Abstract
Identifying genetic variants influencing human brain structures may reveal new biological mechanisms underlying cognition and neuropsychiatric illness. The volume of the hippocampus is a biomarker of incipient Alzheimer’s disease1,2 and is reduced in schizophrenia3, major depression4 and mesial temporal lobe epilepsy5. Whereas many brain imaging phenotypes are highly heritable6,7, identifying and replicating genetic influences has been difficult, as small effects and the high costs of magnetic resonance imaging (MRI) have led to underpowered studies. Here we report genome-wide association meta-analyses and replication for mean bilateral hippocampal, total brain and intracranial volumes from a large multinational consortium. The intergenic variant rs7294919 was associated with hippocampal volume (12q24.22; N = 21,151; P = 6.70 × 10−16) and the expression levels of the positional candidate gene TESC in brain tissue. Additionally, rs10784502, located within HMGA2, was associated with intracranial volume (12q14.3; N = 15,782; P = 1.12 × 10−12). We also identified a suggestive association with total brain volume at rs10494373 within DDR2 (1q23.3; N = 6,500; P = 5.81 × 10−7).
The hippocampal formation is a key brain structure for learning, memory8,9 and stress regulation10 and is implicated in many neuropsychiatric disorders. Further, overall brain and head sizes are altered in many disorders and are significantly correlated with general cognitive ability11–13. Hippocampal, total brain and intracranial volumes are highly heritable in non-human primates14,15 and in humans6,7. Finding loci that influence these measures may lead to the identification of genes underlying susceptibility for neuropsychiatric diseases. Here we sought to identify common genetic polymorphisms influencing hippocampal, total brain and intracranial volumes in a large multinational consortium.
Our discovery sample comprised 17 cohorts of European ancestry from whom genome-wide SNPs and structural MRI data were collected (Supplementary Tables 1–3). Unselected population samples and case-control studies were included, with cases ascertained for neuropsychiatric disorders including depression, anxiety, Alzheimer’s disease and schizophrenia. To distinguish whether putative effects at these loci varied with disease status, analyses were run in the full sample (N = 7,795) and in a healthy subsample (N = 5,775). To help disentangle overall brain size effects from those specific to hippocampal volume, associations were assessed with and without controlling for total brain and intracranial volumes (Online Methods). As the initial goal of the study was to explore associations with hippocampal volume, total brain and intracranial volumes were analyzed in healthy subjects only.
Phenotypes were computed from three-dimensional anatomical T1-weighted magnetic resonance images, using validated automated segmentation programs16–18 (Supplementary Fig. 1 and Supplementary Tables 4 and 5). Extensive quality control analysis of segmentation was performed on sample outliers; subjects with poorly delineated brain volume phenotypes were removed (Supplementary Figs. 2–6). The mean bilateral hippocampal volume across the discovery cohorts was 3,917.4 mm3 (s.d. = 441.0 mm3).
Heritability of structural brain phenotypes was estimated in a sample of Australian monozygotic and dizygotic twins and their siblings (Queensland Twin Imaging (QTIM) study; N = 646, including ungenotyped participants; age range = 20–30 years) for hippocampal volume (h2 = 0.62), total brain volume (h2 = 0.89) and intracranial volume (h2 = 0.78). Hippocampal volume was also highly heritable in an extended pedigree cohort of Mexican-Americans from the United States (Genetics of Brain Structure and Function (GOBS); N = 605; age range = 18–85; h2 = 0.74), as were total brain volume (h2 = 0.77) and intracranial volume (h2 = 0.84). All heritability estimates were highly significant (P < 0.001).
To enable consortium-wide comparison of ancestry and to adjust appropriately for population stratification, each site conducted multidimensional scaling (MDS) analyses comparing their data to the HapMap 3 reference populations (Supplementary Fig. 7). All subsequent analyses included the following covariates: sex, linear and quadratic effects of age, interactions of sex with age covariates, MDS components and dummy covariates for different magnetic resonance acquisitions. Analyses were filtered for genotyping and imputation quality (Supplementary Fig. 8 and Supplementary Table 6); distributions of test statistics were examined at the cohort level through Manhattan and quantile-quantile plots (Supplementary Figs. 9–24). We conducted fixed-effects meta-analysis with METAL, applying genomic control19 (Supplementary Figs. 25–32). For completeness and to account for heterogeneity across sites, a random-effects meta-analysis was also performed20 (Supplementary Figs. 33–40). We attempted in silico replication of the top five loci for each trait within the combined CHARGE Consortium discovery set and 3C replication sample21 (N = 10,779), as well as in two cohorts of European ancestry (imputed to the Utah residents of Northern and Western European ancestry (CEU) and/or Toscani in Italy (TSI) HapMap cohorts; N = 449) and in two additional cohorts (imputed to combined CEU and Yoruba in Ibadan, Nigeria (YRI), and to Mexican ancestry in Los Angeles, California (MEX); N = 842). We also undertook custom genotyping of the two most promising SNPs in two additional samples of European ancestry (BIG replication and Trinity College Dublin/National University of Ireland, Galway (TCD/NUIG); N = 1,286).
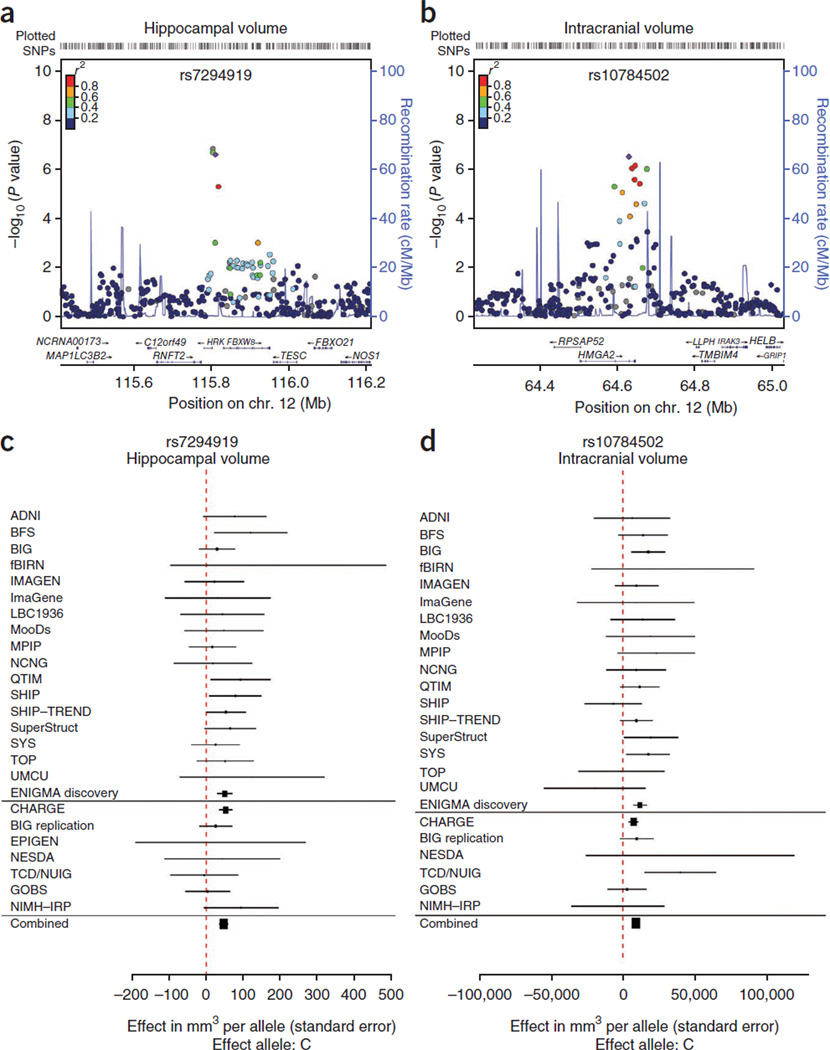
In general, previously identified polymorphisms associated with hippocampal volume showed little association in our meta-analysis (BDNF, TOMM40, CLU, PICALM, ZNF804A, COMT, DISC1, NRG1, DTNBP1; Supplementary Table 7), nor did SNPs previously associated with schizophrenia22 and bipolar disorder23 (Supplementary Table 8). The most significant SNPs in each analysis from the discovery sample (P ≤ 5 × 10−5) are listed (Supplementary Tables 9–16). No markers reached genome-wide significance (P < 1.25 × 10−8; Online Methods) in the discovery sample alone. However, the strongest associations for hippocampal and intracranial volumes were replicated, yielding results at genome-wide significance (Fig. 1 and Table 1; see Supplementary Tables 17–25 for additional results and gene-based tests24).
Figure 1.
