Abstract
The outcome of the interactions between native commensal microorganisms and opportunistic pathogens is crucial to the health of the coral holobiont. During the establishment within the coral surface mucus layer, opportunistic pathogens, including a white pox pathogen Serratia marcescens PDL100, compete with native bacteria for available nutrients. Both commensals and pathogens employ glycosidases and N-acetyl-glucosaminidase to utilize components of coral mucus. This study tested the hypothesis that specific glycosidases were critical for the growth of S. marcescens on mucus and that their inhibition by native coral microbiota reduces fitness of the pathogen. Consistent with this hypothesis, a S. marcescens transposon mutant with reduced glycosidase and N-acetyl-glucosaminidase activities was unable to compete with the wild type on the mucus of the host coral Acropora palmata, although it was at least as competitive as the wild type on a minimal medium with glycerol and casamino acids. Virulence of the mutant was modestly reduced in the Aiptasia model. A survey revealed that ∼8% of culturable coral commensal bacteria have the ability to inhibit glycosidases in the pathogen. A small molecular weight, ethanol-soluble substance(s) produced by the coral commensal Exiguobacterium sp. was capable of the inhibition of the induction of catabolic enzymes in S. marcescens. This inhibition was in part responsible for the 10–100-fold reduction in the ability of the pathogen to grow on coral mucus. These results provide insight into potential mechanisms of commensal interference with early colonization and infection behaviors in opportunistic pathogens and highlight an important function for the native microbiota in coral health.
Keywords: coral microbiology, coral diseases, coral mucus, Serratia marcescens
Introduction
The foundation of reef ecosystems is the coral holobiont: a dynamic symbiosis comprised of the coral polyp, the dinoflagellate Symbiodinium spp. and their associated microbial communities. Interactions within this tripartite symbiosis determine the overall health of the coral colony and its resistance to opportunistic pathogens (Reshef et al., 2006; Rosenberg et al., 2007; Teplitski and Ritchie, 2009). The subject of this investigation is Serratia marcescens PDL100, one of the strains that causes white pox disease in Acropora palmata and is associated with anthropogenic sources (Patterson et al., 2002; Sutherland et al., 2011). To colonize and infect the coral, this multi-host opportunistic pathogen must first establish within the coral surface mucus layer.
Coral mucus is a polymer made in specialized mucocytes of the polyp from the photosynthate produced by their endosymbiotic dinoflagellates and then excreted onto the coral surface (Brown and Bythell, 2005; Bythell and Wild, 2011). The chemical structure of coral mucus has been determined for only few species (Ducklow and Mitchell, 1979; Meikle et al., 1987, 1988; Coddeville et al., 2011). The mucus polymer of Acroporid corals is a glycoprotein, which consists of a serine and threonine-rich polypeptide backbone with sulfated oligosaccharide side chains attached to it through O-glycosidic linkages (Meikle et al., 1987). The major sugar residues in the oligosaccharide side chains of A. formosa mucus are 𝒟-arabinose, 𝒟-mannose, N-acetyl-𝒟-glucosamine and 𝒟-galactose (Meikle et al., 1988). The patterns of induction of glycosidase activities in S. marcescens and coral commensals, when grown on coral mucus, are consistent with its chemical structure (Krediet et al., 2009a, 2009b).
Coral mucus supports growth of bacteria up to 106–108 cfu ml−1 (Sharon and Rosenberg, 2008; Krediet et al., 2009b; Garren and Azam, 2010). Nutrients and chemicals from the host excreted with the mucus determine the composition and structure of the associated microbial communities (Ritchie, 2006; Garren and Azam, 2012). The ability to efficiently utilize carbon and nitrogen sources within mucus likely directly affects competitiveness of the microorganisms in this environment.
Aside from the need to be able to grow on available substrates, successful establishment within a niche requires that microorganisms efficiently spread to and colonize the sites where nutrients are available. Coordinated, multi-cellular spreading over semi-solid surfaces (known as ‘swarming') is one such mechanism of expansion and surface colonization (Verstraeten et al., 2008). Swarming is often coregulated with certain metabolic pathways (Toguchi et al., 2000; Wang et al., 2004). Global regulatory networks (GacS/GacA-Csr, FlhDC-FliA and EnvZ/OmpR) contribute to the control of both the motility and catabolism of specific carbon sources (Park and Forst, 2006; Jones et al., 2008a; Timmermans and Van Melderen, 2010). This coregulation may be instrumental for the regulatory switch that contributes to the efficient establishment within a particular niche.
A better understanding of the interactions between coral commensals and opportunistic pathogens led to the appreciation of the fact that commensals have evolved to protect their nutrient niche (and the coral hosts) from the invading opportunists (Reshef et al., 2006). Coral commensals produce antibiotics, inhibitors of cell-to-cell communication, and currently unknown compounds capable of inhibiting swarming and biofilm formation (Ritchie, 2006; Nissimov et al., 2009; Shnit-Orland and Kushmaro, 2009; Rypien et al., 2010; Tait et al., 2010; Alagely et al., 2011). When coral commensals capable of interfering with swarming and biofilm formation in a pathogen were pre-colonized onto a model polyp, they completely inhibited the progression of a disease caused by S. marcescens (Alagely et al., 2011). This is similar to the ability of the commensal microbiota of vertebrates to act as a barrier to colonization and infection by pathogens (Leatham et al., 2009).
With this study we aimed to test whether the ability of the native commensal microbiota to inhibit specific catabolic enzymes in the coral pathogen S. marcescens disrupts the ability of the pathogen to establish within coral mucus. S. marcescens PDL100 possesses a broad suite of catabolic enzymes. Although many of the tested glycosidases are produced apparently constitutively, β-𝒟-galactosidase, N-acetyl-β-glucosaminidase, and α-𝒟-glucopyranosidase were differentially accumulated when the pathogen was grown on mucus (Krediet et al., 2009a). The over-arching hypothesis was tested with a three-step experiment: first, a Serratia mutant defective in mucus utilization was identified to test the role of specific catabolic enzymes in growth on mucus; second, coral commensals capable of inhibiting specific enzymatic activities were identified; third, interactions of the wild-type S. marcescens and its isogenic mutant defective in mucus utilization with the commensals were tested in mesocosm experiments.
Materials and Methods
Bacterial strains, media and growth conditions
Bacterial strains used in this study are listed in Table 1. Coral-associated bacteria were isolated from mucus of A. palmata by dilution plating onto Glycerol Artificial Sea Water medium as in Ritchie, 2006. The identities of the marine isolates were confirmed by PCR-amplifying and sequencing fragments of their 16S rRNA genes as previously described (Alagely et al., 2011). Data are deposited in GenBank (Accession numbers JQ954975; JQ954976; JQ954977; JQ954978; JQ954979; JQ954980).
