Abstract
The substrate partitioning of sympatric populations of freshwater bacterioplankton was studied via microautoradiography and fluorescence in situ hybridization. Fourteen radiolabeled tracers were used to assess microbial acquisition spectra of low-molecular-weight (LMW) organic compounds. The most abundant group, ac1 Actinobacteria, were highly active in leucine, thymidine and glucose assimilation, whereas Alphaproteobacteria from the LD12 lineage (the freshwater sister clade of SAR11) only weakly incorporated these tracers, but exhibited a distinct preference for glutamine and glutamate. Different Bacteroidetes showed contrasting uptake patterns: Flavobacteriales did not incorporate significant amounts of any LMW compound, and Cyclobacteriaceae were clearly specialized on leucine, glucose and arginine. Betaproteobacteria represented the most active and versatile bacterioplankton fraction and >90% of them could be assigned to eight species- to genus-like populations with contrasting substrate specialization. Limnohabitans sp. were the most abundant and active Betaproteobacteria, incorporating almost all tracers. While three closely related betaproteobacterial populations substantially differed in their uptake spectra, two more distantly related lineages had very similar preferences, and one population did not incorporate any tracer. The dominant phototrophic microorganism, the filamentous cyanobacterium Planktothrix rubescens, assimilated several substrates, whereas other (pico)cyanobacteria had no heterotrophic activity. The variable extent of specialization by the studied bacterial taxa on subsets of LMW compounds contrasts theoretical considerations about non-selective microbial substrate assimilation at oligotrophic conditions. This physiological niche separation might be one explanation for the coexistence of freshwater bacterioplankton species in a seemingly uniform environment.
Keywords: Betaproteobacteria , freshwater bacterioplankton, low-molecular-weight DOM/MAR-FISH, substrate uptake
Introduction
The free-living bacterial assemblages in freshwater lakes are mainly composed of small Actinobacteria (esp. the acI cluster; Warnecke et al., 2005; Allgaier and Grossart, 2006), of members of the LD12 cluster of Alphaproteobacteria (Salcher et al., 2011b), of Betaproteobacteria (Glöckner et al., 1999; Newton et al., 2011), and diverse Flavobacteria and Sphingobacteria (Eiler and Bertilsson, 2007; Newton et al., 2011). Improved techniques such as filtration-acclimatization or dilution to extinction have allowed for the cultivation of numerically important freshwater Betaproteobacteria (Limnohabitans sp. (Kasalický et al., 2010) and Polynucleobacter sp. (Hahn et al., 2009, 2012a)), of Flavobacteria (for example, F. dankookense, F. aquatile (Lee et al., 2011)), and of Sphingobacteria (for example, Algoriphagus aquatilis (Liu et al., 2009)), while the most abundant freshwater bacteria (acI Actinobacteria and LD12 Alphaproteobacteria) still lack cultivated representatives. Therefore, cultivation-independent approaches are essential in order to gain more information about the in situ (eco)physiology of these microbes.
However, it is challenging to link the identity of microbes with a specific set of metabolic processes in order to shed light on their potential role in the environment. Microautoradiography coupled to fluorescence in situ hybridization (MAR-FISH) is a powerful single-cell tool to study particular physiological properties of different microbes. Short incubation times with low concentrations of radiolabeled tracers allow an assessment of the uptake of defined substrates at close to in situ conditions. Since MAR-FISH sample preparation and evaluation is rather laborious, most studies so far have focused on a small number of tracers and/or microbial populations (Pérez et al., 2010; Salcher et al., 2010), which does not allow to draw conclusions about substrate partitioning by sympatric bacterioplankton taxa. Moreover, as FISH-based studies depend on the availability of specific rRNA-targeted oligonucleotide probes, many investigations using MAR-FISH have only analyzed uptake of broad phylogenetic lineages rather than of species- to genus-like bacterial populations (Pérez et al., 2010; Salcher et al., 2010). Leucine (Leu) and thymidine (TdR) are widely used as precursors for biomass synthesis and cell growth (Kirchman, 2001; Cottrell and Kirchman, 2003; Pérez et al., 2010). However, bacterial production cannot be solely assessed by using Leu or TdR, as only a minor proportion of obviously active bacteria is able to take up TdR (Pedros-Alio and Newell, 1989; Pérez et al., 2010), probably due to a lack in TdR transport systems or preferred de novo synthesis. Moreover, substantial differences in the incorporation of TdR and Leu were found for different freshwater microbial populations (Pérez et al., 2010).
Substrate-based niche partitioning of freshwater bacterioplankton populations has also been proposed for acetate, glucose and Leu incorporation (Buck et al., 2009), as well as for N-acetyl-glucosamine and Leu uptake (Eckert et al., 2012). Such a specialization is in contrast to the idea of non-selective microbial substrate assimilation (‘mixed-substrate-growth') under oligotrophic conditions (Egli, 2010). By contrast, the ‘paradox of the plankton' concept (Hutchinson, 1961) suggests niche differentiation of coexisting species in an seemingly unstructured environment such as freshwaters where competitive exclusion could otherwise not explain the high observed microbial diversity.
We compared the substrate uptake spectra of prominent bacterioplankton populations in an oligomesotrophic prealpine lake (Lake Zurich, Switzerland). Specifically, we tested if these microbial lineages mainly competed for the same substrates or if their co-occurrence might be also related to specific substrate niches. Fourteen monomeric radiolabeled tracers were used to determine microbial acquisition of various low-molecular-weight (LMW) organic compounds and substrate specialization of different co-occurring microbial populations. As Betaproteobacteria were found to incorporate a wide range of the applied LMW tracers, we designed new oligonucleotide probes targeting the 16S rRNA of species-like betaproteobacterial populations to study the most active microbes in more detail.
Materials and methods
Study site and sampling
Lake Zurich is a large, deep (136 m), prealpine, oligomesotrophic lake, characterized by persistent annual blooms of the toxic cyanobacterium Planktothrix rubescens (Posch et al., 2012). Sampling took place on 16 September 2009 at the deepest part of the lake (136 m). Vertical profiles of temperature, conductivity, turbidity, oxygen and chlorophyll a content were recorded using a YSI multiprobe (Yellow Springs Instruments, model 6600, Yellow Springs, OH, USA) and a bbe fluoroprobe (TS-16-12, bbe Moldaenke GmbH, Schwentinental, Germany), respectively. The fluoroprobe was calibrated to distinguish different phytoplankton groups (that is, diatoms, cryptophytes, and green algae) and P. rubescens according to their respective pigment fluorescence spectra (Beutler et al., 2002; Salcher et al., 2011a). Water samples were taken from 0, 5, 10, 20, 30, 40, 60, 80 and 100 m depth for flow cytometry and FISH analyses. In all, 40 ml of water was fixed with formaldehyde (2% final concentration) for flow cytometry, and 5–10 ml was fixed with freshly prepared buffered paraformaldehyde (pH 7.4, 2% final concentration) for CARD (catalyzed reporter deposition)-FISH analyses. Chemical parameters (dissolved organic carbon, dissolved phosphorus, nitrate and ammonium concentrations) were determined by the Zurich Water Supply using standard techniques.
Sequence analysis of 16S rRNA genes, and probe design
In all, 250 ml of unfixed water samples from 10 m, 12.5 m and 15 m depth from 1 October 2007 were used for the construction of 3 16S rRNA gene clone libraries (see Salcher et al., 2011b; and Van den Wyngaert et al., 2011 for more details). Phylogenetic analyses were performed with the ARB software package (Ludwig et al., 2004) using the SILVA database SSU Ref release 106 (Pruesse et al., 2007). Sequences affiliated with different clades of Betaproteobacteria (>1200 nucleotides and with good quality scores) were selected for bootstrapped maximum likelihood analyses (RAxML, 100 repetitions) on a web server (Stamatakis et al., 2005). Nodes with bootstrap values <50% were collapsed into multifurcations. Oligonucleotide probes were designed for four species- to genus-like betaproteobacterial populations with the respective ARB tools. The probes were checked in silico against the database of the ribosomal database project (http://rdp.cme.msu.edu), and tested on environmental samples at different formamide concentrations until highest specificity was achieved (Table 1). A set of several helper and competitor oligonucleotides was designed to enhance the accessibility and specificity of probes bet VI-994 and GKS98-584, respectively (Table 1; Amann and Fuchs, 2008).
