Abstract
Objective
Systematic review and meta-analysis to investigate the association between maternal AGTR1 gene single nucleotide polymorphisms (SNPs) and preeclampsia (PE).
Methods
A systematic literature search was performed using PubMed, EMBASE, Scopus, and HuGE Literature Finder databases. The review was conducted according to PRISMA guidelines. Summary odds ratios (ORs) for the allelic and genotypic contrasts were calculated and compared to indicate the most appropriate genetic model for the polymorphism of interest. Among-study heterogeneity was assessed using the I2 statistic and publication bias was evaluated visually using funnel plots.
Results
Seven maternal SNPs investigated with PE were found, but only AGTR1 +1166A>C accumulated sufficient evidence for meta-analysis. Summary ORs calculated from eight studies (10 populations involving 845 PE cases and 1150 controls) did not reveal an association between the +1166A>C polymorphism and PE (allelic OR = 1.19, 95% CI: 0.96–1.47). No evidence of publication bias and among-study heterogeneity was detected.
Conclusions
Meta-analysis findings did not support AGTR1 +1166A>C as a susceptibility locus for PE. Other AGTR1 SNPs require more study.
Keywords: angiotensin II receptor type 1, genetic association study, pregnancy, single nucleotide polymorphism
INTRODUCTION
Preeclampsia (PE), a life-threatening and multi-system disorder of pregnancy, is a leading cause of maternal-fetal morbidity and mortality worldwide. PE complicates 2–7% of pregnancies among healthy nulliparous women and causes 16% of pregnancy-related deaths in the United States [1, 2]. Maternal symptoms may manifest as gestational hypertension and proteinuria with other multi-system abnormalities [2]. The only known treatment for PE is delivery of fetal membranes.
The complex etiology of PE is influenced by maternal and fetal genetic and environmental factors [3]. Alterations in the maternal renin-angiotensin system (RAS), a hormone system involved in blood pressure (BP) regulation, vascular remodeling, and fluid balance, have been implicated in the pathogenesis of PE [4, 5]. The RAS plays a crucial role in maintaining a normal pregnancy, where plasma levels of renin, angiotensin II, and aldosterone are up-regulated [6, 7]. Angiotensin II is the major signaling molecule of the RAS and its major cardiovascular effects are mediated by angiotensin II receptor type 1 (gene denoted as AGTR1), including vasoconstriction, vascular growth promotion, anti-natriuresis, aldosterone synthesis, and inhibition of renin synthesis and release [8]. In PE, this equilibrium is disrupted and plasma levels of these proteins decrease toward the normal non-gravid range. The specific reasons for this imbalance remain unknown, but the genes encoding the components of the RAS, particularly AGTR1, may be plausible candidates [9]. Molecular studies have identified AGTR1 autoantibodies as potential contributors to PE [10].
The AGTR1 gene is highly polymorphic [11]. Seven single nucleotide polymorphisms (SNPs) and a dinucleotide repeat polymorphism have been reported with PE: −810T>A, −713T>G, −521C>T, and −153A>G in the 5’-flanking region; +573T>C and +1162A>G in exon 3 of the coding region; +1166A>C in the 3’-untranslated region (3’-UTR); and a dinucleotide (CA) repeat polymorphism in the 3’-flanking region [9, 12]. The transversion in the 3’-UTR (+1166A>C) of this gene is the best characterized SNP. The association between the maternal AGTR1 +1166A>C SNP and PE has been investigated in several studies with inconsistent findings [9]. The lack of reproducibility may be due to issues relating to study design, sample size, and true variability between populations [13]. The discrepancies may also be due to the inadequate statistical power of individual studies to detect small or moderate effects. A meta-analysis can be performed to address some of these obstacles by increasing statistical power and assessing generalizability of findings across populations [13].
The purpose of the present study is to assess the relationship between the maternal AGTR1 SNPs and PE by conducting a systematic review and meta-analysis. Subgroup analysis was planned to evaluate the consistency of genetic effects across different ethnic populations.
