Abstract
The cellular reaction to genomic instability includes a network of signal transduction pathways collectively referred to as the DNA damage response (DDR). Activated by a variety of DNA lesions, the DDR orchestrates cell cycle arrest and DNA repair, and initiates apoptosis in instances where damage cannot be repaired. As such, disruption of the DDR increases the prevalence of DNA damage secondary to incomplete repair, and in doing so, enhances radiation-induced cytotoxicity. This article describes the molecular agents and their targets within DDR pathways that sensitize cells to radiation. Moreover, it reviews the therapeutic implications of these compounds, provides an overview of relevant clinical trials and offers a viewpoint on the evolution of the field in the years to come.
Keywords: apoptosis, cell cycle, DNA damage response, ionizing radiation, radioprotection, radiosensitization
Ionizing radiation damages numerous sub-cellular structures extending from the plasma membrane to the cell nucleus. Multiple lines of evidence indicate that among these, radiation-induced cytotoxicity is closely linked to DNA damage. Mammalian cells experience in excess of 10,000 DNA lesions per day, secondary to chemical decay, replication errors and environmental radiation, and employ a network of highly conserved molecular machinery to minimize or repair genomic stress. Consequently, the amount of DNA damage induced by radiation varies, not only with the number of ionizations and their proximity to the double helix, but also with the cellular capacity to scavenge free radicals and the efficiency of DNA damage repair pathways [1].
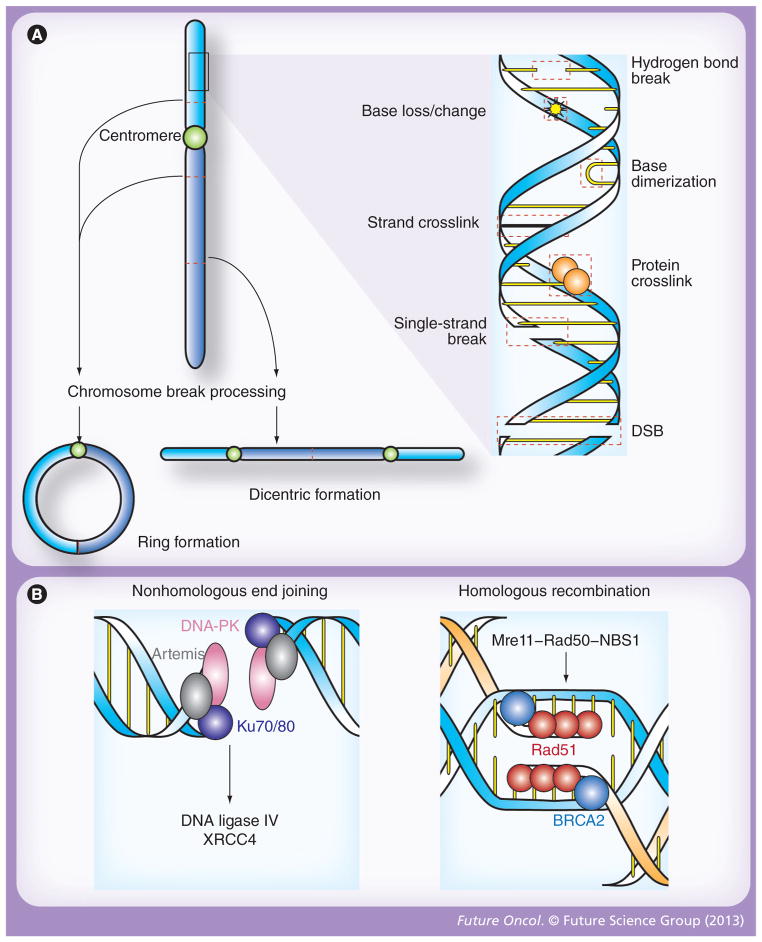
The most common forms of radiation-induced DNA injury are damage to individual nitrogenous bases and single-strand breaks (SSBs) (Figure 1A) [2]. In general, these lesions are easily repaired and merely result in nonlethal mutations if perpetuated through DNA replication. By contrast, numerous lines of evidence indicate that the rate of ionizing radiation-associated apoptosis closely correlates with the incidence of double-strand breaks (DSBs) [3]. Larger radiation-associated chromosomal aberrations, such as dicentric changes, ring formation and anaphase bridges, are also cytotoxic, but these changes occur far less frequently than DSBs (Figure 1A) [4,5]. Therefore, DSBs are considered to be the primary mechanism of cell death in response to radiation.
Figure 1. Patterns of DNA damage and double-stranded break repair following ionizing radiation.
(A) Radiation induces molecular damage, both directly and through the formation of radical oxygen species. At the level of the double helix, these changes including loss, alteration or dimerization of nitrogenous bases; crosslinkage of associated proteins or opposing DNA strands; and breakage of hydrogen bonds as well as one or both strands of DNA (right). While DSBs are the primary cytotoxic lesion associated with radiation, each of these changes activates distinct repair mechanisms, and, if significant, may result in chromosomal aberrations that are also processed by unique molecular repair machinery (left). In this regard, two break points within a single chromatid may fuse to form a ring structure (bottom left), while the union of opposing broken chromatids may result in dicentric formations (bottom right). Stable translocations, deletions and other nonlethal chromosomal aberrations may also occur after exposure to ionizing radiation, but similar to DSBs, ring and dicentric chromosomes lead to cell death via mitotic catastrophe. (B) DSBs are the principle cytotoxic lesion induced by ionized radiation, and their repair is primarily governed by nonhomologous end joining and homologous recombination [9]. In nonhomologous end joining (left), broken ends are recognized by the Ku70/80 heterodimer, which then recruits DNA-PK and artemis. This protein complex serves as a docking site for DNA ligase IV, and its cofactor XRCC4, which catalyze end joining. Homologous recombination (right) is initiated when the Mre11–Rad50–NBS1 complex recognizes the ends of a DSB. BRCA2 then loads Rad51 onto the processed ends, which triggers homologous strand invasion and subsequent ligation of the broken ends. DSB: Double-strand break.
Along with non-DSB oxidative clustered DNA lesions, DSBs are considered to be a form of complex DNA damage that results from clustering of multiple types of damage [6]. DSBs are defined by the presence of two SSBs in close proximity, and while these lesions may be independently created by separate high linear energy transfer events from external radiation, repair enzymes often generate additional SSBs when excising missing nitrogenous bases and oxidative lesions within a single DNA damage cluster [7,8]. Once processed by repair machinery, DSBs are either rejoined in their original confirmation or inappropriately attached to another broken DNA strand to generate a chromosome aberration. The mechanisms of DNA repair are numerous, and include base and nucleotide excision repair, mismatch repair, DNA damage bypass, nonhomologous end joining (NHEJ), homologous recombination, single-strand annealing and crosslink repair [9]. However, among these, NHEJ and homologous recombination are the primary mechanisms of DSB repair (Figure 1B).
