Abstract
The transcription factor Foxp3 is critical to the suppressive phenotype of CD4+ regulatory T cells. Studies have clearly shown that numerous autoimmune diseases are marked by the presence of activated CD4+ T cells within the setting of chronic inflammation. Therefore drugs capable of inducing Foxp3 expression in activated CD4+ T cells could be of great therapeutic interest. We have previously shown that the small molecule G-1, an agonist directed against the membrane-bound estrogen receptor GPER, can induce IL10 expression in naïve CD4+ T cells. In addition, we and others have demonstrated that G-1 attenuates disease in an animal model of experimental autoimmune encephalomyelitis. Using ex vivo cultures of purified CD4+ T cells, we show that G-1 can elicit Foxp3 expression under TH17 polarizing conditions, which mimic the in situ inflammatory milieu of several autoimmune diseases. These findings build upon previous results demonstrating the immunosuppressive properties of the novel estrogenic small molecule G-1.
Keywords: GPR30, GPER, estrogen receptor, G-1, Foxp3, T cells
INTRODUCTION
The immune response is largely coordinated by CD4+ T cells, which upon T cell receptor (TCR) antigen recognition undergo clonal expansion and differentiation into one of three main effector lineages; TH1, TH2, or TH17 cells. These divergent populations coordinate distinct immune responses through the expression of unique mediators and signaling molecules, including canonical transcription factors; T-bet (TH1), GATA3 (TH2), or RORγt (TH17). While highly effective at limiting infection and neoplastic disease, an inherent risk to this process is the generation of clones that recognize self-antigens (eg. autoimmune disease) or otherwise benign environmental contaminants (eg. allergies). To mitigate this danger, the immune system has evolved mechanisms aimed at eliminating self-reactive cells and limiting the extent of inflammation in situ. One such mechanism is the induction of regulatory T (TREG) cell populations.
CD4+ TREG cells are a diverse population of immunosuppressive T cells that function via a variety of mechanisms. The most well defined class of TREG cells expresses the transcription factor Foxp3 (1–3). This protein is critical to immune homeostasis as loss of Foxp3 function in both humans and mice precipitates a fatal multi-organ autoimmune condition marked by the inability to control T cell responses (4, 5). Experimentally, Foxp3+ TREG cells suppress numerous animal models of autoimmune disease (6). Conversely, excessive TREG activity has been shown to inhibit protective anti-tumor immune responses (7, 8) and immunity against infection (9). Thus, the regulation of Foxp3+ cell populations is a critical point of control in many disease settings.
Foxp3 expression can be elicited during thymic development in a TCR dependent fashion (10), leading to “natural” or nTREG cells. However, conventional T cells activated in the periphery can be driven towards a suppressive phenotype when stimulated in the presence of TGFβ (11), IL2 (12), IL10 (13), IL35(14), and/or retinoic acid (15), or when antigen recognition occurs in the absence of appropriate costimulatory ligands such as CD80 and CD86 (16). This leads to numerous types of inducible regulatory T (iTREG) cell populations (17), many of which express Foxp3.
The female sex steroid hormone estrogen (E2) has several well-documented immunosuppressive properties, including induction of Foxp3 in T cells (18) and expression of the cytokine IL10 (19). Although the effects of E2 are traditionally thought to be mediated by the classical nuclear estrogen receptors, ERα and ERβ, recent studies have suggested that the G protein-coupled estrogen receptor (GPER, previously termed GPR30) may contribute to many of the physiological effects of E2 (reviewed in (20, 21)). GPER is a member of the 7-transmembrane spanning superfamily of G protein-coupled receptors (GPCRs) and has been shown to function as an estrogen-binding and-responsive protein (22, 23). The identification and characterization of GPER presented a novel therapeutic target relative to the classical estrogen receptors and provided the opportunity to identify receptor class-specific regulators of estrogen activity.
In 2006, we identified a non-steroidal GPER agonist G-1 that displays high selectivity against the classical estrogen receptors permitting the selective activation of GPER in cellular and animal models (24). G-1 binds to GPER with similar affinity to E2 (G-1 exhibits about 2-fold lower affinity) and initiates multiple signaling pathways, similar to E2, including calcium mobilization and ERK, PI3K and adenylyl cyclase activation (24, 25). However, unlike E2, G-1 exhibits minimal reproductive effects on the uterus, such as imbibition and epithelial proliferation (26). Nevertheless in animal models, like E2, G-1 can attenuate multiple pathophysiological conditions that exhibit an important immune/inflammatory component such as stroke (27), cardiac ischemia-reperfusion injury (28) and experimental autoimmune encephalomyelitis (EAE, a murine model of multiple sclerosis) (29, 30). Although the effects of estrogen in ameliorating symptoms and immune cell infiltration in EAE have been well documented (31), a critical role for GPER in the protective anti-inflammatory effects of E2 and oral ethinyl estradiol in the EAE model has only recently been demonstrated using GPER knockout mice (19). A potential mechanism involves the direct or indirect regulation of pro-inflammatory cells and cytokines or the conversion of pro-inflammatory immune cells into regulatory or anti-inflammatory cells. We have recently shown that G-1 can induce IL10 expression in T cells both in vitro and in in vivo (32). In the current study, we show that G-1 can induce Foxp3 expression in cultured CD4+ T cells, even under pro-inflammatory TH17-polarizing conditions. Our findings are significant as numerous disease processes are associated with chronic inflammation characterized by TH17-polarizing conditions. Therefore, G-1’s effects on Foxp3 expression, and its immunosuppressive properties in additional autoimmune models, warrant further exploration.
