Abstract
Large-scale mortality of marine invertebrates is a major global concern for ocean ecosystems and many sessile, reef-building animals, such as sponges and corals, are experiencing significant declines through temperature-induced disease and bleaching. The health and survival of marine invertebrates is often dependent on intimate symbiotic associations with complex microbial communities, yet we have a very limited understanding of the detailed biology and ecology of both the host and the symbiont community in response to environmental stressors, such as elevated seawater temperatures. Here, we use the ecologically important sponge Rhopaloeides odorabile as a model to explore the changes in symbiosis during the development of temperature-induced necrosis. Expression profiling of the sponge host was examined in conjunction with the phylogenetic and functional structure and the expression profile of the symbiont community. Elevated temperature causes an immediate stress response in both the host and symbiont community, including reduced expression of functions that mediate their partnership. Disruption to nutritional interdependence and molecular interactions during early heat stress further destabilizes the holobiont, ultimately leading to the loss of archetypal sponge symbionts and the introduction of new microorganisms that have functional and expression profiles consistent with a scavenging lifestyle, a lack virulence functions and a high growth rate. Previous models have postulated various mechanisms of mortality and disease in marine invertebrates. Our study suggests that interruption of symbiotic interactions is a major determinant for mortality in marine sessile invertebrates. High symbiont specialization and low functional redundancy, thus make these holobionts extremely vulnerable to environmental perturbations, including climate change.
Keywords: community stability, functional redundancy, marine disease, microbial symbionts, ocean warming
Introduction
Environmental factors, including climate change, anthropogenic pollution, introduced species and nutrient enrichment have all been linked to stress and disease in marine organisms (Lafferty et al., 2004). Large-scale mortality events and disease outbreaks are being observed in marine invertebrates globally (Lafferty et al., 2004; Plowright et al., 2008) and many of these events have been linked with increasing sea surface temperatures (Hoegh-Guldberg et al., 2007; Webster, 2007; Cebrian et al., 2011). Despite the prevalence of these large-scale mortality events, the biological and ecological mechanisms behind them remain unclear.
Sponges, corals and many other marine invertebrates form obligate symbiotic associations with microorganisms (Taylor et al., 2007; Bourne et al., 2009; Rosenberg et al., 2009; Webster and Taylor, 2011; Fan et al., 2012b). Changes in microbial community structure correlate with the development of ‘disease-like' symptoms (Webster, 2007; Webster et al., 2008; Bourne et al., 2009; Rosenberg et al., 2009), however, the role that microbial symbionts or potential pathogens have in the health outcomes of the host are difficult to define. This is highlighted in the study of coral symbiosis, where bleaching can be caused by thermal impairment of zooxanthellae symbionts (Warner et al., 1999) or temperature-induced invasion of bacterial pathogens encoding virulence-related genes (Toren et al., 1998; Banin et al., 2003; Ben-Haim et al., 2003; Kimes et al., 2012). In addition, many studies have reported shifts in microbial community composition before disease development, thereby questioning the importance or involvement of specific pathogens (Bourne, 2005; Bourne et al., 2008; Sussman et al., 2008). These observations illustrate the complex interplay within symbiotic systems and the difficulty in dissecting cause and effect for stress-related syndromes. Systematic studies that simultaneously assess the composition and function of the host and the symbiont community under controlled stress conditions will clarify these issues.
In this study, we monitor an ecologically important Great Barrier Reef (GBR) sponge (Rhopaloeide odorabile) under controlled temperature conditions to systematically investigate the mechanisms that contribute to the decline in a marine holobiont (host and associated symbionts). Community fingerprinting, metagenomics and metaproteomics were used to characterize the community structure and function of symbionts, and gene expression analysis was used to define the host state. We show that the sponge and its microbial symbionts are highly interdependent with respect to their metabolism, cellular recognition and molecular interactions. Elevated seawater temperature suppresses these symbiotic functions in both the host and the symbionts, causing the decline of the holobiont. The disturbed holobiont then offers niches that are rapidly occupied by a new set of bacteria, which lack the capacity for symbiotic interactions, but instead opportunistically scavenge on the decaying host. These findings indicate that interruption of symbiotic functions by environmental perturbation may be a major cause for the decline of marine sessile invertebrates. Highly specialized symbioses and low levels of functional redundancy make marine invertebrates particularly vulnerable to environmental perturbations.
Materials and methods
Sampling and experimental setup
Three individual R. odorabile sponges were collected from Davies Reef on the GBR, Australia (18°82′S, 147°65′E) in July 2009 by scuba diving. The donor sponges were cut into 48 clones. Each of these was ∼15 cm3 in size weighing ∼30 g. They were secured in plastic racks and left on the reef to heal for 16 weeks. Clones were then transported to indoor aquaria at the Australian Institute of Marine Science, Townsville, Australia. The water temperature of the aquaria was set to 27±0.5 °C to correspond to the ambient water temperature at Davies Reef and clones were acclimatized for 7 days. They were then randomly separated into six 60-L tanks, each of which held eight clones. Tanks were supplied with 5 μm filtered seawater at a flow rate of 600 ml per minute and illuminated at 80 μmol quanta m−2 s−1 (consistent with the light intensity at 15 m depth on the GBR) using 3 ft overhead compact fluorescent tubes under a diel cycle (12:12 h). Three additional individuals of R. odorabile (wild-types) were collected from the same site of the GBR and directly processed for microbial community analysis (see below).
The thermal stress experiment was commenced at 0800 hours on 18 November 2009 (day 0). The six tanks were randomly assigned to two temperature treatments (T27 and T32) with three replicate tanks (a, b and c) in each treatment. Experimental temperatures were selected to incorporate ambient summer seawater temperature at Davies Reef (http://www.aims.gov.au/docs/research/monitoring/weather/weather.html) and a 2 °C increase predicted by Intergovernmental Panel on Climate Change B1 and A2 scenarios for sea surface temperature by 2070 (32 °C). Tanks in the T27 treatment were maintained at 27 °C throughout the experiment, while seawater in the T32 treatment tanks was gradually heated (0.2 °C h−1) to the final treatment temperature of 32 °C and then maintained at this temperature until the end of the experiment. Sampling was conducted at the time points listed in Table 1. One clone was randomly selected from each tank and immediately processed for microbial community analysis (see below).
Table 1. Sample collections in the temperature-shifting experiment.
| Timepoint |
Temperature |
|
|---|---|---|
| 27 °C | 32 °C | |
| Day 0 (18 Nov 2009) | Day0-a (H), Day0-b (H), Day0-c (H) | |
| Day 1 (20 Nov 2009) | T27-Day1-a (H), T27-Day1-b (H), T27-Day1-c (H) | T32-Day1-a (H), T32-Day1-b (H), T32-Day1-c (H) |
| Day 3 (22 Nov 2009) | T27-Day3-a (H), T27-Day3-b (H), T27-Day3-c (H) | T32-Day3-a (H), T32-Day3-b (H), T32-Day3-c (N) |
| Day 4 (23 Nov 2009) | T27-Day4-a (H), T27-Day4-b (H), T27-Day4-c (H) | T32-Day4-a (N), T32-Day4-b (I), T32-Day4-c (I) |
Abbreviations: H, the healthy group. I, the, intermediate group. N, the necrotic group.
