Abstract
The goals of this study were to better understand the ecology of oral subgingival communities in health and periodontitis and elucidate the relationship between inflammation and the subgingival microbiome. Accordingly, we used 454-pyrosequencing of 16S rRNA gene libraries and quantitative PCR to characterize the subgingival microbiome of 22 subjects with chronic periodontitis. Each subject was sampled at two sites with similar periodontal destruction but differing in the presence of bleeding, a clinical indicator of increased inflammation. Communities in periodontitis were also compared with those from 10 healthy individuals. In periodontitis, presence of bleeding was not associated with different α-diversity or with a distinct microbiome, however, bleeding sites showed higher total bacterial load. In contrast, communities in health and periodontitis largely differed, with higher diversity and biomass in periodontitis. Shifts in community structure from health to periodontitis resembled ecological succession, with emergence of newly dominant taxa in periodontitis without replacement of primary health-associated species. That is, periodontitis communities had higher proportions of Spirochetes, Synergistetes, Firmicutes and Chloroflexi, among other taxa, while the proportions of Actinobacteria, particularly Actinomyces, were higher in health. Total Actinomyces load, however, remained constant from health to periodontitis. Moreover, an association existed between biomass and community structure in periodontitis, with the proportion of specific taxa correlating with bacterial load. Our study provides a global-scale framework for the ecological events in subgingival communities that underline the development of periodontitis. The association, in periodontitis, between inflammation, community biomass and community structure and their role in disease progression warrant further investigation.
Keywords: bacterial load, community structure, inflammation, periodontitis, subgingival microbiome
Introduction
Caries and periodontitis, the main oral diseases of humans are a consequence of alterations in the ecology of resident microbial communities. The oral cavity represents an easily accessible niche to study such community dynamics and the community-host interactions that result in dysbiosis. Periodontitis affects the supporting structures of teeth and it is characterized by accumulation of bacterial deposits at the gingival margin with formation of an inflammatory infiltrate resulting in destruction of connective tissue attachment to the tooth, alveolar bone resorption and tooth loss (Hernandez et al., 2011). Periodontitis is diagnosed clinically by measuring the level of attachment of periodontal tissues to the tooth root, together with radiographic evidence of bone loss (Armitage, 1995). Apart from determining the depth of periodontal pockets to measure periodontal destruction, clinicians also evaluate whether a pocket bleeds upon probing as this parameter is considered the main clinical indicator of periodontal inflammation. Presence of bleeding on probing (BoP) positively correlates with the size of the inflammatory infiltrate in adjacent gingival tissues (Greenstein et al., 1981), with risk for periodontitis progression (Tanner et al., 2007) and with levels in gingival exudates of proinflammatory mediators such as interleukin-1β, interleukin-8 and destruction markers such as matrix metalloproteinase-8 (Teles et al., 2010).
Decades of investigations have demonstrated that the microbial communities associated with periodontitis differ from those in health (Socransky and Haffajee, 2005). Studies using DNA-DNA hybridization techniques showed that microbial counts for 14 species were generally higher in disease than in health, with species proportions also differing between conditions (Socransky et al., 1991). Culture-based studies and molecular surveys targeting specific species, however, offered limited information on the total diversity present in the subgingival environment. A recent molecular survey performed via pyrosequencing of 16S rRNA gene amplicons provided a much broader picture of overall differences in relative abundances of ∼700 subgingival species-level taxa, confirming the association of Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia with periodontitis, and revealing new species, among them Filifactor alocis, strongly associated with disease (Griffen et al., 2011). This study was conducted in a USA cohort, with inclusion of smokers in the periodontitis group. As smoking has been shown to influence microbiome composition (Shchipkova et al., 2010), further investigation of differences in health and periodontitis is necessary in a homogeneous population. Moreover, although geography does not determine overall structure of oral microbial communities, differences in relative abundances of individual taxa exist among locations around the globe (Nasidze et al., 2009). Further study of the subgingival microbiome in health and periodontitis in populations at geographic locations other than the USA could reveal whether reported shifts in community structure in periodontitis are generalizable at a global scale.
Although it is clear that sites with periodontal destruction harbor a microbiome of distinct structure, the relationship between the subgingival microbiome and clinical inflammation is less understood. Previous studies have reported an association between BoP and certain taxa. For example, Armitage et al. (1982) detected a positive correlation between the percentage of Spirochetes and bleeding tendency, while Socransky et al. (1991) found that the relative proportion of Prevotella intermedia positively correlated with BoP. Furthermore, a comparison of a large number of samples showed the levels of 15 out of 40 bacterial species to be higher in sites with BoP (Socransky and Haffajee, 2005). These findings, however, are confounded by the fact that sites with different degrees of periodontal destruction were compared. This is problematic as it is known that the composition of the subgingival microbiome varies according to probing depths possibly because of dissimilar ecological parameters such as oxygen tension (Loesche et al., 1983). Therefore, associations between inflammation and the subgingival microbiome need to be investigated at ecologically similar niches, that is, sites with similar amounts of periodontal destruction.
This study aimed to clarify the relationship between the subgingival microbiome and clinical inflammation in periodontitis by comparing the microbiome composition and microbial load of sites with a similar amount of periodontal destruction but differing in the presence of BoP. The subgingival microbiome of subjects with periodontitis was also compared with that of healthy subjects to gain a better in-depth understanding of the ecological events leading to periodontitis.