MATERIALS AND METHODS
Mice
Wild type and Foxp3-IRES-EGFP knockin (Foxp3egfp) mice (33) (7–11 weeks of age) were used in this study for collection of purified T cell populations by fluorescence-activated cell sorting (FACS). All mice were on the C57BL/6 genetic background and were purchased from Jackson Laboratory. Animals were housed, bred, and cared for according to the institutional guidelines in the Animal Resource Facility at the University of New Mexico, and studies were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) under approved protocols. Only male mice were used in this study.
Purification of T cell populations
T cells were obtained from single cell suspensions following homogenization of spleens and lymph nodes by mechanical disruption and passage through a 70μm nylon filter. Suspensions were stained with anti-CD4, anti-CD62L, and anti-CD44 antibodies (Biolegend). Enriched populations of CD4+CD62Lhi and CD4+CD44loCD62Lhi naïve T cells were collected by flow cytometric cell sorting on a MoFlo cell sorter (Cytomation). Purity was regularly >96%.
Culture conditions
All experiments and cell purification were carried out in RPMI 1640 medium supplemented with fetal bovine serum (FBS), penicillin/streptomycin, L-glutamine, HEPES, sodium pyruvate, and 2-mercaptoethanol. Phenol red-free buffers and charcoal-stripped FBS were used to minimize exposure to estrogens or phyto/xenoestrogens that could confound results. Cells were stimulated in culture with soluble anti-CD3ε (1.0 μg/mL) and anti-CD28 (2.5 μg/mL) antibodies (Biolegend), and supplemented with various combinations of TGFβ (0.5–5.0 ng/mL, 0.5 ng/mL was used unless otherwise indicated), IL6 (20 ng/mL), and IL23 (20 ng/mL) as described (Biolegend and eBiosciences). Where indicated, cultures were supplemented with 100nM G-1 (a concentration based on previous studies (32)).
Flow cytometry
Cells were collected from single cell suspensions of homogenized tissue or from purified cultures of T cells as indicated. For surface staining, cells were resuspended in 100μl 50% PBS + 50% medium with appropriate antibodies (including the appropriate isotype matched control antibodies) diluted 1:100. Cells were stained for 30 minutes at room temperature, after which 500μl of PBS/medium was added to dilute the antibody, and incubated for an additional 5 minutes before being harvested by centrifugation. Cells were then fixed with Fixation Buffer (FB, Biolegend). Alternatively, for intracellular cytokine staining, cultures were then treated with PMA (50 ng/mL) and ionomycin (500 ng/mL) for 4–5 hours in the presence of Brefeldin A (Biolegend) followed by fixation in FB prior to staining with antibodies diluted 1:50. Immediately after staining, data were collected on a FACScalibur (Becton Dickinson). Data analysis was performed using FlowJo software (TreeStar).
RT-PCR
For RNA collection, cells were homogenized with QIAshredder tubes (Qiagen) and RNA was extracted using the RNeasy mini kit (Qiagen) following manufacturer instructions. RNA was then quantitated using a Nanodrop spectrophotometer (Thermo Scientific). Reverse transcription was performed in a 20ul reaction volume using 100 ng RNA and Applied Biosystems High Capacity cDNA Reverse Transcription kit with RNase inhibitor (Applied Biosystems). For end-point PCR, 2 ul RT reaction was amplified with Taq DNA polymerase (Applied Biosystems) according to manufacturers instructions. Resulting amplicons were separated on agarose gels and visualized using ethidium bromide. For quantitative PCR (qRT-PCR), samples were prepared using Applied Biosystems SYBR Green Master Mix. Reactions were carried out in a 20 ul reaction volume containing 10 ul 2X SYBR Green master mix, 0.5 uM forward and reverse primer, and 2 ul (10 ng) cDNA template. Quantitative PCR was performed on an Applied Biosystems 7500 Fast Real-time PCR system under standard conditions consisting of 50°C for 2 min followed by 40 cycles of 95° C for 15 sec, 60° C for 1 min. GAPDH was used as a loading control for all samples. 7500 Fast software was used for data collection. Data were analyzed using the standard ΔΔCT method (34).