Association results and meta-analysis of effects in individual and combined analyses. (a) The strongest association with hippocampal volume was found for rs7294919. Fixed-effects meta-analysis P values are shown41 after controlling for intracranial volume using all subjects in the discovery sample. (b) The strongest association with intracranial volume was found for rs10784502. Fixed-effects meta-analysis P values are shown in healthy subjects only. (c,d) The effect within each sample contributing to the meta-analysis is shown in forest plots for hippocampal volume (c) and intracranial volume (d). Association data using intracranial volume as a phenotype were not available for the EPIGEN sample. Head size was not controlled for in the CHARGE Consortium association analyses.
Table 1.
Results from the genome-wide association meta-analyses of mean hippocampal, intracranial and total brain volumes
| Sample | N | Freq. of the effect allele |
β (mm3) | S.E. (mm3) | P value | Heterogeneity P value |
Variance explained (%)h |
|---|---|---|---|---|---|---|---|
| Mean bilateral hippocampal volumea | |||||||
| rs7294919b | |||||||
| Discovery Fixed-effects model | 7,795 | 0.104 | 50.27 | 9.71 | 2.42 × 10−7 | 0.913 | 0.242 |
| Random-effects model | 50.12 | 9.65 | 4.43 × 10−7 | 0.910 | 0.241 | ||
| ENIGMA CEU and TSI replication | 1,735 | 0.101 | 22.05 | 19.00 | 0.246 | 0.924 | 0.042 |
| ENIGMA CEU and YRI or MEX replication | 842 | 0.125 | 27.77 | 25.96 | 0.285 | 0.127 | 0.095 |
| Discovery and replication | 10,372 | 0.106 | 42.74 | 8.22 | 1.99 × 10−7 | 0.347 | 0.177 |
| CHARGE in silico replication | 10,779 | 0.093 | 52.70 | 8.45 | 3.40 × 10−10 | 0.442 | 0.458 |
| ENIGMA and CHARGE | 21,151 | 0.099 | 47.58 | 5.89 | 6.70 × 10−16 | 0.419 | 0.265 |
| Intracranial volumec | |||||||
| rs10784502d | |||||||
| Discovery Fixed-effects model | 5,778 | 0.488 | 11860.73 | 2319.00 | 3.14 × 10−7 | 0.783 | 0.281 |
| Random-effects model | 11841.80 | 2270.07 | 3.93 × 10−7 | 0.771 | 0.280 | ||
| ENIGMA CEU and TSI replicatione | 1,130 | 0.525 | 15758.59 | 5244.69 | 0.003 | 0.065 | 0.468 |
| ENIGMA CEU and YRI or MEX replication | 699 | 0.348 | 1928.43 | 6215.31 | 0.756 | 0.710 | 0.008 |
| Discovery and replication | 7,607 | 0.479 | 11395.74 | 2007.27 | 1.37 × 10−8 | 0.217 | 0.261 |
| CHARGE in silico replication | 8,175 | 0.501 | 7429.56 | 1630.92 | 5.23 × 10−6 | NA | 0.110 |
| ENIGMA and CHARGE | 15,782 | 0.491 | 9006.71 | 1265.78 | 1.12 × 10−12 | 0.145 | 0.166 |
| Total brain volumef | |||||||
| rs10494373g | |||||||
| Discovery Fixed-effects model | 5,778 | 0.082 | 13693.29 | 3187.51 | 1.74 × 10−5 | 0.688 | 0.198 |
| Random-effects model | 13562.00 | 3114.17 | 2.69 × 10−5 | 0.728 | 0.194 | ||
| ENIGMA CEU and TSI replication | 117 | 0.107 | 8435.89 | 20256.09 | 0.678 | NA | 0.001 |
| ENIGMA MEX replication | 605 | 0.097 | 26883.36 | 8608.20 | 0.001 | NA | 0.964 |
| Discovery and replication | 6,500 | 0.085 | 14778.23 | 2957.14 | 5.81 × 10−7 | 0.182 | 0.240 |
Freq., frequency. CEU, TSI YRI and MEX refer to the HapMap 3 reference panels most representative of the sample and used for imputation; NA, not applicable.
Mean bilateral hippocampal volume association results were corrected for intracranial volume, sex, age, age2, sex × age, sex × age2 and four MDS components, and individuals with disease were included in the analysis.
rs7294919 is located at 12q24.22: position 115,811,975. Effect allele, C; non-effect allele, T. Genomic positions are based on the NCBI36/hg18 (March 2006) genome assembly.
Association results for intracranial volume were corrected for sex, age, age2, sex × age, sex × age2 and four MDS components, and individuals with disease were excluded from this analysis.
rs10784502 is at 12q14.3: position 64,630,077. Effect allele, C; non-effect allele, T.
Intracranial volume and total brain volume were available for two participants in MPIP and one participant in the BIG cohort who did not have hippocampal volume measures. The proxy SNP rs8756 was genotyped in the TDC/NUIG cohort.
Analysis for total brain volume was corrected for sex, age, age2, sex × age, sex × age2 and four MDS components, and individuals with disease were excluded. Total brain volume was not available for the ENIGMA replication cohorts. Within the CHARGE Consortium, a normalized version of total brain volume was analyzed and defined as total brain volume intracranial volume, and, because of this, the results are not comparable between consortia.
rs10494373 is at 1q23.3: position 160,885,986. Effect allele, C; non-effect allele, A.
Calculated as 2pq × β2 / (s.d.)2, where p and q are the minor and major allele frequencies, β is the unstandardized regression coefficient and s.d. is from the phenotype in the absence of covariate corrections. Intracranial volume phenotypic variance from the ENIGMA discovery sample was used to calculate percent variance explained in the CHARGE in silico replications, as this information was not available from the CHARGE consortium.
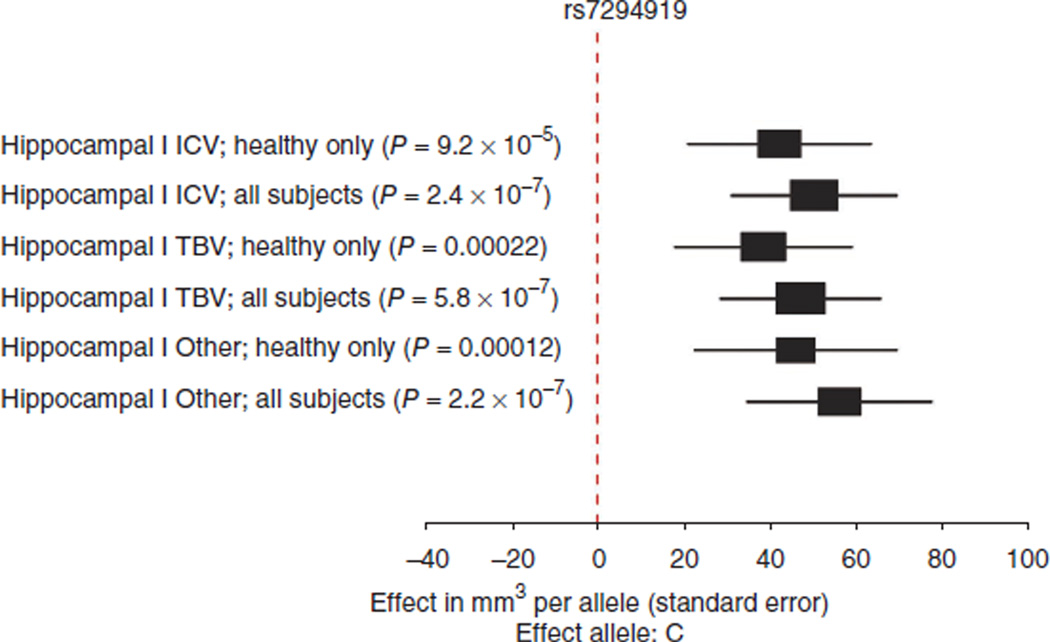
In our discovery sample, two SNPs in the same linkage disequilibrium (LD) block showed strong associations with hippocampal volume after controlling for intracranial volume (rs7294919 and rs7315280; r2 = 0.81, CEU 1000 Genomes Pilot 1). A random-effects analysis of the discovery sample, conducted to examine heterogeneity between cohorts, reduced significance only slightly for rs7294919 (P = 4.43 × 10−7) compared to the primary fixed-effects analysis (P = 2.42 × 10−7). The association was consistent, although stronger, in the full sample compared to the healthy subset (Fig. 2). Notably, the association was robust to the effects of head and brain size (Fig. 2), and the locus was not significantly associated with intracranial volume (P = 0.54) or total brain volume (P = 0.41). This suggests an effect at the level of the hippocampus rather than on brain size in general. The direction of the effect was consistent across samples and ages (Fig. 1). Haplotype analysis of directly genotyped variants near rs7294919 in two samples confirmed that the association was present across the haplotype and that the causal variant was well marked by rs7294919 (Supplementary Note). rs7294919 was also significantly associated with hippocampal volume in the cohorts from the CHARGE Consortium, which are composed of elderly subjects. Meta-analysis of the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) discovery and replication samples with those from the CHARGE Consortium yielded a highly significant association for rs7294919 (P = 6.70 × 10−16; N = 21,151).