Multiplexed reverse transcription quantitative PCR analysis
To investigate the expression profiles of 23 selected host genes in sponges exposed to thermal stress, we undertook a multiplexed reverse transcription quantitative PCR using the GenomeLab GeXP Genetic Analysis System (Beckman Colter, Fullerton, CA, USA). The set of 23 genes were selected following screening of a previously prepared R. odorabile complementary DNA clone library (Pantile and Webster, 2011) on the basis of their known or putative role in the cell stress response and cellular homeostasis-related processes (Supplementary Table S1). The kanA gene conferring kanamycin resistance was used as the internal control. Gene sequences and detailed protocols for multiplex design, mRNA extraction, complementary DNA preparation, PCR, electropherogram analysis and normalization were described by Webster et al. (In Review). Coefficient of variation among technical replicate reactions was within a maximum of 20%, matching values reported in other studies (Rai et al., 2009; Souter et al., 2011). The optimal pairs of reference genes were identified as Prolifin/YWHAQ and their geometric means were used as the normalization factor.
Microbial community analysis
Microbial cell enrichment, DNA and protein extraction and preparation were performed as described by Thomas et al. (2010) and Liu et al. (2012). Details of the terminal restriction fragment length polymorphism analysis are described in Supplementary Information. Metagenomic shotgun sequencing was performed on the Roche 454 Titanium platform (J. Craig Venter Institute, Rockville, USA). Reconstruction of 16S rRNA genes from metagenomic shotgun data and phylogenetic analysis was performed as described by Fan et al. (2012a). The shotgun sequencing is available through the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis website (http://camera.calit2.net/) under project accession ‘CAM_PROJ_BotanyBay'. Metagenomic read processing, assembly, functional annotation, metaproteomic analysis by tandem mass spectrometry, MS/MS data analysis and protein identification were performed as described by Liu et al. (2012). Mass spectrometry spectra for the metaproteomic analysis are available through the PRIDE database (www.ebi.ac.uk/pride; accession numbers: 20986–20998) and raw peptide and protein identifications are given in Supplementary Table S2. Functional abundance matrices were generated by counting the number of genes (for metagenomic data) or proteins (for metaproteomic data) in each sample that had been assigned to a particular function (for example, Clusters of Orthologous Group (COG) (Tatusov et al., 2003), protein family (Pfam-A) (Finn et al., 2010) and Subsystem (Aziz et al., 2008)). The abundance was weighted by the coverage of the genes (calculated during assembly) or proteins (number of peptides identified) assigned to this function. Metagenomic data were normalized using single copy genes (see Supplementary Information) and metaproteomic data were standardized to the relative proportion of all functions detected.
Data analysis
Pairwise statistical comparisons between the healthy/intermediate, intermediate/necrotic and healthy/necrotic samples were conducted using a modified R script for MetaStats (White et al., 2009). Significantly different functions annotated by COG, Pfam, and Subsystem, respectively, were used for sample clustering using Primer 6 (PRIMER-E Ltd, Lutton, UK). Heatmaps were generated using Cluster 3.0 (de Hoon et al., 2004) and Java TreeView (Saldanha, 2004). Multi-dimensional scaling plots of the GeXP, terminal restriction fragment length polymorphism, metagenomic and metaproteomic results were generated with Primer 6.
Results and discussion
Elevated temperature results in sponge necrosis and changes in stress-related gene expression
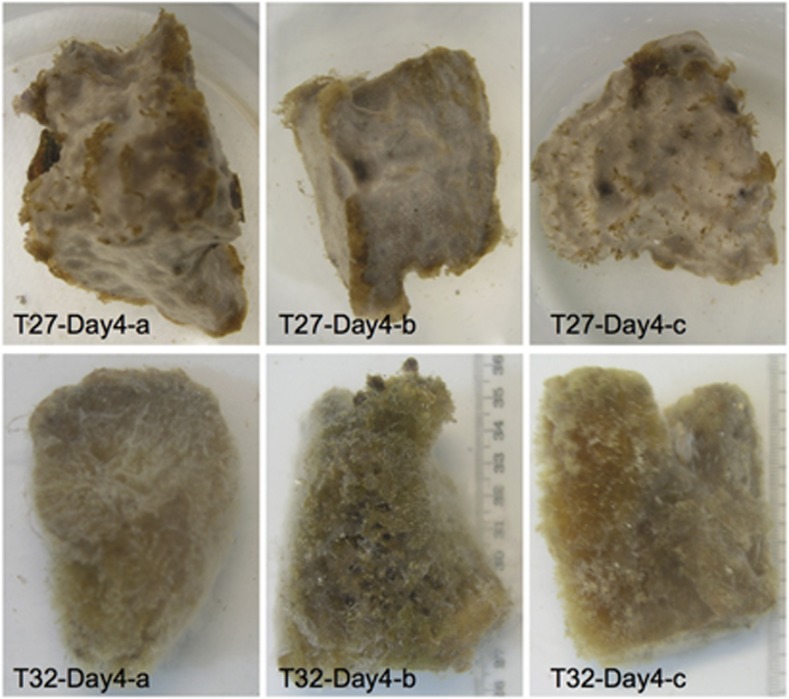
The experimental design used 21 R. odorabile clones that were maintained in aquaria at 27 °C and 32 °C (Table 1, see Supplementary Information for details). All sponge clones from the 27 °C treatment remained healthy throughout the experiment, whereas some clones from the 32 °C treatment suffered substantial tissue necrosis (decolorization and appearance of exposed skeletal fibers) after 3–4 days. Samples collected on day 4 were photographed (Figure 1). Although T32-Day4-a had lost most of its pinacoderm and appeared to be dead, two other samples (T32-Day4-b and T32-Day4-c) still retained a visible amount of healthy tissue (Figure 1). These observations are consistent with previous observations, where R. odorabile began to eject small amounts of cellular material and exhibited surface necrosis after 24 h at 33 °C and major tissue necrosis, resulting in the protrusion of skeletal fibers, after three days (Webster et al., 2008). On the basis of such morphological observations, we could define three health stages in the 21 collected clones: fully necrotic (T32-Day3-c and T32-Day4-a), intermediate/partially necrotic (T32-Day4-b and T32-Day4-c), and healthy (that is, all other clones) (Table 1).
Figure 1.
Morphological changes observed in sponge R. odorabile clones taken on day 4. T27-Day4-a, T27-Day4-b and T27-Day4-c present the healthy sponges. T32-Day4-b and T32-Day4-c present the sponges in intermediate health state. T32-Day4-a presents the necrotic sponges.
