Abstract
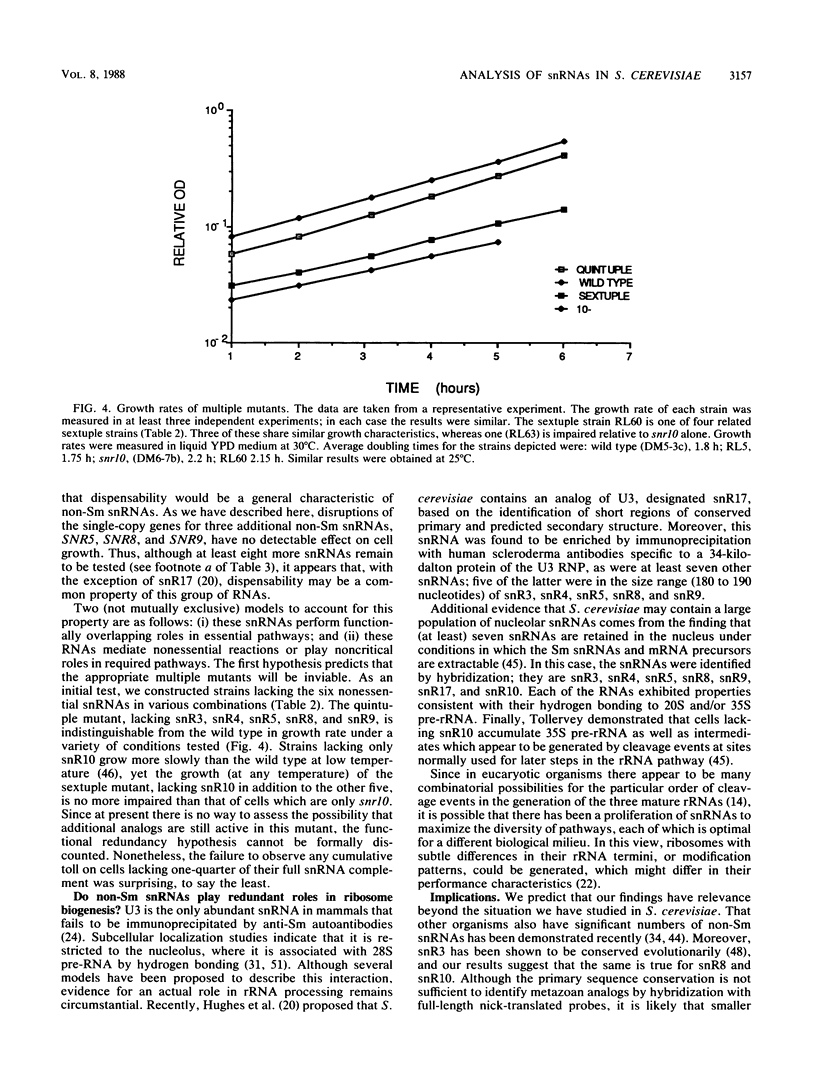
Saccharomyces cerevisiae contains at least 24 distinct small nuclear RNAs (snRNAs), several of which are known to be essential for viability and to participate in the splicing of pre-mRNAs; the RNAs in this subset contain binding sites for the Sm antigen, a hallmark of metazoan snRNAs involved in mRNA processing. In contrast, we showed previously that the single-copy genes for three other snRNAs (snR3, snR4, and snR10) are not required for viability, although cells lacking snR10 are growth impaired at low temperature. None of these RNAs associates with the Sm antigen. To assess this apparent correlation, we cloned and sequenced the genes encoding three additional non-Sm snRNAs. Comparison of these genes with nine additional yeast snRNA genes revealed a highly conserved TATA box located 92 +/- 8 nucleotides 5' of the transcriptional start site. By using the technique of gene replacement with null alleles, each of these three single copy genes was shown to be completely dispensable. We constructed multiple mutants to test the hypothesis that, individually, each of these snRNAs is nonessential because the snRNAs play functionally overlapping roles. A mutant lacking five snRNAs (snR3, snR4, snR5, snR8, snR9) was indistinguishable from the wild type, and growth of the sextuple mutant was no more impaired than that in strains lacking only snR10. This widespread dispensability of snRNAs was completely unexpected and forces us to reconsider the possible roles of these ubiquitous RNAs.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986 Oct 10;47(1):49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y. C. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. The metabolic role of RNases. Trends Biochem Sci. 1988 Apr;13(4):136–139. doi: 10.1016/0968-0004(88)90070-9. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Stunnenberg H. G., Birnstiel M. L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3' histone mRNA termini. Cell. 1983 Oct;34(3):823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987 Dec 24;51(6):887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. A small nuclear ribonucleoprotein associates with the AAUAAA polyadenylation signal in vitro. Cell. 1986 May 23;45(4):581–591. doi: 10.1016/0092-8674(86)90290-4. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hotta Y., Stern H. Small nuclear RNA molecules that regulate nuclease accessibility in specific chromatin regions of meiotic cells. Cell. 1981 Dec;27(2 Pt 1):309–319. doi: 10.1016/0092-8674(81)90414-1. [DOI] [PubMed] [Google Scholar]
- Hughes J. M., Konings D. A., Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987 Jul;6(7):2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. C., Sirdeshmukh R., Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci U S A. 1984 Jan;81(1):185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Rymond B. C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987 Aug 14;50(4):593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987 Jun 5;49(5):613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Primary and secondary structure of U8 small nuclear RNA. J Biol Chem. 1985 Sep 15;260(20):10930–10935. [PubMed] [Google Scholar]
- Riedel N., Wise J. A., Swerdlow H., Mak A., Guthrie C. Small nuclear RNAs from Saccharomyces cerevisiae: unexpected diversity in abundance, size, and molecular complexity. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8097–8101. doi: 10.1073/pnas.83.21.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel N., Wolin S., Guthrie C. A subset of yeast snRNA's contains functional binding sites for the highly conserved Sm antigen. Science. 1987 Jan 16;235(4786):328–331. doi: 10.1126/science.2948278. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Brow D. A., Roiha H., Guthrie C. An essential snRNA from S. cerevisiae has properties predicted for U4, including interaction with a U6-like snRNA. Cell. 1987 Aug 14;50(4):585–592. doi: 10.1016/0092-8674(87)90031-6. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Jones M. H., Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987 Sep 18;237(4821):1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Guthrie C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985 Dec 30;4(13B):3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. High level of complexity of small nuclear RNAs in fungi and plants. J Mol Biol. 1987 Jul 20;196(2):355–361. doi: 10.1016/0022-2836(87)90696-6. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Mattaj I. W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987 Feb;6(2):469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Wise J. A., Guthrie C. A U4-like small nuclear RNA is dispensable in yeast. Cell. 1983 Dec;35(3 Pt 2):753–762. doi: 10.1016/0092-8674(83)90108-3. [DOI] [PubMed] [Google Scholar]
- Wise J. A., Tollervey D., Maloney D., Swerdlow H., Dunn E. J., Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983 Dec;35(3 Pt 2):743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]






