Abstract
A wide array of microorganisms survive and thrive in extreme environments. However, we know little about the patterns of, and controls over, their large-scale ecological distribution. To this end, we have applied a bar-coded 16S rRNA pyrosequencing technology to explore the phylogenetic differentiation among 59 microbial communities from physically and geochemically diverse acid mine drainage (AMD) sites across Southeast China, revealing for the first time environmental variation as the major factor explaining community differences in these harsh environments. Our data showed that overall microbial diversity estimates, including phylogenetic diversity, phylotype richness and pairwise UniFrac distance, were largely correlated with pH conditions. Furthermore, multivariate regression tree analysis also identified solution pH as a strong predictor of relative lineage abundance. Betaproteobacteria, mostly affiliated with the ‘Ferrovum' genus, were explicitly predominant in assemblages under moderate pH conditions, whereas Alphaproteobacteria, Euryarchaeota, Gammaproteobacteria and Nitrospira exhibited a strong adaptation to more acidic environments. Strikingly, such pH-dependent patterns could also be observed in a subsequent comprehensive analysis of the environmental distribution of acidophilic microorganisms based on 16S rRNA gene sequences previously retrieved from globally distributed AMD and associated environments, regardless of the long-distance isolation and the distinct substrate types. Collectively, our results suggest that microbial diversity patterns are better predicted by contemporary environmental variation rather than geographical distance in extreme AMD systems.
Keywords: acid mine drainage, biogeography, contemporary environmental variation, geographical distance, microbial diversity, pyrosequencing
Introduction
Microbial biogeography is increasingly becoming an exciting topic in microbial ecology and a growing number of researches are addressing the spatial scaling and distribution pattern of microorganisms in the environment (Martiny et al., 2006; Green et al., 2008). Despite their tremendous potential for global dispersal, there is accumulating evidence that free-living microorganisms exhibit nonrandom distribution patterns across diverse habitats at various spatial scales. Niche-based processes have been implied as the primary drivers for the widely observed environment-dependent diversity patterns and environmental variables such as salinity (Lozupone and Knight, 2007; Auguet et al., 2010), pH (Fierer and Jackson, 2006; Lauber et al., 2009; Rousk et al., 2010; Griffiths et al., 2011) and C:N ratio (Bates et al., 2011) identified as the major determinants of microbial community composition. However, there is also evidence that spatial distance, which may be seen as a proxy variable that represents differential community dynamics related to the past historical events and disturbances (Ramette and Tiedje, 2007), have a role in structuring natural microbial assemblages (Cho and Tiedje, 2000; McAllister et al., 2011; Martiny et al., 2011). These studies of biogeography have provided initial insights into the processes that generate diversity patterns and improved our understanding of why organisms live where they do and how they will respond to environmental change. However, systematically exploring the microbial geographical patterns by considering both contemporary environmental variations and spatial distance simultaneously is limited (Ramette and Tiedje, 2007; Ge et al., 2008), and the relative importance of these factors in shaping microbial communities in natural environments remains largely unsolved.
The Earth's extreme environments harbor a wide array of extraordinary microorganisms that are, in some ways or others, similar to the ancient life forms (Amaral-Zettler et al., 2011). Analyzing the dynamic changes of these microbial communities coupled with physical and geochemical factors will reveal how microbes can adapt and tolerate to different kinds of environmental extremes and increase our understanding of microbial ecology and evolution. Acid mine drainage (AMD) is a widespread environmental problem primarily resulting from the oxidative dissolution of pyrite (FeS2) and other sulfide minerals exposed to oxygen and water during metal ore mining (Nordstrom and Alpers, 1999). Although typically low in overall microbial diversity, these unique environments harbor metabolically active, acidophilic microorganisms that are well adapted to the multiple environmental stresses encountered and are mainly responsible for the generation of these hot, sulfuric acid- and toxic metals-rich solutions (Baker and Banfield, 2003). While Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans (the two iron-oxidizing species most commonly isolated from acidic drainage waters) are widely implicated to be the microorganisms that control the rate of AMD generation, more recent molecular-based investigations have revealed that other less known organisms (for example, Ferroplasma spp. in the Archaea and Leptospirillum group III within the Nitrospira) are dominant in certain specific mine environments and they probably have important roles in the pyrite dissolution in situ (Bond et al., 2000; Tan et al., 2007; Huang et al., 2011). Because of their biological and geochemical simplicity, AMD environments have the potential as model systems for quantitative analysis of microbial ecology and evolution and community function (Baker and Banfield, 2003; Denef et al., 2010). The first 16S rRNA gene-based microbial diversity surveys of AMD systems date back to the mid-1990s (Goebel and Stackebrandt, 1994). Further molecular diversity inventories of AMD microbes have been conducted in a number of acidic environments in diverse geographical locations, including Iron Mountain in California, USA (Bond et al., 2000; Druschel et al., 2004) and the Rio Tinto (RT) in southwestern Spain (Gonzalez-Toril et al., 2003; Garcia-Moyano et al., 2007). Although expanding our knowledge of the biodiversity of extremely acidic systems, these studies have typically examined a limited number of samples from a single mining environment, and the sequencing depth provided by a standard clone library analysis is relatively limited. Consequently, a global understanding of the pattern of AMD microbial diversity has not been available, and it is not clear how communities are shaped by the prevailing geochemical factors in these extreme environments and whether the major environmental determinants of microbial community composition differ from those working in ‘normal' environments. The advent of high-throughput pyrosequencing technology now affords new opportunities to address these knowledge gaps by comprehensively characterizing microbial communities in large numbers of ecological samples to examine broad trends of microbial distribution in AMD environments.
Here, we applied a massively parallel tag pyrosequencing of the V4 region of the 16S rRNA gene to examine in-depth microbial communities from diverse AMD sites across Southeast China to gain insight into the ecological characteristics of these extraordinary microorganisms. We wanted to determine whether AMD microbes exhibit specific biogeographic patterns and which abiotic factors (contemporary environmental factors versus spatial distance) are more important in relating their diversity and composition across a broad range of physical and geochemical gradients. A meta-analysis based on previous molecular inventory studies of AMD environments from diverse geographical locations was subsequently conducted to determine if the patterns observed in our pyrosequence data set are applicable at broader (global) scales.