Methods
Sample collection from human subjects
Twenty-two subjects with chronic periodontitis and 10 periodontally healthy individuals participated in this study. Subjects were recruited from the dental clinic of the School of Dentistry at University of Chile under an approved protocol. All subjects were non-smokers. Subjects with periodontitis were sampled at two non-adjacent tooth sites with a similar amount of periodontal destruction (probing depths of 5 mm), but differing in the presence of BoP. Two subgingival plaque samples from sites without BoP were also collected from healthy subjects.
DNA isolation, 16S rRNA gene library preparation and sequencing
DNA was isolated and amplicon libraries of the 16S rRNA gene V1-V2 hypervariable regions were sequenced using 454 titanium chemistry, as previously described (Diaz et al., 2012). Sequences are available at the Short Reads Archive (Accession number SRA051864).
Quantification of total load for specific genera and total bacterial load
Total bacterial load was determined via real-time PCR using universal primers and Fusobacterium nucleatum ATCC 10953 genomic DNA as a standard. The total load of Actinomyces, Streptococcus and Veillonella was determined using genus-specific primers.
Sequencing data processing
Sequencing data were processed using MOTHUR (Schloss et al., 2009) and the pipeline of Schloss et al. (2011), as previously described (Diaz et al., 2012). Template taxonomies included the RDP reference dataset and the Human Oral Microbiome Database, a curated dataset for oral taxa (Dewhirst et al., 2010).
Statistical analysis
Clinical and demographic data were compared via t-tests. Differences in α-diversity were evaluated using paired t-test for bleeding and non-bleeding and unpaired t-test for healthy and periodontitis groups. Significant separation of clusters after principal coordinate analysis was evaluated via analysis of molecular variance (Amova) (Excoffier et al., 1992). Differences in relative abundances of individual operational taxonomic units (OTUs), genera and phyla were determined with LEfSe (Segata et al., 2011), using an alpha value of 0.01 for the Kruskal−Wallis test and a threshold of 2.0 for logarithmic linear discriminant analysis scores. Differences in taxon prevalence between bleeding and non-bleeding sites were tested via McNemar and between health and periodontitis via χ2. For comparison of taxa between health and periodontitis, samples originating from the same subject were averaged and the subject was used as the unit of analysis. Real-time data were compared between bleeding and non-bleeding sites using Wilcoxon Signed Rank test and between healthy and periodontitis groups with the Mann−Whitney test. Correlations between relative abundance of taxa and total bacterial load were calculated by Pearson correlation coefficients after data transformation using the inverse hyperbolic sine method (Burbidge et al., 1988). The significance threshold for statistical tests when comparing individual taxa (McNemar, χ2 and Pearson) was adjusted using the Benjamini−Hochberg false discovery rate method.
Detailed methods appear as Supplementary material.
Results
Presence of periodontal destruction, but not increased inflammation (BoP), is associated with higher α-diversity of subgingival bacterial communities
The demographic and clinical characteristics of subjects are depicted in Table 1. We used 454-pyrosequencing to analyze 44 samples from subjects with periodontitis and 17 samples from healthy individuals. Three samples from the healthy group did not yield a PCR product. After processing, our dataset included 179 239 sequences with an average of 3110±2279 (range 796–11 224) sequences per sample and an average sequence length of 263 nt. We found a total of 750 OTUs across all samples with a range of 30–194 OTUs present at individual sites. Richness coverage in individual samples according to CatchAll ranged from 32.2%−79.8%.
Table 1. Demographic and clinical characteristics of studied subjects.
| Characteristics | Periodontitis (n=22) | Health (n=10) |
|---|---|---|
| Women (%) | 54.5 | 80.0 |
| Age (years±s.d.) | 42.5±3.3 | 34.1±5.5 |
| PD (mean±s.d.) | 3.0±0.7 | 1.5±0.1* |
| CAL (mean±s.d.) | 3.9±0.9 | 1.4±0.2§ |
| % sites with BoP (mean±s.d.) | 44.5±18.6 | 4.4±4.3* |
| % surfaces with plaque (mean±s.d.) | 82.4±9.6 | 16.1±13.4§ |
| Periodontal diagnosis | Chronic severe periodontitisa | Periodontal health |
Abbreviations: BoP, bleeding on probing; CAL, clinical attachment levels; PD, probing depths. CAL and PD were measured in mm and represent the mean for all sites in the oral cavity of studied subjects. *P<0.001, §P<0.0001.
Based on criteria defined by Page and Eke (2007).
