Abstract
Because psychotropic drugs affect behavior, we can use changes in behavior to discover psychotropic drugs. The original prototypes of most neuroactive medicines were discovered in humans, rodents and other model organisms. Most of these discoveries were made by chance, but the process of behavior based drug discovery can be made more systematic and efficient. Fully automated platforms for analyzing the behavior of embryonic zebrafish capture digital video recordings of animals in each individual well of a 96-well plate before, during, and after a series of stimuli. To analyze systematically the thousands of behavioral recordings obtained from a large-scale chemical screen, we transform these behavioral recordings into numerical barcodes, providing a concise and interpretable summary of the observed phenotypes in each well. Systems-level analysis of these behavioral phenotypes generate testable hypotheses about the molecular mechanisms of poorly understood drugs and behaviors. By combining the in vivo relevance of behavior-based phenotyping with the scale and automation of modern drug screening technologies, systematic behavioral barcoding represents a means of discovering psychotropic drugs and provides a powerful, systematic approach for unraveling the complexities of vertebrate behavior.
I. Introduction
The prototypes of most modern psychiatric medicines were discovered due to their unexpected behavioral phenotypes in humans and other animals (Kokel and Peterson, 2008). Despite the efficacy of behavioral assays for drug discovery, systematic approaches to behavior-based chemical screening have been difficult to develop. Most model organisms (including mice, rats, and other rodents) are simply too large to be efficiently exposed to the small amounts of compounds in modern chemical libraries. Unlike larger vertebrates, zebrafish embryos are very well suited for high-throughput (HT) chemical screens. Thus, chemical screens in zebrafish may be useful for discovering novel psychotropic molecules to improve our understanding of the brain and behavior.
In theory, almost any behavioral phenotype could be used to identify psychotropic compounds. Embryonic and larval zebrafish exhibit a wide variety of behavioral phenotypes (Granato et al., 1996) including the optomotor response (Orger et al., 2000), optokinetic response (Emran et al., 2008), spontaneous activity (Saint-Amant and Drapeau, 1998), prey capture (Gahtan et al., 2005), sleep (Prober et al., 2006; Zhdanova et al., 2001), response to touch (Low et al., 2010), thermoregulation, and habituation (Best et al., 2008; Burgess and Granato, 2007). Although the small size of young zebrafish is essential for their utility in microwell plate screening assays, it is important to remember that differences may exist between developing and adult nervous systems. For example, although associative learning readily occurs in adult zebrafish, it has not (yet) been described in larvae.
Multidimensional approaches to complex phenotyping can uncover relationships among diseases, genes, and drugs (Lamb et al., 2006). Traditionally, such approaches have used gene expression profiling and high-content cell-based imaging (Perlman et al., 2004; Sørlie et al., 2001). Recently, zebrafish behavioral phenotypes have also been used to link small molecules to their target signaling pathways (Kokel et al., 2010; Rihel et al., 2010). Here, we describe how one such behavior, the photomotor response (PMR), can be used to identify novel psychotropic compounds and their targets.
II. The PMR Behavior
Between 30 and 42 h post fertilization (hpf), zebrafish embryos normally exhibit a low level of basal motor activity (Saint-Amant and Drapeau, 1998). During this phase of development, presentation of an intense photic stimulus elicits a series of robust and reproducible behaviors that we refer to as the photomotor response. Following the stimulus, animals remain motionless for a 1–2 s latency period before entering an excitation phase characterized by vigorous high-frequency body flexions and tail oscillations. After 5–7 s, the excitation behaviors cease abruptly, and are followed by a refractory period (lasting >15 s) during which basal activity is suppressed, and animals fail to respond to a second pulse of light. The neuronal circuitry that underlies the PMR is poorly understood. However, despite its novelty, the PMR offers several advantages as a chemical screening assay.