DNA damage response
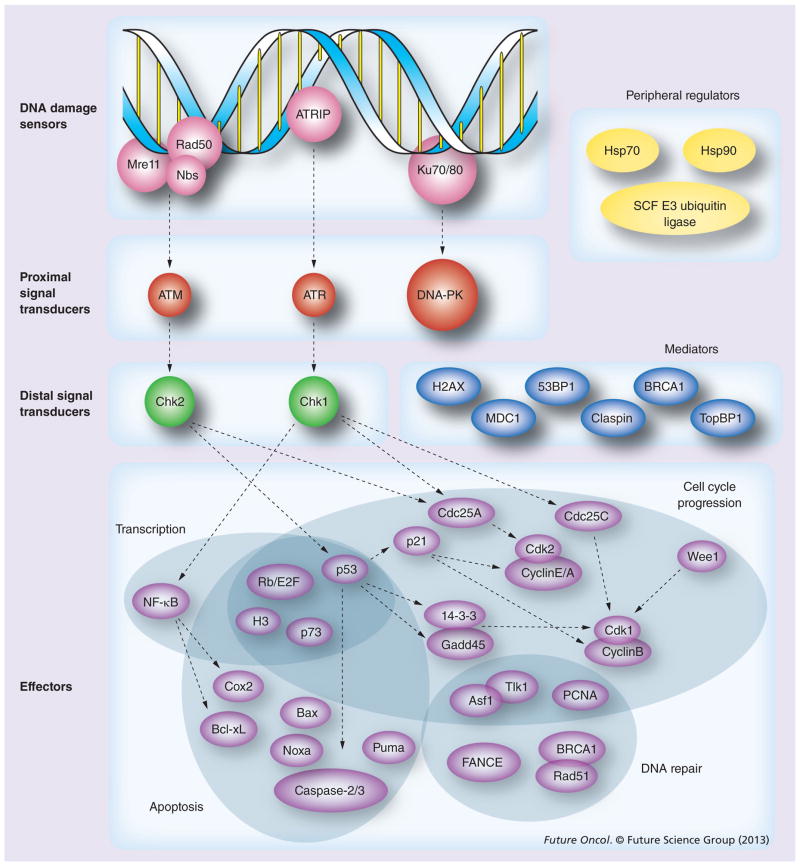
The DNA damage response (DDR) is an evolutionarily conserved signal transduction network that is activated in response to genomic instability [10,11]. Individual DNA lesions trigger distinct yet overlapping pathways to regulate the cell cycle and influence a myriad of distal functions including DNA repair, transcription, cell cycle progression, apoptosis and senescence. Initially, DNA damage is identified by sensor proteins, such as the Mre11–Rad50–Nbs (MRN) complex, ATRIP and Ku70/80, which rapidly activate the proximal signal transduction kinases ATM, ATR and DNA-PK, respectively (Figure 2). While the exact mechanisms of DNA damage recognition and signal transduction continue to be elucidated, several canonical pathways have been identified. ATM is activated in response to MRN association with DSBs [12] and, in turn, catalyzes H2AX phosphorylation to recruit DNA repair proteins through successive histone ubiquitination events [13–15]. By contrast, the ATRIP–ATR pathway is classically associated with SSB processing [16], and both Ku70 and DNA-PK mediate NHEJ [17]. However, in spite of these classic signal transduction pathways, multiple lines of evidence suggest that abundant overlap exists within the DDR. For instance, ATR is also activated by the MRN complex in response to DSBs [18] and NHEJ can also be initiated by the MRN complex [17,19].
Figure 2. DNA damage response pathways.
While numerous feedback pathways exist within the DNA damage response (DDR), the canonical flow of signaling proceeds from sensors of DNA damage through proximal and distal signal transducers, and on to effector proteins. Shaded regions represent unique effector functions, although much like upstream transduction kinases, many DDR effectors fulfill multiple roles. Mediator proteins – many of which are activated by proximal signal transducers – and peripheral regulators influence multiple levels of DDR signaling. Dashed arrows indicate classical signaling pathways within the DDR, but are not specific for either activating or inhibitory signaling events [10,35,36,105].
Downstream of DDR sensor proteins, the PI3-like proximal transduction kinases ATM, ATR and DNA-PK are similarly known to phosphorylate numerous overlapping targets in response to ionizing radiation [20]. Nevertheless, ATM, ATR and DNA-PK retain distinct sub-cellular functions, and among their substrates, several key regulators of the cell cycle and apoptosis have been shown to be essential for tumor progression and the response to radiation. ATM, a heavily phosphorylated 370-kDa kinase, is the primary transducer of DSB repair. Through phosphorylation of checkpoint kinase Chk2, and subsequent activation of the tumor suppressor p53 [21], ATM has been shown to regulate a multitude of cellular process including DNA repair, cell cycle progression, oxidative stress levels and mitochondrial homeostasis [22,23]. The myriad of downstream effector proteins influenced by ATM include the apoptotic factors Bax, Noxa, Puma, caspase-2 and -3, as well as cell cycle regulators, such as p21, 14-3-3, and Gadd45 [24–26]. p21, which functions downstream of p53, plays an especially critical role in mediating the effect of ATM on cell cycle progression through interaction with the Cdk complex, Cdk2/cyclin E/A, to control the G1 checkpoint (Figure 2) [27]. More than 50% of human malignancies bear a mutation or deletion in the gene that encodes for p53, and consequently the G1 checkpoint is thought to have particular relevance for cancer development [28]. Notably, disruption of the G2 checkpoint in the absence of G1 arrest profoundly enhances the cytotoxic effects of ionizing radiation, and has, therefore, proven to be an effective therapeutic strategy for cancer treatment. Premature cell cycle progression in the absence of both G1 and G2 checkpoints leads to segregation of aberrant chromosomes and mitotic catastrophe, a salient mechanism of lethality induced by ionizing radiation through delayed cell death. While the determination between cell cycle arrest and DNA repair versus cell death is poorly understood, numerous mediators and peripheral regulators of DDR activity have been shown to influence the ultimate cellular response to DNA damage (Figure 2) [29–32].
Much like ATM, ATR is activated in response to DSBs and can phosphorylate p53 in an ATM-dependent manner to influence the transcription of anti-apoptotic and cell cycle effector proteins [33]. ATR is constitutively bound by ATRIP and, unlike ATM, can also be activated by virtue of ATRIP interaction with RPA-coated SSBs [16]. Structurally distinct from either ATM or ATR, DNA-PK is classically activated in conjunction with Ku70 during NHEJ. Among other functions, DNA-PK recruits the endonuclease Artemis and phosphorylates the DNA ligase IV complex to facilitate DNA end joining [11]. While often considered to be of ancillary importance due to functional redundancy with other PI3-like kinases, expression of DNA-PK lacking activatable phosphorylation sites leads to bone marrow failure, suggesting that further investigation is needed to characterize the role of DNA-PK beyond the DDR [34].
Many of the downstream targets of the DDR pathway are regulated by distal serine/threonine checkpoint kinases, and numerous lines of evidence indicate that Chk1 and Chk2 are critical for maintenance of genomic integrity and tumor inhibition [35]. Chk1 is classically activated by ATR and is critical for DNA repair through regulation of Cdc proteins (Figure 2) [36]. Under basal conditions, Cdc25 phosphatases activate Cdks to facilitate cell cycle progression, but phosphorylation by Chk1 in response to DNA damage leads to ubiquitination and degradation of Cdc25, thus halting the cell cycle to allow for DNA repair [27]. Chk2, a substrate of ATM, activates p53 both through direct phosphorylation and removal of the negative inhibitor MDM2 to facilitate p53-dependent transcription of target genes including p21 [24,26]. However, as is the case elsewhere within the DDR, distal transduction kinases have overlapping functions, and Chk2 can similarly phosphorylate Cdc25 to trigger G1 arrest [37].
Radioresistance & radiosensitization through the DDR pathway
Upregulation of DDR activity protects cells from genomic instability by increasing the capacity for DNA repair. As such, many malignancies overexpress DDR proteins due to the high rate of replication errors associated with increased cell division. For instance, Chk1 is upregulated in colorectal cancer [38] as well as triple-negative breast carcinoma, where transduction kinase expression correlates strongly with histological tumor grade [39]. Similarly, atypical activation of DNA damage checkpoints and enhanced DNA repair in glioma stem cells results in tumoral radioresistance [40]. In contrast, downregulation or loss of DDR components sensitizes cells to the cytotoxic effects of radiation. Indeed, some of the earliest evidence in support of the DDR as a radioprotective mechanism was derived from heritable radiosensitivity syndromes associated with DDR dysfunction (Table 1) [41–50]. Characterized by numerous germ-line and somatic defects, including a profound sensitivity to environmental radiation, ataxia telangiectasia (A-T) is an autosomal recessive disorder resulting from loss of ATM [51]. A-T patients have approximately a 25% increased lifetime risk of developing cancer due to dysregulation of the cellular response to DSBs [52,53]; in particular, women with A-T have a twofold increased risk of developing breast cancer [54,55]. Atm knockout mice bear similar phenotypic characteristics to A-T patients, including radiosensitivity and cancer predisposition [56,57]. However, two recently reported murine models that express physiologic levels of catalytically inactive ATM on an Atm−/− background, demonstrate that inactive ATM expression results in embryonic lethality [58,59]. Notably, kinase-dead ATM is still recruited to DNA damage foci, but expression of this mutant leads to much greater levels of genomic instability due to decreased homologous recombination repair than is observed in ATM-null mice. These data suggest that ATM may also play a structural role in recruiting or stabilizing clusters of repair proteins at sites of DNA damage. Moreover, kinase-dead ATM expression resulting in embryonic lethality is consistent with the observation that human A-T patients rarely express catalytically inactive ATM, but instead often exhibit complete ATM loss.
Table 1.