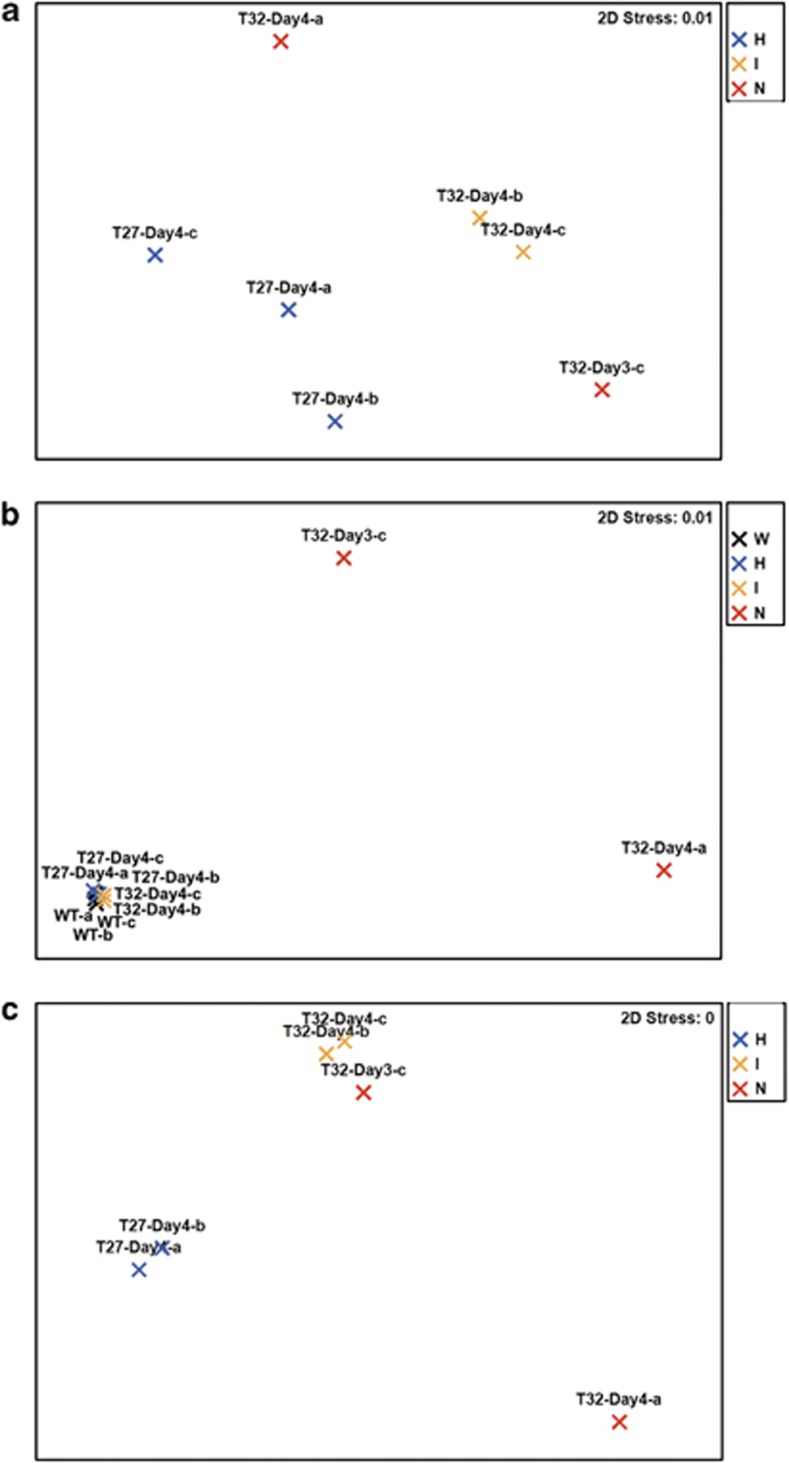
To further define the physiological state of sponges in these three groups, we investigated the expression profiles of 23 selected host genes using multiplexed reverse transcription quantitative PCR (see Materials and methods). Three samples on day 4 at 27 °C were used as representatives of the healthy group (Figure 2a). Ordination analysis of the overall expression profiles showed a clear distinction between the intermediate and necrotic samples, which were all exposed to 32 °C, and the healthy sponge clones at 27 °C (Figure 2a). The high-temperature samples were characterized by a statistically significant decrease in the expression of genes involved in cytoskeletal/skeletal structures (α-, β-tubulin, B-thymosin, radial spoke protein) and protein synthesis/degradation (cyclophilin NIMA-interacting 4, elongation factor Tu, ribosomal protein S9), which is consistent with the morphological changes observed (Supplementary Table S1, Figure 1). Genes involved in signal transduction (calmodulin) and detoxification (glutathione-S-transferase) also showed lower expression, which indicates stress-induced changes in gene regulation and sponge metabolism. Expression of the Hsp 70 and apoptosis-linked genes increased in sponges at 32 °C when compared with healthy sponges, demonstrating a direct molecular response to heat stress (Supplementary Table S1). These results are consistent with our recent studies of thermal stress responses in R. odorabile during different life stages based on quantitative PCR (Pantile and Webster, 2011) and multiplexed reverse transcription quantitative PCR (Webster et al., In Review). In these two studies, the immediate cellular response to thermal stress was also an increased production of Hsp transcripts, which, due to the energetic cost of expression, might compromise normal cellular functions (Feder and Hofmann, 1999).
Figure 2.
Sample multi-dimensional scaling clustering based on host expression, community functional composition and community expression. (a) GeXP. (b) Metagenome annotated by COG. (c) Metaproteome annotated by COG. Black: wild-type samples (W); blue: healthy samples (H); yellow: intermediate samples (I); red: necrotic samples (N).
Together our experiments showed reproducible changes in morphology and gene expression for R. odorabile in response to elevated temperature. Sponges exposed to 32 °C (that is, the intermediate group and the necrotic group) showed a clear molecular response related to heat stress (for example, Hsp expression) and ultimately experience cellular apoptosis and tissue-wide necrosis.
