Abstract
The response of microbial communities to long-term environmental change is poorly understood. Here, we study bacterioplankton communities in a unique system of coastal Antarctic lakes that were exposed to progressive long-term environmental change, using 454 pyrosequencing of the 16S rDNA gene (V3–V4 regions). At the time of formation, most of the studied lakes harbored marine-coastal microbial communities, as they were connected to the sea. During the past 20 000 years, most lakes isolated from the sea, and subsequently they experienced a gradual, but strong, salinity change that eventually developed into a gradient ranging from freshwater (salinity 0) to hypersaline (salinity 100). Our results indicated that present bacterioplankton community composition was strongly correlated with salinity and weakly correlated with geographical distance between lakes. A few abundant taxa were shared between some lakes and coastal marine communities. Nevertheless, lakes contained a large number of taxa that were not detected in the adjacent sea. Abundant and rare taxa within saline communities presented similar biogeography, suggesting that these groups have comparable environmental sensitivity. Habitat specialists and generalists were detected among abundant and rare taxa, with specialists being relatively more abundant at the extremes of the salinity gradient. Altogether, progressive long-term salinity change appears to have promoted the diversification of bacterioplankton communities by modifying the composition of ancestral communities and by allowing the establishment of new taxa.
Keywords: Antarctica, Bacteria, environmental change, long-term, pyrosequencing, salinity
Introduction
Unveiling the mechanisms that determine the spatio-temporal structuring of microbial communities and how these respond to environmental change are major challenges in microbial ecology. It has been shown that environmental filtering is one of the main mechanisms of community assembly in prokaryotes (for example, Jones and McMahon, 2009; Lindström and Langenheder, 2012). Yet, in some cases, microbial community assembly dynamics also follow predictions made by the neutral model (NM; Hubbell, 2001; Sloan et al., 2006; Östman et al., 2010). Hubbell's NM assumes that organisms with the same fitness can occupy a given environment depending on migration, compositional drift and extinction (but not on environmental filtering). As there is evidence that both environmental filtering and neutral dynamics shape bacterial communities simultaneously (Ofiteru et al., 2010; Langenheder and Szekely, 2011), both processes need to be considered when investigating the structure of microbial communities.
Environmental change is likely to have an effect on abundances and taxonomic composition of microbial communities (Allison and Martiny, 2008). Most communities are composed of a few abundant taxa and a great many rare ones (Pedrós-Alió, 2006, 2012). So far, both groups have been usually treated without distinction, although recent studies suggest that rare taxa may present characteristic metabolic activities (Jones and Lennon, 2010; Campbell et al., 2011), carry out particular metabolic functions (Pester et al., 2010) and show distinct spatial distribution patterns (Galand et al., 2009). Hence, a relevant question is whether abundant and rare taxa show similar or different responses to environmental change (considering ‘no reaction' as a possible response).
Changes in the environment may promote habitat diversification, which is likely to lead to an increase in habitat specialists (that is, taxa preferring specific habitats). Previous models and field studies indicate that there tend to be more habitat specialists than generalists in ecological communities, with generalists usually showing greater abundances than specialists (Guo et al., 2000; Kolasa and Romanuk, 2005; van der Gast et al., 2011). In general, habitat specialists seem to be mostly affected by environmental factors, while habitat generalists appear to respond predominantly to spatial factors (for example, geographical distance; Pandit et al., 2009).
To date, most studies exploring changes in microbial community composition in response to environmental change have focused on short-term (months to years) effects (Allison and Martiny, 2008). As a consequence, the effects of long-term (decades to thousands of years) environmental change on microbial communities are poorly understood. In the present work, we investigated aquatic microbial communities that have been exposed to strong long-term environmental change. Specifically, we studied Antarctic bacterioplankton communities inhabiting a unique set of lakes which, in most cases, have been progressively isolated from the sea over the past 20 000 years. Over this time period, the studied marine-derived lakes have gradually developed into waterbodies spanning a salinity gradient that now ranges from freshwater (0) to hypersaline (100), reaching salinities of about 300 in other lakes not included in the current analysis. Consequently, the associated marine-derived bacterioplankton communities were exposed to strong changes in salinity, a factor well known to be one of the most important environmental variables affecting the spatio-temporal distribution and evolution of aquatic microbes (Lozupone and Knight, 2007; Logares et al., 2009; Tamames et al., 2010). Moreover, most studied lakes are unconnected by rivers or streams and thus can be depicted as islands for aquatic microbes (Rengefors et al., 2012). For comparison purposes, we also included a set of 15 Scandinavian freshwater lakes that are fairly well connected and that present highly similar physicochemical conditions between each other.
Overall, the progressive long-term salinity change has most likely shaped the lacustrine bacterioplankton communities observed today by modifying the composition of ancestral communities and by promoting the establishment of immigrants. Considering these factors, we aim at answering the following main questions: (a) What roles do environmental filtering and neutral processes have in shaping actual communities? (b) To what extent do salinity changes affect community composition? (c) How do rare and abundant taxa respond towards salinity change? And (d) How are habitat specialists and generalists distributed along the observed environmental gradient?
Materials and methods
Study site
Our study system consists of 15 coastal lakes in Eastern Antarctica (Vestfold Hills, 68° S; 78° E; Pickard, 1986) and a coastal marine reference site (Supplementary Table S1, Supplementary Figure S1). These lakes range from freshwater to hypersaline conditions (Supplementary Table S1) with their present-day salinity depending on the particular history of each lake (for example, length of isolation from the sea and historical freshwater or marine introgressions). Most lakes were formed during the past 10 000 years (Bird et al., 1991; Zwartz et al., 1998) and a few could be at least 20 000 years, perhaps as much as 125 000 years old (Gibson et al., 2009). In addition, several lakes have developed strong vertical physicochemical gradients, particularly with regard to salinity, oxygen and temperature (Gibson, 1999), rendering them permanently stratified. Additional details on the studied lakes are presented in Supplementary Methods. Details on the analyzed Scandinavian lakes can be found in Supplementary Table S2 and in Logue et al. (2012). The concentrations of total phosphorus (Tot-P), total nitrogen (Tot-N) and dissolved organic carbon (DOC) measured in the studied Antarctic lakes and the sea are presented in Supplementary Table S3 (see Supplementary Methods for a description of nutrient analyses).