Radiosensitivity syndromes linked to the DNA damage response.
| Disease | Deficiency | Gene(s) | Estimated incidence | Primary inheritance pattern | Characteristics |
|---|---|---|---|---|---|
| A-T† | Proximal signal transduction | ATM (11q22-23) | 1 in 40,000 | Autosomal recessive | Cancer predisposition, immunodeficiency, skin changes, ataxia and neurodegeneration |
| A-T-like disorder | DNA damage recognition | Mre11 (11q21) | Unknown | Hypomorphic mutations | Similar features to A-T but milder presentation |
| Bloom syndrome†‡ | Homologous recombination | BLM (15q26) | Unknown; 1 in 48,000 (Ashkenazi Jewish populations) | Autosomal recessive | Cancer predisposition, immunodeficiency, skin changes and congenital abnormalities |
| Cockayne syndrome‡ | Transcription- coupled repair | ERCC6 (10q11.23) or ERCC8 (5q12.1) | 1 in 500,000 | Autosomal recessive | Cutaneous and congnenital abnormalities and leukodystrophy |
| DNA ligase IV deficiency† | Nonhomologous end joining | LIG4 (13q33-34) | Unknown | Hypomorphic mutations | Cancer predisposition, pancytopenia and congenital abnormalities |
| FA† | DNA damage recognition and homologous recombination | FA complex members (multiple loci) | 1 in 350,000 | Autosomal recessive | Cancer predisposition, aplastic anemia and congenital abnormalities |
| Nijmegen breakage syndrome† | DNA damage recognition | NBS1 (8q21) | 1 in 100,000 | Autosomal recessive | Cancer predisposition, immunodeficiency and congenital abnormalities |
| Radiosensitive severe combined immunodeficiency | Nonhomologous end joining | Artemis (10p13) | Unknown | Hypomorphic mutations | Cancer predisposition and immunodeficiency |
| Trichothiodystrophy‡ | Nucleotide excision repair | ERCC2 (19q13.3) or ERCC3 (2q21) | 1 in 1,000,000 | Autosomal recessive | Cutaneous and congenital abnormalities |
| XRCC4-like factor severe combined immunodeficiency | Nonhomologous end joining | NHEJ1 (2q35) | Unknown | Hypomorphic mutations | Immunodeficiency and congenital abnormalities |
| XP‡ | Nucleotide excision repair | XP genes (multiple loci) | 1 in 250,000 | Autosomal recessive | Cutaneous cancer predisposition |
Documented therapeutic radiation sensitivity.
Sunlight sensitivity.
A-T: Ataxia telangiectasia; FA: Fanconi anemia; XP: Xeroderma pigmentosum.
Mutations in other DDR genes have also been reported in human cancer predisposition and radiosensitivity syndromes. For instance, loss of NBS1 and Mre11 result in Nijmegan breakage syndrome and A-T-like disorder, respectively (Table 1) [44], and the combination of cancer predisposition and sensitivity to genotoxic agents has proven to be a great challenge in the treatment of malignancy for these patients. However, investigation of the molecular mechanisms underlying clinical radiosensitivity has lead to the development of numerous agents to disrupt the DDR for therapeutic purposes. For the remainder of the article we will review many of the therapies that target DDR pathways to enhance radiation-associated cytotoxicity, with a particular emphasis on those chemicals and procedures that have allowed the transition from the laboratory to clinical use. These agents not only offer a means to boost the anti-tumor effects of therapeutic radiation and genotoxic chemotherapy, but, through abrogation of the innately enhanced capacity for tumoral DNA repair, may also be useful as monotherapy.
Radiosensitization within DDR pathways
DNA damage sensors
Hyperthermia was among the first mechanisms of cellular radiosensitization that was investigated for clinical use, and it was initially assumed that heat indiscriminately denatured DNA repair proteins. Since then, several particularly heat-labile proteins have been identified within the DDR, and it is now thought that inhibition of specific factors leads to hyperthermia-induced radiosensitization [60–62]. However, conflicting evidence indicates that hyperthermia also activates ATM and Hsp 70 (a molecular chaperone that maintains protein stability and is known to contribute to radioresistance) [63]. Hyperthermia continues to be the subject of numerous clinical investigations as an adjuvant to radiation therapy and other genotoxic agents [64].
Early in vitro experiments aimed at elucidating the mechanisms of hyperthermia-associated radiosensitization demonstrated that Ku70 is heat labile and suggested that elevated temperature may impede recognition of DNA damage [65]. More recent studies have confirmed this hypothesis by demonstrating that hyperthermia induces MRN complex translocation from the nucleus to the cytoplasm in vivo [66,67], in addition to disrupting interactions between Mre11, Rad50 and Nbs1 [68]. Hyperthermic treatment may also inactivate DNA-PK, yet the extent of DNA-PK inhibition following hyperthermia does not correlate with the degree of impaired DSB repair [69]. These data suggest that while hyperthermia may affect the DDR at multiple levels, the primary mechanism of radio-sensitization occurs through impaired recognition of damaged DNA, as opposed to ineffectual downstream signal transduction.
Proximal transduction kinases
While essential for the initial characterization of DDR pathways, many of the first-generation PI3-like kinase inhibitors have numerous off-target effects that preclude clinical implementation. Caffeine and other methylxanthine-derived drugs are potent abrogators of the G2 checkpoint that sensitize cells to radiation-induced cytotoxicity [70]. Mechanistic studies reveal that these compounds inhibit both ATM and ATR, thereby preventing downstream activation of p53- and p21-dependent cell cycle arrest [71,72].
Similar to caffeine, the sterol-like molecule wortmannin is a potent radiosensitizer that inhibits DDR transduction kinases [73]. This fungal metabolite irreversibly inhibits DNA-PK and ATM [74], but is highly unstable in solution and causes acute hepatic toxicity secondary to nonspecific binding [75]. In addition to DNA-PK and ATM, wortmannin also inhibits myosin light-chain kinase, MAPK, mTOR, some phosphoinositide 4 kinases and polo-like kinase, and has, therefore, not been useful as a therapeutic agent [76–78].
Structurally unrelated and less potent than either caffeine or wortmannin, LY294002 is a radiosensitizer derived from the flavonoid quercetin [79,80]. LY294002, and the related compounds rutin and quercitrin inhibit DNA-PK without significantly altering ATM or ATR activity [81,82], but nevertheless, they cause excessive toxicity when administered systemically [83]. Recent studies have attempted to circumvent LY294002 toxicity through dose-reduction chemotherapy regimens and intraperitoneal infusion. Using these strategies in animal models of pancreatic and ovarian cancer, LY294002 has been shown to potentiate the anti-tumor effects of cisplatin and carboplatin, and moreover, is not associated with significant side effects [84,85].
Many of the more recent pharmacologic investigations of DDR transduction kinase-targeted molecules have focused on synthetic derivates of known PI3-like kinase inhibitors [9]. PX-866, a structural analog of wortmannin, demonstrates increased selectivity for PI3-like kinases relative to mTOR binding, and not only has reduced hepatic toxicity but also displays a greater reduction of tumor cell growth compared with wortmannin [75]. While not yet assessed in conjunction with radiotherapy, PX-866 is currently under evaluation in Phase I and II clinical trials as monotherapy for solid tumors. KU-60019, another recently reported DDR transduction kinase inhibitor, was derived from a small-molecule library based on the structure of LY294002. Like PX-866, KU-60019 is extraor- for ATM in dinarily selective and has an IC50 the nanomolar range [86]. KU-60019 sensitizes tumor cells to radiation in vitro and in vivo, and also attenuates tumor cell motility and invasion through downstream inhibition of prosurvival signaling pathways. Moreover, KU-60019 is highly soluble in water, demonstrates few non-specific effects up to the micromolar range [86] and is compatible with the standard chemotherapy regimen for glioblastoma [87]. These findings suggest that KU-60019 has significant therapeutic promise, especially in combination with genotoxic agents, such as external beam radiation and systemic chemotherapy.
While not yet investigated in conjunction with radiotherapy in vivo, a new generation of potent ATR inhibitors similarly hold significant therapeutic potential given their selectivity and synergistic lethality when used in combination with other genotoxic agents. VE-821, a recently reported highly specific ATR inhibitor, sensitizes pancreatic cancer cells to both radiation and gemcitabine [88,89]. The pyrimidine NU6027, which was originally designed to block Chk2 signaling, potently inhibits ATR in the micromolar range, and not only enhances both hydroxy urea and cisplatin toxicity but leads to synergistic cell killing when applied to cells with impaired SSB repair [90].