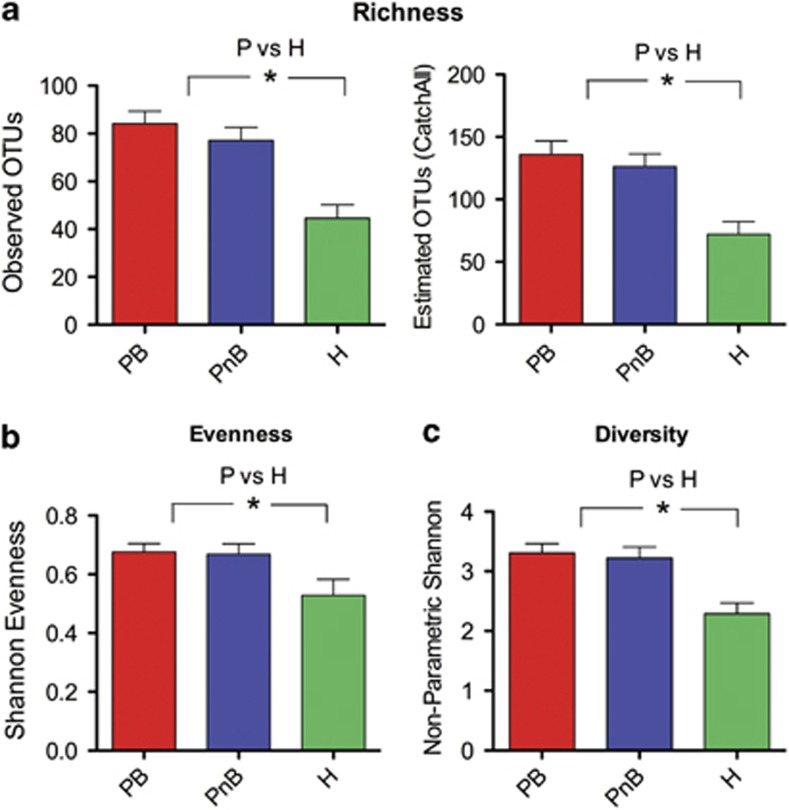
First, we investigated whether periodontal health status or increased inflammation at sites with periodontitis were related to diversity metrics. A comparison of subgingival communities from bleeding and non-bleeding sites revealed no differences in α-diversity, while communities from subjects with periodontitis showed higher α-diversity than those in health (Figure 1).
Figure 1.
Periodontal destruction, but not increased periodontal inflammation, is associated with greater α-diversity. (a) shows community richness (observed and estimated) for subjects with periodontitis (P) in sites with bleeding (PB) and those without bleeding (PnB) and for communities from healthy subjects (H). In (b) evenness of communities was compared, while (c) shows comparisons of diversity. *indicates a P<0.05. To compare P and H, communities from bleeding and non-bleeding sites were combined. α-diversity metrics were calculated after subsampling to obtain equal number of sequences per library.
Presence of periodontal destruction, but not increased inflammation (BoP), is associated with a shift in the composition of subgingival communities
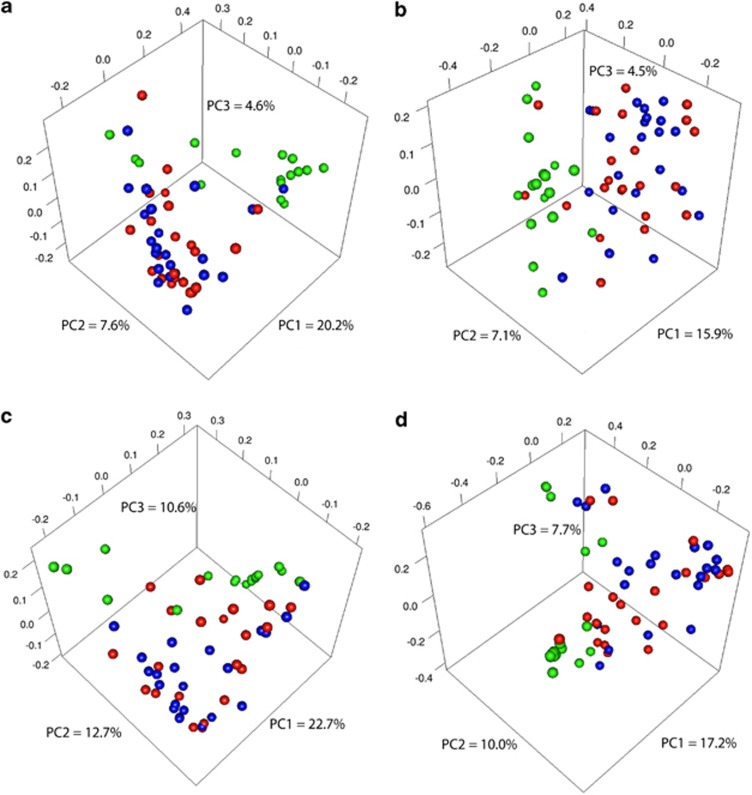
Principal coordinate analysis plots depicting distances among communities based on qualitative community metrics showed that increased inflammation at periodontitis sites was not a strong driver of community composition (Figure 2a and b). However, healthy communities tended to cluster apart from periodontitis communities (Amova P<0.001). Community distance metrics that take into account structure yielded similar results (Amova P<0.001) (Figure 2c and d).
Figure 2.
Communities from periodontitis cluster apart from those in health, while bleeding does not drive community clustering. Graphs are principal coordinate analysis plots based on community membership metrics [(a) is based on unweighted UNIFRAC and (b) on the Jaccard Index] or community structure metrics [(c) is based on weighted UNIFRAC, while (d) is based on the θYC distance]. Communities from healthy subjects appear in green. For the periodontitis group, communities from sites with BoP appear in red, while communities from sites without BoP appear in blue.