Changes in structure of the microbial community
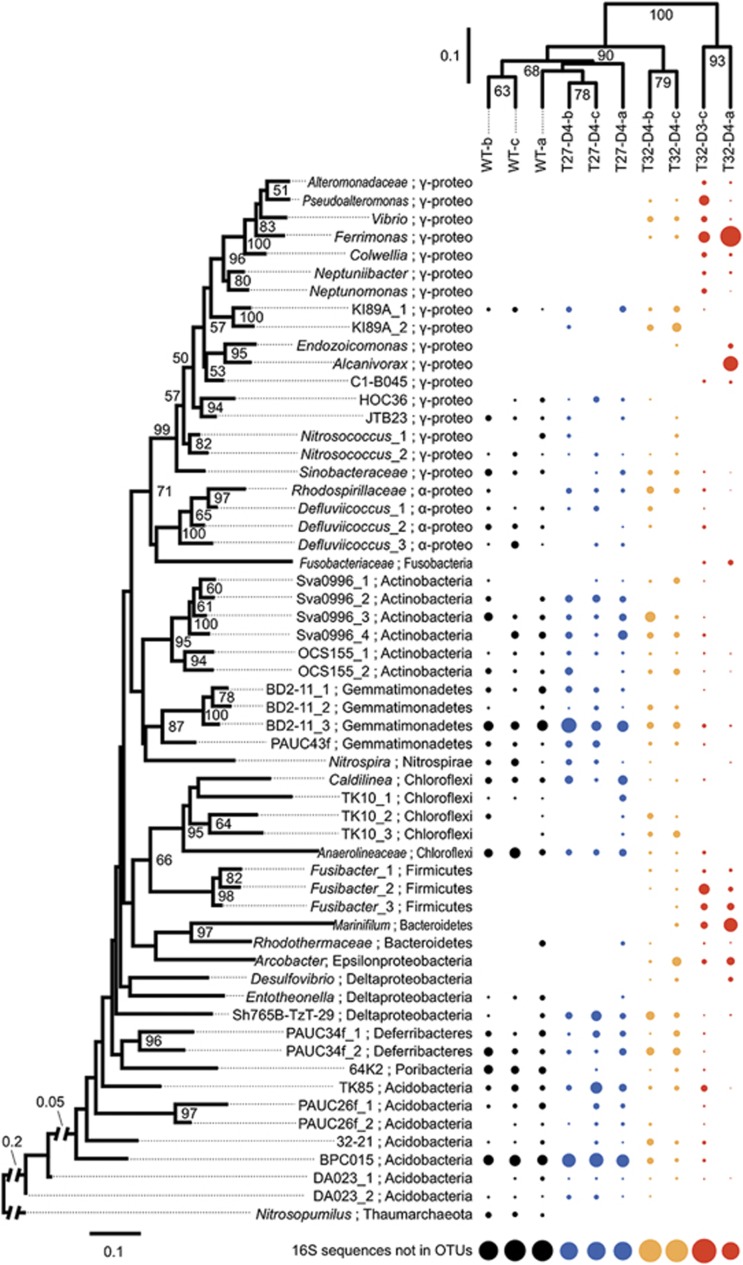
To investigate if the stress response of R. odorabile occurred in conjunction with a shift in the microbial population, we investigated the community structure with terminal restriction fragment length polymorphism of the 16S rRNA gene for all the 21 samples listed in Table 1 (see Supplementary Information, Supplementary Figure S1), and shotgun sequencing of metagenomic DNA for seven representatives in the three groups and three wild-type samples (see Figure 3, Materials and methods). Archaeal and bacterial community profiles based on the 16S rRNA gene were reconstructed from the metagenomic data sets and generated 131 operational taxonomic units of the 16S rRNA genes with phylogenetic distance cut-off of 0.03 (Materials and methods). A proportion of metagenomic shotgun reads for the 16S rRNA gene did not assemble into sufficiently long contigs for phylogenetic analysis and hence, was not included in our analysis. These 16S rRNA gene containing reads come from low-abundance organisms (Fan et al. 2012a) and therefore, our analysis mainly describes the changes in the abundant microbial community members in each sample.
Figure 3.
Shift in the bacterial community of R. odorabile. The relative abundance (dot size) of the 58 most abundant operational taxonomic units (OTUs) (according to the sum of the relative abundance across all samples) is illustrated. Phylogenetic distance cut-off for OTU generation is 0.03. Maximum-likelihood tree of the OTUs (left) is supported by 1 000 replications of bootstrapping (percentage greater than 50% are given) and rooted with the archaeon Nitrosopumilus. Samples are clustered based on the phylogenetic relationships of all their OTUs using the weighted Unifrac algorithm with 1000 rounds of Jackknife (percentage values shown in nodes). ‘16S rRNA sequences not in OTUs' reflects those reads that fail to assemble into contigs used for OTU generation. Black, wild-type group; blue, healthy group; yellow, intermediate group; red, necrotic group.
Comparison of healthy in vitro samples with R. odorabile samples taken in the field showed high similarity between microbial communities in the laboratory and the natural environment (Figure 3). This is consistent with previous observations on the short-term maintenance of this sponge in aquaria (Webster et al., 2011). Only minor differences in operational taxonomic unit abundance (for example, Defluviicoccus, Nitrosopumilus) were observed, hence our experimental sponges reflect a typical symbiont-system in nature.
The clustering of samples based on the abundance of operational taxonomic units was consistent with the grouping into healthy, intermediate and necrotic sponges, and was supported by high Jackknife confidence values (Figure 3). The same grouping profile was observed for the terminal restriction fragment length polymorphism analysis (see Supplementary Information, Supplementary Figure S1). Microbial community composition of the intermediate samples showed a large overlap with that of the healthy samples, but also showed some similarity with the microbial community of the necrotic samples. The abundance of certain sponge–associated phylotypes decreased (for example, BD2-11, Gp9 (BPC015), Gp10 (TK85), and Anaerolineaceae) in sponges at the intermediate health state, while other phylotypes (for example, some proteobacteria) increased in abundance or could be newly detected (Figure 3). These novel and low-abundance phylotypes in intermediate samples eventually dominated the communities of necrotic sponges and belonged to the taxa of Vibrionaceae, Pseudoalteromonas, Colwelliaceae, Ferrimonas, Oceanospirillaceae 2, Endozoicomonas, the BD107 clade, Arcobacter, Marinifillum and Fusibacter. These taxonomic shifts resulted in the microbial communities of necrotic sponges being clearly distinct from those of healthy and intermediate samples, and in the case of the necrotic sample T32-Day4-a, also lead to a community with lower operational taxonomic unit richness (see Supplementary Information). A recent investigation of thermal stress effects on the total and/or active bacteria in R. odorabile found that sublethal temperature (31 °C) had no effect on the community, but a shift consistent with the findings of this study occurred in necrotic sponges at 32 °C (Simister et al., 2012). Interestingly, Simister et al. found that whilst two-thirds of the DNA and RNA sequences derived from healthy sponges could be assigned to previously described clusters of sponge-specific sequences (Taylor et al., 2007), only 12% of the sequences originating from necrotic sponges resided within sponge-specific clusters (Simister et al., 2012).
Overall, exposure to elevated sea surface temperature increased the microbial diversity within R. odorabile in the intermediate health state as newly colonizing or low-abundance bacteria are able to grow with the community of existing symbionts, which are still present but at lower abundance (Figure 3, Supplementary Figure S5). Diversity then decreased in the necrotic state as sponge symbionts were lost or outcompeted by other bacteria that benefited from the necrotic sponge environment (see below).
Symbiotic functions are lost during temperature-induced shift in community composition
The observations above show that microbial phylotypes, which are typically associated with R. odorabile, were lost during temperature stress and consequently replaced by a new set of microorganisms. Work on zooxanthella species in corals (Rowan et al., 1997; Baker, 2001) and microbial communities in soil (Girvan et al., 2005) has shown that perturbation can simply cause a replacement of resident microorganisms with functionally equivalent species, which are resistant or adapted to the stress. In other cases, stress was suspected to allow for the introduction of pathogenic strains in the microbial communities of marine invertebrates (Webster, 2007; Rosenberg et al., 2009). To investigate if functional changes accompany the taxonomic shift observed for R. odorabile and to characterize the newly introduced taxa, we conducted functional annotation and comparison for the metagenomic samples (Supplementary Table S3).