RESULTS
GPER expression in CD4+ T cells
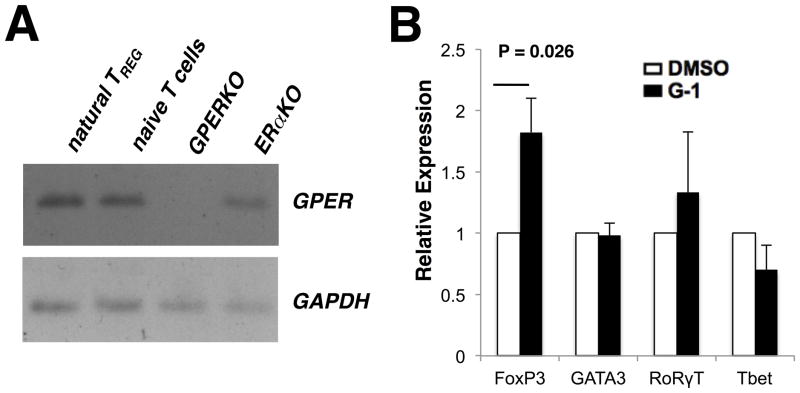
It has been reported that human regulatory T cells (29) and murine splenocytes (35) express GPER. However, no reports investigating GPER expression in murine CD4+ T cells have been published to date. To begin our studies, we sought to determine if GPER is expressed within various CD4+ T cell populations from C57BL/6 mice. Hence CD4+Foxp3+ TREGs and CD4+CD44loCD62LhiFoxp3− naïve T cells were sorted by FACS from Foxp3egfp mice. Due to a lack of antibody sensitivity, expression of GPER mRNA was determined by endpoint RT-PCR, with GPERKO and ERαKO splenocytes serving as controls. We detected GPER expression in both TREG cells and naïve T cells (Figure 1A).
Figure 1. G-1 drives Foxp3 expression in CD4+ T cells.
(A) CD4+GFP+ natural Treg and CD4+CD62LhiCD44loGFP- naïve T cells (NTC) were isolated by FACS from Foxp3egfp, GPERKO, and ERαKO mice. RNA was collected and reverse transcription used to make cDNA that was then used as the template for end-point PCR. PCR products confirmed the presence of GPER in nTREG, NTC, and ERαKO but not GPERKO cells. GAPDH was used as a loading control. (B) Naïve T-cells (CD4+CD62Lhi ) were collected from spleen and inguinal lymph nodes of mice by FACS. Quantitative PCR was performed using GAPDH as an internal control and data were analyzed using the 2ΔΔCT method. Average relative expression over 4 independent experiments is shown for the Transcription factors Foxp3, GATA3, RORγT, and T-bet. P values determined by Student’s t-test. Error bars = S.D. If no P value is indicated, P > 0.05.
G-1 induces Foxp3 mRNA expression
We next examined the impact of G-1 stimulation on the expression of lineage-specific transcription factors responsible for programming the various helper T cell subsets. To determine if G-1 could affect the expression of the canonical transcription factors by direct action on CD4+ T cells, naïve T cells were collected by FACS from Foxp3egfp mice and expanded ex vivo with antiCD3 and antiCD28 antibody treatment (to mimic stimulation by antigen-presenting cells) under TH0 conditions (without the addition of exogenous cytokines or neutralizing antibodies), in the presence of 100 nM G-1 or equivalent concentrations of DMSO. Samples were collected after 4 days in culture and analyzed for mRNA expression of T-bet (TH1), GATA3 (TH2), RORγt (TH17), and Foxp3 (TREG) by qRT-PCR. G-1 treatment led to an increase in the expression of Foxp3, with no change in the expression of the other three effector T cell transcription factors (Figure 1B). The increase in overall Foxp3 RNA expression within the population of cells correlated with an increase in the number of cells expressing Foxp3, as determined by GFP expression (see Figure 2, first column). This result was interesting given the findings from previous reports which showed that estrogen could expand the Foxp3 population in vivo (18), while G-1-mediated suppression of EAE was associated with an increase in the expression of PD-1 on Foxp3+ cells with no corresponding increase in the number of Foxp3+ cells under disease conditions (30).
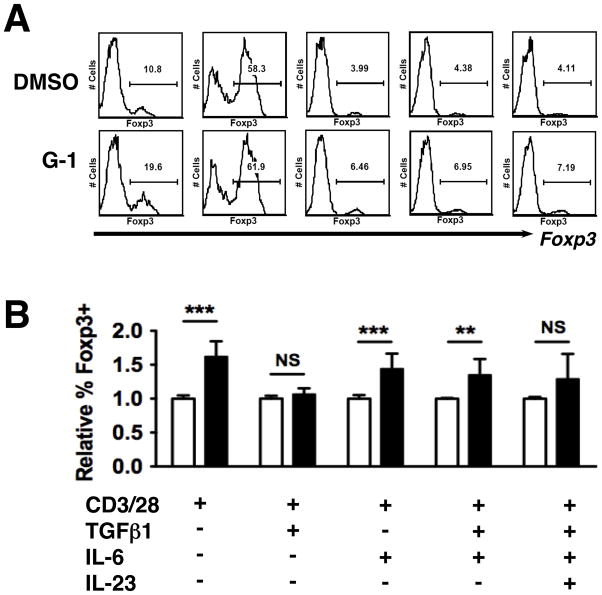
Figure 2. G-1 induces Foxp3 under T(H)17-polarizing conditions.