Field sampling, DNA extraction, PCR and pyrosequencing
In the Antarctic summer of 2008/9 (December—February), water samples were collected from lakes and a marine coastal site (Supplementary Table S1). Water samples were filtered through a 0.2-μm polycarbonate filters (Supor-200, 47 mm; PALL Corporation, East Hills, NY, USA) from which whole community DNA was extracted using the Power Soil kit (MO BIO Laboratories Inc., Carlsbad, CA, USA). The bacterial hypervariable domains V3-V4 of the16S rDNA gene were amplified using the primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′ Herlemann et al., 2011). Pyrosequencing of amplicons, using unidirectional Lib-L chemistry, was carried out on a 454 GS FLX Titanium system (454 Life Sciences, Branford, CT, USA) installed at the Norwegian High Throughput Sequencing Centre (NSC, Oslo, Norway, http://www.sequencing.uio.no). See further details in Supplementary Methods.
Data analysis
Reads between 200–550 bp were used for downstream analysis. They were checked for quality (sliding window Phred average (50 bp)>25) and denoised using DeNoiser (v 0.851; Reeder and Knight, 2010). Archaea and Chloroplast reads were removed. Chimeras were detected using ChimeraSlayer (Haas et al., 2011) and subsequently removed. The total number of clean reads (Antarctica and Scandinavia) amounted to 618 888, which were deposited at MG-RAST (http://metagenomics.anl.gov/; identification number 4487042.3).
All sequences were run through QIIME v. 1.3.0 (Caporaso et al., 2010). Sequences were clustered into Operational Taxonomic Units (OTUs) using UCLUST v1.22q (Edgar, 2010) with a 97% threshold of sequence similarity. OTU tables were constructed for both Antarctic and Scandinavian (OTUANT-SWE), as well as independently for Antarctic (OTUANT) and Scandinavian (OTUSWE) sites. In addition, randomly subsampled OTU tables (2800 reads per sample for all samples) were generated for the Antarctic–Scandinavian (OTUANT-SWE2800=4022 OTUs), the Antarctic (OTUANT2800=2586 OTUs) and the Scandinavian (OTUSWE2800=1557 OTUs) sites, to correct for possible biases introduced by unequal sequencing efforts. Unless stated otherwise, singletons were included in the subsampled OTU tables, as their abundances were higher in the original tables. See Supplementary Methods for further details.
Fit to the neutral model of community assembly
We used equation (14) from Sloan et al. (2006), which considers OTU detection frequency and regional relative abundance (that is, average of local relative abundances for each OTU, including zero values). This model is an adaptation of Hubbell's neutral community model (Hubbell, 2001) adjusted to large microbial populations analyzed with molecular tools (Sloan et al., 2006). In this equation, the parameter Nm determines the relationship between detection frequency and regional relative abundance, with N being metacommunity size and m immigration rate (likelihood of an individual being replaced by an immigrant). Assuming a relatively constant metacommunity size (N), Nm is an estimate of dispersal between communities (Sloan et al., 2006). For this analysis, we used separately the data sets OTUANT2800 and OTUSWE2800. In addition, for Antarctic lakes only (excluding marine samples), we calculated the fit to the NM for surface (<5 m depth) and deep (>5 m) samples separately. This subdivision was carried out as several lakes present strong and permanent vertical physicochemical gradients. In all cases, the detection limit parameter d was 1/2800. All computations were done in R (R-Development-Core-Team, 2008).
Analysis of beta diversity
Beta diversity indices that take into account relative abundances were chosen as they tend to provide better estimates than presence–absence indices (Kuczynski et al., 2010). Bray–Curtis (Bray and Curtis, 1957) and Unifrac (phylogeny-based; Lozupone and Knight, 2005) indices were used. Unifrac calculates the amount of unique branch length that is captured by different communities in a phylogeny. We used the weighted normalized Unifrac version, as it considers relative abundances (that is, number of reads mapping to a specific OTU in a tree) as well as different branch lengths in a tree. The normalized data set used for calculating beta diversity indices was OTUANTSWE2800. Singletons were included, as their removal did not modify beta diversity patterns. Data were explored using NMDS (Non-Metric Multidimensional Scaling) analyses and UPGMA clustering dendrograms. Environments were arbitrarily categorized in relation to salinity: freshwater (salinity 0), low-brackish (1–6), high-brackish (7–30), marine salinity (31–35), hypersaline (61–75) and high-hypersaline (76–100); see Supplementary Table S1. Differences between categories were tested with ANOSIM (ANalysis Of SIMilarity; Clarke, 1993), performing 10 000 permutations. To determine which taxa generated most differences between categories, we used SIMPER (SIMilarity PERcentage; Clarke, 1993) analyses. NMDS, ANOSIM and SIMPER were either run under the R environment (R-Development-Core-Team, 2008) with the package VEGAN (Oksanen et al., 2008) or in PAST (Hammer et al., 2001). UPGMA dendrograms were generated with QIIME and VEGAN, and support values were calculated using Jackknifing (in QIIME) or with the R package PVCLUST v1.2–2 (Suzuki and Shimodaira, 2006). Jackknife support was calculated using 100 permutations, and values>70% were considered significant. Using PVCLUST and 1000 permutations, we calculated Approximately Unbiased P-values as well as bootstrap probability values. Approximately Unbiased values>95% and bootstrap probability values >70% indicate statistical significance. Unifrac distance metrics were calculated with QIIME and both the Unifrac-test (Lozupone and Knight, 2005; Hamady et al., 2010) and the P-test (Martin, 2002) were computed online using FastUnifrac (http://bmf.colorado.edu/fastunifrac/) with 1000 permutations. Agreement between both tests indicate stronger patterns. See further details of these tests in Martin (2002); Lozupone and Knight (2005) and Hamady et al. (2010).
Relationships between community composition, environment and geographical distance
To explore which environmental factors explained most variability in Antarctic bacterioplankton community composition, we ran PERMANOVA (R, VEGAN) and Redundancy Analysis (CANOCO (Leps and Smilauer, 2003)). The OTU table used for these analyses was OTUANT2800. Environmental data included nutrient composition (Supplementary Table S3) and salinity. Stepwise selection coupled to Monte Carlo tests was applied in CANOCO to assess the usefulness of each variable.