Figure 2.
Association of rs7294919 with hippocampal volume stratified by disease and covariates. Effects are consistent in the discovery sample regardless of whether individuals with disease (N = 7,795) or only healthy subjects (N = 5,775) were included. The effect is also consistent whether accounting for intracranial volume (ICV), total brain volume (TBV) or without a measure of head size (Other).
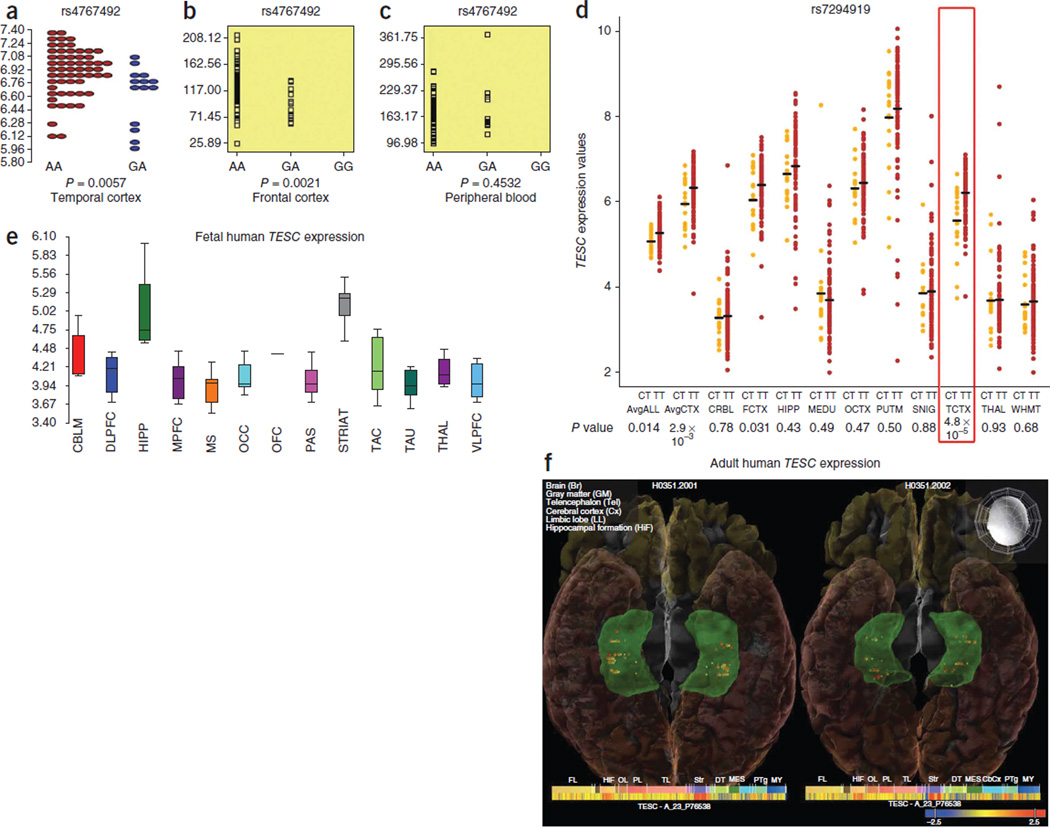
rs7294919 lies between HRK and FBXW8 (12q24.22; Fig. 1) and is not in LD with any SNPs within coding sequences, UTRs or splice sites within 500 kb (r2 > 0.4) in the CEU sample from the 1000 Genomes Project Phase 1. To determine whether the observed association is related to a regulatory mechanism, we examined potential cis effects of this variant on expression levels of genes within a 1-Mb region. In temporal lobe tissue resected from 71 individuals with mesial temporal lobe epilepsy and hippocampal sclerosis in the University College London (UCL) epilepsy cohort, we examined association between rs4767492 (a proxy for rs7294919, which was not directly genotyped; r2 = 0.636 in 1000 Genomes Project Phase 1) and expression levels. This analysis suggested an association (P = 0.006, controlling for age) with expression of the TESC gene, which lies 3′ to FBXW8 (149 kb; Fig. 3). To corroborate this finding, we used the publicly available SNPExpress database (see URLs), which includes data on gene expression in post-mortem frontal cortex from 93 subjects. In this independent sample, expression levels of TESC again significantly differed by genotype (rs4767492; P = 0.0021). Additional replication came from the UK Brain Expression Database, where TESC expression in post-mortem brain tissues from 134 individuals free from neurological disorders showed a strong difference by genotype in temporal cortex (rs7294919; P = 9.7 × 10−4 for gene and 4.8 × 10−5 for exon 8). Given the small sample sizes and low minor allele frequency of this SNP (MAF = 0.099), no homozygotes for the minor allele were observed in any brain tissue sample, limiting the inferences we can draw regarding mode of action. Expression of HRK showed little evidence of association with the proxy genotype in the UCL epilepsy cohort (P = 0.11) or SNPExpress (P = 0.16) but was associated with rs7294919 in temporal cortex within the UK Brain Expression Database (P = 0.0051). Additional associations were observed in peripheral blood mononuclear cells (PBMCs; Supplementary Note).
Figure 3.
Regulatory effects of hippocampal-associated variant and expression of TESC within the hippocampus. (a–d) The locus most associated with hippocampal volume was also associated with mRNA expression of the TESC gene in brain in three independent samples, the UCL epilepsy cohort (a), the SNPExpress database (b), where a proxy SNP was used, and the UK Brain Expression Database (d), where differences in TESC expression of the directly genotyped hippocampal variant (rs7294919) were strongest in the temporal cortex (TCTX) (red box) but also found in the average expression of all cortex (AvgCTX) and average expression of all brain structures tested (AvgALL). Symbol color represents genotype in a and d. These regional gradients in expression support the hypothesis that the SNP may associate with hippocampal but not total brain volume. No effects were detected in PBMCs from the SNPExpress database (c). CRBL, cerebellar cortex; FCTX, frontal cortex; HIPP, hippocampus; MEDU, medulla (specifically the inferior olivary nucleus); OCTX, occipital cortex; PUTM, putamen; SNIG, substantia nigra; THAL, thalamus; WHMT, intralobular white matter. (e) TESC is differentially expressed within the fetal human brain (P = 1.33 × 10−12), with the highest expression in striatum (STRIAT) and hippocampus (HIPP)42. Box plots represent median and 25th to 75th percentiles. Upper and lower lines show minimum and maximum values, respectively. CBLM, cerebellum; DLPFC, dorsolateral prefrontal neocortex; MPFC, medial prefrontal neocortex; MS, motor-somatosensory neocortex; OCC, occipital visual neocortex; OFC, orbital prefrontal neocortex; PAS, parietal association neocortex; TAC, temporal association neocortex; TAU, temporal auditory neocortex; THAL, mediodorsal thalamus; VLPFC, ventrolateral prefrontal neocortex. (f) TESC has moderate to high gene expression throughout the adult human hippocampus (shown in green), as visualized in the Allen Institute Human Brain Atlas using Brain Explorer 2 software. An inferior view of the brain is shown in two subjects; the anterior portion of the brain is at the top. The colors of spheres within the hippocampus indicate the Z-scores of TESC expression normalized within each subject across brain structures. Heat maps show that expression of TESC is higher in the hippocampus (HiF) and striatum (Str) than in other brain structures.
The expression results in brain tissue suggest that TESC is a primary positional candidate for our quantitative trait locus (QTL). Studies of mouse and chicken embryos show that TESC is expressed throughout the brain during development, with the strongest expression in the developing telencephalon and mesencephalon and near the developing ventricles25. TESC also has moderate expression in the human hippocampus during adulthood (Allen Institute Brain Atlas, see URLs; Fig. 3). Its protein product, tescalcin, interacts with the Na+/H+ exchanger (NHE1)26, which is involved in the regulation of intracellular pH21, cell volume and cytoskeletal organization27. TESC expression is strongly regulated during cell differentiation in a cell lineage–specific fashion28,29. Our data suggest that this role in cell proliferation and differentiation is relevant for hippocampal volume and brain development.