CD4+CD62LhiCD44lo naive T cells from Foxp3egfp mice were collected by FACS and cultured for 4 days under the conditions indicated. (A) Representative histograms showing gating for GFP (Foxp3) expression analysis. (B) Summary of data from three to four independent experiments, with conditions for all panels indicated at the bottom of the figure. Individual wells were supplemented with either 100nM G-1 (Black bars) or DMSO (White bars). P values determined by Student’s t-test. *** = P < 0.0005, ** = P< 0.005, * = P < 0.05, N.S. = not significant. Error bars = S.D.
G-1 increases the number of Foxp3+ cells under T(H)17 polarizing conditions
We hypothesized that the difference between our observations in Figure 2B and the previous report discussed above (30) was due to the presence of a TH17-polarizing inflammatory milieu in the EAE mice. This would not be unexpected as it is known that IL6, one of the cytokines implicated in TH17 differentiation, can inhibit Foxp3 expression (36). If there were a high concentration of IL6 during the preclinical stages of EAE development it is possible that this would mask the effects of G-1 in terms of Foxp3 induction, even without the development of overt disease. To determine if any of the key TH17-polarizing cytokines could alter G-1-mediated increases in Foxp3, naïve T cells were collected by FACS from Foxp3egfp mice and stimulated in cultures supplemented with various combinations of TGFβ, IL6, and IL23. The cytokine IL23 is important in the stabilization of the TH17 lineage (37) and is thought to be a critical factor in establishing the balance between TH17 and regulatory T cells in chronic inflammatory diseases such as Crohn’s Disease (38–40). Following 4 days in culture, cells were analyzed for the expression of GFP (a marker for Foxp3+ cells). Representative histograms are shown in Figure 2A. As mentioned above, we observed that G-1 treatment resulted in an increase in the number of Foxp3+ cells under TH0 conditions (Figure 2B). We also observed that G-1 led to an increase in the number of cells expressing Foxp3 in cultures supplemented with IL6 and IL6 + TGFβ. No effect was observed in cultures supplemented with TGFβ alone, though this may reflect the fact that TGFβ is a potent inducer of Foxp3 on its own, which has the potential to mask any additional effects of G-1. While we observed a trend towards increased numbers of Foxp3+ cells in G-1-treated cultures supplemented with IL23, the results were sufficiently variable between experiments that statistical significance was not reached (Figure 2B). However, the latter result does suggest that G-1-induced upregulation of Foxp3 continues to be physiologically regulated as IL23 opposes this upregulation.
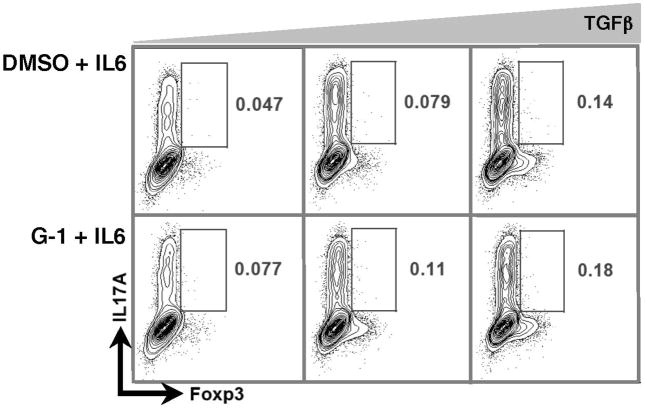
To further delineate G-1’s effects under TH17-polarizing conditions (IL6 + TGFβ), we asked whether Foxp3+ cells induced in this setting express the TH17 cytokine IL17A, and if so, was there was a difference between control and G-1-treated cultures? This was pertinent as we have previously shown that cells expressing Foxp3 under these experimental conditions are entirely of the hybrid T cell category (Foxp3+RORγt+) (32). Thus in theory they could secrete IL17A. If G-1-induced Foxp3+ cells were in fact secreting IL17A, they may exacerbate local inflammation rather than attenuate it. However, we detected very few IL17A+ cells in the Foxp3+ population (<0.2%, Figure 3), consistent with evidence showing Foxp3 can inhibit the function of RORγt (41).
Figure 3. Foxp3+ cells induced under T(H)17-polarizing conditions do not express IL17.
CD4+CD62LHI naive T cells from Foxp3egfp mice were collected by FACS and cultured for 4 days with IL6 + varying concentrations of TGFβ (0.5, 1.0, or 5.0 ng/mL). Cultures were supplemented with either DMSO or 100nM G-1, as indicated. Cells were subsequently stained for IL17A. Representative plots demonstrating IL17A and Foxp3 staining are shown. Data are from one of two independent experiments performed in triplicate.