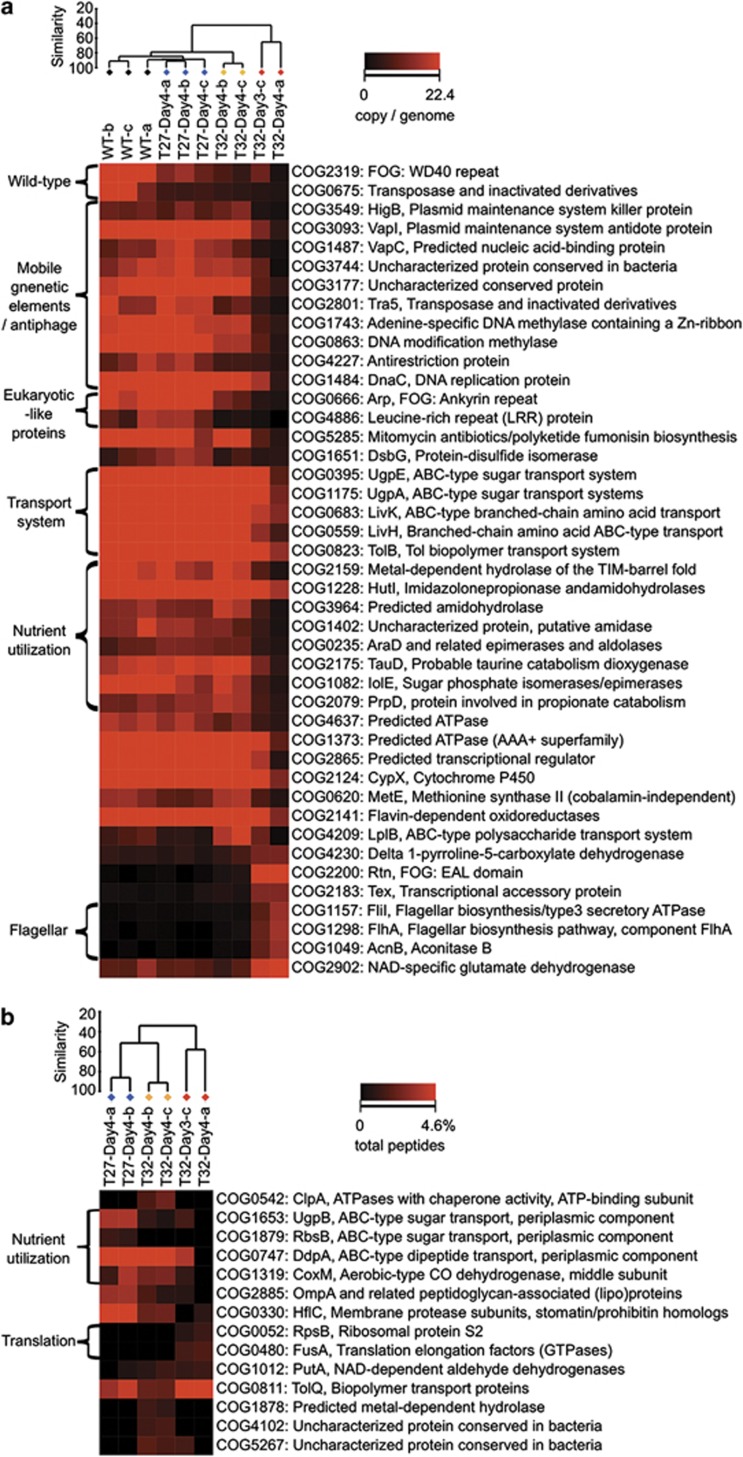
Multi-dimensional scaling shows that the functional profiles of wild-type and healthy sponges from the 27 °C treatment were highly similar (Figure 2b, Supplementary Figures S2AB). They only differed in the abundance of WD40 repeat proteins (COG2319, PF00400) and a transposase (COG0675, PF01385) (Figure 4a, Supplementary Figures S3AB). This reiterates that the aquarium-maintained R. odorabile are representative of wild-type sponges. Microbial communities of these native and healthy samples are abundant in functions that have been also described to be overrepresented in the microbiomes of the sponges Cymbastela concentrica, C. coralliophila, Stylissa sp. 445, Scopalina sp. and Tedania anhelans (Thomas et al., 2010; Fan et al., 2012b). Among these are mobile genetic elements (plasmids and transposases), restriction-modification systems and clustered regularly interspaced short palindromic repeats, which might control genetic exchange in the symbiont community, and eukaryotic-like proteins, which can potentially be used by symbionts to manipulate their host (Thomas et al., 2010; Fan et al., 2012b). Other functions that are often found in sponge symbionts are represented by genes involved in membrane transport, substrate utilization, cell signaling, regulation, stress response mechanisms and cell–cell adhesion. All of these specific functions were still present in sponges at the intermediate health state, resulting in tight clustering of these samples with the healthy group (Figure 4a, Supplementary Figures S2AB, S3AB). In contrast, functions related to symbiosis were dramatically reduced in the communities from necrotic sponges (Figure 4a, Supplementary Figures S3AB) demonstrating that they are not composed of ‘typical' symbionts. A few functions increased in abundance, and specifically, genes encoding for EAL domain proteins (COG2200) were enriched in necrotic sponges (Figure 4a). The EAL domain may degrade cyclic diguanylate, which is an important intracellular signaling molecule that regulates the switch between a motile and surface-associated lifestyle as well as virulence traits in many free-living bacteria (Hengge, 2009). Also overrepresented in the necrotic samples was the function of flagellar biosynthesis, (COG1298, COG1157, COG1049, PF00771) indicating that many of the newly colonizing bacteria were motile. Other pathogenic functions or virulence proteins, including those identified in temperature-stressed corals (Vega Thurber et al., 2009), were not detected in the necrotic sponge samples (Figure 4a, Supplementary Figures S3AB).
Figure 4.
Abundance of specific functions (annotated by COG) in the metagenomes (a) and metaproteomes (b) of microbial communities in normal and stressed sponge. Samples are clustered using Bray–Curtis similarity and group averages. The heatmap is plotted according to the abundance of each function (copy per genome for metagenome and percentage in all peptides) per sample. Black, the wild-type group. Blue, the healthy group. Orange, the intermediate group. Red, the necrotic group.
Our data demonstrates that temperature stress changes the microbial community from one with predominantly symbiotic functions to one that is characterized by motile bacteria capable of using cyclic diguanylate as a gene expression regulator. Importantly, the genetic potential for functions that may be crucial for sponge symbiosis was still largely present in the intermediate samples, yet the sponge host had already undergone a stress-related gene expression response (Figure 2a). For this reason, we predicted that the microbial communities of sponges in the intermediate health state, while being genetically similar to the communities in healthy sponges, would nevertheless have altered their expression pattern. In particular, we hypothesized altered expression of functions related to symbiosis or temperature stress, and this was investigated with metaproteomics (Supplementary Table S4).
