Abstract
The meiosis-specific MER3 protein of Saccharomyces cerevisiae is required for crossing over, which ensures faithful segregation of homologous chromosomes at the first meiotic division. The predicted sequence of the MER3 protein contains the seven motifs characteristic of the DExH-box type of DNA/RNA helicases. The purified MER3 protein is a DNA helicase, which can displace a 50-nucleotide fragment annealed to a single-stranded circular DNA. MER3 was found to have ATPase activity, which was stimulated either by single- or double-stranded DNA. The turnover rate, kcat, of ATP hydrolysis was ~500/min in the presence of either DNA. MER3 was able to efficiently displace relatively long 631-nucleotide fragments from single-stranded circular DNA only in the presence of the S. cerevisiae single-stranded DNA-binding protein, RPA (replication protein A). It appears that RPA inhibits re-annealing of the single-stranded products of the MER3 helicase. The MER3 helicase was found to unwind DNA in the 3′ to 5′ direction relative to single-stranded regions in the DNA substrates. Possible roles for the MER3 helicase in meiotic crossing over are discussed.
During meiosis, two successive rounds of chromosome segregation occur following a single round of DNA replication. The first meiotic division, meiosis I, is unique in that homologous chromosomes synapse and then segregate to opposite poles. Crossing over, but not gene conversion, provides a physical connection between homologous chromosomes and ensures their proper segregation at meiosis I (for review, see Refs. 1 and 2). The distribution of crossovers along a chromosome is regulated. When multiple crossovers occur on a chromosome, they are further apart than predicted from the frequency of individual crossovers, a phenomenon called crossover interference. This regulation takes place so that every pair of homologous chromosomes sustains at least one crossover, and it underscores the importance of crossing over for faithful segregation of chromosomes. An extensive body of evidence has accumulated indicating that crossing over is under the control of a specific set of proteins that are required for crossing over but not gene conversion (3–11).
The initiation of meiotic recombination involves the programmed induction of DNA double strand breaks (DSBs)1 (12). The ends of DSBs are rapidly resected to produce 3′ -overhangs of about 600 nucleotides (nt) long (13). The 3′ -single strands then participate in strand invasion reactions catalyzed by protein complexes containing RAD51 (14, 15) and/or DMC1 (13, 16), the yeast counterparts of the Escherichia coli strand exchange protein, RecA (17). Double-Holliday junctions are the prominent intermediates formed during homologous recombination initiating from DSBs (18–20). It should be noted that both of the two 3′ -single-strand ends originating from a single DSB must invade the same chromatid to form a double-Holliday junction. It is possible that the two different outcomes of recombination, crossing over and gene conversion, result from alternate resolutions of double-Holliday junctions. However, it is also possible that crossing over and gene conversion events are differentiated from each other before or during the formation of the joint molecules. The observation that mutations in the genes that are specifically required for crossing over show a defect in the transition from DSBs to the joint molecules supports this latter view (8, 21). However, the determination of the stage at which recombination crossover control is imposed remains elusive.
The MER3 gene of Saccharomyces cerevisiae is expressed only in meiosis (8, 22). In the absence of MER3, the frequency of crossing over is reduced, and the distribution of crossovers along a chromosome is randomized because of an apparent defect in crossover interference. The predicted sequence of the MER3 protein contains the seven motifs characteristic of the DExH-box type of DNA/RNA helicases (23). Interestingly, the MER3 sequence shows significant homology to S. cerevisiae SGS1 (24) and Homo sapiens BLM (25) helicases, which are involved in DNA replication, recombination, and cell cycle regulation (26–31). Consistent with the presence of the helicase motifs, an initial study has demonstrated that the purified MER3 protein can displace 50-nt DNA fragments annealed to single-stranded circular DNA.2 The MER3 protein binds to both single- and double-stranded DNA, with a slight preference for single-stranded DNA. The mer3G166D mutation, which causes an amino acid substitution changing an invariable glycine to an aspartic acid in a putative nucleotide-binding domain of MER3, decreases crossing over and impairs crossover interference. The spore viability of the mer3GD mutant is also decreased, which is likely to be due to a high incidence of non-disjunction of homologous chromosomes at meiosis I. The mutant MER3GD protein is defective for DNA helicase activity but has DNA binding activity that is indistinguishable from that seen for wild-type MER3. These genetic and biochemical data suggest that the DNA helicase activity of MER3 is required for meiotic crossing over and, in turn, for faithful segregation of homologous chromosomes.
To better understand the role of the MER3 protein in meiotic recombination, we have extensively characterized the ATPase and DNA helicase activity of MER3. The MER3 protein was found to have an ATPase activity that was stimulated by either poly (dA) or M13mp18 replicative form I (M13 RF) DNA. The MER3 protein could displace 50-, 100-, and 631-nt single strands from M13mp18 single-stranded DNA, although high concentrations of MER3 were required to displace the longer fragments. The displacement of 631-nt-long fragments was stimulated by either the single-stranded DNA-binding protein of S. cerevisiae, RPA (33), or that of E. coli, SSB (34). These single-stranded DNA-binding proteins appear to stimulate the activity of MER3 by preventing the re-annealing of the DNA region once it is unwound by the MER3 helicase. The polarity of the MER3 helicase was determined to be 3′ to 5′ with regard to the single-stranded region present in the DNA substrates. Possible roles for a processive 3′ to 5′ DNA helicase like MER3 in crossover control are discussed.
MATERIALS AND METHODS
Preparation of MER3 Protein
The MER3 and MER3G166D proteins tagged with a FLAG epitope at their C termini were over-expressed under the control of GAL10 promoter in yeast cells and were purified by binding to FLAG affinity gel (Sigma) followed by sequential chromatography on MonoQ HR5/5, HiTrap heparin, and MonoS HR5/5 (Amersham Pharmacia Biotech) columns as previously described.2 The final protein preparations were more than 98% pure MER3.
Construction of Yeast RPA Overexpressors
The yeast RPA1gene (yRPA1) was amplified using primers 10786 (5′ -CGACCGCTCGAGACACCATGAGCAGTGTTCAACTTTCG-3′), 10787 (5′ -GCTCCCAAGCTTAAGAGATGCTGAACCGCCC-3′), and pRPA1 (35) as template. The resulting PCR product of 2.0 kb was used to replace the XhoI to HindIII region of pRDK249 (2 µ, GAL10, ampr, URA3) (36). The EcoRV to HindIII region of this construct was then replaced by a 1.8-kb EcoRVHindIII fragment derived from pRPA1 yielding plasmid pRDK273. yRPA2 was amplified using primers 10994 (5′ -CGACCGCTCGAGACACCATGGCAAGTATGTATTAGTGCTAGG-3′), 10993 (5′ -GCTCCCAAGCTTATGTACACCTAGAACCAGATAC-3′) and a clone containing the yRPA2 open reading frame isolated from a YEp213 yeast genomic library as the template. The PCR product of 1.3 kb was inserted between the XhoI and HindIII sites of plasmid pRDK249. A 2.0-kb BamHI-HindIII fragment from the pRDK249 derivative, which contained the GAL10 promoter and yRPA2, was introduced between the BamHI and HindIII sites of pRS425 (2 µ, ampr, LEU2) creating plasmid pRDK274. yRPA3 was amplified using primers 10788 (5′ -CGACCGCTGCAGCTCGAGACACCATGGCCAGCGAAACACCAAG-3′), 10789 (5′ -GGCCTTGGGCCCGCGGAAGGCGTTAAGGCAGC-3′), and a YEP213 yeast library-derived clone that contained the yRPA3 gene as template. The PCR product of 560 bp was cloned between the PstI and ApaI sites of pRS424 (2 µ, ampr, TRP1). A 750-bp BamHI-XhoI fragment from pRDK249, which contained the GAL10 promoter, was introduced upstream of the yRPA3 gene generating plasmid pRDK275. Correct clones were confirmed by DNA sequencing.