G-1 induces small changes in PD-1 & CTLA-4 on stimulated CD4+ T cells
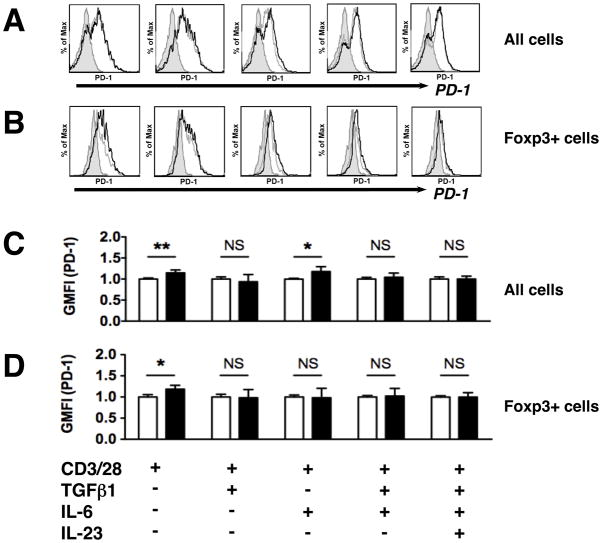
As mentioned previously, G-1 treatment has been reported result in increased surface expression of the inhibitory molecule programmed death 1 (PD-1) on Foxp3+ TREG cells within the draining lymph nodes of EAE mice (30). Thus we next examined the surface expression of PD-1, and another inhibitory receptor cytotoxic T-lymphocyte antigen 4 (CTLA-4), following G-1 treatment of purified T cells stimulated in culture. Like PD-1, CTLA-4 inhibits T cell activation and is upregulated on TREG populations. As before, we utilized naïve T cells purified by FACS and cultured with various combinations of TH17-polarizing cytokines TGFβ, IL6, and IL23, in addition to either G-1 or DMSO. Analysis of the entire culture population collectively showed that, under TH0 conditions, G-1 treatment led to an increase in the expression of PD-1 (Figure 4A/C) and CTLA-4 (Suppl. Figure S1A/C), as measured by geographic mean fluorescence intensity (GMFI). Similarly, PD-1 expression was increased in cultures supplemented with IL6 (Figure 4C). CTLA-4 was increased in cultures supplemented with either IL6 or TGFβ, but not when both were added together (Suppl. Figure S1C). Overall, no changes in either PD-1 or CTLA-4 expression were detected under either of the TH17-polarizing conditions (TGFβ + IL6 ± IL23). When gated exclusively on the Foxp3+ population, increases in PD-1 (Figure 4B/D) and CTLA-4 (Suppl. Figure S1B/D) were detected solely under non-polarizing conditions (TH0). Thus, there was no change in the surface expression of either inhibitory molecule on Foxp3+ cells when exogenous cytokines were added to the culture medium. All increases in PD-1 (Figure 4) and CTLA-4 (Suppl. Figure S1) were modest, in the range of 10 – 20%.
Figure 4. G-1 has a modest effect on PD-1 surface expression.
CD4+CD62LhiCD44lo naive T cells from Foxp3egfp mice were collected by FACS and cultured for 4 days under the conditions indicated, supplemented with either DMSO or 100nM G-1, as indicated. Surface expression of PD-1 was determined by flow cytometry. (A, B) Representative histograms showing gating for analysis of PD-1 surface expression, quantified using geographic mean fluorescence intensity (GMFI), on DMSO- (grey line) or G-1- (black lines) treated cells, and isotype (rat IgG2b,κ) controls (shaded region). (C, D) Summary of data from three to four independent experiments showing relative GMFI for G-1 treated cells (black bars) relative to DMSO treated cells (white bars). Treatment conditions for all panels (AD) are indicated at the bottom of the figure. P values were determined by Student’s t-test. *** = P < 0.0005, ** = P< 0.005, * = P < 0.05, N.S. = not significant. Error bars = S.D.
DISCUSSION
In this study, we have built upon our previous work delineating the effects of G-1 treatment on helper T cell lineages. Our previous report showed that G-1 induces IL10 expression within and secretion from CD4+ T cell undergoing clonal expansion and differentiation in culture (32), including under conditions known to drive TH17 differentiation. In that same report we also showed that splenocytes extirpated from naïve mice treated with subcutaneous G-1 demonstrated increased production of IL10 upon T cell activation in culture. Here we report that G-1 treatment of naïve T cells cultured under non-polarizing (TH0) conditions increases expression of the canonical regulatory T cell transcription factor Foxp3, while not affecting the expression of the effector transcription factors T-bet, GATA3, or RORγt, as determined by qRT-PCR. To determine whether increased Foxp3 mRNA reflected an increase in the number of Foxp3+ cells, we cultured naïve T cells from Foxp3egpf knock-in mice, which express GFP under the control of the Foxp3 promoter. Flow cytometric analysis of FACS purified naïve T cells stimulated under TH0 conditions demonstrated that G-1-treated cultures contained increased numbers Foxp3+ cells, consistent with the qRT-PCR data.