Distal transduction kinases
Cells that harbor p53 mutations are frequently deficient in the G1 cell cycle checkpoint, and as a result, many tumors exclusively rely on Chk1-mediated pathways for G2 cell cycle arrest to repair damaged DNA (Figure 2) [44]. The importance of Chk1 for genomic integrity has been well documented and multiple lines of evidence suggest that, when combined with loss of p53, Chk1 inhibition profoundly sensitizes cancer cells to genomic stress [91,92]. Cancers that are deficient in the tumor suppressor genes BRCA1 and BRCA2 are similarly susceptible to so-called ‘synthetic lethality’ when treated with inhibitors of the DNA repair enzyme PARP1 [93]. Given the clinical success of PARP inhibitors, a myriad of pharmacological checkpoint kinase inhibitors have been identified and are in various stages of preclinical and clinical testing as radiosensitizers and adjuvants to genotoxic chemotherapy.
Initially isolated from the bacteria Streptomyces, the staurosporine analogue UCN-01 was the first Chk1 inhibitor to be evaluated in human patients and has since been the subject of numerous Phase I and II clinical trials. Many mechanistic studies have shown that UCN-01 sensitizes p53-deficient cells to the cytotoxic effects of ionizing radiation and other DNA damaging agents through abrogation of the G2 checkpoint [94,95]. While UCN-01 has been clinically tested both as monotherapy and in combination with several radiomimetic chemotherapies, dose-limiting toxicities have resulted in the cessation of most clinical investigations [96–100]. Nevertheless, treatment with UCN-01 has resulted in partial tumor responses when combined with either cisplatin or topotecan, and these results have fueled extensive efforts to identify second-generation Chk1 inhibitors with reduced toxicity and more favorable pharmacodynamics.
Structurally distinct and more specific than UCN-01, CEP-3891 reduces Cdc25 phosphorylation and prevents the inhibition of DNA synthesis by inhibiting Chk1. Studies in osteo-sarcoma cell lines demonstrate that CEP-3891 abrogates G2 arrest and potentiates mitotic catastrophe after ionizing radiation [101]. In spite of these promising preclinical data, there have been no reports of CEP-3891 in other cancer cell lines, animal models, or clinical trials. Similarly, no clinical data have been reported for the specific Chk1 inhibitors CHIR-124 or for the marine sponge alkaloid debromohymenialdisine, both of which enhance the effects of radiation in vivo [102–104].
Among the second-generation Chk1 inhibitors tested in conjunction with radiation, the thiophene carboxamide urea AZD7762 features prominently in clinical investigations [105]. The subject of parallel Phase I clinical trials for advanced solid malignancies in combination with either gemcitabine or irinotecan, this non-specific Chk1/Chk2 inhibitor abrogates the G2 checkpoint, potentiates cytotoxicity from genotoxic agents in wild-type cells and enhances cell killing in the absence of p53 [106]. These effects are mediated through interference with Rad51 focus formation that prevents DNA repair through homologous recombination (Figure 2) [107]. These results, which are corroborated by studies with the distinct Chk1 inhibitor PD-321852 [108], suggest that similarly to the loss of p53, Rad51 overexpression may be a prognostic marker for tumor response to Chk1 inhibition.
Many other small-molecule Chk1 inhibitors have been identified, but the vast majority of these have not been assessed in conjunction with radiation. PF-00477736 [109], EXEL-9844 [110], CBP501 [111], PD-321852 [108], Go6976 [112], 17-AAG [113], MK-8776 [114], antisense Chk1 cDNA delivered by oncolytic viruses [102] and other Chk1 inhibitors are particularly efficacious in p53-deficient cell lines [105,115]. More Chk1 inhibitors can be found in the patent literature [105,116], and while each of these agents represents a presumptive radiosensitizer, there have been no direct investigations reported to date.
Evolutionarily unrelated to Chk1, Chk2 is classically activated downstream of MRN and ATM in response to DSBs. Despite their overlapping substrates, Chk1 and Chk2 have drastically different cellular functions and significance, and Chk2 is nonessential for genomic maintenance [105]. As such, Chk2 has often been portrayed as an amplifier kinase [37], and its potential as a therapeutic target for radiosensitization is unclear. Many Chk1 inhibitors also inhibit Chk2 in a comparable concentration range and the specific contributions of Chk2 to tumor progression have remained elusive. Moreover, there is conflicting evidence concerning the synthetic lethality of Chk2 inhibition in p53-deficient cells, and until recently, no selective Chk2 inhibitors were available to address these inconsistent data. Notably, each of the specific Chk2 inhibitors described thus far, including 2-arylbenzimidazole, PV1019, VRX0466617 and CCT241533, either attenuate or fail to enhance apoptotic cell death following ionizing radiation in untransformed cells [117–120]. Indeed, Chk2-deficient mice are resistant to the effects of radiation, suggesting that Chk2 is not a viable target for radiosensitization [121]. However, PV1019 enhances the antiproliferative effects of radiation and several genotoxic agents in ovarian cancer and glioma cell lines [119], and CCT241533 potentiates the cytotoxicity of PARP inhibitors [120]. These results not only suggest that Chk2 inhibitors may represent valuable therapeutic agents if prognostic markers can be identified to guide patient selection, but also highlight the need for further mechanistic investigation into the DDR itself.
Cell cycle effectors
Much like cell cycle checkpoint kinases, Cdks are often dysregulated in cancer. Numerous Cdk inhibitors have been investigated as potential anti-tumor agents, but these studies have been complicated by functional redundancies among Cdk family members [115]. Moreover, recent evidence has suggested that Cdks may not be essential for cell cycle or tumor progression, and may therefore not be effective targets for anticancer therapeutics [122,123]. While a complete review of the agents that target cyclin/Cdk complexes is beyond the scope of this article, it is nevertheless notable that Cdks and their substrates function downstream of the DDR, and therefore represent potential targets for therapeutic radiosensitization. Toward that goal, multiple Cdk inhibitors have been shown to potentiate the cytotoxic effects of radiation and several compounds are currently undergoing clinical evaluation [124,125].
Beyond direct regulators of the cell cycle, numerous studies have demonstrated that inhibition of the cyclin/Cdk complex inhibitor Wee1 results in potent radiosensitization [126]. At baseline, Wee1 prevents mitotic catastrophe by inhibiting Cdk1 phosphorylation to negatively regulate the G2 checkpoint (Figure 2). Consequently, Wee1 inhibition in p53-deficient cells leads to synthetic lethality by a mechanism similar to Chk1 inhibition [127]. Consistent with the G2 checkpoint-dependence of tumors, over-expression of Wee1 has been reported in glioblastoma, hepatocellular carcinoma, non-small-cell lung carcinoma, seminoma and colonic carcinoma [128,129]. Adding to the therapeutic potential of Wee1 inhibitors, Wee1-targeted molecules do not enhance radiation-induced cytotoxicity in untransformed cells, and several studies have suggested that Wee1 inhibition is an effective radiosensitizer, irrespective of p53 status [129]. Indeed, preclinical data indicates that the Wee1 inhibitor PD0166285 abrogates the G2 checkpoint and radiosensitizes multiple radioresistance tumors, including melanoma, osteosarcoma and glioblastoma cell lines [129,130]. Another potent and selective small-molecule inhibitor of Wee1, MK-1775, enhances the cytotoxicity of multiple genotoxic therapies and is currently under clinical investigation, both as monotherapy and in multidrug anti-tumor regimens, for patients with advanced solid tumors [131,132]. Preliminary results indicate that MK-1775 is associated with minimal toxicity, has favorable pharmacodynamics in vivo and displays promising anti-tumor activity [133]. Moreover, pharmacodynamic biomarkers for response to Wee1 inhibition have been incorporated into ongoing clinical trials with MK-1775, and have the potential to guide patient selection for optimization of therapy [134].
Transcription factors
Many of the distal functions of the DDR require transcriptional regulation to enhance the expression of effector proteins or reduce the prevalence of inhibitors. DDR-associated transcription factors are therefore numerous, and include Rb, E2F, H3 and p73 (Figure 2). In keeping with the critical role of new protein synthesis for the DDR, several studies have demonstrated that inhibition of specific transcription factors, such as NF-κB [135] and p53 [136], can radiosensitize cells.