Table 1. Oligonucleotide probes designed in this study.
| Probe name | Specificity | No. target sequences/outgroup hits | Sequence (5′ to 3′) | FA % |
|---|---|---|---|---|
| bet I-B | Uncultured bet I-B tribe (Rhodoferax sp. relatives) | 3 | GAG TAG GCC GTT TCG TTC | 35 |
| bet I-C-449 | Uncultured bet I-C tribe (Curvibacter sp. relatives) | 7 | ATT AGC CAA GAC TTT TTC G | 20 |
| bet VI-994 | Uncultured bet VI tribe | 4 | GCA GAA TTC CGA ACA TGT | 35 |
| bet VI-H1 | Helper for bet VI-994 | GAG CAC TTC CGC ATC TCT | ||
| bet VI-H2 | Helper for bet VI-994 | CAA GGG TAG GTA AGG TTT | ||
| bet VI-H3 | Helper for bet VI-994 | CAC CTG TGT TCC GGT TC | ||
| GKS98-584 | Uncultured GKS98 (bet III) tribe | 72/2 | ACC TCT CTT TCC GAA CCG CC | 35 |
| GKS98-C | Competitor for GKS98-584 | AYC TYT CTT TCC GAA CCG CC | ||
| GKS98-H1 | Helper for GKS98-584 | TGC GCA CGC TTT ACG CCC | ||
| GKS98-H2 | Helper for GKS98-584 | TAA GCC CTG GGA TTT CAC | ||
| GKS98-H3 | Helper for GKS98-584 | CGC TTT CGT GCA TGA GCG |
Abbreviation: FA%, formamide concentration for stringent conditions.
Abundances of auto- and heterotrophic prokaryotes
Abundances of bacteria and autotrophic picocyanobacteria were determined by an inFlux V-GS cell sorter (Becton Dickinson, Frankling Lakes, NJ, USA) equipped with a UV (355 nm) and blue (488 nm) laser. All samples were stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg ml−1 final concentration), and scatter plots of DAPI fluorescence vs 90° light scatter were analyzed with the software FlowJo 7.2.2. (Tree Star Inc., Ashland, OR, USA).
Abundances of different bacteria
CARD-FISH with fluorescein-labeled tyramides was carried out as previously described (Sekar et al., 2003) with slight modifications for probe LD12-121 (Salcher et al., 2011b). Horseradish peroxidase-labeled oligonucleotide probes used for community analysis were: EUB I–III (all Bacteria), HGC69a (Actinobacteria), BET42a (Betaproteobacteria), ALF968 (Alphaproteobacteria), CF319a (mostly Flavobacteriales, phylum Bacteroidetes), GAM42a (Gammaproteobacteria) (for details see Amann and Fuchs, 2008), Ver47 (Verrucomicrobia (Arnds et al., 2010)), ac1-853 (acI cluster of Actinobacteria (Warnecke et al., 2005)), LD12-121 (uncultured LD12 Alphaproteobacteria (Salcher et al., 2011b)), Cyc715 (Cyclobacteriaceae, Sphingobacteriales, Bacteroidetes (Eckert et al., 2012)), LD2-739 (LD2 cluster of Saprospiraceae, Sphingobacteriales, Bacteroidetes (Pernthaler et al., 2004)), R-BT065 (Limnohabitans sp., Betaproteobacteria (Šimek et al., 2001)), PnecB-23S-166 (Polynucleobacter acidiphobus and P. difficiles, Betaproteobacteria), PnecC-445 (P. necessarius spp. asymbioticus, Betaproteobacteria (Hahn et al., 2005)), LD28-1017 (uncultured LD28 tribe, Betaproteobacteria (Salcher et al., 2011a)), and the newly designed probes listed in Table 1. CARD-FISH stained filters were analyzed by fully automated high-throughput microscopy (Zeder and Pernthaler, 2009). All images were analyzed with the freeware image analysis software ACMEtool (technobiology.ch), and interfering autofluorescent cyanobacteria or debris were individually subtracted from hybridized cells. At least 10 high quality images or >1000 DAPI-stained bacteria were analyzed per sample.
Tracer incubations and MAR-FISH
MAR assays and bulk uptake measurements with 14 radiolabeled tracers (Table 2, all purchased from Amersham, GE Healthcare, Pittsburgh, PA, USA; and American Radiolabeled Chemicals, Saint Louis, MO, USA) were performed on water samples from 5 m depth. The amino-acid mixture (AA) is composed of 15 individual AAs (that is, Ala, Arg, Asp, Glu, Gly, His, Ile, Leu, Lys, Phe, Pro, Ser, Thr, Tyr and Val). Each substrate was added to a subsample of 6 ml at a final concentration of 10 nmol l−1, and subsamples were incubated for 2 h at in situ temperature in the dark in triplicates plus 2 prefixed controls. Bulk uptake rates were determined from 5 ml of these samples with a scintillation counter (Tri-Carb 3170TR/SL, Perkin-Elmer, Waltham, MA, USA) according to the cold TCA method described in Kirchman (2001). The remaining 1 ml of subsample was filtered onto white polycarbonate filters (Millipore, Billerica, MA, USA), processed as described in Salcher et al. (2011b) and manually evaluated with a Zeiss microscope (Carl Zeiss, Jena, Germany) at bright field illumination and blue excitation. At least 100 hybridized cells were inspected per sample. Filamentous bacteria, picocyanobacteria and P. rubescens filaments were determined from several preparations and counts for a single tracer were pooled.
Table 2. Radiolabeled tracers, their specific activities and bulk incorporation rates (±s.e.) used in this study.
| Tracer | Abbreviation | Specific activity (Ci mmol−1) | Bulk incorporation rates (pmol l−1 h−1) |
|---|---|---|---|
| [3H]-Thymidine | TdR | 77 | 60.2 (±1.3) |
| [3H]-Glucose | Glc | 29 | 590.0 (±17.1) |
| [14C]-Fructose | Fru | 0.316 | 384.7 (±9.7) |
| [14C]-Acetate | Ace | 56 | 5.7 (±0.1) |
| [3H]-Amino-acid mixture | AA | 45 | 829.0 (±45.9) |
| [3H]-Lysine | Lys | 73 | 1.5 (±0.2) |
| [3H]-Arginine | Arg | 49 | 5.4 (±0.3) |
| [14C]-Aspartate | Asp | 0.207 | 1150.7 (±8.5) |
| [3H]-Glutamate | Gln | 45 | 43.3 (±1.2) |
| [3H]-Glutamine | Glu | 53 | 18.8 (±3.3) |
| [14C]-Serine | Ser | 0.255 | 723.5 (±38.8) |
| [14C]-Glycine | Gly | 0.101 | 1200.2 (±57.9) |
| [14C]-Alanine | Ala | 0.154 | 1122.2 (±115.4) |
| [3H]-Leucine | Leu | 61 | 351.6 (±31.1) |
Statistical analysis
Agglomerative hierarchical clustering was done to identify significant clusters of either tracers or populations based on Pearson's correlation coefficients using the unweighted pair-group average. Data were normalized by log(x+1) transformation. A mantel test (1000 permutations) was used to test for a significant relationship between depth distribution and tracer uptake of different bacterial populations. Only species- to family-like bacterial populations and all tracers except AA were included in cluster computation. Clustering was done with XLSTAT ADA (Addinsoft, New York, NY, USA).