Table 1. Bacterial strains and plasmids.
| Isolate or strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | F-ϕ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk-, mk+) phoA supE44 λ- thi-1 gyrA96 relA1 | Invitrogen |
| DH5α λ-pir | DH5α lysogenized with a bacteriophage λ434 derivative containing the pir gene | (Macinga et al., 1995) |
| Top10 | F mcrA (mrr-hsdRMS-mcrBC) 80lacZ M15 lacX74 deoR recA1 araD139 (ara-leu)7697 galU galK | Invitrogen |
| W3110 | rpsL endA1 nupG | (Hayashi et al., 2006) |
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | (Baba et al., 2006) |
| BW25113 ΔmalG15::kan | Single knockout deletion mutant of malG | (Baba et al., 2006) |
| BW25113 ΔmalF16::kan | Single knockout deletion mutant of malF | (Baba et al., 2006) |
| BW25113 ΔmalE17::kan | Single knockout deletion mutant of malE | (Baba et al., 2006) |
| W3110 ΔmalG15 | ΔmalG transduced into W3110 by P1 | This study |
| W3110 ΔmalF16 | ΔmalF transduced into W3110 by P1 | This study |
| W3110 ΔmalE17 | ΔmalE transduced into W3110 by P1 | This study |
| Serratia marcescens strains | ||
| Serratia marcescens PDL100 | Coral pathogen isolated from white pox lesion on Acropora palmata, ATCC BAA-632, TetR | ATCC, (Patterson et al., 2002) |
| Serratia marcescens CK2A4 | Mariner transposon mutant deficient in β-galactosidase activity | This study |
| Serratia marcescens MG1 | Isolated from rotten cucumber | (Givskov et al., 1997) |
| Serratia marcescens ATCC 43422 | Human throat isolate, pigmented | ATCC, (Krediet et al., 2009b) |
| Coral commensal strains | ||
| Photobacterium sp. 33G2 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954975 | This study, (Ritchie, 2006) |
| Photobacterium damselae 33G4 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954976 | This study, (Ritchie, 2006) |
| Photobacterium leiognathi 33E3 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954977 | This study, (Ritchie, 2006) |
| Photobacterium leiognathi 33C4 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954978 | This study, (Ritchie, 2006) |
| Exiguobacterium sp. 33G8 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954979 | This study, (Ritchie, 2006) |
| Vibrio harveyi 34B3 | Isolated from A. palmata mucus; identity confirmed by 16S rRNA gene sequencing; JQ954980 | This study, (Ritchie, 2006) |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector; KmR | Invitrogen |
| pRK600 | ColE1 replicon with RK2 tra genes, conjugation helper plasmid | (Karunakaran et al., 2005) |
| pBT20 | Mariner transposon delivery vector, GmR, R6K ori | (Kalivoda et al., 2008) |
| pCP20 | Temperature-sensitive replicon, thermal induction of FLP synthesis, CmR, ApR | (Datsenko and Wanner, 2000) |
| pBBR1-MCS2-kan | Cloning vector, KmR | (Kovach et al., 1995) |
| pBAD18-kan | Cloning vector, KmR, under arabinose inducible araBAD promoter | (Guzman et al., 1995) |
| pCJK16 | malEFG from PDL100 PCR amplified and cloned into pCR2.1-TOPO, ApR, KmR | This study |
| pCJK17 | malEFG from pCJK16 subcloned into EcoRI site of pBAD18-kan, KmR | This study |
| pCJK18 | malEFG from pCJK17 subcloned into EcoRI site of pBBR1-MCS2-kan, KmR | This study |
Abbreviations: Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Tetr, tetracycline resistance.
S. marcescens PDL100 and coral commensals were grown at 30 °C on the Glycerol Artificial Sea Water medium or marine broth (Difco-Becton, Dickinson and Company, Franklin Lakes, NJ, USA); Escherichia coli strains were grown at 37 °C in Luria-Bertani broth (Fisher Scientific, Atlanta, GA, USA) and when necessary on 1.5% agar plates. Unless otherwise specified, S. marcescens cultures were supplemented with tetracycline 10 μg ml−1 (Tc10), to which S. marcescens PDL100 is spontaneously resistant.
Coral mucus was collected from apparently healthy colonies of A. palmata at Looe Key Reef, Florida (24° 32.764' N: 81° 24.304' W) in July 2011 and prepared as previously described (Ritchie, 2006; Krediet et al., 2009a).
Mariner transposon mutagenesis of Serratia marcescens PDL100 and characterization of the mutants
Tri-parental conjugation was conducted using E. coli SM10-λ pir pBT20, S. marcescens PDL100 and the helper strain E. coli DH5α pRK600 as previously described (Kalivoda et al., 2008). One thousand colonies from each of the three independent matings were compiled into a library. This library was then screened for β-galactosidase activity (see below). To characterize mutants of interest, inverse PCR of DNA sequences flanking the transposon insertion was performed (Ochman et al., 1988; Supplementary Material).
For the experiments with E. coli, individual E. coli BW25113 mutants were obtained through the Keio Collection (Baba et al., 2006) and each of the mutants was transduced into strain W3110 (lac+) using phage P1 (Lennox, 1955; Ikeda and Tomizawa, 1965).
β-Galactosidase assays
Independent screens were conducted either with a library of transposon mutants or with coral commensals. The ability of the commensals to inhibit β-galactosidase in S. marcescens PDL100 was tested by coinoculating coral commensals with PDL100 in 0.3% marine agar in 96-well plates supplemented with 5-bromo-4-chloro-indolyl-β-𝒟-galactopyranoside (X-gal) at 40 μg ml−1 (Gold Biotechnology, St Louis, MO, USA). Plates were incubated at 30 °C overnight, and were monitored for blue color. Candidates were retested in cross-streaks on marine agar. Inhibitory capabilities of the coral commensals were tested against PDL100 in β-galactosidase assays through coincubation of S. marcescens in the spent cell-free culture supernatants of coral commensals. The ability of the cell-free supernatants to inhibit activity of the purified E. coli β-galactosidase enzyme (Sigma-Aldrich, St Louis, MO, USA) was also tested (Supplementary Material).
Individual transposon mutants were assayed in soft marine agar with X-gal and were monitored for blue color development. Mutants deficient in β-galactosidase activity (compared to the wild type) were isolated to single colony on X-gal containing media and then assayed quantitatively against the wild type using ONPG (Miller, 1972).
Competitive fitness assays
Overnight cultures of S. marcescens PDL100 and CK2A4 were grown from glycerol stock in marine broth to an approximate OD600 of 1.7 and cultures were serially diluted and mixed in 1:1 ratio. Strains were inoculated individually or as a mix at starting concentrations of 102 cfu ml−1 into filter sterilized coral mucus, high-molecular-weight fraction of coral mucus from A. palmata, and an artificial seawater-based casamino acids/glycerol media (in artificial seawater: 0.05% Casamino acids, 0.4% glycerol, 10 mℳ HEPES, pH 7.1). All cultures were grown in triplicates. For enumeration, cultures were dilution plated at 0, 12, 24, 48, 72 and 96 h on marine agar supplemented with Tc10. All assays on each medium were repeated at least twice. To distinguish between the wild type and the mutant, individual colonies were patched onto selective media with Tc10 and gentamycin (Gm) 50 μg ml−1.
For the three-strain competition assay, S. marcescens PDL100, CK2A4 and commensals with β-galactosidase inhibitory properties, strains were grown individually in marine broth overnight and diluted as above. S. marcescens PDL100 was inoculated at 102 cfu ml−1 and the cocktail of the six inhibitory coral commensals was inoculated at 104 cfu ml−1 into high-molecular-weight coral mucus from A. palmata. Competitive indices were calculated for each treatment using the formula (Mout/WTout)/(Min/WTin), where M is the proportion of mutant cells and WT is the proportion of wild-type cells in the inoculum (in) or in the recovered samples (out). Statistical significance of each competitive index was established by comparing log values of the competitive indices using a two-tailed t-test.
Preliminary characterization of inhibitory compounds in Exiguobacterium sp. 33G8
Potential antibiotic production by the coral commensal strains was tested by filling agar cores of the commensal strains into semi-solid marine agar (0.5%) seeded with PDL100. The coinoculated strains were incubated at 30 °C overnight and monitored for inhibition of growth in PDL100.