The PMR is very well suited for HT chemical screens because it is robust, reproducible, and scalable to a fully automated 96-well screening format. The PMR assay is performed on ~1.5-day-old animals that have not yet hatched from their chorions. The chorion serves as a natural testing chamber, isolating individual animals even when many of them are grouped together. Due to their chorions, large numbers of animals (eight to ten) can be assayed in a small volume without confounding interactions between individuals.
Automated phenotyping is incredibly useful for high-throughput screens. For example, we developed a fully automated system capable of tracking and quantifying zebrafish behaviors in high-throughput, 96-well format. This behavioral phenotyping platform is built upon a high-content imaging system that combines robotic scanning of microtiter plates and sophisticated image processing software. Digital video microscopy records movements in groups of zebrafish (up to 10 animals per microtiter well) before and after a series of light stimuli. Image analysis software extracts PMR activity data and plots the movements. This generates a behavioral profile for zebrafish exposed to any genetic manipulation or small molecule treatment. This automated approach can assess behavioral effects of hundreds or thousands of experimental conditions per day. Because the behavioral phenotypes integrate inputs from the intact nervous system, it should be possible to identify genes or small molecules that act through a broad range of mechanisms. Automated screening technologies are objective, quantitative, and indefatigable. Such technologies are essential for analyzing large-scale data sets and can be creatively designed for custom needs.
III. Methods
A. Aquaculture and Multiwell Plates
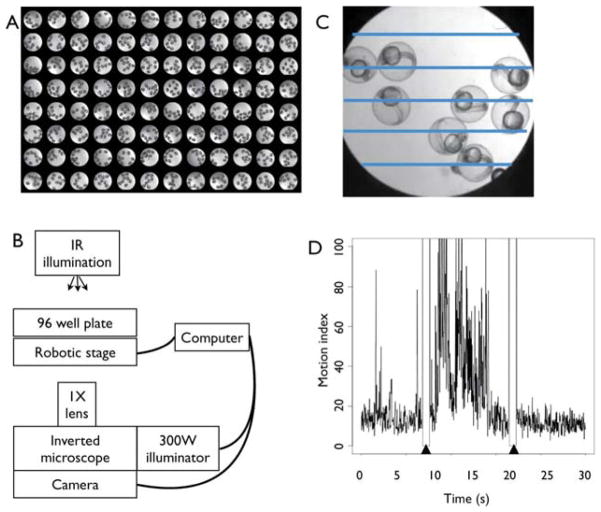
The PMR assay can be scaled to accommodate from tens to thousands of embryos. Embryos are collected from group matings of well-fed zebrafish. Embryos are sorted, at 6–12 hpf, to identify and eliminate any animals with gross development defects or that are substantially older or younger than expected. Embryos are raised in HEPES (10 mM) buffered E3 medium in a dark incubator at 28 degrees until 30 hpf. At 30 hpf, embryos are transferred to a slightly cooler (25 degree) incubator to slow their development, thereby prolonging the developmental window when the PMR can be elicited. Groups of 8–10 animals are manually distributed into the wells of flat bottom black 96-well plates (Fig. 1A). Opaque plates with transparent bottoms allow photic stimuli to be restricted to individual wells. Round bottom plates can also be used, however they hold fewer animals in the same visual plane.
Fig. 1.
The PMR assay and imaging system. (A) Composite photograph of a 96-well plate. Each well holds approximately 10 zebrafish embryos. (B) Schematic representation of the imaging hardware configuration. Infrared (IR) wavelengths provide illumination for the digital camera without affecting the PMR. A computer controls the robotic stage, stimulus shutter, and digital camera. We use a 300 W illuminator to deliver white light stimuli to the individual wells of the 96-well plate through the 1X objective lens. (C) Photograph of a single well. The five parallel lines represent regions of interests (ROIs) used to measure motor activity in the well. (D) A representative plot of a normal PMR in wild-type animals. The y-axis represents the motor index and the x-axis represents time. Arrowheads at 10 s and 20 s indicate stimulus artifacts from the light pulse. (For color version of this figure, the reader is referred to the web version of this book.)