Results
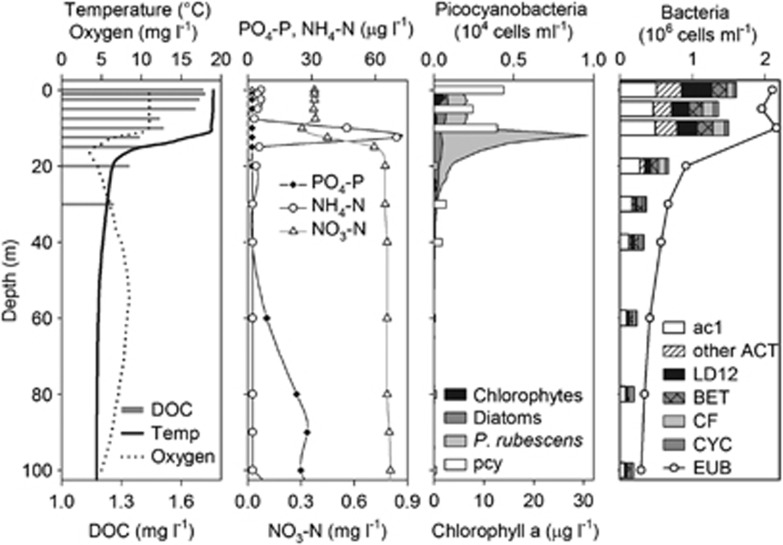
Lake Zurich was thermally stratified with a metalimnetic oxygen minimum (<4 mg O2 l−1) between 16–18 m depth in 16 September 2009 (Figure 1). Dissolved organic carbon was present in very low concentrations (1.3–1.7 mg l−1) with a slight tendency to decrease with depth, while dissolved phosphorus (PO4-P) was below the limit of detection (2 μg l−1) until 40 m depth and increased with depth. Nitrate (NO3-N) was depleted in the epilimnion and increased in the hypolimnion, whereas ammonia (NH4-N) showed a pronounced peak (70 μg l−1) in 12 m depth. The dominant primary producer, the filamentous cyanobacterium P. rubescens, had a distinct maximum in 12 m depth, while small picocyanobacteria (presumably Synechococcus sp.) were abundant in the water layers above. Diatoms and chlorophytes were present in low numbers as samples were taken at the decline of a diatom summer bloom usually observed in August (data not shown). The highest abundances of heterotrophic bacteria were found in the warm, light penetrated epilimnion (0–10 m depths) with a slight decline in 5 m depth. The majority of microbes belonged to the low nucleic acid fraction (77% of all bacteria, data not shown), whereas high nucleic acid cells became more abundant in the deep hypolimnion (that is, 30% in 40–100 m vs 17% in the epilimnion). Thus, the epilimnion was dominated by small-sized cells.
Figure 1.
Vertical profiles of physical, chemical and biological parameters in Lake Zurich at the sampling date. DOC, dissolved organic carbon; PO4-P, phosphate-P; NH4-N, ammonium-N; NO3-N, nitrate-N; pcy, picocyanobacteria; ac1, ac1 tribe of Actinobacteria; ACT, Actinobacteria; LD12, LD12 tribe of Alphaproteobacteria; BET, Betaproteobacteria; CF, Flavobacteriales; CYC, Cyclobacteriaceae; EUB, Bacteria.
Very small bacteria (that is, ultramicrobacteria with a cell length of ≤1 μm) affiliated with the ac1 clade and other lineages of Actinobacteria were more frequent in the epi- than in the hypolimnion (Figure 1). Moreover, LD12 ultramicrobacteria (Alphaproteobacteria) were only present in high numbers in 0–10 m depths (up to 0.4 × 106 cells ml−1 in the surface), and 10 times less abundant between 20–100 m depth. Other Alphaproteobacteria, as well as Gammaproteobacteria and Verrucomicrobia contributed <1% to the total assemblage and were therefore excluded from further analyses. Betaproteobacteria and Bacteroidetes (as hybridized with probes CF319a and Cyc715) could be detected throughout the whole water column (between 4–17% of Bacteria).
Incorporation profiles of different bacterial populations
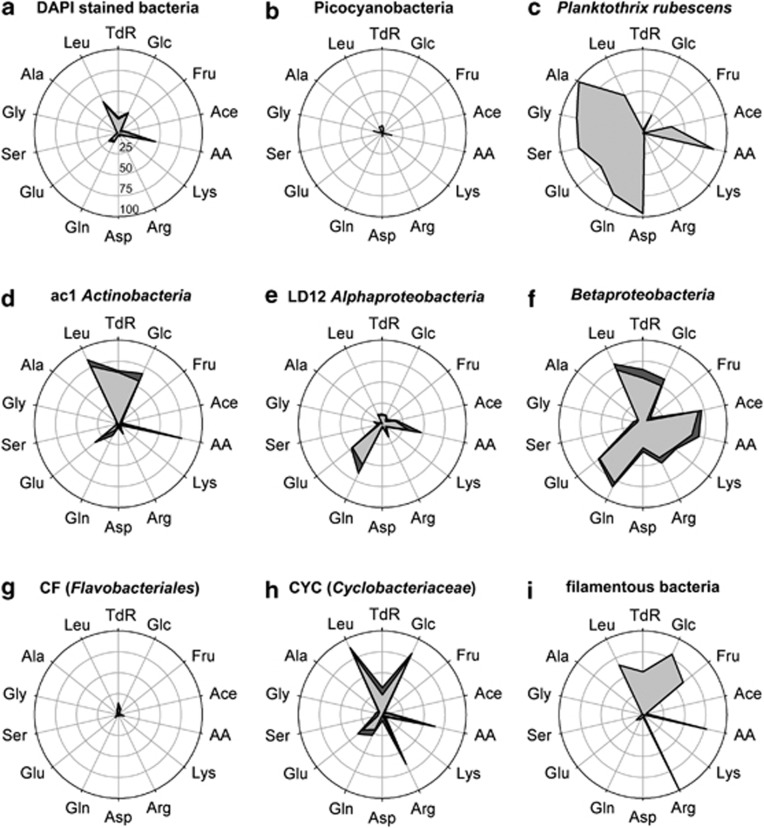
The different radiolabeled tracers were incorporated by 1–44% of all DAPI-stained microbes, respectively (Figure 2a). While Fru, Lys, Arg, Asp, Ser, Gly and Ala (abbreviations see Table 2) were incorporated by <4% of all cells, Ace, TdR, Glc, Gln and Glu were more frequently taken up (8–26%). Highest incorporation was detected for Leu (40%) and AA (44%). Picocyanobacteria did not take up any of the offered substrates (Figure 2b), while P. rubescens was highly efficient in the incorporation of Asp, Gln, Glu, Ser, Gly and Ala (64–98% of filaments, Figure 2c). Ace and Glc were also incorporated by one-third and one-quarter of all P. rubescens filaments, respectively (Figure 2c).
Figure 2.
Ecophysiological MAR-FISH assay with 14 radiolabeled substrates. Means and standard errors (n=3) of the percentage of all DAPI-stained bacteria (a), picocyanobaceria (b), the cyanobacterium P. rubescens (c), ac1 Actinobacteria (d), LD12 Alphaproteobacteria (e), Betaproteobacteria (f), Flavobacteriales targeted by probe CF319a (g), Cyclobacteriaceae (h) and filamentous bacteria (i) with active tracer incorporation are shown. See Table 2 for abbreviations of tracers. Scaling in (a) applies to all populations.