The strongest association with intracranial volume was observed at rs10784502 (Table 1), an intronic SNP near the 3′ UTR of the HMGA2 gene (12q14.3; Fig. 1). This locus was associated with intracranial volume across lifespan, as shown by the strong replication in samples from healthy elderly individuals in the CHARGE Consortium. The combined analysis resulted in the identification of a highly significant association (P = 1.12 × 10−12). Of note, rs10784502 has been reliably associated with increased adult height (P = 3.636 × 10−32; effect allele: C)30. The genetic correlation between height and intracranial volume within the QTIM sample was significant (rg = 0.31; P = 1.34 × 10−7), as was that observed in the GOBS sample (rg = 0.20; P = 0.026), suggesting modest overlap of shared genetic determinants. rs10784502 also had an effect on total brain volume in the discovery sample (P = 9.49 × 10−5). When considering the results from the intracranial volume meta-analysis in SNPs previously associated with height31–33 (NSNPs = 175; Supplementary Fig. 41), a clear inflation of the test statistic was observed (λ = 1.44), indicating that SNPs associated with height are also associated with intracranial volume. This enrichment, which was not observed for hippocampal volume (Supplementary Figs. 42 and 43), was due to a systematically higher degree of association throughout the candidate SNP set rather than a small number of large effects. Structural equation modeling showed that the effect of rs10784502 on intracranial volume could not completely be accounted for by the indirect effects of this SNP on height or by the correlation between height and intracranial volume (Supplementary Fig. 44).
Examining correlations between rs10784502 and expression levels of genes within a 1-Mb region, we identified a significant effect on the expression of HMGA2 (P = 0.0077) as the single significant result in the GOBS transcriptional profile data. Additionally, HMGA2 expression levels in PBMCs were significantly negatively genetically correlated with intracranial volume (rg = −0.49; P = 0.016) in this cohort. These results support HMGA2 as a positional candidate gene underlying our observed QTL. HMGA2 encodes the high-mobility group AT-hook 2 protein, which is a chromatin-associated protein that regulates stem cell renewal during development34. It is implicated in human growth through genetic association studies and the presence of rare mutations35 and also has known roles in neural precursor cells36. Whether both functions are due to the same underlying mechanisms warrants further study.
To test for pleiotropic effects of rs7294919 and rs10784502, we examined the influence of these variants on cognition in the Brisbane Adolescent Twin Study37 (N = 1642). The C allele of rs10784502, which was associated with increased intracranial volume, was also associated with increased full-scale IQ, as measured via the Multidimensional Aptitude Battery38 (effect size (β) = 1.29, standard error (S.E.) = 0.47; P = 0.0073; phenotypic correlations are shown in Supplementary Table 26). This effect was driven by performance (PIQ; β = 1.74, S.E. = 0.61; P = 0.0044) rather than by verbal subtests (VIQ; P = 0.103). rs7294919 was not associated with full-scale IQ (P = 0.139) or PIQ (P = 0.489) but showed nominal association with VIQ (effect allele: C; β = 0.126, S.E. = 0.062; P = 0.043).
No associations at genome-wide significance were detected for total brain volume. Following inclusion of the replication samples, the strongest evidence for association was detected at rs10494373 within DDR2 (1q23.3; P = 5.81 × 10−7) (Table 1), which encodes a receptor tyrosine kinase involved in cell growth and differentiation39.
The current study identified and replicated two quantitative trait loci for hippocampal and intracranial volumes across lifespan in a large sample including both healthy subjects and those with neuropsychiatric diagnoses. The rs7294919 variant was associated with decreased hippocampal volume of 47.6 mm3 or 1.2% of the average hippocampal volume per risk allele. Although further work is necessary to confirm the causal variant(s) and functional mechanisms, this QTL influencing hippocampal volume differences may act by regulating expression of TESC specifically within the brain. In addition, the C allele of rs10784502 is associated, on average, with 9,006.7 mm3 larger intracranial volume, or 0.58% of intracranial volume per risk allele and is weakly associated with increased general intelligence by approximately 1.29 IQ points per allele.
It has previously been hypothesized that brain imaging endophenotypes would have large effect sizes; however, this has proven not to be the case for the specific volumetric traits measured here, which had comparable effect sizes to those observed in other genome-wide association studies of complex traits40. Notably, the discovery sample had 99.92% power to detect variants with effect sizes of 1% of the variance for MAF ≥ 0.05. It remains to be determined whether specific genetic variations linked to volumetric brain differences are also associated with other neuropsychiatric disorders, brain function and other cognitive traits. If this is the case, neuroimaging genetics may also discover new treatment targets related to the neurobiology of these disorders, in addition to improving phenomenologically based diagnostic criteria.
ONLINE METHODS
All participants provided written informed consent, and studies were approved by the respective Local Research Ethics committees or Institutional Review Boards. MRI scans came from previously collected data. Suggested protocols for imaging analysis are publicly available on the ENIGMA Consortium website (see URLs); however, any validated segmentation software was permitted. Accuracy of segmentation programs is influenced by scanner and head-coil type and scanner sequences and by participant characteristics, such as age. Each site was permitted to use any validated automated segmentation algorithm that worked most accurately in their data set. The two most commonly used hippocampal segmentation packages were the FMRIB’s Integrated Registration and Segmentation Tool (FIRST)16 from the FMRIB Software Library (FSL) package of tools43 and FreeSurfer17. Brain volume, the sum of gray and white matter excluding ventricles and cerebrospinal fluid (CSF), was calculated using the FSL FMRIB’s Automated Segmentation Tool (FAST)44 package or FreeSurfer. Estimated total intracranial volume was calculated through registration of each MRI scan to a standard brain image template18, using either FSL FLIRT45 or FreeSurfer (exceptions referenced in Supplementary Table 2). To calculate intracranial volume, the inverse of the determinant of the transformation matrix was multiplied by the template volume (1,948,105 mm3). Extensive quality control analysis on phenotype segmentations included manual examination of phenotype volume histograms (Supplementary Figs. 2–6) and box plots of all volumetric measures. Outliers were manually evaluated by overlaying the automated segmentations on the original MRI scan. Subjects were excluded from the analysis if structures were poorly segmented.
As assessed previously, the correlation in volumes between automatic and manually segmented hippocampi was high; the accuracy was reported to be higher with FreeSurfer than with FIRST in one study (FreeSurfer r = 0.82; FIRST r = 0.66)46 and similar between the two in another (FreeSurfer r = 0.73; FIRST r = 0.71)47. Scan-rescan reliability was also high for both methods (FreeSurfer intraclass correlation (ICC) = 0.98; FIRST ICC = 0.93)48. We undertook a large-scale assessment to determine the correspondence between segmentations from both FSL and FreeSurfer in the same subjects. Correspondence was found to be reasonably high for average bilateral hippocampal segmentation (r = 0.75; N = 6,093; Supplementary Table 4). This is close to the agreement between different human raters, as quantified by interrater reliability (ICC = 0.73–0.85)49,50, which may be a reasonable upper bound on the accuracy of automated segmentation. Brain volume and intracranial volume were delineated with high correspondence between the two methods (r = 0.95, r = 0.90, respectively; N = 4,321).
Heritability estimates for trait measures were calculated in two family-based samples, QTIM and GOBS. Estimates for the QTIM sample used a twin and sibling analysis within Mx. An extended family analysis in Sequential Oligogenic Linkage Analysis Routines (SOLAR)51 was used for the GOBS sample.
Given sample size and the heritability of hippocampal volume, power calculations were performed using the Genetic Power Calculator52. We had 99.92% power to detect variants with effect sizes of 1% of the variance and 71.16% power to detect variants with effect sizes of 0.5% of the variance for MAF ≥ 0.05.
All cohorts were genotyped using commercially available arrays. Genetics protocols were developed to standardize the filtering, imputation and association of genome-wide genotype data (see ENIGMA protocols in URLs). SNPs were filtered out of samples on the basis of standard quality control criteria, including low MAF (<0.01), poor genotype calling (call rate of <95%) and deviations from Hardy-Weinberg equilibrium indicating possible errors in genotyping (P < 1 × 10−6). Genotyping methods and exceptions to these thresholds are summarized in Supplementary Table 3.
Genetic homogeneity within each sample was assessed through MDS plots (Supplementary Fig. 7). Ancestry outliers were excluded through visual inspection. A standardized population template from HapMap 3 representing those sampled was selected for imputation. Performance of software for imputation is generally similar between the most used methods53,54 for common variants (MACH55, IMPUTE56 and BEAGLE57); the protocols provided included use of the MACH tool. As raw genotype data were not directly transferred to the meta-analysis site, a histogram of allele frequency differences between each contributing group and the HapMap 3 CEU population was generated for each group (Supplementary Fig. 8) to further examine genotyping and imputation quality. A simulation to determine the effect of varying quality control thresholds on imputation quality (Supplementary Table 6) showed that the minor variation in quality control thresholds and imputation reference panels between sites was unlikely to have influenced imputation accuracy.