METHODS
Identification of eligible studies
Complete study methods are in Supplemental Digital Content 1. This review was performed according to PRISMA guidelines [14]. PubMed, EMBASE, and Scopus literature databases were searched using a sensitive search strategy to identify relevant studies. The HuGE Literature Finder database was consulted for its listing of articles under the PE phenotype and AGTR1. A manual review of the reference lists of retrieved full-text articles and existing review articles on the topic [9, 12] was conducted for report of other relevant studies. No search restrictions were applied. The last search was conducted on December 4, 2011.
Article titles and abstracts were screened and excluded based on the following exclusion criteria: ineligible phenotype, review article, conference abstract, basic science research, or animal research. The full-text of studies passing initial screening was retrieved and reviewed for eligibility based on the aforementioned and following criteria: not case-control or nested case-control study design, subjects included in another study, or consanguineous subjects. For multiple publications based on related data sets, the study with the greatest number of subjects was included. For studies with multiple geographical or ethnic populations, each population was considered separately.
Phenotype specification
PE was defined as having new onset hypertension (systolic BP ≥ 140 mmHg or diastolic BP ≥ 90 mmHg on at least one occasion after 20 weeks of gestation) and proteinuria (24-hour urine collection with protein ≥ 300 mg or dipstick protein test value of 1+ or greater) [15].
Statistical analysis
A meta-analytic approach not assuming an a priori genetic model was used, as prior evidence did not favor one specific model over others for the polymorphism under study. Data analysis was only performed on AGTR1 SNPs investigated in at least five studies using Review Manager Version 5.1.2 (Nordic Cochrane Centre, Cochrane Collaboration, Copenhagen, Sweden) and MetaAnalyst Version Beta 3.13 (Tufts Medical Center, Boston, MA).
First, Hardy-Weinberg equilibrium (HWE) was checked in normotensive controls using the chi-square (χ2) goodness of fit test. Second, the pooled frequency of the minor allele was estimated in various ethnic groups using the inverse variance method. Third, among-study heterogeneity was assessed using the I2 statistic [16] separately for the summary odds ratios (ORs) across studies for the allelic comparison (m vs. M) and genotypic comparisons of co-dominant (mm vs. MM and Mm vs. MM), dominant (Mm + mm vs. MM), and recessive (mm vs. MM + Mm) models, where “M” is the major allele and “m” is the minor allele. For all comparisons, the minor allele was considered to be the putative risk allele. The inverse variance method was used to estimate the pooled OR and 95% confidence interval (CI), assuming a fixed effects model, in the absence of significant heterogeneity (I2 < 25%). Otherwise, a random effects model was assumed. Comparisons of the summary ORs indicated the most appropriate genetic model for the polymorphism. The significance of the pooled OR was determined by the Z-test with a p-value of < 0.05 considered statistically significant.
To evaluate the impact of HWE-deviated studies on the summary effect estimates, pooled ORs and corresponding 95% CIs were recalculated based on HWE-predicted genotype frequencies in controls rather than using the observed frequencies [17]. Sensitivity analysis was performed including and excluding studies with HWE-deviated and HWE-predicted controls.
Planned subgroup analysis was conducted to examine the effect of ethnicity on the association. Influence analysis was conducted to allow identification of studies excessively perturbing the summary estimate by recalculating the pooled estimate omitting one study at a time. Publication bias was assessed visually using funnel plots.