Actinobacteria of the ac1 tribe mainly incorporated TdR, Glc, AA and Leu (Figure 2d), as opposed to microbes affiliated with the LD12 cluster (Alphaproteobacteria), which showed only a weak incorporation of these tracers (except AA, Figure 2e). These microbes were on the other hand highly active in Gln and Glu uptake. Betaproteobacteria in general incorporated most of the offered tracers to a high extent; only for Fru, Ser, Gly and Ala no incorporation could be detected (Figure 2f). Bacteroidetes showed a contrasting pattern: while microbes targeted by probe CF319a (mainly Flavobacteriales) were seemingly inactive (Figure 2g), bacteria affiliated with Cyclobacteriaceae (Sphingobacteriales) positively selected Arg, Glc and Leu (65–85%), and to a lesser extent TdR, Gln and Glu (24–34%, Figure 2h). A comparable pattern was detected for filamentous DAPI-stained microbes (Figure 2i): Almost all filaments (99%) took up Arg and 51–80% were positive for Leu, TdR, Glc and Fru. A large proportion (44.4%) of these filaments was affiliated with the ubiquitous LD2 cluster of Saprospiraceae (Sphingobacteriales, Bacteroidetes, data not shown).
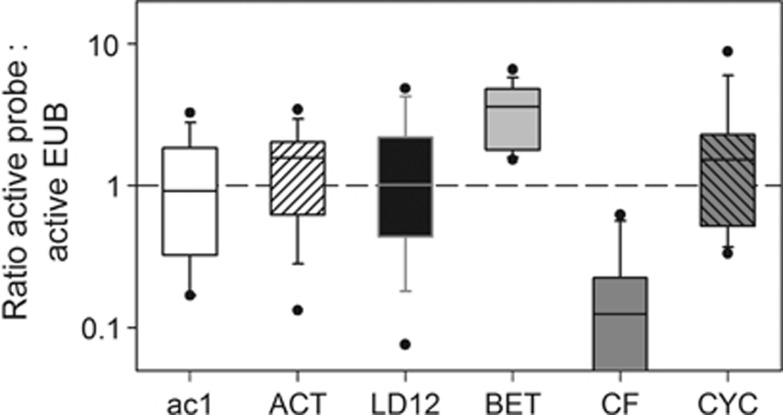
Ratios of active cells in populations to all active bacteria
A ratio of population-specific activity vs activity of all bacteria (as hybridized by probe EUB) was calculated by summing up the abundances of MAR+ cells of all 14 tracers (Figure 3). The most abundant microbes (that is, Actinobacteria and therein the ac1 cluster and LD12 bacteria) mirrored the activity of the total assemblage, while the active cell fraction within Betaproteobacteria was 2.5 times above the community average. Microbes hybridized by probe CF319a were 8 times less active, while Cyclobacteriaceae were slightly above the community average.
Figure 3.
Ratio of active populations to active Bacteria (as hybridized with probe EUB I–III). All tracers were pooled for calculation. Abbreviations see Figure1.
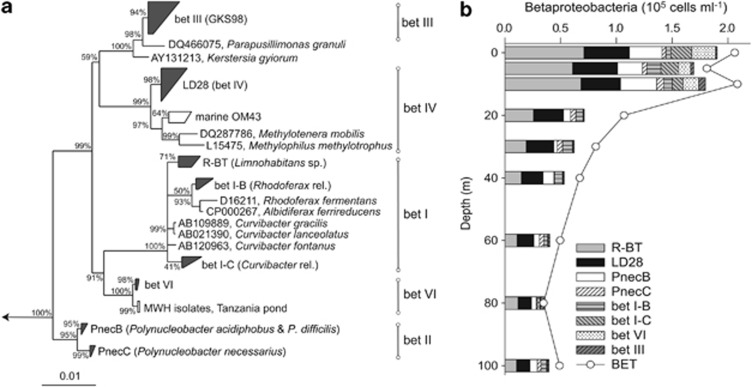
Betaproteobacterial populations
By applying eight species- to genus-specific probes (that is, targeting genotypes with >98% and >96.6% 16S rRNA gene sequence similarity), we succeeded to identify the vast majority (84±11%) of all Betaproteobacteria along the depth profile (Figure 4). In the sampling depth for the MAR-FISH analysis (5 m), 94% of all Betaproteobacteria were covered by the specific probes. The most abundant Betaproteobacteria were affiliated with Limnohabitans sp. (R-BT), followed by the species-like tribe LD28. Polynucleobacter acidiphobus and P. difficiles (PnecB) mainly populated the epilimnion, while Polynucleobacter necessarius ssp. asymbioticus (PnecC) were only present in low densities. The four newly designed probes target small clusters within bet I, bet VI and the whole tribe bet III (GKS98). The species-like population bet I-B, closely related to Rhodoferax sp. and Albidiferax sp., was present throughout the water column, while microbes of the species-like cluster bet I-C, relatives of Curvibacter sp., were restricted to the epilimnion. A similar spatial pattern was found for a species-like cluster within the bet VI lineage. Microbes affiliated with the genus-like (that is, >96.6% 16S rDNA sequence similarity) bet III cluster, related to Parapusillimonas granuli, were present in all depths, albeit in very low abundances. These microbes, as well as PnecC were not further analyzed by MAR-FISH due to their very low numbers.
Figure 4.
Maximum likelihood tree of the 16S rDNA of Betaproteobacteria and their ubiquitous freshwater lineages (a). Clusters in gray mark populations subjected to CARD-FISH with newly designed or already published probes. (b) Vertical distribution of betaproteobacterial populations.
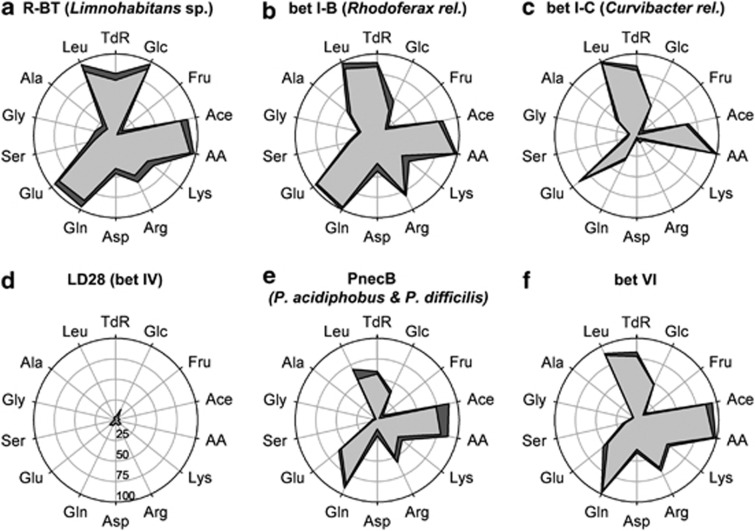
Five of the six analyzed betaproteobacterial subpopulations shared several preferred substrates, that is, TdR, Ace, AA, Gln and/or Glu, and Leu (Figure 5). On the other hand, Glc, Lys, Arg, Asp, Ser and Ala were variable for each population. Fru and Gly were not or only slightly taken up by any of the populations.
Figure 5.
Ecophysiological MAR-FISH assay with 14 radiolabeled substrates. Means and standard errors (n=3) of the percentage of different betaproteobacterial populations with active tracer incorporation are shown (a–f). See Table 2 for abbreviations of tracers and Figure 4 for phylogeny and abundance of populations. Scaling in (d) applies to all populations.
Nine of the 14 substrates were incorporated by >50% of all R-BT (54–95%), and four more by at least one sixth of the population. Only Fru was not incorporated by R-BT (Figure 5a). Microbes affiliated with bet I-B incorporated all substrates to a certain extent (Figure 5b). High uptake (>80%) was observed for seven tracers, medium uptake (22–44%) for six more tracers, and low uptake for Fru (15%). Very high uptake of five radiolabeled tracers was observed for the bet I-C population, while five other substrates were not incorporated at all (Figure 5c). Microbes of the LD28 tribe did not take up any of the offered tracers (Figure 5d), while PnecB and bacteria affiliated with bet VI incorporated 10–12 substrates (Figures 5e and f). The latter two populations showed a comparable pattern with high uptake of Ace, AA, Arg, Gln, Glu, Leu, TdR and Glc.