Metaproteomic analysis reveals expression changes related to stress and symbiosis function
Comparison of the metaproteomic data sets showed that the microbial communities of sponges in the intermediate health state had expression profiles much more closely related to necrotic samples than to healthy sponges (Figure 2c, Supplementary S2CD). This confirms that the microbial communities of the intermediate samples had undergone substantial changes in protein expression consistent with the trend observed in the sponge host (Figure 2a). Annotation of the expressed proteins based on COG classification revealed that overall functions, like amino acid transport and metabolism were relatively overrepresented in the healthy group, while post-translational modification was overrepresented in the intermediate group. In contrast, translation, transcription and intracellular trafficking were overrepresented in the necrotic sponges (Supplementary Figure S4).
Further detailed analysis revealed clear support for a heat stress in the microbial communities of intermediate samples (Figure 4b, Supplementary Figures S3CD). Specifically, expression of the ClpA protein (COG0542, PF02861, Subsystem: proteolysis in bacteria, ATP-dependent), belonging to the heat-shock protein family Hsp100, was upregulated. Clp acts as a chaperone to stabilize and refold proteins (Zavilgelsky et al., 2002) or to deliver unfolded proteins to the peptidases for degradation (Weber-Ban et al., 1999). Expression of elongation factor Tu domain 2 was also increased in intermediate samples (Supplementary Figure S3C), and this protein has an established chaperone-like function (Caldas et al., 1998), in addition to its chain-elongation role in translation. Other potential chaperone activities were also detected (PF08406). Small increases in physiological temperature can cause protein unfolding, entanglement and unspecific aggregation, and the microbial community of heat-treated R. odorabile seemed to have experienced and responded to this thermal stress.
Elevated temperature also decreased functions related to the metabolic interactions between symbionts and the host. Nutritional interdependence between partners in symbiosis often occurs during coevolution (Taylor et al., 2007; Fan et al., 2012b) and is, for example, reflected in the high abundance and diversity of transport proteins found in the metagenomes of healthy R. odorabile (Figure 4a, Supplementary Figures S3AB) and other sponges (Fan et al., 2012b), the metaproteome the sponge C. concentrica (Liu et al., 2012) and the metatranscriptome of the sponge Geodia barretti (Radax et al., 2012). Consistent with these observations, we found transporters involved in the uptake of sugars, peptides and other substrates, to be highly expressed in healthy R. odorabile (Figure 4b, Supplementary Figures S3CD). Specifically, this included UgpB and RbsB for sugar utilization, ABC-type transporters for dipeptides, oligopeptides, branched-chain amino acids and trehalose. Expression of these transporters was largely decreased in sponges with intermediate health, clearly reflecting the shutdown of metabolic processes and interactions within the sponge holobiont. For example, trehalose is often accumulated in the cytoplasm of animal cells, including the sponge Suberites domuncula (Bachinski et al., 1997), to create an osmotic equilibrium with the aqueous surrounding (Eleutherio et al., 1993). Bacteria can potentially benefit from trehalose as a carbon or energy source as well as an osmolyte (Jensen et al., 2002). Reduced trehalose utilization by temperature-stressed symbionts may reflect a lack of production by the host, which would inevitably lead to a reduced nutrient availability for the microbial community.
Interactions between microbial symbionts and eukaryotic cells may also require specific proteins involved in cell–cell contact (Thomas et al., 2010; Fan et al., 2012b). In the microbial community of healthy sponges, the expression of fibronectin type III domains (PF00041), domains related to collagen binding (PF05738, PF01391) and membrane proteins (COG2885, PF00691) may be related to cell–cell interactions (Figure 4b, Supplementary Figures S3CD). Expression of these eukaryotic-like proteins was reduced in intermediate and necrotic samples, which could negatively affect the interactive ability of symbionts and their host. The expression of peroxiredoxin (PF10417) and the eukaryotic-like proteins NIPSNAP (PF07978) with antimacrophage activity (Lee et al., 2002) may be related to symbiont protection, and their lower abundance in intermediate samples potentially reduces protection against host oxidative stress and phagocytosis.
Overall, these observations support a scenario whereby symbionts still persist in the sponges during the intermediate health state, but no longer carry out normal symbiotic functions. The disruption of these features likely unbalances a well-tuned association based on nutritional interdependence and molecular interactions and ultimately leads to a collapse of the holobiont and necrosis of the sponge. Other aspects of the expression changes are discussed in the Supplementary Information.
Opportunistic scavengers dominated the necrotic sponges
Consistent with the metagenomic results, no known virulence-related proteins were expressed in intermediate or fully necrotic samples (Figure 4b, Supplementary Figures S3CD). It is therefore, unlikely that the decreased health of the intermediate or necrotic sponges is a direct result of pathogenic bacteria or specific virulence mechanisms. Instead, the new taxa found in the necrotic samples appeared to have a high growth rate and metabolic activity compared with native symbionts. This is supported by the high abundance of RNA polymerase (PF00562, PF04997), ribosomal protein S2 (for example, COG0052) and translation elongation factors (for example, COG0480) indicating a high rate of transcription and translation, which is generally related to high growth rates. Numerous studies have shown that microbial symbionts derive energy from the oxidation of ammonium that is secreted by sponges (for example, (Mohamed et al., 2010; Radax et al., 2012)). However, in the necrotic samples, ammonium appeared to be preferentially assimilated through the expression of NADP-specific glutamate dehydrogenase (Subsystem: arginine and ornithine degradation) (Bonete et al., 1996), presumably by the new taxa. This enzyme requires a lower energy cost than the ubiquitous glutamine/glutamate synthetase pathway and is often used under conditions of nitrogen excess (Belanger and Hatfull, 1999).
A stressed sponge exhibiting minor necrosis (as observed in the intermediate samples) would be a rich source of nutrients. Bacteria that can quickly move towards the nutrient source, colonize the tissue, rapidly grow and assimilate key nutrients will rapidly outcompete the native symbiont community. Our observations on the functional gene content and protein expression are consistent with such a scavenging tactic. Bursts of nutrients also occur in the marine environment during algal blooms, (Loureiro et al., 2011) and some of newly colonizing sponge taxa, such as Pseudoalteromonas and Vibrio, are often associated with decaying matter and blooms (Kelly and Chistoserdov, 2001; Alderkamp et al., 2007).
Conclusion
Substantial research effort is currently being directed towards understanding the increasing disease outbreaks in the world's oceans, especially for corals and sponges (Reshef et al., 2006; Webster, 2007; Bourne et al., 2009; Rosenberg et al., 2009). Several hypotheses have been proposed to explain large-scale mortality events, disease outbreaks and bleaching in corals and other marine invertebrates. These include models based on photoinhibition (Warner et al., 1999; Cebrian et al., 2011), energetic constraints (Coma et al., 2009) and antibiotic production (Mao-Jones et al., 2010). The mathematical modeling proposed by Mao-Jones et al. predicted that a temperature-induced decrease in antibiotic production in corals would cause a general shift in microbial communities and the invasion of pathogens that effectively compete for nutrients (Mao-Jones et al., 2010). Our observations for R. odorabile support some aspects of this model, however, they also indicate that a decline in symbiotic interactions is likely to be a key factor in the loss of holobiont function. Specifically, an intermediate state exists, where elevated temperature causes changes in gene expression in both the host and the symbiont community. This is characterized by the expression of a heat-shock response and a loss of symbiosis function. Changes in the community composition occur secondarily to this stress process. Given the prevalence of microbial symbiosis in marine invertebrates, the mechanism we propose here would be broadly applicable.