RESULTS
Study inclusion and characteristics
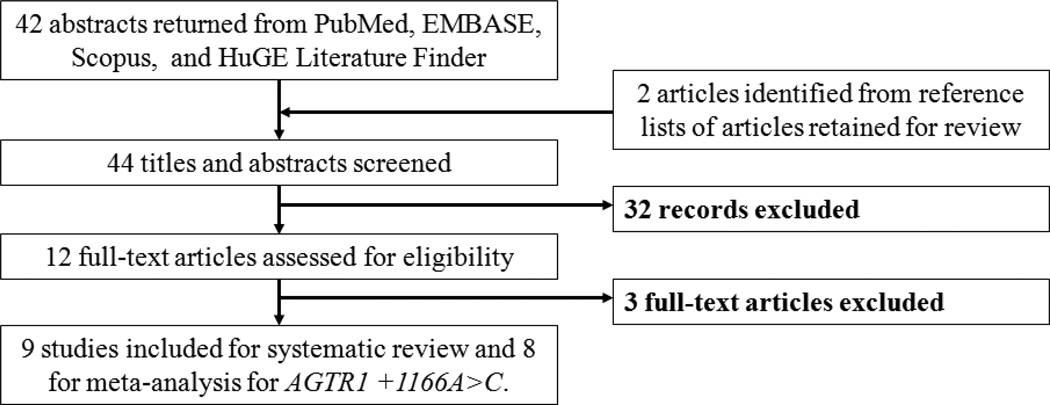
Literature search identified 44 potentially relevant articles. Initial screening of titles and abstracts excluded 32 studies not meeting the eligibility criteria. The full-text of the remaining 12 articles was retrieved for review and three additional articles were excluded for shared subjects. Multiple publications were discovered for two data sets: a United Kingdom (UK) population [18–20] and a Romanian population [21, 22]. For the UK population, the study with the largest number of subjects was retained [20]. AGTR1 +1162A>G genotype frequency data for this population, which was not assessed by Plummer et al. [20], was abstracted from the related paper with shared subjects by Morgan et al. [18]. The study with the larger number of case subjects was retained for the Romanian population [21]. The study by Akbar et al. [23] included three ethnic populations (black Caribbean, Pakistani, and white). Nine studies reporting a SNP association for the +1166A>C polymorphism were included for review. One population (black Zulu) was monomorphic at this site and excluded from meta-analysis [24]. Polymorphisms at −810, −713, −521, −153, +573, and +1162 were each investigated in only one study [18, 20] and excluded from meta-analysis. Therefore, this review yielded 10 populations from eight studies published in the English language [20, 21, 23, 25–29] for meta-analysis of the AGTR1 +1166A>C polymorphism and PE. The study selection process is presented in Figure 1.
Figure 1. Flow diagram of systematic review and meta-analysis literature search results.
HuGE is the Human Genome Epidemiology Network. Nine studies reporting a SNP association for the +1166A>C polymorphism were included for review. One population (black Zulu) was monomorphic at this site and was not included for meta-analysis [24]. Polymorphisms at −810, −713, −521, −153, +573, and +1162 were each reported in only one study [18, 20].
All studies retained for review used a case-control design. Of the 10 populations examining the +1166A>C polymorphism with diagnosed PE for meta-analysis, one included only mild PE (140/90 ≤ BP ≤ 160/110 mmHg) [28], a Pakistani population included a proportion of eclampsia cases [23], and another included severe PE superimposed to chronic hypertension [21]. Six European populations [20, 21, 23, 25, 26, 28] and one each of black Caribbean [23], Chinese [27], Iranian [29], and Pakistani [23] populations were investigated. Most studies defined exclusion criteria for cases, but not for controls. Although PE diagnostic criteria was not uniform across all studies, all studies met diagnostic criteria defined in the Methods section, with the exception of Akbar et al. [23] who recruited subjects presenting with clinical signs of PE prior to 20 weeks of gestation. Genotyping methods were judged appropriate in all studies. However, only two studies reported application of some form of genotyping quality control and none reported that genotyping was blinded to case-control status. Deviation from HWE was detected in the controls of one study [29]. All studies used unique samples and a total of 845 genotyped PE cases and 1150 genotyped normotensive controls were included in the meta-analysis. Study characteristics are summarized in Table S1, Supplemental Digital Content 2, and AGTR1 +1166A>C allele and genotype frequencies are listed in Table S2, Supplemental Digital Content 3.
Pooled AGTR1 +1166C allele frequency in controls
Pooled AGTR1 +1166C allele frequency was 24.9% (95% CI: 22.5–27.4%) for European populations using the inverse variance fixed effects model. The +1166C allele frequencies were 10.1% (95% CI: 6.9–14.6%) in a black Caribbean population, 5.7% (95% CI: 3.3–9.8%) in a Chinese population, and 9.0% (95% CI: 6.5%–12.3%) in a Pakistani population.