Clustering of populations and tracers
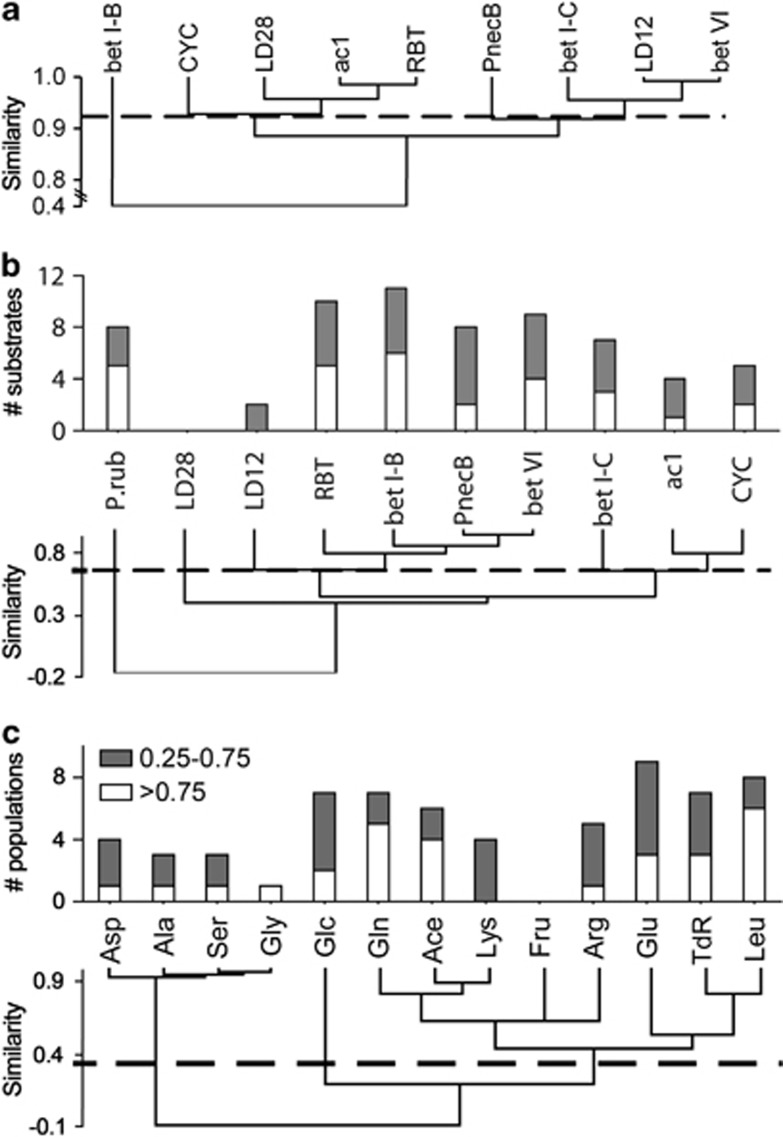
Agglomerative hierarchical clustering based on Pearson's correlation coefficients identified four significant clusters of bacterial populations according to their depth distribution (Figure 6a). Populations with a clear surface preference (LD12, bet VI and bet I-C) were separated from others with a more homogeneous distribution over depth. Clustering based on similarities of microbial uptake spectra (Figure 6b) resulted also in four significant clusters, albeit with a significantly different grouping (that is, the absence of a significant correlation between the data sets in a Mantel test, P=0.131). P. rubescens was clearly separated from all heterotrophic microbes in its substrate uptake pattern, and incorporation patterns of LD28 Betaproteobacteria were different from all other investigated bacterial groups. Alphaproteobacteria of the LD12 clade clustered weakly with the betaproteobacterial tribes R-BT, bet I-B, PnecB and bet VI. Microbes affiliated with bet I-C differed from all other bacteria, and a cluster of ac1 Actinobacteria and Cyclobacteriaceae could be distinguished. Specialized bacteria with an incorporation of only some tracers were affiliated with LD12, ac1, Cyclobacteriaceae and bet I-C. On the other hand, all other populations affiliated with Betaproteobacteria (except for LD28) and P. rubescens incorporated a wide range of substrates, pointing to a more generalist lifestyle. Three significant clusters of substrates that tended to be consumed together could be identified (Figure 6c). The first deeply branching cluster represented tracers, which were incorporated to a high extent exclusively by P. rubescens (that is, Asp, Ala, Ser and Gly). The second cluster included the remaining tracers. Therein, Glc was clearly separated from all other substrates. Glu was incorporated by almost all analyzed microbial populations, followed by Leu, Glc, Gln and TdR. On the other hand, Lys, Asp, Ala, Ser and Gly appeared to be more specialized substrates, and Fru was never incorporated by >25% of any of the analyzed populations.
Figure 6.
(a) Agglomerative hierarchical clustering of bacterial populations based on the depth distribution of their abundances in Lake Zurich. (b) Upper panel: Number of substrates incorporated by a specific population to a very high (>0.75, white) or medium (0.25–0.75, gray) extent. Lower panel: Agglomerative hierarchical clustering of bacterial populations based on substrate incorporation. (c) Upper panel: Number of populations incorporating a specific tracer to a very high (>0.75, white) or medium (0.25–0.75, gray) extent. Lower panel: Agglomerative hierarchical clustering of tracers. Agglomerative hierarchical clustering was based on Pearson's correlation coefficients using unweighted pair-group average. Only species- to family-like bacterial populations and all tracers except AA were computed.
Discussion
Phytoplankton as major sources of LMW substances
Dissolved organic matter (DOM) in freshwaters is composed of a multitude of different substances. The fraction of LMW DOM mainly consists of amino acids (AAs), carbohydrates (CHs) and carboxylic acids (CAs), molecules that can be rapidly utilized by microbes with turnover rates of several hours (Sundh, 1992; Rosenstock and Simon, 1993). Dissolved free AAs form a substantial pool (that is, 12–32% in Lake Constance) of the labile DOM in freshwaters with strong seasonal variations (Weiss and Simon, 1999). Ace is often the most important CA in lakes, while Glc and Fru were found to be equally abundant CHs (Bertilsson and Tranvik, 1998; Zotina et al., 2003; Berggren et al., 2010). Previous studies on Lake Zurich aimed to quantify the concentrations of AAs (that is, ∼10–25 nℳ of individual AAs, ∼100 nℳ in total), CHs (∼20 nℳ) and CAs (∼1 μℳ) (Meon and Jüttner, 1999; Zotina et al., 2003). We added all tracers in final concentrations of 10 nℳ which corresponds approximately to the in situ conditions, except for acetate, which was added in tracer amounts in our assays.
Phytoplankton blooms are a main source of labile DOM readily available for microbes, as a substantial fraction of extracellular phytoplankton products (EPP) such as AAs, CHs or CAs is released by algae (Hama and Handa, 1987). Although the majority of EPP are polymers (Giroldo and Vieira, 2005), bacteria were found to utilize algal LMW exudates more rapidly and to a much higher extent (Giroldo et al., 2007). The exudation of for example, CHs is both, species- and growth phase-dependent, with highest amounts usually released during late stationary phase (Giroldo and Vieira, 2005). Our experiments were conducted at the late phase of a phytoplankton summer bloom dominated by chlorophytes, chrysophytes and diatoms; therefore, substantial EPP release can be expected. The observed high number of active cells within different microbial populations (Figures 2 and 5) might thus be also a result of microbes already being pre-adapted to increased availability of LMW substrates. Glc, besides mannose/xylose, released by a freshwater diatom was found to be the principle source of CHs supporting bacterial growth (Giroldo et al., 2007). In our study, Glc was mainly incorporated by ac1 Actinobacteria, Cyclobacteriaceae, R-BT and filamentous bacteria (Figures 2 and 5), microbes that are typically associated with algal blooms (Šimek et al., 2008; Salcher et al., 2010; Eckert et al., 2012).