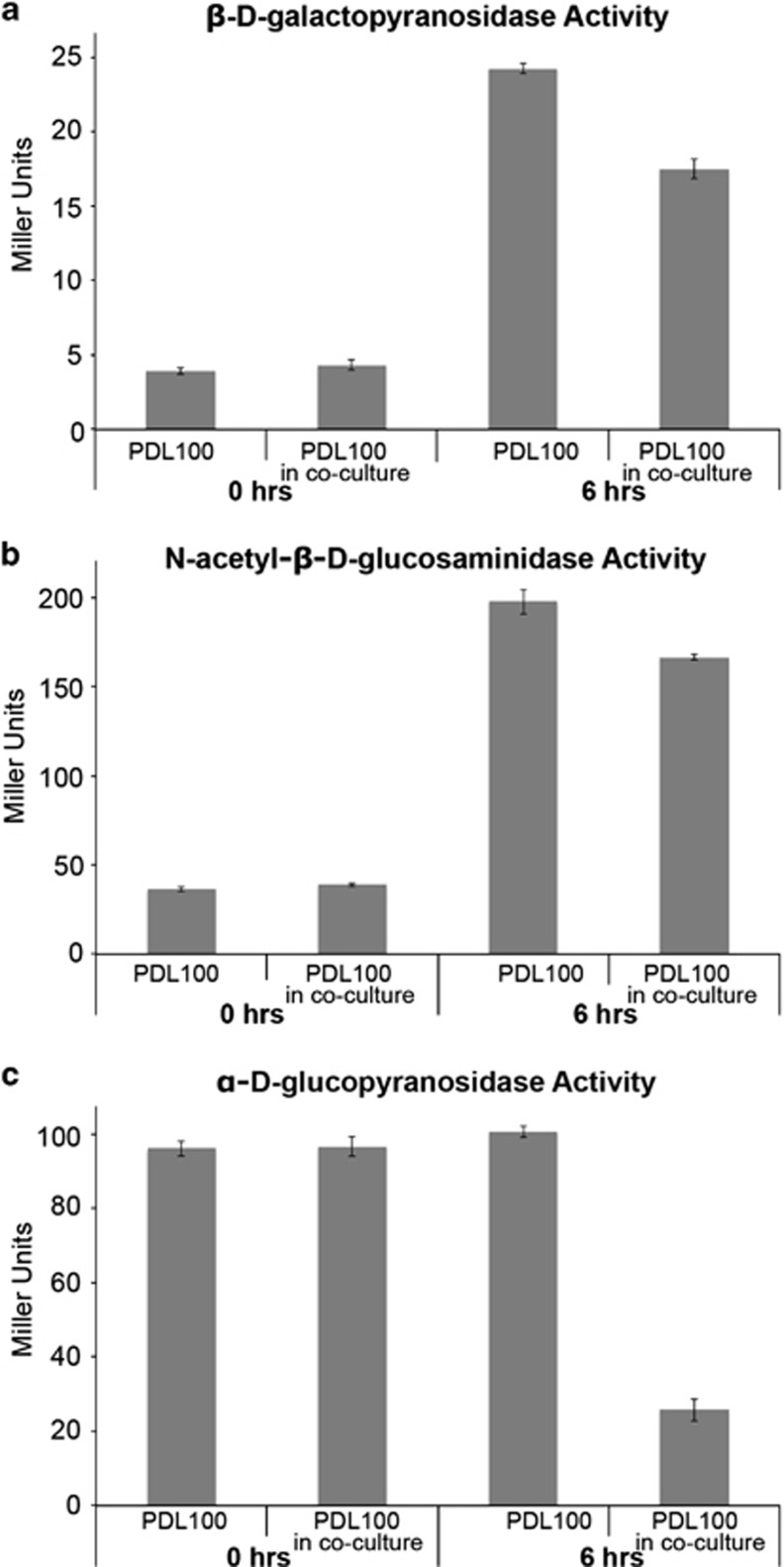
Inhibition of β-galactopyranosidase, N-acetyl-β-𝒹-glucosaminidase and α-𝒹-glucopyranosidase enzymatic activies in PDL100 by coral commensal bacteria were tested through coincubation of the strains using dialysis pouches. Cellulose ester dialysis membrane (MWCO: 1 000 000) stored in 0.05% sodium azide (Spectrum Laboratories, Inc., Rancho Dominguez, CA, USA) was cut into 10-cm strips and washed in sterile DI water (10 water changes) at 4 °C. S. marcescens PDL100 was grown from glycerol stock in 5 ml marine broth and then subcultured into 100 ml marine broth. Exiguobacterium sp. 33G8 was grown from glycerol stock in 5 ml of marine broth overnight at 30 °C and subcultured into 100 ml of sterile marine broth. After overnight incubation, the OD600 of PDL100 was 0.7 and Exiguobacterium 33G8 was at 1.8. The PDL100 culture was concentrated to an OD600 of 1.2 and 5 ml of these cells were transferred into the dialysis pouches. The entire 100 ml culture of Exiguobacterium 33G8 was transferred into 500 ml of sterile marine broth. A dialysis tube containing PDL100 cells was placed into the 33G8 culture and into a flask of sterile marine broth as a control. The cocultures were incubated at 30 °C for 6 h and enzymatic activities of all three enzymes in PDL100 were measured at the beginning of the experiment and 6 h later using nitrophenyl substrates as in Miller, 1972.
To begin to determine the type of inhibitory activity produced by Exiguobacterium sp. 33G8 in coculture, cell-free supernatant from the Exiguobacteriu-Serratia coincubation experiment was subjected to flash ion-exchange chromatography (DOWEX 50-W, Sigma-Aldrich) and the flow-through was then subjected to reverse-phase Silica-C18 chromatography (Alltech Associates, Inc., Deerfield, IL, USA) (Supplementary Material).
Each of the eluted fractions was dried, re-extracted as described in Materials and methods, serially diluted (threefold) and then subjected to a bioassay. The highest dilution contained ∼1.5 ml of the ‘culture equivalent'. The dilutions and solvent controls were evaporated in the chamber of the Centrivap Concentrator (Labconco, Kansas City, MO, USA) at 50 °C for 30 min. Six hundred and fifty microliters of S. marcescens PDL100 culture (OD600=0.922) that had been washed in equal volume of sterile marine broth were added to each sample. Cells were incubated with the diluted fractions for 6 h at 30 °C and assayed for α-𝒟-glucopyranosidase activity as described by Miller, 1972.
Swarming assays
Twenty microliters of cultures of individual commensal strains grown overnight in marine broth were spotted onto a sterile glass fiber GF/C filter disk and placed on a marine agar plate. Five microliters of S. marcescens cultures were spotted onto AB swarm agar plate ∼2 cm from the disk containing commensals as described previously (Alagely et al., 2011). For swimming motility, 5 μl of each culture were stab inoculated into marine broth 0.3% agar plates 2 cm apart. Plates were incubated at 30 °C overnight and photographed with a Canon EOS Rebel Xsi digital camera (Canon USA Inc., San Jose, CA, USA). Images were corrected for auto levels in Adobe Photoshop.
Virulence in a sea anemone model
Individual polyp infections with PDL100 wild type and CK2A4 were performed as described previously (Krediet et al., in review). The polyps were monitored for disease signs and Kaplan–Meier survivorship analyses were performed for each infection using JMP 9.0 Pro statistical software (SAS Institute Inc., Cary, NC, USA) and mortality across treatments was compared with a Cox proportional hazards χ2 model (Hosmer and Lemeshow, 1999).