Association between AGTR1 +1166A>C and preeclampsia
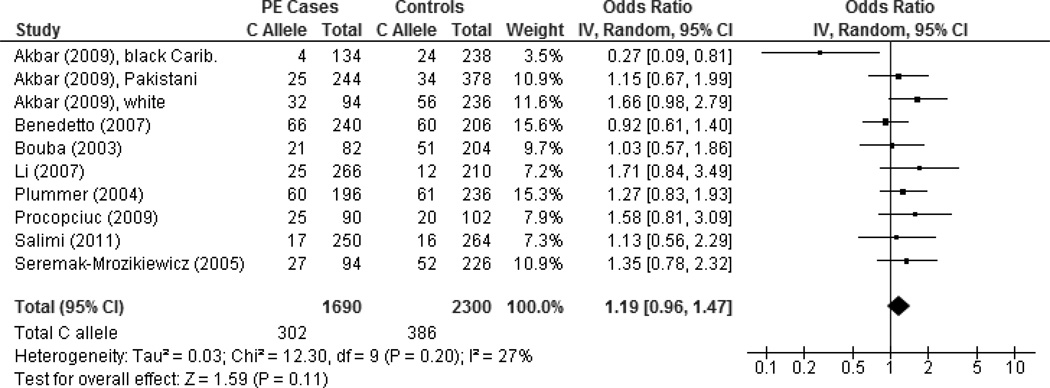
Summary ORs for the allelic and genotypic contrasts and corresponding I2 statistics are presented in Table I. For all studies, no to low heterogeneity were detected for the comparisons, revealing an absence of among-study heterogeneity. Meta-analysis findings did not reveal an association between the AGTR1 +1166A>C polymorphism and PE (allelic OR = 1.19, 95% CI: 0.96–1.47, p = 0.11; I2 = 27%) (Figure 2). The funnel plot did not show evidence of publication bias (Figure S1, Supplemental Digital Content 4). Sensitivity analyses excluding a black Caribbean population in the UK with no CC carriers among cases [23] or excluding the HWE-deviated and including the HWE-predicted study [29] did not meaningfully change the meta-analysis results. Further sensitivity analyses excluding a study that did not fully conform to the review’s phenotype specification [23] or excluding studies with combined PE and non-PE (i.e. eclampsia or PE superimposed to chronic hypertension) outcomes [21, 23] obtained similar non-substantial changes. Additionally, statistically similar results were obtained by the exclusion of any one particular study in the influence analysis, indicating stability of the combined effect.
Table I.
Summary ORs and 95% CIs of the association between AGTR1 +1166A>C polymorphism and preeclampsia
| Comparison | Subgroup | No. of studies (populations) a |
OR (95% CI) | p-value b | I2 (%) c | Statistical model |
|---|---|---|---|---|---|---|
| Allelic | ||||||
| C vs. A | Overall | 8 (10) | 1.19 (0.96–1.47) | 0.11 | 27 | Random |
| European ethnicity | 6 (6) | 1.23 (1.00–1.51) | 0.05 | 0 | Fixed | |
| Non-Euro. ethnicity | 3 (4) | 0.99 (0.55–1.79) | 0.98 | 62 | Random | |
| Codominant model | ||||||
| CC vs. AA | Overall | 8 (10) | 1.48 (0.95–2.30) | 0.08 | 0 | Fixed |
| AC vs. AA | Overall | 8 (10) | 1.14 (0.91–1.43) | 0.26 | 2 | Fixed |
| Dominant model | ||||||
| AC + CC vs. AA | Overall | 8 (10) | 1.18 (0.95–1.46) | 0.13 | 16 | Fixed |
| Recessive model | ||||||
| CC vs. AA + AC | Overall | 8 (10) | 1.42 (0.92–2.18) | 0.11 | 0 | Fixed |
Abbreviations: CI, confidence interval; NS, non-significant; OR, odds ratio.
Number of studies (number of populations).
Z-test for overall effect.
Guideline for interpretation of the I2 statistic: I2 = 0% no heterogeneity, I2 = 25% low heterogeneity, I2 = 50% moderate heterogeneity, and I2 = 75% high heterogeneity [16].
Figure 2. Forest plot of AGTR1 +1166 C versus A alleles for all studies.
The forest plot displays the meta-analysis results of all studies included in the review. Meta-analysis was conducted using an inverse variance (IV), random effects model. For each study in the forest plot, the area of the black square is proportional to study weight and the horizontal bar represents the 95% confidence interval (CI). Preeclampsia is abbreviated as PE.