Materials and methods
Sample collection, physicochemical analyses and DNA extraction
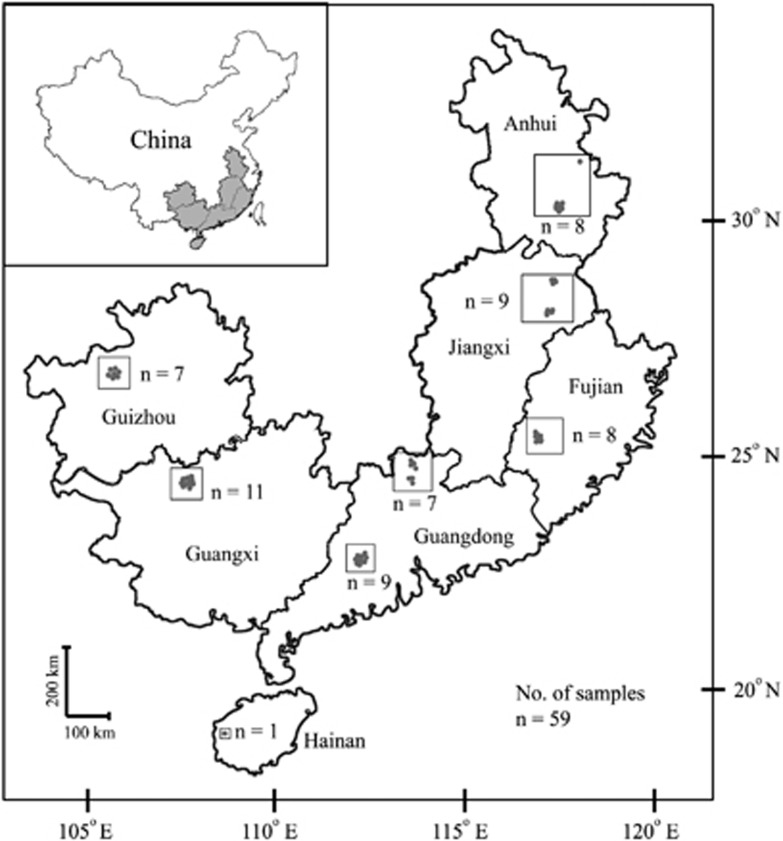
A total of 59 AMD samples were collected from 14 mining areas (12 active and two abandoned) across Southeast China (19.24°–31.64°N, 105.71°–118.62oE; Figure 1 and Supplementary Table 1) with different mineralogy (for example, copper, lead–zinc, pyrite and polymetallic) and representing a broad variety of environmental conditions. Site locations were recorded by global positioning system and the geographical distances between sampling sites ranged from about 10 m to over 1600 km. Water samples were taken from acidic streams, runoff ponds and AMD collection ponds (for storage before treatment) using sterile serum bottles and immediately kept on ice for transport to the laboratory. For DNA extraction, a 500 ml aliquot of each sample was coarse filtered through a 3 μm fiber filter (Type A/D Glass; Pall Corporation, Port Washington, NY, USA) and then filtered through a 0.22 μm polyethersulfone membrane filter (Supor-200; Pall) using a peristaltic pump. The cell pellets on the polyethersulfone membranes were stored at −40 °C before nucleic acid extraction, and the filtrates were temporarily stored at 4 °C for the chemical analyses within 10 days. Temperature, solution pH, dissolved oxygen (DO) and electrical conductivity (EC) were measured on-site by use of specific electrodes. Ferric and ferrous irons were measured by ultraviolet colorimetric assay with 1,10-phenanthroline at 530 nm (Hill et al., 1978). Total organic carbon (TOC) was measured by high-temperature catalytic oxidation and infrared detection with a TOC analyzer (TOC-VCSH; Shimadzu, Kyoto, Japan) and sulfate determined by a BaSO4-based turbidimetric method (Chesnin and Yien, 1951). The element analysis was performed by inductively coupled optical emission spectrometry (Optima 2100DV; Perkin-Elmer, Waltham, MA, USA) after the filtrates were digested at 180 °C with conc. HNO3 and HCl (1:3, v v−1). Genomic DNA was extracted from the filters by following the protocol described by Frias-Lopez et al. (2008). As an additional step to facilitate cell lysis, the membranes were placed into the bead tubes and homogenized by shaking with a Fast Prep-24 Homogenization System equipped with QuickPrep Adapter (MP Biomedicals, Seven Hills, NSW, Australia) for 40 s at maximum speed.
Figure 1.
Location of sampling sites of AMD across Southeast China. Detailed site characteristics are listed in Supplementary Tables 1 and 2.
Amplification and bar-coded pyrosequencing of bacterial and archaeal 16S rRNA genes
PCR amplification, purification, pooling and pyrosequencing of a region of the 16S rRNA gene were performed following the procedure described by Fierer et al. (2008). We used the primer set F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACVSGGGTATCTAAT-3′) that was designed to amplify the V4 hypervariable region and demonstrated in silico to be universal for nearly all bacterial and archaeal taxa (Bates et al., 2011). This short targeted gene region (∼300 bp) can provide sufficient resolution for the accurate taxonomic classification of microbial sequences (Liu et al., 2007). An 8-bp error-correcting tag (Hamady et al., 2008) was added to the forward primer. Samples were amplified in triplicate following the thermal cycling described previously (Fierer et al., 2008). Replicate PCR reactions for each sample were pooled and purified using a QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA, USA). A single composite sample for pyrosequencing was prepared by combining approximately equimolar amounts of PCR products from each sample. Sequencing was carried out on a 454 GS FLX Titanium pyrosequencer (Roche 454 Life Sciences, Branford, CT, USA) at Macrogen (Seoul, Korea).
Processing of pyrosequencing data
Raw data generated from the 454-pyrosequencing run were processed and analyzed following the pipelines of Mothur (Schloss et al., 2009) and QIIME (Caporaso et al., 2010). Pyrosequences were denoized using the commands of ‘shhh.flows' (translation of PyroNoise algorithm; Quince et al., 2009) and ‘pre.cluster' (Huse et al., 2010) in Mothur platform. Chimeric sequences were identified and removed using UCHIME with de novo method (Edgar et al., 2011). Quality sequences were subsequently assigned to samples according to their unique 8-bp barcode and binned into phylotypes using average clustering algorithm (Huse et al., 2010) at the 97% similarity level. Representative sequences were aligned using NAST (DeSantis et al., 2006) and then used to build the neighbor-joining phylogenetic trees using FastTree (Price et al., 2009). Taxonomic classification of phylotypes was determined based on the Ribosomal Database Project at the 80% threshold (Wang et al., 2007).
We estimated the relative abundance (%) of individual taxa within each community by comparing the number of sequences assigned to a specific taxon versus the number of total sequences obtained for that sample. We also calculated the number of phylotypes (richness) and the Faith's index of phylogenetic diversity (Faith's PD, sums of the total branch length in a phylogenetic tree that leads to each member of a community) to compare the community diversity across all 59 AMD samples. Weighted UniFrac analyses (Lozupone and Knight, 2005; Lozupone et al., 2006) were applied to calculate the pairwise distance between microbial assemblages. Calculations of diversity indices and UniFrac dissimilarity were based on a randomly selected subset of 540 sequences per sample. Normalizing the number of sequences per sample allowed us to control the effects of survey effort at same level in comparing the diversity indices and lineage-specific UniFrac distances across the samples (Lauber et al., 2009).
