Abstract
Pulmonary surfactant, a lipoprotein complex, maintains alveolar integrity and plays an important role in lung host defense, and control of inflammation. Altered inflammatory processes and surfactant dysfunction are well described events that occur in patients with acute or chronic lung disease that can develop secondary to a variety of insults. Genetic variants of surfactant proteins, including single nucleotide polymorphisms, haplotypes, and other genetic variations have been associated with acute and chronic lung disease throughout life in several populations and study groups. The hydrophilic surfactant proteins SP-A and SP-D, also known as collectins, in addition to their surfactant-related functions, are important innate immunity molecules as these, among others, exhibit the ability to bind and enhance clearance of a wide range of pathogens and allergens. This review focuses on published association studies of human surfactant proteins A and D genetic polymorphisms with respiratory, and non-respiratory diseases in adults, children, and newborns. The potential role of genetic variations in pulmonary disease or pathogenesis is discussed following an evaluation, and comparison of the available literature.
Keywords: SFTPA1, SFTPA2, SP-D, Polymorphism, Single Nucleotide, collectins, lung disease, review
2. INTRODUCTION
Disease susceptibility is influenced by a number of overlapping genetic and non genetic factors, most of which may have a different level of impact at different stages of life. Genetic components that determine susceptibility to acute and chronic lung disease have been studied within different biological contexts, and correlated with environmental factors, such as pollutants, concurrent diseases, or particular conditions such as prematurity or need for mechanical ventilation (1–11). For example, genetic variation in genes involved in pulmonary adaptation may contribute, under certain conditions, to differences in disease susceptibility, or disease severity among individuals. Study of the genetics of pulmonary surfactant components, and in particular of the surfactant proteins, have revealed correlation with lung disease in neonates, children, and adults (12–14). Genetic variations and mutations of the surfactant proteins have been correlated with disease susceptibility or pathogenesis (12). In the present review, we expand on the most recent review (12) and summarize associations of single nucleotide polymorphisms (SNPs), and genetic variants or haplotypes of the two innate immunity molecules, surfactant proteins A (SP-A) and D (SP-D), with neonatal, pediatric, and adult disease.
2.1. Pulmonary surfactant and surfactant proteins
Pulmonary surfactant, a lipoprotein complex, is synthesized by the alveolar epithelial type II cells in the lung, and its key function is to reduce the surface tension at the alveolar air-liquid interface, and consequently prevent alveolar collapse at low lung volumes. As a consequence of its surface tension lowering function, and the role of its components in innate immunity, pulmonary surfactant is not only essential for life, but is also critical for lung health, and normal lung function throughout life (14, 15). Pulmonary surfactant is composed by approximately 90% of lipids, and 10% of proteins. The lipid fraction is primarily composed by phospholipids, most of which is phosphatidylcholine (PC) and dipalmitoylphosphatidylcholine (DPPC) in particular, a key component involved in the reduction of surface tension. The second most abundant phospholipid is phosphatidylglycerol (PG), and the remainder consists of phosphatidylethanolamine (PE), phosphatidylSerine (PS), and other phospholipids, as well as non-phosphorylated lipids such as cholesterol and triglycerides (14). The protein component consists of serum proteins, and surfactant proteins that were originally collectively thought to be specific to the lung or surfactant. However, this is no longer the case. Surfactant proteins, and especially the hydrophilic proteins (SP-A and SP-D) have been found in several other tissues (16–27).
Surfactant proteins (SP-A, SP-B, SP-C, and SP-D) are divided into two groups, by their hydrophobicity properties. The hydrophobic, surfactant protein B (SP-B), and C (SP-C) are primarily involved in the prevention of alveolar collapse at low lung volumes by lowering surface tension. SP-C stabilizes surfactant at low lung volumes, and since it has the ability to bind LPS, a role in innate immunity has been proposed for SP-C (28, 29). SP-A and SP-D are hydrophilic proteins that belong to the C-type lectin family (collectins), and are primarily host defense proteins (30, 31). SP-A has been shown to play an important role in the structure of the extracellular form of surfactant, tubular myelin, and other surfactant-related functions (32–34), and SP-D has been shown to play an important role in surfactant homeostasis (31, 35–37).
Members of the collectin family are characterized by an N-terminal collagen-like domain and a C-terminal carbohydrate recognition domain (CRD) that allows binding to various types of macromolecules, including carbohydrates, phospholipids, and proteins, as well to a number of pathogens and allergens (30). SP-A and SP-D are found in large oligomeric structures that bind viruses, bacteria and fungi in a calcium-dependent, and carbohydrate-specific manner (2, 38, 39), and it has been proposed that the oligomerization pattern may affect binding (40). The mature SP-A, a 248 amino acid (aa) protein (35kDa), associates in a trimeric structural subunit (105kDa), and six of these trimers assemble in a hexameric (630kDa) oligomeric bouquet-like structure that contains a total of eighteen SP-A1 and SP-A2 monomers. Both hetero-oligomers (i.e. consisting of both SP-A1, and SP-A2 monomers (41)), and homo-oligomers (i.e. consisting of SP-A1 or SP-A2 monomers) are functional (33, 42–46). SP-D oligomers are 540kDa cruciform tetrameric structures, comprised by four subunits (130kDa) of three 43kDa (375 aa) SP-D monomers each (39).
2.2. SP-A and SP-D functions in innate immunity
SP-A and SP-D are important components of the innate immune system (30). These molecules combat infections caused by bacteria, viruses, fungi, and other pathogens by mechanisms that may involve, among others, binding, aggregation, agglutination, inhibition of their growth, and promotion of their phagocytosis by immune cells (2, 30, 40, 47–50). Interaction between SP-A and the phagocytic cells, such as alveolar macrophages has also been demonstrated, and shown to promote phagocytosis (43, 51, 52). Studies of interactions between SP-A or SP-D and pathogens, and between collectins and immune cells have been previously reviewed (36, 39, 53–59).
A number of soluble and membrane receptors have been shown to interact with collectins. SP-A is known to interact with CD35 (CR1) (60), C1qR (CD93) (61), CD14 (62), CD91/calreticulin complex (63), signal inhibitory protein α (SIRPα) (64, 65), SP-R210 (66), glycoprotein 340 (gp-340) (67), Toll-like receptors TLR-2 (68) and TLR-4 (69), and others (51, 66–68, 70–72). Some SP-A receptors have been identified in alveolar type II cells, but not in alveolar macrophages cell surface, indicating their potential role in surfactant function (73, 74), whereas others are ubiquitous (CD91/calreticulin, (64)). Most of the receptor molecules that interact with SP-A have also been shown to bind SP-D (SIRPα, CD91/calreticulin, gp-340, TLR-2, TLR-4, and CD14), and promote phagocytosis, clearance of apoptotic cells, modulate cytokine production, and/or act as opsonins, stimulating alveolar macrophage migration (39, 63, 65, 75–78).
Collectins facilitate destruction of various bacteria, viruses, and fungi, by at least two different mechanisms that involve either direct interaction with the pathogen, and/or activation of the immune cells (2, 36, 79–81). Both SP-A and SP-D can stimulate chemotaxis and enhance phagocytosis of alveolar macrophages, an important cellular component of the first line of defense of the lung. The influence of collectins on the interactions of alveolar macrophages with pathogens varies depending on the microorganism (57, 82). There is evidence that human SP-A binds and enhances phagocytosis of Klebsiella pneumoniae (83–85), Escherichia coli, Staphylococcus aureus (45, 86), and Pseudomonas aeruginosa (43, 52) by alveolar macrophages. SP-D has been shown to bind LPS from Klebsiella pneumoniae, and other bacteria (2). Furthermore, SP-A has also been found to promote macrophage production of pro- and anti-inflammatory cytokines (87).
SP-A and SP-D not only regulate the function of innate immune cells, but also interact and modulate the functions of dendritic cells, and other antigen-presenting cells, as well as T cells, providing a link between the innate and adaptative immune systems, in order to alleviate infection and inflammation in an attempt to restore tissue homeostasis (88).
2.3. Genetics of surfactant proteins
The two hydrophobic surfactant proteins, SP-B and SP-C, are encoded by a single gene, located in chromosomes 2 and 8, respectively. Their genetic complexities and polymorphisms associations with disease have been studied and reviewed elsewhere (12, 15, 89–94). The focus here is on SP-A and SP-D associations with disease, and therefore the hydrophobic surfactant proteins will not be discussed any further.
Human SP-A and SP-D are encoded by three genes (SFTPA1: SP-A1, SFTPA2: SP-A2, and SFTPD: SP-D), that have been mapped to chromosome 10q21–q23 (95–97). The human SP-A locus consists of two functional, highly homologous genes (SP-A1, and SP-A2) in opposite transcriptional orientation and a pseudogene being located between the two genes. These are found in a cluster along with the SP-D gene (Figure 1). The genetic complexity of SP-A1, SP-A2, and SP-D genes has been extensively studied, and reviewed (12, 98–103). SP-A1, SP-A2, and SP-D have all been found to be polymorphic (103–105). A particular locus is considered to be polymorphic if the less frequent allele has a population frequency of no less than 1%, and a heterozygosity frequency of at least 2%. Single nucleotide polymorphisms occur when a single nucleotide (purine or pyrimidine) in a DNA sequence is substituted with a different nucleotide. A SNP may either result in a synonymous or non-synonymous aa substitution, where the aa coded for is the same or different, respectively. The probability of recombination occurring within a haplotype partially depends on the physical distance between the SNP loci. Closely spaced loci are therefore less likely to be separated and are described as being in linkage disequilibrium. Consequently, if the genotype of one SNP is known, the genotype of another SNP may be predicted if there is a high level of linkage disequilibrium between the two SNPs.
Figure 1. SP-A and SP-D loci on chromosome 10.
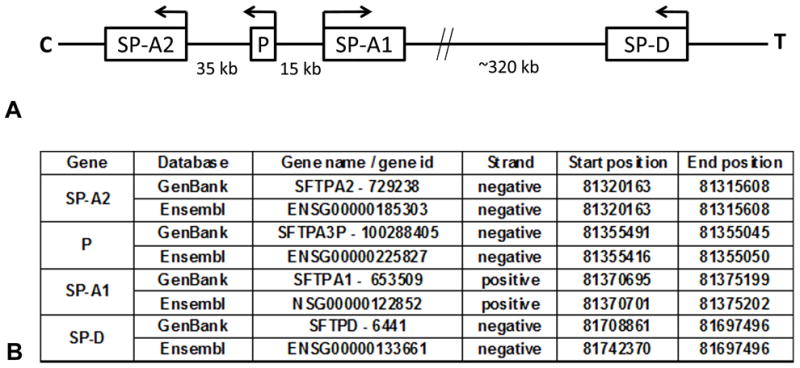
Diagrammatic representation of the 10q22–23 region of the human chromosome 10 (not to scale). The human surfactant protein A locus consists of two functional genes (SP-A1, SP-A2) located in opposite transcriptional orientation, and a pseudogene (P). The SP-D locus is located closer to the telomere (T) in the negative strand, as are SP-A2 and P (Panel A). The information with regards to the specific locations of these genetic loci, available at the GenBank (www.ncbi.nlm.nih.gov/genbank/) and Ensembl (www.ensembl.org/) databases is shown in Panel B. Positive strand refers to the DNA orientation (5′-3′) from C to T, whereas negative strand refers to the opposite orientation.
SP-A1 and SP-A2 genes are in linkage disequilibrium, and exhibit a similar genomic organization. The structure of both genes consists of four coding exons (I-IV) that show coding nucleotide differences that result in aa changes within the collagen-like domain of the protein, and these can distinguish between SP-A1 and SP-A2 gene products, and between their corresponding variants. Multiple SNPs have been identified in SP-A1 and SP-A2 coding regions, and UTRs (103, 106, 107). Nucleotide/aa changes at the coding region that determine the identity of SP-A1 and SP-A2 are shown in Table 1. In brief, both SP-A1 and SP-A2 protein molecules consist of 248 aa, and differ at the following residues: Met66, Asp73, Ile81, and Cys85 for SP-A1, and Thr66, Asn73, Val81, and Arg85 for SP-A2 (44, 102, 103, 108) (Table 1).
Table 1.
Amino acid differences that distinguish between human SP-A1 and SP-A2
| Amino acid position1 | ||||
|---|---|---|---|---|
| 66 | 73 | 81 | 85 | |
| SP-A1 | ATG (Met) | GAT (Asp) | ATC (Ile) | TGT (Cys) |
| SP-A2 | ACA (Thr) | AAT (Asn) | GTC (Val) | CGT (Arg) |
Numbering of amino acid position is based on the precursor molecule that includes the signal peptide
Several coding variants have been identified and characterized for each gene (98, 100, 102, 103, 108). These are combinations of several SNPs, and are summarized in Table 2. Variants 6A, 6A2, 6A3, and 6A4 are combinations of five biallelic SNPs within the exons of SP-A1, corresponding to aa positions 19, 50, 62, 133, and 219, and determined by combinations of SNPs rs1059047, rs1136450, rs1136451, rs1059057, and rs4253527, respectively. Two of these SNPs are silent (aa62 and aa133), whereas the remainder result in non-conservative aa substitutions. Similarly, SP-A2 variants 1A, 1A0, 1A1, 1A2, 1A3, and 1A5 involve four exonic SNPs at aa9, aa91, aa140, and aa223 (rs1059046, rs17886395, rs1965707, rs1965708, respectively), most of which result in a non-synonymous aa change, except for aa140, where a synonymous change occurs (103) (Table 2). These nine variants have been found in the population with different frequencies (103,104). The expression of SP-A1 and SP-A2 appears to differ among individuals as a function of age and lung health status (e.g. healthy vs. cystic fibrosis, culture positive vs. culture negative), as assessed by differences in the protein ratio of SP-A1 to total SP-A in human BAL samples (109).
Table 2.
SNP and/or amino acid variation within the coding region of SP-A1 and SP-A2, that distinguish among the most frequently observed SP-A1 and SP-A2 variants or intragenic haplotypes
| SP-A1 variants | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP id | Nucleotide | amino acid substitutiona | 6A | 6A2 | 6A3 | 6A4 | ||
| rs1059047 | T/C | aa19: GTG (Val) > GCG (Ala) | C (Ala) | T (Val) | T (Val) | T (Val) | ||
| rs1136450 | C/G | aa50: CTC (Leu) > GTC (Val) | C (Leu) | G (Val) | C (Leu) | C (Leu) | ||
| rs1136451 | A/G | aa62: CCA (Pro) > CCG (Pro) | G (Pro) | A (Pro) | A (Pro) | G (Pro) | ||
| rs1059057b | A/G | aa133: ACA (Trp) > ACG (Trp) | G (Trp) | A (Trp) | A (Trp) | A (Trp) | ||
| rs4253527 | C/T | aa219: CGG (Arg) > TGG (Trp) | C (Arg) | C (Arg) | C (Arg) | T (Trp) | ||
| SP-A2 variants | ||||||||
| SNP id | Nucleotide | amino acid substitutiona | 1A | 1A0 | 1A1 | 1A2 | 1A3 | 1A5 |
| rs1059046 | A/C | aa9: ACC (Thr) > AAC (Asn) | C (Thr) | A (Asn) | C (Thr) | C (Thr) | A (Asn) | C (Thr) |
| rs17886395 | G/C | aa91: GCT (Ala) > CCT (Pro) | C (Pro) | G (Ala) | G (Ala) | G (Ala) | G (Ala) | C (Pro) |
| rs1965707b | C/T | aa140: TCC (Ser) > TCT (Ser) | C (Ser) | C (Ser) | T (Ser) | C (Ser) | T (Ser) | T (Ser) |
| rs1965708 | C/A | aa223: CAG (Gln) > AAG (Lys) | C (Gln) | C (Gln) | A (Lys) | C (Gln) | A (Lys) | C (Gln) |
Numbering of amino acid (aa) position is based on the precursor molecule that includes the signal peptide,
These SNPs have been identified in SP-D but have not been associated with any disease described in the present review.
In addition, splice and sequence differences are also found at the 5′ and 3′ untranslated regions (UTRs) of SP-A1 and SP-A2 genes (107, 108, 110, 111). At the 5′UTR, several exons (A, B, B′, C, C′, D, D′) splice in different configurations to give rise to a number of different 5′UTR variants for SP-A1 and SP-A2 (108). These have been shown to differentially impact SP-A regulation of gene expression (112, 113). Similarly, SNPs and other sequence variations located at the 3′UTR of SP-A1 and SP-A2 variants have also been shown to play a role in SP-A regulation (114).