Subgroup analysis of studies by ethnicity (European versus non-European) did not meaningfully change the gene effects (see Table I). Based on the heterogeneity analysis, non-European studies (I2 = 62%) appear to account for the among-study heterogeneity observed in the overall allelic contrast (I2 = 27%). No substantial heterogeneity was detected among European studies (I2 = 0%). Funnel plots did not provide any indication of possible publication bias (figures not shown).
DISCUSSION
The present systematic review and meta-analysis examined the relationship between AGTR1 gene polymorphisms and susceptibility to PE. Only one AGTR1 SNP at position +1166 with sufficiently accumulated published evidence was included in the meta-analysis. AGTR1 is one of the most plausible candidate genes given the current understanding of the biological processes involved in the pathogenesis of PE. Recent functional genetic studies have demonstrated that microRNA-155 attenuates the expression of +1166A, but not +1166C allele [30, 31]. Nonetheless, no association was found between AGTR1 +1166A>C and PE when all evidence (eight case-control studies and 10 corresponding populations) was considered.
The pooled AGTR1 +1166C allele frequencies showed large differences across ethnicities (European populations: 24.9%; non-European populations: black Caribbean, 10.1%; Chinese, 5.7%; and Pakistani, 9.0%). The C allele was not detected in a black Zulu population [24]. In comparison, allele frequencies reported in reference white, black American, Chinese, and Japanese populations were 25.0%, 5.0%, 4.0%, and 3.0%, respectively [32]. Although different ethnicities were combined in the meta-analysis, the gene effects were consistent across the broad ethnic categorizations (European and non-European) in the subgroup analysis. Unfortunately, non-European studies are limited in number, rendering more specific stratification by ethnicity impossible.
AGTR1 SNPs other than +1166A>C have been investigated with PE in only one population [18, 20]. Allele or genotype distributions for polymorphisms at positions −810, −713, −521, −153, +573, and +1162 were statistically similar between cases and controls in this population as was the distribution of dinucleotide (CA) repeat alleles.
Several caveats should be noted when interpreting the results of this review. First, although efforts were made to obtain data from multiple sources and overt publication bias was not detected, the completeness of evidence may be impeded by the file drawer effect, where positive rather than negative findings tend to be published [33]. Second, completeness of evidence may also be limited by language bias. Language bias is a tendency for studies conducted in non-English speaking countries to publish significant results in international journals, which are usually indexed in major international bibliographic databases, and non-significant results in domestic journals, many of which are not indexed [34]. Selective publication of polymorphisms in the human genome and diseases may obscure true relationships between the polymorphisms under investigation and the outcomes of interest. An additional issue influenced by language bias is ethnicity. Information on different allele frequencies associated with different ethnicities with possible relation to the outcome may be excluded unintentionally if studies are being published in non-indexed journals. Third, related to the last point, studies examining the +1166A>C polymorphism were primarily performed in European populations and the association with PE in other ethnic populations should be further investigated due to the ethnic differences of this SNP. Last, most reviewed studies did not report genotypes for cases by severity (e.g. mild versus severe PE). It was demonstrated previously that restricting analysis to studies with more precisely defined case-control phenotypes may improve meta-analysis efficiency [35].
Despite weaknesses inherent to the systematic review and meta-analysis process, the present study provides a comprehensive examination of the available evidence on the association between maternal AGTR1 polymorphisms and PE. The larger number of cases and controls compared to individual studies included in the AGTR1 +1166A>C meta-analysis allowed a more precise risk estimate to be obtained. The results of this meta-analysis demonstrated that AGTR1 +1166A>C is not a likely susceptibility locus for PE. Given the limited investigation of other AGTR1 SNPs, additional studies are warranted to better elucidate their roles in PE pathogenesis.
Supplementary Material
Acknowledgments
DECLARATION OF INTEREST
This work was supported by grants AI41040 and DA05484 from the National Institutes of Health.