Data collection and beta diversity of microbial communities from global AMD and associated environments
To reveal broader patterns in the distribution of microorganisms among acidic environments globally distributed, we searched papers in Web of Science and reviewed molecular inventory studies that explored microbial communities in natural AMD and associated environments (such as acidic biofilm, sediment and tailings) from diverse geographical locations. 16S rRNA clone sequences were identified and recovered from GenBank for samples with detailed information of operational taxonomic units and their relative abundance, and environmental parameters were extracted and summarized. Community composition and weighted UniFrac dissimilarity were calculated for the subsequent meta-analysis (see detailed methods in Supplementary Information).
Statistical analyses
All statistical analyses were implemented using various packages within the R statistical computing environment. Aggregated boosted tree analysis (ABT) (De'ath, 2007) was carried out using the gbmplus package (with 5000 trees used for the boosting, 10-folds cross-validation and three-way interactions) to evaluate quantitatively the relative influence of environmental variables to the community diversity. A sum of squares multivariate regression tree (MRT) (De'ath, 2002) was performed using the mvpart package (with default parameters) to relate relative abundance of lineages to the site characteristics. Multiple linear regression (MLR) with stepwise method and Mantel test were conducted within the vegan package (Oksanen et al., 2010) to test the significance between diversity indices and the site properties. For data sets of PD, phylotypes richness and relative abundance of individual taxa in the subsequent MLR analyses, independent variables, including pH, EC, DO, TOC, SO42−, Fe3+, Fe2+, latitude and longitude, were input into the MLR model, while for the weighted UniFrac dissimilarity data set Bray–Curtis dissimilarities of these environmental variables and geographical distance were used. ABT and MRT analyses are statistical techniques that fundamentally aim to perform accurate prediction and explanation between complex ecological data (as we used diversity indices and dissimilarity matrices in ABT and relative lineage abundance in MRT) and environmental characteristics (De'ath, 2002, 2007). More importantly, the application of these methods allowed us to quantify and visualize the different contribution of environmental variables and geographical distance to the community diversity.
Results
Site characteristics and environmental conditions
The AMD samples captured a wide range of physical and geochemical gradients (Supplementary Tables 1 and 2) and were characterized by extremely acidic pH values ranging from 1.9 to 4.1 (2.6±0.45, mean±s.d.) and high concentrations of dissolved solids (measured as EC) ranging from 134 to 20 000 (4528±3786) μS cm−1. Concentrations of sulfate and ferric and ferrous irons were highly variable across the samples, averaging at 3787±2129, 1317±3915 and 130±354 mg l−1, respectively. Additionally, DO (3.3±3.6 mg l−1) and TOC (8.9±12.4 mg l−1) were also subjected to considerable fluctuations.
Composition and diversity of AMD microbial communities
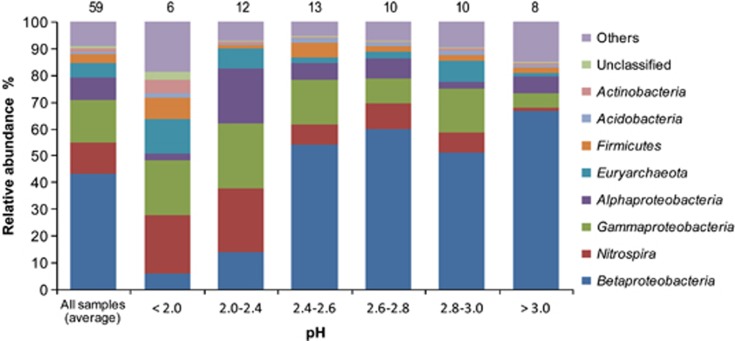
The bar-coded pyrosequencing generated 131 720 quality sequences from the 59 AMD samples, with an average of 2234±1756 sequences and a range of 542 to 9263 sequences per sample. All but 436 of the 131 720 sequences could be classified at the domain level (Bacteria or Archaea) by the RDP classifier (80% threshold). A total of 2198 phylotypes were defined at the 97% similarity level, with the majority (∼54% of the phylotypes) represented by a single sequence, whereas all of these singletons could be assigned to the taxa that were identified in the whole pyrosequence data set. The number of phylotypes detected in each sample ranged from 10 to 244, with an average of 61±49 according to a subset of 540 randomly selected sequences. Of the classifiable sequences, 18 phyla were identified, with Proteobacteria, Nitrospira and Euryarchaeota representing the most dominant lineages and accounting for 72%, 12% and 5.1% of all sequences, respectively. Some other phyla were less abundant but still detected in most of the samples; these included Firmicutes (3.4%), Actinobacteria (1.1%) and Acidobacteria (1.1%). Down to the genus level, the most abundant phylotypes were affiliated with the ‘Ferrovum' (59 333 sequences), Acidithiobacillus (13 744 sequences), Acidiphilium (9461 sequences) and Leptospirillum (7756 sequences); these collectively accounted for 69% of the total sequences. Specifically, the Acidithiobacillus sequences were composed predominantly (>95%) of A. ferrooxidans-like organisms, with an approximately 4.3% of A. caldus. The largest portion of the Leptospirillum reads was affiliated with Leptospirillum ferrodiazotrophum (60%), with the remaining being phylogenetically affiliated with L. ferrooxidans (>39%) and L. ferriphilum (0.34%). Additionally, almost all of the Acidiphilium sequences (>98%) were affiliated with Acidiphilium cryptum. The relative abundance of different lineages varied considerably across the AMD communities (Figures 2 and 3 and Supplementary Table 3a, also see the variances of the measured averages of individual lineages comprising the defined pH levels in Supplementary Tables 3b and 4).
Figure 2.
Relative abundances (%) of dominant lineages (phylum level) in overall communities and in different groups of AMD samples along the gradient of pH levels. The numbers above the columns indicate the number of samples in each group. Others include 12 phyla: Bacteroidetes, Chlamydiae, Chloroflexi, Crenarchaeota, Cyanobacteria, Deinococcus-Thermus, Gemmatimonadetes, OD1, OP11, Planctomycetes, TM7 and Verrucomicrobia; and two subphyla for Proteobacteria: Deltaproteobacteria and Epsilonproteobacteria.