Substrate niche differentiation of the dominant microbes
The most abundant microbes in Lake Zurich (that is, ac1 Actinobacteria and LD12 Alphaproteobacteria, Figure 1) showed strikingly different substrate preferences (Figures 2d and e). Therefore, these apparently highly specialized ultramicrobacterial populations might coexist in the water column by largely avoiding competition for LMW DOM compounds. The uptake pattern of ac1 Actinobacteria suggested adaptation to phytoplankton exudates (Glc and Leu) in combination with rapid cell division (TdR). A nearly complete genome was recently obtained from a single sorted cell of the ubiquitous ac1-B1 tribe (Garcia et al., 2012). This streamlined genome included several genes encoding for AA transporters (for example, ABC transporters for branched-chain AAs such as Leu), but lacked a transporter for Glc. This is in contrast to our results (Figure 2d), as well as to previous findings of Glc uptake by ac1-B1 (Buck et al., 2009). Other successful planktonic microorganisms with reduced genomes such as the marine SAR11 clade or Polynucleobacter sp. are highly diversified, with physiologically distinct ecotypes adapted to specific environmental conditions (Schwalbach et al., 2010; Hahn et al., 2012b). Such ecotype diversification might also be the case for ac1 Actinobacteria.
In contrast, LD12 Alphaproteobacteria only weakly incorporated Leu, TdR and Glc, but were instead specialized on Gln and Glu (Figure 2e; Salcher et al., 2011b). These AAs are rich in nitrogen and their α-amino groups can be used for the biosynthesis of other AAs (Lengeler et al., 1999). Therefore, LD12 microbes seemed to be well adapted to nitrogen depletion in Lake Zurich during summer (Salcher et al., 2011b). LD12 Alphaproteobacteria can be regarded as typical oligocarbophils (Salcher et al., 2011b), comparable to their marine sister group SAR11 (Giovannoni et al., 2005). Since these bacteria were almost three times more abundant 2 weeks earlier (Salcher et al., 2011b), the population likely was already in decline at the sampling date. As their level of AA incorporation was found to be tightly linked to their seasonal population development (Salcher et al., 2011b), they might not have been at the peak of their activity. A recent study conducted in brackish waters of the Gulf of Gdańsk (Baltic Sea) revealed that up to one-quarter of LD12 bacteria actively incorporated TdR, particularly at times of freshwater influx (Piwosz et al., accepted).
Differences within Bacteroidetes
Three lineages affiliated with Bacteroidetes were analyzed in our assay, that is, Flavobacteriales targeted by probe CF319a, Cyclobacteriaceae (Sphingobacteriales) (Eckert et al., 2012), and filamentous microbes most of which were affiliated with the ubiquitous LD2 tribe of Saprospiraceae (Sphingobacteriales) (Schauer et al., 2006). Microbes targeted by probe CF319a did not incorporate any of the offered substrates (Figure 2g). This does not imply that these microbes were metabolically inactive. Marine and brackish Flavobacteriales preferably incorporated high-molecular-weight DOM such as proteins or chitin (Cottrell and Kirchman, 2000). Freshwater Flavobacteriales were repeatedly reported to be only marginally involved in AA turnover (Pérez and Sommaruga, 2006; Salcher et al., 2010), but could be experimentally enriched with chitin (Beier and Bertilsson, 2011) and incorporated the mono- and dimeric chitin units N-acetyl-glucosamine and di-N-acetyl-glucosamine (Beier and Bertilsson, 2011; Eckert et al., 2012). The freshwater bacterium Flavobacterium johnsoniae strain A3 preferentially utilized oligopeptides and proteins over individual AAs and had a higher specific affinity for di- and polysaccharides than for Fru (Sack et al., 2011). Our results support the hypothesis of Flavobacteriales being specialized on substrates other than LMW DOM (Figure 2g).
Cyclobacteriaceae (Sphingobacteriales), on the other hand, were specialized on Glc, Arg and Leu incorporation (Figure 2h). The applied FISH probe targets almost the complete family and was designed to quantify Cyclobacteriaceae that were not detected by the general probe CF319a during a phytoplankton spring bloom in Lake Zurich (Eckert et al., 2012). These microbes were most abundant during the late bloom phase and were highly involved in turnover of N-acetyl-glucosamine (Eckert et al., 2012). Cultivated members of this lineage (Algoriphagus sp.) were able to grow on Glc as sole carbon source (Liu et al., 2009), pointing to a dependency on phytoplankton exudates.
Filamentous microbes, predominantly affiliated with the LD2 tribe of Saprospiraceae (Schauer et al., 2006) specialized on the uptake of Arg, Leu, TdR, Glc and Fru (Figure 2i). Moreover, high TdR incorporation indicated rapid cell division. Filaments often reach maximal densities at late phases of algal blooms when protistan grazing pressure is high (Pernthaler et al., 2004; Eckert et al., 2012), as this morphology protects from flagellate predation (Pernthaler, 2005; Schauer et al., 2006). However, the intense uptake of AAs and sugar monomers suggested that filaments might also have directly profited from algal exudates.
Active betaproteobacterial populations
Using a set of eight species- to genus-like probes we were able to identify almost all Betaproteobacteria (Figure 4), which were the most active microbial group with respect to LMW assimilation (Figure 3). Cryptophytes and chlorophytes, both of which were present at the sampling date, are known to release EPP that support the growth of Betaproteobacteria (R-BT, PnecB and others), especially during late bloom phases (Šimek et al., 2011). Closely related populations (that is, R-BT, bet I-B and bet I-C, all belonging to the bet I lineage, Figure 4) differed drastically in their incorporation patterns (Figure 5), and thus seemed to largely avoid competition for the same substrates. On the other hand, microbes affiliated with PnecB and bet VI showed similar uptake whereas they are not phylogenetically closely related (Figures 4 and 5). In general, R-BT, PnecB, and microbes of the uncultured bet I-B and bet VI tribes had very broad tracer uptake spectra (Figures 5 and 6), pointing to a more generalist life style and the preparedness to grow on a wide range of substrates. An ‘opportunistic' life strategy has been previously postulated for R-BT, based on their fast growth that is tightly linked to phosphorus and algal-derived carbon sources (Šimek et al., 2005, 2008, 2011), and their sensitivity to grazing by protists (Jezbera et al., 2006). Bet I-C on the other hand seemed to be more specialized, for example, Leu and Glu were the only preferred AAs. Bacteria of the uncultured LD28 (bet IV) tribe of Betaproteobacteria are close relatives of the marine OM43 lineage and other methylotrophs, for example, Methylotenera sp. (Figure 4). Marine OM43 are commonly associated with diatom blooms (Sekar et al., 2004; Morris et al., 2006) and utilize C1 substrates such as methanol, formate and formaldehyde (Giovannoni et al., 2008). High numbers of LD28 were detected in deep water beneath the chlorophyll a maxima in Lake Zurich (Salcher et al., 2011a) and a small prealpine lake (Salcher et al., 2008). In the present study, LD28 did not incorporate any of the offered substrates (Figure 5d), probably due to a methylotrophic life style.
Photoheterotrophy of cyanobacteria—powerful competitors for heterotrophs
Picocyanobacteria (presumably Synechococcus sp.) did not incorporate any of the offered substrates (Figure 2b) and therefore likely were predominantely phototrophic. This differs from observations in marine waters where they could assimilate AAs and ATP (Zubkov et al., 2003; Michelou et al., 2011). By contrast, the filamentous cyanobacterium P. rubescens, the dominant primary producer in Lake Zurich (Posch et al., 2012), took up a variety of AAs and even incorporated Glc and Ace (Figure 2c), as also observed previously (Zotina et al., 2003; Walsby and Jüttner, 2006; Horňák et al., 2012). In particular, nitrogenous organic compounds may fuel most of the nitrogen demand of this non-N2-fixing cyanobacterium (Zotina et al., 2003). While most of the preferentially incorporated AAs (esp. Ala, Gly, Ser and Asp) were not or only marginally taken up by heterotrophic bacteria (Figures 2, 5 and 6), Gln and Glu were highly incorporated by both microbial guilds. Therefore, P. rubescens can be a potent competitor of heterotrophic microbes for DOM. This competition might be even more significant when considering biomass specific substrate uptake, as P. rubescens represents 10 times more biomass than bacteria in the epilimnion of Lake Zurich (Van den Wyngaert et al., 2011). Moreover, unlike other phototrophs, this cyanobacterial species has only very low DOM excretion rates (Feuillade et al., 1988), which makes it doubtful that it may also represent a source of dissolved substrates for heterotrophic bacteria in the photic zone of the lake.