Standard and partial Mantel tests were used to investigate correlations between salinity (identified as one of the most important environmental variables in our system), geographical distance between lakes and community composition (Antarctic lakes only). The used data set was OTUANT2800, excluding marine samples. Correlations with salinity would indicate environmental filtering, while correlations with geographical distance would indicate unequal dispersal among sites. The Mantel statistic r(A × B) estimates the correlation between two matrices (A and B). The partial Mantel statistic (A × B|C) estimates the correlation between A and B controlling for the effects of a third matrix, C. Standard and partial Mantel tests were run in R (VEGAN) using Bray–Curtis distances between communities. Salinities were square-root transformed and Euclidean distances between samples were calculated. Geographical distances between lakes were calculated from maps provided by the Australian Antarctic Division. The significance of the Mantel statistic was obtained after 1000 permutations. Tests were done considering all Antarctic samples together as well as samples from the surface of lakes (<5 m) and deeper samples (>5 m).
Rare and abundant taxa
Rare and abundant OTUs were identified in the data set considering Antarctic and Scandinavian lakes (OTUANT-SWE2800). Abundant OTUs were arbitrarily defined as those containing >100 reads/OTU considering the entire data set (mean relative abundance >0.06% ). Rare OTUs were defined as those containing 2–10 reads/OTU (mean relative abundance >0.001% and <0.006%, respectively). OTUs encompassing 11–99 reads were considered to be in an ambiguous zone between rare and abundant OTUs and were not used in our analyses. Singletons were not used to avoid possible biases. Separate OTU tables containing abundant and rare OTUs were constructed and used for UPGMA clustering (based on Bray–Curtis distances).
Analysis of habitat specialists and generalists
To measure habitat specialization, we used the ‘niche breadth' approach (Levins, 1968) described by the formula:
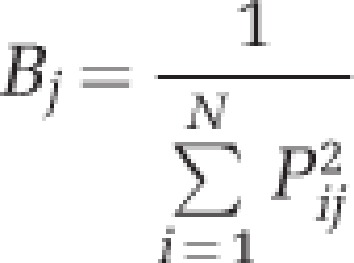 |
where Bj indicates niche breadth and Pij is the proportion of individuals belonging to species j present in a given habitat i. OTUs that are present, and more evenly distributed, along a wider range of habitats will have a higher B-value and can be considered habitat generalists (Pandit et al., 2009). Similarly, OTUs with a lower B-value can be regarded as habitat specialists. The considered habitats were: freshwater, low-brackish, high-brackish, marine salinity, hypersaline, and high-hypersaline. Niche breadth was measured only for the Antarctic samples using the OTUANT2800 data set. OTUs with mean relative abundances <2 × 10−5 were not considered, as they could erroneously indicate specialized taxa (Pandit et al., 2009). The considered OTUs after the removal of rare taxa were 1272. Niche breadth ranged between 1 and 5.59 (with a theoretical maximum of 6). OTUs with B>3 were arbitrarily defined as generalists, whereas those with B<1.5 were regarded as specialists. B>3 was chosen because this value lies within the outlier area of the B distribution (see Supplementary Figure S2). B<1.5 was selected as it is close to 1, the smallest possible B-value.
Identification of strict habitat specialists
Species that characterize a given habitat are known as indicator species. Good indicator species should be found mostly in a single habitat and be present in most sites or samples from that habitat (Legendre and Legendre, 1998). Thus, indicator species can be considered as strict habitat specialists. Niche breadth (B) and indicator species (strict specialists) complement each other. Using B-values, it is possible to identify different levels of specialization. However, this approach will not indicate which taxa are associated with each investigated environment. For the latter purpose, we used the INDVAL (INDicator VALues; Dufrene and Legendre, 1997) analysis, which identifies indicator species based on OTU fidelity and relative abundance. Analyses were run using the data set OTUANT2800 and the package labdsv (http://ecology.msu.montana.edu/labdsv/R/) within the R environment. Only OTUs with significant (P<0.05) INDVAL values that were>0.3 were considered, as this latter value can be regarded as a good threshold for habitat specialization (see Dufrene and Legendre, 1997). Environments were categorized by salinity (freshwater (salinity 0; 5 samples), low-brackish (1–6; 16 samples), high-brackish (7–30; 4 samples), marine salinity (31–35; 3 samples), hypersaline (61–75; 4 samples) and high-hypersaline (76–100; 4 samples)).
Results
Fit to the neutral model of community assembly
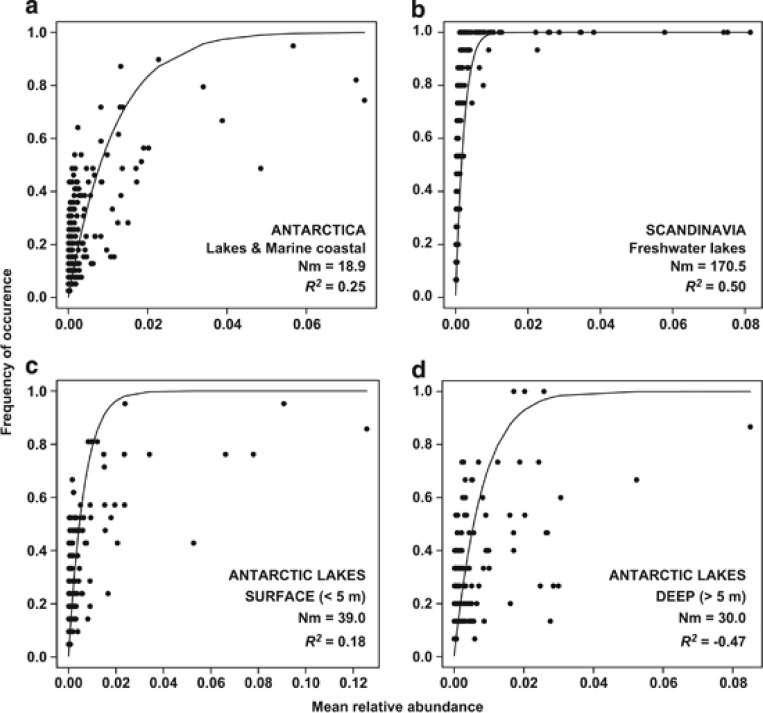
The NM estimated a low fraction of the variation in the frequency of occurrence of different OTUs in Antarctic lakes (Figure 1a, R2=0.25). By contrast, the NM explained a larger fraction of the variation in frequency of occurrence in Scandinavian lakes (Figure 1b R2=0.50), and most departures from the NM originated from rare taxa that tended to be more frequent than expected (Figure 1b). The Nm-value was much higher for Scandinavian (Nm=170.5) than for Antarctic communities (Nm=18.9). As some of the studied Antarctic lakes present strong and permanent vertical physicochemical gradients, we also tested the fit to the NM in surface (<5 m depth) and deep (>5 m depth) communities (Figure 1c and d). In surface samples, the NM also explained a low proportion of the variability in occurrence frequency (Figure 1c, R2=0.18), while deep communities showed no fit to the NM (Figure 1 d, R2=−0.47, note that negative R2 values can occur when there is no fit to the model).