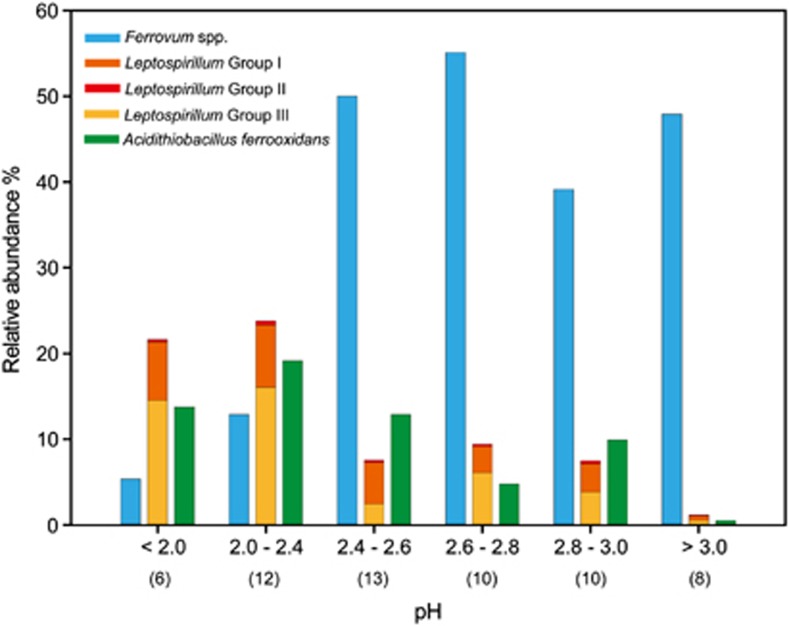
Figure 3.
Relative abundances of Ferrovum spp., Leptospirillum groups and A. ferrooxidans in different groups of microbial assemblages along the gradient of pH levels in AMD. The numbers within the parentheses indicate the number of samples in each group.
Relative influence of environmental conditions on microbial diversity
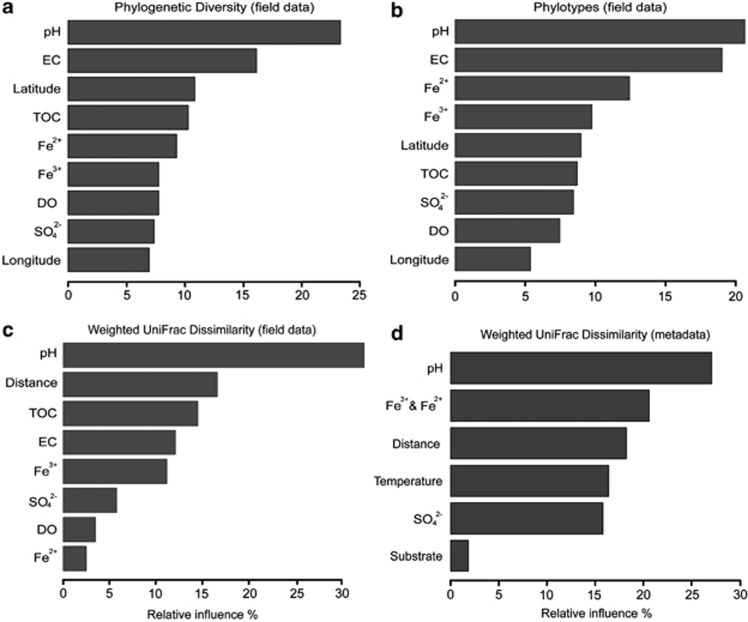
ABT models were conducted to interpret the relative importance of environmental conditions and spatial isolation to the diversity patterns of AMD microbial communities in Southeast China. ABT analysis indicated that pH was the major factor for the patterns of both PD and phylotypes, accounting for approximately 23% and 21% of the relative influence, respectively (Figures 4a and b). Partial dependency plots of pH from the fitted model revealed that high values of diversity index were most likely to be observed at higher pH conditions (Supplementary Figures 1a and b). These results were in good agreement with the significantly positive correlations between solution pH and overall diversity determined by MLR analysis (Faith's PD: r=0.349, P=0.008; Phylotypes: r=0.359, P=0.006; for both PD and Phylotypes, environmental variables other than pH were all eliminated in the predicted MLR model, see Statistical analyses). Additionally, the ABT analysis also revealed a moderate effect for EC and weaker effects for other environmental variables such as TOC and DO on both diversity estimates. In comparison, the spatial isolation represented by the gradients of latitude and longitude made less contributions, indicating that there is no obvious effect of geographical distance on the AMD microbial diversity. When considering the pairwise community distances between microbial assemblages, the variation of pH calculated by Bray–Curtis distance again revealed the most important influence on the weighted UniFrac dissimilarity (Figure 4c). This relationship was corroborated by the Mantel test (Spearman's r=0.329, P<0.001) and MLR analysis (r=0.337, P<0.001, with Bray–Curtis dissimilarity of pH as the only variable remained in the MLR model), with higher divergence of solution pH likely leading to higher UniFrac dissimilarity (Supplementary Figure 1c). In contrast, no significant correlation between geographical distance and the UniFrac distance was detected by the Mantel test (Spearman's r=0.106, P=0.072), implying that the contribution of spatial isolation to the community dissimilarity was limited (Figure 4c).
Figure 4.
Relative influence (%) of environmental properties and spatial distance for phylogenetic diversity (a), phylotypes (b), weighted UniFrac dissimilarity of field data (pyrosequecing) (c) and weighted UniFrac dissimilarity of metadata (meta-analysis) (d) evaluated by ABT models.