Genome-wide association analyses were performed that included and excluded individuals with disease. Including individuals with disease (all subjects) offers advantages of greater sample size and wider phenotype distribution, which may provide greater power to detect genetic effects58–60. We reanalyzed phenotypes after we excluded individuals with disease to confirm that the observed associations were not due to confounds relating to disease, medication or the possibly altered environments and experiences of these persons. To aid in the interpretation of results, we reanalyzed hippocampal volume after controlling for intracranial volume and total brain volume in two separate analyses. This helped to determine whether the observed associations were caused by direct effects on hippocampal volume or were attributable to more global associations with head size. In addition, genome-wide association analyses of intracranial volume and brain volume were conducted in the healthy controls to clarify whether observed associations were specific to hippocampal volume or influenced brain size in general. Participating sites were asked to conduct five genome-wide association analyses (three analyses of hippocampal volume, intracranial volume and brain volume). In addition, cohorts with groups of individuals with disease were asked to perform hippocampal analyses including data from these individuals.
Evidence for association was assessed using the allelic dosage of each SNP (accounting for familial relationships in the GOBS, QTIM and SYS samples). SNP-derived covariates were tested as fixed effects, while explicitly modeling the genetic relationships between family members in these pedigree-based studies51,61,62. Analyses used multiple linear regression with the phenotype of interest as a dependent variable and the additive dosage of each SNP as an independent variable of interest, controlling for covariates of population stratification (four MDS components), age, age2, sex and the interactions between age and sex and age2 and sex. Dummy covariates were used to control for different scanner sequences or equipment within a site when needed. We refer to these covariates as ‘other covariates’, and these were included in all analyses. The extensive regression model was used to statistically control for factors known to affect hippocampal volume that are not specific genetic influences. Recommended protocols for association were provided to the studies based on those used in mach2qtl software (see ENIGMA protocols).
To combine information from multiple studies, we generated a secure web-accessible upload site for participants to upload their association results. An automated system parsed the uploaded results files (see URLs). This parser was designed to read raw results files from a variety of association software packages (mach2qtl, PLINK, SOLAR, SNPTEST, QUICKTEST, Merlin-offline and ProbABEL), perform a series of tests on the incoming data to ensure quality, correctly assign the effect allele (dependent on both the imputation and association programs used) and correctly scale the β values and standard errors from association into the same units. Quality control was performed on imputed SNPs to filter out SNPs with low frequency (MAF of <0.01) or poor imputation quality (estimated R2 of <0.3). Result files and summary statistics from each group were pooled for meta-analysis. Meta-analysis was undertaken for each SNP across all groups based on a fixed-effects model using an inverse standard error–weighted meta-analysis protocol implemented in METAL19. Genomic control was applied at the level of each study and at the meta-analysis level to adjust for population stratification or cryptic relatedness not accounted for by MDS components63. To account for heterogeneity across samples, a random-effects meta-analysis20 was also conducted via the program METASOFT without using genomic control. Using KGG64 we performed gene-based tests on the double genome–controlled meta-analysis results, using the extended Simes test24 to obtain an overall P value for association of the entire gene with a 50-kb boundary on either side. Results from genes with P ≤ 1 × 10−4 are presented (Supplementary Tables 18–25).
Meta-analysis was performed separately on the discovery sample, the CEU and TSI replication sample and the CEU and YRI or MEX replication sample. These results were then pooled to form the combined meta-analysis statistics for discovery and replication. The in silico replication results from the CHARGE Consortium were added to this, and a final meta-analysis was conducted. The location of Manhattan and quantile-quantile plots is specified in Supplementary Table 27.
To appropriately account for the multiple comparisons conducted, we first sought to determine the effective number of independent phenotypes among the eight highly correlated genome-wide association analyses. This was calculated by creating an 8 × 8 matrix derived from cross-correlations of meta-analytic t statistics of association for each SNP across phenotypes. The resulting correlation matrix provided an estimate of the similarity between phenotypes after adjusting for covariates of interest and appropriately controlling for family structure. The effective number of tests was then calculated by summing eigenvalues of the correlation matrix, weighted by a formula that appropriately controls false positive rates in simulation65. The effective number of tests was determined to be 4 and an overall genome-wide significance threshold of 5 × 10−8/4 = 1.25 × 10−8 was used throughout the manuscript.
Regulatory potential of SNPs identified in the genome-wide association analysis was examined in three samples. In the UCL epilepsy cohort, tissue was obtained from resection material from affected individuals who had undergone surgery for drug-resistant mesial temporal lobe epilepsy with hippocampal sclerosis, according to established clinical protocols. Total RNA from the middle temporal cortex (Brodmann areas 20 and 21) from 86 subjects was isolated and randomly hybridized to Affymetrix Human Exon 1.0 ST arrays, and quality control analysis was performed using standard methods. The effects of several methodological (day of expression hybridization, RNA integrity number (RIN)) and biological covariates (sex, age and medication) on exon–gene expression relationships were tested for significance. Of these individuals, 71 had participated in a published epilepsy genome-wide association study, and, therefore, genotyping data were available. Details of sample collection and genotyping quality control steps have been published previously66. These samples were assayed with Illumina HumanHap550v3 (N = 44) and Illumina Human610-Quadv1 (N = 27) arrays.
In the UK Brain Expression database, post-mortem brain tissues from 134 individuals free from neurological disorders were obtained from the MRC Sudden Death Brain Bank in Edinburgh and Sun Health Research Institute67. Genotype information was obtained using Illumina HumanOmni 1M arrays and standard quality control methods. Expression profiling was conducted in up to ten separate brain regions for each individual brain using the Affymetrix GeneChip Human Exon 1.0 ST array. Expression levels were normalized using the Robust Multi-array Analysis (RMA) algorithm restricting to probe sets containing more than three probes, unique hybridization target (probes that map to a single position within the genome) and supported by evidence from EntrezGene. The average signals for all neocortex (AvgCTX) and all brain regions (AvgAll) were tested, as were individual cortical and subcortical regions. Any significant association where the probe set contained the SNP or a SNP in high LD (r2 > 0.50) was removed from further analysis.
SNPExpress, a publically available database, was also used for replication of the findings. The SNPExpress database68 used autopsy-collected frontal cortex brain tissue in 93 samples from human subjects with no neuropsychiatric conditions and PBMCs in 80 samples. In this database, transcript expression levels were measured on Affymetrix Human ST 1.0 exon arrays, and genome-wide genotyping was performed using Illumina HumanHap550K arrays.
Raw gene expression data from human fetal brain were gathered from a published study42. Post-mortem specimens from four late mid-fetal human brains (18, 19, 21 and 23 weeks of gestation) were collected from the Human Fetal Tissue Repository at the Albert Einstein College of Medicine. Details of specimens, tissue processing, microdissection and neuropathological assessment have been described elsewhere42.
Supplementary Material
ACKNOWLEDGMENTS
Some authors received commercial funding unrelated to the topic of this paper. N.J.v.d.W. received speaking fees from Eli Lilly & Company and Wyeth and served on advisory panels of Eli Lilly & Company, Pfizer, Wyeth and Servier. A.A. received an investigator-initiated unrestricted research grant from Bristol-Myers Squibb and speaker’s fees from AstraZeneca, Bristol-Myers Squibb and GlaxoSmithKline. H.J.G. received external research support from the German Research Foundation, the Federal Ministry of Education and Research Germany, speaker’s honoraria from Bristol-Myers Squibb, Eli Lilly & Company, Novartis, Eisai, Boehringer Ingelheim and Servier and travel funds from Janssen-Cilag, Eli Lilly & Company, Novartis, AstraZeneca, Lundbeck and the SALUS–Institute for Trend-Research and Therapy Evaluation in Mental Health. M.N. received research grants from the Federal Ministry of Education and Research, Germany, the German Research Foundation, BioRad Laboratories, Siemens AG, Zeitschrift für Laboratoriumsmedizin, Bruker Daltronics, Abbott, Jurilab Kuopio, Roche Diagnostics, Instand and Becton Dickinson. H.V. received external research support via research grants from Hofmann La Roche, the Humboldt Foundation, the Federal Ministry of Education and Research (Germany) and the German Research Foundation. M.W. is on the following scientific advisory boards: Lilly, Araclon and Institut Catala de Neurociencies Aplicades, the Gulf War Veterans Illnesses Advisory Committee, VACO, Biogen Idec and Pfizer. M.W. received funding for consulting from Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, Pfizer and Janssen; for travel from NeuroVigil, CHRU–Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, the University of California, San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, the University of New Mexico School of Medicine, Ipsen, Clinical Trials on Alzheimer’s Disease (CTAD), Pfizer, AD PD Meeting, Paul Sabatier University, Novartis and Tohoku University; and research support from: Merck, Avid, DoD, VA. M.W. received honoraria from PMDA/ the Japanese Ministry of Health, Labour, and Welfare, Tohoku University, Neuro Vigil, Insitut Catala de Neurociencies Aplicades. M.W. owns stock options for Synarc, Elan. Organizations contributing to the Foundation for the US NIH and thus to the National Institute on Aging (NIA)-funded Alzheimer’s Disease Neuroimaging Initiative included Abbott, the Alzheimer’s Association, the Alzheimer’s Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica (ADNI 2), Bristol-Myers Squibb, the Cure Alzheimer’s Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck, Novartis, Pfizer, Roche, Schering Plough, Synarc and Wyeth.