Previous studies have indicated that the invasion of pathogenic microorganisms is responsible for the declining health in some marine invertebrate holobionts (Webster et al., 2002; Rosenberg et al., 2009; Vega Thurber et al., 2009), while other studies found no support for this (Bourne, 2005; Lesser et al., 2007; Sussman et al., 2008; Angermeier et al., 2011). Our characterization of the temperature-induced decay of R. odorabile showed no evidence for virulence or pathogens, but instead found support for a community shift towards motile and nutrient-scavenging bacteria with high growth activity. These bacteria would opportunistically take advantage of the niches provided by the interruption of symbiosis. We propose that these opportunists might thus have lower host specificity than the actual symbionts. This prediction is consistent with the broad similarity of disease-related microorganisms found in various marine hosts, including sponges, gorgonian sea fans and corals (Webster et al., 2008; Ein-Gil et al., 2009; Negandhi et al., 2010).
The mechanisms underpinning the assembly and stability of microbial communities have received recent attention by ecologists (Allison and Martiny, 2008; Little et al., 2008; Jones et al., 2009). A general conclusion is that community diversity and functional redundancy is often positively correlated to community stability (Girvan et al., 2005; Allison and Martiny, 2008; Robinson et al., 2010) and our data give experimental support for this. In obligate symbiotic systems, such as sponge holobionts, microbial community members are highly interdependent and potentially very specialized to particular niches (Thomas et al., 2010; Fan et al., 2012b; Liu et al., 2012). This high degree of interdependence and specialization would limit functional redundancy within the community, making it unlikely that the loss of an essential symbiont would be compensated by the presence or introduction of a functionally equivalent species (Allison and Martiny, 2008; Mao-Jones et al., 2010), even when species richness is high. In contrast, communities with many functionally redundant members, but low total species richness, may still maintain function when exposed to stressful conditions. Therefore, obligate symbiotic systems are likely to be very sensitive to environmental perturbation, highlighted here in the R. odorabile holobiont, which collapses at 32 °C, only 2–3 °C above the average annual maximal temperature of their habitat on the GBR (Webster et al., 2008).
Environmental perturbations, including the effects of climate change and urbanization, are likely to continue to negatively impact on marine species and the ocean's ecosystem (Harvell et al., 2004; Lafferty et al., 2004; Plowright et al., 2008). Although flow-through experimental systems cannot be expected to perfectly mimic reef situations, they have a major role to play in understanding cause-effect relationships and sensitivity to pressures such as elevated sea temperatures. Controlled experiments that simultaneously analyze the structure and function of all symbiosis partners have the potential to reveal, which aspects of the holobiont are most sensitive to the environmental change. The multifaceted approach described here would help to discern the mechanisms behind the many models proposed for a range of marine species and their disease syndromes (Reshef et al., 2006; Webster, 2007; Bourne et al., 2009; Coma et al., 2009; Rosenberg et al., 2009; Mao-Jones et al., 2010; Cebrian et al., 2011). The impact of elevated temperature on a model sponge holobiont suggests that in invertebrates with similar highly interdependent microbial partnerships, environmental change may irreversibly disrupt the symbiosis with significant implications for host health.
Acknowledgments
We thank Florita Flores, Emmanuelle Botte and Raffaella Pantile (AIMS) and Kerensa McElroy, Martin Thompson and Ling Zhong (UNSW) for technical advice and support. We also acknowledge the J. Craig Venter Institute‘s Joint Technology Center, under the leadership of Yu-Hui Rogers and the assistance of Matt Lewis for producing the sequencing data. This work was funded by the Australian Research Council, the Gordon and Betty Moore Foundation, the Center for Marine Bio-Innovation and the JCVI, USA.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alderkamp AC, van Rijssel M, Bolhuis H. Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol. 2007;59:108–117. doi: 10.1111/j.1574-6941.2006.00219.x. [DOI] [PubMed] [Google Scholar]
- Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105 (Suppl 1:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermeier H, Kamke J, Abdelmohsen UR, Krohne G, Pawlik JR, Lindquist NL, et al. The pathology of sponge orange band disease affecting the Caribbean barrel sponge Xestospongia muta. FEMS Microbiol Ecol. 2011;75:218–230. doi: 10.1111/j.1574-6941.2010.01001.x. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachinski N, Koziol C, Batel R, Labura Z, Schröder HC, Müller WE. Immediate early response of the marine sponge Suberites domuncula to heat stress: reduction of trehalose and glutathione concentrations and glutathione S-transferase activity. J Exp Mar Bio Ecol. 1997;210:129–141. [Google Scholar]
- Baker AC. Reef corals bleach to survive change. Nature. 2001;411:765–766. doi: 10.1038/35081151. [DOI] [PubMed] [Google Scholar]
- Banin E, Vassilakos D, Orr E, Martinez RJ, Rosenberg E. Superoxide dismutase is a virulence factor produced by the coral bleaching pathogen Vibrio shiloi. Curr Microbiol. 2003;46:418–422. doi: 10.1007/s00284-002-3912-5. [DOI] [PubMed] [Google Scholar]
- Belanger AE, Hatfull GF. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J Bacteriol. 1999;181:6670–6678. doi: 10.1128/jb.181.21.6670-6678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonete MJ, Perez-Pomares F, Ferrer J, Camacho ML. NAD-glutamate dehydrogenase from Halobacterium halobium: inhibition and activation by TCA intermediates and amino acids. Biochim Biophys Acta. 1996;1289:14–24. doi: 10.1016/0304-4165(95)00134-4. [DOI] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne DG. Microbiological assessment of a disease outbreak on corals from Magnetic Island (Great Barrier Reef, Australia) Coral Reef. 2005;24:304–312. [Google Scholar]
- Bourne DG, Garren M, Work TM, Rosenberg E, Smith GW, Harvell CD. Microbial disease and the coral holobiont. Trends Microbiol. 2009;17:554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Caldas TD, El Yaagoubi A, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- Cebrian E, Uriz MJ, Garrabou J, Ballesteros E. Sponge mass mortalities in a warming Mediterranean Sea: are cyanobacteria-harboring species worse off. PLoS One. 2011;6:e20211. doi: 10.1371/journal.pone.0020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coma R, Ribes M, Serrano E, Jiménez E, Salat J, Pascual J. Global warming-enhanced stratification and mass mortality events in the Mediterranean. Proc Natl Acad Sci USA. 2009;106:6176–6181. doi: 10.1073/pnas.0805801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ein-Gil N, Ilan M, Carmeli S, Smith GW, Pawlik JR, Yarden O. Presence of Aspergillus sydowii, a pathogen of gorgonian sea fans in the marine sponge Spongia obscura. ISME J. 2009;3:752–755. doi: 10.1038/ismej.2009.18. [DOI] [PubMed] [Google Scholar]