Radiomimetic DNA alkylators are widely used in cancer treatment and in general, they induce O6-methylguanine (O6MeG) lesions that mispair with deoxythymidine and lead to cell killing via a p53-independent pathway. Despite the dispensability of p53, the importance of the DDR for DNA alkylation-mediated cell death is underscored by clinical studies demonstrating that hyperthermia potentiates the cytotoxic effects of ifosfamide and carboplatin [137]. Notably, NF-κB is known to be involved in the cellular response to O6MeG [138], and recent evidence demonstrates that Chk1 specifically phosphorylates the p50 subunit of NF-κB in response to temozolomide to regulate DNA binding and gene transcription (Figure 2) [139]. This modification decreases expression of NF-κB-sensitive anti-apoptotic factors including Cox2 and Bcl-xL, thereby lowering the cytotoxic threshold to ionizing radiation. Notably, p50-dependent radiosensitization in response to DNA alkylation can be mimicked by treatment of glioma cells with exogenous DNA oligonucleotides that contain O6MeG–T mismatched pairs. These synthetic oligonucleotides are nontoxic in the absence of adjuvant radiation [139], and represent promising clinical agents in combination with tumor-specific genotoxic therapies.
Peripheral regulators
DDR activity is influenced at many levels by ancillary factors that augment or temper the cellular response to genomic instability (Figure 2). As many of these proteins participate in oncogenesis, cancer cells often display abnormal expression patterns of peripheral DDR regulators. For instance, Hsps, a family of functionally related molecular chaperones, are often overexpressed in malignancy where they contribute to tumor progression and chemotherapy resistance [140]. In particular, Hsp90 has been implicated in a variety of pathways that favor cell proliferation and survival, including the DDR. Indeed, inhibition of Hsp90 with the glendanamycin analog 17-AAG abrogates the G2 checkpoint through depletion of Chk1 and Wee1 [113,141]. Studies with the related molecule 17-DMAG demonstrate that Hsp90 inhibition similarly decreases the activities of DNA-PK, ATM and the MRN complex [142]. Given the widespread disruption of the DDR achieved by Hsp90 inhibition, it is not surprising that 17-AAG and 17-DMAG sensitize cells to the cytotoxic effects of a variety of genotoxic agents, including radiation. Hsp90 inhibitors have, therefore, been the subjects of more than a dozen clinical trials either alone or in combination with standard chemotherapy regimens [143]. Early reports from several Phase I studies indicate that Hsp90 inhibition, in combination with irinotecan, can be administered safely to patients with advanced solid tumors [144,145], and is especially efficacious in p53-deficient tumors [141,144], as well as in cells that express EGF [143].
Skp1/Cullins/F-box (SCF) E3 ubiquitin ligases also peripherally regulate the DDR by targeting cell cycle and apoptotic proteins for proteosome-mediated degradation [146]. Similar ly to Hsps, SCF E3 ubiquitin ligase components, such as RBX1 and SAG, are overexpressed in a variety of human tumors [147,148]. Notably, silencing of either RBX1 or SAG sensitizes cells to radiation-induced cytotoxicity and profoundly increases apoptosis of tumor cells, but has no effect on normal cell growth [147]. While the mechanism of radiosensitization through SCF E3 ubiquitin ligase inhibition is unclear, several studies demonstrate an accumulation of downstream DDR effectors including p21 and the proapoptotic protein Noxa. MLN4924, a specific in inhibitor of the SCF E3 ubiquitin ligase activator NEDD8, radiosensitizes breast and pancreatic cancer cell lines, and multiple clinical trials for the evaluation of MLN4924 are underway [149–151].
Conclusion
The cellular response to genomic instability involves a highly coordinated cascade of DNA damage sensors, proximal and distal signal transducers, and effectors including cell cycle regulators, transcription factors, mediators of apoptosis and DNA repair proteins. While regulated internally, a network of peripheral proteins and mediators, including molecular chaperones and the ubiquitin proteasome among others, also influence the activity of the DDR. As such, the DDR contains a myriad of targets for radiosensitization and numerous molecular therapies have been developed to disrupt the radioprotective features of this signal transduction pathway. Some of these agents have transitioned to clinical investigation, but further studies are needed to fully elucidate the molecular mechanisms activated in response to radiation-induced DNA damage. Moreover, additional in vivo experiments and clinical trials of targeted radiation in combination with molecular radiosensitizers are required to sufficiently evaluate the effects of these agents in the absence of confounding systemic toxicities from chemotherapy.
Future perspective
The ongoing elucidation of signal transduction pathways within the DDR continues to reveal novel therapeutic targets for radiosensitization. ATR was recently shown to suppress NF-κB in response to replication stress, but the implications of these findings on the cellular response to DNA damage are unclear [152]. Similarly, combined Chk1 and PARP inhibition in pancreatic cancer cells dramatically decreases the cytotoxic threshold for ionizing radiation, suggesting that new avenues of synthetic lethality have yet to be characterized [153]. Furthermore, emerging evidence indicates that synthetic lethality may also be achieved through the simultaneous inhibition of more distal signaling pathways within the DDR, such as interference of base excision repair in conjunction with abrogation of DSB repair [154]. Furthermore, there is debate surrounding the mechanism of action for many DDR-directed radiosensitizers already in clinical use. It has been hypothesized that Chk1 inhibitors induce cell death in p53-deficient cells, not through the synergism of combined G1 and G2 checkpoint abrogation, but rather, via enhanced replicative stress [33]. This model suggests that other cancer-associated mutations that ease entry into S phase may also render cells susceptible to checkpoint kinase inhibition, and is consistent with numerous lines of data demonstrating that Chk1 inhibitors are effective in cells with wild-type p53. Similarly, the observation that ATM may play a structural role for homologous recombination repair, in addition to a catalytic one, suggests that the currently available collection of ATM inhibitors may be both more potent and more toxic than previously estimated. Future mechanistic studies will almost certainly shed light on these controversies, and perhaps provide guidance for the implementation of existing and novel therapies alike.
Continued biomarker identification is similarly likely to enhance the efficacy of DDR-directed radiosensitizers. For instance, the discovery of preferential DDR overexpression in malignancies for which no selective molecular therapies exist, such as triple-negative breast cancer, has revealed new populations of patients for which radiosensitizers may be of particular utility [39]. Appropriate patient selection through biomarker screening will also reduce the unnecessary risk of secondary malignancies associated with induced genomic instability for patients who are unlikely to benefit from DDR-directed agents.
With the exception of hyperthermia and inhibitors of Hsps and SCF E3 ubiquitin ligases, the vast majority of currently available DDR-directed radiosensitizers are small molecules that preferentially affect the activity or stability of a single target. However, future advances in basic cancer biology and drug delivery are likely to diversify the structures and mechanisms of action in the next generation of radiosensitizers. DNA-based agents either alone or via tumor-specific viruses have already been used as radiosensitizers through modulation of NF-κB and Chk1 [102,139]. Furthermore, combination therapies targeting both the DDR and other mediators of cell survival have already transitioned to clinical trials. A study evaluating the safety and action of a dual DNA-PK and mTOR inhibitor is currently underway for patients with advanced solid tumors, non-Hodgkin’s lymphoma or multiple myeloma [201]. These and other molecular therapies may not only improve on the selectivity of small-molecule regulators of the DDR, but also enhance the potency of radiosensitization and tumor cell death.
Executive summary.
DNA damage response
The cellular response to genomic stress consists of highly conserved signal transduction pathways that identify damaged regions, arrest the cell cycle, repair DNA and initiate cell senescence or apoptosis in instances where damage cannot be removed.
The DNA damage response (DDR) is composed of DNA damage sensors, proximal and distal transduction kinases, cell cycle effectors, transcription factors, mediators and peripheral regulators, such as Hsps and ubiquitinases.
Cancer cells that harbor p53 mutations are deficient in the G1 checkpoint, and are therefore particularly susceptible to the radiosensitizing effects of Chk1 inhibitors through abrogation of the G2 checkpoint and synthetic lethality.
Radioresistance & radiosensitization
Components of the DDR are selectively overexpressed in a variety of malignancies with radioresistant phenotypes.
Heritable cancer syndromes involving dysregulation of the DDR are characterized by an enhanced sensitivity to the cytotoxic effects of radiation and genotoxic chemotherapeutic agents.
Radiosensitization within DDR pathways
DNA damage sensors: hyperthermia impairs recognition of DNA damage.