Figure 1.
Frequency of occurrence of different Operational Taxonomic Units as a function of mean relative abundance for Antarctic and Scandinavian communities (2800 reads per sample in all data sets). Lines indicate the best fit to the neutral model as in Sloan et al. (2006). Nm indicates metacommunity size times immigration. R2 indicates the fit to the neutral model, and negative R2 values indicate no fit to this model. The same detection limit d was used in all analyses (d=1/2800). In panel (a), only Antarctic samples were considered, while in (b) only Scandinavian samples were used. Results from surface (<5 m depth) and deep (>5 m depth) Antarctic samples only are shown in panels (c) and (d), respectively.
Salinity explained most of the community variability between lakes
In the PERMANOVA analysis, salinity explained between 44% and 51% of the variability (data sets OTUANT-SWE2800 and OTUANT2800), followed by Tot-P, which explained about 12% (P<0.05 for both variables). Redundancy analysis indicated that salinity explained 43% of the total variability followed by both Tot-P and Tot-N (18% each; P<0.05 for all variables).
Large heterogeneity in community composition among Antarctic samples
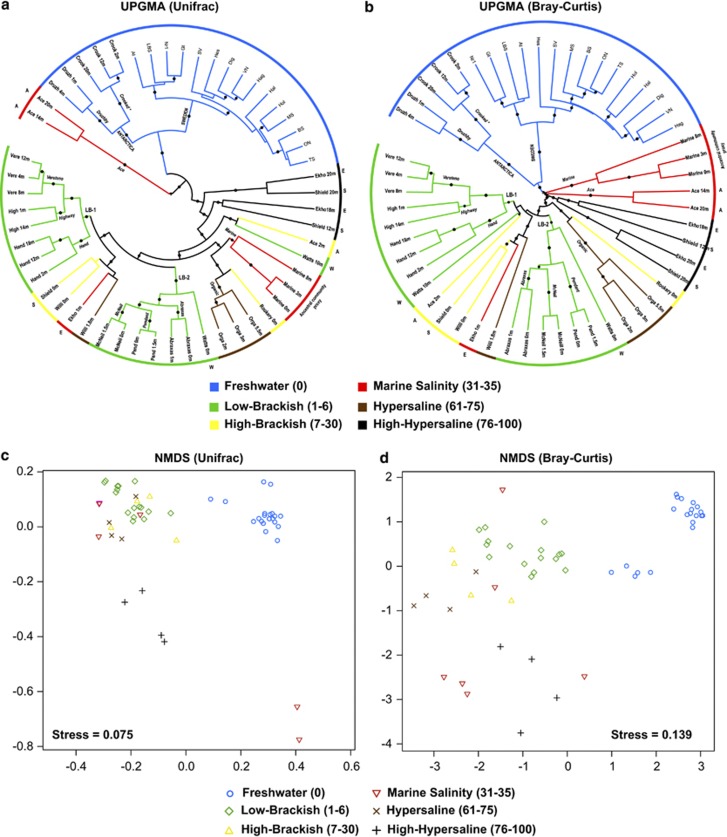
Freshwater communities were most distinct from all other communities (Figure 2; Supplementary Tables S4 and S5). Most marine-derived lake communities differed in composition from marine communities (Figure 2a and b, note that marine and lake samples normally do not cluster together; Supplementary Tables S4 and S5). Community composition changed abruptly with increasing salinity in stratified lakes (Figure 2a and b). For example, the Ace Lake community at 14 m (more saline) was significantly different from the community at the surface (less saline; Figure 2a and b).
Figure 2.
Panels (a) and (b) show UPGMA dendrograms based on Weighted-Normalized Unifrac and Bray–Curtis distances. Dots over branches indicate a Jackknife support>70%. The salinity classification of samples is shown in colors. For the Antarctic data set, the name of the sample and the depth are indicated (using the format: site, depth (in meters)). Most sites refer to lakes, except the ones indicated as ‘Marine'. Samples originating from the same site and clustering together are indicated. Samples from the same lake that do not cluster together are indicated with a letter outside the main circle. LB-1 and LB-2 indicate the two low-brackish clusters. Panels (c) and (d) show Non-metric Multidimensional scaling (NMDS) charts based on Weighted-Normalized Unifrac (panel c) and Bray–Curtis (panel d) distances. Note that Antarctic freshwater lakes cluster with Scandinavian lakes.
There was a large variation in taxonomic composition between lakes (Supplementary Figure S3). Flavobacteria (Bacteroidetes) were predominant in several saline lakes, except in the highly hypersaline ones. Actinobacteria were abundant in communities from freshwater and most low-salinity lakes. Gammaproteobacteria were mostly present in marine communities and a number of saline lakes. Betaproteobacteria predominated in Scandinavian freshwater lakes, being present only in very low numbers in Antarctic lakes. Sphingobacteria (Bacteroidetes) were present in saline Antarctic lakes as well as in Scandinavian (freshwater) lakes, pointing to a halotolerant group. Alphaproteobacteria were moderately abundant in a number of lakes and marine environments. Other groups were abundant in specific samples, for example, Epsilonproteobacteria, which were only present at 12 m in Lake Shield as well as the group Chlorobia, which abounded only in the deeper waters of Ace Lake. See Supplementary Figure S3 for a detailed description.
Similar communities in different lakes
Bacterioplankton communities from different lakes, which experience similar salinity, tended to be similar in composition. In particular, samples from specific layers in different lakes, but with similar salinity, featured communities that were more similar to each other than to communities from other depth layers within the same lake, but with different salinity (Figure 2a and b). For example, communities from lakes Ekho and Shield taken at >12 m depth were significantly more similar to each other than to surface communities within each of the lakes, respectively (Figure 2a and b). Similar results were found in surface communities of lakes Shield and Williams when compared with deeper communities in each lake (Figure 2a and b).