Next, we compared relative abundances and prevalence of individual taxa between bleeding and non-bleeding sites of subjects with periodontitis to investigate whether increased inflammation is associated with specific changes in the microbiome. OTU-level analysis showed no statistically significant differences in relative abundances and prevalence of OTUs between bleeding and non-bleeding sites. Similar analyses performed with sequence data binned at genus or phylum levels also revealed no differences. Taken together, these results confirm that increased inflammation at sites with periodontitis is not associated with a distinct microbiome.
We then investigated, which taxa were responsible for overall community differences between health and disease. Analysis of relative abundances showed 60 significantly different OTUs, 14 of which were increased in health and 46 in periodontitis (Supplementary Figure S1). Supplementary Figure S2 shows the 40 most abundant OTUs in periodontitis (A) and health (B). Among the most abundant OTUs in periodontitis and over-represented with respect to health were bacterial species classically associated with this disease such as Treponema denticola, Tannerella forsythia and Porphyromonas gingivalis (Socransky et al., 1998). However, a broad range of taxa beyond these three species are associated with periodontitis.
Differences in prevalence of individual OTUs between health and periodontitis were also explored. We found 102 OTUs with significantly different prevalence, 41 of which were more frequently detected in health and 61 in periodontitis (Supplementary Figure S3). Supplementary Figure S4 shows the most prevalent OTUs under each condition.
Analyses at the genus level showed four genera with increased relative abundance in health and 25 with higher relative abundance in periodontitis (Supplementary Figure S5). These results largely agreed with OTU analysis, showing Treponema as the genus over-represented in periodontitis with the highest linear discriminant analysis score, while Rothia showed the highest effect size from the genera over-represented in health, closely followed by Actinomyces. Supplementary Figure S6 depicts the most abundant genera in periodontitis and health, illustrating the change in Treponema from a mean relative abundance of ∼1% in health to ∼20% of sequence reads in periodontitis, becoming the most abundant genus. Rothia and Actinomyces displayed an opposite behavior dominating communities in health and dramatically decreasing their abundance in periodontitis.
Prevalence analysis at the genus level showed 36 genera as significant, with 32 genera more frequently detected in periodontitis and 4 in health (Supplementary Figure S7). Most genera, however, were detected under both conditions, with the exception of Eubacterium (11)(G-5), Peptostreptococcaceae(13)(G-1), Bacteroidetes (G-6), Bacteroidetes (G-5) and Sneathia, which were only detected in periodontitis. Supplementary Figure S8 shows the most prevalent genera in health and disease, indicating that the shift in communities is mainly due to an increase in frequency of detection of genera in periodontitis.
Supplementary Figure S9 shows phylum level results. Periodontitis communities were characterized by increased relative abundance of Firmicutes, Spirochetes, Synergistetes and Chloroflexi, while healthy communities were characterized by increased relative abundance of Actinobacteria. Spirochetes, Synergistetes and Chloroflexi were also more prevalent in periodontitis.
Quantification of total load for specific genera in health and periodontitis
As 454-pyrosequencing only reveals relative proportions of species, we also used quantitative real-time PCR to investigate total load for selected, highly prevalent genera to better understand changes in the subgingival microbiome from health to disease. Supplementary Figure S10 shows that although the relative abundance of Actinomyces decreased from health to disease (Supplementary Figures S5 and S6), its total load did not change between the two states. This finding suggests that, changes in relative abundance of this genus in periodontitis are due to an increase in biomass of other community members rather than a loss of Actinomyces total biomass. In contrast, genera with equal relative abundance in health and disease, such as Streptococcus and Veillonella (Supplementary Figures S5 and S6), showed an increase in total abundance from health to periodontitis (Supplementary Figure S10), which confirms that although these genera are not specifically associated with periodontitis, their load increases as the disease develops.
Defining the core subgingival microbiome in health and periodontitis
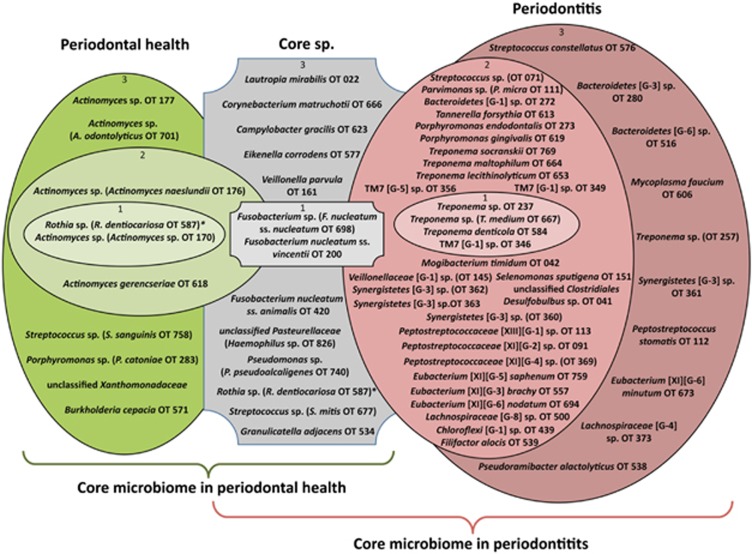
Our next goal was to develop a model framework for changes in the subgingival microbiome from health to disease. Despite high inter-individual variability, we found some OTUs present in most subjects, representing the core subgingival microbiome, that is, taxa best adapted to the living conditions of the subgingival environment. Some of these highly prevalent OTUs were preferentially associated with health or periodontitis, while some others were equally prevalent and abundant under both conditions (Figure 3). The core subgingival OTUs were defined as those present in a majority of subjects, with equal prevalence and relative abundance in health or disease (Figure 3, gray shapes). Two OTUs closely related to F. nucleatum were the most abundant core members, while 11 other OTUs were less abundant but present in most subjects regardless of the health status of the site. We then identified those OTUs present in the majority of subjects in health and appearing at increased prevalence and relative abundance compared with disease (Figure 3, green shapes). The health-associated OTUs included five Actinomyces spp., a Streptococcus spp. closely related to Streptococcus sanguinis, two Proteobacteria and a Porphyromonas spp. closely related to Porphyromonas catoniae. Thus, the core subgingival microbiome in health are the health-associated OTUs (green shapes) and the core subgingival OTUs (gray shapes). It is worth noting that an OTU closely related to Rothia dentocariosa was found to numerically dominate the health-associated communities showing increased abundance in health and therefore it is depicted in the inner green shape in Figure 3. This OTU, however, could also be considered a subgingival core species as its prevalence was similar in health and disease.