The SP-D gene contains a total of eight exons, seven of which are coding. Exon I, and part of exon II correspond to the 5′UTR, and the last part of exon 8 corresponds to the 3′UTR. Coding and non-coding SNPs (rs721917, rs6413520, rs2243639, rs3088308, rs1051246, rs1923537, rs2245121, rs911887, rs2255601, and rs7078012) are also found within the SP-D gene (Table 3); most of these have been associated with a number of diseases (115–117). In addition, evidence indicates that serum SP-D levels are genetically influenced (106, 118–120).
Table 3.
Human SP-D SNPs
| SNP id | Nucleotide | amino acid substitution1 |
|---|---|---|
| rs721917 | C/T | aa11: ATG (Met) > ACG (Thr) |
| rs6413520 | T/C | aa25: AGT (Ser) > AGC (Ser) |
| rs2243639 | A/G | aa160: ACA (Thr) > GCA (Ala) |
| rs30883082 | A/T | aa270: TCT (Ser) > ACT (Thr) |
| rs10512462 | C/T | aa286: GCT (Ala) > GCC (Ala) |
| rs1923537 | T/C | 3′UTR |
| rs2245121 | A/G | intron |
| rs911887 | A/G | Intron |
| rs22556012 | G/A | Intron |
| rs7078012 | C/T | Intron |
Numbering of amino acid (aa) position is based on the mature protein and does not include signal peptide,
These SNPs have been identified in SP-D but have not been shown to associate with any disease at present.
Overall, the literature indicates that associations exist between SP-A or SP-D genetic variants that may include single nucleotide polymorphisms and haplotypes and the development of both chronic and acute lung diseases, as well as some non-pulmonary diseases. In the following paragraphs we discuss the clinical evidence of these associations.
3. ASSOCIATION OF SP-A AND SP-D POLYMORPHISMS WITH DISEASE
Variants of SP-A1, SP-A2, and SP-D genes have been found to associate with a range of pulmonary and non pulmonary diseases. A summary of the available information that significantly associated gene variants, SNPs and haplotypes of SP-A1, SP-A2, and SP-D with adult and pediatric disease is described below, and summarized in Table 4.
Table 4.
SP-A and SP-D SNP associations with adult and pediatric disease
| Disease studied | Gene(s) | Population | SNP or haplotype | nucleotide (amino acid) | susceptibility | cases | controls | Reference |
|---|---|---|---|---|---|---|---|---|
| ABPA | SP-A1 | Indian | rs1136454 (G/A) | G (Arg) | risk | 22 | 23 | (132) |
| ABPA | SP-A1/SP-A2 | Indian | rs1136454-rs1136452 (C/G) | G(Arg)-G(Ala) | risk | 22 | 23 | (132) |
| ABPA | SP-A2 | Indian | rs1650223 (intron) | risk | 10 | 11 | (132) | |
| AR | SP-A2 | Chinese | rs1965708 | A (Lys) | risk | 216 | 84 | (128) |
| AR | SP-A2 | Chinese | 1A2 | protective | 216 | 84 | (128) | |
| AR | SP-D | Chinese | rs721917 | C (Thr) | risk | 216 | 84 | (127) |
| Asthma | SP-A1 | mixed | 6A | risk | 221 | 355a | (129) | |
| Asthma | SP-A1/SP-A2 | mixed | 6A/1A | risk | 221 | 355a | (129) | |
| Asthma | SP-D | German | no association | 322 | 270 | (126) | ||
| Asthma | SP-D | Black | rs721917 | T (Met) | risk | 162 | 97 | (130) |
| Cardiovascular Disease (CS) | SP-D | Norwegian | rs721917 | C/C (Thr/Thr) | risk | 130 | 100 | (174) |
| CF | SP-A1 | Caucasian | 6A3 | risk | 135b | n/a | (139) | |
| CF | SP-A2 | Caucasian | 1A1 | risk | 135b | n/a | (139) | |
| CF | SP-A1/SP-A2 | Caucasian | 6A3/1A1 | risk | 135b | n/a | (139) | |
| COPD | SP-D | Caucasian | rs2245121 | A | risk | 389 | 472 | (115) |
| COPD | SP-D | Caucasian | rs911887 | G | risk | 389 | 472 | (115) |
| COPD | SP-D | Caucasian | rs6413520 | C (Ser) | risk | 389 | 472 | (115) |
| COPD | SP-D | Caucasian | rs721917 | C (Thr) | risk | 389 | 472 | (115) |
| COPD | SP-D | Caucasian | rs7078012 | C | risk | 389 | 472 | (115) |
| COPD | SP-A1 | Mexican | rs1136451 | A (Pro) | risk | 101 | 81 | (258) |
| HAPE | SP-A1 | Indian | rs1059047 | C (Ala) | risk | 27 | 19 | (134) |
| HAPE | SP-A2 | Indian | rs17880902 (T/C) | T (Asp) | risk | 27 | 19 | (134) |
| HAPE | SP-A2 | Indian | rs17096771 (T/C) | T (Pro) | risk | 27 | 19 | (134) |
| HAPE | SP-A2 | Indian | rs1965708 | C (Gln) | risk | 27 | 19 | (134) |
| IPF | SP-A1 | Mexican | 6A4 | risk | 84 | 194 | (143) | |
| IPF | SP-A1 | Mexican | rs1136450 | C (Leu) | risk | 84 | 194 | (143) |
| IPF | SP-A1 | Mexican | rs1136451 | G (Val) | risk | 84 | 194 | (143) |
| IPF | SP-A1 | Mexican | rs4253527 | T (Trp) | risk | 84 | 194 | (143) |
| Lung Cancer (SCC) | SP-A1 | German | 6A4 | risk | 35 | 110c | (145) | |
| Lung Cancer (SCC) | SP-A1 | German | 6A4 | risk | 35 | 99d | (145) | |
| Lung Cancer (NSCLC) | SP-A1 | German | 6A11 | risk | 68 | 110c | (145) | |
| Lung Cancer (NSCLC) | SP-A2 | German | 1A9 | risk | 68 | 110c | (145) | |
| Lung Cancer (AC) | SP-A1 | German | 6A11 | risk | 23 | 110c | (145) | |
| Lung Cancer (AC) | SP-A2 | German | 1A9 | risk | 23 | 99d | (145) | |
| Meningococcal Disease | SP-A2 | English | rs1965708 | A (lys) | risk | 303 | 222 | (159) |
| Meningococcal Disease | SP-A2 | English | 1A1/1A1 | risk | 303 | 222 | (159) | |
| Meningococcal Disease | SP-A2 | English | 1A5 | protective | 303 | 222 | (159) | |
| Otitis Media | SP-A1/SP-A2 | Finnish | 6A2/1A0 | risk | 47 (acute) | 228 | (164) | |
| Otitis Media | SP-A1/SP-A2 | Finnish | 6A4/1A5 | risk | 47 (acute), 147 (recurrent) | 228 | (164) | |
| Otitis Media | SP-A1 | Finnish | 6A2/6A2 | risk | 147 (recurrent) | 228 | (164) | |
| Otitis Media | SP-A1 | mixed | 6A4 | protective | 258 | 355a | (163) | |
| Otitis Media | SP-A1/SP-A2 | mixed | 6A4/1A5 | protective | 258 | 355a | (163) | |
| Otitis Media | SP-A1 | mixed | rs1059047 | C (Ala) | risk | 258 | 355a | (163) |
| rUTI | SP-A1 | Chinese | rs1059047 | C (Ala) | risk | 32 | 30 | (170). |
| rUTI | SP-A2 | Chinese | rs1965708 | C (Gln) | risk | 32 | 30 | (170). |
| RSV | SP-A2 | mixed | rs1059046 | A (Asn) | risk | 277 | n/a | (153) |
| RSV | SP-A2 | mixed | 1A0/1A0 | protective | 277 | n/a | (153) | |
| RSV | SP-A2 | mixed | 1A2 | protective | 277 | n/a | (153) | |
| RSV | SP-A2 | mixed | 1A2 | protective | 148 | n/a | (152) | |
| RSV | SP-D | mixed | rs2243639 | A (Thr) | risk (possible) | 148 | n/a | (152) |
| RSV | SP-D | mixed | rs721917-rs2243639 | T(Met)-G(Ala) | protective | 148 | n/a | (152) |
| RSV | SP-A1/SP-A2/SP-D | mixed | 6A2-1A0-rs2243639 | G (Ala) | protective | 148 | n/a | (152) |
| RSV | SP-A2 | Finnish | 1A3 | risk | 86 | 95 | (151) | |
| RSV | SP-A2 | Finnish | 1A | protective | 86 | 95 | (151) | |
| RSV | SP-A1 | Finnish | 6A | protective | 86 | 95 | (151) | |
| RSV | SP-A1/SP-A2 | Finnish | 6A/1A | protective | 86 | 95 | (151) | |
| RSV | SP-A1 | Finnish | rs1059047 | C (Ala) | protective | 86 | 95 | (151) |
| RSV | SP-A2 | Finnish | rs17886395 | C (Pro) | protective | 86 | 95 | (151) |
| RSV | SP-A2 | Finnish | rs1965708 | A (Lys) | risk | 86 | 95 | (151) |
| RSV | SP-A2 | Finnish | 1A1/1A1 | risk | 86 | 95 | (151) | |
| RSV | SP-A2 | Finnish | 1A0/1A3 | risk | 86 | 95 | (151) | |
| RSV | SP-A1/SP-A2 | Finnish | 6A2/1A3 | risk | 86 | 95 | (151) | |
| RSV | SP-D | Finnish | rs721917 | T (Met) | risk | 84 | 93 | (150) |
| TB | SP-D | Indian | G459A (exon 7)e | risk | 30 | 30 | (124) | |
| TB | SP-D | Mexican | rs721917 | C (Thr) | risk | 178 | 101 | (117) |
| TB | SP-A1 | Mexican | 6A4 | risk | 178 | 101 | (117) | |
| TB | SP-A2 | Mexican | 1A3 | risk | 178 | 101 | (117) | |
| TB | SP-A2 | Ethiopian | 1A3 | risk | 226 (181 families) | n/a | (123) | |
| TB | SP-A2 | Ethiopian | 1A5 | risk | 226 (181 families) | n/a | (123) | |
| TB | SP-A1 | Ethiopian | rs1136451 | A (Pro) | risk | 226 (181 families) | n/a | (123) |
| TB | SP-A1 | Ethiopian | rs4253527 | T (Trp) | risk | 226 (181 families) | n/a | (123) |
| TB | SP-A2 | Ethiopian | rs17886395 | C (Pro) | risk | 226 (181 families) | n/a | (123) |
| TB | SP-A2 | Ethiopian | rs1965708 | C (Gln) | risk | 226 (181 families) | n/a | (123) |
| UC | SP-D | Japanese | rs911887 | G | risk | 296 | 394 | (22) |
| UC | SP-D | Japanese | rs2243639-rs911887 | A (Thr)-G | risk | 296 | 394 | (22) |
ABPA: allergic bronchopulmonary aspergillosis; AR: allergic rhinitis; CS: coronary stenosis; CF: cystic fibrosis; COPD: chronic obstructive pulmonary disease; HAPE: high altitude pulmonary edema; IPF: idiopathic pulmonary fibrosis; SCC: squamous cell carcinoma; NSCLC: non-small cell lung cancer; AC: adenocarcinoma; rUTI: recurrent urinary tract infection; RSV: respiratory syncytial virus infection; TB: tuberculosis; UC: ulcerative colitis.
infants at risk,
cases were stratified according to pulmonary outcomes (e.g. predicted FEV1, etc), and the risk variants were associated with cases of poor pulmonary outcome,
healthy controls,
clinical controls,
SNP id not available.
3.1. Respiratory pediatric and adult disease associations with SP-A1, SP-A2, and SP-D variants, SNPs and intragenic haplotypes
Since the lung is one of the major sites of surfactant protein synthesis, and because most surfactant proteins are involved collectively in surfactant-related functions, host defense, and regulation of inflammatory processes in the lung, it is expected that collectins play a role in clinical situations where surfactant homeostasis, and/or host defense mechanisms are affected. Therefore, it is not surprising that SP-A, and SP-D (as well as the hydrophobic surfactant proteins, which are not discussed in this review) are involved in the development of, or protection from, various pulmonary diseases (Table 4). In this section, we discuss SP-A and SP-D polymorphisms associations with adult and pediatric respiratory disease, and discuss potential interactions of collectins with viral, bacterial, fungal, and other disease-causing pathogens and molecules, as well as with the immune cells involved in pathogen clearance, and control of infection. It is likely that mechanisms involving function and regulation of collectins in human disease may overlap or differ entirely, depending on the physiologic context or derangement associated with each particular disease.
Different effects of SP-A and SP-D interactions with the same and/or different pathogens do occur. SP-A has been shown to enhance binding of alveolar macrophages to Mycobacterium tuberculosis (121), whereas SP-D binds to the bacterial surface causing a reduction in its uptake by alveolar macrophages (122). Interestingly, several SP-A and SP-D polymorphisms have been found to associate with risk for tuberculosis (117, 123–125). These include variants SP-A1 6A4 (117), and SP-A2 1A3 (117, 123) and 1A5 (123). Also, SP-A1 SNPs rs1136451 and rs4253527 (123), SP-A2 rs17886395 and rs1965708 (123), and SP-D rs721917 (117) and G459A (124) variation have been associated with TB.
Allergic diseases such as asthma and allergic rhinitis (AR) are very common multifactorial diseases, and polymorphisms in SP-A and SP-D genes have been associated with these diseases in several populations in three different clinical studies (126–130). Innate immunity collectins play a critical role in preventing damage and injury to nasal mucosa, which is constantly exposed to inhaled pollutants, microbes, and allergens. SP-A2 rs1965708 SNP was associated with increased risk for AR in a Chinese population (124). In addition, the SP-A2 1A2 variant was found to be protective for AR (128), and the SP-A1 6A variant, and the 6A/1A haplotype were shown to associate with risk for asthma (129). For SP-D, a Threonine at aa11 (rs721917) was associated with increased risk for AR in the Chinese population (125). In contrast, a Methionine at aa11 (rs721917) was found to associate with risk for asthma in a black population (127). A study performed in a German population did not find any association of this SNP and susceptibility to asthma (123). The SP-D Met11Thr variant has been found to associate with assembly, function, and concentration of SP-D, with the Thr variant having a negative impact on these (131).
A small study conducted in an Indian population with allergic bronchopulmonary aspergillosis (ABPA), detected an association of the SP-A1 rs1136454, the SP-A2 rs1136452 SNPs (located at the collagen-like region), and an intronic SNP (rs1650223), with disease susceptibility, and severity of clinical markers of ABPA (total IgE levels and eosinophilia) (132). ABPA is caused by the pathogenic fungus Aspergillus fumigatus, and SP-A binds to various glycosylated allergens-antigens and glycoproteins from the fungus. In addition, SP-A inhibits the Aspergillus fumigatus-induced histamine release from sensitized basophils (133), and enhances macrophage- and neutrophil-mediated clearance of the pathogen (133). Another small association study in an Indian population found the SP-A1 rs1059047, and SP-A2 rs17880902, rs17096771, and rs1965708 SNPs as risk determinants of high altitude pulmonary edema (HAPE), a disease characterized by increased capillary permeability due to exaggerated inflammation, and free radical-mediated lung injury (134). Together, these data indicate that SP-A polymorphisms may play a role in allergy control.
Chronic obstructive pulmonary disease (COPD) is characterized by chronic bronchitis, and/or emphysema. Elevated serum levels of SP-D are a biomarker for COPD (115), and a recent study associated the SP-D SNPs rs2245121, rs911887, rs6413520, rs721917, rs7078012, as well as the combination of rs1051246, rs2245121, rs911887, rs225601, rs6413520, and rs721917, with risk to develop COPD, and with SP-D serum levels, in independent populations, and multiple study designs (115). The authors proposed that the SP-D genetic variants may differentially modulate mechanisms involved in inflammatory signaling functions (135) and SP-D-mediated clearance of apoptotic cells (63), and that these may underline COPD pathogenesis (136, 137). Moreover, SP-A has been found to bind and enhance alveolar macrophage IFN-γ-mediated phagocytosis of Mycoplasma pulmonis, a pathogen involved in pneumonia and exacerbation of asthma, and COPD (138).