Substrate specialization: a common feature of freshwater microbes?
We identified two general strategies of LMW DOM acquisition by planktonic bacteria: For one, there was uptake of a broad range of substrates by several generalist and ecophysiological versatile betaproteobacterial populations (Figures 5 and 6). Such preparedness for growth on mixed substrates has been postulated as a general rule for microbes in oligotrophic environments (Egli, 2010). However, our results also indicated a high degree of specialization, as the most abundant microbes (that is, ac1 Actinobacteria and LD12 Alphaproteobacteria), both of which are known for their oligotrophic life style (Salcher et al., 2011b; Garcia et al., 2012), showed no overlap in their LMW substrate acquisition spectra (Figure 2). Moreover, substrate selectivity was also found for Bacteroidetes (CF, Cyclobacteriaceae, and filaments) as well as for two populations of Betaproteobacteria. This suggests that any generalized estimate of the in situ activity of freshwater microbes (or their active fraction) by single radiolabeled tracers such as Leu or TdR (Cottrell and Kirchman, 2003; Pérez et al., 2010) may be biased. For example, at least one of the tested substrates was taken up by >90% of cells from the various betaproteobacterial populations (except for LD28, Figure 5), but the preferred substrates differed (that is, Glc for R-BT, Gln for PnecB, bet I-B, and bet VI, Leu for bet I-C). Therefore, all these populations must be regarded as highly active.
The observed physiological niche separation might also be an important reason for the coexistence and considerable diversity of planktonic bacterial taxa in freshwater lakes. Interestingly, microbial populations with similar substrate uptake patterns also had different vertical distributions in the lake (Figures 6a and b), hinting at other niche separation factors such as oxygen content or temperature. Our results nevertheless provide an example for coexistence mediated by substrate-based ecological diversification, as was recently suggested for the degradation of complex polysaccharides (Teeling et al., 2012). Such an ecophysiological niche separation might moreover be regarded as a (partial) solution for the classical ‘paradox of the plankton' (Hutchinson, 1961) with respect to heterotrophic pelagic bacteria.
Acknowledgments
We thank captain E Loher for help during sampling and the Zurich Water Supply (WVZ) for the provision of limnochemical data. Three anonymous reviewers are acknowledged for their helpful comments. This study was supported by the Swiss National Science Foundation (SNF, Project 31003A-117765) and, as part of the European Science Foundation EUROCORES Programme EuroEEFG, by the SNF Project 31EE30-132771.
The authors declare no conflict of interest.
References
- Allgaier M, Grossart H. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol. 2006;72:3489–3497. doi: 10.1128/AEM.72.5.3489-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- Arnds J, Knittel K, Buck U, Winkel M, Amann R. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst Appl Microbiol. 2010;33:139–148. doi: 10.1016/j.syapm.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Beier S, Bertilsson S. Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol Oceanogr. 2011;56:1179–1188. [Google Scholar]
- Berggren M, Laudon H, Haei M, Strom L, Jansson M. Efficient aquatic bacterial metabolism of dissolved low-molecular-weight compounds from terrestrial sources. ISME J. 2010;4:408–416. doi: 10.1038/ismej.2009.120. [DOI] [PubMed] [Google Scholar]
- Bertilsson S, Tranvik LJ. Photochemically produced carboxylic acids as substrates for freshwater bacterioplankton. Limnol Oceanogr. 1998;43:885–895. [Google Scholar]
- Beutler M, Wiltshire K, Meyer B, Moldaenke C, Lüring C, Meyerhöfer M, et al. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynth Res. 2002;72:39–53. doi: 10.1023/A:1016026607048. [DOI] [PubMed] [Google Scholar]
- Buck U, Grossart HP, Amann R, Pernthaler J. Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a humic lake. Environ Microbiol. 2009;11:1854–1865. doi: 10.1111/j.1462-2920.2009.01910.x. [DOI] [PubMed] [Google Scholar]
- Cottrell M, Kirchman D. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacteria cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell M, Kirchman D. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol Oceanogr. 2003;48:168–178. [Google Scholar]
- Eckert EM, Salcher MM, Posch T, Eugster B, Pernthaler J. Rapid successions affect microbial N-acetyl-glucosamine uptake patterns during a lacustrine spring phytoplankton bloom. Environ Microbiol. 2012;14:794–806. doi: 10.1111/j.1462-2920.2011.02639.x. [DOI] [PubMed] [Google Scholar]
- Egli T. How to live at very low substrate concentration. Water Res. 2010;44:4826–4837. doi: 10.1016/j.watres.2010.07.023. [DOI] [PubMed] [Google Scholar]
- Eiler A, Bertilsson S. Flavobacteria blooms in four eutrophic lakes: Linking population dynamics of freshwater bacterioplankton to resource availability. Appl Environ Microbiol. 2007;73:3511–3518. doi: 10.1128/AEM.02534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillade M, Bohatier J, Bourdier G, Dufour P, Feuillade J, Krupka H. Amino-acid uptake by a natural population of Oscillatoria rubescens in relation to uptake by bacterioplankton. Arch Hydrobiol. 1988;113:345–358. [Google Scholar]
- Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R, et al. 2012Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton ISME Je-pub ahead of print 19 July 2012, doi:2010.1038/ismej.2012.2086. [DOI] [PMC free article] [PubMed]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Giroldo D, Ortolano PIC, Vieira AAH. Bacteria–algae association in batch cultures of phytoplankton from a tropical reservoir: the significance of algal carbohydrates. Freshwater Biol. 2007;52:1281–1289. [Google Scholar]
- Giroldo D, Vieira AAH. Polymeric and free sugars released by three phytoplanktonic species from a freshwater tropical eutrophic reservoir. J Plankton Res. 2005;27:695–705. [Google Scholar]
- Glöckner F, Fuchs B, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Pöckl M, Wu Q. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol. 2005;71:4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T. Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int J Syst Evol Microbiol. 2009;59:2002–2009. doi: 10.1099/ijs.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Minasyan A, Lang E, Koll U, Spröer C. Polynucleobacter difficilis sp. nov., a planktonic freshwater bacterium affiliated with subcluster B1 of the genus Polynucleobacter. Int J Syst Evol Microbiol. 2012a;62:376–383. doi: 10.1099/ijs.0.031393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Scheuerl T, Jezberova J, Koll U, Jezbera J, Šimek K, et al. The passive yet successful way of planktonic life: genomic and experimental analysis of the ecology of a free-living Polynucleobacter population. PLoS ONE. 2012b;7:e32772. doi: 10.1371/journal.pone.0032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama T, Handa N. Pattern of organic-matter production by natural phytoplankton population in a eutrophic lake. 2. Extracellular products. Arch Hydrobiol. 1987;109:227–243. [Google Scholar]
- Horňák K, Zeder M, Blom JF, Posch T, Pernthaler J. Suboptimal light conditions negatively affect the heterotrophy of Planktothrix rubescens but are beneficial for accompanying Limnohabitans spp. Environ Microbiol. 2012;14:765–778. doi: 10.1111/j.1462-2920.2011.02635.x. [DOI] [PubMed] [Google Scholar]
- Hutchinson GE. The paradox of the plankton. Am Nat. 1961;95:137–145. [Google Scholar]