Figure 3.
The core subgingival microbiome in health and periodontitis. OTUs that are part of the core microbiome were first defined as those present in at least 50% of subjects in both the healthy and periodontitis groups (gray), in at least 50% of healthy subjects (green) or in at least 50% of subjects with periodontitis (red). Prevalence and relative abundance data were used for further filtering. OTUs in gray represent the core subgingval OTUs, present at equal prevalence and relative abundance in health and periodontitis. OTUs in green represent the core health-associated species, appearing at increased prevalence and relative abundance in health compared to disease. OTUs in red represent the core periodontitis-associated species, present at increased prevalence and relative abundance in periodontitis compared to health. Inner circles labeled with 1, contain highly prevalent and highly abundant OTUs, that is, present in at least 2/3 of samples from each group (core, health or periodontitis) and numerically dominant with a mean relative abundance of ⩾2% of total sequences. Middle circles labeled with 2 contain OTUs highly prevalent but present in low abundance, that is present in at least 2/3 of samples from the specific group but with a mean relative abundance of <2% of total sequences. Outer circles labeled with 3 contain OTUs moderately prevalent and present in low abundance, that is OTUs present in 1/2 to 2/3 of samples from each group and with a mean relative abundance of <2% of total sequences.
Red shapes in Figure 3 show OTUs that appeared in a majority of samples from periodontitis subjects and had higher prevalence and abundance than in health. Among these, three Treponema spp. and an OTU from the phylum TM7 numerically dominated the communities. However, a phylogenetically diverse range of taxa were also associated with periodontitis and present in most subjects, including other Treponema spp., TM7 spp., several Bacteroidetes, Synergistetes, one Chloroflexi and a great number of OTUs from the Clostridia class of the phylum Firmicutes. The core subgingival microbiome in periodontitis are thus the OTUs in red shapes plus the core OTUs appearing in gray shapes in Figure 3.
Core species could be considered those capable of a cosmopolitan lifestyle in the subgingival environment. However, we also observed endemic OTUs appearing in only a few samples, some of which numerically dominated a particular community (at least 5% of sequence reads). These endemic but locally abundant OTUs are depicted in Supplementary Figure S11. Although these OTUs were not widespread (present in less than 25% of samples), particular assembly rules may have allowed them to thrive locally.
Increased inflammation at sites with periodontal destruction is associated with higher bacterial load
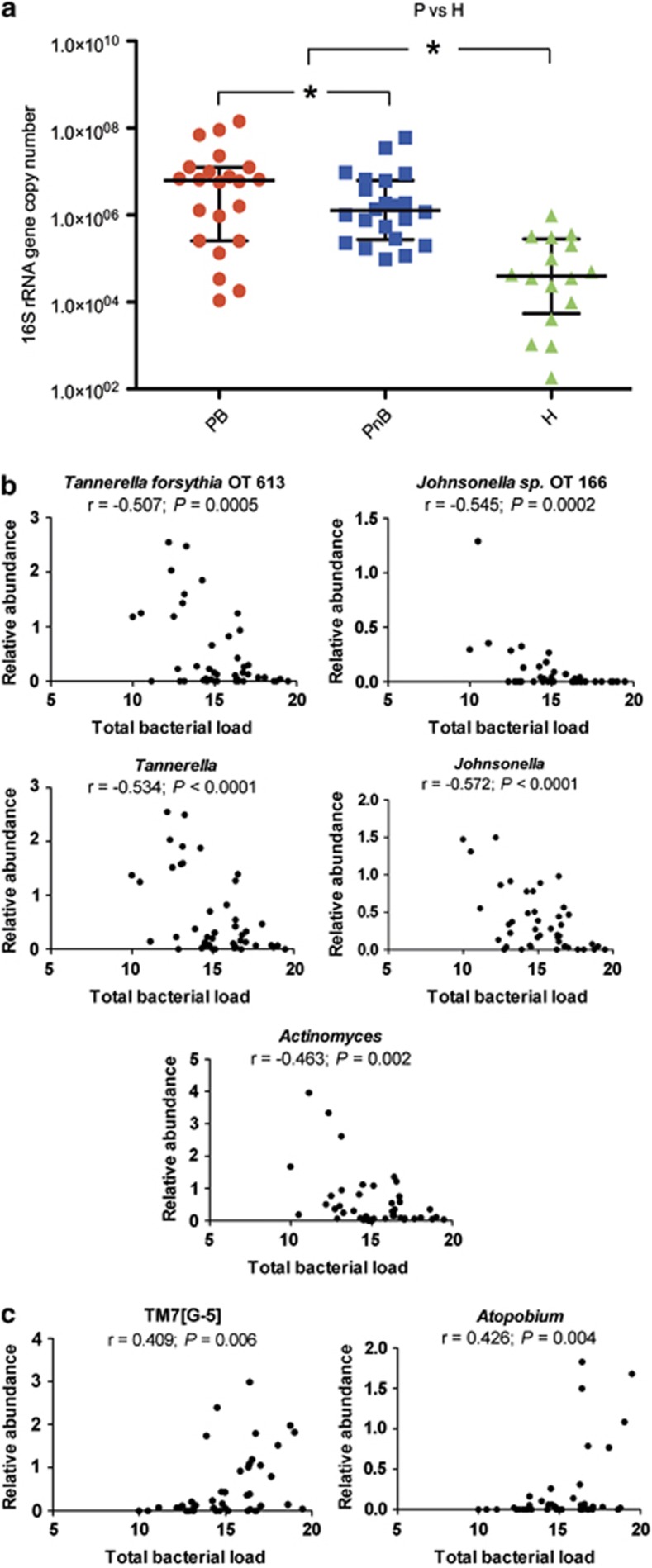
Next, the relationship between increased inflammation in periodontitis, as evidenced by BoP, and subgingival bacterial load was investigated. We found higher number of 16S rRNA gene copies in bleeding than in non-bleeding sites (Figure 4a). Additionally, as expected from previous reports (Moore and Moore, 1994; Socransky and Haffajee, 2005), communities in periodontitis had higher bacterial load than those in health.
Figure 4.
Bacterial load is higher in periodontitis than in health, is also higher in bleeding than in non-bleeding sites and correlates with the relative abundance of specific taxa within periodontitis samples. (a) shows bacterial load measurements in periodontitis (P) in sites with bleeding (PB) or without bleeding (PnB) and in health (H). *indicates a P<0.05. To compare P and H, communities from bleeding and non-bleeding sites were combined. (b) shows taxa from periodontitis communities that showed a significant negative correlation between their relative abundance and bacterial load, while (c) shows taxa from periodontitis communities with a significant positive correlation between their relative abundance and bacterial load. Relative abundance (%) and bacterial load (number of 16S rRNA copies) data were transformed (see methods).