Overexpression and Purification of yRPA
The yRPA overexpressor strain (RDKY2275) was created by transforming plasmids pRDK273, pRDK274, and pRDK275 into yeast strain RDKY1293 (MATα, ura3–52, trp1, leu2Δ1, his3Δ200, pep4::HIS3, prb1Δ1.6R, can1, GAL). The over-producer strain was grown at 30 °C with vigorous shaking in 2 liters of Leu− Ura− Trp− minimal drop-out medium supplemented with 2% (w/v) raffinose. The expression from GAL10 promoter was induced by the addition of a final concentration of 2% (w/v) galactose at an A600 of 2.0 and incubation was continued until late log phase. Cells were collected by centrifugation and washed with ice-cold water followed by two washes with buffer A [25 mm Tris·HCl (pH 7.5), 1 mm EDTA, 0.01% Nonidet P-40, 10% glycerol, 1 mm dithiothreitol, 10 mm benzamidine, 1 µg/ml pepstatin A, 2 µm leupeptin, 1 µm aprotinin, and 0.1 mm phenylmethylsulfonyl fluoride). The wet cell pellet (22 g) was squirted into liquid nitrogen through a 60-ml syringe, and the frozen cell “noodles” were ground to a fine powder under liquid nitrogen in a motorized mortar grinder (Retsch). Purification was carried out at 4 °C based on a previously described procedure (33). The yeast powder was resuspended with buffer A containing 0.5 m NaCl. The crude cell lysate was centrifuged in a Sorvall SA600 rotor at 15,000 rpm for 60 min, and the cleared lysate (100 ml, 660 mg) was adjusted to 0.5 m NaCl and loaded onto an Affi-gel blue (Bio-Rad) column (1.8 cm2 × 7 cm). The column was washed with 120 ml of buffer A containing 0.8 m NaCl, and the protein was eluted with 120 ml of buffer A containing 2.5 m NaCl and 40% (w/v) ethylene glycol. Peak fractions were pooled (40 ml, 95.8 mg), diluted to a conductivity equal to buffer A containing 0.5 m NaCl, and loaded onto a single-stranded DNA cellulose column (3.1 cm2 × 8 cm). The column was then washed with 125 ml of buffer A containing 0.75 m NaCl, and the protein was eluted with 125 ml of buffer A containing 1.5 m NaCl and 50% ethylene glycol. Peak fractions were pooled (8 ml, 1.8 mg), dialyzed against 2 changes of 500 ml of buffer A containing 0.1 m NaCl, and loaded onto a DEAE-Sepharose column (0.8 cm2 × 1.5 cm), and the protein was eluted with a linear gradient of 100–500 mm NaCl in buffer A. Peak fractions of RPA eluting at 200 mm NaCl were pooled and stored at −80 °C. Purity was similar to that observed previously (33). The yield was 0.6 mg/liter of cell culture.
Nucleotides and DNA
M13mp18 single-stranded circular and M13mp18 RF I were from New England Biolabs and Life Technologies, Inc., respectively. Poly (dA) and ATP were from Amersham Pharmacia Biotech. [γ-32P]ATP was from PerkinElmer Life Sciences. High pressure liquid chromatography-purified oligonucleotides were from CyberSyn, Inc. (Aston, PA). E. coli single-stranded DNA-binding protein (SSB) was from the United States Biochemical Corp. (Cleveland, OH).
ATPase Assays
Reactions containing 20 mm Tris·HCl (pH 7.6), 50 mm NaCl, 5 mm MgCl2, 2 mm dithiothreitol, 100 µg/ml bovine serum albumin, 0.05 Ci/ml [γ-32P]ATP, 1.5 µg/ml DNA, and 1 mm ATP (unless otherwise indicated) were pre-incubated at 30 °C for 5 min, and the reaction was initiated by the addition of 5 nm MER3 protein. For time course reactions, aliquots (5 µl) were withdrawn at the indicated time points and mixed with 2 µl of 0.2 m EDTA. 1 µl of each reaction was spotted onto polyethyleneimine cellulose thin layer chromatography plates (Sigma). The plates were developed in 1 m formic acid, 0.5 m LiCl and dried, and the amounts of 32Pi and [γ-32P]ATP in the reaction mixture were determined using a PhosphorImager (445 SI, Molecular Dynamics).
DNA Helicase Substrates
DNA helicase substrates were prepared essentially as described before (37). An 0.6-kb ClaI-BamHI fragment from M13mp18 RF I was introduced between the ClaI and BamHI sites of pBluescript II KS+ (Stratagene) to create pRDK4182. pRDK4182 was digested with ClaI, treated with calf alkaline phosphatase (New England Biolabs), purified using a PCR Purification Kit (Qiagen), and digested with BamHI, and then the resulting 0.6-kb ClaI-BamHI fragment was separated by electrophoresis through an 0.7% agarose gel and purified from the gel slice using a Gel Extraction Kit (Qiagen). The 0.6-kb Cla-BamHI fragment and the M13–100 oligonucleotide (100-nt), which is complementary to nucleotide coordinates 6230–6329 of M13mp18 single-stranded circular DNA, were 5′ -end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs), and unincorporated nucleotides were removed using a Nucleotide Removal Kit (Qiagen). In separate reactions, each labeled DNA was mixed with an equal molar amount of M13mp18 single-stranded circular DNA in an annealing buffer (10 mm Tris·HCl (pH 7.6), 1 mm EDTA, 50 mm NaCl), heated to 95 °C for 10 min, and then slowly cooled to 25 °C in a PCR machine at a rate of temperature decrease of −1 °C/2 min. Each annealed DNA product was purified by electrophoresis through an 0.7% agarose gel as described above and then by chromatography through an 0.12 cm2 × 25 cm Bio-Gel A 5-m (Bio-Rad) column equilibrated and run in annealing buffer containing 100 mm NaCl. The DNA substrate containing the M13–50 oligonucleotide (50-nt) annealed to M13mp18 single-stranded circular DNA at nucleotides 6230–6279 were prepared as described previously.2 The concentration of the purified DNA was determined using a spectrophotometer (DU 640B, Beckman).