ADNI: The ADNI study was supported by the US NIH (U01 AG024904) and the Foundation for the NIH for genotype and phenotype data collection, the NIH (RC2 AG036535-01) for data analysis, the NIA (R01 AG019771-09) for additional data analysis and NCRAD (U24AG021886) for DNA used in part for the GWAS. Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (see URLs). As such, the investigators within ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. The ADNI sample wishes to acknowledge the investigators who contributed to the design and implementation of ADNI (see URLs). Data collection and sharing for this project were funded by ADNI (NIH grant U01 AG024904). ADNI is funded by the NIA, the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and through generous contributions from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck and Cocpany, Novartis AG, Pfizer, F. Hoffman–La Roche, Schering-Plough and Synarc, as well as from nonprofit partners at the Alzheimer’s Association and the Alzheimer’s Drug Discovery Foundation, with participation from the US Food and Drug Administration (FDA). Private sector contributions to ADNI are facilitated by the Foundation for the NIH (see URLs). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants (P30 AG010129 and K01 AG030514) and by the Dana Foundation. ADNI was launched in 2003 by the NIA, the NIBIB, the FDA, private pharmaceutical companies and nonprofit organizations as a 5-year public- private partnership. The primary goal of ADNI has been to test whether serial MRI), positron emission tomography (PET), other biological markers and clinical and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease. Determination of sensitive and specific markers of very early Alzheimer’s disease progression is intended to aid researchers and clinicians in developing new treatments and monitoring their effectiveness, as well as lessening the time and cost of clinical trials. The Principal Investigator of this initiative is M.W. Weiner. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the United States and Canada. The initial goal of ADNI was to recruit 800 adults ages 55 to 90 to participate in the research—approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early Alzheimer’s disease to be followed for 2 years. For up-to-date information, please visit the ADNI website (see URLs).
BIG: The BIG study wishes to acknowledge S. Kooijman for coordination of sample collection and A. Heister, M. Naber, R. Makkinje, M. Hakobjan and M. Steehouwer for genotyping. The BIG study was supported by a Biobanking and Biomolecular Resources Research Infrastructure Netherlands (BBMRI-NL) complementation grant for brain segmentation and the Netherlands Organisation for Scientific Research (NWO) Horizon Breakthrough grant (grant number 93511010 (to A.A.V.).
Bipolar Family Study: The Bipolar Family Study wishes to thank the Scottish Mental Health Research Network for research assistant support, the Brain Research Imaging Centre Edinburgh (see URLs), a center in the Scottish Funding Council Scottish Imaging Network–A Platform for Scientific Excellence (SINAPSE) Collaboration (see URLs), for image acquisition and the Wellcome Trust Clinical Research Facility for genotyping. Genotyping was supported by the National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Award (to A.M.M.), and data collection was supported by the Health Foundation Clinician Scientist Fellowship.
fBIRN: fBIRN wishes to acknowledge D.B. Keator for leading fBIRN neuroinformatics development, B.A. Mueller for image calibration and quality assurance and A. Belger, V.D. Calhoun, G.G. Brown, J.M. Ford, G.H. Glover, R. Kikinis, K. Lim, J. Laurriello, J. Bustillo, G. McCarthy, D.S. O’Leary, B. Rosen, A.W.T. and J.T. Voyvodic for their leadership contributions to fBIRN scanner and sequence calibration, tool development and data collection efforts. The fBIRN study was supported by the US NIH (U24 RR21992) for phenotypic data collection. Genotyping was performed with the support of the grant RBIN04SWHR to F.M. from the Italian Ministry of University and Research.
GOBS: The GOBS study was supported by the US NIH (MH0708143 and MH083824 to D.C.G., MH078111 and MH59490 to J.B., C06 RR13556 and C06 RR017515). P.K. was also supported by an NIH grant (EB006395).
IMAGEN: IMAGEN is funded by the European Commission Framework Programme 6 (FP-6) Integrated Project IMAGEN (PL037286), the European Commission Framework Programme 7 (FP-7) Project Alzheimer’s Disease, Alcoholism, Memory, Schizophrenia (ADAMS), the FP-7 Innovative Medicine Initiative Project European Autism Interventions (AIMS), the UK Department of Health National Institute of Health Research (NIHR)–Biomedical Research Centre Mental Health program and the MRC programme grant Developmental Pathways into Adolescent Substance Abuse (93558).
ImaGene: ImaGene wishes to acknowledge J. Lee and J. Lane for processing the blood samples, The Easton Consortium for Alzheimer’s Disease Drug Discovery and Biomarker Development and the Alzheimer’s Disease Research Center (ADRC) funded by the NIA at the University of California, Los Angeles (AG16570).
LBC1936: We thank the participants in LBC1936. We thank C. Murray, A.J. Gow, S.E. Harris, M. Luciano, P. Redmond, E. Sandeman, I. Gerrish, J. Boyd-Ellison, N. Leslie, A. Howden and C. Scott for data collection and preparation. This project is funded by the Age UK’s Disconnected Mind programme and also by Research Into Ageing (251 and 285). The entire genome association part of the study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/F019394/1). Analysis of brain images was funded by UK MRC grants (G1001401 and 8200). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Imaging was performed at the Brain Research Imaging Centre, Edinburgh, a center in the SINAPSE Collaboration. Funding from BBSRC, the Engineering and Physical Sciences Research Council (EPSRC), the Economic and Social Research Council (ESRC) and the MRC and Scottish Funding Council through the SINAPSE Collaboration is gratefully acknowledged. L.M.L. is the beneficiary of a postdoctoral grant from the AXA Research Fund.
MooDS: This work was funded by the German Federal Ministry of Education and Research (BMBF) in the National Genome Research Network (NGFN-plus) through the MooDs grant Molecular Causes of Major Mood Disorders and Schizophrenia (coordinator M.M.N.). Additional funding for genotyping was provided by a NARSAD Distinguished Investigator award to A.M.-L.
MPIP: The MPIP Munich Morphometry Sample comprises images acquired as part of the Munich Antidepressant Response Signature Study and the Recurrent Unipolar Depression (RUD) Case-Control Study performed at the MPIP and control subjects acquired at the Department of Psychiatry at the Ludwig-Maximilians-University. We wish to acknowledge A. Olynyik and radiographers R. Schirmer, E. Schreiter and R. Borschke for image acquisition and data preparation. We thank D.P. Auer for local study management in the initial phase of the RUD study. We are grateful to GlaxoSmithKline for providing the genotypes of the RUD Case-Control Sample. We thank the staff of the Center of Applied Genotyping (CAGT) for generating the genotypes of the MARS cohort. The study is supported by a grant from the Exzellenz-Stiftung of the Max Planck Society. This work has also been funded by the BMBF in the framework of the National Genome Research Network (NGFN) (FKZ 01GS0481).
NCNG: We would like to thank the personnel involved in recruitment and data collection and, in particular, P. Due-Tønnessen for clinical assessment of the MRI images. The NCNG study was supported by Research Council of Norway grants (154313/V50 and 177458/V50). The NCNG GWAS was financed by grants from the Bergen Research Foundation, the University of Bergen, the Research Council of Norway (FUGE; Psykisk Helse), Helse Vest Regionalt Helseforetak (RHF) and the Dr Einar Martens Fund.
NESDA-NTR: Funding was obtained from the NWO (MagW/ZonMW 904-61-090; 985-10-002; 904-61-193; 480-04-004; 400-05-717, Addiction-31160008; 911-09-032; SPI 56-464-14192 and Geestkracht Program, 10-000-1002), the Center for Medical Systems Biology (CMSB; NWO Genomics), NBIC/BioAssist/RK/2008.024, BBMRI-NL, Biobanking and Biomolecular Resources Research Infrastructure, the VU University, the EMGO Institute for Health and Care Research and Neuroscience Campus Amsterdam, the European Science Foundation (EU/QLRT-2001-01254), the European Community’s FP7 (HEALTH-F4-2007-201413), the European Science Council (ERC) Genetics of Mental Illness (230374), Rutgers University Cell and DNA Repository (cooperative agreement NIMH U24 MH068457-06), the US NIH (R01D0042157-01A) and the Genetic Association Information Network (a public-private partnership between the NIH and Pfizer, Affymetrix and Abbott Laboratories).
NIMH-IRP: This study was supported by funding from the Intramural Research Program of the National Institute of Mental Health (NIMH) from the NIH and the US Department of Health and Human Services (K99 MH085098 to G.L., 1ZIA MH002810 to F.J.M. and 1ZIA MH002790 to W.C.D.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government.