Proximal transduction kinases: nonselective, first-generation PI3-like kinase inhibitors have given rise to a collection of relatively specific small-molecule agents that are under ongoing clinical investigation.
Distal transduction kinases: the highly toxic checkpoint kinase inhibitor UCN-01 has been replaced by selective second-generation agents that inhibit Chk1 and Chk2 with more favorable pharmacodynamics.
Cell cycle effectors: Wee1 inhibition facilitates cell cycle progression through disinhibition of Cdk/cyclin complexes and enhances mitotic catastrophe following ionizing radiation.
Transcription factors: mismatched oligonucleotides mimic the radiosensitizing effect of DNA alkylating agents by decreasing NF-κB-mediated transcription of anti-apoptotic factors.
Peripheral regulators: small-molecule inhibition of Hsp90 and SCF E3 ubiquitin ligase interferes with multiple levels of the DDR to potentiate radiation-induced cytotoxicity.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ward JF. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int J Radiat Biol. 1990;57(6):1141–1150. doi: 10.1080/09553009014551251. [DOI] [PubMed] [Google Scholar]
- 2.Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156(5 Pt 2):577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int J Radiat Biol. 1994;66(5):427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 4.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 5.Olive PL. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat Res. 1998;150(Suppl 5):S42–S51. [PubMed] [Google Scholar]
- 6.Hada M, Georgakilas AG. Formation of clustered DNA damage after high-LET irradiation: a review. J Radiat Res. 2008;49(3):203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 7.Hanai R, Yazu M, Hieda K. On the experimental distinction between SSBs and DSBs in circular DNA. Int J Radiat Biol. 1998;73(5):475–479. doi: 10.1080/095530098142013. [DOI] [PubMed] [Google Scholar]
- 8.Datta K, Jaruga P, Dizdaroglu M, Neumann RD, Winters TA. Molecular analysis of base damage clustering associated with a site-specific radiation-induced DNA double-strand break. Radiat Res. 2006;166(5):767–781. doi: 10.1667/RR0628.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪▪.Lord CJ, Garrett MD, Ashworth A. Targeting the double-strand DNA break repair pathway as a therapeutic strategy. Clin Cancer Res. 2006;12(15):4463–4468. doi: 10.1158/1078-0432.CCR-06-1269. Extensive review of DNA repair mechanisms and the molecular agents that disrupt these pathways. [DOI] [PubMed] [Google Scholar]
- 10▪▪.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. Review of the evolution of knowledge concerning the DNA damage response (DDR) signaling network. [DOI] [PubMed] [Google Scholar]
- 11.Lawson JD, Kahle KT, Ng K, Carter B, Kesari S, Chen CC. DNA damage response and repair: insights into strategies for radiation sensitization. In: Chen CC, editor. Selected Topics in DNA Repair. InTech; Rijeka, Croatia: 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪▪.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. Landmark study demonstrating ATM activation by the Mre11–Rad50–Nbs1 complex in response to DNA double-strand breaks. [DOI] [PubMed] [Google Scholar]
- 13.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 14.Huen MSY, Grant R, Manke I, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doil C, Mailand N, Bekker-Jensen S, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.Goedecke W, Eijpe M, Offenberg HH, Van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23(2):194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 18.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281(14):9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinkelmann M, Spehalski E, Stoneham T, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16(8):808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20▪▪.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. Describes overlapping yet distinct roles for DDR proximal signal transduction kinases. [DOI] [PubMed] [Google Scholar]
- 21.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 22.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 23.Valentin-Vega YA, Kastan MB. A new role for ATM: regulating mitochondrial function and mitophagy. Autophagy. 2012;8(5):840–841. doi: 10.4161/auto.19693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- 25.Peters M, Deluca C, Hirao A, et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc Natl Acad Sci USA. 2002;99(17):11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Q, Chen J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle. 2010;9(3):472–478. doi: 10.4161/cc.9.3.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guardavaccaro D, Pagano M. Stabilizers and destabilizers controlling cell cycle oscillators. Mol Cell. 2006;22(1):1–4. doi: 10.1016/j.molcel.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama KI, Nakayama K. Ubiquitin ligases: cell cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 30.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19(2):238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 31▪.Al-Hakim A, Escribano-Diaz C, Landry MC, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9(12):1229–1240. doi: 10.1016/j.dnarep.2010.09.011. Reviews the numerous roles played by ubiquitin ligases throughout the DDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ETH. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011;585(18):2891–2896. doi: 10.1016/j.febslet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol. 2011;5(4):368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Yajima H, Huynh H, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193(2):295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin Cancer Res. 2007;13(7):1955–1960. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 36.Stracker TH, Usui T, Petrini JHJ. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8(9):1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3(5):421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 38.Madoz-Gúrpide J, Cañamero M, Sanchez L, Solano J, Alfonso P, Casal Ji. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics. 2007;6(12):2150–2164. doi: 10.1074/mcp.M700006-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Verlinden L, Vanden Bempt I, Eelen G, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res. 2007;67(14):6574–6581. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 40▪.Bao S, Wu Q, Re Mclendon, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. DDR hyperactivation in tumor stem cells protects against radiation damage. [DOI] [PubMed] [Google Scholar]
- 41.Buck D, Malivert L, De Chasseval R, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124(2):287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 42.Gatti RA. The inherited basis of human radiosensitivity. Acta Oncol. 2001;40(6):702–711. doi: 10.1080/02841860152619115. [DOI] [PubMed] [Google Scholar]
- 43.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom’s Syndrome Registry. Hum Mutat. 2007;28(8):743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 44.Kastan MB, Bartek J. Cell cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 45.Moshous D, Li L, Chasseval R, et al. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum Mol Genet. 2000;9(4):583–588. doi: 10.1093/hmg/9.4.583. [DOI] [PubMed] [Google Scholar]
- 46.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42(1):68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 47.O’Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 2004;3(8–9):1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 48.Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Phys. 2009;74(5):1323–1331. doi: 10.1016/j.ijrobp.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor AMR, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD) – its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3(8–9):1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Winkelstein JA, Marino MC, Lederman HM, et al. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine (Baltimore) 2006;85(4):193–202. doi: 10.1097/01.md.0000229482.27398.ad. [DOI] [PubMed] [Google Scholar]
- 51.Rotman G, Shiloh Y. ATM: from gene to function. Hum Mol Genet. 1998;7(10):1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 52.Morgan SE, Kastan MB. p53 and ATM: cell cycle, cell death, and cancer. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 53.Reiman A, Srinivasan V, Barone G, et al. Lymphoid tumours and breast cancer in ataxia telangiectasia; substantial protective effect of residual ATM kinase activity against childhood tumours. Br J Cancer. 2011;105(4):586–591. doi: 10.1038/bjc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97(11):813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 55.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 56.Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86(1):159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10(19):2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto K, Wang Y, Jiang W, et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198(3):305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daniel JA, Pellegrini M, Lee BS, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198(3):295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Hur E, Elkind MM, Bronk BV. Thermally enhanced radioresponse of cultured Chinese hamster cells: inhibition of repair of sublethal damage and enhancement of lethal damage. Radiat Res. 1974;58(1):38–51. [PubMed] [Google Scholar]