To determine the polarity of translocation by the MER3 helicase, two kinds of DNA substrates were constructed using essentially the same methods as described above. To prepare the DNA substrate shown in Fig. 6A, oligonucleotides T75 (5′ -GACGCTGCCGAATTCTGGCTTGCTAGGACA-3′, 30-nt) and T82 (5′ -CCCATAAACAAACTTCGTTAACTGAACTTGCCTGTACGATTCGTC-3′, 45-nt) were 5′ -end-labeled with [γ-32P]ATP and then annealed to T81 (5′ -GACGAATCGTACAGGCAAGTTCAGTTAACGAAGTTTGTTTATGGGTACTCTCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGTC, 100-nt). The annealed DNA was separated by 10% non-denaturing polyacrylamide gel electrophoresis (60:1 acrylamide/bisacrylamide) in TBE (90 mm Tris·borate, 2 mm EDTA). The DNA was recovered by soaking the excised band in elution buffer (0.5 m NaCl, 0.6 m sodium acetate, 1 mm EDTA) overnight at 4 °C followed by phenol/chloroform extraction and ethanol precipitation. The recovered DNA pellet was dissolved in suspension buffer (10 mm Tris·HCl (pH 7.6), 250 mm NaCl). The concentration of purified DNA was determined using a DNA DipStick™ (Invitrogen). To prepare the DNA substrate shown in Fig. 6D, the M13–100 oligonucleotide was 5′ -end-labeled with [γ-32P]ATP and annealed to M13mp18 single-stranded circular DNA in extension buffer (40 mm Tris·HCl (pH 7.6), 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol). The 3′ -end of M13–100 was then labeled with [α-32P] dGTP using Klenow fragment (New England Biolabs). After removal of unincorporated nucleotides and digestion with HincII, the DNA substrate was purified using Bio-Gel A 5-m (Bio-Rad) as described above.
Fig. 6. Unwinding of DNA duplexes flanking a single-stranded region.
A, the 100-nt DNA substrate containing annealed 5′ -end-labeled 45- and 30-nt fragments used to determine the polarity of MER3 helicase is illustrated. B, standard helicase reactions (20 µl each) containing 2 nm (in molecules) DNA substrate, illustrated in A, and 10 mm NaCl were started by the addition of 1, 2, 3, 4, or 5 nm MER3, and incubated at 30 °C for 30 min. The DNA product formed was monitored by electrophoresis through 10% polyacrylamide gels. C, the average value of the amount of 45- and 30-nt fragments displaced in three independent experiments is plotted. The error bars indicate the standard deviation. D, the DNA substrate containing an ~7,100-nt-long single-stranded region flanked by double-stranded regions containing end-labeled 64- and 37-nt fragments used to determine the polarity of MER3 helicase is illustrated. E, standard helicase reactions (20 µl each) containing 1 µm (in nucleotides) DNA substrate having a long single-stranded region, illustrated in panel D, and 100 mm NaCl were started by the addition of 0.2, 0.4, 0.8, 1.6, 3.2, or 6.4 nm of MER3 and incubated at 30 °C for 30 min. The percentage of 64- or 37-nt fragment displaced is plotted.
DNA Helicase Assays
The indicated amount of protein and DNA substrate (indicated concentrations are in moles of nucleotides) were incubated in 20-µl volumes containing DNA helicase buffer (20 mm Tris·HCl (pH 7.6), 50 mm NaCl, 2 mm dithiothreitol, 100 µg/ml bovine serum albumin, 2 mm MgCl2, 2 mm ATP) unless otherwise indicated. All reactions were pre-incubated at 30 °C for 5 min, started by the addition of MER3 protein, and then incubation was continued for 30 min. Reactions were stopped by the addition of 5 µl of stop buffer (50 mm Tris·HCl (pH 7.6), 50 mm EDTA, 2.5% SDS) and 0.5 µl of 25 mg/ml proteinase K followed by incubation at 37 °C for 10 min. The DNA products were analyzed by electrophoresis through non-denaturing polyacrylamide gels run in TBE. The gels were dried, and the radiolabeled DNA was visualized using a PhosphorImager.
RESULTS
MER3 Is an ATPase That Is Stimulated by Single- and Double-stranded DNA