Possible correlations between bacterial load and relative abundance of individual taxa within periodontitis samples were then evaluated to understand differences in structure of communities with high or low biomass. This analysis showed that the relative abundance of Actinomyces, a health-associated genus, and that of the periodontitis-associated genera Tannerella and Johnsonella negatively correlated with bacterial load (Figure 4b). These findings were also consistent at an OTU-level, where we found a negative correlation between bacterial load and the relative abundance of the known periodontal pathogen Tannerella forsythia and that of an uncultured Jonhsonella spp. (Figure 4b), both previously shown to be periodontitis-associated (Supplementary Figure S1). Interestingly, other genera associated with periodontitis such as Filifactor, Porphyromonas, Synergistetes (G-3) and Peptostreptococcaceae (11)(G-4) also showed a trend for a negative correlation with total bacterial load in periodontitis samples (data not shown). Figure 4c shows taxa that positively correlated with bacterial load, which included the periodontitis-associated genus TM7(G-5) and Atopobium, a genus that did not differ between health and disease. These data suggest that the productivity of subgingival communities is related to community structure. Moreover, high levels of some periodontitis-associated microorganisms are not necessarily an indication of greater community biomass, which suggests that their presence in low abundance may actually ensure higher community productivity than when they are dominant community members.
Discussion
Periodontitis is caused by ecological disturbances in subgingival communities (Marsh et al., 2011; Diaz, 2012). In this study, we confirmed previous reports finding higher total bacterial load and richness in periodontitis (Socransky et al., 1991; Griffen et al., 2011). Moreover, periodontitis is also associated with increased evenness and thus contrasts with other mucosal diseases of the gastrointestinal tract, such as inflammatory bowel disease, in which dysbiosis is associated with a decrease in bacterial diversity (Walker et al., 2011).
Our findings suggest that periodontitis communities result mostly from ecological shifts in community structure, rather than shifts in membership. Despite differences in frequency of detection of taxa between health and periodontitis, most disease-associated taxa were also present in health, albeit detected in a small number of samples and in low proportion. Conversely, most health-associated taxa were commonly seen in periodontitis (Supplementary Figures S3, S4, S7 and S8). As detection of a taxon is dependent on its abundance and sampling effort, it is likely that deeper sampling would have increased the membership resemblance of health and periodontitis communities. However, a great number of sequences (∼100-fold more than those obtained) would have been necessary to cover all richness, particularly in the unevenly distributed health-associated communities (Diaz et al., 2012). It is then likely that the ecological shifts from health to periodontitis are a consequence of the rise of low-abundance community members as dominant species, rather than a result of the disappearance of health-associated species. Measurements of total load for specific genera also support this concept. For example, the load of health-associated Actinomyces did not change from health to disease, while that of Streptococcus and Veillonella, neither health- nor disease-associated, increased in unison with overall community biomass. These findings are consistent with the concept of microbial succession events as drivers of the maturation of communities with pathogenic potential. Ecological succession, however, is not experienced as replacement of primary species, but rather as the emergence of newly dominant community members as biomass accumulates.