Results
Characterization of the S. marcescens mutant defective in glycosidase and N-acetyl-glucosaminidase activities
To test the importance of β-galactosidase, N-acetyl-β-𝒟-glucosaminidase and α-𝒟-glucopyranosidase for the ability of S. marcescens to utilize components of coral mucus, a transposon mutant, S. marcescens CK2A4, deficient in these activities was isolated. To characterize the mutant, inverse PCR was carried out, and a single PCR band was detected and then sequenced. The sites flanking the transposon insertion were identified as homologous to the malF gene (Spro_4473; base pairs 283–715) of Serratia proteamaculans. However, attempts to complement the transposon mutant with the entire predicted 4.8-kb ‘malEFG' operon amplified from S. marcescens PDL100 genomic DNA were not successful (data not shown).
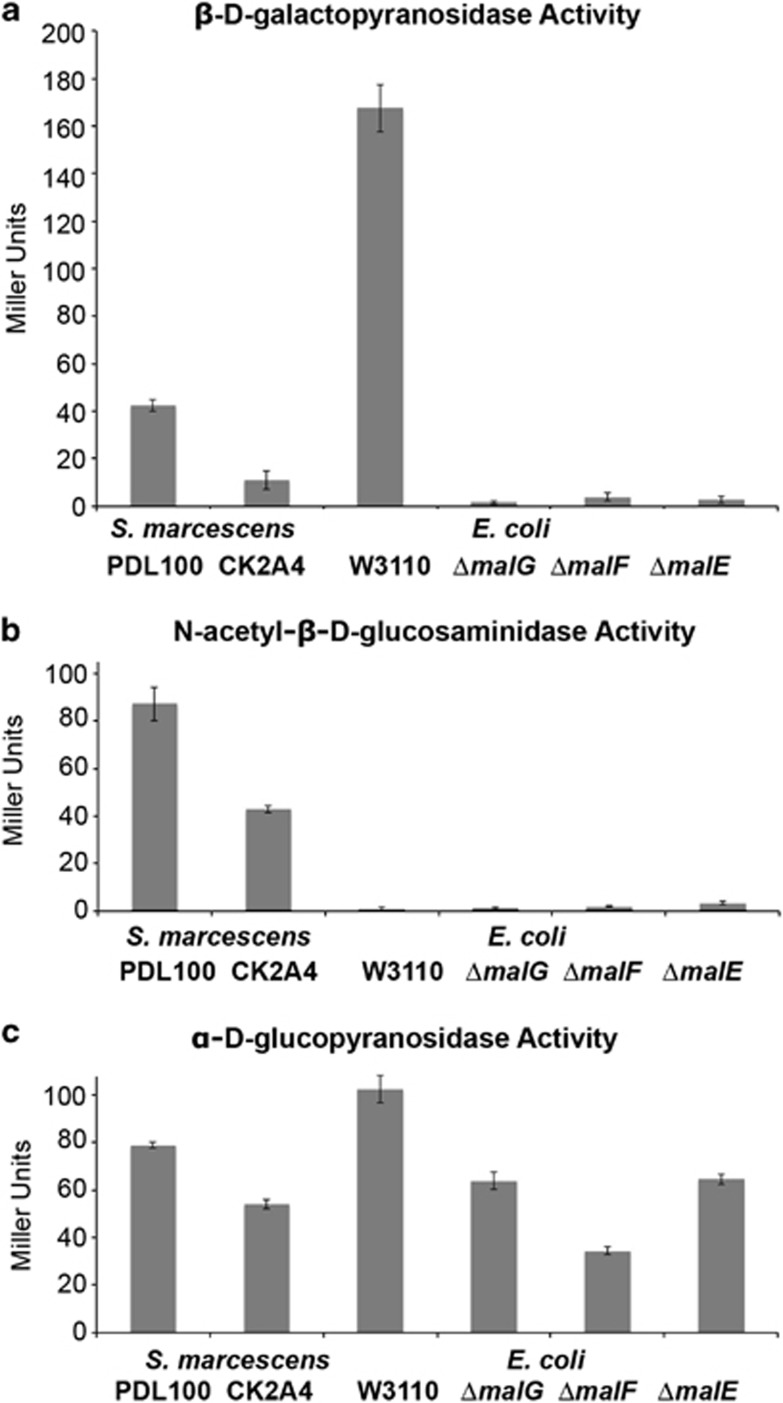
Because the complementation of the CK2A4 mutant was unsuccessful, glycosidase and N-acetyl-glucosaminidase activities of the defined malE, malF and malG mutants were tested in E. coli W3110 to corroborate the identification of CK2A4 as a malF mutant to establish whether the phenotype of the S. marcescens mutant is consistent with the mutation in the mal operon. Enzymatic activities in the defined E. coli mutants and in CK2A4 were similarly decreased (Figure 1). Note, that E. coli W3110 does not produce N-acetyl-β-𝒟-glucosaminidase (Figure 1). Therefore, even though the malF mutation in CK2A4 could not be complemented, its phenotype is consistent with the phenotypes of three defined E. coli mutants in the malEFG operon.
Figure 1.
Enzymatic activities in mutants of S. marcescens and E. coli. Overnight cultures of wild-type S. marcescens PDL100 and the transposon mutant (CK2A4) were tested for activities of (a) β-𝒟-galactopyranosidsase, (b) N-acetyl-β-𝒟-glucosaminidase and (c) α-𝒟-glucopyranosidase using a Miller assay (Miller, 1972) with appropriate p-nitrophenyl-glycoside substrates. E. coli W3110 and individual in-frame single gene knockouts of malE, malF and malG were also tested to compare phenotypes of CK2A4 with those of the defined well-characterized E. coli mutants in the malEFG operon. All cultures were grown overnight before the assays; S. marcescens was cultured in Marine broth, E. coli was grown in LB. Averages of three biological and four technical replications are shown. Error bars are standard error. There was a significant reduction in the enzymatic activities between wild-type S. marcescens and CK2A4 (β-𝒟-galactopyranosidsase: t=13.55, dF=6, P<0.0001; N-acetyl-β-𝒟-glucosaminidase: t=12.22, dF=3.3, P=0.0008; α-𝒟-glucopyranosidase: t=20.86, dF=4.87, P<0.0001). In E. coli, β-𝒟-galactopyranosidsase and α-𝒟-glucopyranosidase activities were reduced in malE, malF and malG mutants.