REFERENCES
- 1.Chang J, Elam-Evans LD, Berg CJ, Herndon J, Flowers L, Seed KA, Syverson CJ. Pregnancy-related mortality surveillance--United States, 1991--1999. MMWR Surveill Summ. 2003;52(2):1–8. [PubMed] [Google Scholar]
- 2.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365(9461):785–799. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 3.Williamson C. Molecular biology related to pre-eclampsia. Int Congr Ser. 2005;1279:282–289. [Google Scholar]
- 4.Shah DM. The role of RAS in the pathogenesis of preeclampsia. Curr Hypertens Rep. 2006;8(2):144–152. doi: 10.1007/s11906-006-0011-1. [DOI] [PubMed] [Google Scholar]
- 5.Vitoratos N, Hassiakos D, Iavazzo C. Molecular mechanisms of preeclampsia. J Pregnancy. 2012;2012:298343. doi: 10.1155/2012/298343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham F, Leveno K, Bloom S, Hauth J, Rouse D, Spong C. Williams Obstetrics. 23rd ed. New York: McGraw-Hill; 2010. Pregnancy hypertension; pp. 706–756. [Google Scholar]
- 7.Irani RA, Xia Y. The Functional Role of the Renin-Angiotensin System in Pregnancy and Preeclampsia. Placenta. 2008;29(9):763–771. doi: 10.1016/j.placenta.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senbonmatsu T, Saito T, Landon EJ, Watanabe O, Price E, Jr, Roberts RL, Imboden H, Fitzgerald TG, Gaffney FA, Inagami T. A novel angiotensin II type 2 receptor signaling pathway: possible role in cardiac hypertrophy. EMBO J. 2003;22(24):6471–6482. doi: 10.1093/emboj/cdg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutze S, Rudnik-Schoneborn S, Zerres K, Rath W. Genes and the preeclampsia syndrome. J Perinat Med. 2008;36(1):38–58. doi: 10.1515/JPM.2008.004. [DOI] [PubMed] [Google Scholar]
- 10.Hubel CA, Wallukat G, Wolf M, Herse F, Rajakumar A, Roberts JM, Markovic N, Thadhani R, Luft FC, Dechend R. Agonistic angiotensin II type 1 receptor autoantibodies in postpartum women with a history of preeclampsia. Hypertension. 2007;49(3):612–617. doi: 10.1161/01.HYP.0000256565.20983.d4. [DOI] [PubMed] [Google Scholar]
- 11.Baudin B. Polymorphism in angiotensin II receptor genes and hypertension. Exp Physiol. 2005;90(3):277–282. doi: 10.1113/expphysiol.2004.028456. [DOI] [PubMed] [Google Scholar]
- 12.Medica I, Kastrin A, Peterlin B. Genetic polymorphisms in vasoactive genes and preeclampsia: a meta-analysis. Eur J Obstet Gynecol Reprod Biol. 2007;131(2):115–126. doi: 10.1016/j.ejogrb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Salanti G, Sanderson S, Higgins JP. Obstacles and opportunities in meta-analysis of genetic association studies. Genet Med. 2005;7(1):13–20. doi: 10.1097/01.gim.0000151839.12032.1a. [DOI] [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner LK. Diagnosis and management of preeclampsia. Am Fam Physician. 2004;70(12):2317–2324. [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163(4):300–309. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 18.Morgan L, Crawshaw S, Baker PN, Edwards R, Broughton Pipkin F, Kalsheker N. Functional and genetic studies of the angiotensin II type 1 receptor in pre-eclamptic and normotensive pregnant women. J Hypertens. 1997;15(12 Pt 1):1389–1396. doi: 10.1097/00004872-199715120-00004. [DOI] [PubMed] [Google Scholar]
- 19.Morgan L, Crawshaw S, Baker PN, Brookfield JF, Broughton Pipkin F, Kalsheker N. Distortion of maternal-fetal angiotensin II type 1 receptor allele transmission in pre-eclampsia. J Med Genet. 1998;35(8):632–636. doi: 10.1136/jmg.35.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plummer S, Tower C, Alonso P, Morgan L, Baker P, Broughton-Pipkin F, Kalsheker N. Haplotypes of the angiotensin II receptor genes AGTR1 and AGTR2 in women with normotensive pregnancy and women with preeclampsia. Hum Mutat. 2004;24(1):14–20. doi: 10.1002/humu.20050. [DOI] [PubMed] [Google Scholar]