- Jezbera J, Horňák K, Šimek K. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol. 2006;8:1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- Kasalický V, Jezbera J, Šimek K, Hahn MW. Limnohabitans planktonicus sp. nov., and Limnohabitans parvus sp. nov., two novel planktonic Betaproteobacteria isolated from a freshwater reservoir and emended description of the genus limnohabitans. Int J Syst Evol Microbiol. 2010;60:2710–2714. doi: 10.1099/ijs.0.018952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Meth Microbiol. 2001;30:227–237. [Google Scholar]
- Lee S, Weon H-Y, Han K, Ahn T-Y. Flavobacterium dankookense sp. nov., isolated from a freshwater reservoir and emended descriptions of Flavobacterium cheonanense, F. chungnamense, F. koreense and F. aquatile. Int J Syst Evol Microbiol. 2011;62:2378–2382. doi: 10.1099/ijs.0.032698-0. [DOI] [PubMed] [Google Scholar]
- Lengeler J, Drews G, Schlegel H. Biology of the Prokaryotes. Georg Thieme Verlag; 1999. [Google Scholar]
- Liu Y, Li H, Jiang J-T, Liu Y-H, Song X-F, Xu C-J, et al. Algoriphagus aquatilis sp. nov., isolated from a freshwater lake. Int J Syst Evol Microbiol. 2009;59:1759–1763. doi: 10.1099/ijs.0.005215-0. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acid Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meon B, Jüttner F. Concentrations and dynamics of free mono- and oligosaccharides in a shallow eutrophic lake measured by thermospray mass spectrometry. Aquat Microb Ecol. 1999;16:281–293. [Google Scholar]
- Michelou VK, Lomas MW, Kirchman DL. Phosphate and adenosine-5'-triphosphate uptake by cyanobacteria and heterotrophic bacteria in the Sargasso Sea. Limnol Oceanogr. 2011;56:323–332. [Google Scholar]
- Morris RM, Longnecker K, Giovannoni SJ. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol. 2006;8:1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedros-Alio C, Newell SY. Microautoradiography study of thymidine uptake in brackish waters around Sapelo Island, Georgia, USA. Mar Ecol Prog Ser. 1989;55:83–94. [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Pernthaler J, Zollner E, Warnecke F, Jurgens K. Bloom of filamentous bacteria in a mesotrophic lake: Identity and potential controlling mechanism. Appl Environ Microbiol. 2004;70:6272–6281. doi: 10.1128/AEM.70.10.6272-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwosz K, Salcher MM, Zeder M, Ameryk A, Pernthaler J.(accepted). Seasonal dynamics and activity of typical freshwater bacteria in brackish waters of the Gulf of Gdańsk Limnol Oceanogr
- Posch T, Köster O, Salcher MM, Pernthaler J. Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nature Clim Change. 2012;2:809–813. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez MT, Hörtnagl P, Sommaruga R. Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ Microbiol. 2010;12:74–82. doi: 10.1111/j.1462-2920.2009.02043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez MT, Sommaruga R. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol Oceanogr. 2006;51:2527–2537. [Google Scholar]
- Rosenstock B, Simon M. Use of dissolved combined and free amino-acids by planktonic bacteria in Lake Constance. Limnol Oceanogr. 1993;38:1521–1531. [Google Scholar]
- Sack ELW, van der Wielen PWJJ, van der Kooij D. Flavobacterium johnsoniae as a Model Organism for Characterizing Biopolymer Utilization in Oligotrophic Freshwater Environments. Appl Environ Microbiol. 2011;77:6931–6938. doi: 10.1128/AEM.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Frater N, Posch T. Vertical and longitudinal distribution patterns of differnt bacterioplankton populations in a canyon-shaped, deep prealpine lake. Limnol Oceanogr. 2011a;56:2027–2039. [Google Scholar]
- Salcher MM, Pernthaler J, Posch T. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol Oceanogr. 2010;55:846–856. [Google Scholar]
- Salcher MM, Pernthaler J, Posch T. Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria 'that rule the waves' (LD12) ISME J. 2011b;5:1242–1252. doi: 10.1038/ismej.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol. 2008;10:2074–2086. doi: 10.1111/j.1462-2920.2008.01628.x. [DOI] [PubMed] [Google Scholar]
- Schauer M, Jiang J, Hahn M. Recurrent seasonal variations in abundance and composition of filamentous SOL cluster bacteria (Saprospiraceae, Bacteroidetes) in oligomesotrophic lake Mondsee (Austria) Appl Environ Microbiol. 2006;72:4704–4712. doi: 10.1128/AEM.02935-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol. 2010;12:490–500. doi: 10.1111/j.1462-2920.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- Sekar R, Fuchs BM, Amann R, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Horňák K, Jezbera J, Masin M, Nedoma J, Gasol J, et al. Influence of top-down and bottom-up manipulation on the R-BT065 subcluster of beta-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl Environ Microbiol. 2005;71:2381–2390. doi: 10.1128/AEM.71.5.2381-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Horňák K, Jezbera J, Nedoma J, Znachor P, Hejzlar J, et al. Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat Microb Ecol. 2008;51:249–262. [Google Scholar]
- Šimek K, Kasalický V, Zapomelova E, Horňák K. Alga-derived substrates select for distinct betaproteobacterial lineages and contribute to niche separation in Limnohabitans strains. Appl Environ Microbiol. 2011;77:7307–7315. doi: 10.1128/AEM.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Pernthaler J, Weinbauer MG, Horňák K, Dolan JR, Nedoma J, et al. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl Environ Microbiol. 2001;67:2723–2733. doi: 10.1128/AEM.67.6.2723-2733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-II: a program for sequential, parallel and distributed inference of large phylogenetic. Concurr Comput-Pract Exp. 2005;17:1705–1723. [Google Scholar]
- Sundh I. Biochemical composition of dissolved organic carbon derived from phytoplankton and used by heterotrophic bacteria. Appl Environ Microbiol. 1992;58:2938–2947. doi: 10.1128/aem.58.9.2938-2947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- Van den Wyngaert S, Salcher MM, Pernthaler J, Zeder M, Posch T. Quantitative dominance of seasonally persistent filamentous cyanobacteria (Planktothrix rubescens) in the microbial assemblages of a temperate lake. Limnol Oceanogr. 2011;56:97–109. [Google Scholar]
- Walsby AE, Jüttner F. The uptake of amino acids by the cyanobacterium Planktothrix rubescens is stimulated by light at low irradiances. FEMS Microbiol Ecol. 2006;58:14–22. doi: 10.1111/j.1574-6941.2006.00143.x. [DOI] [PubMed] [Google Scholar]
- Warnecke F, Sommaruga R, Sekar R, Hofer JS, Pernthaler J. Abundances, identity, and growth state of Actinobacteria in mountain lakes of different UV transparency. Appl Environ Microbiol. 2005;71:5551–5559. doi: 10.1128/AEM.71.9.5551-5559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Simon M. Consumption of labile dissolved organic matter by limnetic bacterioplankton: the relative significance of amino acids and carbohydrates. Aquat Microb Ecol. 1999;17:1–12. [Google Scholar]
- Zeder M, Pernthaler J. Multispot live-image autofocusing for high-throughput microscopy of fluorescently stained bacteria. Cytometry Part A. 2009;75A:781–788. doi: 10.1002/cyto.a.20770. [DOI] [PubMed] [Google Scholar]
- Zotina T, Köster O, Jüttner F. Photoheterotrophy and light-dependent uptake of organic and organic nitrogenous compounds by Planktothrix rubescens under low irradiance. Freshwater Biol. 2003;48:1859–1872. [Google Scholar]
- Zubkov M, Fuchs B, Tarran G, Burkill P, Amann R. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl Environ Microbiol. 2003;69:1299–1304. doi: 10.1128/AEM.69.2.1299-1304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]