Competitive fitness of S. marcescens PDL100 and CK2A4
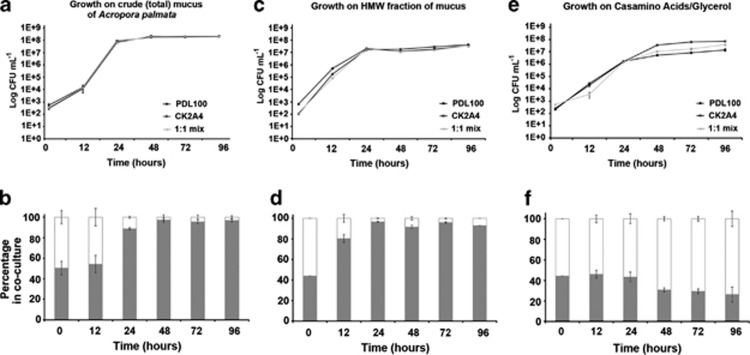
The ecological importance of β-galactosidase and other catabolic enzymes for utilization of coral mucus from A. palmata was tested by growing wild-type PDL100 and the CK2A4 mutant on total coral mucus and high-molecular-weight fraction of mucus both individually and in a 1:1 coculture. Individual growth curves for both strains showed the same growth trend on total mucus and high-molecular-weight coral mucus (Figures 2a and c). Importantly, however, within the coculture, CK2A4 was not competitively fit on either total coral mucus or on high-molecular-weight fraction of the mucus (Figures 2b and d; log competitive indices −1.64 and −1.23, respectively), consistent with the predicted role of the β-galactosidase, N-acetyl-β-𝒟-glucosaminidase and α-𝒟-glucosidase enzymes in mucus degradation (Krediet et al., 2009a, 2009b). After 12–24 h, the wild type dominated the culture and after 48 h the wild type represented ∼95% of the culture. However, in the absence of the substrates for glycosidases or N-acetyl-glucosaminsidases (that is, when grown on a minimal medium with casamino acids and glycerol), the CK2A4 mutant had a modest competitive advantage (Figure 2e; log competitive index 0.480). This indicates that the reduced fitness of the mutant on coral mucus is not due to a general reduction in the metabolism, but rather is due to a disruption of the specific catabolic activities required for the utilization of coral mucus.
Figure 2.
Competitive fitness of S. marcescens PDL100 and CK2A4. To test the importance of catabolic enzymes for growth of S. marcescens on coral mucus, both wild type and CK2A4 were grown both individually and in coculture on mucus of A. palmata and in the HEPES-buffered (10 mℳ, pH 7.1) artificial seawater with casamino acids (0.5 g l−1) and glycerol (0.4% vol/vol). For competition experiments, overnight inocula were serially diluted and mixed (1:1 ratio). Cultures in (a) crude (total) mucus of Acropora palmata, (c) high-molecular-weight (>5 kDa) fraction of A. palmata mucus or (e) casamino acids/glycerol were inoculated at 102 cfu ml−1. Cultures were incubated at 30 °C with shaking. To enumerate cells, at 12, 24, 48, 72 and 96 h cultures were dilution plated onto marine agar supplemented with Tc10. Averages of three biological replications of the experiment (three independent cultures) are shown, error bars are standard error. In monocultures, all strains grew similarly and reached the same final population densities. To estimate the percentage of each strain within each coculture (competitive fitness), colonies were patched onto marine agar supplemented with Tc10 and Gm50 to differentiate between the wild type and the transposon mutant. The relative proportion of the wild type is shown as the gray portion of the stacked column. Competitive fitness experiments were repeated at least twice, and averages are shown (error bars are standard errors). On total mucus (b) and high-molecular-weight fraction of the mucus (d), the mutant was not competitive against the wild type. CK2A4 had a fitness advantage on the casamino acid/glycerol defined medium and grew to higher population densities than wild type and the 1:1 coculture (f).
Virulence of S. marcescens PDL100 and CK2A4 in a polyp model
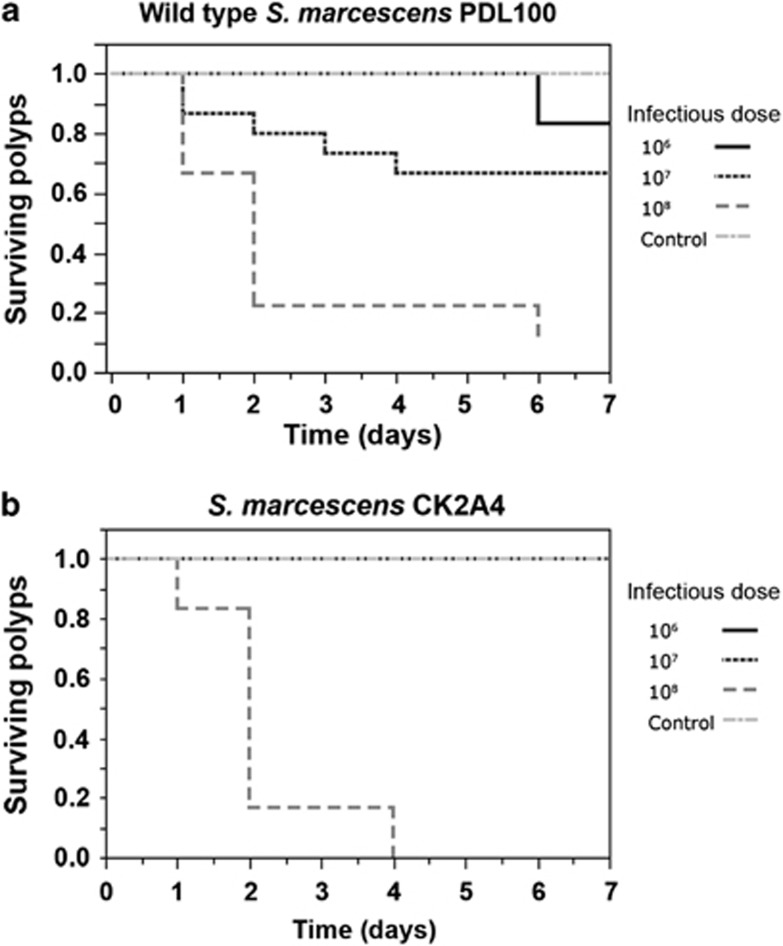
Sixty-nine Aiptasia pallida polyps were used to test the virulence of S. marcescens PDL100 and CK2A4 over a 7-day period. After 24 h of incubation at the highest concentration (108 cfu ml−1) the polyps compressed their stalk length, retracted their tentacles and exhibited darkening of the tissue. These responses were seen to a lesser degree at the 107 cfu ml−1 treatment and rarely at the 106 cfu ml−1 concentration in the wild type but no mortality was observed at the lower concentrations in CK2A4. The survivorship of the infected polyps is plotted in Figure 3. The differences in survivorship between the different treatments were statistically significant (PDL100: χ2=38.60; dF=4; P<0.0001; CK2A4: χ2=28.25; dF=3; P<0.0001).
Figure 3.
Survivorship of Aiptasia pallida polyps infected with S. marcescens. Ten-fold dilutions of overnight cultures of S. marcescens PDL100 and CK2A4 were inoculated into microtiter plate wells with A. pallida. Infections were maintained at room temperature (22–24 °C) with illumination. Polyps were scored daily. Each line represents the proportion of polyps surviving at each concentration (106 cfu ml−1, 107 cfu ml−1, 108 cfu ml−1, or control as shown in the key). Survivorship decreased significantly with increasing pathogenic concentration for polyps infected with (a) S. marcescens PDL100 (χ2=38.604; dF=4; P<0.0001) and (b) S. marcescens CK2A4 (χ2=28.249; dF=3; P<0.0001).