In addition, freshwater lake communities from Antarctica tended to be more similar to Scandinavian freshwater communities than to saline neighboring communities (Figure 2). Yet, freshwater communities from Antarctica and Scandinavia showed significant differences (Figure 2; ANOSIM RBrayCurtis/Unifrac=1/0.99, P<0.001) when analyzed independently from the saline samples. Such differences and similarities in bacterioplankton community composition between Antarctic and Scandinavian freshwater habitats are caused by those taxa that are restricted to either Antarctica or Scandinavia or shared between both zones (see Supplementary Tables S6 and S7). Some of the shared OTUs were highly similar (>99%) to lacustrine freshwater OTUs with an apparently worldwide distribution (Supplementary Table S7).
In limited cases, communities from different lakes experiencing similar salinity featured different compositions. For instance, different low-brackish (LB) communities clustered into two groups (Figure 2a and b, see LB-1 and LB-2). Statistical tests are inconclusive on whether LB-1 and LB-2 are different in terms of phylotype composition. The P-test indicated that LB-1 and LB-2 could be considered as two different groups (P<0.05), whereas the Unifrac-test indicated the opposite (P>0.05).
Only a few abundant OTUs were shared between marine and lacustrine Antarctic habitats
Out of a total of 2586 OTUs (data set OTUANT2800), 173 (6.7%) were found only in marine sites, 2246 OTUs (86.8%) were found in only lakes, and 167 OTUs (6.5%) were shared between lakes and marine sites. OTUs restricted to lakes accounted for the 54.0% of the total number of reads (OTUANT2800=109,200 reads), while OTUs restricted to marine sites were only 0.5% of the reads in the data set. Shared OTUs represented 45.5% of the reads in the data set. In total, in the Antarctic data set OTUANT2800, 100 800 reads (92.3%) originated from lake samples and 8400 reads (7.7%) from marine samples.
Correlations between environment and geographical distance with community composition
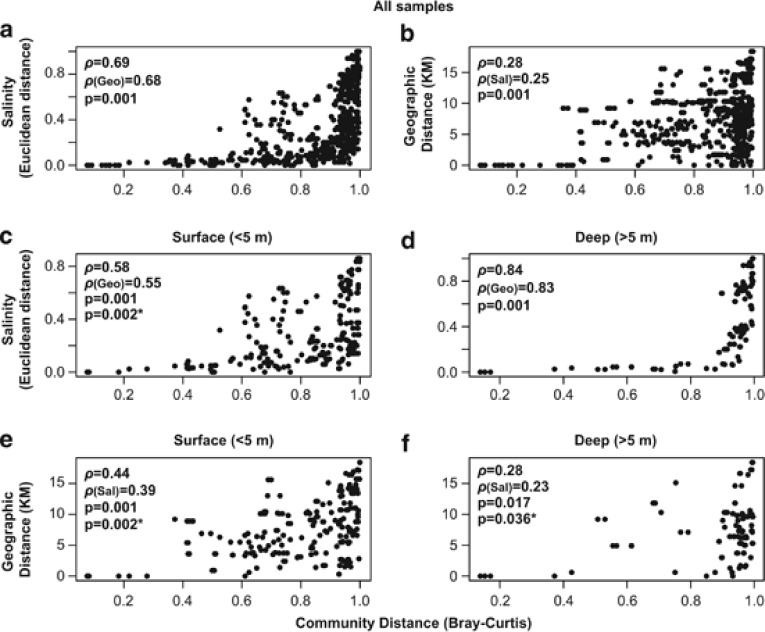
Salinity showed significant correlations to community composition in all Mantel tests (regular and partial), although the correlation coefficient was higher in deeper samples than in surface ones (Figure 3). Geographical distance between lakes was moderately correlated to community composition in surface samples and displayed a weak correlation in deeper ones (Figure 3). In all the cases, salinity was more strongly correlated to community composition than geographical distance. The Spearman correlation coefficient was used as the dissimilarity matrices were skewed (Figure 3; see Legendre and Legendre, 1998).
Figure 3.
Correlations between bacterioplankton community composition with salinity and geographical distance between lakes (only Antarctic lakes are considered). In panels (a) and (b), all samples were used, while in panels c–f, samples were divided into surface (<5 m depth) and deep (>5 m depth) categories. Spearman correlation coefficient (ρ) values are shown for regular (first value) and partial Mantel (second) tests. P-values are indicated for each test (*P-values of partial Mantel tests). Only one P-value is shown in cases where both regular and partial Mantel tests presented identical values.
Abundant and rare communities showed comparable dissimilarity patterns
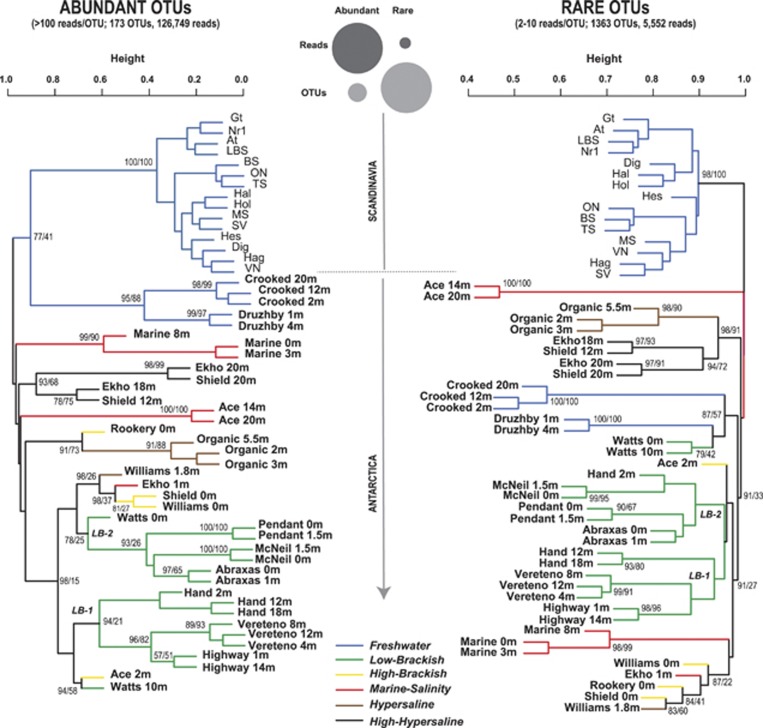
Similar community differentiation patterns were observed in abundant and rare taxa when analyzing the data set OTUANT-SWE2800 (Figure 4; Mantel test Abundant|Rare(Bray–Curtis) r=0.82, P<0.01). However, one difference was that abundant freshwater communities from Antarctica and Scandinavia clustered together, while rare Antarctic freshwater communities clustered with rare communities from brackish Antarctic lakes (Figure 4). Furthermore, rare communities from Hypersaline and High-Hypersaline lakes clustered together, which did not happen with abundant communities. Abundant OTUs (173) contributed 126 749 reads, while rare OTUs (1363) contributed 5552 reads (Figure 4). In general, the clustering patterns received significant statistical support (Figure 4).