A significant association between SP-A1 6A3, SP-A2 1A1, or the combined haplotype 6A3/1A1 with poor pulmonary outcomes in cystic fibrosis (CF) patients has been reported (139). Cystic fibrosis is an autosomal-recessive disease characterized by multi-organ disorders and decreased life expectancy, and recurrent or chronic airway infections with bacteria, including Haemophilus influenzae, Staphylococcus aureus, and Pseudomonas aeruginosa (140) Of relevance, differences in the phagocytic index of alveolar macrophages between SP-A1 and SP-A2 variants have been observed with regards to Pseudomonas aeruginosa and Staphylococcus aureus (43–45, 52). Moreover, pulmonary function deterioration is listed as one of the primary complications of CF, and surfactant proteins have been identified as candidates to mediate pathogenesis of pulmonary disease in CF (141). Decreased levels of SP-A and SP-D were detected in BAL from CF patients (141, 142) and attributed to persistent inflammation, which in turn increases degradation of collectins, and may also affect collectin synthesis (140). Of relevance, the SP-A2 1A1 variant has the lowest activity for enhancement of TNF-α in THP-1 cells. Furthermore, both activities (phagocytosis and cytokine production) are negatively affected by oxidative stress (42, 45), a situation that may exist in the CF microenvironment, and may explain the compromised pulmonary innate immunity commonly seen in CF cases.
The SP-A1, 6A4 variant, and three SNPs: rs1136450, rs1136451, and rs4253527 were associated with risk to develop idiopatic pulmonary fibrosis (IPF) in a nonsmoker Mexican population (143). Derangement in pulmonary surfactant or its individual components and alveolar collapse are common findings in IPF, a progressive lung disorder characterized by sequential acute lung microinjuries, and fibroblastic foci formation, scarring, and end-stage, usually lethal, lung disease (144). The 6A4 variant exhibited differences in self-aggregation when compared with other SP-A variants that differ at aa219, in the CRD region (143). The authors proposed a Tryptophan at aa219 instead of Arginine (rs4253527) to be responsible for biochemical differences between 6A4 and other SP-A variants, and that these differences may affect SP-A1 ability to maintain the function, stability, and structure of surfactant.
Common SP-A1 (6A4), and rare SP-A1 and SP-A2 (6A11, 1A9) variants have been associated with risk for various types of lung cancer (145), when compared to normal and clinical controls. This study concluded that SP-A gene variants may be involved in mechanisms that influence susceptibility to lung cancer of a particular histological type. These mechanisms may include modulation of inflammatory processes and host defense, defense against toxic gases, cigarette smoke, and other environmental factors, NF-kB activity (146, 147), modulation of cytokine production (42, 148, 149), and other determinants of cancer pathogenesis.
Polymorphisms in SP-A and SP-D genes have been linked to susceptibility to infection with respiratory syncytial virus (RSV) (150–153). RSV infection is the most common cause of hospitalization in infants, and the major cause of bronchiolitis during early childhood (154, 155). The risk of RSV infection was associated with the SP-A2 1A3 variant, the 1A1/1A1, and 1A1/1A3 genotypes, and with the 6A2/1A3 haplotype in a Finnish population (151). Asparagine at aa9 (rs1059046), Lysine at aa223 (rs1965708) of SP-A2 (151), as well as the SP-D SNP rs721917 (150) were also associated with risk. In contrast, Alanine at SP-A1 aa9 (rs1059047), and Proline at SP-A2 aa91 (rs17886395), were found to be protective (151). Other protective associations included the SP-A1 6A variant (151), SP-A2 variants 1A2 (152, 153), and 1A (151), 1A0/1A0 genotype (153), and haplotypes, SP-A1/SP-A2 6A2/1A3 (151), SP-D Met11 (rs721917)/Ala160 (rs2243639) (152), and SP-A1/SP-A2/SP-D 6A2/1A0/Ala160 (rs2243639) (152). SP-A has the ability to bind RSV virion glycoproteins and enhance its uptake by immune cells (156, 157). SP-D has also been found to bind the RSV fusion glycoprotein and decrease RSV infectivity (158).
3.2. Non-respiratory pediatric and adult disease associations with SP-A1, SP-A2, and SP-D variants, SNPs and intragenic haplotypes
The literature provides evidence of associations between SNPs and haplotypes of SP-A and SP-D genes with a number of non-respiratory diseases, either caused by diverse microorganisms, or in which the inflammatory/immune response is altered. In this section, we describe associations of polymorphisms in the collectins genes with diseases that occur outside the respiratory tract.
A study performed in an English population found that an SP-A2-specific, non synonymous SNP, rs1965708, was associated with susceptibility to infection, and increased risk of death by meningococcal disease (159). This sickness is caused by a bloodstream infection of Neisseria meningitides after a period of nasopharyngeal colonization (160). The risk allele exhibits a Lysine residue at aa223, located at the SP-A2 domain. Although, SP-A and SP-D are expressed at the site of initial meningococcal colonization, there is no evidence of SP-A binding to meningococci. The authors proposed other factors contributing to disease susceptibility, such as the ability of SP-A2 to bind SIRPα (64), altered inflammatory response, or inefficient handling of prior upper respiratory infections, which is known to affect susceptibility to meningococcal infection (161). The SP-A2 1A1 variant that also contains Lysine at aa223 was associated with disease susceptibility, and the 1A5 variant that contains a Glutamine at the 223 residue was found to be protective. The 1A1 protein has a lower ability to stimulate TNF-α release when compared to 1A, 1A0, and 1A2 (148). The authors proposed a role of the lower TNF-α responses in the increased risk of death (162). Other common and rare SP-A2 variants (1A3, 1A8) encode Lysine at aa223, but these occurred at low frequency in this and other studies.
Two independent groups analyzed associations of SP-A polymorphisms with susceptibility to otitis media, one of the most common infections of early childhood (163, 164), caused by bacterial pathogens (Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis (165), and Staphylococcus aureus), or secondary to respiratory syncitial virus (RSV) infection (166). SP-A and SP-D are expressed in the Eustachian tube, and alterations in the expression or regulation of these molecules may also be the major risk factor for otitis media (167). The Finnish group identified the SP-A1/SP-A2 haplotypes 6A2/1A0 as risk for acute, and 6A4/1A5 as risk for acute and recurrent infection, and the SP-A1 6A2/6A2 genotype for recurrent otitis. Since SP-A binds to, and increases phagocytosis of Streptococcus pneumoniae and Haemophilus influenzae, the most common pathogens in acute otitis media (168), the authors proposed a distinct role of SP-A variants/haplotypes in disease pathogenesis (164). This study took place in Finland, and the distribution of SP-A haplotypes in this population has been shown to differ from frequencies found within the United States (169). This, along with potential differences in patient stratification, is probably one of the reasons why their results differ from those obtained by the American group, whose study population were infants at risk for asthma (163), with no distinction between acute and recurrent otitis media. In this study, the 6A4 variant, and the 6A4/1A5 haplotype were protective for otitis media in white infants. SNP rs1059047, which is located within the N-terminal domain, was also associated with otitis media risk in infants with an Alanine at aa19, whereas infants with a valine at codon 19 were more likely to have otitis media during their first year of life. To date, the role of this SP-A1 domain in infection is unknown.
Recently, the SP-A1 rs1059047 (Alanine at aa19), and the SP-A2 rs1965708 (Glutamine at aa223) SNPs were found to associate with susceptibility to recurrent urinary tract infection (rUTI) (170), a disease caused by Escherichia coli and other microorganisms (171). Host genetic factors have been proposed to play an important role in the pathogenesis of rUTI, and polymorphisms within TLR-2 and TLR-4 genes were also identified to associate with rUTI (172). In addition, lower serum SP-A and SP-D levels correlated with these haplotypes and rUTI, and it is possible that different SP-A haplotypes result in variable SPA levels (99), and contribute to disease susceptibility.
In a recent study, two SP-D SNPs (rs2243639, and rs911887) were associated with ulcerative colitis (UC), a chronic inflammation of the colon, in a Japanese population. Like Crohn’s disease, UC is caused by abnormal activation of the immune system in the intestines. Authors hypothesize that SP-D, by affecting dendritic cell and T-helper cell functions (70), modulates the inflammatory response. The SP-D association with UC is likely related to SP-D involvement the regulation of innate and adaptive immunity against bacteria in the colon (173). A study performed in a Norwegian population, subjects homozygous for the allele encoding Threonine at aa11 of SP-D (SNP rs721917) were found to exhibit higher risk for coronary artery stenosis, a type of cardiovascular disease, in which inflammation has been shown to play an important role (174). The C/C genotype (Thr/Thr) has been shown to correlate with lower serum SP-D levels, smaller oligomeric structures, lower affinity to pathogens, and susceptibility to lung disease (131). However, although it is known that SP-D plays an important role in lipid homeostasis, and removal of dying host cells, the authors conclude that SP-D variants are weakly associated with the atherogenic process. Interestingly, a polymorphism in the TLR-4 gene was also associated with coronary disease in this study; SP-D has been shown to bind TLR-4 via the CRD (175).
3.3. Newborn lung disease associations with SP-A1, SP-A2, and SP-D variants (SNPs and haplotypes)
Genetic associations with newborn disease, as well as their relationship with other factors (e.g. prematurity, use of pre-and post- partum treatment, such as antenatal corticoids, or mechanical ventilation, etc.) are of special interest, because of the potential that these associations hold to predict susceptibilities, and to help on decisions about the use of appropriate treatment. In this section, we focus on studies of SP-A and SP-D gene polymorphisms, and their association with two neonatal diseases, respiratory distress syndrome (RDS) and bronchopulmonary dysplasia (BPD) that reflect, respectively, acute and chronic lung injury. A comparison of the main findings obtained in 13 different studies (11 for RDS, 2 for BPD), is shown in Table 5.
Table 5.
SP-A and SP-D SNP associations with neonatal disease
| Disease studied | Gene(s) | Population | SNP or haplotype | nucleotide (amino acid) | susceptibility | cases | controls | Reference |
|---|---|---|---|---|---|---|---|---|
| BPD | SP-A1 | German | 6A6 | risk | 23 | 23 | (199) | |
| BPD | SP-A2/SP-D | Greek | 1A2-rs2243639 | G (Ala) | protective | 71 (60 families) | (198) | |
| BPD | SP-A/SP-D | Greek | 1A2-rs721917-rs2243639 | C(Thr)-G(Ala) | protective | 71 (60 families) | (198) | |
| RDS | SP-A1 | Chinese | 6A2 | risk | 18 | 28 | (191) | |
| RDS | SP-D | German | rs1923537 (3′UTR) | G/G | protective | 202 | 68 | (116) |
| RDS | SP-A2/SP-D | mixed | 1A1-rs2243639 | A (Thr) | protective | 132 families | (197) | |
| RDS | SP-A2/SP-D | mixed | 1A1-rs721917-rs2243639 | T(Met)-A(Thr) | protective | 132 families | (197) | |
| RDS | SP-A1/SP-A2/SP-D | mixed | 6A4-1A2-rs2243639 | A (Thr) | protective | 132 families | (197) | |
| RDS | SP-A1/SP-A2/SP-D | mixed | 6A3-1A1-rs721917-rs2243639 | T(Met)-A(Thr) | protective | 132 families | (197) | |
| RDS | SP-A1/SP-A2/SP-D | mixed | 6A4-1A2-rs721917-rs2243639 | T(Met)-A(Thr) | protective | 132 families | (197) | |
| RDS | SP-A1 | Finnish | 6A2 | protectived | 198a | (187) | ||
| RDS | SP-A1 | Finnish | 6A2/6A2 | protectived | 198a | (187) | ||
| RDS | SP-A1/SP-A2 | Finnish | 6A2/1A0 | protectived | 198a | (187) | ||
| RDS | SP-A1 | Finnish | 6A2 | riskb | 441/480c | (190) | ||
| RDS | SP-A1 | Finnish | 6A2/6A2 | riskb | 441/480c | (190) | ||
| RDS | SP-A1/SP-A2 | Finnish | 6A2/1A0 | risk | 86 (76 families) | 35 (31 families) | (184) | |
| RDS | SP-A1 | Finnish | 6A2 | risk | 88 (76 families) | 35 (31 families) | (184) | |
| RDS | SP-A1 | Finnish | 6A3 | protective | 88 (76 families) | 35 (31 families) | (184) | |
| RDS | SP-A2 | Finnish | 1A0 | risk | 93 (76 families) | 38 (31 families) | (184) | |
| RDS | SP-A1 | Finnish | 6A2 | risk | 46 | 43 | (185) | |
| RDS | SP-A1 | Finnish | 6A3 | protective | 46 | 43 | (185) | |
| RDS | SP-A1/SP-A2 | Finnish | 6A2/1A0 | risk | 88 | 88 | (185) | |
| RDS | SP-A1 | mixed | 6A2 | risk | 122 (32 families) | (186) | ||
| RDS | SP-A1 | mixed | 6A4 | protective | 122 (32 families) | (186) | ||
| RDS | SP-A2 | mixed | 1A0 | risk | 122 (32 families) | (186) | ||
| RDS | SP-A2 | Mixed | 1A5 | protective | 122 (32 families) | (186) | ||
| RDS | SP-A1/SP-A2 | Mixed | 6A2/1A0 | risk | 122 (32 families) | (186) | ||
| RDS | SP-A1/SP-A2 | Mixed | 6A4/1A5 | protective | 122 (32 families) | (186) | ||
| RDS | SP-A1 | Finnish | 6A2 | riske | 184 | 500 | (188) | |
| RDS | SP-A1 | Finnish | 6A3 | protectivee | 184 | 500 | (188) | |
| RDS | SP-A1/SPA2 | Finnish | 6A2/1A0 | riske | 184 | 500 | (188) | |
| RDS | SP-A1/SP-A2 | Finnish | 6A3/1A2 | protectivee | 184 | 500 | (188) | |
| RDS | SP-A1 | Black | 6A3 | protectiveg | 40 | 38 | (192) | |
| RDS | SP-A1 | Caucasian | 6A2 | riskf | 203 | 331 | (192) | |
| RDS | SP-A2 | Caucasian | 1A0 | riskf | 203 | 331 | (192) | |
| RDS | SP-A2 | Caucasian | 1A0 | risk | 106 | 86 | (196) |
RDS: respiratory distress syndrome; BPD: bronchopulmonary dysplasia;
twin pairs;
singletons;
twin or multiple infants;
protective if twins are discordant for RDS, but risk if twins are concordant for RDS;
together with SP-B Thr/Thr genotype at Ile131Thr polymorphism;
together with the SP-B genotype 9306(A/G) or intron 4 (del/*);
together with the SP-B genotype 1580(T/T).
Deficiency of surfactant can result in RDS in prematurely born infants. Insufficient amounts of surfactant proteins, particularly SP-A, as well as absence of tubular myelin, an extracellular structural form of surfactant, have been shown to occur in RDS (176–180). Numerous studies have explored the relationship among SP-A1 and SP-A2 gene variants, and expression of SP-A in in vitro and in vivo models (32, 110, 114, 181, 182), however no clinical correlations among SP-A1, and/or SP-A2 haplotypes or variants, and SP-A levels in the lung have yet been fully described to date (109, 183). A specific SP-A1/SP-A2 haplotype, 6A2/1A0, associated with RDS risk in most studies (184–186). In a particular study, this haplotype was only found to be a risk factor if twins were concordant for RDS (187). Another study identified an interaction of this haplotype with an SP-B polymorphism (Ile131Thr) (188), and the same interaction was found for 6A3/1A2 and SP-B in protection (188).
The SP-A1 6A2 variant has been associated with risk in the majority of the studies (184, 186, 189–191). These associations may be also dependent on SP-B polymorphisms (188, 192, 193), and influenced by multiple birth and birth order (187, 190), antenatal glucocorticoid therapy (11, 194, 195), size of the uterus and length of gestation (2). The SP-A1 6A2/6A2 genotype association with RDS susceptibility was also found to be influenced by multiple birth, being associated with risk when twins were concordant for RDS, or protective when twins were discordant for RDS (187, 190).
Variants 6A3 of SP-A1 (184, 185, 188, 192), and 1A0 of SP-A2 (184, 186, 192, 196) and other less frequent SP-A1 (6A4) and SP-A2 (1A5) variants (186) were associated with protection for RDS in several studies. In a separate study, the haplotype 6A4/1A5 was found to be protective (186).
For SP-D, a non-coding SNP (rs1923537) was protective for RDS in a German study (116), and a different study identified several haplotypes of SP-A1, SP-A2, and SP-D that also associate with protection. These include haplotypes SP-A2/SP-D: 1A1-Thr160 (rs2243639), 1A1-Met11 (rs721917)-Thr160 (rs2243639), and SP-A1/SP-A2/SP-D: 6A4-1A2-Tr160 (rs2243639), 6A3-1A1-Met11 (rs721917)-Thr160 (rs2243639), and 6A4-1A2-Met11 (rs721917)-Thr160 (rs2243639) (197).