B. Chemical Treatment
Chemical stocks dissolved in DMSO are added to each well such that the final concentration of DMSO is <1%. Low levels of DMSO do not affect the PMR, but higher concentrations may have detrimental effects on the animals. Each well is thoroughly mixed using a multi channel pipette. We find that many psychotropic compounds show activity at final concentrations between 10 and 100 μM. Typically, compounds lose activity at lower concentrations and become toxic at higher concentrations. Embryos are allowed to incubate in chemical solutions for 1–10 h prior to the PMR assay.
C. Dark Adaptation
Exposure to ambient light disrupts the PMR. Animals must be kept in the dark for at least 10 min prior to running the PMR assay. Following dark adaptation, intense white will elicit the PMR. Other wavelengths (~400–500 nm) will also elicit the response. Infrared wavelengths (>650 nm) do not affect the PMR, and can be safely used for imaging purposes.
D. Stimulus Presentation
We use a 300 W xenon bulb housed in a Sutter Lambda LS illuminator to deliver light stimuli (lasting 1 s) to the individual wells of a 96-well plate. Light stimuli can be restricted to single wells using a field diaphragm or pinhole mask. If a 300 W illuminator is unavailable, the PMR can be elicited using a variety of common light sources, including the standard 100 W halogen bulbs used on many dissecting microscopes.
E. Digital Video Capture
In a typical assay, 1000 frames of digital video are recorded at 33 fps using a Hamamatsu ORCA-ER camera mounted on a Nikon TE200 microscope with a 1X objective (Fig. 1B). We use Metamorph Software (Universal Imaging) to automate the robotic stage, digital video camera, and stimulus presentation. The entire PMR assay, including video recording and data processing, takes less than 1 min per well, enabling the routine screening of hundreds of wells per microscope per day. Uncompressed video files can be ~200 MB per 30 s video, so it is important to plan for enough hard drive space to store the files.
F. Measuring Motor Activity
We use Metamorph Software to analyze the digital video recordings and quantify the total motor activity in each well. The software is programmed to draw automatically a number of evenly spaced regions of interest (ROIs) across each well such that each embryo is likely to be crossed by one of the ROIs (Fig. 1C). The software then tracks the average intensity of the pixels for each ROI on each frame for the duration of the movie. As the embryos move, the average pixel intensity (API) of each ROI will change, providing a measure of motor activity in the well. Taking the absolute value of the difference in API for adjacent time points and then summing over the ROIs forms a motion index. This motion index correlates with the overall amount of motion in the well, both in terms of contraction frequency and number of animals in motion. Plotting the motion index over time provides a graphical representation of movement in the well (Fig. 1D). If Metamorph software is unavailable, similar types of data analysis can be made using freely available software such as ImageJ (Abramoff et al., 2004).
G. Behavioral Barcoding, Hit Identification and Phenoclustering
For small data sets, compounds that induce strong behavioral phenotypes can be identified and sorted manually. But for the purposes of large-scale chemical screening, software is used to identify wells that are statistically distinct from control wells. To facilitate the analysis of complex behaviors, the motion index profile for each well is first transformed into a “behavioral barcode.” First, salient features of the behavior are identified and extracted from the profile. For example, several phases of the PMR appear to be differentially modulated by specific small molecules. These phases include: background (prior to the stimulus), latency (0–2 s following the stimulus), excitation (2–5 s following the stimulus), and the refractory phases (10–30 s following the stimulus). Together, these measurements can be thought of as a behavioral barcode. Small molecule barcodes that are statistically distinct from controls are considered to be potential hits (Fig. 2A). It is important to retest and confirm the reproducibility of any putative hits. We typically retest and confirm behavioral phenotypes using at least 5–10 replicate wells on multiple days. Finally, freely available software packages, including Cluster (Eisen et al., 1998), and R (Team, 2008), can be used to cluster the behavioral barcodes based on their phenotypic similarities (Fig. 2B).