We next sought to further delineate the conditions under which G-1 could elicit Foxp3 expression, in particular the ability of G-1 to act as an immunosuppressive mediator in the setting of chronic inflammation, using T cell populations exposed to a TH17-like milieu, common in diseases such as rheumatoid arthritis (42) and inflammatory bowel disease (43). Thus we assessed the impact of G-1 on Foxp3 expression in cultures supplemented with various combinations of TH17-polarizing cytokines. Our data demonstrate that G-1 can elicit Foxp3 expression in cultures treated with IL6 and IL6 + TGFβ. Collectively, these data suggest that G-1-mediated Foxp3 expression resulting from direct action on the T cell populations can occur in a variety of inflammatory milieux. However, as IL23-treated cultures did not exhibit a similar extent of Foxp3 induction following G-1 treatment, it is possible that stabilization of the TH17 lineage following prolonged exposure to IL23 (37, 44–46) may limit G-1-mediated Foxp3 expression. This would indicate that G-1 is acting on differentiating but uncommitted TH17 cells to drive Foxp3 expression, a concept that warrants further investigation.
Two previous reports demonstrated that G-1 suppresses disease in the MS-like animal model EAE (29, 30). In one study, the authors found that G-1’s protective effects correlated with increased PD-1 expression on Foxp3+ TREG cells, and were dependent on intact PD-1 expression in the host animal as PD-1KO mice were not protected from disease by G-1 (30). These experiments were based on in vivo administration of G-1, and analysis was based on experiments with cells from the draining lymph nodes of diseased animals. Thus, it is not clear whether these observations reflect a direct effect of G-1 driving PD-1 expression within the Foxp3+ population itself, or are the result of G-1 effects on another cell type, perhaps leading to the induction of other mediators that indirectly led to the increase in PD-1 expression. We were able to detect increased expression of both PD-1 and CTLA-4 in some of the conditions tested ex vivo, but these effects were much smaller than those reported by Wang et. al. following in vivo G-1 treatment of EAE mice, wherein the percent of Foxp3+ cells expressing PD-1 nearly doubled (30). Additionally, we detected no changes under TH17-polarizing conditions (TGFβ + IL6 ± IL23), suggesting that G-1-mediated induction of PD-1 is likely the result of a distinct mechanism from that responsible for G-1-mediated Foxp3 expression.
The above data provide evidence that GPER is involved in estrogen-induced regulation of Foxp3 expression, building on data previously demonstrating a role for the “classical” estrogen receptor ERα. It has been reported that estrogen treatment induces Foxp3+ expression within the draining lymph nodes of EAE mice in an ERα-dependent fashion as E2 treatment did not increase Foxp3 expression in ERα−/− mice (47). Moreover, these findings correlate with clinical outcomes as E2-mediated protection from EAE is attenuated, but not eliminated, in ERα−/− mice (47, 48). It is notable that a similar reduction in E2-mediated protection that has been shown in GPER−/− mice (30). It is possible that ERα and GPER act through distinct mechanisms, which would be consistent with the data showing that G-1’s protective effects in the setting of EAE are completely lost in GPERKO mice while estrogens effects are only partly attenuated in the same mice. The direct relationships between GPER and ERα signaling pathways within immune populations are likely complex and warrant further investigation.
There are several caveats to our work that deserve further discussion. We chose to use male mice in our studies to eliminate the confounding effects of surgery (ovariectomy) and/or cycling levels of endogenous estrogens found in female mice. Thus, an important caveat to this approach is that the findings herein may not recapitulate in ovary-intact female mice and some of the differences in the data discussed above may in fact represent a sexual dimorphism of GPER signaling. Additionally, the data presented here reflect the direct effects of G-1 on T cell populations, but it is unclear how readily these findings will translate to the disease context in vivo where complex networks of cytokine signaling may have a profound impact on the final outcome of GPER and/or ERα activation. Moreover, previous studies have focused on Foxp3 expression within the setting of EAE in female mice, and in vivo data outside this model are currently lacking. Further studies investigating GPER involvement in estrogen-induced Foxp3 expression and immune regulation in a variety of contexts are warranted.
Supplementary Material
CD4+CD62LhiCD44lo naive T cells from Foxp3egfp mice were collected by FACS and cultured for 4 days under the conditions indicated, supplemented with either DMSO or 100nM G-1, as indicated. Surface expression of CTLA-4 was determined by flow cytometry. (A, B) Representative histograms showing gating for analysis of CTLA-4 surface expression, quantified using GMFI, on DMSO- (grey line) or G-1- (black lines) treated cells, and isotype controls (shaded region). (C, D) Summary of data from three to four independent experiments showing relative GMFI for G-1-treated cells (black bars) relative to DMSO-treated cells (white bars). Conditions for all panels (A–D) are indicated at the bottom of the figure. P values were determined by Student’s t-test. *** = P < 0.0005, ** = P< 0.005, * = P < 0.05, N.S. = not significant. Error bars = S.D.
Acknowledgments
This work was supported by NIH grants R01 CA116662, CA127731 and CA163890 (ERP). Data was generated in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center. The authors would like to thank Drs. Rick Lyons and Mary Lipscomb for insightful discussions, and Dr. Helen Hathaway for her expertise in the care and use of mice.
Footnotes
Financial Disclosure: RLB and KSO declare no conflicts of interest. ERP declares US patent #7,875,721 for G-1.
AUTHORSHIP CONTRIBUTIONS
RLB, KSO, and ERP designed and interpreted experiments. RLB and KSO carried out experiments. RLB, KSO, and ERP wrote the manuscript.