BPD is the most common chronic lung disease in infants. A number of antenatal and postnatal risk factors influence susceptibility to BPD. SP-A2/SP-D haplotypes were found to protect against BPD: 1A2-Ala160 (rs2243639), and 1A2-Thr11 (rs721917)-Ala160 (rs2243639) (198), and an association between the rare SP-A1 6A6 variant and risk for BPD has also been reported (199).
3.4. SP-A, and SP-D polymorphisms found in more than one study group
Several SP-A1, and SP-A2 genetic variants, as well as, specific SP-A1, SP-A2, and SP-D SNPs were found to associate with lung disease susceptibility in more than one study group. A previous review has described these associations (12). We provide an updated summary of this information, and also included associations with non-respiratory diseases in Tables 6 (SNPs) and Table 7 (gene variants).
Table 6.
Summary of SP-A1, SP-A2, and SP-D SNPs and disease susceptibility
| Gene | SNP | Nucleotide (aa) | Risk | Protection |
|---|---|---|---|---|
| SP-A1 | rs1059047 | C (Ala) | HAPE | RSV |
| Otitis Media | ||||
| rUTI | ||||
| rs1136450 | C (Leu) | IPF | ||
| rs1136451 | A (Pro) | COPD | ||
| TB | ||||
| G (Val) | IPF | |||
| rs4253527 | T (Trp) | IPF | ||
| TB | ||||
| SP-A2 | rs1059046 | A (Asn) | RSV | |
| rs17886395 | C (Pro) | TB | RSV | |
| rs1965708 | A (Lys) | AR | ||
| Meningococcal Disease | ||||
| RSV | ||||
| C (Gln) | HAPE | |||
| rUTI | ||||
| TB | ||||
| SP-D | rs721917 | C (Thr) | AR | BPD1 |
| Cardiovascular (CS) | RDS1 | |||
| COPD | ||||
| TB | ||||
| T (Met) | Asthma | RSV2 | ||
| RSV | ||||
| rs6413520 | C (Ser) | COPD | ||
| rs2243639 | A (Thr) | RSV | RDS1 | |
| UC | ||||
| G (Ala) | RSV1 | |||
| BPD1 | ||||
| RDS3 | ||||
| rs1923537 | G | RDS | ||
| rs2245121 | A | COPD | ||
| rs911887 | G | COPD | ||
| UC | ||||
| rs7078012 | C | COPD |
AR: allergic rhinitis; BPD: bronchopulmonary dysplasia; CS: coronary stenosis; COPD: chronic obstructive pulmonary disease; HAPE: high altitude pulmonary edema; IPF: idiopathic pulmonary fibrosis; rUTI: recurrent urinary tract infection; RDS: respiratory distress syndrome; RSV: respiratory syncytial virus infection; TB: tuberculosis; UC: ulcerative colitis.
haplotypes with SP-A1 and/or SP-A2 polymorphisms;
haplotype with SP-D rs2243639;
both SNP alleles were associated with protection, depending on the population
Table 7.
Summary of SP-A1 and SP-A2 genetic variants and disease susceptibility
| Gene | Variant | Risk | Protection |
|---|---|---|---|
| SP-A1 | 6A | Asthma | RSV |
| CF | |||
| 6A2 | Otitis Media | ||
| RSVa | |||
| RDS | |||
| 6A3 | CF | RDS | |
| 6A4 | IPF | Otitis Mediab | |
| Lung cancer | RDSb | ||
| 6A6 | BPD | ||
| 6A11 | Lung cancer | ||
| SP-A2 | 1A | Asthmaa | RSV |
| 1A0 | RDS | ||
| Otitis Media RSVc | |||
| 1A1 | CF | RDSa | |
| RSV | |||
| 1A2 | AR | ||
| BPDa | |||
| RDSa | |||
| RSV | |||
| 1A3 | RSV | ||
| TB | |||
| 1A5 | Otitis Mediaa | RDS | |
| TB |
AR: allergic rhinitis; BPD: bronchopulmonary dysplasia; CF: cystic fibrosis; IPF: idiopathic pulmonary fibrosis; RDS: respiratory distress syndrome; RSV: respiratory syncytial virus infection; TB: tuberculosis.
only in haplotypes (with SP-A or SP-D),
in Otitis Media, 6A4 is risk if in haplotype with 1A5, but in RDS remains protective if in haplotype with 1A5,
risk in 1A0/1A3, protectivein 1A0/1A0.
4. ASSOCIATION OF SP-A AND SP-D SERUM AND BRONCHOALVEOLAR LAVAGE FLUID (BAL) PROTEIN LEVELS WITH DISEASE
Collectins are considered to be markers of and/or contributors to the pathogenesis of various diseases characterized by inflammation, infection, and/or derangement of pulmonary function or integrity. Although associations of SP-A and SP-D gene polymorphisms and disease have been observed, to date, no human disease has been identified to be the result of one or more SPA or SP-D gene polymorphisms. However, clinical conditions have been identified where a) the amounts of SP-A, SP-D, as well as the SP-A1/SP-A ratio in BAL or serum are altered, and b) host defense and inflammation mechanisms mediated by collectins are deranged. Association studies have correlated clinical outcomes and collectins concentrations (140–142, 200–251). These associations may underline the contribution SP-A and SP-D make to innate host defense by altering cytokine production, enhancing immune cells chemotaxis and function, and regulating cell proliferation and apoptosis, as well as the previously decribed interaction with pathogens (252, 253). In Table 8 we have reviewed and summarized the clinical studies that correlated significant changes in serum and BAL SP-A and SP-D protein levels with pulmonary and non-pulmonary disorders. We present the information as increases/decreases compared to control subjects for each study. The absolute SP-A and SP-D levels are not reported due to potential variation among studies that may arise from the use of different antibodies and standards.
Table 8.
SP-A and SP-D protein levels in disease, and other clinical conditions, compared to control.
| Disease | Serum levels | References | BAL levels | References | ||
|---|---|---|---|---|---|---|
| SP-A | SP-D | SP-A | SP-D | |||
| Asthma | ↑ | (200) | ↓ | (226) | ||
| ↑ | ↑ | (227) | ||||
| BPD | ↓ | (228) | ||||
| Bronchitis | ↑ | (201) | ||||
| CF | ↑ | (202) | ↓ (no infection) | (142) | ||
| ↓ (no infection) | (140, 141) | |||||
| ↑ (infection) | (229) | |||||
| ↓ (infection) | (140, 141) | |||||
| CLE | ↑ | (203) | ||||
| COPD | ↑ | (204) | ↑1 | (230, 231) | ||
| ↑ | (205) | ↓ | (232) | |||
| HP | ↑ | (206) | ↑ | (233, 234) | ||
| IPF | ↑ | (207–212) | ↓ | (235, 236) | ||
| ↑ | (210, 211, 213, 214) | |||||
| Lung trauma | ↓ | (237) | ||||
| Measles | ↑ | (215) | ||||
| PAP | ↑ | (207) | ↑ | (207, 238) | ||
| ↑ | (210) | ↑2 | (239) | |||
| ↑ | (210, 240, 241) | |||||
| Pneumonia | ↑ | (213, 216, 217) | ↓ | (242–244) | ||
| ↑ (HIV +) | (244, 245) | |||||
| ↑ | (217) | |||||
| RA | ↓ | (218, 219) | ||||
| RDS | ↑ | (203, 220–222) | ↓ | (242, 246, 247) | ||
| ↓ | (223) | ↓ | (248, 249) | |||
| ↑ | (211, 221, 224) | ↓2 | ↓2 | (249) | ||
| RSV | ↓ | (251) | ||||
| Sarcoidosis | ↑ | (210, 225) | ↑ | (233) | ||
| ↑ | (210) | |||||
| TB | ↑ | (210) | ||||
BPD: bronchopulmonary dysplasia, CF: cystic fibrosis, CLE: cardiac lung edema, HP: hypersensitivity pneumonitis, IPF: idiopatic pulmonary fibrosis, PAP: pulmonary alveolar proteinosis, RA: rheumatoid arthritis, RDS: respiratory distress syndrome, RSV: respiratory syncytial virus infection, TB: tuberculosis.
Levels measured in induced sputum and lung tissue,
The samples from these studies are from tracheal aspirates
The literature provides evidence that SP-A and SP-D levels are influenced by age, health, and smoking status, circadian rhythm, as well as by genetic factors (106, 109, 118, 120, 232, 254–257). However, very few studies have correlated genetic polymorphisms with collectins serum levels. In one study, an SP-D haplotype revealed a negative association with serum SP-D levels (118). In addition, the SP-D rs721917 SNP has been shown to influence oligomerization, function, and serum concentration of SP-D (131). With regards to SP-A, since the functional activity of SP-A1 and SP-A2 has been shown to differ (33, 43, 44, 52), the overall functional activity of human SP-A cannot be assessed if total SP-A levels are reported. Therefore, the relative SP-A1/SP-A2 ratio is likely to be more informative of the total functional SP-A activity in the lung, and potentially provide a more disease-specific marker, especially if it is further correlated with the specific SP-A1 and SP-A2 genotype. Future studies may focus on potential correlations among SP-A and SP-D genetic polymorphisms, protein levels, and susceptibility to disease, and provide further evidence of collectins as genetic biomarkers for disease.
5. SUMMARY
We have reviewed the available experimental evidence of SP-A1, SP-A2, and SP-D genetic associations with disease susceptibility in adults, children, and newborns. Interactions among these and other genes products, as well as the impact of environmental factors, and other genetic and non-genetic factors are a necessary extension of this work, and will broaden our knowledge about the complexities underlying the role of collectins in respiratory disease. Understanding these complexities, and the impact of genetic variability will help us understand individual disease-susceptibilities, identify risk groups, permit early detection of risk in neonates, and therefore design proper interventions in an attempt to decrease the long-term pulmonary injury.
Acknowledgments
This work was supported by the NIH HL-34788 grant.
Abbreviations
- SNP
single nucleotide polymorphism
- SFTPA
SP-A, surfactant protein A
- SFTPD
SP-D, surfactant protein D
- TNF-α
tumor necrosis factor alpha
- IFNγ
interferon gamma
- CRD
carbohydrate recognition domain
- aa
amino acid
- LPS
lipopolisaccharide
References
- 1.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8:440–7. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kishore U, Bernal AL, Kamran MF, Saxena S, Singh M, Sarma PU, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D in human health and disease. Arch Immunol Ther Exp (Warsz) 2005;53:399–417. [PubMed] [Google Scholar]
- 3.Meyers DA. Genetics of asthma and allergy: what have we learned? J Allergy Clin Immunol. 2010;126:439–46. doi: 10.1016/j.jaci.2010.07.012. quiz 447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie JD. Genetic epidemiology of acute lung injury: choosing the right candidate genes is the first step. Crit Care. 2004;8:411–3. doi: 10.1186/cc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc. 2005;2:188–94. doi: 10.1513/pats.200501-005AC. [DOI] [PubMed] [Google Scholar]
- 6.Leikauf GD, McDowell SA, Wesselkamper SC, Hardie WD, Leikauf JE, Korfhagen TR, Prows DR. Acute lung injury: functional genomics and genetic susceptibility. Chest. 2002;121:70S–75S. doi: 10.1378/chest.121.3_suppl.70s. [DOI] [PubMed] [Google Scholar]
- 7.Villar J, Flores C, Mendez-Alvarez S. Genetic susceptibility to acute lung injury. Crit Care Med. 2003;31:S272–5. doi: 10.1097/01.CCM.0000057903.11528.6D. [DOI] [PubMed] [Google Scholar]
- 8.Lam E, dos Santos CC. Advances in molecular acute lung injury/acute respiratory distress syndrome and ventilator-induced lung injury: the role of genomics, proteomics, bioinformatics and translational biology. Curr Opin Crit Care. 2008;14:3–10. doi: 10.1097/MCC.0b013e3282f42211. [DOI] [PubMed] [Google Scholar]
- 9.Meyer NJ, Garcia JG. Wading into the genomic pool to unravel acute lung injury genetics. Proc Am Thorac Soc. 2007;4:69–76. doi: 10.1513/pats.200609-157JG. [DOI] [PubMed] [Google Scholar]
- 10.Reddy AJ, Kleeberger SR. Genetic polymorphisms associated with acute lung injury. Pharmacogenomics. 2009;10:1527–39. doi: 10.2217/pgs.09.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallman M, Haataja R. Genetic influences and neonatal lung disease. Semin Neonatol. 2003;8:19–27. doi: 10.1016/s1084-2756(02)00196-3. [DOI] [PubMed] [Google Scholar]
- 12.Floros J, Thomas N. Genetic variations of surfactant proteins and lung injury. In: Nakos G, Papathanasiou A, editors. Surfactant Pathogenesis and Treatment of Lung Diasease. K. Research Signpost; India: 2009. [Google Scholar]
- 13.Floros J, Pavlovic J. Genetics of acute respiratory distress syndrome: challenges, approaches, surfactant proteins as candidate genes. Semin Respir Crit Care Med. 2003;24:161–8. doi: 10.1055/s-2003-39015. [DOI] [PubMed] [Google Scholar]
- 14.Floros J, Phelps DS. Pulmonary surfactant. In: Yaksh TL, et al., editors. Anesthesia: Biologic Foundations. Lippincott-Raven Publishers; Philadelphia: 1997. [Google Scholar]
- 15.Floros J, Kala P. Surfactant proteins: molecular genetics of neonatal pulmonary diseases. Annu Rev Physiol. 1998;60:365–84. doi: 10.1146/annurev.physiol.60.1.365. [DOI] [PubMed] [Google Scholar]
- 16.Murray E, Khamri W, Walker MM, Eggleton P, Moran AP, Ferris JA, Knapp S, Karim QN, Worku M, Strong P, Reid KB, Thursz MR. Expression of surfactant protein D in the human gastric mucosa and during Helicobacter pylori infection. Infect Immun. 2002;70:1481–7. doi: 10.1128/IAI.70.3.1481-1487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitoh H, Okayama H, Shimura S, Fushimi T, Masuda T, Shirato K. Surfactant protein A2 gene expression by human airway submucosal gland cells. Am J Respir Cell Mol Biol. 1998;19:202–9. doi: 10.1165/ajrcmb.19.2.3239. [DOI] [PubMed] [Google Scholar]
- 18.Stahlman MT, Gray ME, Hull WM, Whitsett JA. Immunolocalization of surfactant protein-D (SP-D) in human fetal, newborn, and adult tissues. J Histochem Cytochem. 2002;50:651–60. doi: 10.1177/002215540205000506. [DOI] [PubMed] [Google Scholar]
- 19.Bourbon JR, Chailley-Heu B. Surfactant proteins in the digestive tract, mesentery, and other organs: evolutionary significance. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:151–61. doi: 10.1016/s1095-6433(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 20.Bräuer L, Möschter S, Beileke S, Jäger K, Garreis F, Paulsen FP. Human parotid and submandibular glands express and secrete surfactant proteins A, B, C and D. Histochem Cell Biol. 2009;132:331–8. doi: 10.1007/s00418-009-0609-x. [DOI] [PubMed] [Google Scholar]
- 21.Madsen J, Tornoe I, Nielsen O, Koch C, Steinhilber W, Holmskov U. Expression and localization of lung surfactant protein A in human tissues. Am J Respir Cell Mol Biol. 2003;29:591–7. doi: 10.1165/rcmb.2002-0274OC. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Arimura Y, Goto A, Hosokawa M, Nagaishi K, Yamashita K, Yamamoto H, Sonoda T, Nomura M, Motoya S, Imai K, Shinomura Y. Genetic variants in surfactant, pulmonary-associated protein D (SFTPD) and Japanese susceptibility to ulcerative colitis. Inflamm Bowel Dis. 2009;15:918–25. doi: 10.1002/ibd.20936. [DOI] [PubMed] [Google Scholar]
- 23.Paananen R, Glumoff V, Sormunen R, Voorhout W, Hallman M. Expression and localization of lung surfactant protein B in Eustachian tube epithelium. Am J Physiol Lung Cell Mol Physiol. 2001;280:L214–20. doi: 10.1152/ajplung.2001.280.2.L214. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z, deMello D, Phelps DS, Koltun WA, Page M, Floros J. Both human SP-A1 and Sp-A2 genes are expressed in small and large intestine. Pediatr Pathol Mol Med. 2001;20:367–86. [PubMed] [Google Scholar]
- 25.Lin Z, Floros J. Heterogeneous allele expression of pulmonary SP-D gene in rat large intestine and other tissues. Physiol Genomics. 2002;11:235–43. doi: 10.1152/physiolgenomics.00061.2002. [DOI] [PubMed] [Google Scholar]