QTIM: We are extremely grateful to the twins for their participation, the radiographers at the Centre for Advanced Imaging at the University of Queensland for image acquisition and the many research assistants and support staff at the Queensland Institute of Medical Research for twin recruitment and daily management, and we especially thank K. Johnson for MRI scanning and processing, A. Henders for DNA processing and preparation and S. Gordon for quality control and management of the genotypes. Phenotyping was funded by the US National Institute of Child Health and Human Development (R01 HD050735) and the Australian National Health and Medical Research Council (NHMRC) (project grant 496682). Genotyping was funded by the NHMRC (Medical Bioinformatics Genomics Proteomics Program, 389891). G.M. was supported by an NHMRC Fellowship (613667), and G.Z. was supported by Australian Research Council (ARC) Future Fellowship (FT0991634). S.E.M. is funded by an ARC Future Fellowship (FT110100548). J.L.S. was supported by the Achievement Rewards for College Scientists foundation and the US NIMH (F31 MH087061). D.P.H. is partially supported by a National Science Foundation (NSF) Graduate Research Fellowhip Program (GRFP) grant (DGE-0707424). P.T. was also supported by the NIH (grants U01 AG024904, AG040060, EB008432, P41 RR013642, HD050735, AG036535, AG020098 and EB008281).
SYS: The Saguenay Youth Study Group wishes to thank the following individuals for their contribution in acquiring and analyzing the data: N. Arbour, M.-È. Bouchard, A. Houde, A. Gauthier and H. Simard for the recruitment and assessment of participating families, M. Bérubé, S. Masson, S. Castonguay and M.-J. Morin for MRI acquisition and E. Ding and N. Qiu for MR data management. We thank J. Mathieu for the medical follow up of participants in whom we detected any medically relevant abnormalities. We are grateful to all families for participating in the study. The Saguenay Youth Study Group is supported by the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Quebec and the Canadian Foundation for Innovation. For more information, please see the study website (see URLs).
SHIP: The Study of Health in Pomerania (SHIP) is supported by the German Federal Ministry of Education and Research (grants 01ZZ9603, 01ZZ0103 and 01ZZ0403) and the German Research Foundation (DFG; GR 1912/5-1). Genome-wide data and MRI scans were supported by the Federal Ministry of Education and Research (grant 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Center of Knowledge Interchange program of the Siemens AG. We thank all staff members and participants of the SHIP study, as well as all of the genotyping staff for generating the SHIP SNP data set. The genetic data analysis workflow was created using the Software InforSense. Genetic data were stored using the database Caché (InterSystems).
SHIP-TREND: The authors from SHIP are grateful to M. Stanke for the opportunity to use his Server Cluster for SNP Imputation. This cohort is part of the Community Medicine Research net (CMR) of the University of Greifswald, which is funded by the German Federal Ministry of Education and Research and the German Ministry of Cultural Affairs, as well as by the Social Ministry of the Federal State of Mecklenburg–West Pomerania. CMR encompasses several research projects that share data from the population-based Study of Health in Pomerania (SHIP; see URLs). The work is also supported by the German Research Foundation (DFG; GR 1912/5-1) and the Greifswald Approach to Individualized Medicine (GANI_MED) network funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genome-wide data and MRI scans were supported by the Federal Ministry of Education and Research (grant 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Center of Knowledge Interchange program of the Siemens AG.
Superstruct: We thank the investigators and participants who contributed to the brain genomics data collection for Superstruct at Massachusetts General Hospital and Harvard University, with funding from the Simons Foundation, the Howard Hughes Medical Institute and the US NIH (grant MH079799).
TOP: We thank the study participants of TOP and the personnel involved in data collection and logistics, especially T.D. Bjella. This work was supported by the Oslo University Hospital–Ullevål, the Eastern Norway Health Authority (2004-123), the Research Council of Norway (167153/V50, 163070/V50 and 183782/V50), and by Eli Lilly & Company (who covered part of the genotyping costs).
TCD: We wish to express our sincere thanks to all participants and to clinical staff who facilitated patients’ involvement. In particular, we acknowledge colleagues from the Trinity College Institute of Neuroscience A. Bodke, J. McGrath, F. Newell, H. Garavan, and J. O’Doherty for their support in sample collection. Collection and analysis of these samples were funded by the Wellcome Trust (072894/z/03/z-Gill) and the Science Foundation Ireland (08/IN.1/B1916_Corvin).
EPIGEN: Work from the London Cohort was supported by research grants from the Wellcome Trust (grant 084730 to S.M.S.), University College London (UCL)/University College London Hospitals (UCLH) Comprehensive Biomedical Research Centre/Specialist Biomedical Research Centres (CBRC/SBRC) (grant 114 to S.M.S.), the European Union Marie Curie Reintegration (to M. Matarin and S.M.S.), the UK NIHR (08-08-SCC), the Comprehensive Local Research Network (CLRN) Flexibility and Sustainability Funding (FSF) (grant CEL1300 to S.M.S.), The Big Lottery Fund, the Wolfson Trust and the Epilepsy Society. This work was undertaken at UCLH/UCL, which received a proportion of funding from the UK Department of Health’s NIHR Biomedical Research Centres funding scheme. Work from the Royal College of Surgeons in Ireland was supported by research grants from the Science Foundation Ireland (Research Frontiers Programme award 08/RFP/GEN1538) and Brainwave–the Irish Epilepsy Association. The collection of Belgian subjects was supported by the Fonds National de la Recherche Scientifique (grant FC 63574 / 3.4.620.06 F) and the Fonds Erasme pour la Recherche Médicale at the Université Libre de Bruxelles.
UCL Institute of Neurology Control Brain Tissue Collection: Funding was provided by the UK MRC (grant G0901254), the MRC Sudden Death Brain and Tissue Bank and the Sun Health Research Institute Brain Bank.
UMCU: The UMCU study was supported by the Netherlands Organization for Health Research and Development ZonMw (917.46.370 to H.E.H.) and the US NIMH (MH078075 to R.A.O.).
Footnotes
URLs. Allen Institute Brain Atlas, http://human.brain-map.org/; SNPExpress database, http://compute1.lsrc.duke.edu/softwares/SNPExpress/index.php; ADNI database, http://adni.loni.ucla.edu/; ADNI acknowledgements, http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf; the Foundation for the NIH, http://www.fnih.org/; ADNI information, http://www.adni-info.org/; Brain Research Imaging Centre Edinburgh, http://www.bric.ed.ac.uk/; SINAPSE Collaboration, http://www.sinapse.ac.uk/; fBIRN, http://www.birncommunity.org/; SYS, http://www.saguenay-youth-study.org/; SHIP, http://ship.community-medicine.de/; ENIGMA Consortium protocols, http://enigma.loni.ucla.edu/protocols/; Mx, http://www.vcu.edu/mx/; SOLAR, http://solar.txbiomedgenetics.org/; Genetic Power Calculator, http://pngu.mgh.harvard.edu/~purcell/gpc/; HapMap, http://hapmap.ncbi.nlm.nih.gov/; Data upload site for participating studies, http://enigma.loni.ucla.edu/; METAL, http://www.sph.umich.edu/csg/abecasis/Metal/; METASOFT, http://genetics.cs.ucla.edu/meta/; matSpD, http://gump.qimr.edu.au/general/daleN/matSpD/.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
The ENIGMA support group designed the project, established the consortium, determined the analysis and quality control procedures, offered analytical support and performed and coordinated cross-site and replication analyses. This group included J.L.S., S.E.M., A.A.V., D.P.H., M.J.W., B.F., N.G.M. and P.M.T. The imaging protocols group determined and refined protocols for computing brain measures from the MRI scans and helped sites implement them as needed. This group included J.L.S., R.T., A.M.W., T.E.N., M.J. and M. Rijpkema. The genetics protocols group created analysis methods for imputation, quality control and association testing of genome-wide data and helped to ensure that protocols were implemented consistently across all sites. This group included S.E.M., J.L.S., A.A.V. and D.P.H. The meta-analysis was carried out by the meta-analysis group, consisting of S.E.M., R.E.S., J.L.S., D.P.H., A.A.V., M.J.W., N.G.M., B.F. and P.M.T. The first draft was written by J.L.S., S.E.M., A.A.V., D.P.H., M.J.W., B.F., N.G.M. and P.M.T. Local image processing, involving statistical analysis and analysis of the data, was performed by J.L.S., A.M.W., D.P.H., R.B., Ø.B., M.M.C., O.G., M. Hollinshead, A.J.H., S.M.M., A.C.N., M. Rijpkema, N.A.R., M.C.V.H., T.G.M.v.E., S.W., D.G.B., S.L.R., J.L.R., M.-J.v.T., S.E., P.T.F., P.K., J.L.L., R.M., G.B.P., J. Savitz, H.G.S., K.S., A.W.T., M.V.d.H., N.J.v.d.W., N.E.M.V.H., H.W., A.M.D., C.R.J., D.J.V., E.J.C.d.G., G.I.d.Z., T.E., G.F., P.H., H.E.H.P., K.L.M., A.J.S., L.S., J.B., D.C.G., K.N., E.L., A.M.