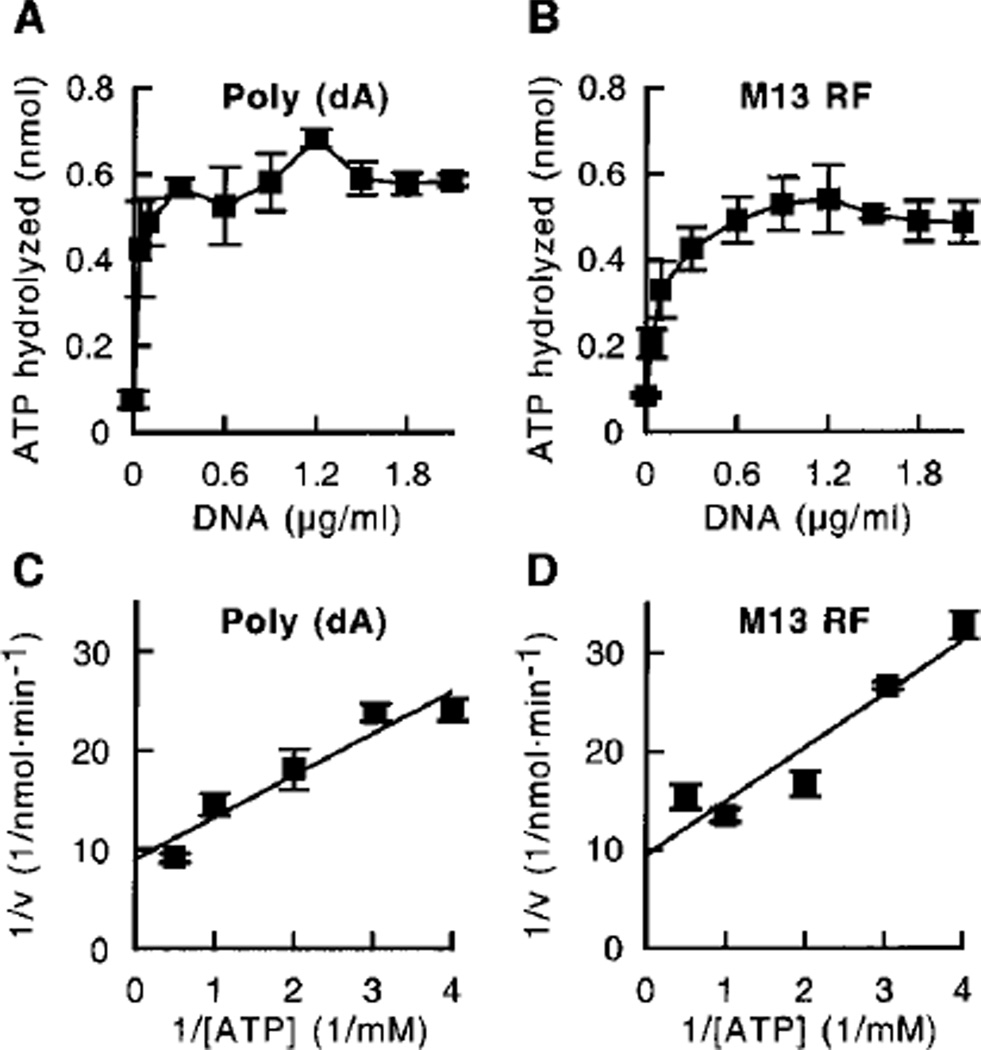
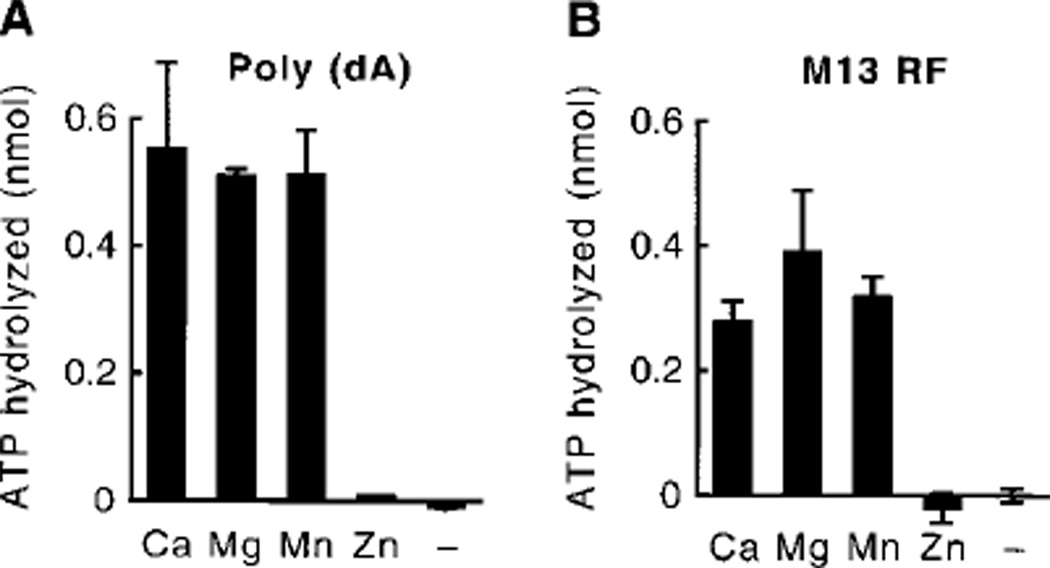
In a previous study, we demonstrated that the MER3 protein has ATPase activity and displace oligonucleotide fragments annealed to M13 single-stranded circular DNA in an ATP-dependent manner.2 To better characterize the MER3 ATPase activity, MER3 protein was incubated with [γ-32P] ATP, and the formation of 32Pi was measured using thin layer chromatography. MER3 hydrolyzed ATP, and the addition of increasing concentrations of poly (dA) or M13 RF DNA increased the amount of ATP hydrolyzed by up to 5–6-fold (Fig. 1, A and B), showing that the MER3 ATPase activity is stimulated by either single- or double-stranded DNA. Stimulation of the ATPase activity by the addition of M13mp18 single-stranded circular DNA was also observed, indicating that single-stranded ends are not required to stimulate the MER3 ATPase activity (data not shown). In addition, M13 RF DNA linearized by digestion with HindIII stimulated the MER3 ATPase activity as well as undigested M13 RF DNA (at a DNA concentration of 1.5 µg/ml; data not shown), excluding the possibility that single-stranded regions that might appear in super-coiled DNA stimulate the ATPase activity. The ATPase activity reached maximal levels at poly (dA) and M13 RF DNA concentrations of 0.3 and 0.6 µg/ml, respectively, which is equivalent to 180 nt of poly (dA) or 180 bp of M13 RF DNA per MER3 molecule. To determine the turnover rate of the MER3 ATPase, the initial velocity of ATP hydrolysis was determined at different concentrations of ATP in the presence of an excess amount of DNA (10 µg/ml) (Fig. 1, C and D). Shown in Table I are the Km, Vmax, and kcat values obtained from analyses of the data presented in Fig. 1, C and D. The turnover rate, kcat was found to be about 500 ATP/min in the presence of either poly (dA) or M13 RF DNA, although the Km value was slightly lower in the presence of poly (dA) as compared with M13 RF DNA. The MER3 ATPase activity required a divalent cation. Maximal activity could be observed in the presence of either Ca2+, Mg2+, or Mn2+, whereas Zn2+ did not support the ATPase activity (Fig. 2, A and B).
Fig. 1. Poly (dA) and M13 RF DNA stimulate the MER3 ATPase activity.
Reactions (20 µl each) containing different concentrations (0, 0.04, 0.1, 0.3, 0.6, 0.9, 1.2, 1.5, 1.8, and 2.1 µg/ml) of poly (dA) (A) or M13mp18 RF (B) were initiated by the addition of MER3, incubated at 30 °C for 15 min, and terminated by the addition of 2 µl of 0.5 m EDTA, and the amount of ATP hydrolyzed was measured as described under “Materials and Methods.” C and D, the initial velocity of ATP hydrolysis in reactions (40 µl each) was measured at different concentrations of ATP (0.25, 0.33, 0.5, 1, and 2 mm) in the presence of 10 µg/ml poly (dA) (C) or M13mp18 RF DNA (D); the data are presented as a Lineweaver-Burk plot. The average of ATP hydrolyzed in three independent experiments is plotted, and the error bars show the standard deviation.
Table I.
ATPase activity of the MER3 protein
| DNA | Km | Vmax | kcat |
|---|---|---|---|
| µM | pmol · min−1 | min−1 | |
| Poly(dA) | 470 | 111 | 550 |
| M13mp18 RF | 580 | 106 | 530 |
Fig. 2. Requirement of divalent cations for the MER3 ATPase activity.
Reactions (20 µl each) were carried out as described in the legend to Fig. 1, A and B, except that EDTA was present at a final concentration of 0.1 mm in order to remove contaminating divalent cations; and then CaCl2, MgCl2, MnCl2, or ZnCl2 was added to a final concentration of 5 mm prior to the pre-incubation step. The amount of ATP hydrolyzed in the presence of poly (dA) (A) or M13 RF (B) is presented. The average of two independent experiments is shown, and the error bars indicate the standard deviation.