Relationship between relative abundance of dominant lineages and environmental conditions
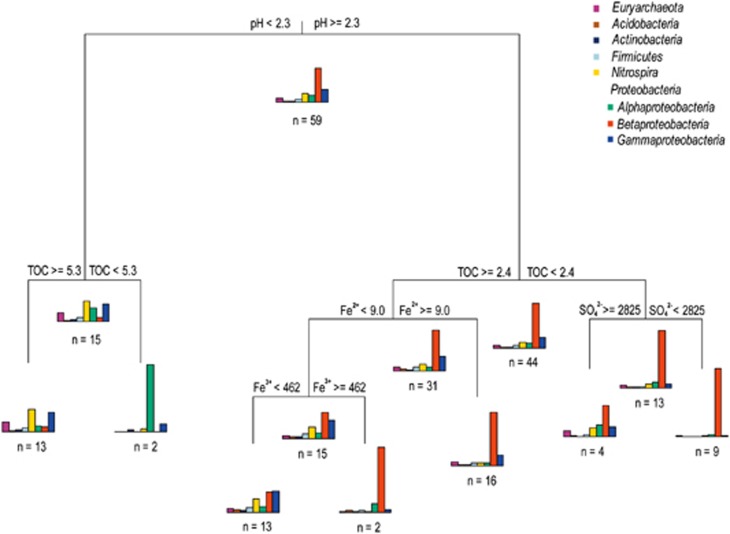
Taxonomy-supervised analysis has the advantages of less computation requirement and more tolerance of sequencing errors (Sul et al., 2011). We further conducted an MRT analysis using our AMD field data, which interpreted the relationship between the relative abundance of dominant lineages and environmental conditions by providing a tree with seven terminal nodes based on pH, TOC and concentrations of sulfate, ferric and ferrous irons, collectively explaining 70% of the standardized abundance variance (Figure 5). The results suggested that spatial isolation represented by sampling location (measuring longitude and latitude as two variables in the MRT model) was less of a factor than environmental variables in explaining the variation in microbial community composition, and pH appeared to be a strong predictor of relative lineage abundance with samples with low pH levels (pH <2.3) clustering separately from those with moderate pH values. Betaproteobacteria, the most abundant lineage (44±34%) across all communities, showed an intense response to solution pH, with low relative abundance (4.2±9.1%, n=15) in extremely acidic environments, but explicitly predominant (48±32%, n=44) in the microbial assemblages under moderate pH conditions. Such a trend was largely attributed to the distribution pattern of ‘Ferrovum'-related organisms (Figure 3). In contrast, other lineages such as Alphaproteobacteria, Euryarchaeota, Gammaproteobacteria and Nitrospira exhibited a distinct adaptation to more acidic environments with an increase of relative abundance. These results were coincident with the overall dynamics of community composition along the pH gradient (Figure 2). Similar patterns were found for other environmental variables in the MRT analysis, with groups uniquely dominated by Betaproteobacteria generally separating from those with notable increase of relative abundance of other lineages (Figure 5), implying that the optimal conditions for the growth of Betaproteobacteria (mainly ‘Ferrovum' spp.) were apparently different from those for the other lineages.
Figure 5.
Multivariate regression tree analysis of the relation between relative abundance of dominant lineages and environmental parameters in microbial communities of AMD. The bar plots show the mean relative abundance of specific lineages at each terminal nodes and the distribution patterns of relative abundance represent the dynamics of community composition among each split. The numbers under the bar plots indicate the number (n) of samples within each group. All values are in mg l−1, except pH, which is in standard units.
Global distribution patterns of microbial diversity in AMD and associated environments
From the 25 molecular inventory studies that met our literature searching criteria, 66 samples with overall community composition were extracted for the taxon-based analysis in MRT model, and 45 of them (which had detailed information of OTUs and their relative abundance) were retained for the phylogenetic-based analysis in ABT model (Supplementary Tables 5a and b). It should be noted, however, that some parameters were missing from individual studies, but this problem could be overcome in the ABT and MRT model analyses as these models can deal with different types of response variables (for example, numeric or categorical) with missing values (De'ath, 2002, 2007).
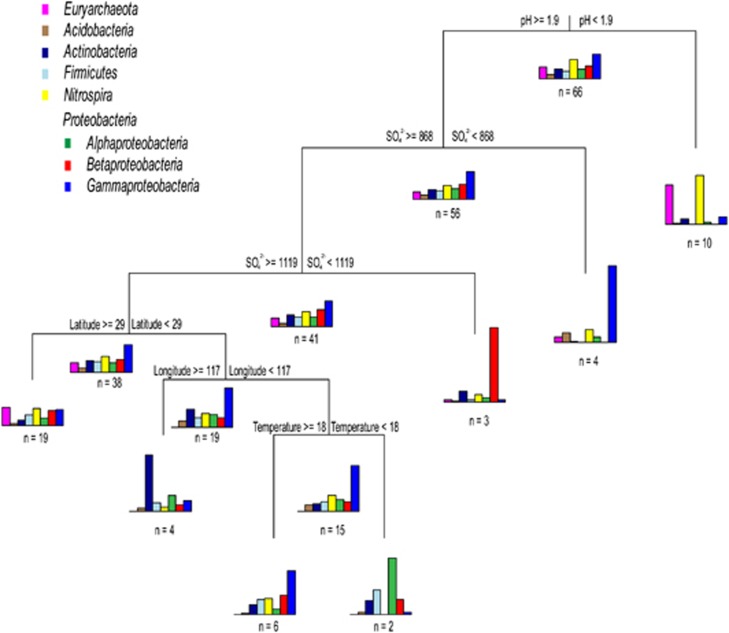
Overall, similar patterns of microbial composition were found in the global AMD and associated systems, with Proteobacteria, Nitrospira and Euryarchaeota as the major groups despite considerable fluctuations in their relative recovery in the 16S rRNA gene libraries (Supplementary Table 6). Most strikingly, pairwise UniFrac distances between the 45 samples were still largely affected by environmental pH as revealed by the ABT model (Figure 4d), implying that microbial assemblages from different substrates may have similar community composition under a similar pH condition. Furthermore, the geographical distance between the globally distributed samples (up to 18 000 km) still had less influence to the community dissimilarity than pH, supporting environmental variation as the major factor relating microbial communities as observed in our pyrosequence data set. Likewise, the MRT analysis using the metadata of 66 samples indicated that microbial community composition was mainly shaped by pH level, with relatively little influence from spatial isolation (Figure 6). However, in comparison with the significant contribution of Betaproteobacteria to the overall microbial distribution in the Chinese AMD environments, the global-scale pH-dependent pattern was largely attributed by the predominant distribution of Euryarchaeota and Nitrospira under relatively low pH conditions (pH <1.9).
Figure 6.
Multivariate regression tree analysis of the relation between relative abundance of dominant lineages and environmental parameters in microbial communities of global AMD and associated systems using metadata. pH, latitude and longitude are in standard units. Temperature is in °C and sulfate concentration is in mg l−1.
Discussion
A comprehensive survey of AMD microbial diversity
We characterized the diversity and composition of microbial communities from diverse and geographically separated acidic mining environments in Southeast China. The large number of samples surveyed and the sequencing depth provided by the bar-coded pyrosequencing generated an unprecedented number of AMD microbial 16S rRNA gene sequence data that far exceed the total number of sequences reported in previous clone library studies, significantly expanding our knowledge of the broad trends of microbial distribution in extremely acidic environments. Although most of the AMD communities have been sufficiently sampled by the pyrosequencing (as suggested by the rarefaction analyses; Supplementary Figure 2), the full extent of microbial diversity in a few samples has not been captured. It is not likely that this is due to inflation of biodiversity estimate by sequencing errors generated by noise introduced during pyrosequencing and the PCR amplification stage (Reeder and Knight, 2010), as such bias should have been limited after our stringent denoizing of data. Similar results have been reported in other extreme habitats such as hydrothermal chimneys (Brazelton et al., 2010) and acidic hot spring (Bohorquez et al., 2012), where numerous rare taxa account for most of the observed diversity, implying that microbial diversity could be higher than expected in some specific sites with complex interactions among environmental variables and microorganisms. Interestingly, although the microbial diversity (Faith's PD and Phylotypes) generally increased along the solution pH gradient, a moderately higher diversity and relatively uniform distribution pattern were found in the lowest pH level (pH <2.0; Figure 2 and Supplementary Table 7). This may be related to the significantly higher organic carbon contents in a few samples in this pH group, as high carbon contents with heterogeneous resource condition have previously been found to promote high species diversity in soil (Zhou et al., 2002) and marine sediments (Stach et al., 2003).