References
- 1.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 3.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nature immunology. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 4.Patey-Mariaud de Serre N, Canioni D, Ganousse S, Rieux-Laucat F, Goulet O, Ruemmele F, Brousse N. Digestive histopathological presentation of IPEX syndrome. Mod Pathol. 2008 doi: 10.1038/modpathol.2008.161. [DOI] [PubMed] [Google Scholar]
- 5.Clark LB, Appleby MW, Brunkow ME, Wilkinson JE, Ziegler SF, Ramsdell F. Cellular and molecular characterization of the scurfy mouse mutant. J Immunol. 1999;162:2546–2554. [PubMed] [Google Scholar]
- 6.Yuan Q, Bromley SK, Means TK, Jones KJ, Hayashi F, Bhan AK, Luster AD. CCR4-dependent regulatory T cell function in inflammatory bowel disease. J Exp Med. 2007;204:1327–1334. doi: 10.1084/jem.20062076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boissonnas A, Scholer-Dahirel A, Simon-Blancal V, Pace L, Valet F, Kissenpfennig A, Sparwasser T, Malissen B, Fetler L, Amigorena S. Foxp3+ T cells induce perforin-dependent dendritic cell death in tumor-draining lymph nodes. Immunity. 2010;32:266–278. doi: 10.1016/j.immuni.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nature reviews. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 9.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annual review of immunology. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 10.Nunes-Cabaco H, Ribot JC, Caramalho I, Serra-Caetano A, Silva-Santos B, Sousa AE. Foxp3 induction in human and murine thymus precedes the CD4+ CD8+ stage but requires early T-cell receptor expression. Immunol Cell Biol. 2010;88:523–528. doi: 10.1038/icb.2010.4. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nature immunology. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 13.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nature immunology. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kochetkova I, Golden S, Holderness K, Callis G, Pascual DW. IL-35 stimulation of CD39+ regulatory T cells confers protection against collagen II-induced arthritis via the production of IL-10. J Immunol. 2010;184:7144–7153. doi: 10.4049/jimmunol.0902739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elias KM, Laurence A, Davidson TS, Stephens G, Kanno Y, Shevach EM, O’Shea JJ. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–1020. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. The Journal of experimental medicine. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nature reviews. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polanczyk MJ, Hopke C, Huan J, Vandenbark AA, Offner H. Enhanced FoxP3 expression and Treg cell function in pregnant and estrogen-treated mice. Journal of neuroimmunology. 2005;170:85–92. doi: 10.1016/j.jneuroim.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 19.Yates MA, Li Y, Chlebeck PJ, Offner H. GPR30, but not estrogen receptor-alpha, is crucial in the treatment of experimental autoimmune encephalomyelitis by oral ethinyl estradiol. BMC Immunol. 2010;11:20. doi: 10.1186/1471-2172-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prossnitz ER, Arterburn JB, Smith HO, Oprea TI, Sklar LA, Hathaway HJ. Estrogen signaling through the transmembrane G protein-coupled receptor GPR30. Annu Rev Physiol. 2008;70:165–190. doi: 10.1146/annurev.physiol.70.113006.100518. [DOI] [PubMed] [Google Scholar]
- 21.Prossnitz ER, Barton M. The G-protein-coupled estrogen receptor GPER in health and disease. Nat Rev Endocrinol. 2011;7:715–726. doi: 10.1038/nrendo.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–1630. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 23.Filardo EJ, Thomas P. GPR30: a seven-transmembrane-spanning estrogen receptor that triggers EGF release. Trends Endocrinol Metab. 2005;16:362–367. doi: 10.1016/j.tem.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Bologa CG, Revankar CM, Young SM, Edwards BS, Arterburn JB, Kiselyov AS, Parker MA, Tkachenko SE, Savchuck NP, Sklar LA, Oprea TI, Prossnitz ER. Virtual and biomolecular screening converge on a selective agonist for GPR30. Nat Chem Biol. 2006;2:207–212. doi: 10.1038/nchembio775. [DOI] [PubMed] [Google Scholar]
- 25.Prossnitz ER, Barton M. The G-protein-coupled estrogen receptor GPER in health and disease. Nat Rev Endocrinol. 2011;7:715–726. doi: 10.1038/nrendo.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis MK, Burai R, Ramesh C, Petrie WK, Alcon SN, Nayak TK, Bologa CG, Leitao A, Brailoiu E, Deliu E, Dun NJ, Sklar LA, Hathaway HJ, Arterburn JB, Oprea TI, Prossnitz ER. In vivo effects of a GPR30 antagonist. Nat Chem Biol. 2009;5:421–427. doi: 10.1038/nchembio.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Subramanian S, Dziennis S, Jia J, Uchida M, Akiyoshi K, Migliati E, Lewis AD, Vandenbark AA, Offner H, Hurn PD. Estradiol and G-1 reduce infarct size and improve immunosuppression after experimental stroke. J Immunol. 