An Amino Acid Substitution, mer3G166D, in a Putative Nucleotide-binding Domain of MER3 Impairs ATPase Activity
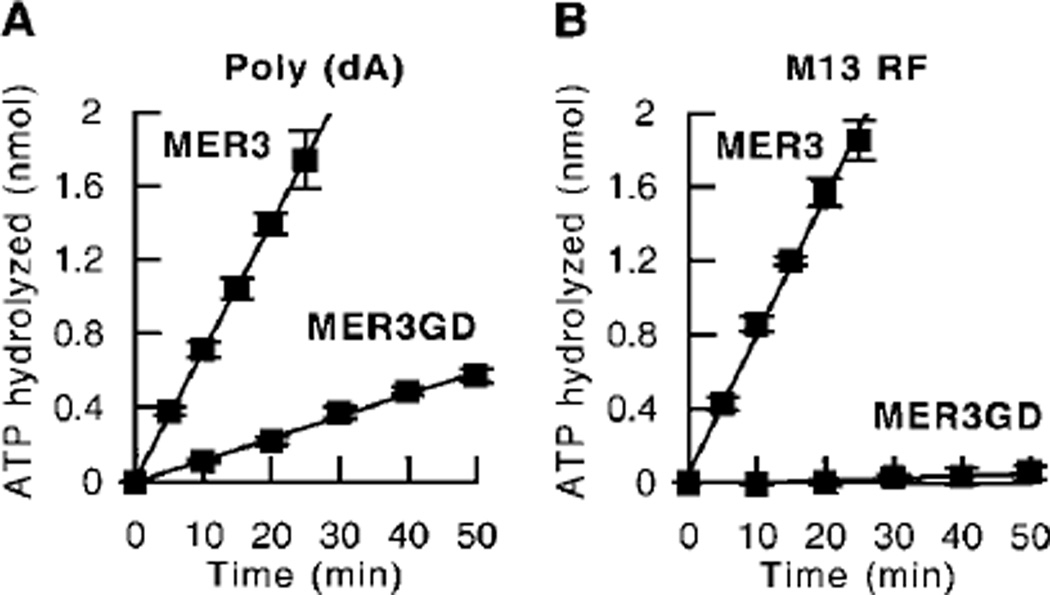
An amino acid substitution changing a conserved glycine to an aspartic acid at amino acid 166, mer3G166D, in a putative nucleotide-binding domain of the helicase motifs of MER3 was previously found to virtually eliminate the MER3 DNA helicase activity.2 However, the mutant MER3GD protein had single-and double-stranded DNA-binding activity that was indistinguishable from that of the wild-type MER3 protein. To analyze the effect of this amino acid substitution on ATPase activity, the time course of ATP hydrolysis by MER3GD was determined (Fig. 3). In the presence of either poly (dA) or M13 RF, the mer3G166D amino acid substitution significantly decreased the MER3 ATPase activity. Interestingly, the residual ATPase activity of MER3GD was different in the presence of different DNA cofactors. In the case of poly (dA), the rate of ATP hydrolysis carried out by MER3GD was about 17% of that of wild-type MER3 (Fig. 3A), whereas no detectable amounts of ATP were hydrolyzed by MER3GD in the presence of M13 RF DNA (Fig. 3B). In summary, mutant MER3GD retains an ability to bind DNA but has defects in ATP hydrolysis and DNA helicase reaction. Thus, it is likely that the glycine in a putative nucleotide-binding domain is important for ATP hydrolysis, which is coupled to the DNA unwinding activity of MER3.
Fig. 3. The mer3G166D mutation impairs ATPase activity.
A time course of ATP hydrolysis by the wild-type MER3 and mutant MER3GD proteins was performed as described under “Materials and Methods.” Reactions (40 µl each) were incubated for the indicated times in the presence of poly (dA) (A) or M13mp18 RF (B). The average of three independent experiments is presented, and the error bars indicate the standard deviation.
MER3 Unwinds Long DNA Duplexes
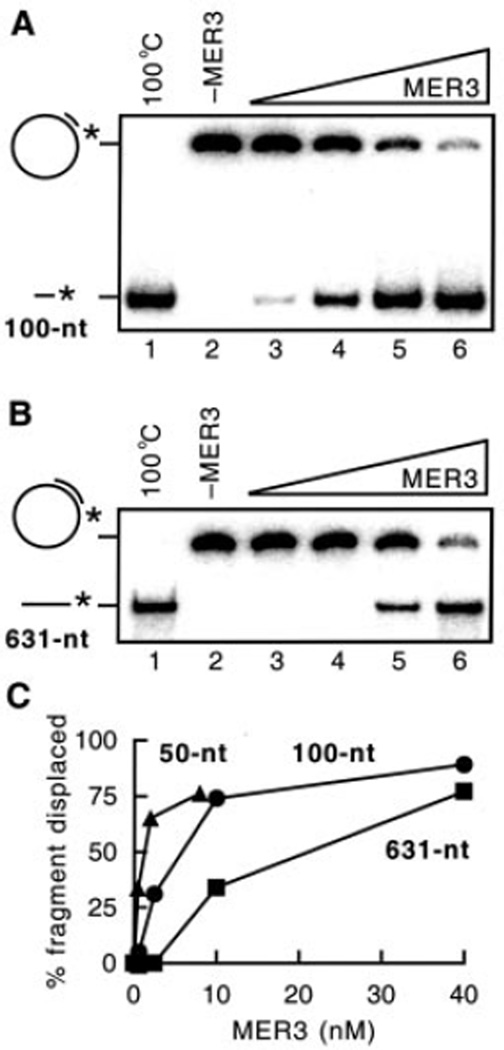
In previous studies, it was observed that the MER3 helicase could displace a 50-nt oligonucleotide annealed to M13mp18 single-stranded circular DNA.2 To determine whether MER3 can unwind longer DNA duplexes, similar DNA substrates were constructed containing 100- or 631-nt fragments annealed to M13mp18 single-stranded circular DNA. DNA helicase assays were performed with each DNA substrate in the presence of increasing amounts of MER3 (Fig. 4, A and B). The MER3 helicase displaced both the 100- and 631-nt fragments, although higher amounts of MER3 were required to displace the 631-nt fragment compared with the 100-nt fragment. The activity of MER3 helicase as a function of protein concentration was quantified using substrates containing 50-, 100-, or 631-nt fragments annealed to M13mp18 single-stranded circular DNA (Fig. 4C). This analysis showed that the amount of MER3 required to displace each annealed fragment increased as a function of the size of the annealed fragment. For example, 0.5, 2.5, and 10 nm MER3 was required to displace ~30% of the fragments from the 50-, 100-, and 631-nt substrates, respectively. These results show that high concentrations of MER3 are required to unwind long DNA duplexes.
Fig. 4. Displacement of different length fragments by MER3 helicase.
Displacement of 5′ -end-labeled fragments annealed to M13mp18 single-stranded circular DNA was examined as described under “Materials and Methods.” The reactions contained 1 µm (in nucleotides) DNA substrates. 6 and 4% non-denaturing polyacrylamide gels were used for the detection of the 100- and 631-nt fragments, respectively. Increasing amounts (0, 0.625, 2.5, 10, and 40 nm) of MER3 were added to the reaction containing the 100-nt fragment substrate (A) or the 631-nt fragment substrate (B). The percentage of fragment displaced by the different amounts of MER3 is plotted (C). To obtain the percentage for the 100- and 631-nt fragments, the gels shown in A and B, respectively, were analyzed using a PhosphorImager. The percentage displacement for 50-nt fragment substrate was measured similarly except that the reactions contained 0, 0.5, 2, or 8 nm MER3.