Figure 4.
Contrasted UPGMA clustering of bacterioplankton samples (Bray–Curtis distances) based on their rare and abundant Operational Taxonomic Unit (OTU) composition. Abundant OTUs were arbitrarily defined as those containing more than 100 reads and rare OTUs as those containing between 2–10 reads. There were 173 abundant OTUs, encompassing a total of 126 749 reads. A total of 1363 rare OTUs were present, which included 5552 reads. The circles on the top-center section of the figure illustrate the proportions of OTUs and reads in both the abundant and rare data sets. For the Antarctic data set, the name of the sample and the depth are indicated (using the format: site, depth (in meters)). Most sites refer to lakes, except the ones indicated as ‘Marine'. Support values for the clustering pattern are indicated on top of the branches. Values indicate the approximately unbiased (AU) P-values as well as the bootstrap probability (BP) using the format AU/BP. AU values>95% and BP>70% indicate statistical significance. The colors of the branches indicate the salinity of the samples. LB-1 and LB-2 indicate the two low-brackish clusters.
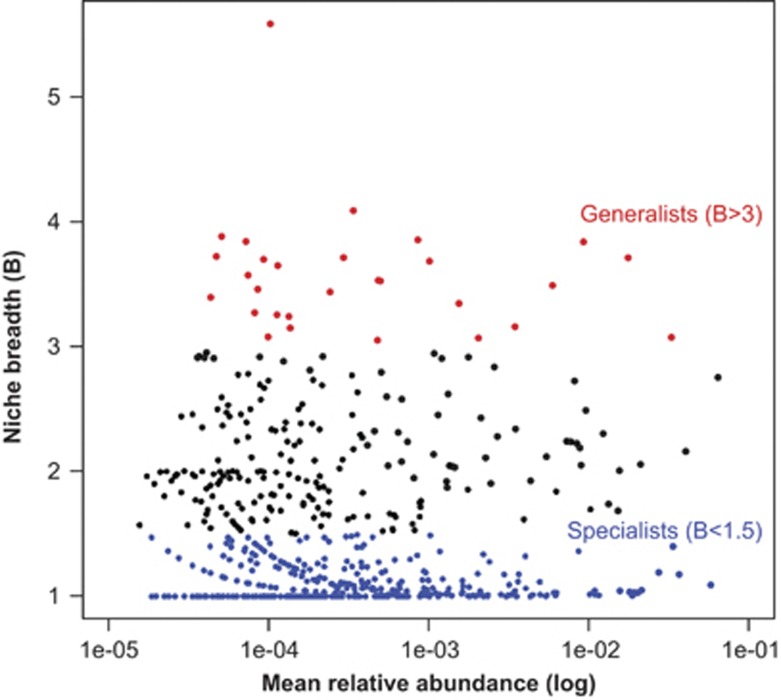
Few habitat generalists and many specialists
Generalists and specialists were present among both rare and abundant taxa (Figure 5). In total, we identified 30 OTUs (out of 1272 OTUs, 2.3%) as generalists and 939 OTUs as specialists (73.8%). Specialists identified in this manner (that is, niche breadth, B) comprise different degrees of habitat specialization. Using INDVAL, strict specialists (that is, OTUs that tend to be present in only one habitat type and in most samples from that habitat type) were identified. We detected 365 OTUs that seemed to be strict specialists (INDVAL >0.3, P<0.05), representing 28.7% of the total OTU data set. The distribution of such strict specialist OTU among the salinity classes was: freshwater (103), low-brackish (25), high-brackish (16), marine salinity (79), hypersaline (37), and high-hypersaline (105).
Figure 5.
Habitat specialists and generalists in relation to salinity in Antarctic lakes. Each dot represents an Operational Taxonomic Unit (OTU). The x axis indicates mean relative abundances (log) and the y axis indicates Niche Breadth (B). Generalists (red; B>3) and Specialists (blue; B<1.5) OTUs are indicated. The considered salinity habitats were: freshwater, low-brackish, high-brackish, marine salinity, hypersaline and high-hypersaline. Note that habitat specialists and generalists were distributed along most of the range of mean relative abundances.
Discussion
Major role for environmental filtering in Antarctic lakes
Compared with other biogeography studies (Lindström and Langenheder, 2012), the fraction of variation in Antarctic community composition that could be explained by environmental variables was high. This means that environmental filtering was a very important mechanism for the generation of spatial distribution patterns in our study system. The dispersal rates via air are probably too low to cause mass effect (Jones and McMahon, 2009; Lindström and Östman, 2011) and as we found similar taxa in Antarctica and Scandinavia, dispersal limitation seems unlikely. The impact of environmental filtering appeared to be more important in deeper Antarctic lacustrine waters than in surface waters, which seemed slightly more affected by spatial processes (dispersal). Similar results were obtained by Barberan and Casamayor (2011) in a study focusing on 12 stratified lakes from diverse geographical locations situated mostly in the Northern Hemisphere.
We tested the neutral model of community assembly in order to determine whether processes unrelated to environmental variability could also explain community composition. The fit to the NM obtained for the physicochemically different Antarctic lakes was contrasted to the fit obtained for the physicochemically similar Scandinavian lakes. The NM explained a minor part (25%) of the variation in Antarctic community composition, supporting the conclusion by Östman et al. (2010) that strong environmental gradients may favor environmental filtering over neutral dynamics. Dispersal and stochastic processes appeared to have more impact in surface-water communities compared with deeper ones, as surface communities showed a limited fit to the NM, while deeper communities showed no fit to this model. In contrast to Antarctic lakes, the NM explained 50% of the variation in community composition in Scandinavian lakes, agreeing with previous results from lakes in the same area using fingerprinting techniques (ARISA and tRFLP; Drakare and Liess, 2010; Östman et al., 2010). Thus, the more homogeneous physicochemical conditions in Scandinavian lakes appear to favor neutral dynamics. In Scandinavian lakes, most departures from the NM observed in our data occurred among rare taxa. In general, the deeper taxonomic coverage achieved by pyrosequencing increases the chances of detecting rare taxa. This may be the reason why rare OTUs that do not follow the NM have been poorly represented in previous works using tRFLP (Östman et al., 2010), ARISA (Drakare and Liess, 2010) or Sanger clone libraries (Sloan et al., 2006); all techniques that normally detect taxa with high or moderate abundances. Such rare OTUs could be slow growing or dormant taxa (Pedrós-Alió, 2006) that accumulate over time in communities, thus departing from the assumptions of the NM (equal growth rates for all taxa). As rivers interconnect several of the analyzed Scandinavian lakes, dispersal and accumulation of rare dormant OTUs over time seems more likely than in the physically unconnected Antarctic lakes. According to the calculated Nm values, dispersal between the analyzed Scandinavian lakes is likely higher than among Antarctic counterparts (assuming similar metacommunity sizes).