Better prediction of microbial diversity patterns by solution pH
Previous molecular investigations have documented spatial and seasonal variations in microbial populations in specific AMD environments, and different major environmental determinants such as conductivity and rainfall (Edwards et al., 1999), solution pH (Lear et al., 2009) and oxygen gradient (Gonzalez-Toril et al., 2011) have been identified across different studies at local scale, which may be due to the site-specific geochemical characteristics. Other factors, in particular resolutions of the different methods used to evaluate community composition (fluorescent in situ hybridization, community fingerprinting, and so on) and the relatively small number of samples examined, should also be taken into account. More recent studies have used extensive genomic sequencing (Denef and Banfield, 2012) and proteomics (Mueller et al., 2010) to investigate how rapid adaptive evolution may have assisted in the maintenance of the dominant populations in local AMD communities and how the physiologies of the dominant and less abundant organisms change along environmental gradients corresponding with their ecological distribution within the Richmond Mine at Iron Mountain in California. Our pyrosequencing survey of microbial diversity in multiple geographically separated AMD sites across Southeast China has provided an initial insight into the large-scale distribution patterns of microbes in these unique habitats, with greater variability in physical and geochemical attributes. Our phylogenetic- and taxon-based analyses (ABT and MRT models) supported the important idea that similar ecological conclusions could be drawn by using these two commonly used clustering approaches (Sul et al., 2011), and consistently suggested that, notably, pH is still the definitive factor structuring AMD communities, regardless of the fact that low pH is a major feature of these extreme environments and the indigenous microorganisms are predominantly acidophiles that are well adapted and tolerant to these prevailing extreme conditions. More strikingly, our subsequent meta-analysis of previous molecular surveys of AMD systems in diverse geographical locations also revealed similar pH-dependent patterns in microbial diversity at a global scale despite the long-distance separation and regardless of distinct substrate types. These consistent results were presumably due to the strong selective pressures with extremely acidic conditions that primarily determine which lineages can survive there. Indeed, optimum pH for growth can vary significantly among cultivated acidophilic species or even between phylogenetically highly similar (as evidenced by 16S rRNA sequence comparison) microorganisms isolated from different acidic mining environments (Edwards et al., 2000; Golyshina et al., 2000). Furthermore, recent quantitative proteomic analyses of the response of acidophilic microbial communities to different pH conditions has suggested pH-specific niche partitioning of prokaryotes and confirmed the importance of pH and related geochemical factors in fine-tuning acidophilic microbial community structure and function (Belnap et al., 2011).
Hot springs have also been targets for the study of microbial diversity and adaptation in extreme environments. Although some studies have suggested that environmental temperature is a primary factor controlling the structure and dynamics of microbial communities (Ward et al., 1998; Miller et al., 2009), significant differences in microbial community composition exist between the alkaline (Miller et al., 2009) and acidic (Mathur et al., 2007) hot springs in Yellowstone National Park (WY, USA). Furthermore, the acidic hot springs with high sulfate concentrations (Stout et al., 2009) and iron-rich conditions (Mathur et al., 2007) were dominated by iron-oxidizing acidophiles such as Acidithiobacillus spp., irrespective of the wide range of temperature gradients. This similar distribution and adaptation of such acidophiles in acidic hot springs and acid mine water may indicate that pH is a common determinant for structuring the microbial communities in the two different extreme environments. These results are potentially important since recent studies have repeatedly identified pH as one of the general selective pressures working in ‘normal' environments like soil (Fierer and Jackson, 2006; Lauber et al., 2009; Rousk et al., 2010; Griffiths et al., 2011).
Other potential factors explaining community differences
Several recent meta-analyses have identified salinity as the primary environmental factor shaping the ecological distribution of prokaryotic taxa along broad environmental gradients and across different habitat types (Lozupone and Knight, 2007; Tamames et al., 2010). It is striking that general environmental properties such as salinity still primarily determine the microbial composition of extreme environments, although these organisms are presumably under strong selective pressures (for example, temperate and pH). Although salinity (as reflected by the EC values) had a weaker effect than solution pH in our ABT models, a significantly negative relationship was detected between EC and pH (Spearman's r=−0.631, P<0.001), and the combination of pH, EC and their interaction was the best MLR model to predict the microbial community diversity (Faith's PD r=0.505, P=0.006 and Phylotypes r=0.478, P=0.002, for MLR model considering the interaction between pH and EC), implying the potential importance of salinity for structuring AMD microbial communities, a feature documented in a previous fluorescent in situ hybridization-based survey of an acid-generating site at Iron Mountain in California (Edwards et al., 1999).
Dispersal limitation and past environmental conditions can lead to genetic divergence of microbial assemblages from geographically separated sites (Martiny et al., 2006). Although our results suggested that the overall microbial diversity patterns were better predicted by contemporary environmental variation, a moderate influence of geographical isolation on the UniFrac dissimilarity of the Acidithiobacillus and Leptospirillum genera could still be found (although no significant correlation was detected by the Mantel test), indicating that observed patterns could be taxa-/lineage-dependent or might vary with the levels of analytical/phylogenetic resolution. It should also be noted that relative influence of historical versus environmental factors could be related to the scale of sampling (Martiny et al., 2006), as exemplified by the inter- and intracontinental surveys of hot spring Synechococcus and Sulfolobus assemblages (Papke et al., 2003; Whitaker et al., 2003).
Taxonomic distribution pattern of dominant species in AMD microbial communities
Taxonomic classification of our pyrosequences revealed that Betaproteobacteria were ubiquitous and dominant across the AMD sites, especially at moderate pH conditions (pH >2.4; Figures 2 and 5). Significantly, the most abundant phylotype, which accounted for approximately 91% of the total Betaproteobacteria reads, was affiliated with the recently discovered genus ‘Ferrovum' (Hallberg et al., 2006). Although less known, ‘Ferrovum' spp. are suggested to be obligately autotrophic and capable of growth only by ferrous iron oxidation with less acid tolerance than the well-studied AMD species L. ferrooxidans and A. ferrooxidans (Rowe and Johnson, 2008; Hallberg, 2010). Our pyrosequencing survey of diverse AMD sites supports the acidic susceptibility of these ‘Ferrovum'-affiliated organisms and their preference for relatively high ferrous iron conditions (Figure 5) (Heinzel et al., 2009b). More importantly, their wide distribution and dominance were conspicuous across geochemically distinct mining environments examined in this study (Figure 3) and other acidic sites (Brockmann et al., 2010; Brown et al., 2011; Kimura et al., 2011), implying their important ecological role in the extreme AMD systems. Notably, although pH was identified as the primary force shaping their large-scale ecological range (across Southeast China), EC was found (r=−0.76, P=0.018, MLR with all other independent variables excluded) to influence their local distribution at the YunFu mine (where a relatively large sample size is available for analyzing the local patterns), implying the potential effects of site-specific geochemical characteristics. The interactions between ‘Ferrovum' spp. and other populations over a continuum of spatial and temporal scales within entire regions merit further study (Ricklefs, 2008).