Coral commensal bacteria capable of inhibiting enzymatic activities limit growth of S. marcescens in coculture
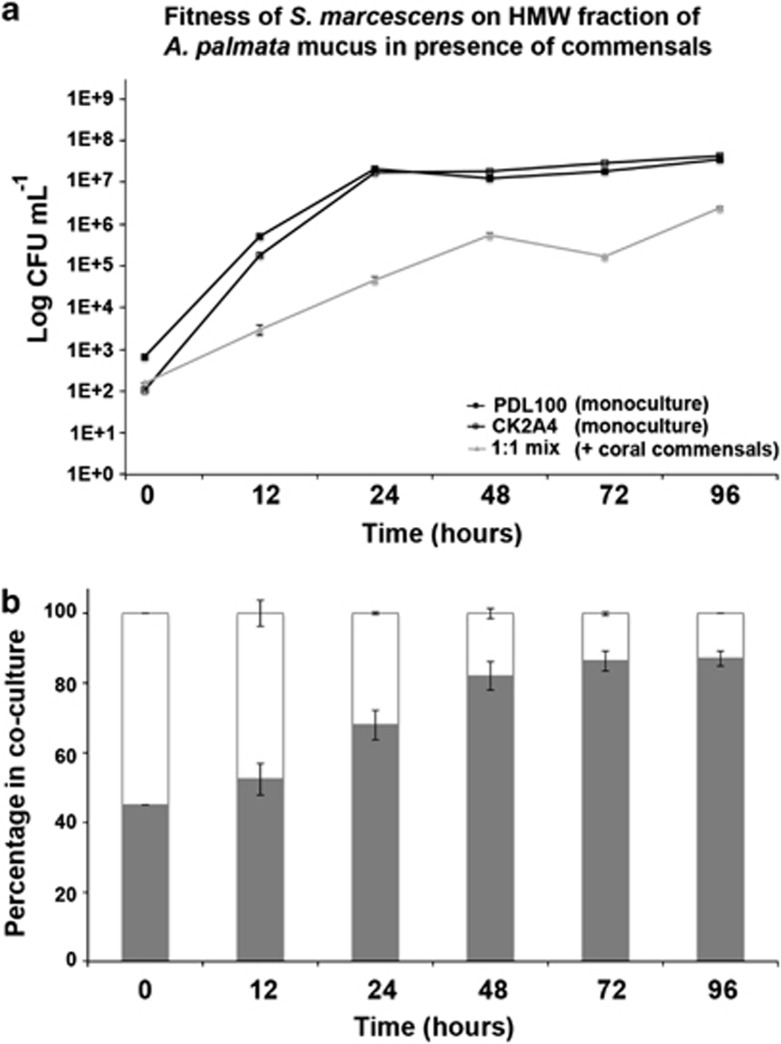
Because the ability to cleave sugar and amino-sugar residues off coral mucus appears important for the successful colonization of the coral mucus surface layer, we next tested whether the native commensals have evolved the mechanisms to inhibit glycosidases in the invading opportunists. Initial screens of a library of commensal isolates collected from the mucus surface layer of a healthy A. palmata (which is the only coral known to be susceptible to S. marcescens infections) revealed that ∼8% of isolates are capable of inhibiting β-galactosidase activity in S. marcescens (Supplementary Figure 1). The competitive fitness of PDL100 and CK2A4 on high-molecular-weight mucus was tested in the presence of these inhibitory coral commensal strains (Photobacterium sp. 33G2, P. damselae 33G4, Exiguobacterium sp. 33G8, P. leiognathi 33C4, and P. leiognathi 33E3, and Vibrio harveyi 34B3). We tested the hypothesis that in the presence of the commensals that inhibit glycosidase and N-acetyl-glucosaminidase activities, the CK2A4 mutant will be as fit as the wild type due to the ability of the commensals to inhibit these enzymatic activities in the wild type. In the coral mucus coculture of Serratia with the commensals, growth of the pathogen was reduced by 10–100-fold (compared with the monoculture of Serratia; Figure 4a). Furthermore, the CK2A4 mutant (defective in the enzymatic activities targeted by the commensals) was more competitive against the wild type compared with the mesocosms without the commensals at 12 and 24 h (Figures 2d vs 4b; log competitive index −0.729 vs −0.129, t3.82=4.769, P=0.0099). These observations suggest that the ability of commensals to specifically inhibit catabolic enzymes (glycosidases and N-acetyl-glucosaminidase) is at least in part responsible for the reduction in growth of S. marcescens on coral mucus in the presence of these commensals. Our subsequent experiments focused on the characterization of the activities produced by the commensals.
Figure 4.
Competitive fitness of S. marcescens with coral commensals. Coral commensal strains shown to inhibit β-𝒟-galactopyranosidase activity in S. marcescens PDL100 were grown in coculture on high-molecular-weight fraction of A. palmata mucus with wild-type PDL100 and transposon mutant CK2A4. To prepare inoculum of S. marcescens strains, overnight cultures were serially diluted and wild type and CK2A4 were mixed in 1:1 ratio and inoculated at 102 cfu ml−1. The coral commensals (Photobacterium sp. 33G2, P. damselae 33G4, Exiguobacterium sp. 33G8, P. leiognathi 33C4, P. leiognathi 33E3 and Vibrio harveyi 34B3) were grown separately, mixed as a cocktail and inoculated at 104 cfu ml−1 concurrently with S. marcescens. (a) In monocultures (black lines) of S. marcescens without coral commensals, the wild type (filled square) and CK2A4 mutant (empty square) reached the same population densities. In the coculture on high-molecular-weight fraction of A. palmata mucus in the presence of the coral commensals, growth of S. marcescens was significantly reduced (gray line). Averages of three biological replicates (three independent cultures) are shown. Error bars are standard errors. (b) Fitness of the mutant versus the wild type in the presence of the commensals was estimated by patching of Serratia colonies onto media with the appropriate antibiotics. The relative proportion of the wild type is shown as the gray portion of the stacked column.
Preliminary characterization of inhibitory compounds in Exiguobacterium sp. 33G8
Exiguobacterium sp. 33G8 strongly inhibited β-galactosidase activity in S. marcescens strains PDL100 and MG1 (Supplementary Figure 1) and was chosen for further investigation. Our experiments aimed to (a) test mode of action of the compound(s) and (b) characterize chemical properties of the activity.
Reduction in enzymatic activity in PDL100 in the coculture with Exiguobacterium sp. was first assayed by growing PDL100 in a dialysis pouch in the presence of Exiguobacterium sp. At the start of the experiment, the enzymatic activities were not significantly different between the control culture and PDL100 inoculated with Exiguobacterium sp. (Figure 5). After the 6 h coculture, the enzymatic activities of PDL100 in coculture were significantly reduced for each enzyme (β-galactosidase: t=9.340, dF=4.2, P=0.0005; N-acetyl-β-𝒟-glucosaminidase: t=4.410, dF=3.27, P=0.0180; α-𝒟-glucopyranosidase: t=22.191; dF=4.34; P<0.0001; Figure 5). We then tested whether the substance(s) produced by the commensal directly inhibit the enzymatic activity or regulation of the enzyme synthesis. Incubation of Serratia in the cell-free supernatant from the monoculture of Exiguobacterium neither reduced the wild-type β-galactosidase activity, nor did the coral commensal cultures reduce activity of a purified β-galactosidase from E. coli (data not shown). This indicates that the substance is not a compound that inhibits the function of the β-galactosidase enzyme. The inhibitory activity was extracellular, but was produced by Exiguobacterium sp. only when it was cultured with S. marcescens.
Figure 5.