- 26.Bräuer L, Johl M, Börgermann J, Pleyer U, Tsokos M, Paulsen FP. Detection and localization of the hydrophobic surfactant proteins B and C in human tear fluid and the human lacrimal system. Curr Eye Res. 2007;32:931–8. doi: 10.1080/02713680701694369. [DOI] [PubMed] [Google Scholar]
- 27.Bräuer L, Kindler C, Jäger K, Sel S, Nölle B, Pleyer U, Ochs M, Paulsen FP. Detection of surfactant proteins A and D in human tear fluid and the human lacrimal system. Invest Ophthalmol Vis Sci. 2007;48:3945–53. doi: 10.1167/iovs.07-0201. [DOI] [PubMed] [Google Scholar]
- 28.Glasser SW, Burhans MS, Korfhagen TR, Na CL, Sly PD, Ross GF, Ikegami M, Whitsett JA. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc Natl Acad Sci U S A. 2001;98:6366–71. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Augusto LA, Synguelakis M, Johansson J, Pedron T, Girard R, Chaby R. Interaction of pulmonary surfactant protein C with CD14 and lipopolysaccharide. Infect Immun. 2003;71:61–7. doi: 10.1128/IAI.71.1.61-67.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crouch E, Wright JR. Surfactant proteins A and D and pulmonary host defense. Annu Rev Physiol. 2001;63:521–54. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- 31.Hawgood S, Poulain FR. The pulmonary collectins and surfactant metabolism. Annu Rev Physiol. 2001;63:495–519. doi: 10.1146/annurev.physiol.63.1.495. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Guo X, Diangelo S, Thomas N, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: Formation of tubular myelin in vivo requires both gene products. J Biol Chem. 2010;285:11998–2010. doi: 10.1074/jbc.M109.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, Bates-Kenney S, Tao J, Phelps D, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. 2004;43:4227–39. doi: 10.1021/bi036023i. [DOI] [PubMed] [Google Scholar]
- 34.Williams MC, Hawgood S, Hamilton RL. Changes in lipid structure produced by surfactant proteins SP-A, SP-B, and SP-C. Am J Respir Cell Mol Biol. 1991;5:41–50. doi: 10.1165/ajrcmb/5.1.41. [DOI] [PubMed] [Google Scholar]
- 35.Ikegami M, Whitsett JA, Jobe A, Ross G, Fisher J, Korfhagen T. Surfactant metabolism in SP-D gene-targeted mice. Am J Physiol Lung Cell Mol Physiol. 2000;279:L468–76. doi: 10.1152/ajplung.2000.279.3.L468. [DOI] [PubMed] [Google Scholar]
- 36.LeVine AM, Whitsett JA, Gwozdz JA, Richardson TR, Fisher JH, Burhans MS, Korfhagen TR. Distinct effects of surfactant protein A or D deficiency during bacterial infection on the lung. J Immunol. 2000;165:3934–40. doi: 10.4049/jimmunol.165.7.3934. [DOI] [PubMed] [Google Scholar]
- 37.Botas C, Poulain F, Akiyama J, Brown C, Allen L, Goerke J, Clements J, Carlson E, Gillespie AM, Epstein C, Hawgood S. Altered surfactant homeostasis and alveolar type II cell morphology in mice lacking surfactant protein D. Proc Natl Acad Sci U S A. 1998;95:11869–74. doi: 10.1073/pnas.95.20.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Floros J, Wang G, Lin Z. Genetic Diversity of Human SP-A, a Molecule with Innate host Defense and Surfactant-Related Functions; Characteristics, Primary Function, and Significance. Current Pharmacogenomics. 2005;3:87–95. [Google Scholar]
- 39.Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43:1293–315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–78. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 41.Voss T, Melchers K, Scheirle G, Schäfer KP. Structural comparison of recombinant pulmonary surfactant protein SPA derived from two human coding sequences: implications for the chain composition of natural human SP-A. Am J Respir Cell Mol Biol. 1991;4:88–94. doi: 10.1165/ajrcmb/4.1.88. [DOI] [PubMed] [Google Scholar]
- 42.Wang G, Umstead T, Phelps D, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. 2002;110:79–84. doi: 10.1289/ehp.0211079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, Floros J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. 2007;75:1403–12. doi: 10.1128/IAI.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. 2007;46:8425–35. doi: 10.1021/bi7004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikerov A, Umstead T, Gan X, Huang W, Guo X, Wang G, Phelps D, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008;294:L121–30. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikerov A, White M, Hartshorn K, Wang G, Floros J. Inhibition of hemagglutination activity of influenza A viruses by SP-A1 and SP-A2 variants expressed in CHO cells. Med Microbiol Immunol. 2008;197:9–12. doi: 10.1007/s00430-007-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crouch EC. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol. 1998;19:177–201. doi: 10.1165/ajrcmb.19.2.140. [DOI] [PubMed] [Google Scholar]
- 48.Crouch E, Hartshorn K, Ofek I. Collectins and pulmonary innate immunity. Immunol Rev. 2000;173:52–65. doi: 10.1034/j.1600-065x.2000.917311.x. [DOI] [PubMed] [Google Scholar]
- 49.Kishore U, Reid KB. Structures and functions of mammalian collectins. Results Probl Cell Differ. 2001;33:225–48. doi: 10.1007/978-3-540-46410-5_12. [DOI] [PubMed] [Google Scholar]
- 50.Ding J, Umstead TM, Floros J, Phelps DS. Factors affecting SP-A-mediated phagocytosis in human monocytic cell lines. Respir Med. 2004;98:637–50. doi: 10.1016/j.rmed.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 51.Schagat TL, Wofford JA, Wright JR. Surfactant protein A enhances alveolar macrophage phagocytosis of apoptotic neutrophils. J Immunol. 2001;166:2727–33. doi: 10.4049/jimmunol.166.4.2727. [DOI] [PubMed] [Google Scholar]
- 52.Mikerov A, Umstead T, Huang W, Liu W, Phelps D, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2005;288:L150–8. doi: 10.1152/ajplung.00135.2004. [DOI] [PubMed] [Google Scholar]
- 53.Kingma PS, Whitsett JA. The pulmonary collectins and respiratory syncytial virus: is there a clinical link? J Pediatr. 2010;156:349–50. doi: 10.1016/j.jpeds.2009.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kingma PS, Whitsett JA. In defense of the lung: surfactant protein A and surfactant protein D. Curr Opin Pharmacol. 2006;6:277–83. doi: 10.1016/j.coph.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 55.McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest. 2002;109:707–12. doi: 10.1172/JCI15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeVine AM, Whitsett JA. Pulmonary collectins and innate host defense of the lung. Microbes Infect. 2001;3:161–6. doi: 10.1016/s1286-4579(00)01363-0. [DOI] [PubMed] [Google Scholar]
- 57.Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. 2001;20:269–92. [PubMed] [Google Scholar]
- 58.Haagsman HP, Hogenkamp A, van Eijk M, Veldhuizen EJ. Surfactant collectins and innate immunity. Neonatology. 2008;93:288–94. doi: 10.1159/000121454. [DOI] [PubMed] [Google Scholar]
- 59.Haagsman HP. Interactions of surfactant protein A with pathogens. Biochim Biophys Acta. 1998;1408:264–77. doi: 10.1016/s0925-4439(98)00072-6. [DOI] [PubMed] [Google Scholar]
- 60.Tenner AJ, Robinson SL, Borchelt J, Wright JR. Human pulmonary surfactant protein (SP-A), a protein structurally homologous to C1q, can enhance FcR- and CR1-mediated phagocytosis. J Biol Chem. 1989;264:13923–8. [PubMed] [Google Scholar]
- 61.Nepomuceno RR, Henschen-Edman AH, Burgess WH, Tenner AJ. cDNA cloning and primary structure analysis of C1qR(P), the human C1q/MBL/SPA receptor that mediates enhanced phagocytosis in vitro. Immunity. 1997;6:119–29. doi: 10.1016/s1074-7613(00)80419-7. [DOI] [PubMed] [Google Scholar]
- 62.Sano H, Chiba H, Iwaki D, Sohma H, Voelker DR, Kuroki Y. Surfactant proteins A and D bind CD14 by different mechanisms. J Biol Chem. 2000;275:22442–51. doi: 10.1074/jbc.M001107200. [DOI] [PubMed] [Google Scholar]
- 63.Vandivier RW, Ogden CA, Fadok VA, Hoffmann PR, Brown KK, Botto M, Walport MJ, Fisher JH, Henson PM, Greene KE. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169:3978–86. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- 64.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 65.Janssen WJ, McPhillips KA, Dickinson MG, Linderman DJ, Morimoto K, Xiao YQ, Oldham KM, Vandivier RW, Henson PM, Gardai SJ. Surfactant proteins A and D suppress alveolar macrophage phagocytosis via interaction with SIRP alpha. Am J Respir Crit Care Med. 2008;178:158–67. doi: 10.1164/rccm.200711-1661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chroneos ZC, Abdolrasulnia R, Whitsett JA, Rice WR, Shepherd VL. Purification of a cell-surface receptor for oysurfactant protein A. J Biol Chem. 1996;271:16375–83. doi: 10.1074/jbc.271.27.16375. [DOI] [PubMed] [Google Scholar]
- 67.Tino MJ, Wright JR. Glycoprotein-340 binds surfactant protein-A (SP-A) and stimulates alveolar macrophage migration in an SP-A-independent manner. Am J Respir Cell Mol Biol. 1999;20:759–68. doi: 10.1165/ajrcmb.20.4.3439. [DOI] [PubMed] [Google Scholar]
- 68.Murakami S, Iwaki D, Mitsuzawa H, Sano H, Takahashi H, Voelker DR, Akino T, Kuroki Y. Surfactant protein A inhibits peptidoglycan-induced tumor necrosis factor-alpha secretion in U937 cells and alveolar macrophages by direct interaction with toll-like receptor 2. J Biol Chem. 2002;277:6830–7. doi: 10.1074/jbc.M106671200. [DOI] [PubMed] [Google Scholar]
- 69.Guillot L, Balloy V, McCormack FX, Golenbock DT, Chignard M, Si-Tahar M. Cutting edge: the immunostimulatory activity of the lung surfactant protein-A involves Toll-like receptor 4. J Immunol. 2002;168:5989–92. doi: 10.4049/jimmunol.168.12.5989. [DOI] [PubMed] [Google Scholar]
- 70.Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. 2005;5:58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 71.Yamada C, Sano H, Shimizu T, Mitsuzawa H, Nishitani C, Himi T, Kuroki Y. Surfactant protein A directly interacts with TLR4 and MD-2 and regulates inflammatory cellular response. Importance of supratrimeric oligomerization. J Biol Chem. 2006;281:21771–80. doi: 10.1074/jbc.M513041200. [DOI] [PubMed] [Google Scholar]
- 72.Kuroki Y, Mason RJ, Voelker DR. Alveolar type II cells express a high-affinity receptor for pulmonary surfactant protein A. Proc Natl Acad Sci U S A. 1988;85:5566–70. doi: 10.1073/pnas.85.15.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stevens PA, Wissel H, Sieger D, Meienreis-Sudau V, Rüstow B. Identification of a new surfactant protein A binding protein at the cell membrane of rat type II pneumocytes. Biochem J. 1995;308:77–81. doi: 10.1042/bj3080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kresch MJ, Christian C, Lu H. Isolation and partial characterization of a receptor to surfactant protein A expressed by rat type II pneumocytes. Am J Respir Cell Mol Biol. 1998;19:216–25. doi: 10.1165/ajrcmb.19.2.3061. [DOI] [PubMed] [Google Scholar]
- 75.Miyamura K, Leigh LE, Lu J, Hopkin J, López Bernal A, Reid KB. Surfactant protein D binding to alveolar macrophages. Biochem J. 1994;300:237–42. doi: 10.1042/bj3000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Restrepo CI, Dong Q, Savov J, Mariencheck WI, Wright JR. Surfactant protein D stimulates phagocytosis of Pseudomonas aeruginosa by alveolar macrophages. Am J Respir Cell Mol Biol. 1999;21:576–85. doi: 10.1165/ajrcmb.21.5.3334. [DOI] [PubMed] [Google Scholar]
- 77.LeVine AM, Whitsett JA, Hartshorn KL, Crouch EC, Korfhagen TR. Surfactant protein D enhances clearance of influenza A virus from the lung in vivo. J Immunol. 2001;167:5868–73. doi: 10.4049/jimmunol.167.10.5868. [DOI] [PubMed] [Google Scholar]
- 78.LeVine AM, Elliott J, Whitsett JA, Srikiatkhachorn A, Crouch E, DeSilva N, Korfhagen T. Surfactant protein-d enhances phagocytosis and pulmonary clearance of respiratory syncytial virus. Am J Respir Cell Mol Biol. 2004;31:193–9. doi: 10.1165/rcmb.2003-0107OC. [DOI] [PubMed] [Google Scholar]
- 79.Eggleton P, Reid KB. Lung surfactant proteins involved in innate immunity. Curr Opin Immunol. 1999;11:28–33. doi: 10.1016/s0952-7915(99)80006-5. [DOI] [PubMed] [Google Scholar]
- 80.Wu H, Kuzmenko A, Wan S, Schaffer L, Weiss A, Fisher JH, Kim KS, McCormack FX. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. 2003;111:1589–602. doi: 10.1172/JCI16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schaeffer LM, McCormack FX, Wu H, Weiss AA. Bordetella pertussis lipopolysaccharide resists the bactericidal effects of pulmonary surfactant protein A. J Immunol. 2004;173:1959–65. doi: 10.4049/jimmunol.173.3.1959. [DOI] [PubMed] [Google Scholar]
- 82.Floros J, Phelps DS. Pulmonary surfactant protein A; structure, expression, and its role in innate host defense. In: Lekka GNaM., editor. Surfactant-Update of Intensive Care Medicine. University of Ioannina; Ioannina, Greece: 2001. [Google Scholar]
- 83.Hickman-Davis JM, O’Reilly P, Davis IC, Peti-Peterdi J, Davis G, Young KR, Devlin RB, Matalon S. Killing of Klebsiella pneumoniae by human alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2002;282:L944–56. doi: 10.1152/ajplung.00216.2001. [DOI] [PubMed] [Google Scholar]
- 84.Mikerov AN, Haque R, Gan X, Guo X, Phelps DS, Floros J. Ablation of SP-A has a negative impact on the susceptibility of mice to Klebsiella pneumoniae infection after ozone exposure: sex differences. Respir Res. 2008;9:77. doi: 10.1186/1465-9921-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mikerov AN, Gan X, Umstead TM, Miller L, Chinchilli VM, Phelps DS, Floros J. Sex differences in the impact of ozone on survival and alveolar macrophage function of mice after Klebsiella pneumoniae infection. Respir Res. 2008;9:24. doi: 10.1186/1465-9921-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Iwaarden JF, Pikaar JC, Storm J, Brouwer E, Verhoef J, Oosting RS, van Golde LM, van Strijp JA. Binding of surfactant protein A to the lipid A moiety of bacterial lipopolysaccharides. Biochem J. 1994;303:407–11. doi: 10.1042/bj3030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Reid K. The immunoregulatory roles of lung surfactant collectins SP-A, and SP-D, in allergen-induced airway inflammation. Immunobiology. 2007;212:417–25. doi: 10.1016/j.imbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 88.Pastva AM, Wright JR, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4:252–7. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mulugeta S, Beers MF. Surfactant protein C: its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006;8:2317–23. doi: 10.1016/j.micinf.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Hawgood S. Surfactant protein B: structure and function. Biol Neonate. 2004;85:285–9. doi: 10.1159/000078169. [DOI] [PubMed] [Google Scholar]