Fig. 2.
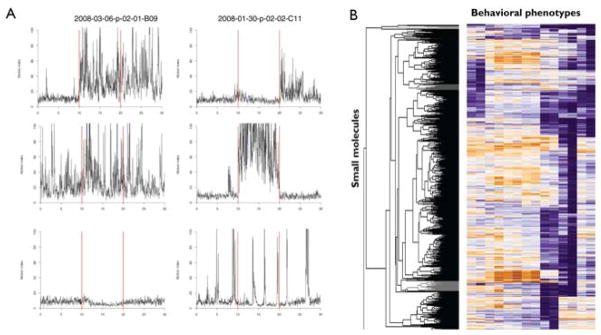
Behavioral phenotypes and phenoclustering. (A) Motor activity plots illustrating examples of six different PMR phenotypes. (B) Heat map showing phenotype-based clustering of small molecules (y-axis) based on behavioral barcode (x-axis) similarity. (See color plate.)
IV. Discussion
The PMR behavior provides a rapid, robust, and reproducible assay of nervous system function in living zebrafish. We have found that different structural and functional classes of neuroactive compounds cause specific and reproducible phenotypes in the PMR assay (Kokel et al., 2010). Compounds causing similar phenotypes may act on common molecular pathways. Thus, behavioral phenotypes can be used to link uncharacterized small molecules, and other perturbagens, to their molecular mechanisms and target pathways.
The PMR is just one of many potential behavioral assays. As more scalable behavioral phenotypes are identified, it may be possible to evaluate the same plates of animals through a battery of behavioral and phenotypic assays. Because different neuronal signaling pathways control different behaviors, additional assays would likely improve the resolution of behavioral barcodes. In the future, it may be possible to combine chemical screens with mutant or morphant phenotypes to identify small molecule suppressors and enhancers of specific pathways and phenotypes.
Acknowledgments
The authors acknowledge members of the Peterson lab for comments on the manuscript. This work was supported by NIH K01 mentored research scientist development award MH091449 (David Kokel) and grants MH085205 (Randall T. Peterson), MH086867 (Randall T. Peterson).
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11(7):36–42. [Google Scholar]
- Best J, et al. Non-associative learning in larval zebrafish. Neuropsychopharmacology. 2008;33:1206–1215. doi: 10.1038/sj.npp.1301489. [DOI] [PubMed] [Google Scholar]
- Burgess HA, Granato M. Sensorimotor gating in larval zebrafish. J Neurosci. 2007;27:4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran F, Rihel J, Dowling JE. A behavioral assay to measure responsiveness of zebrafish to changes in light intensities. J Vis Exp. 2008:20. doi: 10.3791/923. http://www.jove.com/details.php?id=923. [DOI] [PMC free article] [PubMed]
- Gahtan E, Tanger P, Baier H. Visual prey capture in larval zebrafish is controlled by identified reticulospinal neurons downstream of the tectum. J Neurosci. 2005;25:9294–9303. doi: 10.1523/JNEUROSCI.2678-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6:231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D, Peterson RT. Chemobehavioural phenomics and behaviour-based psychiatric drug discovery in the zebrafish. Brief Funct Genom Proteom. 2008;7:483. doi: 10.1093/bfgp/eln040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Low SE, et al. Touché is required for touch-evoked generator potentials within vertebrate sensory neurons. J Neurosci. 2010;30:9359–9367. doi: 10.1523/JNEUROSCI.1639-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orger MB, Smear MC, Anstis SM, Baier H. Perception of Fourier and non-Fourier motion by larval zebrafish. Nat Neurosci. 2000;3:1128–1133. doi: 10.1038/80649. [DOI] [PubMed] [Google Scholar]
- Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin over-expression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in1. The zebrafish embryo. J Neurobiol. 1998;37:622–632. doi: 10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.D.C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. URL http://www.R-project.org. [Google Scholar]
- Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]