2010;184:4087–4094. doi: 10.4049/jimmunol.0902339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bopassa JC, Eghbali M, Toro L, Stefani E. A novel estrogen receptor GPER inhibits mitochondria permeability transition pore opening and protects the heart against ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2010;298:H16–23. doi: 10.1152/ajpheart.00588.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasko E, Haskell CA, Leung S, Gualtieri G, Halks-Miller M, Mahmoudi M, Dennis MK, Prossnitz ER, Karpus WJ, Horuk R. Beneficial role of the GPR30 agonist G-1 in an animal model of multiple sclerosis. J Neuroimmunol. 2009;214:67–77. doi: 10.1016/j.jneuroim.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Dehghani B, Li Y, Kaler LJ, Proctor T, Vandenbark AA, Offner H. Membrane estrogen receptor regulates experimental autoimmune encephalomyelitis through up-regulation of programmed death 1. J Immunol. 2009;182:3294–3303. doi: 10.4049/jimmunol.0803205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matejuk A, Bakke AC, Hopke C, Dwyer J, Vandenbark AA, Offner H. Estrogen treatment induces a novel population of regulatory cells, which suppresses experimental autoimmune encephalomyelitis. J Neurosci Res. 2004;77:119–126. doi: 10.1002/jnr.20145. [DOI] [PubMed] [Google Scholar]
- 32.Brunsing RL, Prossnitz ER. Induction of interleukin-10 in the T helper type 17 effector population by the G protein coupled estrogen receptor (GPER) agonist G-1. Immunology. 2011;134:93–106. doi: 10.1111/j.1365-2567.2011.03471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol. 2007;178:2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods (San Diego, Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Isensee J, Meoli L, Zazzu V, Nabzdyk C, Witt H, Soewarto D, Effertz K, Fuchs H, Gailus-Durner V, Busch D, Adler T, de Angelis MH, Irgang M, Otto C, Noppinger PR. Expression pattern of G protein-coupled receptor 30 in LacZ reporter mice. Endocrinology. 2009;150:1722–1730. doi: 10.1210/en.2008-1488. [DOI] [PubMed] [Google Scholar]
- 36.Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, Saouaf SJ, Wang Q, Hancock WW, Shen Y, Greene MI. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci U S A. 2008;105:14023–14027. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nature immunology. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neurath MF. IL-23: a master regulator in Crohn disease. Nature medicine. 2007;13:26–28. doi: 10.1038/nm0107-26. [DOI] [PubMed] [Google Scholar]
- 39.Izcue A, Hue S, Buonocore S, Arancibia-Carcamo CV, Ahern PP, Iwakura Y, Maloy KJ, Powrie F. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–570. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, Kitazume MT, Nakazawa A, Sugita A, Koganei K, Isobe K, Hibi T. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut. 2008;57:1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 41.Zhou L, Lopes JE, Chong MM, Ivanov, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alzabin S, Abraham SM, Taher TE, Palfreeman A, Hull D, McNamee K, Jawad A, Pathan E, Kinderlerer A, Taylor PC, Williams R, Mageed R. Incomplete response of inflammatory arthritis to TNFalpha blockade is associated with the Th17 pathway. Annals of the rheumatic diseases. 2012 doi: 10.1136/annrheumdis-2011-201024. [DOI] [PubMed] [Google Scholar]
- 43.Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. The Journal of clinical investigation. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nature immunology. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 46.McGeachy MJ, Cua DJ. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin Immunol. 2007;19:372–376. doi: 10.1016/j.smim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Polanczyk MJ, Carson BD, Subramanian S, Afentoulis M, Vandenbark AA, Ziegler SF, Offner H. Cutting edge: estrogen drives expansion of the CD4+CD25+ regulatory T cell compartment. J Immunol. 2004;173:2227–2230. doi: 10.4049/jimmunol.173.4.2227. [DOI] [PubMed] [Google Scholar]
- 48.Polanczyk M, Yellayi S, Zamora A, Subramanian S, Tovey M, Vandenbark AA, Offner H, Zachary JF, Fillmore PD, Blankenhorn EP, Gustafsson JA, Teuscher C. Estrogen receptor-1 (Esr1) and -2 (Esr2) regulate the severity of clinical experimental allergic encephalomyelitis in male mice. Am J Pathol. 2004;164:1915–1924. doi: 10.1016/s0002-9440(10)63752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD4+CD62LhiCD44lo naive T cells from Foxp3egfp mice were collected by FACS and cultured for 4 days under the conditions indicated, supplemented with either DMSO or 100nM G-1, as indicated. Surface expression of CTLA-4 was determined by flow cytometry. (A, B) Representative histograms showing gating for analysis of CTLA-4 surface expression, quantified using GMFI, on DMSO- (grey line) or G-1- (black lines) treated cells, and isotype controls (shaded region). (C, D) Summary of data from three to four independent experiments showing relative GMFI for G-1-treated cells (black bars) relative to DMSO-treated cells (white bars). Conditions for all panels (A–D) are indicated at the bottom of the figure. P values were determined by Student’s t-test. *** = P < 0.0005, ** = P< 0.005, * = P < 0.05, N.S. = not significant. Error bars = S.D.