-L., P.G.S., L.G.A., K.S.H., T.P., M.D., R.P., N.H., K.W., I.A., Ø.B., A.M.D., D.H., M.C., S.A., N.D., C. Depondt, M. Pandolfo, E.J.R., D.M.C., J.C.R., J.R., J.T., R.T., C.L., S.M., A.H., C.D.W., N.J., D.J.H., L.T.W. and M. Hoogman. Local genetics processing, involving statistical analysis and analysis of the data, was performed by J.L.S., S.E.M., A.A.V., A.M.W., D.P.H., M.B., A.A.B., A. Christoforou, G. Davies, J.-J.H., L.M.L., G.L., P.H.L., D.C.L., X.L., M. Mattingsdal, K.N., E. Strengman, K.v.E., T.G.M.v.E., S.W., S.K., L.A., R.M.C., M.A.C., J.E.C., R.D., T.D.D., N.B.F., H.H.H.G., M.P.J., J.W.K., M. Mattheisen, E.K.M., T.W.M., M.M.N., M. Rietschel, V.M.S., A.W.T., J.A.V., S.C., S.D., T.M.F., P.H., S.L.H., G.W.M., O.A.A., H.G.B., R.A.O., B.W.P., A.J.S., L.S., J.B., D.C.G., M.J.W., N.G.M., A.L., E.B.B., C.W., B.P., B.M.-M., G.C., Z.P., G.H., M.N., A.T., D.K., M. Matarin, S.M.S., G.L.C., N.K.H., M.E.R., D.W.M., C.O., A. Corvin, M.G., J.F., J.C.R., A.R., M. Ryten, D.T., N.S., C.S., R.W., J. Hardy, M.E.W. and M.A.A.d.A. Local study oversight and management, involving joint supervision of research, contribution of reagents, materials and/or analysis tools, was carried out by R.L.B., R.D., P.T.F., R.S.K., I.M., R.L.O., I.R., I.A., W.C.D., P.H., F.M., A.M.-L., D.J.P., S.G.P., J.M.S., M.W.W., O.A.A., M.E.B., H.G.B., E.J.C.d.G., I.J.D., G.I.d.Z., T.E., G.F., H.E.H.P., F.J.M., K.L.M., R.A.O., T.P., Z.P., B.W.P., A.J.S., L.S., J.W.S., J.M.W., J.B., D.C.G., M.J.W., B.F., P.M.T., A.M.M., J. Hall, M. Papmeyer, E. Sprooten, J. Sussmann, S.M.L., J.B.P., L.G.A., G.C., D.R., E.M., G.S., K.S.H., P.G.S., E.B.B., D.I.B., H.J.G., H.V., K.A., C.M., G. Donohoe, F.H., A.V.S., V.G., C.T., M.W.V., L.J.L., C. DeCarli, S.S., J.C.B., M.A.I., A.A. and J. Hardy.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Jack CR, Jr, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer’s disease. Alzheimers Dement. 2011;7:474 e4–485 e4. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simić G, Kostovic I, Winblad B, Bogdanovic N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer’s disease. J. Comp. Neurol. 1997;379:482–494. doi: 10.1002/(sici)1096-9861(19970324)379:4<482::aid-cne2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Wright IC, et al. Meta-analysis of regional brain volumes in schizophrenia. Am. J. Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- 4.Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am. J. Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- 5.Keller SS, Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia. 2008;49:741–757. doi: 10.1111/j.1528-1167.2007.01485.x. [DOI] [PubMed] [Google Scholar]
- 6.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum. Brain Mapp. 2007;28:464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kremen WS, et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire EA, et al. Navigation-related structural change in the hippocampi of taxi drivers. Proc. Natl. Acad. Sci. USA. 2000;97:4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess N, Maguire EA, O’Keefe J. The human hippocampus and spatial and episodic memory. Neuron. 2002;35:625–641. doi: 10.1016/s0896-6273(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 10.Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–461. doi: 10.1038/nature10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag CM, et al. Total brain volume and corpus callosum size in medication-naive adolescents and young adults with autism spectrum disorder. Biol. Psychiatry. 2009;66:316–319. doi: 10.1016/j.biopsych.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanfield AC, et al. Towards a neuroanatomy of autism: a systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur. Psychiatry. 2008;23:289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Posthuma D, et al. The association between brain volume and intelligence is of genetic origin. Nat. Neurosci. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- 14.Fears SC, et al. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. J. Neurosci. 2009;29:2867–2875. doi: 10.1523/JNEUROSCI.5153-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers J, et al. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage. 2010;53:1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischl B, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 18.Buckner RL, et al. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bis JC, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat. Genet. doi: 10.1038/ng.2237. published online (15 April 2012; doi:10.1038/ng.2237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripke S, et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sklar P, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful genebased association test using extended Simes procedure. Am. J. Hum. Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao Y, et al. Expression and evolutionary conservation of the tescalcin gene during development. Gene expression patterns. Gene Exp. Patterns. 2009;9:273–281. doi: 10.1016/j.gep.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Baumgartner M, Patel H, Barber DL. Na+/H+ exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am. J. Physiol. Cell Physiol. 2004;287:C844–C850. doi: 10.1152/ajpcell.00094.2004. [DOI] [PubMed] [Google Scholar]
- 27.Slepkov ER, Rainey JK, Sykes BD, Fliegel L. Structural and functional analysis of the Na+/H+ exchanger. Biochem. J. 2007;401:623–633. doi: 10.1042/BJ20061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levay K, Slepak VZ. Tescalcin is an essential factor in megakaryocytic differentiation associated with Ets family gene expression. J. Clin. Invest. 2007;117:2672–2683. doi: 10.1172/JCI27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levay K, Slepak VZ. Up- or downregulation of tescalcin in HL-60 cells is associated with their differentiation to either granulocytic or macrophage-like lineage. Exp. Cell Res. 2010;316:1254–1262. doi: 10.1016/j.yexcr.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 32.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 35.Litterman N, et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond SM, Sharpless NE. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135:1013–1016. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright MJ, Martin NG. Brisbane adolescent twin study: outline of study methods and research projects. Aust. J. Psychol. 2004;56:65–78. [Google Scholar]
- 38.Jackson DN. MAB: Multidimensional Aptitude Battery Manual. Port Huron, Michigan: Research Psychologists Press; 1984. [Google Scholar]
- 39.Vogel W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 1999;13(suppl):S77–S82. doi: 10.1096/fasebj.13.9001.s77. [DOI] [PubMed] [Google Scholar]
- 40.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 45.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 46.Morey RA, et al. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardoe HR, Pell GS, Abbott DF, Jackson GD. Hippocampal volume assessment in temporal lobe epilepsy: how good is automated segmentation? Epilepsia. 2009;50:2586–2592. doi: 10.1111/j.1528-1167.2009.02243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morey RA, et al. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum. Brain Mapp. 2010;31:1751–1762. doi: 10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pantel J, et al. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10:752–758. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Morra JH, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 53.Pei YF, Li J, Zhang L, Papasian CJ, Deng HW. Analyses and comparison of accuracy of different genotype imputation methods. PLoS ONE. 2008;3:e3551. doi: 10.1371/journal.pone.0003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 55.Allen AS, Martin ER, Qin X, Li YJ. Genetic association tests based on ranks (GATOR) for quantitative traits with and without censoring. Genet. Epidemiol. 2006;30:248–258. doi: 10.1002/gepi.20141. [DOI] [PubMed] [Google Scholar]
- 56.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 57.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durston S, et al. Differential effects of DRD4 and DAT1 genotype on fronto-striatal gray matter volumes in a sample of subjects with attention deficit hyperactivity disorder, their unaffected siblings, and controls. Mol. Psychiatry. 2005;10:678–685. doi: 10.1038/sj.mp.4001649. [DOI] [PubMed] [Google Scholar]
- 59.Dick DM, et al. Genome-wide association study of conduct disorder symptomatology. Mol. Psychiatry. 2011;16:800–808. doi: 10.1038/mp.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein JL, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol. Psychiatry. 2011;16:927–937. 881. doi: 10.1038/mp.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 64.Li MX, Sham PC, Cherny SS, Song YQ. A knowledge-based weighting framework to boost the power of genome-wide association studies. PLoS ONE. 2010;5:e14480. doi: 10.1371/journal.pone.0014480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 66.Kasperaviciūte D, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–2147. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trabzuni D, et al. Quality control parameters on a large dataset of regionally-dissected human control brains for whole genome expression studies. J. Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinzen EL, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.