- 61.Dewey WC, Hopwood LE, Sapareto SA, Gerweck LE. Cellular responses to combinations of hyperthermia and radiation. Radiology. 1977;123(2):463–474. doi: 10.1148/123.2.463. [DOI] [PubMed] [Google Scholar]
- 62.Corry PM, Robinson S, Getz S. Hyperthermic effects on DNA repair mechanisms. Radiology. 1977;123(2):475–482. doi: 10.1148/123.2.475. [DOI] [PubMed] [Google Scholar]
- 63.Pandita TK, Pandita S, Bhaumik SR. Molecular parameters of hyperthermia for radiosensitization. Crit Rev Eukaryot Gene Expr. 2009;19(3):235–251. doi: 10.1615/critreveukargeneexpr.v19.i3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Zee J. Heating the patient: a promising approach? Ann Oncol. 2002;13(8):1173–1184. doi: 10.1093/annonc/mdf280. [DOI] [PubMed] [Google Scholar]
- 65.Matsumoto Y, Suzuki N, Sakai K, Morimatsu A, Hirano K, Murofushi H. A possible mechanism for hyperthermic radiosensitization mediated through hyperthermic lability of Ku subunits in DNA-dependent protein kinase. Biochem Biophys Res Commun. 1997;234(3):568–572. doi: 10.1006/bbrc.1997.6689. [DOI] [PubMed] [Google Scholar]
- 66.Zhu WG, Seno JD, Beck BD, Dynlacht JR. Translocation of MRE11 from the nucleus to the cytoplasm as a mechanism of radiosensitization by heat. Radiat Res. 2001;156(1):95–102. doi: 10.1667/0033-7587(2001)156[0095:tomftn]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 67.Seno JD, Dynlacht JR. Intracellular redistribution and modification of proteins of the Mre11/Rad50/Nbs1 DNA repair complex following irradiation and heat-shock. J Cell Physiol. 2004;199(2):157–170. doi: 10.1002/jcp.10475. [DOI] [PubMed] [Google Scholar]
- 68.Gerashchenko BI, Gooding G, Dynlacht JR. Hyperthermia alters the interaction of proteins of the Mre11 complex in irradiated cells. Cytometry A. 2010;77(10):940–952. doi: 10.1002/cyto.a.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng ZC, Jiang GL, Wang GM, Tang ZY, Curran WJ, Iliakis G. DNA-PKcs subunits in radiosensitization by hyperthermia on hepatocellular carcinoma hepG2 cell line. World J Gastroenterol. 2002;8(5):797–803. doi: 10.3748/wjg.v8.i5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rowley R, Zorch M, Leeper DB. Effect of caffeine on radiation-induced mitotic delay: delayed expression of G2 arrest. Radiat Res. 1984;97(1):178–185. [PubMed] [Google Scholar]
- 71.Yao SL, Akhtar AJ, Mckenna KA, et al. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat Med. 1996;2(10):1140–1143. doi: 10.1038/nm1096-1140. [DOI] [PubMed] [Google Scholar]
- 72.Sarkaria JN, Busby EC, Tibbetts RS, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Research. 1999;59(17):4375–4382. [PubMed] [Google Scholar]
- 73.Price BD, Youmell MB. The phosphatidylinositol 3-kinase inhibitor wortmannin sensitizes murine fibroblasts and human tumor cells to radiation and blocks induction of p53 following DNA damage. Cancer Res. 1996;56(2):246–250. [PubMed] [Google Scholar]
- 74.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Research. 1998;58(19):4375–4382. [PubMed] [Google Scholar]
- 75.Wipf P, Minion DJ, Halter RJ, et al. Synthesis and biological evaluation of synthetic viridins derived from C(20)-heteroalkylation of the steroidal PI-3-kinase inhibitor wortmannin. Org Biomol Chem. 2004;2(13):1911–1920. doi: 10.1039/b405431h. [DOI] [PubMed] [Google Scholar]
- 76.Ferby IM, Waga I, Sakanaka C, Kume K, Shimizu T. Wortmannin inhibits mitogen-activated protein kinase activation induced by platelet-activating factor in guinea pig neutrophils. J Biol Chem. 1994;269(48):30485–30488. [PubMed] [Google Scholar]
- 77.Vanhaesebroeck B, Leevers SJ, Ahmadi K, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y, Jiang N, Wu J, Dai W, Rosenblum JS. Polo-like kinases inhibited by wortmannin Labeling site and downstream effects. J Biol Chem. 2007;282(4):2505–2511. doi: 10.1074/jbc.M609603200. [DOI] [PubMed] [Google Scholar]
- 79.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269(7):5241–5248. [PubMed] [Google Scholar]
- 80.Rosenzweig KE, Youmell MB, Palayoor ST, Price BD. Radiosensitization of human tumor cells by the phosphatidylinositol3-kinase inhibitors wortmannin and LY294002 correlates with inhibition of DNA-dependent protein kinase and prolonged G2–M delay. Clin Cancer Res. 1997;3(7):1149–1156. [PubMed] [Google Scholar]
- 81.Izzard RA, Jackson SP, Smith GC. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 1999;59(11):2581–2586. [PubMed] [Google Scholar]
- 82.Fuhrman CB, Kilgore J, Lacoursiere YD, et al. Radiosensitization of cervical cancer cells via double-strand DNA break repair inhibition. Gynecol Oncol. 2008;110(1):93–98. doi: 10.1016/j.ygyno.2007.08.073. [DOI] [PubMed] [Google Scholar]
- 83.Stauffer F, Holzer P, García-Echeverría C. Blocking the PI3K/PKB pathway in tumor cells. Curr Med Chem Anticancer Agents. 2005;5(5):449–462. doi: 10.2174/1568011054866937. [DOI] [PubMed] [Google Scholar]
- 84.Fujiwara M, Izuishi K, Sano T, et al. Modulating effect of the PI3-kinase inhibitor LY294002 on cisplatin in human pancreatic cancer cells. J Exp Clin Cancer Res. 2008;27:76. doi: 10.1186/1756-9966-27-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Westfall SD, Skinner MK. Inhibition of phosphatidylinositol 3-kinase sensitizes ovarian cancer cells to carboplatin and allows adjunct chemotherapy treatment. Mol Cancer Ther. 2005;4(11):1764–1771. doi: 10.1158/1535-7163.MCT-05-0192. [DOI] [PubMed] [Google Scholar]
- 86.Golding SE, Rosenberg E, Valerie N, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009;8(10):2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Golding SE, Rosenberg E, Adams BR, et al. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle. 2012;11(6):1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prevo R, Fokas E, Reaper PM, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13(11):1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pires IM, Olcina MM, Anbalagan S, et al. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br J Cancer. 2012;107(2):291–299. doi: 10.1038/bjc.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peasland A, Wang LZ, Rowling E, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105(3):372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20(51):7453–7463. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 92.Luo Y, Rockow-Magnone SK, Kroeger PE, et al. Blocking Chk1 expression induces apoptosis and abrogates the G2 checkpoint mechanism. Neoplasia. 2001;3(5):411–419. doi: 10.1038/sj.neo.7900175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 94.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88(14):956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 95.Graves PR, Yu L, Schwarz JK, et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275(8):5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 96.Sausville EA, Arbuck SG, Messmann R, et al. Phase I trial of 72-hour continuous infusion UCN-01 in patients with refractory neoplasms. J Clin Oncol. 2001;19(8):2319–2333. doi: 10.1200/JCO.2001.19.8.2319. [DOI] [PubMed] [Google Scholar]
- 97.Kortmansky J, Shah MA, Kaubisch A, et al. Phase I trial of the cyclin-dependent kinase inhibitor and protein kinase C inhibitor 7-hydroxystaurosporine in combination with fuorouracil in patients with advanced solid tumors. J Clin Oncol. 2005;23(9):1875–1884. doi: 10.1200/JCO.2005.03.116. [DOI] [PubMed] [Google Scholar]
- 98.Lara PN, Mack PC, Synold T, et al. The cyclin-dependent kinase inhibitor UCN-01 plus cisplatin in advanced solid tumors: a California cancer consortium Phase I pharmacokinetic and molecular correlative trial. Clin Cancer Res. 2005;11(12):4444–4450. doi: 10.1158/1078-0432.CCR-04-2602. [DOI] [PubMed] [Google Scholar]
- 99.Hotte Sj, Oza A, Winquist Ew, et al. PhaseI trial of UCN-01 in combination with topotecan in patients with advanced solid cancers: a Princess Margaret Hospital Phase II Consortium study. Ann Oncol. 2006;17(2):334–340. doi: 10.1093/annonc/mdj076. [DOI] [PubMed] [Google Scholar]
- 100.Sampath D, Cortes J, Estrov Z, et al. Pharmacodynamics of cytarabine alone and in combination with 7-hydroxystaurosporine (UCN-01) in AML blasts in vitro and during a clinical trial. Blood. 2006;107(6):2517–2524. doi: 10.1182/blood-2005-08-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Syljuåsen RG, Sørensen CS, Nylandsted J, Lukas C, Lukas J, Bartek J. Inhibition of Chk1 by CEP-3891 accelerates mitotic nuclear fragmentation in response to ionizing radiation. Cancer Research. 2004;64(24):9035–9040. doi: 10.1158/0008-5472.CAN-04-2434. [DOI] [PubMed] [Google Scholar]
- 102.Gao Q, Zhou J, Huang X, et al. Selective targeting of checkpoint kinase 1 in tumor cells with a novel potent oncolytic adenovirus. Mol Ther. 2006;13(5):928–937. doi: 10.1016/j.ymthe.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 103.Tse AN, Rendahl KG, Sheikh T, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13(2 Pt 1):591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- 104.Tao Y, Leteur C, Yang C, et al. Radiosensitization by Chir-124, a selective CHK1 inhibitor: effects of p53 and cell cycle checkpoints. Cell Cycle. 2009;8(8):1196–1205. doi: 10.4161/cc.8.8.8203. [DOI] [PubMed] [Google Scholar]