- 91.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest. 2004;125:203–11. doi: 10.1378/chest.125.1.203. [DOI] [PubMed] [Google Scholar]
- 92.Seifart C, Plagens A, Brödje D, Müller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers. 2002;18:129–36. doi: 10.1155/2002/194075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ewis AA, Kondo K, Dang F, Nakahori Y, Shinohara Y, Ishikawa M, Baba Y. Surfactant protein B gene variations and susceptibility to lung cancer in chromate workers. Am J Ind Med. 2006;49:367–73. doi: 10.1002/ajim.20283. [DOI] [PubMed] [Google Scholar]
- 94.Lin Z, deMello DE, Wallot M, Floros J. An SP-B gene mutation responsible for SP-B deficiency in fatal congenital alveolar proteinosis: evidence for a mutation hotspot in exon 4. Mol Genet Metab. 1998;64:25–35. doi: 10.1006/mgme.1998.2702. [DOI] [PubMed] [Google Scholar]
- 95.Hoover RR, Floros J. Organization of the human SP-A and SP-D loci at 10q22–q23. Physical and radiation hybrid mapping reveal gene order and orientation. Am J Respir Cell Mol Biol. 1998;18:353–62. doi: 10.1165/ajrcmb.18.3.3035. [DOI] [PubMed] [Google Scholar]
- 96.Bruns G, Stroh H, Veldman G, Latt S, Floros J. The 35 kd pulmonary surfactant-associated protein is encoded on chromosome 10. Hum Genet. 1987;76:58–62. doi: 10.1007/BF00283051. [DOI] [PubMed] [Google Scholar]
- 97.Kölble K, Lu J, Mole SE, Kaluz S, Reid KB. Assignment of the human pulmonary surfactant protein D gene (SFTP4) to 10q22–q23 close to the surfactant protein A gene cluster. Genomics. 1993;17:294–8. doi: 10.1006/geno.1993.1324. [DOI] [PubMed] [Google Scholar]
- 98.Floros J, Wang G. A point of view: quantitative and qualitative imbalance in disease pathogenesis; pulmonary surfactant protein A genetic variants as a model. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:295–303. doi: 10.1016/s1095-6433(01)00325-7. [DOI] [PubMed] [Google Scholar]
- 99.Floros J, Hoover R. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. 1998;1408:312–22. doi: 10.1016/s0925-4439(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 100.Floros J. Human surfactant protein A (SP-A) variants: why so many, why such a complexity? Swiss Med Wkly. 2001;131:87–90. doi: 10.4414/smw.2001.06136. [DOI] [PubMed] [Google Scholar]
- 101.Crouch E, Rust K, Veile R, Donis-Keller H, Grosso L. Genomic organization of human surfactant protein D (SP-D). SP-D is encoded on chromosome 10q22.2–23.1. J Biol Chem. 1993;268:2976–83. [PubMed] [Google Scholar]
- 102.Floros J, Wang G, Mikerov A. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function. Crit Rev Eukaryot Gene Expr. 2009;19:125–37. doi: 10.1615/critreveukargeneexpr.v19.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, Floros J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers. 1999;15:269–81. doi: 10.1155/1999/961430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu W, Bentley CM, Floros J. Study of human SP-A, SP-B and SP-D loci: allele frequencies, linkage disequilibrium and heterozygosity in different races and ethnic groups. BMC Genet. 2003;4:13. doi: 10.1186/1471-2156-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pantelidis P, Lagan AL, Davies JC, Welsh KI, du Bois RM. A single round PCR method for genotyping human surfactant protein (SP)-A1, SP-A2 and SP-D gene alleles. Tissue Antigens. 2003;61:317–21. doi: 10.1034/j.1399-0039.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- 106.Sorensen GL, Husby S, Holmskov U. Surfactant protein A and surfactant protein D variation in pulmonary disease. Immunobiology. 2007;212:381–416. doi: 10.1016/j.imbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 107.Floros J, DiAngelo S, Koptides M, Karinch A, Rogan P, Nielsen H, Spragg R, Watterberg K, Deiter G. Human SPA locus: allele frequencies and linkage disequilibrium between the two surfactant protein A genes. Am J Respir Cell Mol Biol. 1996;15:489–98. doi: 10.1165/ajrcmb.15.4.8879183. [DOI] [PubMed] [Google Scholar]
- 108.Karinch A, Floros J. 5′ splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol. 1995;12:77–88. doi: 10.1165/ajrcmb.12.1.7811473. [DOI] [PubMed] [Google Scholar]
- 109.Tagaram H, Wang G, Umstead T, Mikerov A, Thomas N, Graff G, Hess J, Thomassen M, Kavuru M, Phelps D, Floros J. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1052–63. doi: 10.1152/ajplung.00249.2006. [DOI] [PubMed] [Google Scholar]
- 110.Wang G, Guo X, Floros J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol. 2003;284:L738–48. doi: 10.1152/ajplung.00375.2002. [DOI] [PubMed] [Google Scholar]
- 111.Krizkova L, Sakthivel R, Olowe SA, Rogan PK, Floros J. Human SP-A: genotype and single-strand conformation polymorphism analysis. Am J Physiol. 1994;266:L519–27. doi: 10.1152/ajplung.1994.266.5.L519. [DOI] [PubMed] [Google Scholar]
- 112.Wang G, Guo X, Silveyra P, Kimball S, Floros J. Cap-independent translation of human SP-A 5′-UTR variants: a double-loop structure and cis-element contribution. Am J Physiol Lung Cell Mol Physiol. 2009;296:L635–47. doi: 10.1152/ajplung.90508.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005;289:L497–508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 114.Silveyra P, Wang G, Floros J. Human SP-A1 (SFTPA1) variant-specific 3′ UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene. Am J Physiol Lung Cell Mol Physiol. 2010;299:L523–34. doi: 10.1152/ajplung.00113.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foreman MG, Kong X, Demeo DL, Pillai SG, Hersh CP, Bakke P, Gulsvik A, Lomas DA, Litonjua AA, Shapiro SD, Tal-Singer R, Silverman EK. Polymorphisms in Surfactant Protein D are Associated with COPD. Am J Respir Cell Mol Biol. 2010 doi: 10.1165/rcmb.2009-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hilgendorff A, Heidinger K, Bohnert A, Kleinsteiber A, König IR, Ziegler A, Lindner U, Frey G, Merz C, Lettgen B, Chakraborty T, Gortner L, Bein G. Association of polymorphisms in the human surfactant protein-D (SFTPD) gene and postnatal pulmonary adaptation in the preterm infant. Acta Paediatr. 2009;98:112–7. doi: 10.1111/j.1651-2227.2008.01014.x. [DOI] [PubMed] [Google Scholar]
- 117.Floros J, Lin H, García A, Salazar M, Guo X, DiAngelo S, Montaño M, Luo J, Pardo A, Selman M. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a Mexican population. J Infect Dis. 2000;182:1473–8. doi: 10.1086/315866. [DOI] [PubMed] [Google Scholar]
- 118.Heidinger K, König IR, Bohnert A, Kleinsteiber A, Hilgendorff A, Gortner L, Ziegler A, Chakraborty T, Bein G. Polymorphisms in the human surfactant protein-D (SFTPD) gene: strong evidence that serum levels of surfactant protein-D (SP-D) are genetically influenced. Immunogenetics. 2005;57:1–7. doi: 10.1007/s00251-005-0775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Husby S, Herskind AM, Jensenius JC, Holmskov U. Heritability estimates for the constitutional levels of the collectins mannan-binding lectin and lung surfactant protein D. A study of unselected like-sexed mono- and dizygotic twins at the age of 6–9 years. Immunology. 2002;106:389–94. doi: 10.1046/j.1365-2567.2002.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sørensen GL, Hjelmborg JB, Kyvik KO, Fenger M, Høj A, Bendixen C, Sørensen TI, Holmskov U. Genetic and environmental influences of surfactant protein D serum levels. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1010–7. doi: 10.1152/ajplung.00487.2005. [DOI] [PubMed] [Google Scholar]
- 121.Gaynor CD, McCormack FX, Voelker DR, McGowan SE, Schlesinger LS. Pulmonary surfactant protein A mediates enhanced phagocytosis of Mycobacterium tuberculosis by a direct interaction with human macrophages. J Immunol. 1995;155:5343–51. [PubMed] [Google Scholar]
- 122.Ferguson JS, Voelker DR, McCormack FX, Schlesinger LS. Surfactant protein D binds to Mycobacterium tuberculosis bacilli and lipoarabinomannan via carbohydrate-lectin interactions resulting in reduced phagocytosis of the bacteria by macrophages. J Immunol. 1999;163:312–21. [PubMed] [Google Scholar]
- 123.Malik S, Greenwood CM, Eguale T, Kifle A, Beyene J, Habte A, Tadesse A, Gebrexabher H, Britton S, Schurr E. Variants of the SFTPA1 and SFTPA2 genes and susceptibility to tuberculosis in Ethiopia. Hum Genet. 2006;118:752–9. doi: 10.1007/s00439-005-0092-y. [DOI] [PubMed] [Google Scholar]
- 124.Vaid M, Kaur S, Taruna M, Singh H, Gupta V, Murthy K, Sarma P. Association of SP-D, MBL and I-NOS genetic variants with pulmonary tuberculosis. Indian Journal of Human Genetics. 2006;12:105–110. [Google Scholar]
- 125.Madan T, Saxena S, Murthy KJ, Muralidhar K, Sarma PU. Association of polymorphisms in the collagen region of human SP-A1 and SP-A2 genes with pulmonary tuberculosis in Indian population. Clin Chem Lab Med. 2002;40:1002–8. doi: 10.1515/CCLM.2002.174. [DOI] [PubMed] [Google Scholar]
- 126.Krueger M, Puthothu B, Gropp E, Heinze J, Braun S, Heinzmann A. Amino acid variants in Surfactant protein D are not associated with bronchial asthma. Pediatr Allergy Immunol. 2006;17:77–81. doi: 10.1111/j.1399-3038.2005.00353.x. [DOI] [PubMed] [Google Scholar]
- 127.Deng YQ, Tao ZZ, Kong YG, Xiao BK, Chen SM, Xu Y, Wang Y, He Q. Association between single nucleotide polymorphisms of surfactant protein D and allergic rhinitis in Chinese patients. Tissue Antigens. 2009;73:546–52. doi: 10.1111/j.1399-0039.2009.01232.x. [DOI] [PubMed] [Google Scholar]
- 128.Deng Y, Chen S, Chen J, Tao Z, Kong Y, Xu Y, Xiao B, He Q. Relationship between surfactant protein A polymorphisms and allergic rhinitis in a Chinese Han population. Mol Biol Rep. 2010 doi: 10.1007/s11033-010-0254-4. [DOI] [PubMed] [Google Scholar]
- 129.Pettigrew MM, Gent JF, Zhu Y, Triche EW, Belanger KD, Holford TR, Bracken MB, Leaderer BP. Respiratory symptoms among infants at risk for asthma: association with surfactant protein A haplotypes. BMC Med Genet. 2007;8:15. doi: 10.1186/1471-2350-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brandt EB, Mingler MK, Stevenson MD, Wang N, Khurana Hershey GK, Whitsett JA, Rothenberg ME. Surfactant protein D alters allergic lung responses in mice and human subjects. J Allergy Clin Immunol. 2008;121:1140–1147.e2. doi: 10.1016/j.jaci.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leth-Larsen R, Garred P, Jensenius H, Meschi J, Hartshorn K, Madsen J, Tornoe I, Madsen HO, Sørensen G, Crouch E, Holmskov U. A common polymorphism in the SFTPD gene influences assembly, function, and concentration of surfactant protein D. J Immunol. 2005;174:1532–8. doi: 10.4049/jimmunol.174.3.1532. [DOI] [PubMed] [Google Scholar]
- 132.Saxena S, Madan T, Shah A, Muralidhar K, Sarma PU. Association of polymorphisms in the collagen region of SP-A2 with increased levels of total IgE antibodies and eosinophilia in patients with allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol. 2003;111:1001–7. doi: 10.1067/mai.2003.1395. [DOI] [PubMed] [Google Scholar]
- 133.Madan T, Kishore U, Shah A, Eggleton P, Strong P, Wang JY, Aggrawal SS, Sarma PU, Reid KB. Lung surfactant proteins A and D can inhibit specific IgE binding to the allergens of Aspergillus fumigatus and block allergen-induced histamine release from human basophils. Clin Exp Immunol. 1997;110:241–9. doi: 10.1111/j.1365-2249.1997.tb08323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saxena S, Kumar R, Madan T, Gupta V, Muralidhar K, Sarma PU. Association of polymorphisms in pulmonary surfactant protein A1 and A2 genes with high-altitude pulmonary edema. Chest. 2005;128:1611–9. doi: 10.1378/chest.128.3.1611. [DOI] [PubMed] [Google Scholar]
- 135.Guo CJ, Atochina-Vasserman EN, Abramova E, Foley JP, Zaman A, Crouch E, Beers MF, Savani RC, Gow AJ. S-nitrosylation of surfactant protein-D controls inflammatory function. PLoS Biol. 2008;6:e266. doi: 10.1371/journal.pbio.0060266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsuji T, Aoshiba K, Nagai A. Alveolar cell senescence in patients with pulmonary emphysema. Am J Respir Crit Care Med. 2006;174:886–93. doi: 10.1164/rccm.200509-1374OC. [DOI] [PubMed] [Google Scholar]
- 137.Park JW, Ryter SW, Choi AM. Functional significance of apoptosis in chronic obstructive pulmonary disease. COPD. 2007;4:347–53. doi: 10.1080/15412550701603775. [DOI] [PubMed] [Google Scholar]
- 138.Hickman-Davis J, Gibbs-Erwin J, Lindsey JR, Matalon S. Surfactant protein A mediates mycoplasmacidal activity of alveolar macrophages by production of peroxynitrite. Proc Natl Acad Sci U S A. 1999;96:4953–8. doi: 10.1073/pnas.96.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi EH, Ehrmantraut M, Foster CB, Moss J, Chanock SJ. Association of common haplotypes of surfactant protein A1 and A2 (SFTPA1 and SFTPA2) genes with severity of lung disease in cystic fibrosis. Pediatr Pulmonol. 2006;41:255–62. doi: 10.1002/ppul.20361. [DOI] [PubMed] [Google Scholar]
- 140.Noah TL, Murphy PC, Alink JJ, Leigh MW, Hull WM, Stahlman MT, Whitsett JA. Bronchoalveolar lavage fluid surfactant protein-A and surfactant protein-D are inversely related to inflammation in early cystic fibrosis. Am J Respir Crit Care Med. 2003;168:685–91. doi: 10.1164/rccm.200301-005OC. [DOI] [PubMed] [Google Scholar]
- 141.Postle AD, Mander A, Reid KB, Wang JY, Wright SM, Moustaki M, Warner JO. Deficient hydrophilic lung surfactant proteins A and D with normal surfactant phospholipid molecular species in cystic fibrosis. Am J Respir Cell Mol Biol. 1999;20:90–8. doi: 10.1165/ajrcmb.20.1.3253. [DOI] [PubMed] [Google Scholar]
- 142.Griese M, Birrer P, Demirsoy A. Pulmonary surfactant in cystic fibrosis. Eur Respir J. 1997;10:1983–8. doi: 10.1183/09031936.97.10091983. [DOI] [PubMed] [Google Scholar]
- 143.Selman M, Lin H, Montaño M, Jenkins A, Estrada A, Lin Z, Wang G, DiAngelo S, Guo X, Umstead T, Lang C, Pardo A, Phelps D, Floros J. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum Genet. 2003;113:542–50. doi: 10.1007/s00439-003-1015-4. [DOI] [PubMed] [Google Scholar]
- 144.Hunninghake GW, Zimmerman MB, Schwartz DA, King TE, Lynch J, Hegele R, Waldron J, Colby T, Müller N, Lynch D, Galvin J, Gross B, Hogg J, Toews G, Helmers R, Cooper JA, Baughman R, Strange C, Millard M. Utility of a lung biopsy for the diagnosis of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2001;164:193–6. doi: 10.1164/ajrccm.164.2.2101090. [DOI] [PubMed] [Google Scholar]
- 145.Seifart C, Lin HM, Seifart U, Plagens A, DiAngelo S, von Wichert P, Floros J. Rare SP-A alleles and the SP-A1-6A(4) allele associate with risk for lung carcinoma. Clin Genet. 2005;68:128–36. doi: 10.1111/j.1399-0004.2005.00470.x. [DOI] [PubMed] [Google Scholar]