In contrast to the high relative abundance of ‘Ferrovum' spp. under moderate pH conditions, A. ferrooxidans and Leptospirillum groups (acidophiles widely implicated for their contribution to AMD production) were more dominant and thus likely to have significant roles in more acidic conditions (Figure 3). A similar distribution of Leptospirillum spp. and A. ferrooxidans was found along the pH gradient, revealing an optimum pH range of 2.0–2.4 and a significant decrease in relative abundance when the pH increases to above 3.0 (Figure 3). However, a relatively high Leptospirillum spp. to A. ferrooxidans ratio was observed at higher Fe3+/Fe2+ (redox potential) (t-test, for independent samples, P<0.01), indicating a potential competition for energy sources (ferrous iron) and supporting the greater Fe2+ affinity and less sensitivity to Fe3+ inhibition of Leptospirillum spp. (Rawlings et al., 1999).
The biogeographic patterns of less abundant AMD taxa and the associated driving forces remain unresolved. While heterotrophic bacteria such as Acidiphilium were widely detected across the AMD samples, they were generally presented in significantly smaller proportions than the acidophilic autotrophs and no clear statistical correlation was found between TOC and relative abundance of the detected heterotrophs, suggesting a limited contribution of external inputs of fixed carbon in the maintenance of the indigenous microbial assemblages. Archaea accounted for a non-negligible proportion (average >5.0%) across the AMD communities; however, only a small fraction of these sequences could be confidently assigned at the genus level, mostly to the Ferroplasma, although recent studies have suggested that acidophilic Archaea related to the ferrous-iron-oxidizing F. acidiphilum are numerically significant and thus ecologically important in some acidic environments in diverse geographical locations (Golyshina and Timmis, 2005; Huang et al., 2011). In general, no obvious distribution patterns of Archaea or Ferroplasma spp. could be observed along either geographical distance or environmental properties in our data set (no independent variables retained in the MLR model). Additionally, the neutrophilic Gallionella spp., which have been commonly found in AMD habitats, were nearly not detected (two phylotypes with 33 reads) among our diverse AMD sites. Acid susceptibility may not be the main reason for their absence in the Chinese AMD environments as these iron oxidizers have been detected in a few other acidic sites with a low pH range of 2.6–3.0 (Hallberg et al., 2006; Heinzel et al., 2009a; Kimura et al., 2011).
Summary and prospect
We report the most comprehensive analysis of the geographical distribution of AMD microbes to date, revealing environmentally dependent patterns at both regional and global scale with less contributions of spatial separation. While pH is identified as the major factor relating microbial communities over the range of acidic habitats examined in this study and ‘‘normal'' environments surveyed in other studies (Lauber et al., 2009; Griffiths et al., 2011), the underlying mechanisms have thus far remained unsolved. Trait-based or functional biogeography assessed by metagenomics (Raes et al., 2011) or comprehensive functional gene arrays such as GeoChip (He et al., 2010) represents a promising strategy to address this issue. We could not distinguish in our pyrosequence data set active community members from the dormant taxa, which may be metabolically inactive because of the unfavorable environmental conditions and thus less sensitive to environmental change. Future investigations specifically focusing on the active members of the community will likely reveal even more pronounced environmentally dependent patterns of microbial diversity. Such knowledge is critical as these active taxa presumably have crucial roles in the functioning of the AMD ecosystems. Additionally, evaluation of microbial population dynamics at fine temporal scales and over relatively long periods of time at a diverse array of AMD sites would bring novel insight and greater predictive power to the microbial diversity patterns in these extreme environments.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 40930212 and 30970548), the Major Science and Technology Project of Ministry of Agriculture of the People's Republic of China (No. 2009ZX08009-002B), the Guangdong Province Key Laboratory of Computational Science and the Guangdong Province Computational Science Innovative Research Team.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R. Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J. 2011;5:42–50. doi: 10.1038/ismej.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet JC, Barberan A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Banfield JF. Microbial communities in acid mine drainage. FEMS Microbiol Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap CP, Pan C, Denef VJ, Samatova NF, Hettich RL, Banfield JF. Quantitative proteomic analyses of the response of acidophilic microbial communities to different pH conditions. ISME J. 2011;5:1152–1161. doi: 10.1038/ismej.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, et al. In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol. 2012;63:103–115. doi: 10.1007/s00248-011-9943-3. [DOI] [PubMed] [Google Scholar]
- Bond PL, Smriga SP, Banfield JF. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol. 2000;66:3842–3849. doi: 10.1128/aem.66.9.3842-3849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen CC, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci USA. 2010;107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann S, Arnold T, Schweder B, Bernhard G. Visualizing acidophilic microorganisms in biofilm communities using acid stable fluorescence dyes. J Fluoresc. 2010;20:943–951. doi: 10.1007/s10895-010-0640-2. [DOI] [PubMed] [Google Scholar]
- Brown JF, Jones DS, Mills DB, Macalady JL, Burgos WD. Application of a depositional facies model to an acid mine drainage site. Appl Environ Microbiol. 2011;77:545–554. doi: 10.1128/AEM.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnin L, Yien CH. Turbidimeteric determination of available sulphates. Proc Soil Sci Soc Am. 1951;15:149–151. [Google Scholar]
- Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De'ath G. Multivariate regression trees: a new technique for modeling species–environment relationships. Ecology. 2002;83:1105–1117. [Google Scholar]
- De'ath G. Boosted trees for ecological modeling and prediction. Ecology. 2007;88:243–251. doi: 10.1890/0012-9658(2007)88[243:btfema]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Banfield JF. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science. 2012;336:462–466. doi: 10.1126/science.1218389. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druschel GK, Baker BJ, Gihring TM, Banfield JF. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem Trans. 2004;5:13–32. doi: 10.1186/1467-4866-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Gihring TM, Banfield JF. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol. 1999;65:3627–3632. doi: 10.1128/aem.65.8.3627-3632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Bond PL, Gihring TM, Banfield JF. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science. 2000;287:1796–1799. doi: 10.1126/science.287.5459.1796. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moyano A, Gonzalez-Toril E, Aguilera A, Amils R. Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Rio Tinto (SW, Spain) Syst Appl Microbiol. 2007;30:601–614. doi: 10.1016/j.syapm.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Ge Y, He JZ, Zhu YG, Zhang JB, Xu Z, Zhang LM, et al. Differences in soil bacterial diversity: driven by contemporary disturbances or historical contingencies. ISME J. 2008;2:254–264. doi: 10.1038/ismej.2008.2. [DOI] [PubMed] [Google Scholar]