- 21.Procopciuc LM, Caracostea G, Iordache G, Olteanu I, Stamatian F. Influence of RAS polymorphisms on the development and perinatal outcome of preeclampsia. Genetic RAS evaluation. Gineco.ro. 2009;5(2):88–93. [Google Scholar]
- 22.Procopciuc LM, Caracostea G, Zaharie G, Puscas M, Iordache G, Popa M, Colcear D, Olteanu I, Stamatian F. Maternal/newborn genotype contribution of the renin-angiotensin system (Met235Thr, Thr174Met, I/D-ACE, A2350G-ACE, A1166C-AT2R1, C3123A- AT2R2, 83A/G-REN) to the risk of pre-eclampsia: a Romanian study. J Renin Angiotensin Aldosterone Syst. 2011;12(4):539–548. doi: 10.1177/1470320311399603. [DOI] [PubMed] [Google Scholar]
- 23.Akbar SA, Khawaja NP, Brown PR, Tayyeb R, Bamfo J, Nicolaides KH. Angiotensin II type 1 and 2 receptors gene polymorphisms in pre-eclampsia and normal pregnancy in three different populations. Acta Obstet Gynecol Scand. 2009;88(5):606–611. doi: 10.1080/00016340902859307. [DOI] [PubMed] [Google Scholar]
- 24.Roberts CB, Rom L, Moodley J, Pegoraro RJ. Hypertension-related gene polymorphisms in pre-eclampsia, eclampsia and gestational hypertension in Black South African women. J Hypertens. 2004;22(5):945–948. doi: 10.1097/00004872-200405000-00016. [DOI] [PubMed] [Google Scholar]
- 25.Benedetto C, Marozio L, Ciccone G, Chieppa G, Quaglia M, Matullo G, Bertola L, Guarrera S, Carturan S, Stratta P. Synergistic effect of renin-angiotensin system and nitric oxide synthase genes polymorphisms in pre-eclampsia. Acta Obstet Gynecol Scand. 2007;86(6):678–682. doi: 10.1080/00016340701415269. [DOI] [PubMed] [Google Scholar]
- 26.Bouba I, Makrydimas G, Kalaitzidis R, Lolis DE, Siamopoulos KC, Georgiou I. Interaction between the polymorphisms of the renin-angiotensin system in preeclampsia. Eur J Obstet Gynecol Reprod Biol. 2003;110(1):8–11. doi: 10.1016/s0301-2115(03)00046-0. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Ma Y, Fu Q, Wang L. Angiotensin-converting enzyme insertion/deletion (ACE I/D) and angiotensin II type 1 receptor (AT1R) gene polymorphism and its association with preeclampsia in Chinese women. Hypertens Pregnancy. 2007;26(3):293–301. doi: 10.1080/10641950701413676. [DOI] [PubMed] [Google Scholar]
- 28.Seremak-Mrozikiewicz A, Dubiel M, Drews K, Breborowicz GH, Mrozikiewicz PM. 1166C mutation of angiotensin II type 1 receptor gene is correlated with umbilical blood flow velocimetry in women with preeclampsia. J Matern Fetal Neonatal Med. 2005;17(2):117–121. doi: 10.1080/14767050500043400. [DOI] [PubMed] [Google Scholar]
- 29.Salimi S, Mokhtari M, Yaghmaei M, Jamshidi M, Naghavi A. Association of Angiotensin-Converting Enzyme Intron 16 Insertion/Deletion and Angiotensin II Type 1 Receptor A1166C Gene Polymorphisms with Preeclampsia in South East of Iran. J Biomed Biotechnol. 2011;2011:941515. doi: 10.1155/2011/941515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282(33):24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3' untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Submitted SNP(ss) details: ss44469395. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss44469395.
- 33.Rosenthal R. The “file drawer problem” and tolerance for null results. Psychol Bull. 1979;86(3):638–641. [Google Scholar]
- 34.Juni P, Holenstein F, Sterne J, Bartlett C, Egger M. Direction and impact of language bias in meta-analyses of controlled trials: empirical study. Int J Epidemiol. 2002;31(1):115–123. doi: 10.1093/ije/31.1.115. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Bracken MB. Association of CD14 −260 (−159) C>T and asthma: a systematic review and meta-analysis. BMC Med Genet. 2011;12(1):93. doi: 10.1186/1471-2350-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.