- 105.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16(2):376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7(9):2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 107.Morgan MA, Parsels LA, Zhao L, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Research. 2010;70(12):4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parsels LA, Morgan MA, Tanska DM, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8(1):45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blasina A, Hallin J, Chen E, et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther. 2008;7(8):2394–2404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- 110.Matthews DJ, Yakes FM, Chen J, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle. 2007;6(1):104–110. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- 111.Sha SK, Sato T, Kobayashi H, et al. Cell cycle phenotype-based optimization of G2-abrogating peptides yields CBP501 with a unique mechanism of action at the G2 checkpoint. Mol Cancer Ther. 2007;6(1):147–153. doi: 10.1158/1535-7163.MCT-06-0371. [DOI] [PubMed] [Google Scholar]
- 112.Kohn EA, Yoo CJ, Eastman A. The protein kinase C inhibitor Gö6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res. 2003;63(1):31–35. [PubMed] [Google Scholar]
- 113.Arlander SJH, Eapen AK, Vroman BT, Mcdonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278(52):52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- 114.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Ther. 2012;11(2):427–438. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115▪.Collins I, Garrett MD. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol. 2005;5(4):366–373. doi: 10.1016/j.coph.2005.04.009. Review of Cdk small-molecule inhibitiors as cancer therapies. [DOI] [PubMed] [Google Scholar]
- 116.Tao ZF, Lin NH. Chk1 inhibitors for novel cancer treatment. Anticancer Agents Med Chem. 2006;6(4):377–388. doi: 10.2174/187152006777698132. [DOI] [PubMed] [Google Scholar]
- 117.Arienti KL, Brunmark A, Axe FU, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005;48(6):1873–1885. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 118.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6(3):935–944. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 119.Jobson AG, Lountos GT, Lorenzi PL, et al. Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide] J Pharmacol Exp Ther. 2009;331(3):816–826. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Anderson VE, Walton MI, Eve PD, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011;71(2):463–472. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takai H, Naka K, Okada Y, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21(19):5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tetsu O, Mccormick F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell. 2003;3(3):233–245. doi: 10.1016/s1535-6108(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 123.Ortega S, Prieto I, Odajima J, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35(1):25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 124.Senderowicz AM. Novel small molecule cyclin-dependent kinases modulators in human clinical trials. Cancer Biol Ther. 2003;2(4 Suppl 1):S84–S95. [PubMed] [Google Scholar]
- 125.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24(11):1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 126.Stathis A, Oza A. Targeting Wee1-like protein kinase to treat cancer. Drug News Perspect. 2010;23(7):425–429. doi: 10.1358/dnp.2010.23.7.1490760. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y, Li J, Booher RN, et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61(22):8211–8217. [PubMed] [Google Scholar]
- 128.Masaki T, Shiratori Y, Rengifo W, et al. Cyclins and cyclin-dependent kinases: comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology. 2003;37(3):534–543. doi: 10.1053/jhep.2003.50112. [DOI] [PubMed] [Google Scholar]
- 129.Mir SE, De Witt Hamer PC, Krawczyk PM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18(3):244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hashimoto O, Shinkawa M, Torimura T, et al. Cell cycle regulation by the Wee1 inhibitor PD0166285, pyrido [2,3-D] pyimidine, in the B16 mouse melanoma cell line. BMC Cancer. 2006;6:292. doi: 10.1186/1471-2407-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hirai H, Iwasawa Y, Okada M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8(11):2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 132.Hirai H, Arai T, Okada M, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9(7):514–522. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 133.Schellens JH, Leijen S, Shapiro GI, et al. A Phase I and pharmacological study of MK-1775, a Wee1 tyrosine kinase inhibitor, in both monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. Presented at: 2009 American Society of Clincal Oncology Annual Meeting.; Orlando, FL, USA. 29 May–2 June 2009; p. Abstract 3510. [Google Scholar]
- 134.Mizuarai S, Yamanaka K, Itadani H, et al. Discovery of gene expression-based pharmacodynamic biomarker for a p53 context-specific anti-tumor drug Wee1 inhibitor. Mol Cancer. 2009;8:34. doi: 10.1186/1476-4598-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim BY, Kim KA, Kwon O, et al. NF-kappaB inhibition radiosensitizes Ki-Ras-transformed cells to ionizing radiation. Carcinogenesis. 2005;26(8):1395–1403. doi: 10.1093/carcin/bgi081. [DOI] [PubMed] [Google Scholar]
- 136.Bristow RG, Benchimol S, Hill RP. The p53 gene as a modifier of intrinsic radiosensitivity: implications for radiotherapy. Radiother Oncol. 1996;40(3):197–223. doi: 10.1016/0167-8140(96)01806-3. [DOI] [PubMed] [Google Scholar]
- 137.Wiedemann GJ, Robins HI, Gutsche S, et al. Ifosfamide, carboplatin and etoposide (ICE) combined with 41.8 degrees C whole body hyperthermia in patients with refractory sarcoma. Eur J Cancer. 1996;32A(5):888–892. doi: 10.1016/0959-8049(95)00622-2. [DOI] [PubMed] [Google Scholar]
- 138.Wu ZH, Miyamoto S. Many faces of NF-kappaB signaling induced by genotoxic stress. J Mol Med. 2007;85(11):1187–1202. doi: 10.1007/s00109-007-0227-9. [DOI] [PubMed] [Google Scholar]
- 139.Schmitt AM, Crawley CD, Kang S, et al. p50 (NF-κB1) is an effector protein in the cytotoxic response to DNA methylation damage. Mol Cell. 2011;44(5):785–796. doi: 10.1016/j.molcel.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Didelot C, Lanneau D, Brunet M, et al. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14(27):2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- 141.Tse AN, Sheikh TN, Alan H, Chou TC, Schwartz GK. 90-kDa heat shock protein inhibition abrogates the topoisomerase I poison-induced G2/M checkpoint in p53-null tumor cells by depleting Chk1 and Wee1. Mol Pharmacol. 2009;75(1):124–133. doi: 10.1124/mol.108.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of Hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66(18):9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 143▪.Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13(15 Pt 1):4326–4330. doi: 10.1158/1078-0432.CCR-07-0632. Review of radiosensitization through Hsp90 inhibition. [DOI] [PubMed] [Google Scholar]
- 144.Tse AN, Klimstra DS, Gonen M, et al. A Phase 1 dose-escalation study of irinotecan in combination with 17-allylamino-17-demethoxygeldanamycin in patients with solid tumors. Clin Cancer Res. 2008;14(20):6704–6711. doi: 10.1158/1078-0432.CCR-08-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pacey S, Wilson RH, Walton M, et al. A Phase I study of the heat shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17(6):1561–1570. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jia L, Yang J, Hao X, et al. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16(3):814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jia L, Bickel JS, Wu J, et al. RBX1 (RING box protein 1) E3 ubiquitin ligase is required for genomic integrity by modulating DNA replication licensing proteins. J Biol Chem. 2011;286(5):3379–3386. doi: 10.1074/jbc.M110.188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2–M arrest, apoptosis, and senescence. Cancer Res. 2009;69(12):4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15(12):3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 150.Wei D, Li H, Yu J, et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Research. 2012;72(1):282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang D, Tan M, Wang G, Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS One. 2012;7(3):e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu ZH, Miyamoto S. Induction of a pro-apoptotic ATM–NF-kappaB pathway and its repression by ATR in response to replication stress. EMBO J. 2008;27(14):1963–1973. doi: 10.1038/emboj.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vance S, Liu E, Zhao L, et al. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle. 2011;10(24):4321–4329. doi: 10.4161/cc.10.24.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sultana R, Mcneill DR, Abbotts R, et al. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int J Cancer. 2012;131(10):2433–2444. doi: 10.1002/ijc.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.ClinicalTrials.gov. [Accessed 1 June 2012];Study to assess safety and tolerability of oral CC-115 for patients with advanced solid tumors, non-Hodgkin’s lymphoma or multiple myeloma. http://clinicaltrials.gov/ct2/show/NCT01353625?term=cc-115&rank=1.