Single-stranded DNA-binding Proteins Stimulate the MER3 Helicase
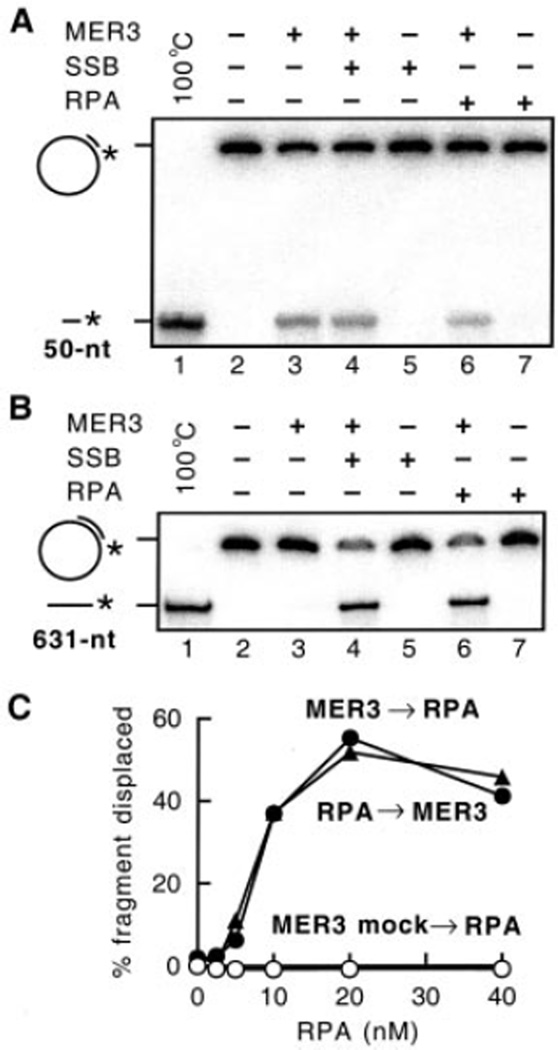
One possible reason for the fact that increasing amounts of MER3 are required to unwind DNA duplexes of increasing length is that binding of MER3 to the displaced single strands is required to prevent re-annealing. If this is the case, the activity of MER3 would be expected to be stimulated by single-stranded DNA-binding proteins. To test this possibility, the activity of the MER3 helicase was measured in the presence of either S. cerevisiae RPA complex or E. coli SSB, the single-stranded DNA-binding proteins that function in DNA replication, recombination, and repair in each organism (Fig. 5). In reactions containing the annealed 50-nt fragment (Fig. 5A), there was no detectable stimulation of strand displacement by the addition of RPA (Fig. 5A, lane 6) or SSB (Fig. 5A, lane 4). Also, no stimulation of MER3 by these SSBs was observed at different concentrations of MER3 (0.1, 0.4, and 0.8 nm; data not shown). However, displacement of the 631-nt fragment was stimulated by either RPA (Fig. 5B, lane 6) or SSB (Fig. 5B, lane 4) at the concentration of RPA or SSB that did not affect the activity of MER3 on the 50-nt substrate (Fig. 5A).
Fig. 5. RPA and SSB stimulate displacement of the 631-nt fragment.
Displacement of 5′ -end-labeled fragments annealed to M13mp18 single-stranded circular DNA was examined in the presence or absence of 20 nm S. cerevisiae RPA or E. coli SSB, essentially as described under “Materials and Methods.” Reactions contained 1 µm (in nucleotides) DNA substrates. Displacement of 50-nt (A) and 631-nt (B) fragments was analyzed in the presence of 0.2 and 5 nm MER3, respectively, and 8 and 4% non-denaturing polyacrylamide gels were used for the detection of 50- and 631-nt fragments, respectively. C, effect of increasing RPA concentration (0, 2.5, 5, 10, 20, and 40 nm) in reactions containing 1 µm (in nucleotides) 631-nt fragment substrates. RPA was added to the reaction 5 min before (closed circles) or 2 min after (closed triangles) the addition of 5 nm MER3. The open circles indicate the reaction to which no MER3 was added.
The helicase activity of MER3 on the substrate containing the 631-nt fragment was examined at different concentrations of RPA (Fig. 5C). The most efficient stimulation of strand displacement occurred at 20 nm RPA. At this concentration of RPA, the amount of 631-nt fragment displaced was increased 25-fold compared with that seen in the absence of RPA. The order of the addition of RPA and MER3 was found not to affect the strand displacement (Fig. 5C, closed triangles and circles). Importantly, at the RPA concentrations examined, RPA alone did not displace any of the 631-nt fragment (Fig. 5C, open circles). Given a binding site size of 90–100 nt for RPA (33), under these reaction conditions of 1 µm (in nucleotides) DNA and 20 nm RPA, there is sufficient RPA present to bind all of the DNA present if it is single-stranded. These results suggest that RPA stimulates MER3 by binding to the unwound regions produced by MER3 and inhibiting their re-annealing. It is also possible that RPA prevents nonspecific binding of MER3 to the single-stranded regions of the substrates, although the lack of an effect of the order of addition makes this latter possibility seem unlikely.
The MER3 Helicase Acts in the 3′ to 5′ Direction
To determine the polarity of the MER3 helicase, a DNA substrate that has a 25-nt single-stranded region flanked by 45- and 30-bp duplex regions was prepared (Fig. 6A). Because both of the 45- and 30-nt fragments were 32P-labeled at their 5′ -ends, displacement of both fragments could be examined in the same reaction. If the MER3 helicase binds to the single-stranded region and translocates in the 5′ to 3′ direction, the 30-nt fragment will be displaced. However, if MER3 translocates in the 3′ to 5′ direction, the 45-nt fragment will be displaced. MER3 preferentially displaced the 45-nt fragment compared with the 30-nt fragment (Fig. 6, B and C). A second DNA substrate was prepared to further examine the polarity of the MER3 helicase (Fig. 6D). This substrate has a long single-stranded region of ~7,100 nt with a 64-nt fragment annealed at one end and a 37-nt fragment annealed at the other end. Each of the blunt ends in this substrate (Fig. 6D) contains a 5′ -phosphate group, whereas only one of the blunt ends contains a 5′ -phosphate in the substrate described in Fig. 6A. Examination of the displacement of 64- and 37-nt fragments revealed that the 64-nt fragment was efficiently displaced by MER3, whereas the 37-nt fragment was not displaced at any concentration of MER3 examined (Fig. 6E). These results indicate that the polarity of the MER3 helicase is 3′ to 5′ relative to the single-stranded region.
DISCUSSION
The MER3 gene is required specifically for crossing over, which ensures faithful segregation of homologous chromosomes at the first meiotic division (8). In the present study, we have extensively characterized the ATPase and DNA helicase activity of the purified MER3 protein. The MER3 protein was found to have potent ATPase activity that was stimulated either by single- or double-stranded DNA. The MER3 protein was able to displace 50-, 100-, and 631-nt fragments annealed to M13mp18 single-stranded circular DNA in a reaction containing ATP and Mg2+. Compared with the displacement of 50-nt fragments, displacement of 100- and 631-nt fragments annealed to M13mp18 single-stranded circular DNA required increasing amounts of MER3. The addition of single-stranded DNA-binding proteins, including S. cerevisiae RPA and E. coli SSB, stimulated unwinding of the longer DNA duplexes and reduced the amount of MER3 required. Using DNA substrates containing two double-stranded regions flanking a single-stranded region, the polarity of MER3 helicase was determined to be in the 3′ to 5′ direction with respect to the single-stranded region. An amino acid substitution in the predicted nucleotide-binding domain of MER3 significantly reduced both the ATPase and DNA helicase activities, indicating that these activities are an intrinsic property of MER3 and that these activities are likely coupled.