This study revealed a broad range of taxa present in health and disease providing a global-scale view of subgingival microbial communities. Some of our results agree with Griffen et al. (2011), the only published study to date comparing the subgingival microbiome in health and disease using 454-pyrosequencing. For example, both studies found similar number of species/OTUs in the subgingival environment, despite different methods used for classification and clustering of sequences. Moreover, in both studies, Fusobacterium and Treponema were among the most abundant subgingival genera and the phyla Spirochetes and Synergistetes were increased in periodontitis. Griffen et al. (2011), however, found Bacteroidetes at increased abundance in periodontitis, reporting high proportions of periodontitis-associated Prevotella, while we found lower Prevotella abundance, unchanged between groups. This discrepancy could be explained by geographic variability (Chile vs USA) or by differences in the depths of the pockets sampled, as the proportion of certain Prevotella spp. has been shown to increase in deeper pockets (with probing depths >6 mm), which were not part of our sampling strategy (Socransky et al., 1991). We also evaluated if variability in Prevotella detection could be due to primer bias as our reverse primer differed from that used by Griffen et al. (2011). This is unlikely, however, as Prevotella denticola, the most abundant periodontitis-associated Prevotella in Griffen et al. (2011), does not have any mismatches with our reverse primer. Discrepancies in relative abundance of other phyla between the two studies were also observed. For instance, contrary to Griffen et al. (2011), we observed Firmicutes as increased in periodontitis, finding Clostridia increased in periodontitis, but the class Bacilli unchanged between groups, with the genus Streptococcus, the most abundant Bacilli, equally represented in health and disease. Interestingly, in our study, some Streptococcus OTUs such as Streptococcus sanguinis were increased in health, sequences related to Streptococcus mitis remained unchanged, whereas Streptococcus constellatus and an uncultured Streptococcus sp. (OT 071) were associated with disease. Physiologic differences that could explain such heterogeneity in niches occupied by streptococci are not currently known. Major differences between Griffen et al. (2011) and our study were also observed for health-associated phyla. Although our study shows Actinobacteria strongly associated with health, Griffen et al. (2011) reports Proteobacteria as the only health-associated phylum. These discrepancies could be explained by increased detection in our study of Actinomyces and Rothia, possibly due to the use of curettes as a sampling method, while Griffen et al. (2011) utilized paper points, which only allow passive translocation of plaque material and fluid into the sampling device. The paper point sampling method is thus likely to represent only the outer biofilm microorganisms, undersampling initial colonizers present in the inner biofilm mass attached to the root surface (Zijnge et al., 2010). Moreover, our study did not find Proteobacteria to differ between health and disease and we did not obtain any sequences from Proteobacteria such as Moraxella, found in great abundance in health by Griffen et al. (2011). Variability in detection of Moraxella could be explained by population differences, as a recent study that sequenced the V3-V5 region of the 16S rRNA gene in a large North American cohort also found it to be abundant in subgingival plaque of healthy subjects (The Human Microbiome Project (HMP) Consortium, 2012). In conclusion, a distinct microbiome was shown to exist in periodontitis by both Griffen et al. (2011) and our study. Future studies, however should reconcile differences by evaluating the subgingival microbiome at locations around the globe using comparable sampling and sequencing methods.
This study also defined the core microbiome associated with periodontal health and disease. Despite the small size of the control group, this initial characterization provides a starting point for future comparisons and will facilitate investigations into the function of subgingival communities, allowing researchers to focus on the microorganisms most commonly found in human hosts. Furthermore, we highlight abundant OTUs that do not change from health to disease (core sp. in Figure 3). These OTUs may have important roles in subgingival communities providing structural support, as suggested for Fusobacterium nucleatum (Kolenbrander et al., 2006), serving as metabolic cornestones or perhaps directing health to disease transitions. Among the species associated with periodontitis, the dominance of Spirochetes requires further attention. Spirochetes may benefit from interactions with other community members and thus autogenic succession may be responsible for their dominance. Indeed, Treponema denticola benefits from succinate produced by Porphyromonas gingivalis (Grenier, 1992), however the nutritional requirements of other Treponema spp. remain for the most part uncharacterized. Our study also highlights the urgency to develop appropriate cultivation technologies for core disease-associated taxa from the phylum TM7, which are understudied due to unrecognized metabolic requirements.