- Goebel BM, Stackebrandt E. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol. 1994;60:1614–1621. doi: 10.1128/aem.60.5.1614-1621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golyshina OV, Pivovarova TA, Karavaiko GI, Kondratéva TF, Moore ER, Abraham WR, et al. Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int J Syst Evol Microbiol. 2000;50:997–1006. doi: 10.1099/00207713-50-3-997. [DOI] [PubMed] [Google Scholar]
- Golyshina OV, Timmis KN. Ferroplasma and relatives, recently discovered cell wall-lacking archaea making a living in extremely acid, heavy metal-rich environments. Environ Microbiol. 2005;7:1277–1288. doi: 10.1111/j.1462-2920.2005.00861.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Toril E, Llobet-Brossa E, Casamayor EO, Amann R, Amils R. Microbial ecology of an extreme acidic environment, the Tinto River. Appl Environ Microbiol. 2003;69:4853–4865. doi: 10.1128/AEM.69.8.4853-4865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Toril E, Aguilera A, Souza-Egipsy V, Lopez Pamo E, Sanchez Espana J, Amils R. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain) Appl Environ Microbiol. 2011;77:2685–2694. doi: 10.1128/AEM.02459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- Hallberg KB, Coupland K, Kimura S, Johnson DB. Macroscopic streamer growths in acidic, metal-rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol. 2006;72:2022–2030. doi: 10.1128/AEM.72.3.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg KB. New perspectives in acid mine drainage microbiology. Hydrometallurgy. 2010;104:448–453. [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- Heinzel E, Hedrich S, Janneck E, Glombitza F, Seifert J, Schlomann M. Bacterial diversity in a mine water treatment plant. Appl Environ Microbiol. 2009a;75:858–861. doi: 10.1128/AEM.01045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel E, Janneck E, Glombitza F, Schlomann M, Seifert J. Population dynamics of iron-oxidizing communities in pilot plants for the treatment of acid mine waters. Environ Sci Technol. 2009b;43:6138–6144. doi: 10.1021/es900067d. [DOI] [PubMed] [Google Scholar]
- Hill AG, Bishop E, Coles LE, McLaughlan EJ, Meddle DW, Pater MJ, et al. Standardized general method for the determination of iron with 1,10-phenanthroline. Analyst. 1978;103:391–396. [Google Scholar]
- Huang LN, Zhou WH, Hallberg KB, Wan CY, Li J, Shu WS. Spatial and temporal analysis of the microbial community in the tailings of a Pb–Zn mine generating acidic drainage. Appl Environ Microbiol. 2011;77:5540–5544. doi: 10.1128/AEM.02458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Bryan CG, Hallberg KB, Johnson DB. Biodiversity and geochemistry of an extremely acidic, low-temperature subterranean environment sustained by chemolithotrophy. Environ Microbiol. 2011;13:2092–2104. doi: 10.1111/j.1462-2920.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear G, Niyogi D, Harding J, Dong Y, Lewis G. Biofilm bacterial community structure in streams affected by acid mine drainage. Appl Environ Microbiol. 2009;75:3455–3460. doi: 10.1128/AEM.00274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Bizzoco RW, Ellis DG, Lipson DA, Poole AW, Levine R, et al. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl Environ Microbiol. 2007;73:2612–2623. doi: 10.1128/AEM.02567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SR, Strong AL, Jones KL, Ungerer MC. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl Environ Microbiol. 2009;75:4565–4572. doi: 10.1128/AEM.02792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SM, Davis RE, McBeth JM, Tebo BM, Emerson D, Moyer CL. Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol. 2011;77:5445–5457. doi: 10.1128/AEM.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RS, Denef VJ, Kalnejais LH, Suttle KB, Thomas BC, Wilmes P, et al. Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol Syst Biol. 2010;6:374. doi: 10.1038/msb.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom DK, Alpers CN. Negative pH, efflorescent mineralogy, and consequences for environmental restoration at the Iron Mountain Superfund site, California. Proc Natl Acad Sci USA. 1999;96:3455–3462. doi: 10.1073/pnas.96.7.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RG, Simpson GL, et al. 2010Vegan: community ecology packageR package version 1.17-0. URL http://CRAN.R-project.org/package=vegan .
- Papke RT, Ramsing NB, Bateson MM, Ward DM. Geographical isolation in hot spring cyanobacteria. Environ Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Noise and the accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol. 2011;7:473. doi: 10.1038/msb.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings DE, Tributsch H, Hansford GS. Reasons why ‘Leptospirillum'-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for the biooxidation of pyrite and related ores. Microbiology. 1999;145:5–13. doi: 10.1099/13500872-145-1-5. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs R. Disintegration of the ecological community. Am Nat. 2008;172:741–750. doi: 10.1086/593002. [DOI] [PubMed] [Google Scholar]
- Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Rowe OF, Johnson DB. Comparison of ferric iron generation by different species of acidophilic bacteria immobilized in packed-bed reactors. Syst Appl Microbiol. 2008;31:68–77. doi: 10.1016/j.syapm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach JE, Maldonado LA, Masson DG, Ward AC, Goodfellow M, Bull AT. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl Environ Microbiol. 2003;69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout LM, Blake RE, Greenwood JP, Martini AM, Rose EC. Microbial diversity of boron-rich volcanic hot springs of St. Lucia, Lesser Antilles. FEMS Microbiol Ecol. 2009;70:402–412. doi: 10.1111/j.1574-6941.2009.00780.x. [DOI] [PubMed] [Google Scholar]
- Sul WJ, Cole JR, Jesus Eda C, Wang Q, Farris RJ, Fish JA, et al. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J, Abellan JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Huang LN. Cultivation-dependent and cultivation-independent characterization of the microbial community in acid mine drainage associated with acidic Pb/Zn mine tailings at Lechang, Guangdong, China. FEMS Microbiol Ecol. 2007;59:118–126. doi: 10.1111/j.1574-6941.2006.00216.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Ferris MJ, Nold SC, Bateson MM. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O'Neill RV, et al. Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol. 2002;68:326–334. doi: 10.1128/AEM.68.1.326-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.