Progressive long-term salinity change promoted the assembly of new communities
Most of the studied lakes separated from the sea at different times (Zwartz et al., 1998) and presumably harbored in their beginnings typical marine-coastal microbial communities. After marine isolation, lakes progressively developed different salinities, exposing organisms (marine derived and immigrant taxa) to strong environmental selection and promoting the assembly of new communities. The array of different communities observed today in marine-derived lakes is most likely the outcome of this process, which has been acting over hundreds to thousands of years.
A relevant question in the studied Antarctic lake system is whether current lake communities harbor marine taxa. We found that a few, normally abundant, OTUs (6.5% of the OTUs comprising 45.5% of the reads) were shared between marine and lacustrine habitats, showing that these environments share at least part of their bacterioplankton community composition. This agrees with previous studies reporting the presence of the ubiquitous marine SAR11 group in some of the studied lakes (Lauro et al., 2010; Logares et al., 2010). These shared taxa could derive from the original marine inoculum of the lakes or from recent colonizations or mass effects related to inputs from the neighboring sea.
Interestingly, most investigated Antarctic OTUs were only detected in lacustrine samples, where they represented 54% of the retrieved reads and 86.8% of the OTUs. Hence, marine-derived lakes contain a distinctive set of taxa that appear to be absent or in very low abundances in neighboring coastal-marine waters. These taxa could have originated from the present or past rare marine biosphere (featuring OTUs that were below our detection threshold) or by long- or short-range dispersal from other saline waterbodies. Some saline Antarctic lakes shared similar taxa with other saline lakes in other continents (for example, Asia), supporting long-range dispersal; previous studies have shown that the influx of biological particles to Antarctica from other continents can be considerable (Marshall, 1996). On the other hand, other lacustrine OTUs were similar to taxa found only in other distant Antarctic lakes (for example, Lake Vida in the Dry Valleys; results not shown), supporting continental dispersal of taxa potentially endemic to the Antarctic.
A similar scenario can be depicted for Antarctic freshwater lakes, which contained taxa shared with freshwater lakes from other continents as well as possible Antarctic endemics. NMDS and clustering analyses based on Bray–Curtis and Unifrac distances indicated that freshwater Antarctic lakes contained mostly freshwater taxa and not saline taxa adapted to low salinity. This agrees with the conclusion of Logares et al. (2009), indicating that freshwater taxa can rarely survive in saline waters and vice versa. Interestingly though, we found contrasting patterns in abundant and rare freshwater Antarctic communities. Abundant communities were more similar to freshwater communities from Scandinavian lakes than to adjacent communities in low saline lakes. Yet, rare freshwater Antarctic communities were more similar to neighboring low-brackish (salinity 1–3) communities (specifically to the brackish Lake Watts community). Thus, the abundant and rare bacterial communities in the studied freshwater Antarctic lakes seem to have different origins and dynamics. Abundant taxa probably encompass several global dispersers, while rare taxa may represent low-saline or freshwater OTUs that are shared with other slightly saline neighboring lakes. Such differences in rare community composition between Antarctic and Scandinavian lakes are most likely due to dispersal limitation or mass effects.
Similar biogeography in abundant and rare saline taxa
Several questions still remain unanswered regarding the rare biosphere; in particular, to what extent the environment affects this rare biosphere and what is the spatial distribution of rare communities (Caron and Countway, 2009; Pedrós-Alió, 2012). Our results indicate that environmental filtering acted similarly on the studied rare and abundant saline communities, generating similar biogeography. Thus, it seems that the rare biosphere contains taxa sensitive to environmental variation, which respond to change in a similar manner to abundant taxa. Altogether, our results and earlier findings (Galand et al., 2009) suggest that, in general, both the rare biosphere and the abundant taxa present similar biogeography.
A few habitat generalists and several specialists
Our results agreed with previous observations and theory indicating few habitat generalists and many specialist in species-rich communities (Guo et al., 2000; Kolasa and Romanuk, 2005; van der Gast et al., 2011). Habitat specialists and generalists were distributed along the entire range of abundances, similar to what has been reported by Pandit et al. (2009) for zooplankton. Strict habitat specialists were more abundant in the extremes of the salinity gradient. Therefore, salinity extremes (freshwater and high-hypersaline environments) appear to have selected more strongly for specific OTUs. In a nutshell, progressive long-term environmental change appears to have generated a variety of niches that were filled by an array of habitat specialists. These were likely in very low abundances or absent in the initial marine communities that populated marine-derived lakes. Some specialist OTU probably arrived via intra-/inter-continental dispersal.
Conclusions
Gradual long-term salinity change seems to have promoted the assembly of an array of different communities in the studied saline Antarctic lakes (that is, marine-derived lakes). Neutral processes appear to have had a minor role in the structuring of the physicochemically heterogeneous Antarctic communities but a more important role in the structuring of the physicochemically homogeneous Scandinavian communities. Most of the studied saline lacustrine communities appear to contain both a marine and a lacustrine component in their taxonomic composition. Still, the studied saline Antarctic lakes contain a large number of taxa not shared with the neighboring marine environment, which could be long-range dispersers or Antarctic continental endemics. Abundant and rare bacterioplankton from saline lakes presented similar biogeography, suggesting that these pools do not present large differences in their environmental sensitivity. Habitat specialists and generalists were present among abundant and rare taxa, with strict specialists being relatively more abundant at the extremes of the salinity gradient. Altogether, our results indicate that gradual long-term environmental change can be a strong force that promotes bacterioplankton community diversification as well as the establishment of habitat specialists.
