mathews WR, Wang F, Eide DJ, Van Doren M. Drosophila fear of intimacy encodes a Zrt/IRT-like protein (ZIP) family zinc transporter functionally related to mammalian ZIP proteins. J. Biol. Chem. 2005;280:787–795. doi: 10.1074/jbc.M411308200. [DOI] [PubMed] [Google Scholar]
- Moore LA, Broihier HT, Van Doren M, Lehmann R. Gonadal mesoderm and fat body initially follow a common developmental path in Drosophila. Development. 1998a;125:837–844. doi: 10.1242/dev.125.5.837. [DOI] [PubMed] [Google Scholar]
- Moore LA, Broihier HT, Van Doren M, Lunsford LB, Lehmann R. Identification of genes controlling germ cell migration and embryonic gonad formation in Drosophila. Development. 1998b;125:667–678. doi: 10.1242/dev.125.4.667. [DOI] [PubMed] [Google Scholar]
- Parsons L, Harris KL, Turner K, Whitington PM. Roundabout gene family functions during sensory axon guidance in the drosophila embryo are mediated by both Slit-dependent and Slit-independent mechanisms. Dev. Biol. 2003;264:363–375. doi: 10.1016/j.ydbio.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: A glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Prasad A, Paruchuri V, Preet A, Latif F, Ganju RK. Slit-2 induces a tumor-suppressive effect by regulating beta-catenin in breast cancer cells. J. Biol. Chem. 2008;283:26624–26633. doi: 10.1074/jbc.M800679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr. Biol. 2005;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Nicolas E, Vivancos V, Berger J, Dickson BJ. Crossing the midline: roles and regulation of Robo receptors. Neuron. 2000a;28:767–777. doi: 10.1016/s0896-6273(00)00152-5. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Vivancos V, Nicolas E, Dickson BJ. Selecting a longitudinal pathway: Robo receptors specify the lateral position of axons in the Drosophila CNS. Cell. 2000b;103:1033–1045. doi: 10.1016/s0092-8674(00)00207-5. [DOI] [PubMed] [Google Scholar]
- Rhee J, Buchan T, Zukerberg L, Lilien J, Balsamo J. Cables links Robo-bound Abl kinase to N-cadherin-bound beta-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat. Cell Biol. 2007;9:883–892. doi: 10.1038/ncb1614. [DOI] [PubMed] [Google Scholar]
- Rhee J, Mahfooz NS, Arregui C, Lilien J, Balsamo J, VanBerkum MF. Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat. Cell Biol. 2002;4:798–805. doi: 10.1038/ncb858. [DOI] [PubMed] [Google Scholar]
- Riechmann V, Irion U, Wilson R, Grosskortenhaus R, Leptin M. Control of cell fates and segmentation in the Drosophila mesoderm. Development. 1997;124:2915–2922. doi: 10.1242/dev.124.15.2915. [DOI] [PubMed] [Google Scholar]
- Riechmann V, Rehorn KP, Reuter R, Leptin M. The genetic control of the distinction between fat body and gonadal mesoderm in Drosophila. Development. 1998;125:713–723. doi: 10.1242/dev.125.4.713. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Artavanis-Tsakonas S. Modularity of the slit protein. Characterization of a conserved carboxy-terminal sequence in secreted proteins and a motif implicated in extracellular protein interactions. J. Mol. Biol. 1992;227:367–370. doi: 10.1016/0022-2836(92)90891-m. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hartley DA, Walther Z, Artavanis-Tsakonas S. slit: an EGF-homologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell. 1988;55:1047–1059. doi: 10.1016/0092-8674(88)90249-8. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Santiago-Martinez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc Natl Acad Sci U S A. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Martinez E, Soplop NH, Patel R, Kramer SG. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. J. Cell Biol. 2008;182:241–248. doi: 10.1083/jcb.200804120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AC, Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004;14:R578–589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Shiau CE, Bronner-Fraser M. N-cadherin acts in concert with Slit1-Robo2 signaling in regulating aggregation of placode-derived cranial sensory neurons. Development. 2009;136:4155–4164. doi: 10.1242/dev.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128:4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O’Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JH, Bland KS, Fetter RD, Goodman CS. Short-range and long-range guidance by Slit and its Robo receptors: a combinatorial code of Robo receptors controls lateral position. Cell. 2000a;103:1019–1032. doi: 10.1016/s0092-8674(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Simpson JH, Kidd T, Bland KS, Goodman CS. Short-range and long-range guidance by slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron. 2000b;28:753–766. doi: 10.1016/s0896-6273(00)00151-3. [DOI] [PubMed] [Google Scholar]
- Starz-Gaiano M, Cho N, Forbes A, Lehmann R. Spatially restricted activity of a Drosophila lipid phosphatase guides migrating germ cells. Development. 2001;128:983–991. doi: 10.1242/dev.128.6.983. [DOI] [PubMed] [Google Scholar]
- Stratmann R, Lehner CF. Separation of Sister Chromatids in Mitosis Requires the Drosophila pimples Product, a Protein Degraded after the Metaphase/Anaphase Transition. Cell. 1996;84:25–35. doi: 10.1016/s0092-8674(00)80990-3. [DOI] [PubMed] [Google Scholar]
- Tei H, Nihonmatsu I, Yokokura T, Ueda R, Sano Y, Okuda T, Sato K, Hirata K, Fujita SC, Yamamoto D. pokkuri, a Drosophila gene encoding an E-26-specific (Ets) domain protein, prevents overproduction of the R7 photoreceptor. Proc Natl Acad Sci U S A. 1992;89:6856–6860. doi: 10.1073/pnas.89.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Mathews WR, Samuels M, Moore LA, Broihier HT, Lehmann R. fear of intimacy encodes a novel transmembrane protein required for gonad morphogenesis in Drosophila. Development. 2003;130:2355–2364. doi: 10.1242/dev.00454. [DOI] [PubMed] [Google Scholar]
- Warrior R. Primordial germ cell migration and the assembly of the Drosophila embryonic gonad. Dev. Biol. 1994;166:180–194. doi: 10.1006/dbio.1994.1306. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, Greenbaum MP, Bellen HJ. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 – Screen Genetics. Mutant lines from our screen were created using the genetic scheme outlined here. See materials and methods for more details.
Supplemental Figure 2 – Balancer chromosome ftz-lacZ expression. Sample embryo from our screening process treated by X-Galactosidase reaction. This embryo carries both the 68-77 enhancer trap (gonad expression denoted by the white arrow) and the ftz-lacZ construct (denoting the presence of the balancer chromosome, black arrow). Compare with embryos in Fig. 1C-E which all lack the ftz-lacZ balancer construct.