- 146.Koptides M, Umstead TM, Floros J, Phelps DS. Surfactant protein A activates NF-kappa B in the THP-1 monocytic cell line. Am J Physiol. 1997;273:L382–8. doi: 10.1152/ajplung.1997.273.2.L382. [DOI] [PubMed] [Google Scholar]
- 147.Antal JM, Divis LT, Erzurum SC, Wiedemann HP, Thomassen MJ. Surfactant suppresses NF-kappa B activation in human monocytic cells. Am J Respir Cell Mol Biol. 1996;14:374–9. doi: 10.1165/ajrcmb.14.4.8600942. [DOI] [PubMed] [Google Scholar]
- 148.Wang G, Phelps D, Umstead T, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol. 2000;278:L946–54. doi: 10.1152/ajplung.2000.278.5.L946. [DOI] [PubMed] [Google Scholar]
- 149.Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. 2004;286:L546–53. doi: 10.1152/ajplung.00267.2003. [DOI] [PubMed] [Google Scholar]
- 150.Lahti M, Lofgren J, Marttila R, Renko M, Klaavuniemi T, Haataja R, Ramet M, Hallman M. Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr Res. 2002;51:696–9. doi: 10.1203/00006450-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 151.Löfgren J, Rämet M, Renko M, Marttila R, Hallman M. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J Infect Dis. 2002;185:283–9. doi: 10.1086/338473. [DOI] [PubMed] [Google Scholar]
- 152.Thomas N, DiAngelo S, Hess J, Fan R, Ball M, Geskey J, Willson D, Floros J. Transmission of surfactant protein variants and haplotypes in children hospitalized with respiratory syncytial virus. Pediatr Res. 2009;66:70–3. doi: 10.1203/PDR.0b013e3181a1d768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.El Saleeby CM, Li R, Somes GW, Dahmer MK, Quasney MW, DeVincenzo JP. Surfactant protein A2 polymorphisms and disease severity in a respiratory syncytial virus-infected population. J Pediatr. 2010;156:409–14. doi: 10.1016/j.jpeds.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 154.Boyce TG, Mellen BG, Mitchel EF, Wright PF, Griffin MR. Rates of hospitalization for respiratory syncytial virus infection among children in medicaid. J Pediatr. 2000;137:865–70. doi: 10.1067/mpd.2000.110531. [DOI] [PubMed] [Google Scholar]
- 155.Glezen WP. The changing epidemiology of respiratory syncytial virus and influenza: impetus for new control measures. Pediatr Infect Dis J. 2004;23:S202–6. doi: 10.1097/01.inf.0000144662.86396.07. [DOI] [PubMed] [Google Scholar]
- 156.Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J Gen Virol. 2002;83:753–7. doi: 10.1099/0022-1317-83-4-753. [DOI] [PubMed] [Google Scholar]
- 157.Hickling TP, Malhotra R, Bright H, McDowell W, Blair ED, Sim RB. Lung surfactant protein A provides a route of entry for respiratory syncytial virus into host cells. Viral Immunol. 2000;13:125–35. doi: 10.1089/vim.2000.13.125. [DOI] [PubMed] [Google Scholar]
- 158.Hickling TP, Bright H, Wing K, Gower D, Martin SL, Sim RB, Malhotra R. A recombinant trimeric surfactant protein D carbohydrate recognition domain inhibits respiratory syncytial virus infection in vitro and in vivo. Eur J Immunol. 1999;29:3478–84. doi: 10.1002/(SICI)1521-4141(199911)29:11<3478::AID-IMMU3478>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 159.Jack DL, Cole J, Naylor SC, Borrow R, Kaczmarski EB, Klein NJ, Read RC. Genetic polymorphism of the binding domain of surfactant protein-A2 increases susceptibility to meningococcal disease. Clin Infect Dis. 2006;43:1426–33. doi: 10.1086/508775. [DOI] [PubMed] [Google Scholar]
- 160.Read RC, Fox A, Miller K, Gray T, Jones N, Borrows R, Jones DM, Finch RG. Experimental infection of human nasal mucosal explants with Neisseria meningitidis. J Med Microbiol. 1995;42:353–61. doi: 10.1099/00222615-42-5-353. [DOI] [PubMed] [Google Scholar]
- 161.Cartwright KA, Jones DM, Smith AJ, Stuart JM, Kaczmarski EB, Palmer SR. Influenza A and meningococcal disease. Lancet. 1991;338:554–7. doi: 10.1016/0140-6736(91)91112-8. [DOI] [PubMed] [Google Scholar]
- 162.Westendorp RG, Langermans JA, Huizinga TW, Elouali AH, Verweij CL, Boomsma DI, Vandenbroucke JP, Vandenbrouke JP. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349:170–3. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
- 163.Pettigrew MM, Gent JF, Zhu Y, Triche EW, Belanger KD, Holford TR, Bracken MB, Leaderer BP. Association of surfactant protein A polymorphisms with otitis media in infants at risk for asthma. BMC Med Genet. 2006;7:68. doi: 10.1186/1471-2350-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rämet M, Löfgren J, Alho OP, Hallman M. Surfactant protein-A gene locus associated with recurrent otitis media. J Pediatr. 2001;138:266–8. doi: 10.1067/mpd.2001.110133. [DOI] [PubMed] [Google Scholar]
- 165.Dudley S, Ashe K, Winther B, Hendley JO. Bacterial pathogens of otitis media and sinusitis: detection in the nasopharynx with selective agar media. J Lab Clin Med. 2001;138:338–42. doi: 10.1067/mlc.2001.119311. [DOI] [PubMed] [Google Scholar]
- 166.Andrade MA, Hoberman A, Glustein J, Paradise JL, Wald ER. Acute otitis media in children with bronchiolitis. Pediatrics. 1998;101:617–9. doi: 10.1542/peds.101.4.617. [DOI] [PubMed] [Google Scholar]
- 167.Lim DJ, Chun YM, Lee HY, Moon SK, Chang KH, Li JD, Andalibi A. Cell biology of tubotympanum in relation to pathogenesis of otitis media - a review. Vaccine. 2000;19:S17–25. doi: 10.1016/s0264-410x(00)00273-5. [DOI] [PubMed] [Google Scholar]
- 168.Tino MJ, Wright JR. Surfactant protein A stimulates phagocytosis of specific pulmonary pathogens by alveolar macrophages. Am J Physiol. 1996;270:L677–88. doi: 10.1152/ajplung.1996.270.4.L677. [DOI] [PubMed] [Google Scholar]
- 169.Rämet M, Haataja R, Marttila R, Hämäläinen A, Knip M, Hallman M. Human surfactant protein--A gene locus for genetic studies in the Finnish population. Dis Markers. 2000;16:119–24. doi: 10.1155/2000/914814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu J, Hu F, Liang W, Wang G, Singhal PC, Ding G. Polymorphisms in the surfactant protein a gene are associated with the susceptibility to recurrent urinary tract infection in chinese women. Tohoku J Exp Med. 2010;221:35–42. doi: 10.1620/tjem.221.35. [DOI] [PubMed] [Google Scholar]
- 171.Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 2010;6:e1001187. doi: 10.1371/journal.ppat.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hawn TR, Scholes D, Li SS, Wang H, Yang Y, Roberts PL, Stapleton AE, Janer M, Aderem A, Stamm WE, Zhao LP, Hooton TM. Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLoS One. 2009;4:e5990. doi: 10.1371/journal.pone.0005990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 174.Berg KK, Madsen HO, Garred P, Wiseth R, Gunnes S, Videm V. The additive contribution from inflammatory genetic markers on the severity of cardiovascular disease. Scand J Immunol. 2009;69:36–42. doi: 10.1111/j.1365-3083.2008.02187.x. [DOI] [PubMed] [Google Scholar]
- 175.Ohya M, Nishitani C, Sano H, Yamada C, Mitsuzawa H, Shimizu T, Saito T, Smith K, Crouch E, Kuroki Y. Human pulmonary surfactant protein D binds the extracellular domains of Toll-like receptors 2 and 4 through the carbohydrate recognition domain by a mechanism different from its binding to phosphatidylinositol and lipopolysaccharide. Biochemistry. 2006;45:8657–64. doi: 10.1021/bi060176z. [DOI] [PubMed] [Google Scholar]
- 176.deMello D, Phelps D, Patel G, Floros J, Lagunoff D. Expression of the 35kDa and low molecular weight surfactant-associated proteins in the lungs of infants dying with respiratory distress syndrome. Am J Pathol. 1989;134:1285–93. [PMC free article] [PubMed] [Google Scholar]
- 177.deMello D, Heyman S, Phelps D, Floros J. Immunogold localization of SP-A in lungs of infants dying from respiratory distress syndrome. Am J Pathol. 1993;142:1631–40. [PMC free article] [PubMed] [Google Scholar]
- 178.Pryhuber GS, Hull WM, Fink I, McMahan MJ, Whitsett JA. Ontogeny of surfactant proteins A and B in human amniotic fluid as indices of fetal lung maturity. Pediatr Res. 1991;30:597–605. doi: 10.1203/00006450-199112000-00023. [DOI] [PubMed] [Google Scholar]
- 179.Chi EY, Lagunoff D. Linear arrays of intramembranous particles in pulmonary tubular myelin. Proc Natl Acad Sci U S A. 1978;75:6225–9. doi: 10.1073/pnas.75.12.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.deMello DE, Chi EY, Doo E, Lagunoff D. Absence of tubular myelin in lungs of infants dying with hyaline membrane disease. Am J Pathol. 1987;127:131–9. [PMC free article] [PubMed] [Google Scholar]
- 181.Karinch AM, Floros J. Translation in vivo of 5′ untranslated-region splice variants of human surfactant protein-A. Biochem J. 1995;307:327–30. doi: 10.1042/bj3070327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Karinch A, Deiter G, Ballard P, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta. 1998;1398:192–202. doi: 10.1016/s0167-4781(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 183.Karinch AM, deMello DE, Floros J. Effect of genotype on the levels of surfactant protein A mRNA and on the SP-A2 splice variants in adult humans. Biochem J. 1997;321:39–47. doi: 10.1042/bj3210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Haataja R, Marttila R, Uimari P, Löfgren J, Rämet M, Hallman M. Respiratory distress syndrome: evaluation of genetic susceptibility and protection by transmission disequilibrium test. Hum Genet. 2001;109:351–5. doi: 10.1007/s004390100574. [DOI] [PubMed] [Google Scholar]
- 185.Rämet M, Haataja R, Marttila R, Floros J, Hallman M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am J Hum Genet. 2000;66:1569–79. doi: 10.1086/302906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Floros J, Fan R, Matthews A, DiAngelo S, Luo J, Nielsen H, Dunn M, Gewolb IH, Koppe J, van Sonderen L, Farri-Kostopoulos L, Tzaki M, Ramet M, Merrill J. Family-based transmission disequilibrium test (TDT) and case-control association studies reveal surfactant protein A (SP-A) susceptibility alleles for respiratory distress syndrome (RDS) and possible race differences. Clin Genet. 2001;60:178–87. doi: 10.1034/j.1399-0004.2001.600303.x. [DOI] [PubMed] [Google Scholar]
- 187.Marttila R, Haataja R, Ramet M, Pokela ML, Tammela O, Hallman M. Surfactant protein A gene locus and respiratory distress syndrome in Finnish premature twin pairs. Ann Med. 2003;35:344–52. doi: 10.1080/07853890310006389. [DOI] [PubMed] [Google Scholar]
- 188.Haataja R, Rämet M, Marttila R, Hallman M. Surfactant proteins A and B as interactive genetic determinants of neonatal respiratory distress syndrome. Hum Mol Genet. 2000;9:2751–60. doi: 10.1093/hmg/9.18.2751. [DOI] [PubMed] [Google Scholar]
- 189.Ramet M, Haataja R, Marttila R, Floros J, Hallman M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am J Hum Genet. 2000;66:1569–79. doi: 10.1086/302906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Marttila R, Haataja R, Guttentag S, Hallman M. Surfactant protein A and B genetic variants in respiratory distress syndrome in singletons and twins. Am J Respir Crit Care Med. 2003;168:1216–22. doi: 10.1164/rccm.200304-524OC. [DOI] [PubMed] [Google Scholar]
- 191.Zhai L, Wu HM, Wei KL, Zhao SM, Jiang H. Genetic polymorphism of surfactant protein A in neonatal respiratory distress syndrome. Zhongguo Dang Dai Er Ke Za Zhi. 2008;10:295–8. [PubMed] [Google Scholar]
- 192.Floros J, Fan R, Diangelo S, Guo X, Wert J, Luo J. Surfactant protein (SP) B associations and interactions with SP-A in white and black subjects with respiratory distress syndrome. Pediatr Int. 2001;43:567–76. doi: 10.1046/j.1442-200x.2001.01474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Floros J, Fan R. Surfactant protein A and B genetic variants and respiratory distress syndrome: allele interactions. Biol Neonate. 2001;80:22–5. doi: 10.1159/000047173. [DOI] [PubMed] [Google Scholar]
- 194.Hallman M, Haataja R, Marttila R. Surfactant proteins and genetic predisposition to respiratory distress syndrome. Semin Perinatol. 2002;26:450–60. doi: 10.1053/sper.2002.37314. [DOI] [PubMed] [Google Scholar]
- 195.Hallman M, Haataja R. Surfactant protein polymorphisms and neonatal lung disease. Semin Perinatol. 2006;30:350–61. doi: 10.1053/j.semperi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 196.Kala P, Ten Have T, Nielsen H, Dunn M, Floros J. Association of pulmonary surfactant protein A (SP-A) gene and respiratory distress syndrome: interaction with SP-B. Pediatr Res. 1998;43:169–77. doi: 10.1203/00006450-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 197.Thomas N, Fan R, Diangelo S, Hess J, Floros J. Haplotypes of the surfactant protein genes A and D as susceptibility factors for the development of respiratory distress syndrome. Acta Paediatr. 2007;96:985–9. doi: 10.1111/j.1651-2227.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 198.Pavlovic J, Papagaroufalis C, Xanthou M, Liu W, Fan R, Thomas N, Apostolidou I, Papathoma E, Megaloyianni E, DiAngelo S, Floros J. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis Markers. 2006;22:277–91. doi: 10.1155/2006/817805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Weber B, Borkhardt A, Stoll-Becker S, Reiss I, Gortner L. Polymorphisms of surfactant protein A genes and the risk of bronchopulmonary dysplasia in preterm infants. Turk J Pediatr. 2000;42:181–5. [PubMed] [Google Scholar]
- 200.Koopmans JG, van der Zee JS, Krop EJ, Lopuhaä CE, Jansen HM, Batenburg JJ. Serum surfactant protein D is elevated in allergic patients. Clin Exp Allergy. 2004;34:1827–33. doi: 10.1111/j.1365-2222.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- 201.Behera D, Balamugesh T, Venkateswarlu D, Gupta A, Majumdar S. Serum surfactant protein-A levels in chronic bronchitis and its relation to smoking. Indian J Chest Dis Allied Sci. 2005;47:13–7. [PubMed] [Google Scholar]
- 202.Krane M, Griese M. Surfactant protein D in serum from patients with allergic bronchopulmonary aspergillosis. Eur Respir J. 2003;22:592–5. doi: 10.1183/09031936.03.00060603. [DOI] [PubMed] [Google Scholar]
- 203.Doyle IR, Nicholas TE, Bersten AD. Serum surfactant protein-A levels in patients with acute cardiogenic pulmonary edema and adult respiratory distress syndrome. Am J Respir Crit Care Med. 1995;152:307–17. doi: 10.1164/ajrccm.152.1.7599839. [DOI] [PubMed] [Google Scholar]
- 204.Kobayashi H, Kanoh S, Motoyoshi K. Serum surfactant protein-A, but not surfactant protein-D or KL-6, can predict preclinical lung damage induced by smoking. Biomarkers. 2008;13:385–92. doi: 10.1080/13547500801903651. [DOI] [PubMed] [Google Scholar]