- Eleutherio EC, Araujo PS, Panek AD. Protective role of trehalose during heat stress in Saccharomyces cerevisiae. Cryobiology. 1993;30:591–596. doi: 10.1006/cryo.1993.1061. [DOI] [PubMed] [Google Scholar]
- Fan L, McElroy K, Thomas T. Reconstruction of ribosomal RNA genes from metagenomic data. PLoS One. 2012a;7:e39948. doi: 10.1371/journal.pone.0039948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, et al. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA. 2012b;109:E1878–E1887. doi: 10.1073/pnas.1203287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan MS, Campbell CD, Killham K, Prosser JI, Glover LA. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol. 2005;7:301–313. doi: 10.1111/j.1462-2920.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- Harvell D, Aronson R, Baron N, Connell J, Dobson A, Ellner S, et al. The rising tide of ocean diseases: unsolved problems and research priorities. Front Ecol Environ. 2004;2:375–382. [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Jensen JB, Peters NK, Bhuvaneswari TV. Redundancy in periplasmic binding protein-dependent transport systems for trehalose, sucrose, and maltose in Sinorhizobium meliloti. J Bacteriol. 2002;184:2978–2986. doi: 10.1128/JB.184.11.2978-2986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Newton RJ, McMahon KD. Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ Microbiol. 2009;11:2463–2472. doi: 10.1111/j.1462-2920.2009.01977.x. [DOI] [PubMed] [Google Scholar]
- Kelly KM, Chistoserdov AY. Phylogenetic analysis of the succession of bacterial communities in the Great South Bay (Long Island) FEMS Microbiol Ecol. 2001;35:85–95. doi: 10.1111/j.1574-6941.2001.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH, et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012;6:835–846. doi: 10.1038/ismej.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty KD, Porter JW, Ford SE. Are diseases increasing in the ocean. Annu Rev Ecol Evol Syst. 2004;35:31–54. [Google Scholar]
- Lee AH, Zareei MP, Daefler S. Identification of a NIPSNAP homologue as host cell target for Salmonella virulence protein SpiC. Cell Microbiol. 2002;4:739–750. doi: 10.1046/j.1462-5822.2002.00225.x. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O. Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Bio Ecol. 2007;346:36–44. [Google Scholar]
- Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- Liu M, Fan L, Zhong L, Kjelleberg S, Thomas T. Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 2012;6:1515–1525. doi: 10.1038/ismej.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro S, Reñé A, Garcés E, Camp J, Vaqué D. Harmful algal blooms (HABs), dissolved organic matter (DOM), and planktonic microbial community dynamics at a near-shore and a harbour station influenced by upwelling (SW Iberian Peninsula) J Sea Res. 2011;65:401–413. [Google Scholar]
- Mao-Jones J, Ritchie KB, Jones LE, Ellner SP. How microbial community composition regulates coral disease development. PLoS Biol. 2010;8:e1000345. doi: 10.1371/journal.pbio.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Saito K, Tal Y, Hill RT. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 2010;4:38–48. doi: 10.1038/ismej.2009.84. [DOI] [PubMed] [Google Scholar]
- Negandhi K, Blackwelder P, Ereskovsky A, Lopez J. Florida reef sponges harbor coral disease-associated microbes. Symbiosis. 2010;51:117–129. [Google Scholar]
- Pantile R, Webster NS. Strict thermal threshold identified by quantitative PCR in the sponge Rhopaloeides odorabile. Mar Ecol Prog Ser. 2011;431:97–105. [Google Scholar]
- Plowright RK, Sokolow SH, Gorman ME, Daszak P, Foley JE, Plowright RK, et al. Causal inference in disease ecology: investigating ecological drivers of disease emergence. Front Ecol Environ. 2008;6:420–429. [Google Scholar]
- Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ Microbiol. 2012;14:909–923. doi: 10.1111/j.1462-2920.2011.02661.x. [DOI] [PubMed] [Google Scholar]
- Rai AJ, Kamath RM, Gerald W, Fleisher M. Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal Bioanal Chem. 2009;393:1505–1511. doi: 10.1007/s00216-008-2436-7. [DOI] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Kushmaro A, Kramarsky-Winter E, Banin E, Yossi L. The role of microorganisms in coral bleaching. ISME J. 2009;3:139–146. doi: 10.1038/ismej.2008.104. [DOI] [PubMed] [Google Scholar]
- Rowan R, Knowlton N, Baker A, Jara J. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature. 1997;388:265–269. doi: 10.1038/40843. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Simister R, Taylor MW, Tsai P, Fan L, Bruxner TJ, Crowe ML, et al. Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ Microbiol. 2012;14:3232–3246. doi: 10.1111/1462-2920.12010. [DOI] [PubMed] [Google Scholar]
- Souter P, Bay LK, Andreakis N, Császár N, Seneca FO, van Oppen MJ. A multilocus, temperature stress-related gene expression profile assay in Acropora millepora, a dominant reef-building coral. Mol Ecol Resour. 2011;11:328–334. doi: 10.1111/j.1755-0998.2010.02923.x. [DOI] [PubMed] [Google Scholar]
- Sussman M, Willis BL, Victor S, Bourne DG. Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS One. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Rusch D, Demaere MZ, Yung PY, Lewis M, Halpern A, et al. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010;4:1557–1567. doi: 10.1038/ismej.2010.74. [DOI] [PubMed] [Google Scholar]
- Toren A, Landau L, Kushmaro A, Loya Y, Rosenberg E. Effect of temperature on adhesion of Vibrio Strain AK-1 to Oculina patagonica and on coral bleaching. Appl Environ Microbiol. 1998;64:1379–1384. doi: 10.1128/aem.64.4.1379-1384.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Warner ME, Fitt WK, Schmidt GW. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc Natl Acad Sci USA. 1999;96:8007–8012. doi: 10.1073/pnas.96.14.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Webster NS, Negri AP, Webb RI, Hill RT. A spongin-boring alpha-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar Ecol Prog Ser. 2002;232:305–309. [Google Scholar]
- Webster NS. Sponge disease: a global threat. Environ Microbiol. 2007;9:1363–1375. doi: 10.1111/j.1462-2920.2007.01303.x. [DOI] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Negri AP. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2008;2:830–842. doi: 10.1038/ismej.2008.42. [DOI] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Soo R, Anthony SL, Battershill CN, Whalan S, et al. Bacterial community dynamics in the marine sponge Rhopaloeides odorabile under in situ and ex situ cultivation. Mar Biotechnol (NY) 2011;13:296–304. doi: 10.1007/s10126-010-9300-4. [DOI] [PubMed] [Google Scholar]
- Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2011;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavilgelsky GB, Kotova VY, Mazhul' MM, Manukhov IV. Role of Hsp70 (DnaK-DnaJ-GrpE) and Hsp100 (ClpA and ClpB) chaperones in refolding and increased thermal stability of bacterial luciferases in Escherichia coli cells. Biochemistry (Mosc) 2002;67:986–992. doi: 10.1023/a:1020565701210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.