MER3 was found to hydrolyze ATP to produce free phosphate and, presumably, ADP. The reaction was stimulated 5–6-fold by the addition of poly (dA) or M13 RF DNA (Fig. 1, A and B), and Ca2+, Mg2+, or Mn2+, but not Zn2+, could serve as a required divalent cation (Fig. 2). The turnover rates, kcat, in the presence of single- and double-stranded DNA were similar, consisting of ~500 ATP molecules hydrolyzed/min. However, the Km value for single-stranded DNA was slightly lower than for double-stranded DNA (Fig. 1, C and D; Table I). At low concentrations, single-stranded DNA stimulated the MER3 ATPase more efficiently than double-stranded DNA (Fig. 1, A and B). These results indicate that the MER3 ATPase has a slight preference for single-stranded DNA. This is consistent with the previous observation that MER3 has a preference for binding to single- compared with double-stranded DNA.2 The mer3G166D mutation, which changes a conserved glycine to aspartic acid in a putative nucleotide-binding domain, impaired the ATPase activity of MER3 (Fig. 3) and the DNA helicase activity,2 consistent with the view that ATP hydrolysis is related to DNA unwinding catalyzed by MER3. No detectable ATP hydrolysis by the mutant MER3GD protein was observed in the presence of double-stranded DNA (Fig. 3B). In contrast, in the presence of single-stranded DNA, MER3GD showed ~17% of the wild-type MER3 ATPase activity (Fig. 3A). This difference and a preference for single-stranded DNA described above suggest that the molecular mechanism underlying ATP hydrolysis catalyzed by MER3 is different in the presence of single- or double-stranded DNA. It is also possible that our MER3 preparations were slightly contaminated with another single-stranded DNA-specific ATPase, although this seems unlikely given the affinity chromatography purification procedure used here and the level of protein purity obtained.
An important property of DNA helicases is their ability to unwind DNA duplexes of various lengths (38). MER3 helicase was observed to displace 50-, 100-, and 632-nt fragments annealed to M13mp18 single-stranded circular DNA, although increasingly higher concentrations of MER3 were required as the length of the annealed fragment increased. The addition of the S. cerevisiae single-stranded DNA-binding protein, RPA, strongly stimulated the displacement of the long fragments by reducing the amount of MER3 required. As similar stimulation was observed upon the addition of E. coli SSB, it is unlikely that a specific interaction between MER3 and RPA is required for stimulation of the MER3 helicase activity. The amino acid sequence of MER3 has significant homology to members of the RecQ family of DNA helicases, S. cerevisiae SGS1, and human BLM, which are involved in DNA replication, repair, recombination, and cell cycle checkpoints (26–31). In contrast to the MER3 helicase, in the absence of single-stranded DNA-binding protein, SGS1 helicase has been shown to displace efficiently 52- and 140-nt but not 588-nt fragments (39), and BLM helicase can displace 30- and 71-nt but not 259-nt fragments using similar DNA substrates (40). Stimulation of displacement of long DNA fragments by single-stranded DNA-binding protein has been also observed for SGS1 and BLM helicases. In the presence of SSB, 588-nt fragments are efficiently displaced by SGS1 (39). In the presence of human or S. cerevisiae RPA, 259-nt fragments are displaced by BLM, although SSB and T4 gp32 failed to stimulate this helicase (40). There are three possible ways for a single-stranded DNA-binding protein to stimulate strand displacement carried out by a DNA helicase. One is to bind to the single-stranded region of the substrate resulting in the disruption of its secondary structures. This would eliminate the need for the helicase to denature such secondary structure as well as possibly reduce nonspecific binding by the helicase. The second way is to bind to the single-stranded DNA produced by unwinding by the helicase and prevent it from re-annealing. The third way is by stimulating the activity of the helicase through specific protein-protein interactions. The observations that the amount of MER3 required for strand displacement increases as the length of the double-stranded region increases, that both RPA and SSB show stimulation of strand displacement only when the length of the double-stranded region increases, and that the order of addition of RPA and MER3 does not affect the extent of unwinding all indicate that RPA prevents the re-annealing of the products produced by MER3 (Fig. 7A). It should be noted that 10–40 MER3 molecules/M13mp18 DNA molecule are required for the efficient unwinding of each substrate examined. As BLM helicase forms homo-oligomers including hexameric rings (41), it is possible that MER3 helicase also functions in an oligomeric form. Consistent with this possibility, analysis by gel filtration indicates that MER3 (137-kDa monomer) chromatographs as oligomeric species ranging from 140 to 750 kDa (data not shown). The relatively high activity level of the MER3 ATPase suggests that MER3 is either continually translocating along DNA or forms a stable MER3-DNA complex capable of hydrolyzing ATP, independent of DNA unwinding. Further studies are required to test these possibilities.
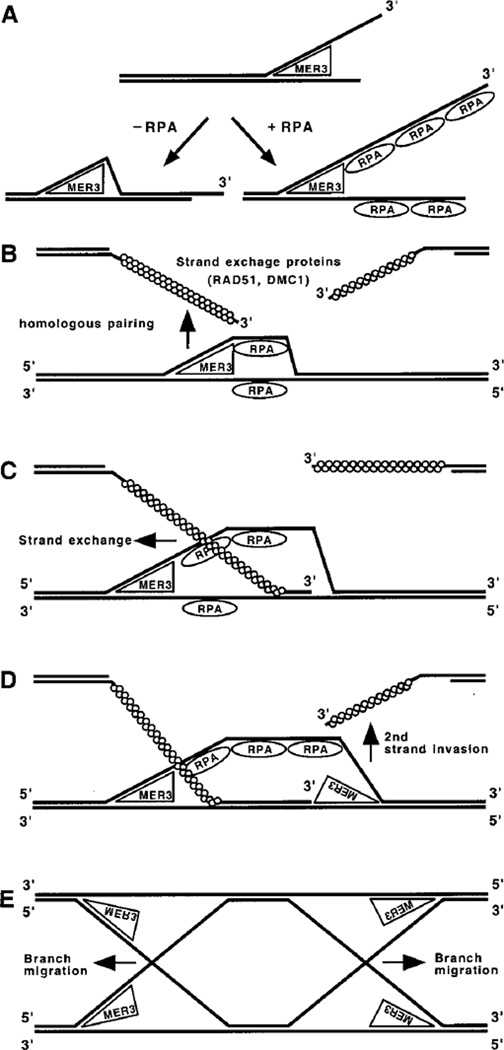
Fig. 7. Roles for MER3 helicase in DNA recombination.