- 205.Lomas DA, Silverman EK, Edwards LD, Locantore NW, Miller BE, Horstman DH, Tal-Singer R E. o. C. L. t. I. P. S. E. s. investigators. Serum surfactant protein D is steroid sensitive and associated with exacerbations of COPD. Eur Respir J. 2009;34:95–102. doi: 10.1183/09031936.00156508. [DOI] [PubMed] [Google Scholar]
- 206.Janssen R, Grutters JC, Sato H, van Velzen-Blad H, Zanen P, Kohno N, Welsh KI, du Bois RM, van den Bosch JM. Analysis of KL-6 and SP-D as disease markers in bird fancier’s lung. Sarcoidosis Vasc Diffuse Lung Dis. 2005;22:51–7. [PubMed] [Google Scholar]
- 207.Kuroki Y, Tsutahara S, Shijubo N, Takahashi H, Shiratori M, Hattori A, Honda Y, Abe S, Akino T. Elevated levels of lung surfactant protein A in sera from patients with idiopathic pulmonary fibrosis and pulmonary alveolar proteinosis. Am Rev Respir Dis. 1993;147:723–9. doi: 10.1164/ajrccm/147.3.723. [DOI] [PubMed] [Google Scholar]
- 208.Takahashi H, Honda Y, Kuroki Y, Imai K, Abe S. Pulmonary surfactant protein A: a serum marker of pulmonary fibrosis in patients with collagen vascular diseases. Clin Chim Acta. 1995;239:213–5. doi: 10.1016/0009-8981(95)06118-w. [DOI] [PubMed] [Google Scholar]
- 209.Honda Y, Kuroki Y, Shijubo N, Fujishima T, Takahashi H, Hosoda K, Akino T, Abe S. Aberrant appearance of lung surfactant protein A in sera of patients with idiopathic pulmonary fibrosis and its clinical significance. Respiration. 1995;62:64–9. doi: 10.1159/000196393. [DOI] [PubMed] [Google Scholar]
- 210.Honda Y, Kuroki Y, Matsuura E, Nagae H, Takahashi H, Akino T, Abe S. Pulmonary surfactant protein D in sera and bronchoalveolar lavage fluids. Am J Respir Crit Care Med. 1995;152:1860–6. doi: 10.1164/ajrccm.152.6.8520747. [DOI] [PubMed] [Google Scholar]
- 211.Greene KE, King TE, Kuroki Y, Bucher-Bartelson B, Hunninghake GW, Newman LS, Nagae H, Mason RJ. Serum surfactant proteins-A and -D as biomarkers in idiopathic pulmonary fibrosis. Eur Respir J. 2002;19:439–46. doi: 10.1183/09031936.02.00081102. [DOI] [PubMed] [Google Scholar]
- 212.Kinder BW, Brown KK, McCormack FX, Ix JH, Kervitsky A, Schwarz MI, King TE. Serum surfactant protein-A is a strong predictor of early mortality in idiopathic pulmonary fibrosis. Chest. 2009;135:1557–63. doi: 10.1378/chest.08-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ohnishi H, Yokoyama A, Kondo K, Hamada H, Abe M, Nishimura K, Hiwada K, Kohno N. Comparative study of KL-6, surfactant protein-A, surfactant protein-D, and monocyte chemoattractant protein-1 as serum markers for interstitial lung diseases. Am J Respir Crit Care Med. 2002;165:378–81. doi: 10.1164/ajrccm.165.3.2107134. [DOI] [PubMed] [Google Scholar]
- 214.Barlo NP, van Moorsel CH, Ruven HJ, Zanen P, van den Bosch JM, Grutters JC. Surfactant protein-D predicts survival in patients with idiopathic pulmonary fibrosis. Sarcoidosis Vasc Diffuse Lung Dis. 2009;26:155–61. [PubMed] [Google Scholar]
- 215.Arai Y, Obinata K, Sato Y, Hisata K, Tadokoro R, Tawa T, Kinoshita K. Clinical significance of the serum surfactant protein D and KL-6 levels in patients with measles complicated by interstitial pneumonia. Eur J Pediatr. 2001;160:425–9. doi: 10.1007/s004310100763. [DOI] [PubMed] [Google Scholar]
- 216.Leth-Larsen R, Nordenbaek C, Tornoe I, Moeller V, Schlosser A, Koch C, Teisner B, Junker P, Holmskov U. Surfactant protein D (SP-D) serum levels in patients with community-acquired pneumonia small star, filled. Clin Immunol. 2003;108:29–37. doi: 10.1016/s1521-6616(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 217.Daimon T, Tajima S, Oshikawa K, Bando M, Ohno S, Sugiyama Y. KL-6 and surfactant proteins A and D in serum and bronchoalveolar lavage fluid in patients with acute eosinophilic pneumonia. Intern Med. 2005;44:811–7. doi: 10.2169/internalmedicine.44.811. [DOI] [PubMed] [Google Scholar]
- 218.Christensen AF, Sørensen GL, Hørslev-Petersen K, Holmskov U, Lindegaard HM, Junker K, Hetland ML, Stengaard-Pedersen K, Jacobsen S, Lottenburger T, Ellingsen T, Andersen LS, Hansen I, Skjødt H, Pedersen JK, Lauridsen UB, Svendsen A, Tarp U, Pødenphant J, Vestergaard A, Jurik AG, Østergaard M, Junker P. Circulating surfactant protein -D is low and correlates negatively with systemic inflammation in early, untreated rheumatoid arthritis. Arthritis Res Ther. 2010;12:R39. doi: 10.1186/ar2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hoegh SV, Lindegaard HM, Sorensen GL, Høj A, Bendixen C, Junker P, Holmskov U. Circulating surfactant protein D is decreased in early rheumatoid arthritis: a 1-year prospective study. Scand J Immunol. 2008;67:71–6. doi: 10.1111/j.1365-3083.2007.02039.x. [DOI] [PubMed] [Google Scholar]
- 220.Greene KE, Ye S, Mason RJ, Parsons PE. Serum surfactant protein-A levels predict development of ARDS in at-risk patients. Chest. 1999;116:90S–91S. [PubMed] [Google Scholar]
- 221.Endo S, Sato N, Nakae H, Yamada Y, Makabe H, Abe H, Imai S, Wakabayashi G, Inada K, Sato S. Surfactant protein A and D (SP-A, AP-D) levels in patients with septic ARDS. Res Commun Mol Pathol Pharmacol. 2002;111:245–51. [PubMed] [Google Scholar]
- 222.Cheng IW, Ware LB, Greene KE, Nuckton TJ, Eisner MD, Matthay MA. Prognostic value of surfactant proteins A and D in patients with acute lung injury. Crit Care Med. 2003;31:20–7. doi: 10.1097/00003246-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 223.Kaneko K, Shimizu H, Arakawa H, Ogawa Y. Pulmonary surfactant protein A in sera for assessing neonatal lung maturation. Early Hum Dev. 2001;62:11–21. doi: 10.1016/s0378-3782(00)00133-x. [DOI] [PubMed] [Google Scholar]
- 224.Eisner MD, Parsons P, Matthay MA, Ware L, Greene K, Network ARDS. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax. 2003;58:983–8. doi: 10.1136/thorax.58.11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Janssen R, Sato H, Grutters JC, Bernard A, van Velzen-Blad H, du Bois RM, van den Bosch JM. Study of Clara cell 16, KL-6, and surfactant protein-D in serum as disease markers in pulmonary sarcoidosis. Chest. 2003;124:2119–25. doi: 10.1378/chest.124.6.2119. [DOI] [PubMed] [Google Scholar]
- 226.van de Graaf EA, Jansen HM, Lutter R, Alberts C, Kobesen J, de Vries IJ, Out TA. Surfactant protein A in bronchoalveolar lavage fluid. J Lab Clin Med. 1992;120:252–63. [PubMed] [Google Scholar]
- 227.Cheng G, Ueda T, Numao T, Kuroki Y, Nakajima H, Fukushima Y, Motojima S, Fukuda T. Increased levels of surfactant protein A and D in bronchoalveolar lavage fluids in patients with bronchial asthma. Eur Respir J. 2000;16:831–5. doi: 10.1183/09031936.00.16583100. [DOI] [PubMed] [Google Scholar]
- 228.Liu DY, Wu J, Zhang XY, Feng ZC. Expression of IL-8, SP-A and TGF-beta1 in bronchoalveolar lavage fluid of neonates with bronchopulmonary dysplasia. Zhongguo Dang Dai Er Ke Za Zhi. 2010;12:444–6. [PubMed] [Google Scholar]
- 229.Hull J, South M, Phelan P, Grimwood K. Surfactant composition in infants and young children with cystic fibrosis. Am J Respir Crit Care Med. 1997;156:161–5. doi: 10.1164/ajrccm.156.1.9609090. [DOI] [PubMed] [Google Scholar]
- 230.Ohlmeier S, Vuolanto M, Toljamo T, Vuopala K, Salmenkivi K, Myllarniemi M, Kinnula VL. Proteomics of Human Lung Tissue Identifies Surfactant Protein A as a Marker of Chronic Obstructive Pulmonary Disease. J Proteome Res. 2008;7:5125–32. doi: 10.1021/pr800423x. [DOI] [PubMed] [Google Scholar]
- 231.Vlachaki EM, Koutsopoulos AV, Tzanakis N, Neofytou E, Siganaki M, Drositis I, Moniakis A, Schiza S, Siafakas NM, Tzortzaki EG. Altered surfactant protein-A expression in type II pneumocytes in COPD. Chest. 2010;137:37–45. doi: 10.1378/chest.09-1029. [DOI] [PubMed] [Google Scholar]
- 232.Moré JM, Voelker DR, Silveira LJ, Edwards MG, Chan ED, Bowler RP. Smoking reduces surfactant protein D and phospholipids in patients with and without chronic obstructive pulmonary disease. BMC Pulm Med. 2010;10:53. doi: 10.1186/1471-2466-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hamm H, Lührs J, Guzman y Rotaeche J, Costabel U, Fabel H, Bartsch W. Elevated surfactant protein A in bronchoalveolar lavage fluids from sarcoidosis and hypersensitivity pneumonitis patients. Chest. 1994;106:1766–70. doi: 10.1378/chest.106.6.1766. [DOI] [PubMed] [Google Scholar]
- 234.Cormier Y, Israël-Assayag E, Desmeules M, Lesur O. Effect of contact avoidance or treatment with oral prednisolone on bronchoalveolar lavage surfactant protein A levels in subjects with farmer’s lung. Thorax. 1996;51:1210–5. doi: 10.1136/thx.51.12.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.McCormack FX, King TE, Jr, Bucher BL, Nielsen L, Mason RJ. Surfactant protein A predicts survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1995;152:751–9. doi: 10.1164/ajrccm.152.2.7633738. [DOI] [PubMed] [Google Scholar]
- 236.McCormack FX, King TE, Voelker DR, Robinson PC, Mason RJ. Idiopathic pulmonary fibrosis. Abnormalities in the bronchoalveolar lavage content of surfactant protein A. Am Rev Respir Dis. 1991;144:160–6. doi: 10.1164/ajrccm/144.1.160. [DOI] [PubMed] [Google Scholar]
- 237.Pison U, Obertacke U, Seeger W, Hawgood S. Surfactant protein A (SP-A) is decreased in acute parenchymal lung injury associated with polytrauma. Eur J Clin Invest. 1992;22:712–8. doi: 10.1111/j.1365-2362.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 238.Honda Y, Takahashi H, Shijubo N, Kuroki Y, Akino T. Surfactant protein-A concentration in bronchoalveolar lavage fluids of patients with pulmonary alveolar proteinosis. Chest. 1993;103:496–9. doi: 10.1378/chest.103.2.496. [DOI] [PubMed] [Google Scholar]
- 239.Masuda T, Shimura S, Sasaki H, Takishima T. Surfactant apoprotein-A concentration in sputum for diagnosis of pulmonary alveolar proteinosis. Lancet. 1991;337:580–2. doi: 10.1016/0140-6736(91)91640-g. [DOI] [PubMed] [Google Scholar]
- 240.Brasch F, Birzele J, Ochs M, Guttentag S, Schoch O, Boehler A, Beers M, Müller K, Hawgood S, Johnen G. Surfactant proteins in pulmonary alveolar proteinosis in adults. Eur Respir J. 2004;24:426–35. doi: 10.1183/09031936.04.00076403. [DOI] [PubMed] [Google Scholar]
- 241.Doyle IR, Davidson KG, Barr HA, Nicholas TE, Payne K, Pfitzner J. Quantity and structure of surfactant proteins vary among patients with alveolar proteinosis. Am J Respir Crit Care Med. 1998;157:658–64. doi: 10.1164/ajrccm.157.2.9701090. [DOI] [PubMed] [Google Scholar]
- 242.Günther A, Siebert C, Schmidt R, Ziegler S, Grimminger F, Yabut M, Temmesfeld B, Walmrath D, Morr H, Seeger W. Surfactant alterations in severe pneumonia, acute respiratory distress syndrome, and cardiogenic lung edema. Am J Respir Crit Care Med. 1996;153:176–84. doi: 10.1164/ajrccm.153.1.8542113. [DOI] [PubMed] [Google Scholar]
- 243.Baughman RP, Sternberg RI, Hull W, Buchsbaum JA, Whitsett J. Decreased surfactant protein A in patients with bacterial pneumonia. Am Rev Respir Dis. 1993;147:653–7. doi: 10.1164/ajrccm/147.3.653. [DOI] [PubMed] [Google Scholar]
- 244.Sternberg RI, Whitsett JA, Hull WM, Baughman RP. Pneumocystis carinii alters surfactant protein A concentrations in bronchoalveolar lavage fluid. J Lab Clin Med. 1995;125:462–9. [PubMed] [Google Scholar]
- 245.Phelps DS, Rose RM. Increased recovery of surfactant protein A in AIDS-related pneumonia. Am Rev Respir Dis. 1991;143:1072–5. doi: 10.1164/ajrccm/143.5_Pt_1.1072. [DOI] [PubMed] [Google Scholar]
- 246.Gregory TJ, Longmore WJ, Moxley MA, Whitsett JA, Reed CR, Fowler AA, Hudson LD, Maunder RJ, Crim C, Hyers TM. Surfactant chemical composition and biophysical activity in acute respiratory distress syndrome. J Clin Invest. 1991;88:1976–81. doi: 10.1172/JCI115523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gerdes JS, Abbasi S, Karp K, Hull W, Whitsett JA. Surfactant protein-A in bronchoalveolar lavage fluid from neonates with RDS on conventional and high-frequency oscillatory ventilation. Pediatr Pulmonol. 1990;9:166–9. doi: 10.1002/ppul.1950090308. [DOI] [PubMed] [Google Scholar]
- 248.Eguchi H, Koyama N, Tanaka T, Kamiya K, Ogawa Y. Surfactant apoprotein A (SP-A) in tracheal aspirates of newborn infants with RDS. Acta Paediatr Jpn. 1991;33:649–54. doi: 10.1111/j.1442-200x.1991.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 249.Wang JY, Yeh TF, Lin YC, Miyamura K, Holmskov U, Reid KB. Measurement of pulmonary status and surfactant protein levels during dexamethasone treatment of neonatal respiratory distress syndrome. Thorax. 1996;51:907–13. doi: 10.1136/thx.51.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Beresford MW, Shaw NJ. Bronchoalveolar lavage surfactant protein a, B, and d concentrations in preterm infants ventilated for respiratory distress syndrome receiving natural and synthetic surfactants. Pediatr Res. 2003;53:663–70. doi: 10.1203/01.PDR.0000054653.89527.F8. [DOI] [PubMed] [Google Scholar]
- 251.Kerr MH, Paton JY. Surfactant protein levels in severe respiratory syncytial virus infection. Am J Respir Crit Care Med. 1999;159:1115–8. doi: 10.1164/ajrccm.159.4.9709065. [DOI] [PubMed] [Google Scholar]
- 252.Davies J, Turner M, Klein N. The role of the collectin system in pulmonary defence. Paediatr Respir Rev. 2001;2:70–5. doi: 10.1053/prrv.2000.0104. [DOI] [PubMed] [Google Scholar]
- 253.Vaandrager AB, van Golde LM. Lung surfactant proteins A and D in innate immune defense. Biol Neonate. 2000;77:9–13. doi: 10.1159/000047051. [DOI] [PubMed] [Google Scholar]
- 254.Christensen AF, Hoegh SV, Lottenburger T, Hølmskov U, Tornoe I, Horslev-Petersen K, Sørensen GL, Junker P. Circadian rhythm and the influence of physical activity on circulating surfactant protein D in early and long-standing rheumatoid arthritis. Rheumatol Int. 2010 doi: 10.1007/s00296-010-1538-7. [DOI] [PubMed] [Google Scholar]
- 255.Honda Y, Takahashi H, Kuroki Y, Akino T, Abe S. Decreased contents of surfactant proteins A and D in BAL fluids of healthy smokers. Chest. 1996;109:1006–9. doi: 10.1378/chest.109.4.1006. [DOI] [PubMed] [Google Scholar]
- 256.Hoegh SV, Sorensen GL, Tornoe I, Lottenburger T, Ytting H, Nielsen HJ, Junker P, Holmskov U. Long-term stability and circadian variation in circulating levels of surfactant protein D. Immunobiology. 2010;215:314–20. doi: 10.1016/j.imbio.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 257.Betsuyaku T, Kuroki Y, Nagai K, Nasuhara Y, Nishimura M. Effects of ageing and smoking on SP-A and SP-D levels in bronchoalveolar lavage fluid. Eur Respir J. 2004;24:964–70. doi: 10.1183/09031936.04.00064004. [DOI] [PubMed] [Google Scholar]
- 258.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. 2001;18:482–90. doi: 10.1183/09031936.01.00043401. [DOI] [PubMed] [Google Scholar]