This study also demonstrates that in periodontitis, clinical evidence of increased inflammation is not associated with a distinct microbiome, but rather corresponds with higher community biomass. Thus, our data do not support previous studies indicating that specific microorganisms are associated with bleeding in established periodontal lesions (Armitage et al., 1982; Socransky et al., 1991). Increased inflammation associated with disease progression may then be the result of disruption of host homeostasis due to an overall greater bacterial challenge. We acknowledge, however, that the cross-sectional nature of this study does not allow the conclusion that increased inflammation is the result of higher bacterial load. Indeed, the relationship between these two factors may be dynamic and reciprocal. Higher microbial load may represent a greater challenge to the host, which responds with increased inflammation, which in turn results in increased biomass via greater supply of host-derived nutrients. Future investigations using in vivo and in vitro models should therefore evaluate the role of inflammation and specific host-derived molecules as modulators of community biomass and conversely, the role of increased biomass, rather than levels of specific microorganisms, in the pathophysiology of periodontitis progression. Moreover, when interpreting these results it is important to take into account that although sampling via curettes is the best available method to obtain subgingival plaque, it is not completely standardized. The large differences in biomass observed between health and periodontitis are consistent with previous findings (Moore and Moore, 1994; Socransky and Haffajee, 2005), however, confirmation of the smaller differences observed in bacterial load between bleeding and non-bleeding sites in a larger cohort could further support our results.
Despite the above-mentioned limitations, we found interesting that in periodontitis-associated communities, the relative abundance of certain taxa correlated with community biomass. Although it appeared logical that Actinomyces, a prominent genus in health, negatively correlated with microbial load in periodontitis, it was unexpected to find lower proportions of periodontitis-associated taxa when microbial load was high. Recent studies in mice demonstrated that, low-levels of colonization by periodontitis-associated microorganisms such as Porphyromonas gingivalis are sufficient to dysregulate innate immunity pathways, facilitate an increase in overall community biomass and trigger periodontal destruction (Hajishengallis et al., 2011). Indeed, in the current study, Porphyromonas was among the genera showing a trend negative correlation with bacterial load in periodontitis. Therefore, it seems possible that certain periodontitis-associated microorganisms may contribute more effectively to an overall increase in community biomass by existing in low abundance, as the growth capacity of the community may be the result of a delicate ecological balance between mutualistic and antagonistic interactions among its members.
In conclusion, we characterized the subgingival microbiome in health and periodontitis providing a comprehensive view of the ecological shifts associated with this disease. Our study suggests that periodontitis communities are the result of microbial succession characterized by emergence of newly abundant taxa concurrent with an increase in bacterial biomass. Furthermore, we demonstrate that in periodontitis, a distinct microbiome signature does not exist for sites with BoP, a sign of increased inflammation. Despite limitations in obtaining fully standardized subgingival samples, we observed that communities from bleeding sites exhibited higher bacterial load than communities from non-bleeding sites and we found a possible relationship between community productivity and microbiome structure, as bacterial load in periodontitis communities correlated with the relative proportion of certain taxa. Our results warrant further investigations regarding the role of increased overall community biomass as the possible trigger for increased periodontal inflammation and the relationship between biomass, community productivity and the proportion of taxa within the highly diverse bacterial communities associated with periodontitis.
Acknowledgments
This work was partly funded by FONDECYT Project 1090046.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Armitage GC, Dickinson WR, Jenderseck RS, Levine SM, Chambers DW. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J Periodontol. 1982;53:550–556. doi: 10.1902/jop.1982.53.9.550. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Clinical evaluation of periodontal diseases. Periodontology 2000. 1995;7:39–53. doi: 10.1111/j.1600-0757.1995.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Burbidge JB, Lonnie M, Robb AL. Alternative transformations to handle extreme values of the dependent variable. J Amer Stat Assoc. 1988;83:123–127. [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI. Microbial diversity and interactions in subgingival biofilm communities. Front Oral Biol. 2012;15:17–40. doi: 10.1159/000329669. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol. 2012;27:182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein G, Caton J, Polson AM. Histologic characteristics associated with bleeding after probing and visual signs of inflammation. J Periodontol. 1981;52:420–425. doi: 10.1902/jop.1981.52.8.420. [DOI] [PubMed] [Google Scholar]
- Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. 1992;60:5298–5301. doi: 10.1128/iai.60.12.5298-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez M, Dutzan N, Garcia-Sesnich J, Abusleme L, Dezerega A, Silva N, et al. Host-pathogen interactions in progressive chronic periodontitis. J Dent Res. 2011;90:1164–1170. doi: 10.1177/0022034511401405. [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project (HMP) Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontology 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- Loesche WJ, Gusberti F, Mettraux G, Higgins T, Syed S. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infect Immun. 1983;42:659–667. doi: 10.1128/iai.42.2.659-667.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontology 2000. 2011;55:16–35. doi: 10.1111/j.1600-0757.2009.00339.x. [DOI] [PubMed] [Google Scholar]
- Moore WEC, Moore LV.1994Periodontal microbiota in different clinical conditions Periodontology 2000 566–77.9673163 [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC, Eke PI. Case definitions for use in population-based surveillance of periodontitis. J Periodontol. 2007;78:1387–1399. doi: 10.1902/jop.2007.060264. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchipkova AY, Nagaraja HN, Kumar PS. Subgingival microbial profiles of smokers with periodontitis. J Dent Res. 2010;89:1247–1253. doi: 10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Kent R, Kanasi E, Lu SC, Paster BJ, Sonis ST, et al. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 2007;34:917–930. doi: 10.1111/j.1600-051X.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Teles R, Sakellari D, Teles F, Konstantinidis A, Kent R, Socransky S, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81:89–98. doi: 10.1902/jop.2009.090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.