A, DNA unwinding reaction carried out by MER3 helicase with or without RPA. The polarity of MER3 helicase is in the 3′ to 5′ direction with respect to a single-stranded region. In the absence of single-stranded DNA-binding protein (−RPA), the region unwound by MER3 will re-anneal to form double-stranded DNA. In the presence of RPA (+RPA), the single-stranded regions produced by MER3 will be bound by RPA. RPA binding to the single-stranded regions prevents them from re-annealing to form double-stranded DNA. Thus, RPA stimulates the unwinding and subsequent displacement of long DNA fragments by MER3 helicase. There are several steps of DNA recombination at which the MER3 helicase might function. B, a possible role for MER3 in homologous pairing by unwinding double-stranded DNA prior to the initiation of DNA strand exchange. At each DSB site, two single-stranded DNA ends having 3′ -overhangs are formed. To create double-Holliday junctions that potentially result in crossovers, both of these ends must invade the same chromatid of homolog. C, once strand exchange has been initiated by RAD51 and/or DMC1, MER3 helicase could bind to the single-stranded region of the resulting D-loop and further unwind the duplex region with the aid of RPA. This would stimulate strand exchange during the first strand invasion. D, MER3 could play a similar role during the second strand invasion by unwinding the duplex region in the opposite direction as illustrated. This would stimulate strand exchange during the second strand invasion. E, it is also possible that MER3 helicase participates in branch migration of Holliday junctions following the first and second strand invasion.
Using two different DNA substrates, the polarity of the MER3 helicase was found to be 3′ to 5′ relative to the single-stranded regions of the helicase substrate. Interestingly, SGS1 and BLM as well as RecQ and WRN also unwind DNA with a 3′ to 5′ polarity (42–45). It is thus likely that a 3′ to 5′ polarity is a general feature for the RecQ family of DNA helicases. There is another similarity among MER3, SGS1, and BLM. The ATPase activity of SGS1 and BLM is also stimulated by either single- or double-stranded DNA (39, 43). These biochemical and sequence similarities among the MER3, SGS1, and BLM helicases suggest a functional similarity consistent with the observation that all of these helicases act in DNA recombination.
What role might MER3 play in meiotic crossing over? It has been observed that meiosis-specific DSBs accumulate in mer3 deletion mutants and that a fraction of these DSBs are hyperresected (8). Thus, MER3 does not appear to be required for DSB formation and subsequent resection of the ends of the DSBs. Mutations in the RAD51 and DMC1 genes, which encode homologous pairing proteins required for the formation of joint molecules during meiotic recombination, cause virtually the same molecular defect during meiosis as that caused by a mer3 deletion mutation. In immunostaining experiments with anti-RAD51 and anti-DMC1 antibodies, RAD51 and DMC1 transiently co-localize as foci in spread meiotic nuclei (46). The RAD51 and DMC1 foci appear to accumulate in a mer3 deletion mutant (8). Two possible but similar roles for MER3 consistent with these observations are that the MER3 helicase promotes the initial unwinding of double-stranded DNA that may be required for the initiation of homologous pairing (Fig. 7B) or that MER3 drives the extensive strand exchange that occurs during recombination promoted by RAD51 and DMC1 (Fig. 7, C and D). Thus, in the absence of MER3 there may be insufficient homologous pairing or strand exchange to produce the joint molecules that are required for crossing over, although pairing and strand exchange sufficient for gene conversion may occur. Because recombination initiates but is not completed in a mer3 mutant, the steady state level of early recombination intermediates likely increases resulting in the apparent accumulation of RAD51 and DMC1 foci. Another possibility is that MER3 helicase catalyzes branch migration of Holliday junctions in the same manner as SGS1 and BLM (47, 48) (Fig. 7E). In this circumstance, it may be that the failure to form a mature crossover intermediate impairs crossover control in a mer3 mutant. Our results are consistent with the idea that crossover control acts during the formation of crossover intermediates rather than during their resolution.
Acknowledgments
We are grateful to the members of the Kolodner laboratory, especially Kyungjae Myung, for helpful discussions.
Footnotes
This work was supported by National Institutes of Health Grant GM26017 (to R. D. K.).
Supported by the Human Frontier Science Program. Present address: Dept. of Biology, Osaka University Graduate School of Science, Machikaneyama, Toyonaka, Osaka 560-0043, Japan.
Supported by the Jane Coffin Childs Memorial Fund for Medical Research.
The abbreviations used are: DSB, double strand break; nt, nucleotide(s); kb, kilobase pair(s); BLM, Bloom syndrome; RPA, replication protein A; SSB, single-stranded DNA-binding protein; PCR, polymerase chain reaction.
T. Nakagawa and R. D. Kolodner, submitted for publication.
References
- 1.Roeder GS. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 2.Kleckner N. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sym M, Roeder GS. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 4.Ross-Macdonald P, Roeder GS. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 5.Hollingsworth NM, Ponte L, Halsey C. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 6.Hunter N, Borts RH. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 7.Chua PR, Roeder GS. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa T, Ogawa H. EMBO J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TF, Kleckner N, Hunter N. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsubouchi H, Ogawa H. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 13.Bishop DK, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 14.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 15.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Golub EI, Gupta R, Radding CM. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington DC: American Society for Microbiology Press; 1995. pp. 407–464. [Google Scholar]
- 18.Schwacha A, Kleckner N. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 19.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 20.Paques F, Haber JE. Microbiol. Mol. Biol. Rev. 1999;63:349–404.7. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storlazzi A, Xu L, Schwacha A, Kleckner N. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa T, Ogawa H. Genes Cells. 1997;2:65–79. doi: 10.1046/j.1365-2443.1997.d01-283.x. [DOI] [PubMed] [Google Scholar]
- 23.Gorbalenya AE, Koonin EV. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 24.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 26.Watt PM, Hickson ID, Borts RH, Louis EJ. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myung K, Datta A, Chen C, Kolodner RD. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 28.Frei C, Gasser SM. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao S, Graham J, Yan H. Genes Dev. 2000;14:2570–2575. doi: 10.1101/gad.822400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleted in proof
- 33.Alani E, Thresher R, Griffith JD, Kolodner RD. J. Mol. Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 34.McEntee K, Weinstock GM, Lehman IR. Proc. Natl. Acad. Sci. U. S. A. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson AW, Kolodner RD. J. Biol. Chem. 1991;266:14046–14054. [PubMed] [Google Scholar]
- 37.Matson SW. J. Biol. Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 38.Matson SW, Bean DW, George JW. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 39.Bennett RJ, Sharp JA, Wang JC. J. Biol. Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 40.Brosh RM, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 41.Karow JK, Newman RH, Freemont PS, Hickson ID. Curr. Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Mullen JR, Brill SJ, Kleff S, Romeo AM, Sternglanz R. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 43.Karow JK, Chakraverty RK, Hickson ID. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 44.Umezu K, Nakayama K, Nakayama H. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 46.Bishop DK. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 47.Bennett RJ, Keck JL, Wang JC. J. Mol. Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 48.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]