Inhibition of enzymatic activities in S. marcescens PDL100 by Exiguobacterium sp. 33G8. Exiguobacterium sp. 33G8 was grown in coculture with PDL100 to test for inhibition of the enzymatic activities that are normally induced in S. marcescens grown on mucus of A. palmata. As detailed in Materials and methods, within the coculture, bacteria were separated with a dialysis membrane (MWCO: 1 000 000) for the ease of recovering cells for enzymatic assays. The dialysis pouch pore size was enough to retain cells but allow compounds produced by both organisms to pass through. Enzymatic assays for PDL100 (a) β-𝒟-galactopyranosidase, (b) N-acetyl-β-𝒟-glucosaminidase and (c) α-𝒟-glucopyranosidase activities were conducted at the time of inoculation and after 6 h of coculture using nitrophenyl substrates as in Miller, 1972. Enzymatic activities of S. marcescens PDL100 in a monoculture or in the coculture with Exiguobacterium sp 33G8 are shown. Experiments were repeated twice, each time with four technical replicates. Data from one replicated experiment are shown. There was no difference in initial activity between the treatments for each enzyme assayed. After coincubation for 6 h, S. marcescens PDL100 showed strongly significantly reduced enzymatic activities as a result of 33G8 for β-𝒟-galactopyranosidase (t4.2=9.340, P=0.0005) and α-𝒟-glucopyranosidase (t4.34=22.191, P<0.0001), and modestly significantly reduced activity of N-acetyl-β-𝒟-glucosaminidase (t3.27=4.410, P=0.0180).
Preliminary characterization of the chemical produced by Exiguobacterium sp. 33G8 was performed by extraction of cell-free supernatant from the 33G8 culture used for the dialysis pouch coculture enzyme assay. Cell-free supernatant was passed through an ion-exchange column and then the flow-through from the ion-exchange column was subjected to reverse-phase Si C18 chromatograpy. Eluent from both columns was dried, re-extracted with two different solvents and the resulting samples were assayed for their ability to reduce of α-𝒟-glucopyranosidase activity in PDL100. This enzyme was chosen out of the three initially tested because of the strong inhibition seen in the coculture experiment (Figure 5). Of the fractions assayed, the ethanol-soluble extract of the eluent from the ion-exchange column showed the greatest reduction in activity (nearly 1.5-fold reduction), as compared with the solvent alone control (Supplementary Figure 2A).
Swarming motility inhibition in S. marcescens PDL100 by coral bacteria
Even though the inhibition of enzymatic activities by the commensals was at least in part responsible for the reduced fitness of S. marcescens within coral mucus, it did not fully account for the observed phenotype. The conclusion that the activities produced by commensals target induction of enzymatic activities, rather than a function of the enzyme, suggests that the compounds may affect a higher-level regulator. Therefore, we explored the possibility that the commensals inhibit behaviors associated with niche colonization and coregulated with metabolism.
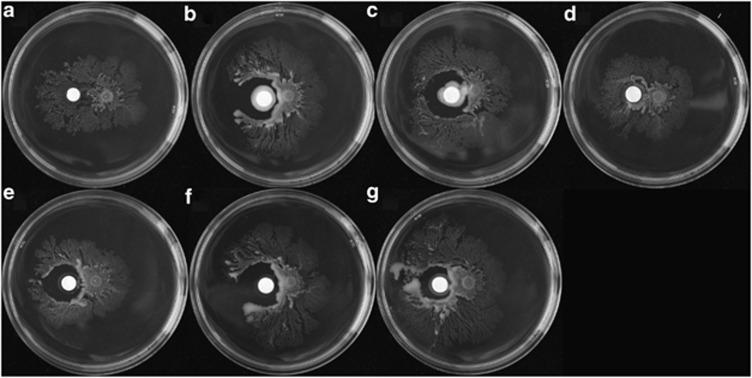
To further characterize the potential coregulation of metabolic enzymes and motility, swarming motility of PDL100 was assayed with and without the commensals as in Alagely et al., 2011. When spotted onto AB swarm agar plate, PDL100 swarms (Figure 6a). Five of the six strains that were capable of inhibiting glycosidases, also inhibited swarming of S. marcescens. Exiguobacterium sp. 33G8 did not inhibit swarming although this strain showed the greatest inhibition of enzyme induction during growth in coculture (Figure 6d). We then tested three hypotheses that explain these observations: commensals produce antibiotics, motility inhibitors or specific inhibitors of swarming.
Figure 6.
Inhibition of swarming of S. marcescens PDL100 by coral commensals. Swarming of S. marcescens PDL100 on AB 0.4% swarm agar next to a sterile glass fiber GF/C disk is shown in (a). Individual coral commensal strains (Photobacterium sp. 33G2 (b), P. damselae 33G4 (c), Exiguobacterium sp. 33G8 (d), P. leiognathi 33C4 (e), P. leiognathi 33E3 (f) and Vibrio harveyi 34B3 (g)) were grown in marine broth and 20 μl were spotted onto a glass fiber GF/C disk on solid marine agar and disks were incubated at 30 °C overnight. Filter disks were placed next to PDL100 (5 μl of overnight culture diluted 1:100 and grown for 2 h). Plates were incubated at 30 °C overnight and all strains, except 33G8, inhibited swarming motility in the pathogen. The experiments were repeated three times, data from one representative experiment are shown.
No growth inhibition of S. marcescens was observed in diffusion assays to test for the antibiotic production by the coral commensals (data not shown), indicating that neither the limitation of growth seen in the competitive fitness coculture test, nor inhibition of swarming is due to antibiotic production by commensals. Five of the six of the commensal strains tested reduced swarming motility in S. marcescens, however, none inhibited swimming within soft agar. These data suggest that the commensal strains may produce inhibitory compounds specific to the coregulation of swarming motility (not flagellar motility in general) and metabolism, perhaps by targeting a higher-level regulator that controls both pathways.
Discussion
Glycosidases and N-acetyl-glucosidase, which correspond to the available substrates in coral mucus, are induced during growth of Serratia marcescens PDL100 on coral mucus (Krediet et al., 2009a, 2009b). This study sought to test the consequences of inhibition of these enzymatic activities in S. marcescens by coral-associated bacteria. A transposon mutant deficient in these activities was identified and tentatively mapped to the operon homologous to the E. coli malEFG maltose/maltodextrin transporter. This high-affinity ABC transporter system consists of a periplasmic maltose-binding protein encoded by malE and a transport complex encoded by malF, malG and malK (Jones et al., 2008b). On the basis of the transport functions of the operon, it was hypothesized that malEFG is necessary for proper colonization and utilization of coral mucus by the pathogen. Although the S. marcescens malEFG mutant showed a threefold decrease in β-galactosidase activity and a twofold decrease in N-acetyl-β-𝒟-glucosaminidase activities compared with the wild type, the mutation did not fully eliminate the ability of the pathogen to utilize and grow on coral mucus. In a monoculture, the mutant grew to the same population density as the wild type, however, was not competitive against the wild type on coral mucus in coculture. It is plausible that the reduced affinity for the substrate resulting from the disruption of the transporters is responsible for the reduced competitive fitness of the strain. The reduction, but not a complete elimination, of the ability of the mutant to utilize mucus is likely due to the fact that bacteria rely on transporters with redundant functions for the uptake of carbon sources. The decrease in the virulence of the mutant in a surrogate host is also likely due to its reduced fitness within the surface mucus layer and its inability to establish population densities that are high enough to effectively invade the host.
Coral pathogens compete with the native coral-associated bacteria to establish within a niche and utilize preferred carbon sources of coral mucus. These coral-associated bacteria interact with and can interfere with the invading pathogen through antibiotic production, inhibition of quorum sensing and secondary metabolite production (Shnit-Orland and Kushmaro, 2009; Teplitski and Ritchie, 2009; Rypien et al., 2010). This study characterized six coral-associated strains capable of interfering with metabolic enzymatic activities and decreasing the overall growth of S. marcescens on coral mucus. Interference of enzymatic activities was cell-associated and dependent on coculturing of commensals and the pathogen.