Acknowledgments
Financial support for this work was provided by the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (FORMAS) via Uppsala Microbiomics Centre (UMC) (granted to SB and LT), the Swedish Research Council (granted individually to SB, ESL and KR), the Marie Curie Intra-European Fellowship (PIEF-GA-2009-235365, EU) and Juan de la Cierva fellowship (JCI-2010-06594, Ministry of Science and Innovation, Spain) (granted to RL), the Australian Antarctic Research Assessment Committee (granted to JL-P and KR), FORMAS (granted to SL) and the Olsson-Borghs and Helge Ax:son Johnsons Foundation (granted to JBL). Computing resources were provided by Bioportal (http://www.bioportal.uio.no/), UPPMAX (http://www.uppmax.uu.se/) as well as the Canarian Institute of Astrophysics (www.iac.es) and MareNostrum (http://www.bsc.es/) through the Barcelona Supercomputing Center and Spanish Network of Supercomputing (Grants BCV-2010-3-0003 and 2011-2-0003/3-0005 to RL). We thank the Australian Antarctic Division staff in Kingston, onboard the research vessel Aurora Australis and at Davis Station, Antarctica, in particular, A. Davidson, T. Bailey and D. Lang. We thank Ö. Östman for comments that helped to improve this work. G. Salazar is thanked for assistance with R analyses. We also thank the three reviewers and the ISME editor who provided useful comments to improve this work.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Martiny JB. Resistance, resilience, and redundancy in microbial communities. Proc Nat Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan A, Casamayor EO. Euxinic freshwater hypolimnia promote bacterial endemicity in continental areas. Microb Ecol. 2011;61:465–472. doi: 10.1007/s00248-010-9775-6. [DOI] [PubMed] [Google Scholar]
- Bird M, Chivas A, Radnell C, Burton HR. Sedimentological and stable-isotope evolution of lakes in the Vestfold Hills, Antarctica. Palaeogeog palaeoclim palaeoecol. 1991;84:109–130. [Google Scholar]
- Pester M, Bittner N, Deevong P, Wagner M, Loy A. A 'rare biosphere' microorganism contributes to sulfate reduction in a peatland. Isme J. 2010;4:1591–1602. doi: 10.1038/ismej.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron D, Countway P. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat Microb Ecol. 2009;57:227–238. [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Drakare S, Liess A. Local factors control the community composition of cyanobacteria in lakes while heterotrophic bacteria follow a neutral model. Freshwater Biol. 2010;55:2447–2457. [Google Scholar]
- Dufrene M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JA, Paterson KS, White CA, Swadling KM. Evidence for the continued existence of Abraxas Lake, Vestfold Hills, East Antarctica during the Last Glacial Maximum. Antarctic Sci. 2009;21:269–278. [Google Scholar]
- Gibson JAE. The meromictic lakes and stratified marine basins of the Vestfold Hills, East Antarctica. Antarctic Sci. 1999;11:175–192. [Google Scholar]
- Guo Q, Brown JH, Valone TJ, Kachman SD. Constraints of seed size on plant distribution and abundance. Ecology. 2000;81:2149–2155. [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electronica. 2001;4:9. [Google Scholar]
- Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell SP. A unified neutral theory of biodiversity and biogeography. Princeton University Press: Princeton, NJ; 2001. [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, McMahon KD. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ Microbiol. 2009;11:905–913. doi: 10.1111/j.1462-2920.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Kolasa J, Romanuk T.2005Assembly of Unequals in the Unequal World of a Rock Pool MetacommunityIn: Holyoak M, Leibold MA, Holt RD, (eds)Metacommunities The University of Chicago Press: Chicago; p. 513 [Google Scholar]
- Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2010;7:813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Szekely AJ. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 2011;5:1086–1094. doi: 10.1038/ismej.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D, et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 2010;5:879–895. doi: 10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Legendre L.1998Numerical Ecology2nd edn. Elsevier Science BV: Amsterdam [Google Scholar]
- Leps J, Smilauer P. Multivariate Analysis of Ecological Data using CANOCO. Cambridge University Press: Cambridge; 2003. [Google Scholar]
- Levins R. Evolution in changing environments. Princeton University Press: Princeton, New Jersey; 1968. [Google Scholar]
- Lindström ES, Langenheder S. Local and regional factors influencing bacterial community assembly. Environ Microbiol Rep. 2012;4:1–9. doi: 10.1111/j.1758-2229.2011.00257.x. [DOI] [PubMed] [Google Scholar]
- Lindström ES, Östman Ö. The importance of dispersal for bacterial community composition and functioning. PLoS One. 2011;6:e25883. doi: 10.1371/journal.pone.0025883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Brate J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Logares R, Brate J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11) Mol Biol Evol. 2010;27:347–357. doi: 10.1093/molbev/msp239. [DOI] [PubMed] [Google Scholar]
- Logue JB, Langenheder S, Andersson AF, Bertilsson S, Drakare S, Lanzen A, et al. Freshwater bacterioplankton richness in oligotrophic lakes depends on nutrient availability rather than on species-area relationships. ISME J. 2012;6:1127–1136. doi: 10.1038/ismej.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WA. Biological particles over Antarctica. Nature. 1996;383:680. [Google Scholar]
- Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Nat Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. 2008. vegan: Community Ecology Package. R package version 1.15-0.
- Östman Ö, Drakare S, Kritzberg ES, Langenheder S, Logue JB, Lindström ES. Regional invariance among microbial communities. Ecol Lett. 2010;13:118–127. doi: 10.1111/j.1461-0248.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- Pandit SN, Kolasa J, Cottenie K. Contrasts between habitat generalists and specialists: an empirical extension to the basic metacommunity framework. Ecology. 2009;90:2253–2262. doi: 10.1890/08-0851.1. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. The rare bacterial biosphere. Annu Rev Marine Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- Pickard J.1986Antarctic Oasis, Davis Station and the Vestfold HillsIn: Pickard J, (eds)Antarctic Oasis Academic Press: Sydney; 1–19. [Google Scholar]
- R-Development-Core-Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria; 2008. [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengefors K, Logares R, Laybourn-Parry J. Polar lakes may act as ecological islands to aquatic protists. Mol Ecol. 2012;21:3200–3209. doi: 10.1111/j.1365-294X.2012.05596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tamames J, Abellan JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwartz D, Bird M, Stone J, Lambeck K. Holocene sea-level change and ice-sheet history in the Vestfold Hills, East Antarctica. Earth Planet Sci Lett. 1998;155:131–145. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.