In addition to inhibiting enzymatic activities in S. marcescens that are involved in coral mucus utilization, the coral-associated bacteria also inhibited swarming motility in the pathogen. We do not yet know whether the same compound is involved in the inhibition of both swarming and catabolism of mucus components. However, if this were the case, it is possible to hypothesize that a higher-level regulatory system (GacS/GacA-Csr or FlhDC) (Romeo, 1998; Pruss et al., 2003; Park and Forst, 2006; Timmermans and Van Melderen, 2010) may be the target. The GacS/GacA-Csr system is not likely to be affected by the activities of these commensals: first, the phenotype of the gacA mutant (Krediet et al., in review) is distinct from what is observed when the wild type is exposed to the commensals tested here, and, second, the commensals did not affect the expression of the gacA::lacZ reporter (Krediet, unpublished data). It is also unlikely that FlhDC is affected by the commensals: even though FlhDC in Enterobacteriacea is known to contribute to the regulation of both motility and metabolism (Pruss et al., 2003), the fact that none of the commensals inhibited swimming motility suggests that FlhDC is also an unlikely target of these activities. Therefore, these observations suggest that either the compounds target another regulatory cascade, or that different compounds disrupt swarming and the induction of enzymatic activities separately.
The interactions between coral-associated mutualistic bacteria and invading pathogens are numerous, complex and still poorly understood. The results of this study demonstrate that coral-associated bacteria disrupt the coregulated metabolic activities and swarming in the invading opportunists, and thus the ability of the pathogens to compete with commensals within the coral mucus environment. Further research will elucidate the exact level of these regulatory pathways at which coral commensal strains interfere. It will be also important to establish whether the bacteria with the ‘beneficial' properties are actively recruited by host (which would be consistent with the Hologenome Theory of Evolution (Rosenberg and Zilber-Rosenberg, 2011)), or whether there are other hypotheses that explain their functions within the coral surface mucus layer.
Acknowledgments
This research was supported by the National Geographic Committee for Research Exploration grant no. 9184–12 and by ‘Protect Our Reefs', a program managed by Mote Marine Laboratory with the proceeds from the state-wide sales of Protect Our Reefs specialty license plates and administered through a peer-reviewed mechanism. KBR's contributions were also funded in part by the Dart Foundation. MT acknowledges funding by Smithsonian Burch Fellowship. We thank Dr Claudio Gonzalez and Ricardo Valladares (UF) for generously providing individual ΔmalE, F, G mutants in E. coli and phage P1 for generalized transductions. We also thank Erich Bartels for field support.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alagely A, Krediet CJ, Ritchie KB, Teplitski M. Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 2011;5:1609–1620. doi: 10.1038/ismej.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BE, Bythell JC. Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser. 2005;296:291–309. [Google Scholar]
- Bythell JC, Wild C. Biology and ecology of coral mucus release. J Exp Mar Biol Ecol. 2011;408:88–93. [Google Scholar]
- Coddeville B, Maes E, Ferrier-Pages C, Guerardel Y. Glycan profiling of gel forming mucus layer from the scleractinian symbiotic coral Oculina arbuscula. Biomacromolecules. 2011;12:2064–2073. doi: 10.1021/bm101557v. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducklow HW, Mitchell R. Composition of mucus released by coral-reef coelenterates. Limnol Oceanogr. 1979;24:706–714. [Google Scholar]
- Garren M, Azam F. New method for counting bacteria associated with coral mucus. Appl Environ Microbiol. 2010;76:6128–6133. doi: 10.1128/AEM.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M, Azam F. New directions in coral reef microbial ecology. Environ Microbiol. 2012;14:833–844. doi: 10.1111/j.1462-2920.2011.02597.x. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol. 2006;2:2006–0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. Applied Survival Analysis. Wiley & Sons: New York; 1999. [Google Scholar]
- Ikeda H, Tomizawa JI. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965;14:85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Jones MK, Warner EB, Oliver JD. csrA inhibits the formation of biofilms by Vibrio vulnificus. Appl Environ Microbiol. 2008a;74:7064–7066. doi: 10.1128/AEM.01810-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, et al. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun. 2008b;76:2531–2540. doi: 10.1128/IAI.00096-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivoda EJ, Stella NA, O'Dee DM, Nau GJ, Shanks RM. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol. 2008;74:3461–3470. doi: 10.1128/AEM.02733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran R, Mauchline TH, Hosie AH, Poole PS. A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology. 2005;151:3249–3256. doi: 10.1099/mic.0.28311-0. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. 4 new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Krediet CJ, Ritchie KB, Cohen M, Lipp EK, Sutherland KP, Teplitski M. Utilization of mucus from the coral Acropora palmata by the pathogen Serratia marcescens and by environmental and coral commensal bacteria. Appl Environ Microbiol. 2009b;75:3851–3858. doi: 10.1128/AEM.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet CJ, Ritchie KB, Teplitski M. Catabolite control in a coral white pox pathogen, Serratia marcescens PDL100. Dis Aquat Organ. 2009a;87:57–66. doi: 10.3354/dao02084. [DOI] [PubMed] [Google Scholar]
- Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox ES. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Macinga DR, Parojcic MM, Rather PN. Identification and analysis of aarP, a transcriptional activator of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1995;177:3407–3413. doi: 10.1128/jb.177.12.3407-3413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle P, Richards GN, Yellowlees D. Structural determination of the oligosaccharide side-chains from a glycoprotein isolated from the mucus of the coral Acropora formosa. J Biol Chem. 1987;262:16941–16947. [PubMed] [Google Scholar]
- Meikle P, Richards GN, Yellowlees D. Structural investigations on the mucus from 6 species of coral. Mar Biol. 1988;99:187–193. [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY; 1972. [Google Scholar]
- Nissimov J, Rosenberg E, Munn CB. Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett. 2009;292:210–215. doi: 10.1111/j.1574-6968.2009.01490.x. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Forst S. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol Microbiol. 2006;61:1397–1412. doi: 10.1111/j.1365-2958.2006.05320.x. [DOI] [PubMed] [Google Scholar]
- Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, Peters EC, et al. The etiology of white pox, a lethal disease of the Caribbean elkhorn coral Acropora palmata. Proc Natl Acad Sci USA. 2002;99:8725–8730. doi: 10.1073/pnas.092260099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol. 2003;185:534–543. doi: 10.1128/JB.185.2.534-543.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Zilber-Rosenberg I. Symbiosis and development: the hologenome concept. Birth Defects Res C. 2011;93:56–66. doi: 10.1002/bdrc.20196. [DOI] [PubMed] [Google Scholar]
- Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- Sharon G, Rosenberg E. Bacterial growth on coral mucus. Curr Microbiol. 2008;56:481–488. doi: 10.1007/s00284-008-9100-5. [DOI] [PubMed] [Google Scholar]
- Shnit-Orland M, Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol. 2009;67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- Sutherland KP, Shaban S, Joyner JL, Porter JW, Lipp EK. Human pathogen shown to cause disease in the threatened elkhorn coral Acropora palmata. PLoS ONE. 2011;6:e23468. doi: 10.1371/journal.pone.0023468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait K, Hutchison Z, Thompson FL, Munn CB. Quorum sensing signal production and inhibition by coral-associated vibrios. Environ Microbiol Reports. 2010;2:145–150. doi: 10.1111/j.1758-2229.2009.00122.x. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Ritchie K. How feasible is the biological control of coral diseases. Trends Ecol Evol. 2009;24:378–385. doi: 10.1016/j.tree.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, et al. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.